Abstract
Methods to assess the impact of goose fecal contamination are needed as the result of the increasing number of Canada geese (Branta canadensis) near North American inland waters. However, there is little information on goose fecal microbial communities, and such data are important for the development of host-specific source-tracking methods. To address this issue, 16S rRNA gene clone libraries for Canada goose fecal samples from Ontario, Canada, and Ohio were analyzed. Analyses of fecal clones from Ontario (447) and Ohio (302) showed that goose fecal communities are dominated by the classes “Clostridia” (represented by 33.7% of clones) and “Bacilli” (38.1% of clones) and the phylum “Bacteroidetes” (10.1% of clones). Sequences not previously found in other avian fecal communities were used to develop host-specific assays. Fecal DNA extracts from sewage plants (10 samples) and different species of birds (11 samples) and mammals (18 samples) were used to test for host specificity. Of all the assays tested, one assay showed specificity for Canada goose fecal DNA. The PCR assay was positive for Canada goose fecal DNA extracts collected from three locations in North America (Ohio, Oregon, and Ontario, Canada). Additionally, of 48 DNA extracts from Lake Ontario waters presumed to be impacted by waterfowl feces, 19 tested positive by the assay, although 10 were positive only after a nested PCR approach was used. Due to the level of host specificity and the presence of signals in environmental waters, the assay is proposed as a part of the toolbox to detect Canada goose contamination in waterfowl-contaminated waters.
Canada goose (Branta canadensis) is one of the most common waterfowl species in inland water areas in North America, especially around the Great Lakes. Indeed, Canada goose populations in the United States have been increasing in the last decade (29). Feces produced by geese congregating around surface water bodies are considered a potential source of fecal bacteria in reservoirs that supply drinking water and in recreational waters (4). More importantly, microbiological studies of Canada geese have shown the incidence of pathogenic bacteria such as Salmonella spp. (19), Escherichia coli (1, 21), and Vibrio spp. (30) and pathogenic protozoa such as Giardia sp. and Cryptosporidium parvum (15, 20).
While the potential impact of waterfowl fecal contamination on public health is relevant to beach closures and zoonosis, unfortunately to date there are no methods that can specifically trace goose fecal pollution in environmental waters. The lack of detection methods is due in part to the limited information on waterfowl fecal microbial community composition, most of which has been obtained using culture-based methods. For example, Hollander (17) studied intestinal microbiota of various wintering goose species and found that the majority of the isolated bacteria belonged to the genera Bacillus and Pseudomonas, while enterobacteria and streptococci were less common. Since culture-based studies can provide only a limited picture of natural microbial communities, it is necessary to rely on alternate methods like the sequence analysis of 16S rRNA gene clone libraries. Thus far, there is no information on the molecular diversity of Canada goose fecal microbiota, although it has been hypothesized that waterfowl gut microbial communities are different from other gut communities (13).
In order to understand the public health impact of Canada goose fecal pollution in natural waters, it is essential to have adequate detection methods. Recently, microbial source-tracking methods based on Bacteroides-Prevotella 16S rRNA gene sequences have been used to differentiate human and cow from other animal fecal sources (2, 22). These methods have taken advantage of the fact that “Bacteroidetes” are abundant in mammalian fecal samples. Since “Bacteroidetes” are less abundant in some avian species, approaches for detecting chicken and gull fecal pollution in water have been based on alternate targets, such as members of the classes “Clostridia” and “Bacilli” (25, 26). The aims of this study were to provide a description of the microbial community composition in goose feces by using 16S rRNA gene sequence analysis and to develop host-specific PCR assays to detect Canada goose fecal pollution in environmental waters.
MATERIALS AND METHODS
Sample collection and DNA extraction.
Fecal samples from Canada geese were collected in Ohio (n = 12), Oregon (n = 3), and Ontario, Canada (n = 24). Water samples were collected from goose-contaminated beaches in Toronto and Hamilton on Lake Ontario. Droppings from healthy animals were aseptically collected immediately after defection by the animals, placed into sterile centrifuge tubes, and stored at −80°C until being used in DNA extractions. Total DNA was extracted from all fecal samples collected in Ontario, Ohio, and Oregon by using the FastDNA kit according to the instructions of the manufacturer (Qbiogene, Irvine, CA) and then eluted in 80 μl of water. The concentration of each fecal DNA extract was measured using an ND-1000 UV-vis spectrophotometer (NanoDrop Technologies, Inc., Berlin, Germany), and aliquots containing equal DNA amounts from each individual fecal sample were mixed to generate a DNA composite of fecal samples collected from each site.
16S rRNA gene clone libraries, DNA sequencing, and data analysis.
Sequence analysis of 16S rRNA gene products was used to describe the phylogenetic affiliations of goose fecal bacterial populations. The 16S rRNA gene was amplified using bacterial primers 27F (positions 8 to 27 [E. coli numbering], 5′-AGAGTTTGATCMTGGCTCAG-3′) and 926R (positions 926 to 945 [E. coli numbering], 5′-CCGTCAATTC[A/C]TTT[A/G]AGTTT-3′) and composite DNA as the template. PCR amplifications were performed in a PTC-240 DNA Engine Tetrad 2 cycler (MJ Research, Inc. Alameda, CA) using the cycling conditions described previously (25). PCR products from five independent reactions were pooled and cloned into pCR4.1 TOPO (Invitrogen). Entire individual clones were sequenced using M13 forward and reverse primers as described by Lu et al. (25). Sequencing was carried out by automated methods using BigDye terminator chemistry and an Applied Biosystems PRISM 3730XL DNA analyzer at the Cincinnati Children's Hospital Medical Center Genomics Core Facility (Cincinnati, OH).
Sequence editing and alignment were completed using Sequencher (Gene Codes Corporation, Ann Arbor, MI). Homology searches were performed using BLASTn and GenBank's nr database (http://www.ncbi.nlm.nih.gov/BLAST/) as described previously (25). Chimeric sequences were identified by employing the Check Chimera program of the Ribosomal Database Project (5) and creating manual alignments of secondary structures. The Bellerophon program (18; http://foo.maths.uq.edu.au/∼huber/bellerophon.pl) was also used to check for chimeras by comparing each sequence against the sequences from the same library. As a final check for chimeras, unique sequences were split into 5′ and 3′ fragments and analyzed separately using BLAST. Sequences for which either the 5′ or 3′ fragment had significantly different closest relatives were considered probable chimeras and were removed from the data set.
Primer design and development of PCR assays.
BLAST results were used to identify sequences that showed less than 90% identity to sequences in GenBank. Closely related sequences, including sequences from other fecal bacteria, were used in alignments to identify unique regions. Sequences were then selected after in silico cross-examination using BLAST. Primers used to develop host-specific assays were designed using Primer Designer (version 2.01) (Scientific and Educational Software, Cary, NC). Prior to host specificity tests, primers were further validated against sequences in GenBank.
Optimal thermal conditions for each primer set were determined using thermal gradients and goose fecal DNA extracts as amplification templates. Assays that produced signals at a wide range of temperatures were used in further studies. The host specificities of the selected assays were evaluated using the same protocol and samples described by Lu et al. (25). In addition, 10 sewage samples from the following locations were included as part of host specificity tests: Waldwick, NJ; Frankfort, KY; West Point, WA; St. Peter, MN; Rutland, VT; Morehead, KY; Buffalo, NY; Milford, OH; Sacramento, CA; and Las Vegas, NV. Assays that showed cross-amplification signals and PCR products of unexpected sizes were not further considered. To confirm the identities of PCR products from Canada goose fecal samples collected in Ontario, clone libraries were developed and analyzed as described above. Promising PCR assays were also tested against individual fecal DNA extracts to determine the distribution of the targeted populations in geese and against DNA extracts from water samples impacted by goose fecal contamination. A nested PCR analysis was also conducted to detect potentially low titers of the targeted sequence. In the nested PCR assays, bacterial primers 27F and 926R (which cover approximately the first 930 positions of the rRNA gene) were first used as described above. An aliquot (2 μl) of the resulting PCR product was diluted 10-fold, and then 2 μl of the dilution was used as the template for the nested PCR assay. The host-specific primers anneal to positions within the first amplification product.
Nucleotide sequence accession numbers.
Representative clone sequences were deposited in GenBank with accession numbers FJ390504 to FJ390840.
RESULTS AND DISCUSSION
Phylogenetic analysis and comparison with other avian fecal microbial communities.
A total of 749 goose fecal 16S rRNA gene sequences (447 from Ontario and 302 from Ohio) were analyzed in this study (Table 1). Most of the sequences clustered within the bacterial classes “Bacilli” (46.3% of those from Ontario and 30.2% of those from Ohio) and “Clostridia” (38.4% of those from Ontario and 28.9% of those from Ohio) (Fig. 1). Twelve genera of “Clostridia” were represented in the clone libraries, although most sequences were closely related to sequences from the Clostridium genus. Among all the clones identified as members of the “Bacilli,” species of the following genera were the most abundant: Lactococcus, Bacillus, Paenibacillus, and Turicibacter. The percentages of Bacillus and Turicibacter sequences in the two different (Ontario and Ohio) libraries were similar. In contrast, most of the Lactococcus sequences were obtained from the Ontario clone library while most of the Paenibacillus sequences were of Ohio origin. Both Lactococcus and Paenibacillus species have been isolated from poultry cecum or feces, so they can be considered normal members of the avian gut microbiota. However, the ecological meaning of the differences in their relative abundance is yet to be determined. Sequences similar to those from Actinobacteria and “Bacteroidetes” were present in samples from both locations, although they were more dominant in the Ohio clone library (accounting for 11.3 and 18.3% of Ohio sequences versus 2.7 and 2.0% of Ontario sequences). Proteobacterial sequences, including sequences closely related to those of potentially pathogenic bacteria such as Campylobacter and Helicobacter spp., were also retrieved from the clone libraries. On average, proteobacterial sequences represented <3% of the total clones analyzed.
TABLE 1.
Distribution of clones obtained in Canada goose fecal clone libraries
| Phylum or class | Division, order, family, or genus | Ontario library
|
Ohio library
|
||
|---|---|---|---|---|---|
| No. of clones | % of total | No. of clones | % of total | ||
| Actinobacteria | Adlercreutzia | 1 | 0.2 | 0 | 0 |
| Arthrobacter | 0 | 0 | 26 | 8.6 | |
| Cryobacterium | 0 | 0 | 1 | 0.3 | |
| Curtobacterium | 1 | 0.2 | 0 | 0 | |
| Frankia | 1 | 0.2 | 0 | 0 | |
| Kineosporia | 1 | 0.2 | 1 | 0.3 | |
| Leifsonia | 0 | 0 | 1 | 0.3 | |
| Microbacterium | 1 | 0.2 | 0 | 0 | |
| Mycobacterium | 1 | 0.2 | 0 | 0 | |
| Olsenella | 1 | 0.2 | 0 | 0 | |
| Patulibacter | 1 | 0.2 | 0 | 0 | |
| Propionibacterium | 1 | 0.2 | 0 | 0 | |
| Rhodoglobus | 1 | 0.2 | 0 | 0 | |
| Sanguibacter | 0 | 0 | 2 | 0.7 | |
| Slackia | 1 | 0.2 | 0 | 0 | |
| Streptomyces | 0 | 0 | 1 | 0.3 | |
| Subtercola | 1 | 0.2 | 2 | 0.7 | |
| “Bacteroidetes” | Bacteroides | 5 | 1.1 | 16 | 5.3 |
| “Bacteroidales” | 0 | 0 | 7 | 2.3 | |
| Chryseobacterium | 1 | 0.2 | 0 | 0 | |
| Flavobacterium | 0 | 0 | 1 | 0.3 | |
| Parabacteroides | 2 | 0.4 | 0 | 0 | |
| Porphyromonas | 0 | 0 | 3 | 1 | |
| Prevotella | 1 | 0.2 | 28 | 9.3 | |
| “Bacilli” | Bacillus | 19 | 4.3 | 18 | 6 |
| Jeotgalibacillus | 1 | 0.2 | 0 | 0 | |
| Lactococcus | 55 | 12.3 | 3 | 1 | |
| Leuconostoc | 6 | 1.3 | 0 | 0 | |
| Paenibacillus | 1 | 0.2 | 20 | 6.6 | |
| Sporosarcina | 1 | 0.2 | 0 | 0 | |
| Streptococcus | 0 | 0 | 1 | 0.3 | |
| Turicibacter | 122 | 27.3 | 49 | 16.2 | |
| Weissella | 1 | 0.2 | 0 | 0 | |
| “Clostridia” | Allisonella | 1 | 0.2 | 0 | 0 |
| Clostridium | 117 | 26.2 | 63 | 20.9 | |
| Clostridiaceae | 8 | 1.8 | 5 | 1.7 | |
| Coprococcus | 0 | 0 | 1 | 0.3 | |
| Clostridiales | 4 | 0.9 | 2 | 0.7 | |
| Dialister | 0 | 0 | 1 | 0.3 | |
| Desulfotomaculum | 2 | 0.4 | 0 | 0 | |
| Eubacterium | 8 | 1.8 | 3 | 1 | |
| Faecalibacterium | 3 | 0.7 | 0 | 0 | |
| “Lachnospiraceae” | 0 | 0 | 1 | 0.3 | |
| Pectinatus | 0 | 0 | 2 | 0.7 | |
| Megamonas | 0 | 0 | 6 | 2 | |
| Peptococcus | 6 | 1.3 | 0 | 0 | |
| Ruminococcus | 11 | 2.5 | 1 | 0.3 | |
| Subdoligranulum | 11 | 2.5 | 2 | 0.7 | |
| Mollicutes | Erysipelothrix | 1 | 0.2 | 0 | 0 |
| “Candidatus Bacilloplasma” | 1 | 0.2 | 0 | 0 | |
| Alphaproteobacteria | Agrobacterium | 1 | 0.2 | 2 | 0.7 |
| Devosia | 1 | 0.2 | 1 | 0.3 | |
| Phyllobacterium | 1 | 0.2 | 0 | 0 | |
| Rhizobium | 0 | 0 | 5 | 1.7 | |
| Rhodobacter | 13 | 2.9 | 0 | 0 | |
| Rhodoplanes | 1 | 0.2 | 0 | 0 | |
| Rhodopseudomonas | 1 | 0.2 | 1 | 0.3 | |
| Sinorhizobium | 1 | 0.2 | 1 | 0.3 | |
| Sphingomonas | 1 | 0.2 | 1 | 0.3 | |
| Betaproteobacteria | Burkholderia | 0 | 0 | 5 | 1.7 |
| Variovorax | 0 | 0 | 1 | 0.3 | |
| Duganella | 0 | 0 | 1 | 0.3 | |
| Epsilonproteobacteria | Campylobacter | 2 | 0.4 | 3 | 1 |
| Helicobacter | 3 | 0.7 | 12 | 4 | |
| Gammaproteobacteria | Acinetobacter | 4 | 0.8 | 0 | 0 |
| Enterobacter | 1 | 0.2 | 0 | 0 | |
| Escherichia | 8 | 1.8 | 0 | 0 | |
| Klebsiella | 1 | 0.2 | 0 | 0 | |
| Pseudomonas | 0 | 0 | 1 | 0.3 | |
| Psychrobacter | 2 | 0.4 | 0 | 0 | |
| Stenotrophomonas | 1 | 0.2 | 0 | 0 | |
| Unknown | Candidate division TM7 | 1 | 0.2 | 0 | 0 |
| Unknown | 4 | 0.9 | 0 | 0 | |
| Total | 447 | 302 | |||
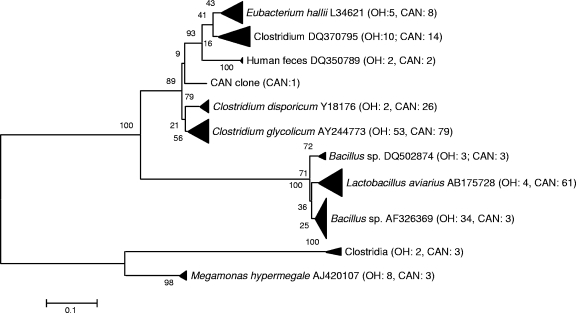
FIG. 1.
Unrooted neighbor-joining tree of 16S rRNA gene sequences from low-G+C-content gram-positive bacteria, obtained from clone libraries. Sequences were aligned and a bootstrap consensus tree was created using MEGA 3.1. The values along branches indicate the percent confidence. The numbers in parentheses indicate numbers of sequences analyzed. CAN, Canada; OH, Ohio.
To further understand the avian fecal microbial community, the sequences obtained in this study were compared to published data from 16S rRNA gene fecal and intestinal clone libraries from gulls (26), chickens (14, 23, 24, 39), and turkeys (27, 31) (Table 2). A total of 11 classes (22 orders) were identified. On average, the majority of the sequences were related to those from “Clostridia” (39.4%), “Bacilli” (24.1%), and “Bacteroidetes” (16.8%). The highest average percentage of clone sequences similar to those from “Clostridia” (i.e., 57.0%) was found in chicken cecum specimens (Table 2), with the dominant sequences being similar to sequences from Clostridium and Ruminococcus spp. On average, sequences similar to those from “Bacilli” were most abundant in waterfowl: Canada goose (37.8%) and gull (37.1%). Based on average results, sequences similar to those from “Bacteroidetes” represent a smaller fraction of the avian fecal community, particularly in waterfowl (7.1%). This finding is relevant to source tracking, as sequences from “Bacteroidetes” are often used to develop host-specific assays. However, it should be noted that “Bacteroidetes” have been found to be abundant in the gastrointestinal tract of turkeys (31).
TABLE 2.
Diversity of microbial communities in avian intestinal or fecal DNA
| Phylum, (sub)class(es), order, or genusb | % in librarya:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CG1 | CG2 | Gull | Chicken1 | Chicken2 | Chicken3 | Chicken4 | Turkey1 | Turkey2 | Turkey3 | |
| Actinobacteria | 2.9 | 11.3 | 6.4 | 8.3 | 0.1 | 0.1 | 5.5 | |||
| Actinomycetales | 1.8 | 11.3 | 6.4 | 2.1 | 5.2 | |||||
| Coriobacteridae | 0.9 | 6.1 | 0.1 | 0.1 | 0.2 | |||||
| Rubrobacterales | 0.2 | 0 | ||||||||
| “Bacteroidetes” | 2.0 | 18.2 | 1.1 | 5.2 | 17.3 | 4.2 | 2.1 | 55.5 | 61.9 | 0.5 |
| “Bacteroidales” | 1.8 | 17.9 | 1.1 | 5.0 | 17.3 | 4.2 | 30.4 | 42.7 | 0.5 | |
| “Flavobacteriales” | 0.2 | 0.3 | 0 | 0.2 | 0.5 | |||||
| “Bacilli” | 45.5 | 30.1 | 37.1 | 11.3 | 5.1 | 28.7 | 1.3 | 0.9 | 3.0 | 77.9 |
| Bacillales | 31.8 | 28.8 | 1.8 | 3.1 | 3.7 | 0.1 | 0 | 30.9 | ||
| “Lactobacillales” | 13.7 | 1.3 | 35.3 | 8.2 | 5.1 | 24.4 | 0.7 | 3.0 | 47.1 | |
| “Clostridia” | 38.9 | 29.1 | 17.3 | 65.6 | 44.9 | 65.2 | 52.3 | 35.0 | 34.8 | 11.3 |
| Clostridiales | 38.9 | 29.1 | 17.3 | 65.6 | 44.9 | 65.2 | 52.3 | 35.0 | 34.8 | 11.3 |
| “Fusobacteria” | 1.1 | 13.9 | ||||||||
| “Fusobacteriales” | 1.1 | 13.9 | ||||||||
| Mollicutes | 0.4 | 8.8 | 0.1 | |||||||
| Anaeroplasmatales | 0.4 | 8.8 | 0.1 | |||||||
| Alphaproteobacteria | 4.4 | 3.6 | 6.7 | 0.8 | 0.1 | |||||
| Rhizobiales (%) | 1.3 | 3.3 | 2.8 | 0.8 | 0.1 | |||||
| Rhodobacterales | 2.9 | 3.2 | ||||||||
| Sphingomonadales | 0.2 | 0.3 | 0.7 | |||||||
| Betaproteobacteria | 4.2 | 2.3 | 4.3 | 0.7 | 0.6 | 4.2 | 1.0 | |||
| Burkholderiales | 4.2 | 2.3 | 3.9 | 0.7 | 4.2 | 1.0 | ||||
| Rhodocyclales | 0.4 | 0.6 | 0 | |||||||
| Deltaproteobacteria | 0.4 | 3.1 | 0.4 | 0.8 | ||||||
| Desulfovibrionales | 0.4 | 3.1 | 0.4 | 0.8 | ||||||
| Epsilonproteobacteria | 1.1 | 5.0 | 0.4 | |||||||
| Campylobacterales | 1.1 | 5.0 | 0.4 | |||||||
| Gammaproteobacteria | 3.8 | 0.3 | 11.3 | 1.3 | 11.2 | 1.2 | 35.8 | 0.6 | 0.2 | 4.7 |
| Aeromonadales | 0.1 | 1.1 | ||||||||
| Pseudomonadales | 1.4 | 0.3 | 4.9 | 1 | ||||||
| “Enterobacteriales” | 2.2 | 6.4 | 1.3 | 11.2 | 1.2 | 34.5 | 0.6 | 0.1 | ||
| Xanthomonadales | 0.2 | 0.1 | 3.7 | |||||||
| Acinetobacter | 0.1 | |||||||||
| Other classes | 4.7 | 8.4 | 2.4 | |||||||
| Unknown | 1.1 | 3.2 | 18.4 | 7.2 | 10.1 | |||||
CG1, Canada goose fecal library (n = 447) for samples from Toronto, Canada (this study); CG2, Canada goose fecal library (n = 302) for samples from Ohio (this study); gull, gull fecal library (n = 282) for samples from West Virginia (26); chicken1, chicken cecal library (n = 616) for samples from Georgia (24); chicken2, chicken cecal library (n = 98) for samples from Guelph, Canada (14); chicken3, chicken cecal library (n = 164) for samples from Hanoi, Vietnam (23); chicken4, chicken cecal library (n = 1,656) for samples from Delaware (39); turkey1, domestic-turkey cecal library (n = 685) for samples from Iowa (31); turkey2, wild-turkey cecal library (n = 627) for samples from Iowa (31); turkey3, turkey fecal library (n = 382) for samples from Ohio (26).
Phylum and (sub)class names are in bold.
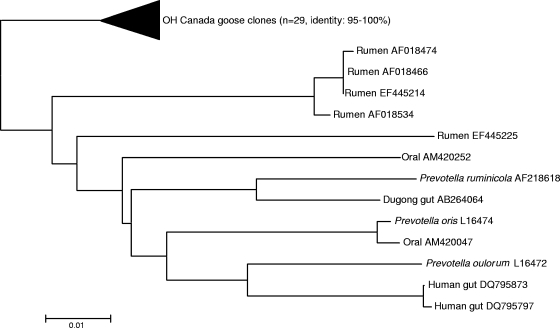
While at the bacterial class level, the compositions of the microbial communities in feces from Canada geese are similar to those in feces from other avian species, there were some differences at the species level. For example, different species of “Bacilli” were dominant in the two waterfowl species studied thus far, Catellicoccus marimammalium in gull and Turicibacter sanguinis in Canada goose. Assays specific to C. marimammalium have been used recently to identify gull fecal contamination in surface waters (26). Although the presence of T. sanguinis in other avian feces has not been reported, similar sequences have been retrieved from swine manure pits and environmental samples. Additionally, Clostridium glycolicum was numerically dominant in the goose feces. In summary, approximately 27% of the Ohio and 41% of the Ontario goose fecal clones were closely related to C. glycolicum and T. sanguinis. Interestingly, in only a few instances have both of these species been isolated from the gastrointestinal tracts of animals. The results from this study suggest that C. glycolicum and T. sanguinis bacteria are among the numerically dominant bacterial populations in goose feces and, therefore, are potential targets for host-specific assays. In contrast, 29 sequences similar to Prevotella sequences (identity, 86 to 90%) were present only in the Ohio clone library. As they formed a unique clade away from sequences retrieved from other gut environments (rumens, human oral tract, dugong gut, and human gut) (Fig. 2), they were also treated as potential targets for source-tracking assay development.
FIG. 2.
Unrooted neighbor-joining tree of 16S rRNA gene sequences (Ohio Canada goose clones including Prevatella-like sequences) compared to other closely related clone library sequences retrieved from GenBank. Sequences were aligned and a bootstrap consensus tree was created using MEGA 3.1.
Design of PCR assays and test for specificity.
Sequences identified as C. glycolicum and T. sanguinis were selected for assay development, as they represented novel sequences that were among the most abundant fecal clones. Additionally, Prevotella-like sequences were selected because they were unique, although they were not as abundant in the clone library as sequences from the two other bacterial species. Sequence alignments identified unique regions for each bacterial target. BLAST searches of potential species-specific sequences confirmed the specificities of the Prevotella group and T. sanguinis primer sequences. In contrast, in silico tests of the selected C. glycolicum primer sequences indicated the presence of sequences from environmental clones retrieved from several fecal sources, suggesting that these may not be good targets for host-specific studies.
A total of eight primers sets were designed (see Table S1 in the supplemental material). Five were developed to target Prevotella sequences from geese, and three were developed to detect T. sanguinis. The T. sanguinis assays showed cross-amplification with pig, cow, chicken, and human (sewage) feces, suggesting that these assays may not be useful in tracking specific fecal sources. Originally isolated from the blood of a febrile patient with acute appendicitis (3), T. sanguinis has also been found in the insect gut (12, 33), dairy waste (28), and cheese and milk samples (6). However, the results from this study suggest that T. sanguinis is more ubiquitous in the animal gut than previously known. While the T. sanguinis assays developed in this study are not fecal source specific, future studies should be conducted to determine if the occurrence of this species in environmental waters can be used as an indication of fecal pollution events. In general terms, the T. sanguinis assays developed in this study will also help in better understanding the ecology of this fecal bacterial species.
Of the PCR assays designed to target the Prevotella goose sequences obtained in this study, only two were host specific (see Table S1 in the supplemental material). One of the host-specific assays (CG199-Prev f2) showed poor host distribution in the host, which was compatible with the fact that no signals were obtained when the assays were challenged against DNA extracts from water. The CG-Prev f5 PCR assay showed no positive amplification signals from nontarget fecal DNA samples, including 10 sewage samples from different states in the Great Lakes region, further validating the host specificity of this assay (Table 3). Sequence analyses of PCR products from goose fecal samples from Ontario confirmed the identities of fecal PCR signals. Specifically, 29 clones from Ontario fecal DNA extracts were analyzed, all of which were ≥97% identical to the Ohio clones used to develop the host-specific assays.
TABLE 3.
Host specificity of Canada goose assay (CGf5r1) tested against feces from various animals
| Sample source | Location of sample collection | No. of samples tested/no. of different composites tested | No. of samples positive by:
|
|
|---|---|---|---|---|
| Normal PCR | Nested PCR | |||
| Pig | DE | 10/2 | 0 | 0 |
| Cow | WV | 17/3 | 0 | 0 |
| Cow | DE | 11/1 | 0 | 0 |
| Human | WV | 16/3 | 0 | 0 |
| Goat | DE | 10/2 | 0 | 0 |
| Sheep | DE | 11/3 | 0 | 0 |
| Horse | WV | 5/1 | 0 | 0 |
| House cat | WV | 11/1 | 0 | 0 |
| Domestic dog | WV | 13/1 | 0 | 0 |
| Coyote | TX | 10/1 | 0 | 0 |
| Gray squirrel | TX | 4/1 | 0 | 0 |
| Deer | WV | 6/1 | 0 | 0 |
| Possum | TX | 2/1 | 0 | 0 |
| Black vulture | TX | 1/1 | 0 | 0 |
| Raccoon | TX | 1/1 | 0 | 0 |
| Hedgehog | WV | 1/1 | 0 | 0 |
| Bobcat | TX | 1/1 | 0 | 0 |
| Red ape | OH | 1/1 | 0 | 0 |
| Asia elephant | OH | 1/1 | 0 | 0 |
| Turkey | DE | 11/1 | 0 | 0 |
| Turkey | OH | 8/8 | 0 | 0 |
| Pigeon | WV | 2/1 | 0 | 0 |
| Pigeon | OH | 3/3 | 0 | 0 |
| Duck | GA | 21/21 | 0 | 0 |
| Duck | OH | 4/4 | 0 | 0 |
| Chicken | WV | 14/1 | 0 | 0 |
| Penguin | OH | 3/3 | 0 | 0 |
| Parrot | OH | 4/4 | 0 | 0 |
| Dove | OH | 2/2 | 0 | 0 |
| Pelican | OH | 1/1 | 0 | 0 |
| Ibis | OH | 1/1 | 0 | 0 |
| Seagull | WV | 8/1 | 0 | 0 |
| Sewage plant in: | ||||
| Waldwick | NJ | 1/1 | 0 | 0 |
| Frankfort | KY | 1/1 | 0 | 0 |
| West Point | WA | 1/1 | 0 | 0 |
| St. Peter | MN | 1/1 | 0 | 0 |
| Rutland | VT | 1/1 | 0 | 0 |
| Morehead | KY | 1/1 | 0 | 0 |
| Buffalo | NY | 1/1 | 0 | 0 |
| Milford | OH | 1/1 | 0 | 0 |
| Sacramento | CA | 1/1 | 0 | 0 |
| Las Vegas | NV | 1/1 | 0 | 0 |
| Canada goose | OH | 12/1 | 1 | 1 |
| Canada goose | Ontario, Canada | 24/1 | 1 | 1 |
Detection limits and marker distribution in the host for the CG-Prev f5 PCR assay.
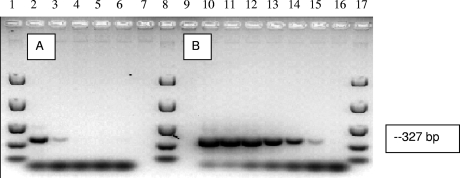
The detection limit of the CG-Prev f5 PCR assay for Canada goose fecal DNA was 2 ng of fecal DNA/reaction. Similar detection limits based on fecal DNA template concentrations have been reported for human- and swine-specific assays (34, 35), although lower detection limits have been reported for chickens (25). Since the detection limits obtained using plasmid constructs (i.e., PCR inserts) showed that the assay is capable of detecting 2 plasmid copies per reaction (Fig. 3), these results suggest that the targeted populations are not dominant members of the microbial community in goose feces. This deduction is consistent with the relatively low abundance of Prevotella-like clones obtained in this study and clones of “Bacteroidetes” in other avian fecal clone libraries (26, 27).
FIG. 3.
Detection limits of the CGf5r1 assay. (A) Lanes: 2 to 5, 10-fold dilutions of Canada goose fecal DNA ranging from 20 to 0.02 ng per PCR assay mixture; 6, negative control. (B) Lanes: 10 to 15, plasmid DNA (10-fold dilutions of plasmid DNA ranging from 2 × 105 to 2 copies per PCR mixture) containing a targeted insert; 16, negative control. Lanes 1, 8, and 17 contain molecular size markers.
Experiments with individual Canada goose fecal samples collected in Ohio and Ontario, Canada (Table 4), showed that the CG-Prev f5 PCR assay produced relatively weak signals from many of the individual samples. Similar results were obtained with three additional samples collected in Oregon. A nested PCR approach using universal primers followed by the species-specific assay was then tested to further determine the occurrence of goose-specific Prevotella (see Fig. S1 in the supplemental material). Approximately 38% of the individual fecal samples were positive by the conventional PCR approach, while half (i.e., 51%) tested positive by the nested approach. Specifically, three times more positive signals from Ohio fecal samples were detected by the nested PCR approach than by normal PCR. This result is in agreement with the fact that only the Ohio clone library contains goose-specific Prevotella-like sequences. Moreover, several Ontario fecal samples as well as all Oregon fecal samples that were negative for the conventional PCR signals were positive by the nested PCR approach. Altogether, these results suggest relatively low abundance of goose-specific Prevotella species in goose feces and potential regional differences in abundance in fecal samples.
TABLE 4.
Performance of Canada goose feces-specific PCR assay with Canada goose fecal and water samples
| Sampling location | No. of samples | Sample type | Normal PCR result (no. of positive signals) | Nested PCR result (no. of positive signals) |
|---|---|---|---|---|
| Mason, OH | 12 | Feces | 3 | 10 |
| Oregon | 3 | Feces | 0 | 3 |
| Ontario, Canada | 24 | Feces | 12 | 6 |
| Ontario, Canada | 48 | Water | 9 | 19 |
Experiments to detect goose-specific PCR signals in waters with a history of waterfowl contamination were also conducted (10, 11). Nearly 19% of the water samples collected in Lake Ontario showed positive PCR signals in the goose-specific assay (Table 4). When a nested PCR approach was used, the total proportion of water samples that tested positive increased to 38%. Evidence of waterfowl fecal contamination at Lake Ontario beaches was detected previously using E. coli antibiotic resistance and repetitive element PCR fingerprinting methods (10, 11). However, these methods were not capable of fully discriminating among waterfowl species. Considering the relatively small populations of goose-specific Prevotella species in fecal samples, the high level of host specificity of the Prevotella marker, and the variable distribution of this marker in Canada goose feces, and assuming that, as anaerobic bacteria, Prevotella-like species are relatively poor survivors in environmental waters, the results from this study suggest that goose feces are an important source of fecal bacteria at some Lake Ontario beaches.
A limited number of host-specific waterfowl assays have been described in the scientific literature. These assays have used different approaches to retrieve host-specific genetic targets. For example, Hamilton et al. (16) used a subtractive hybridization approach to develop goose- and duck-specific macroarray assays based on E. coli gene fragments. By combining different markers, they were able to identify 76 and 73% of goose and duck E. coli isolates, respectively. When these assays were applied to water samples, 51% of the E. coli isolates were classified as being of waterfowl origin. However, these assays showed high degrees of regional specificity and cross-hybridization with approximately 5 to 10% of E. coli strains isolated from human and other animal hosts. Culture-independent methods have also been developed, albeit for a limited number of waterfowl species. Devane et al. (7) developed a nested PCR assay targeting the 16S rRNA gene of a Desulfovibrio sp. strain originally isolated from mallard ducks. The assay was positive for most (76%) of the duck fecal samples tested and for smaller portions (20 and 15%, respectively) of swan and Canada goose fecal samples. Cross-reactivity was detected only with goat fecal samples. Approximately half (i.e., 55%) of the water samples examined were positive in this specific assay, although the correlations between signal detection and total coliforms and E. coli counts were not statistically significant. Another recently developed method to detect gull feces based on 16S rRNA gene sequences targets C. marimammalium (26), a member of the family “Enterococcaceae.” The assay was positive for water samples with known histories of gull contamination but negative for waters impacted by Canadian geese. Metagenomic approaches have also been used to develop PCR assays specific to poultry. While thus far none of the assays are specific to waterfowl, a couple of assays have produced positive signals only with feces from avian species (i.e., chicken, turkey, Canada goose, and pigeon) (25). Bioinformatic analyses suggested that the sequences used for assay development are similar to Bacteroides fragilis and Lactobacillus acidophilus genes encoding proteins. Their potential value in waterfowl source-tracking studies has not been tested, but clearly their application would be restricted to areas in which nonpoultry species are not suspected to be important sources of fecal pollution.
The importance of free-living bird populations as reservoirs for human waterborne pathogens is becoming increasingly evident (19, 21, 32, 36, 38). Due to their migratory character, waterfowl populations can amplify and eventually transmit infectious microbes to humans by directly contaminating agricultural fields or surface waters used for potable water, recreation, or crop irrigation (8). The fact that the microbial composition of waterfowl feces remains poorly studied has become a barrier for developing methods to distinguish goose fecal sources from other animal fecal sources. Recently developed source-tracking methods have provided more direct evidence of the importance of gulls, ducks, and geese in the microbial quality of recreational water, particularly in the Great Lakes (10, 37). Our results suggest that the goose CG-Prev f5 PCR assay developed in this study, in conjunction with a recently developed gull-specific assay (26), can also be used to detect the presence of each waterfowl source without the need for cultivating indicator bacteria or further processing environmental isolates. While these PCR assays may have utility in environmental monitoring, in order to realize their full potential in fecal source-tracking applications, several issues need to be addressed, such as the persistence of target DNA molecules in water, the relevance of the PCR assay to current regulatory fecal indicator methods used to monitor water quality (such as the detection of E. coli and enterococci), and the link between the prevalence of a specific DNA target sequence in the environment and relevant public health risks. The fact that different methods have been based on different bacterial species and different host-specific genetic targets underscores the possibility of developing a robust toolbox for the identification of primary sources of waterfowl fecal pollution that could be used in a variety of environmental scenarios.
Supplementary Material
Acknowledgments
This research was funded in part by a New Start Award from the National Center for Computational Toxicology of the U.S. Environmental Protection Agency to J.W.S.D. and a grant from Environment Canada's STAGE genomics program to T.A.E.
The U.S. Environmental Protection Agency, through its Office of Research and Development, funded and managed, or partially funded and collaborated in, the research described herein. This work has been subjected to the agency's administrative review and has been approved for external publication. Any opinions expressed in this paper are those of the authors and do not necessarily reflect the views of the agency; therefore, no official endorsement should be inferred. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Published ahead of print on 24 July 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alderisio, K. A., and N. DeLuca. 1999. Seasonal enumeration of fecal coliform bacteria from the feces of ring-billed gulls (Larus delawarensis) and Canada geese (Branta canadensis). Appl. Environ. Microbiol. 65:5628-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernhard, A. E., and K. G. Field. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosshard, P. P., R. Zbinden, and M. Altwegg. 2002. Turicibacter sanguinis gen. nov., sp. nov., a novel anaerobic, Gram-positive bacterium. Int. J. Syst. Evol. Microbiol. 52:1263-1266. [DOI] [PubMed] [Google Scholar]
- 4.Cole, D., D. J. V. Drum, D. E. Stallknecht, D. G. White, M. D. Lee, S. Ayers, M. Sobsey, and J. J. Maurer. 2005. Free-living Canada geese and antimicrobial resistance. Emerg. Infect. Dis. 11:935-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delbès, C., L. Ali-Mandjee, and M. C. Montel. 2007. Monitoring bacterial communities in raw milk and cheese by culture-dependent and -independent 16S rRNA gene-based analyses. Appl. Environ. Microbiol. 73:1882-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devane, M. L., B. Robson, F. Nourozi, P. Scholes, and B. J. Gilpin. 2007. A PCR marker for detection in surface waters of faecal pollution derived from ducks. Water Res. 41:3553-3560. [DOI] [PubMed] [Google Scholar]
- 8.Dieter, R. A., Jr., R. S. Dieter, R. A. Dieter III, and G. Gulliver. 2001. Zoonotic diseases: health aspects of Canadian geese. Int. J. Circumpolar Health 60:676-684. [PubMed] [Google Scholar]
- 9.Reference deleted.
- 10.Edge, T. A., and S. Hill. 2007. Multiple lines of evidence to identify the sources of fecal pollution at a freshwater beach in Hamilton Harbour, Lake Ontario. Water Res. 41:3585-3594. [DOI] [PubMed] [Google Scholar]
- 11.Edge, T. A., S. Hill, G. Stinson, P. Seto, and J. Marsalek. 2007. Experience with the antibiotic resistance analysis and DNA fingerprinting in tracking faecal pollution at two lake beaches. Water Sci. Technol. 56:51-58. [DOI] [PubMed] [Google Scholar]
- 12.Egert, M., B. Wagner, T. Lemke, A. Brune, and M. W. Friedrich. 2003. Microbial community structure in midgut and hindgut of the humus-feeding larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae). Appl. Environ. Microbiol. 69:6659-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fogarty, L. R., and M. A. Voytek. 2005. Comparison of Bacteroides-Prevotella 16S rRNA genetic markers for fecal samples from different animal species. Appl. Environ. Microbiol. 71:5999-6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong, J., R. J. Forster, H. Yu, J. R. Chambers, P. M. Sabour, R. Wheatcroft, and S. Chen. 2002. Diversity and phylogenetic analysis of bacteria in the mucosa of chicken ceca and comparison with bacteria in the cecal lumen. FEMS Microbiol. Lett. 208:1-7. [DOI] [PubMed] [Google Scholar]
- 15.Graczyk, T. K., R. Fayer, J. M. Trout, E. J. Lewis, C. A. Farley, I. Sulaiman, and A. A. Lal. 1998. Giardia sp. cysts and infectious Cryptosporidium parvum oocysts in the feces of migratory Canada geese (Branta canadensis). Appl. Environ. Microbiol. 64:2736-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamilton, M. J., T. Yan, and M. J. Sadowsky. 2006. Development of goose- and duck specific DNA markers to determine sources of Escherichia coli in waterways. Appl. Environ. Microbiol. 72:4012-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollander, R. 1982. The aerobic bacterial intestinal flora of various wintering geese species. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 252:394-400. (In German.) [PubMed] [Google Scholar]
- 18.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 19.Hudson, C. R., C. Quist, M. D. Lee, K. Keyes, S. V. Dodson, C. Morales, and J. J. Maurer. 2000. Genetic relatedness of Salmonella isolates from nondomestic birds in the southeastern United States. J. Clin. Microbiol. 38:1860-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kassa, H., B. J. Harrington, and M. S. Bisesi. 2004. Cryptosporidiosis: a brief literature review and update regarding Cryptosporidium in feces of Canada geese (Branta canadensis). J. Environ. Health 66:34-40. [PubMed] [Google Scholar]
- 21.Kullas, H., M. Coles, J. Rhyan, and L. Clark. 2002. Prevalence of Escherichia coli serogroups and human virulence factors in faeces of urban Canada geese (Branta canadensis). Int. J. Environ. Health Res. 12:153-162. [DOI] [PubMed] [Google Scholar]
- 22.Lamendella, R., J. W. Santo Domingo, D. Oerther, J. Vogel, and D. Stoeckel. 2007. Assessment of fecal pollution sources in a small northern-plains watershed using PCR and phylogenetic analyses of Bacteroidetes 16S rDNA. FEMS Microbiol. Ecol. 59:651-660. [DOI] [PubMed] [Google Scholar]
- 23.Lan, P. T. N., H. Hayash, M. Sakamoto, and Y. Benno. 2002. Phylogenetic analysis of cecal microbiota in chicken by the use of 16S rDNA clone libraries. Microbiol. Immunol. 46:371-382. [DOI] [PubMed] [Google Scholar]
- 24.Lu, J., U. Idris, B. G. Harmon, C. Hofacre, J. J. Maurer, and M. D. Lee. 2003. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 69:6816-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu, J., J. W. Santo Domingo, and O. C. Shanks. 2007. Identification of chicken-specific fecal microbial sequences using a metagenomic approach. Water Res. 41:3561-3574. [DOI] [PubMed] [Google Scholar]
- 26.Lu, J., J. W. Santo Domingo, R. Lamendella, T. Edge, and S. Hill. 2008. Phylogenetic diversity and molecular detection of gull feces. Appl. Environ. Microbiol. 74:3969-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu, J., and J. W. Santo Domingo. 2008. Turkey fecal microbial community structure and functional gene diversity revealed by 16S rRNA gene and metagenomic sequences. J. Microbiol. 46:469-477. [DOI] [PubMed] [Google Scholar]
- 28.McGarvey, J. A., W. G. Miller, R. Zhang, Y. Ma, and F. Mitloehner. 2007. Bacterial population dynamics in dairy waste during aerobic and anaerobic treatment and subsequent storage. Appl. Environ. Microbiol. 73:193-202. 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng, J., I. Pavlasek, and U. Ryan. 2006. Identification of novel Cryptosporidium genotypes from avian hosts. Appl. Environ. Microbiol. 72:7548-7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlater, L. K., B. O. Blackburn, R. Harrington, Jr., D. J. Draper, J. Van Wagner, and B. R. Davis. 1981. A non-O1 Vibrio cholerae isolated from a goose. Avian Dis. 25:199-201. [PubMed] [Google Scholar]
- 31.Scupham, A. J., J. Jones, and I. V. Wesley. 2007. Comparison of DNA extraction methods for analysis of turkey cecal microbiota. J. Appl. Microbiol. 102:401-409. [DOI] [PubMed] [Google Scholar]
- 32.Simpson, V. R. 2002. Wild animals as reservoirs of infectious diseases in the UK. Vet. J. 163:128-146. [DOI] [PubMed] [Google Scholar]
- 33.Thongaram, T., Y. Hongoh, S. Kosono, M. Ohkuma, S. Trakulnaleamsai, N. Noparatnaraporn, and T. Kudo. 2005. Comparison of bacterial communities in the alkaline gut segment among various species of higher termites. Extremophiles 9:229-238. [DOI] [PubMed] [Google Scholar]
- 34.Ufnar, J. A., S. Y. Wang, J. M. Christiansen, H. Yampara-Iquise, C. A. Carson, and R. D. Ellender. 2006. Detection of the nifH gene of Methanobrevibacter smithii: a potential tool to identify sewage pollution in recreational waters. J. Appl. Microbiol. 101:44-52. [DOI] [PubMed] [Google Scholar]
- 35.Ufnar, J. A., D. F. Ufnar, S. Y. Wang, and R. D. Ellender. 2007. Development of a swine specific fecal pollution marker based on host differences in methanogen mcrA genes. Appl. Environ. Microbiol. 73:5209-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waldenström, J., S. L. W. On, R. Ottvall, D. Hasselquist, and B. Olsen. 2006. Species diversity of campylobacteria in a wild bird community in Sweden. J. Appl. Microbiol. 102:424-432. [DOI] [PubMed] [Google Scholar]
- 37.Yan, T., M. J. Hamilton, and M. J. Sadowsky. 2007. High-throughput and quantitative procedure for determining sources of Escherichia coli in waterways by using host-specific DNA marker genes. Appl. Environ. Microbiol. 73:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou, L., H. Kassa, M. L. Tischler, and L. Xiao. 2004. Host-adapted Cryptosporidium spp. in Canada geese (Branta canadensis). Appl. Environ. Microbiol. 70:4211-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu, X. Y., T. Zhong, Y. Panya, and R. D. Joerger. 2002. 16S rRNA-based analysis of microbiota from the cecum of broiler chickens. Appl. Environ. Microbiol. 68:124-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.