Abstract
O island 48 (OI-48) of Escherichia coli consists of three functional gene clusters that encode urease, tellurite resistance (Ter), and putative adhesins Iha and AIDA-1. The functions of these clusters in enterohemorrhagic E. coli (EHEC) O157:H7 infection are unknown. Deletion mutants for these three regions were constructed and evaluated for their ability to adhere to epithelial cells in vitro and in ligated pig ileal loops. Deletion of the Ter gene cluster reduced the ability of the organism to adhere to and form large clusters on IPEC-J2 and HEp-2 cells. Complementation of the mutation by introducing the wild-type ter genes restored adherence and large-cluster formation. Tests in ligated pig ileal loops showed a decrease in colonization by the Ter-negative mutant, but the difference was not significant compared to colonization by the wild type (26.4% ± 21.2% versus 40.1% ± 19.1%; P = 0.168). The OI-48 aidA gene deletion had no effect on adherence in vitro or in vivo. Deletion of the iha and ureC genes had no effect on adherence in vitro but significantly reduced the colonization of EHEC O157:H7 in the ligated pig intestine. These data suggest that Ter, Iha, and urease may contribute to EHEC O157:H7 pathogenesis by promoting adherence of the pathogen to the host intestinal epithelium.
The genome of enterohemorrhagic Escherichia coli (EHEC) O157:H7 strain EDL933 contains unique blocks of sequences called O islands (OIs), which are absent from the E. coli K12 MG1655 genome. The OIs contain 1,387 genes, some of which encode virulence factors (21), thus making these OIs pathogenicity islands (PAIs). EHEC pathogenicity is mainly determined by factors encoded in these PAIs, such as OI-148 (locus of enterocyte effacement [LEE] PAI) which causes attaching and effacing (AE) lesions, and OI-45 and OI-93, which contain lambdoid phages encoding verotoxins that are required for bloody diarrhea and the hemolytic uremic syndrome. However, only 40% of the OI genes in O157:H7 strain EDL933 have been assigned a function.
OI-43 and OI-48 are duplicates, encoding tellurite resistance (Ter) and the putative adhesin Iha and are therefore called Ter- and adherence-conferring islands (TAIs) (28). EHEC O157:H7 strain EDL933 contains both OI-43 and OI-48, while the Sakai strain contains only one of these OIs. We used PCR with OI junction primers to show that O157:H7 strain 86-24 contains only OI-48, not OI-43 (data not shown). The TAI can be divided into three functional gene clusters, one encoding urease, a second encoding Ter, and the third encoding the putative adhesins Iha and AIDA-1 (Fig. 1) (21, 28). The urease genes are similar to the genes in Klebsiella aerogenes (9) while the Ter gene cluster is similar to the genes in Serratia marcesens (28). The pattern of homology indicates that the TAI is a mosaic of functional regions that were acquired on separate occasions (1). Iha is an IrgA (iron-regulated gene A) homolog adhesin (25). Because of its presence in eae (E. coli AE)-negative verotoxigenic E. coli (VTEC) strains, it has been suggested that Iha may function as an adhesin in these organisms in place of intimin (23, 25). AIDA-I, encoded by aidA, is an autotransporter membrane protein with a β-barrel structure, and it confers on enteropathogenic E. coli the ability to diffusely adhere to epithelial cells. The AIDA-I adhesin from OI-48 has 68% homology to the AIDA-I of enteropathogenic E. coli (20, 21). However, the roles of Iha and AIDA-I-like adhesins in EHEC pathogenesis have yet to be determined.
FIG. 1.
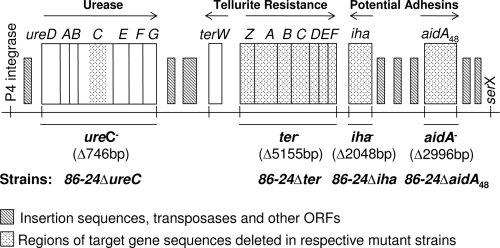
Schematic representation of the three functional regions of OI-48 of EHEC O157:H7 strain EDL933. The island is 87,547 bp, with 92 open reading frames; only genes of interest are shown. The sections in the four mutants that were deleted are shaded, and the sizes of the deletions are shown.
The ure gene cluster consists of three structural genes, ureA, ureB, and ureC, and four accessory genes, ureD, ureE, ureF, and ureG, required for transport and processing of urease (5, 15). Interestingly, ure genes are more frequently present in eae-positive VTEC strains (113 of 132, or 85.6%) than in eae-negative VTEC strains (4 of 70, or 5.7%) although there is no physical linkage of the ure gene and eae, which is present on the LEE (17). These observations suggest that there may be a functional relationship between the ure and LEE genes (17).
Ter imparts resistance against tellurium in bacteria; however, tellurium is rare in nature, and the advantage of Ter to pathogenic bacteria is not understood. The Ter genes have been extensively studied in plasmids pMER610 and R478 and appear to confer phage inhibition, resistance to bacteriophage (such as λ and T5) infection, and pore-forming colicins (27). Tellurite salts are strong oxidative agents, and it is possible that Ter confers a selective advantage in the host environment by aiding the bacteria in a general stress response. Introduction of Ter into nonpathogenic or uropathogenic E. coli (UPEC) results in a significant increase in their survival inside macrophages (29). Ter in EHEC strain EDL933 is encoded by terZABCDEF located in OI-48 and OI-43, and questions have been asked as to whether the operon that encodes Ter in EHEC O157 provides a selective advantage to this pathogen and, if so, how it is associated with bacterial pathogenesis.
To investigate the roles of the putative OI-48 virulence determinants in the pathogenesis of EHEC O157:H7, deletion mutants of the three functional regions of OI-48 were constructed and evaluated for adherence to tissue culture cells and to enterocytes in pig ileal loops.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are described in Table 1. Mutant strains were constructed in EHEC O157:H7 strain 86-24.
TABLE 1.
Bacterial strains and plasmids
| E. coli strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| 86-24 | EHEC O157:H7 strain 86-24 | 26 |
| SM10(λpir) | Kmr RP4-2-Tc::Mu; broad host-range conjugation strain; Tra functions conferred by chromosomal RP4 | 24 |
| DH5α λpir | F−hsdR17 thi-1 gyrA Δ(lacZYA-argF) supE44 recA1 φ80dΔ(lacZ)M15 relA λpir | 4 |
| 86-24Δter | 86-24Δter::Kan (OI-48) | This study |
| 86-24Δter(pTER) | 86-24Δter containing pCR2.1-terZABCDEF | This study |
| 86-24Δiha | 86-24Δiha::Kan (OI-48) | This study |
| 86-24Δiha(pIHA) | 86-24Δiha containing pBAD-iha | This study |
| 86-24ΔaidA48 | 86-24ΔaidA48::Kan (OI-48) | This study |
| 86-24ΔescN | 86-24ΔescN::Gm (OI-148) | This study |
| 86-24ΔureC | 86-24ΔureC::Kan (OI-48) | This study |
| 86-24ΔureC(pUDG) | 86-24ΔureC containing pBAD-ureDABCEFG | This study |
| Plasmids | ||
| pUC-Kan | Template plasmid for kanamycin resistance | 13 |
| pKM208 | pMAK700 derivative with red and gam expressed from Ptac; AprlacI; IPTG inducible | 16 |
| pBAD | pBAD-TOPO; expression vector PBAD; Apr | Invitrogen |
| pCR2.1 | pCR2.1-TOPO, TA cloning vector, Apr | Invitrogen |
| pTER | pCR2.1-TOPO containing terZABCDEF (OI-48) | This study |
| pIHA | pBAD-TOPO containing iha | This study |
| pUDG | pBAD-TOPO containing ureDABCEFG (OI-48) | This study |
| pRE107 | Mobilizable suicide vector; AprsacB | 4 |
| pUreC | pRE107 containing ureC deletion kan cassette | This study |
Creation of mutants. (i) Construction of Ter, iha, aidA48, and escN isogenic mutants.
The OI-48 gene encoding an AIDA-I-like protein is named aidA48. Deletions of the Ter gene cluster, iha, and aidA48 of OI-48 and escN of the LEE PAI were made through the phage λ-Red-mediated recombination system (16). Briefly, competent EHEC O157:H7 strain 86-24 cells containing pKM208 were electroporated with PCR products. The PCR products were generated with primers containing 42-nucleotide overhangs at the 5′ end homologous to the gene to be replaced and 20 nucleotides at the 3′ end targeting the drug resistance genes derived from plasmid pUCKan (carrying a kanamycin resistance cassette) (Tables 1 and 2). After electroporation, cells were incubated at 37°C overnight with shaking and plated on medium containing 50 μg/ml kanamycin. The resulting colonies for each of the target genes or gene clusters were confirmed for absence of the genes by PCR using primers flanking the deleted region (data not shown). Clones with gene deletions were tested for ampicillin sensitivity for the loss of plasmid pKM208. The resultant ampicillin-sensitive mutant clones were named 86-24Δter, 86-24Δiha, 86-24ΔaidA48, and 86-24ΔescN (Fig. 1 and Table 1). To complement the mutant 86-24Δter, the wild-type ter gene cluster was amplified by PCR with primers ter-CF and ter-CR and cloned into the plasmid pCR2.1, resulting in pTER. This plasmid was then electroporated into 86-24Δter, yielding 86-24Δter(pTER). Similarly, complemented strain 86-24Δiha(pIHA) was generated by introduction of pIHA into 86-24Δiha. pIHA was generated by cloning the PCR amplicon with primers iha-CF and iha-CR from the wild-type gene.
TABLE 2.
Primers used for PCR and PCR-mediated mutagenesisa
| Primer target and name (restriction enzyme) | Sequence (5′ to 3′)d |
|---|---|
| 86-24Δter | |
| ter-KF | CAGACGGTATCACTCAGCAAAGAATCATCTGCATTAAGCCAGTGACTAACTAGGAGGAATAA |
| ter-KR | ATCAATGACAACGGTGATCGCAATTTTACTGACGTTCGCCGGTCATTATTCCCTCCAGGTAC |
| ter-CFb | TTGTCAGAGAACTTCATCATG |
| ter-CRb | ATAAGTCAATTGCACCTTCTC |
| 86-24Δihac | |
| iha-KF | CGTATTCTACCGTCAGTGATAGCGTTTTGTTATTATCAGAACTGACTAACTAGGAGGAATAA |
| iha-KR | CGGAGATTAGTAATATGCGAATAACCACTCTGGCTTCCGTAGTCATTATTCCCTCCAGGTAC |
| iha-F | TGAGATAGCACTGGATCGTG |
| iha-R | CAGTTCCCAGGCAATAATGC |
| iha-CF | AGACAGGACTTTGAGATGACG |
| iha-CR | CACCGACATACGGAAAGATACCACG |
| 86-24ΔaidA48 | |
| aidA48-KF | GAATCTCTTCATCATGCAGAACGGAATTGCACACAACAGACTGACTAACTAGGAGGAATAA |
| aidA48-KR | GAAATCGTATTTCCGGGATACCGTATAATCAGAAAGTCATATTCATTATTCCCTCCAGGTAC |
| aidA48-LF | TGATGAGCGCCAGACCAATC |
| aidA48-RR | TAATATGCGCCTGTAGTGACTG |
| 86-24ΔescN | |
| escN-GF | ACGAATAGATAAAATTCTGTCCAACATACTCAGGCAACCACTCGTTGTGACAATTTACCGAA |
| escN-GR | AATATCGAACTTAAAGTATTAGGAACGGTAAATGATTTCAGACGAGCTCGAATTGACATAAG |
| escN-Fm | GATAAAATTCTGTCCAACATAC |
| escN-Rm | TCGAACTTAAAGTATTAGGAAC |
| 86-24ΔureCc | |
| ure-1F (XbaI) | AAAAAATCTAGAGCATGTCCTGAGTCGCGAGCA |
| ure-1R(KpnI) | AAAAAAGGTACCAGGTTGTGGCGTTAGAACC |
| ure-2F (KpnI) | AAAAAAGGTACCTGAAGTCGGCTCGATTGAAGT |
| ure-2R(SacI) | AAAAAAGAGCTCTGCTCCTGATCGTGATTATGG |
| ureC-F | CAGAAGGCAGGATCGTTACC |
| ureC-R | CGAACATTGGGCGATAGTGC |
| ure-CF | CACCTTTGCAGACCAGTCAGTGAC |
| ure-CR | CGGCATCTGGTAAGTACAAC |
All the primers were designed in the present study and were used for generation and confirmation; primers were used for complementation where noted.
Used for both confirmation and complementation.
Primers were also used for complementation.
Restriction sites are in boldface.
(ii) Construction of ureC isogenic mutant by allelic exchange.
A ureC deletion mutant of EHEC O157:H7 strain 86-24 was created by allelic exchange using the suicide vector pRE107, as described by Gunzer et al. (7). First, the suicide plasmid pUreC containing a ureC deletion cassette was constructed by the following steps. Up- and downstream fragments of the EHEC ureC regions were amplified using Expand High Fidelity DNA polymerase (Roche Diagnostics, Montreal, Canada) and the primer pair ure-1F (XbaI) and ure-1R (KpnI) and the pair ure-2F (KpnI) and ure-2R (SacI) (Table 2), generating PCR fragments of 951 bp and 921 bp, respectively. The two PCR fragments were ligated successively into their specific restriction enzyme sites in pRE107. A kanamycin resistance cassette derived from pUC-Kan was then inserted into the KpnI site near the middle of the cloned PCR fragments in pRE107, creating pUreC. Second, the pUreC in E. coli DH5α λpir was isolated and transferred into EHEC O157:H7 strain 86-24 by electroporation. Colonies were screened for the first crossover (plasmid integration) with ampicillin and the second crossover with sucrose (7). Ampicillin-sensitive, sucrose-resistant colonies were screened by PCR with primers ureC-F and ureC-R to confirm that the wild-type ureC had been replaced with the mutated ureC. The PCR yielded a 1,227-bp amplicon for the mutated gene and a 1,070-bp amplicon for the wild-type gene. The resultant urease-negative mutant (86-24ΔureC) (Fig. 1) had a 746-bp deletion in ureC replaced by a 903-bp kanamycin cassette and was confirmed by sequencing. To complement the mutant 86-24ΔureC, the wild-type ure gene cluster was amplified by PCR with primers ure-CF and ure-CR and cloned into the plasmid pBAD, resulting in pUDG. This plasmid was then electroporated into 86-24ΔureC, yielding 86-24ΔureC(pUDG).
In vitro adherence assay.
Effects of the mutations on adherence to HEp-2 and IPEC-J2 cells were evaluated by comparing adherence of the wild-type O157:H7 strain 86-24 with that of the mutant. The bacteria were grown at 37°C for 16 to 18 h in 3 ml of brain heart infusion (BHI) broth containing 44 mM NaHCO3 (BHIN broth) in tightly capped 12-ml sterile plastic tubes (Fisher Scientific, Nepean, ON, Canada) without shaking. The density of all bacterial cultures was adjusted photometrically to contain approximately 5 × 108 CFU/ml prior to their use in the assay.
HEp-2 (ATCC CCL23) cells were maintained in Eagle's minimal essential medium (EMEM) (Invitrogen, Carlsbad, CA). The IPEC-J2 pig jejunal epithelial cells (a gift from Joshua Gong of Agriculture and Agri-Food Canada) were maintained in Dulbecco's minimal essential medium (Invitrogen). Both media were supplemented with 10% fetal bovine serum, penicillin (100 IU/ml), and streptomycin (100 μg/ml). HEp-2 and IPEC-J2 cell adherence assays were conducted as described previously (3), with some modifications. Briefly, approximately 2 × 105 HEp-2 or IPEC-J2 cells per well were dispensed in six-well cell culture plates (Corning, NY) and grown overnight in EMEM or Dulbecco's minimal essential medium, respectively, in the presence of 5% CO2. For the adherence assay, cell monolayers at ∼50% confluence were washed and reconstituted with fresh EMEM (800 μl per well) without antibiotics. A 20-μl volume (approximately 107 bacteria) of an overnight culture of each strain was added individually to sets of duplicate wells. After incubation for 6 h at 37°C in 5% CO2 with a medium change at 3 h, the plates were washed with phosphate-buffered saline to remove unbound bacteria, fixed with 70% methanol, stained 1:40 with Giemsa (Sigma), and examined by light microscopy. Adherence was quantified by examining 100 consecutive cells per well and recording the percentage of HEp-2 or IPEC-J2 cells with clusters of 5 to 9, 10 to 19, and ≥20 bacteria. The percentage of cells with at least five adherent bacteria per cell was calculated as a measure of total adherence. Data are expressed as the means of at least three independent experiments ± standard deviations (SDs).
Pig ileal loop experiments.
The mutants were tested in ligated ileal loops of pigs. The experimental protocol and the care of the pigs were approved by the University of Guelph Animal Care Committee. Bacteria were grown at 37°C overnight in BHIN broth without shaking. Bacteria were then concentrated by centrifugation and resuspended to a concentration of 5 × 1010 CFU/ml in EMEM containing 10% fetal bovine serum.
A total of 63 12- to 14-day-old female pigs were used, with two or three pigs from the same litter being used at one time. The pigs were fed only electrolytes in warm water (Vetoquinol, Lavaltrie, Quebec, Canada) for 24 h before surgery. The pigs were premedicated with a mixture of ketamine (50 mg/ml), xylazine (10 mg/ml), and butorphenol (Wyeth Canada, Saint-Laurent, Quebec, Canada) (1 mg/ml), given intramuscularly at 0.2 ml/kg of body weight. About 10 min later anesthesia was achieved by slow intravenous injection of sodium pentobarbital (55 mg/100 ml). Following cleaning and disinfection of the abdomen, a ventral midline laparotomy was performed aseptically, and the distal ileum was exteriorized. Six to eight ligated loops (each about 10 cm long) were created with nylon ligatures in the distal ileum, beginning approximately 10 cm from the ileocecal junction. Each loop was followed by a short intervening segment (2 to 3 cm) that was not inoculated. A 2-ml volume of inoculum containing 1011 CFU of the test organisms was injected into the lumen of the ileal loops with a 25-gauge needle. In each pig, the treatments were assigned randomly; one loop received the positive control wild-type O157:H7 strain 86-24, and one loop received the negative control mutant 86-24ΔescN. After inoculation, the ileum was replaced in the abdomen, and the laparotomy incision was closed. Immediately following the surgery and at 4-h intervals thereafter, the pigs were injected intramuscularly with butorphenol at 0.4 mg/kg of body weight. The pigs were euthanized by an overdose of pentobarbital at 15 to 16 h after inoculation of the loops, and pieces of the ligated intestine were quickly excised from each loop for histopathology, electron microscopy, and bacteriology. Fluid accumulation in the ileal loops was measured as the volume per loop.
Comparison of the adherence of the mutants and the wild type was based on tests conducted in the same pigs, and data were considered valid when the positive control showed adherent bacterial clusters and the negative control showed no adherent bacterial clusters. Bacterial adherence was evaluated semiquantitatively by examining Giemsa-stained sections of the intestine (30) as described below.
Histological examination, immunoperoxidase staining with anti-O157 antibody, and electron microscopy.
Tissues taken from the loops were fixed immediately in 10% neutral buffered formalin for at least 24 h at room temperature. Additional 4 mm by 4 mm pieces were immediately immersed in cacodylate-HCl-buffered glutaraldehyde for electron microscopy. The fixed tissues were cut into smaller pieces, and every second piece of the tissue was chosen, for a total of four pieces from each loop that were processed by routine methods. After the tissue was embedded in paraffin, 1-μm-thick sections were cut and stained with Giemsa and with hematoxylin and eosin dyes. All the villi in these sections were examined by light microscopy. The percentage of villi that had clusters of five or more adherent bacteria was calculated from four consecutive sections of a single loop from each animal, and the score for each treatment was calculated as the mean percentage (± SD) of villi, with adherent bacterial clusters for all loops subjected to the treatment. Bacteria seen in these sections were tested for the O157 antigen by indirect immunoperoxidase staining of adjacent sections with anti-O157 antibody (Difco, Detroit, MI) using Histostain (Zymed Laboratories, San Francisco, CA), according to the manufacturer's instructions. Selected fixed tissues containing adherent bacterial clusters, as identified by the light-microscopic examination, were processed for electron microscopy. Thin sections were stained with uranyl acetate and lead citrate and examined with a 100S transmission electron microscope (Joel, Japan).
RNA isolation.
To determine the expression of the plasmid-encoded genes in the complemented strains, reverse transcription-PCR (RT-PCR) was performed from bacterial total RNA that was isolated using a RiboPure-Bacteria kit protocol (Ambion, TX). Overnight bacteria were harvested and treated with RNAlater (Ambion). Subsequent RNA isolation and purification and DNase I treatment steps were followed according to the manufacturer's protocol. After DNase I treatment, total RNA was quantified by a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE), and RNA quality was visualized by agarose gel electrophoresis. RNAs were confirmed to be free of DNA by PCR using RNA as a template.
RT-PCR.
First-strand cDNA was synthesized from the DNase I-treated bacterial total RNA using SuperScript II-RT with 100 ng of random primer pd(N)9 according to the procedures recommended by the supplier (Invitrogen). The cDNAs obtained were amplified by PCR in a final reaction mixture of 50 μl containing 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphate mixture, 10 pmol of each primer (Table 2), and 2.5 units of Taq DNA polymerase (New England Biolabs, Boston, MA). An initial denaturation of 94°C for 4 min was followed by 35 cycles of PCR (denaturation at 94°C for 30 s, primer annealing at 55 to 58°C [depending on the primers used] for 30 s, and extension at 72°C for 120 s, with a final amplification step of 10 min at 72°C).
Statistical analysis.
All analyses were performed with SAS for Windows, version 8.02 (SAS Institute Inc., Cary, N.C.). The in vitro adherence of various bacterial strains was compared by analysis of variance using the percent adherence of clusters with 5 to 9, 10 to 19, and ≥20 bacteria per cell, as well as the total percent adherence (≥5 adherent bacteria per cell). Similarly, in vivo adherence of the tested strains was compared by analysis of variance of the mean percentage of villi with adherent bacteria (≥5 bacteria per villus) for the total number of loops that were tested. P values of <0.05 were considered significant.
RESULTS
Effect of a ter deletion on adherence of O157:H7 strain 86-24 to HEp-2 and IPEC-J2 cells.
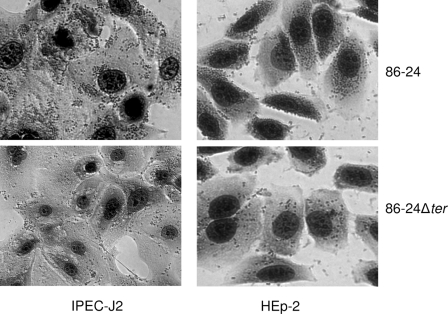
The ter deletion mutant strain 86-24Δter was tellurite sensitive, failing to grow in potassium tellurite at a concentration of 1 μg/ml, as determined by the method of Taylor et al. (28). Adherence of the mutant strain 86-24Δter to the IPEC-J2 and HEp-2 cells was compared with that of the wild-type parent strain 86-24. The patterns of adherence by the wild-type strain and its isogenic mutant to IPEC-J2 and HEp-2 cells are shown in Fig. 2. With both cell types, the mutant 86-24Δter adhered less frequently and in much smaller clusters of bacteria.
FIG. 2.
Effects of the 86-24Δter mutation on the adherence of EHEC O157:H7 strain 86-24 to IPEC-J2 and HEp-2 cells. As described in Materials and Methods, adherence assays were performed for 6 h with bacteria grown in BHIN broth without shaking, and cells were fixed, stained with Giemsa, and then photographed. The 86-24Δter mutant showed decreased adherence compared to the wild-type parent strain 86-24. Magnification, ×200.
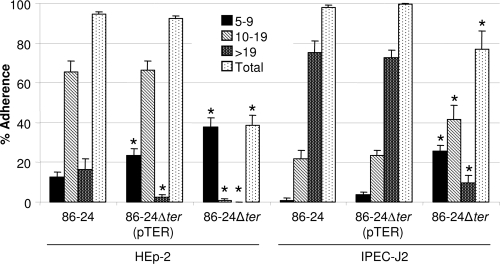
With HEp-2 cells, the mutant strain 86-24Δter caused a marked reduction in the formation of medium clusters (10 to 19 bacteria) and large clusters (>19 bacteria) compared to the wild type (0.83% and 0% versus 65.5% and 16.33%, respectively; P < 0.001) and in the total percentage of the epithelial cells with adherent bacteria (38.8% versus 94.5%; P < 0.001) (Fig. 3). Complementation of the mutant restored the Ter phenotype with a MIC of 128 μg/ml of potassium tellurite. As shown in Fig. 3, total adherence of the complemented mutant strain 86-24Δter(pTER) to HEp-2 cells returned to wild-type levels as did medium-sized cluster formation; however, the complementation only partially restored the formation of large clusters to the wild-type level. The mutant strain transformed with the plasmid vector, 86-24Δter(pCR2.1), adhered to HEp-2 cells at a level similar to that of the mutant strain 86-24Δter (data not shown), affirming the specificity of the complementation. The data for IPEC-J2 cells were similar to those for HEp-2 cells; the total percentage of IPEC-J2 cells with adherent 86-24Δter bacteria was 76.8%, with 9.5% in large clusters, and these values were significantly lower than the corresponding values of 97.8% and 75.1% for the wild type (P < 0.001). There was a reduction in the number of large clusters and an increase in the number of medium clusters of the mutant compared to the wild type (Fig. 3). With IPEC-J2 cells, the complemented mutant strain 86-24Δter(pTER) restored the wild-type adherence phenotype (Fig. 3). The mutant 86-24ΔescN, used as a negative control in the adherence assay, caused no adherence to either cell type (data not shown). It is also noteworthy that EHEC O157:H7 strain 86-24 adhered significantly more to IPEC-J2 cells than to HEp-2 cells (data not shown).
FIG. 3.
Adherence to HEp-2 and IPEC-J2 cells by wild-type strain 86-24, mutant 86-24Δter, and complemented mutant 86-24Δter(pTER) containing a cloned copy of the Ter gene cluster. Bacteria were grown in BHIN broth without shaking and subjected to a 6-h adherence assay. Adherence was quantified as described in Materials and Methods and was compared to the wild-type 86-24. The number of adherent bacteria per cell (5 to 9, 10 to 19, and >19) and the total percentage of cells with clusters of ≥5 adherent bacteria per cell are shown, as indicated. *, P < 0.05.
To determine whether the reduced adherence of the 86-24Δter mutant might be due to a decrease in the growth rate, both the wild-type and the mutant strains were grown under the same conditions as in the adherence assay, excluding the epithelial cells. The total number of bacteria was determined every hour for 6 h. The growth rates for the wild type and mutant were similar, with a doubling time of 36 min (data not shown). A time course study of adherence to HEp-2 cells was also carried out over the 6-h time period. Small clusters of the wild-type EHEC began to appear from 0 to 3 h, medium-sized clusters appeared from 3 to 5 h, and large-sized clusters appear from 5 to 6 h postinfection. The frequency of each category of the clusters gradually increased with increasing time. The pattern is consistent with a conversion of small clusters into large ones and the formation of new clusters after the first 3 h of infection. Relative to the wild type, the Ter mutant showed a slow increase in the formation of small clusters over the initial 3 h. However, after the first 3 h, there was no increase in total adherence or in formation of large clusters with the mutant (data not shown).
Adherence of the ter deletion mutant in ligated pig ileal loops.
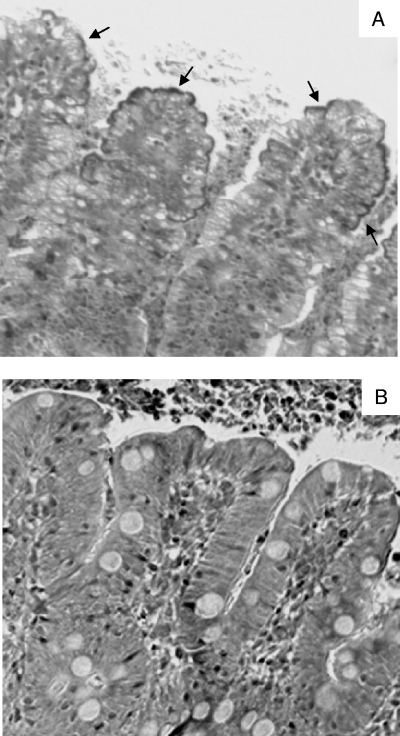
Adherence of the ter mutant was compared with that of the wild-type O157:H7 strain 86-24 in pig ileal loops. Ileal loops were used instead of colon loops because O157:H7 strains grown in BHIN broth without rotation adhered more extensively to the ileal epithelium than to the colon epithelium of pigs (data not shown). The typical appearance of the bacterial clusters on the ileal villi as seen under the light microscope is shown in Fig. 4. Examination of a sample of the bacterial clusters by electron microscopy showed that this pattern of adherence represented characteristic AE lesions, and the adherent bacteria were confirmed to be O157 by immunohistochemistry (data not shown). With nine pairs of valid loops, the mutant strain 86-24Δter adhered to 26.4% ± 21.2% of the villi, whereas the wild type adhered to 40.1% ± 19.1% of the villi (P = 0.168) (Table 3). The mutant 86-24ΔescN, used as a negative control in the pig ileal loops, caused virtually no intimate adherence (Table 3).
FIG. 4.
Light-microscopic appearance of adherence on villi in ligated pig ileal loops inoculated with wild-type EHEC O157:H7 strain 86-24 grown in BHIN broth without shaking (A). Arrows point to clusters of bacteria. Villi in the control loop inoculated with the mutant strain 86-24ΔescN show no adherence (B). The sections were stained with Giemsa. Magnification, ×200.
TABLE 3.
Comparison of adherence in pairs of ileal loops in pigs infected with mutant and wild-type EHEC O157:H7 86-24 strainsa
| Strain pair | No. of loops testedb | No. of loops with bacterial clustersc | % of villi with bacterial clusters (mean ± SD) | P value |
|---|---|---|---|---|
| Wild type 86-24 | 10 | 9 | 40.1 ± 19.1 | 0.168 |
| 86-24Δter | 10 | 9 | 26.4 ± 21.2 | |
| Wild type 86-24 | 10 | 5 | 22.4 ± 7.0 | 0.0347 |
| 86-24Δiha | 10 | 5 | 10.1 ± 8.3 | |
| Wild type 86-24 | 6 | 6 | 32.9 ± 18.9 | 0.8204 |
| 86-24ΔaidA48 | 6 | 6 | 30.4 ± 18.4 | |
| Wild type 86-24 | 6 | 5 | 35.9 ± 8.5 | 0.0129 |
| 86-24ΔureC | 6 | 5 | 17.1 ± 10.0 | |
| Wild type 86-24 | 10 | 6 | 44.8 ± 25.0 | 0.0022 |
| 86-24ΔescN | 10 | 6 | 3.0 ± 2.5 |
Cells were grown in BHIN broth. All inocula consisted of 1011 CFU of bacteria.
Mutant and wild-type bacteria were tested in pairs of loops in the same pigs.
Number of loops with valid data.
Contributions of Iha, AIDA48, and urease to adherence by O157:H7 strain 86-24 in the ligated pig ileal loops.
The deletion mutant strains 86-24Δiha and 86-24ΔureC did not express iha and ureC, respectively, as assayed by RT-PCR (Fig. 5C), and aidA48 was not expressed by the mutant 86-24ΔaidA48 (data not shown). Data from in vitro adherence assays showed that adherence by these mutant strains to IPEC-J2 and HEp-2 cells was not significantly different from that of the wild type (data not shown). When tested in pig ileal loops, mutant 86-24Δiha was impaired in its ability to adhere to pig ileal loops (Table 3); in five loops, the mutant adhered intimately to only 10.1% ± 8.3% of villi, which is significantly lower than 22.4% ± 7.0% for the wild-type parent strain (P = 0.035). In separate ileal loop experiments, the complemented mutant strain 86-24Δiha(pIHA) was evaluated together with the wild type. Data from six loops showed that complementation failed to restore adherence to the level of the wild type (Fig. 5A). RT-PCR showed that transcripts for the iha gene in the complemented mutant strain 86-24Δiha(pIHA) were similar to the level of the wild type (Fig. 5C). Deletion of aidA48 (strain 86-24ΔaidA48) (Fig. 1) made no significant difference in adherence to enterocytes in ligated pig ileal loops compared to adherence with the wild-type strain (Table 3).
FIG. 5.
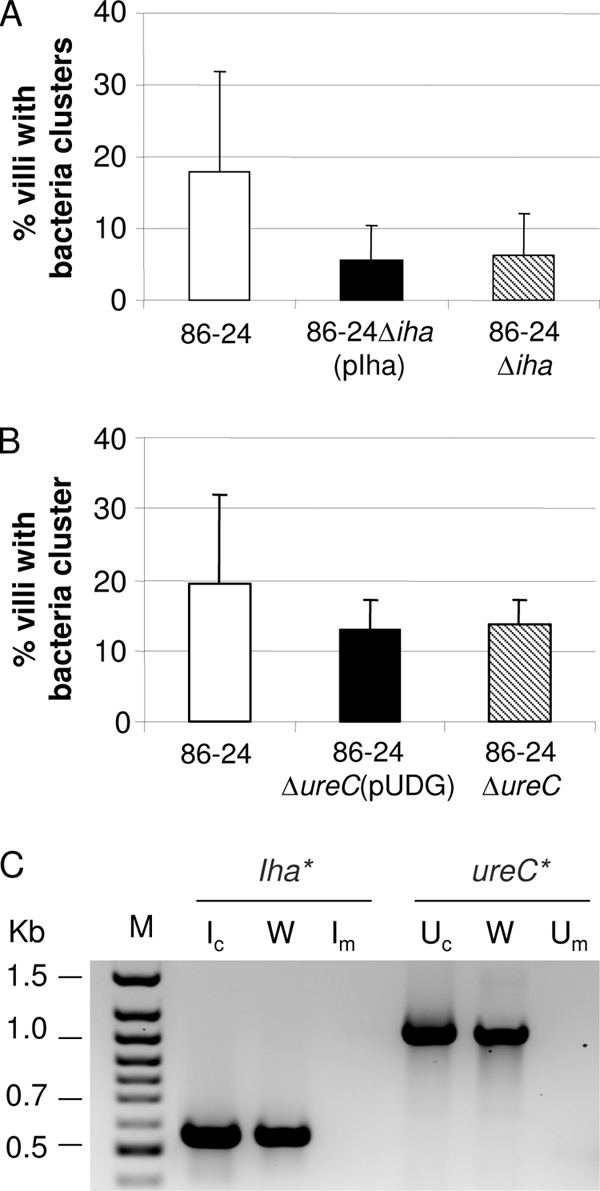
Tests of complemented mutants in pig ileal loops inoculated with bacteria grown in BHIN broth without shaking. Adherence was quantified as the percentage of villi with clusters of ≥5 adherent bacteria per villus (mean + SD). (A) Adherence of wild type (86-24), mutant (86-24Δiha), and complemented mutant [86-24Δiha(pIHA)]. (B) Adherence of wild type (86-24), mutant (86-24ΔureC), and complemented mutant [86-24ΔureC(pUDG)]. (C) Transcripts detected by RT-PCR for complemented mutant strains. Ic, complemented strain 86-24Δiha(pIHA); Im, mutant strain 86-24Δiha; Uc, complemented strain 86-24Δure(pUDG); Um, mutant strain 86-24Δure; W, wild-type EHEC O157:H7 strain 86-24. M, molecular size marker.
Urease activity of the mutant strain 86-24ΔureC, like that of the wild type, was negative on Christensen's urea agar. This is consistent with reports that although ure genes are functional, urease is not expressed in vitro in O157:H7 strains (9). When the plasmid pUDG, carrying a wild-type ure gene cluster and used for complementing the ure-negative mutant, was transformed into E. coli DH5α, it conferred on the bacteria the ability to produce urease, which was detected by Christensen's urea agar. RT-PCR confirmed that the ureC gene transcripts in the complemented mutant strain were produced at a level similar to that of the wild type (Fig. 5C). In pig ileal loops, strain 86-24ΔureC caused adherence in 17.1% ± 10.0% of the villi, which was significantly lower (P = 0.0129) than the 35.9% ± 8.6% for the wild-type parent strain 86-24 (Table 3). Complemented mutant strain 86-24ΔureC(pUDG), tested in ileal loops of four pigs in separate experiments, did not restore the frequency of adherence to that of the wild-type organism (Fig. 5B).
Contribution of mutants to fluid accumulation in ligated pig ileal loops.
In comparison with the wild type, the iha deletion mutant was associated with a nonsignificant decrease in average fluid accumulation (0.5 ± 0.5 ml/loop for the mutant versus 3.9 ± 2.7 ml/loop for the wild type). The mutants 86-24Δter, 86-24Δaid48, and 86-24ΔureC and the control strain 86-24ΔescN also showed no significant change in average fluid accumulation in the ileal loops (data not shown).
DISCUSSION
The role of Ter genes in EHEC O157:H7 infection remains unknown. Ter genes are present in most EHEC O157:H7 strains (2, 18) and are hypothesized to assist EHEC O157:H7 in establishing infection. In the present study, the mutant strain 86-24Δter adhered less to the epithelial cells and formed smaller clusters than the wild-type parent. These results support the hypothesis that Ter plays a role in EHEC O157:H7 infection by enhancing attachment of the organism to the host epithelial cells. The Ter genes might encode an adhesin or promote attachment via enhancing activities of existing adhesins or receptors. Although the Ter gene cluster has no homology with known adhesins (27, 31), products of the Ter genes may control the expression of adhesins or host receptors. Interestingly, plasmid R478 in S. marcescens contains a Ter locus and an Iha homolog colicin I receptor and confers on E. coli J62 the ability to attach to cultured human epithelial cells (14). Alternatively, the Ter genes may have additional functions that are not yet recognized (27, 28), such as enhancing survival of the organisms in the host (29) or indirectly affecting LEE activity.
In the pig ileal loops, the ter-null mutation reduced the ability of EHEC O157:H7 to adhere to the intestinal epithelium although the difference was not significant due to the large pig-to-pig variation. Genetic background or differences in intestinal microbiota of these pigs might contribute to this variation. Studies in gnotobiotic- or antibiotic-treated pigs may be needed to reduce the variation. Taken together, these data support a role of Ter in EHEC O157:H7 infection. The Ter gene cluster is absent from most sorbitol-fermenting O157:NM strains (2, 18), but the role of the Ter genes in adherence in these bacteria may be replaced by other genes. For example, the Ter phenotype is present in the sorbitol-fermenting O157:NM strain 4180/97, which lacks ter genes (2, 18), implying that genes responsible for Ter may be redundant in strains of this serotype.
Iha was identified by Tarr et al. (25) as an adherence-conferring protein of EHEC O157:H7, and it was later demonstrated that iha of UPEC strain CFT073 (O6:K2:H1) is a urovirulence factor (10). The iha of EHEC O157:H7 displays near identity to the UPEC strain CFT073 iha (8, 10, 11). Whether iha functions as a virulence factor in EHEC O157:H7 is unknown. In the present study, 86-24Δiha had a significantly (P < 0.05) reduced capacity to adhere to pig enterocytes, supporting the hypothesis that Iha is a virulence factor and functions as an adhesin. Iha is also a catecholate siderophore receptor in UPEC (12) and may therefore be a dually functional virulence factor of pathogenic E. coli. This provides an advantage to the bacteria for growth and colonization in the iron-restricted milieu of the host intestine by increasing the uptake of iron and promoting in vivo persistence. The iha deletion mutant adhered to HEp-2 and IPEC-J2 cells as well as the wild type in the present in vitro study, which might be explained by the fact that iha is iron regulated and shows decreased expression in rich medium (10, 22). Growth in BHI broth for the adhesion assay might have compromised Iha expression and therefore diminished the role of iha in adherence in vitro. Failure of complementation of the iha deletion mutant might be due to the toxicity of Iha (10) produced by the plasmid, which was supported by our observation that the complemented mutant 86-24Δiha(pIHA) caused even less adherence than the mutant 86-24Δiha to IPEC-J2 cells (data not shown).
Urease in O157:H7 VTEC may contribute to the general metabolic fitness in the human intestine (28), thereby promoting colonization of the intestine. In the present study, a urease-null mutant formed significantly fewer bacterial clusters than the wild-type EHEC O157:H7 parent strain, supporting a role for urease in virulence. Although urease is not expressed in vitro by most VTEC strains (5, 6), it is likely that it is expressed in vivo, thereby contributing to bacterial survival in the intestine (6, 19). This suggestion is supported by our RT-PCR data that show that ureC gene transcripts from the wild-type 86-24 strain present in pig ileal loops were detected at levels similar to those of the bacteria grown in BHIN broth static cultures in vitro (unpublished data). Complementation of the urease-null mutant by introduction of the wild-type ure gene cluster on a plasmid failed to restore the wild-type adherence phenotype in the ligated pig ileal loops. It is possible that regulation of urease synthesis might have prevented the plasmid-encoded urease from expression in the host intestine as regulation of ure expression appears complicated by unknown trans-acting factors (9, 19). On the other hand, it might be the negative effect from overexpression of urease in the complemented strain as it was also observed that the complemented mutant 86-24ΔureC(pUDG) caused even less adherence to IPEC-J2 cells than its parent mutant (data not shown).
The deletion mutant 86-24ΔaidA48 did not appear to contribute to the adherence of O157:H7 strain 86-24 to the cultured cells or to pig intestinal epithelial cells. It is possible that AIDA48 plays only a small role in adherence, and deletion of aidA48 was compensated by the presence of redundant AIDA-like proteins. The LEE PAI, encoding the major adherence mechanism, was intact in all the mutants, and minor effects associated with a single gene deletion might have been difficult to detect unless they affected LEE expression or function. Evaluating mutants with deletions of several genes with potential adherence functions may be one strategy to overcome this difficulty.
In conclusion, the present study demonstrated that the Ter gene cluster is involved in adherence to HEp-2 and IPEC-J2 cells and the formation of large adherent bacterial clusters. Deletion mutation in genes encoding urease and Iha reduced adherence in the ligated pig ileal loops. These data suggest that these genes of OI-48 are involved in adherence of O157:H7 VTEC to epithelial cells.
Acknowledgments
We thank Hai Yu for assistance with statistical analysis and Cornelis Poppe and Abdul Lone for critical reviews of the manuscript.
The research was supported by the Canadian Institutes of Health Research. X.Y. was financially supported by Agriculture and Agri-Food Canada. B.L. and J.Z. were visiting scholars supported by the China Scholarship Council.
Footnotes
Published ahead of print on 24 July 2009.
REFERENCES
- 1.Benson, A. 2003. Molecular and population genetics of virulence traits in Escherichia coli O157:H7, p. 155-166. In M. E. Torrence and R. E. Isaacson (ed.), Microbial food safety in animal agriculture: current topics. Iowa State Press, Ames.
- 2.Bielaszewska, M., P. I. Tarr, H. Karch, W. Zhang, and W. Mathys. 2005. Phenotypic and molecular analysis of tellurite resistance among enterohemorrhagic Escherichia coli O157:H7 and sorbitol-fermenting O157:NM clinical isolates. J. Clin. Microbiol. 43:452-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cravioto, A., R. J. Gross, S. M. Scotland, and B. Rowe. 1979. An adhesive factor found in strains of Escherichia coli belonging to traditional infantile enteropathogenic serotypes. Curr. Microbiol. 3:95-99. [Google Scholar]
- 4.Edwards, R. A., L. H. Keller, and D. M. Schifferli. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149-157. [DOI] [PubMed] [Google Scholar]
- 5.Friedrich, A. W., R. Kock, M. Bielaszewska, W. Zhang, H. Karch, and W. Mathys. 2005. Distribution of the urease gene cluster among and urease activities of enterohemorrhagic Escherichia coli O157 isolates from humans. J. Clin. Microbiol. 43:546-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedrich, A. W., R. Lukas, A. Mellmann, R. Kock, W. Zhang, W. Mathys, M. Bielaszewska, and H. Karch. 2006. Urease genes in non-O157 Shiga toxin-producing Escherichia coli: mostly silent but valuable markers for pathogenicity. Clin. Microbiol. Infect. 12:483-486. [DOI] [PubMed] [Google Scholar]
- 7.Gunzer, F., U. Bohn, S. Fuchs, I. Mühldorfer, J. Hacker, S. Tzipori, and A. Donohue-Rolfe. 1998. Construction and characterization of an isogenic slt-ii deletion mutant of enterohemorrhagic Escherichia coli. Infect. Immun. 66:2337-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guyer, D. M., J. S. Kao, and H. L. Mobley. 1998. Genomic analysis of a pathogenicity island in uropathogenic Escherichia coli CFT073: distribution of homologous sequences among isolates from patients with pyelonephritis, cystitis, and catheter-associated bacteriuria and from fecal samples. Infect. Immun. 66:4411-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heimer, S. R., R. A. Welch, N. T. Perna, G. Posfai, P. S. Evans, J. B. Kaper, F. R. Blattner, and H. L. Mobley. 2002. Urease of enterohemorrhagic Escherichia coli: evidence for regulation by fur and a trans-acting factor. Infect. Immun. 70:1027-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, J. R., S. Jelacic, L. M. Schoening, C. Clabots, N. Shaikh, H. L. Mobley, and P. I. Tarr. 2005. The IrgA homologue adhesin Iha is an Escherichia coli virulence factor in murine urinary tract infection. Infect. Immun. 73:965-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kao, J. S., D. M. Stucker, J. W. Warren, and H. L. Mobley. 1997. Pathogenicity island sequences of pyelonephritogenic Escherichia coli CFT073 are associated with virulent uropathogenic strains. Infect. Immun. 65:2812-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leveille, S., M. Caza, J. R. Johnson, C. Clabots, M. Sabri, and C. M. Dozois. 2006. Iha from an Escherichia coli urinary tract infection outbreak clonal group A strain is expressed in vivo in the mouse urinary tract and functions as a catecholate siderophore receptor. Infect. Immun. 74:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ménard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mignatti, P., L. Pagani, M. Perduca, and E. Romero. 1985. R factor-mediated adhesiveness to mammalian cells in E. coli K12. Microbiologica 8:101-111. [PubMed] [Google Scholar]
- 15.Mobley, H. L., M. D. Island, and R. P. Hausinger. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59:451-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy, K. C., and K. G. Campellone. 2003. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orth, D., K. Grif, M. P. Dierich, and R. Wurzner. 2006. Prevalence, structure and expression of urease genes in Shiga toxin-producing Escherichia coli from humans and the environment. Int. J. Hyg. Environ. Health 209:513-520. [DOI] [PubMed] [Google Scholar]
- 18.Orth, D., K. Grif, M. P. Dierich, and R. Wurzner. 2007. Variability in tellurite resistance and the ter gene cluster among Shiga toxin-producing Escherichia coli isolated from humans, animals and food. Res. Microbiol. 158:105-111. [DOI] [PubMed] [Google Scholar]
- 19.Parreira, V. R., J. H. Liao, S. H. Kim, and C. L. Gyles. 2008. A homolog of the O157 urease-encoding O island 48 is present in porcine O149:H10 enterotoxigenic Escherichia coli. Vet. Res. 39:38. [DOI] [PubMed] [Google Scholar]
- 20.Perna, N. T., J. D. Glasner, V. Burland, and G. Plunkett III. 2002. The genomes of Escherichia coli K-12 and pathogenic E. coli, p. 3-53. In M. S. Donnenberg (ed.), Escherichia coli: virulence mechanisms of a versatile pathogen. Academic Press, San Diego, CA.
- 21.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 22.Rashid, R. A., P. I. Tarr, and S. L. Moseley. 2006. Expression of the Escherichia coli IrgA homolog adhesin is regulated by the ferric uptake regulation protein. Microb. Pathog. 41:207-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt, H., W. L. Zhang, U. Hemmrich, S. Jelacic, W. Brunder, P. I. Tarr, U. Dobrindt, J. Hacker, and H. Karch. 2001. Identification and characterization of a novel genomic island integrated at selC in locus of enterocyte effacement-negative, Shiga toxin-producing Escherichia coli. Infect. Immun. 69:6863-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 25.Tarr, P. I., S. S. Bilge, J. C. Vary, Jr., S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarr, P. I., M. A. Neill, C. R. Clausen, J. W. Newland, R. J. Neill, and S. L. Moseley. 1989. Genotypic variation in pathogenic Escherichia coli O157:H7 isolated from patients in Washington, 1984-1987. J. Infect. Dis. 159:344-347. [DOI] [PubMed] [Google Scholar]
- 27.Taylor, D. E. 1999. Bacterial tellurite resistance. Trends Microbiol. 7:111-115. [DOI] [PubMed] [Google Scholar]
- 28.Taylor, D. E., M. Rooker, M. Keelan, L. K. Ng, I. Martin, N. T. Perna, N. T. Burland, and F. R. Blattner. 2002. Genomic variability of O islands encoding tellurite resistance in enterohemorrhagic Escherichia coli O157:H7 isolates. J. Bacteriol. 184:4690-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vavrova, S., D. Valkova, H. Drahovska, J. Kokavec, J. Mravec, and J. Turna. 2006. Analysis of the tellurite resistance determinant on the pNT3B derivative of the pTE53 plasmid from uropathogenic Escherichia coli. Biometals 19:453-460. [DOI] [PubMed] [Google Scholar]
- 30.Vlisidou, I., M. Lyte, P. M. van Diemen, P. Hawes, P. Monaghan, T. S. Wallis, and M. P. Stevens. 2004. The neuroendocrine stress hormone norepinephrine augments Escherichia coli O157:H7-induced enteritis and adherence in a bovine ligated ileal loop model of infection. Infect. Immun. 72:5446-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walter, E. G., and D. E. Taylor. 1992. Plasmid-mediated resistance to tellurite: expressed and cryptic. Plasmid 27:52-64. [DOI] [PubMed] [Google Scholar]