Abstract
In this study, we evaluated the specificity, distribution, and sensitivity of Prevotella strain-based (PF163 and PigBac1) and methanogen-based (P23-2) PCR assays proposed to detect swine fecal pollution in environmental waters. The assays were tested against 222 fecal DNA extracts derived from target and nontarget animal hosts and against 34 groundwater and 15 surface water samples from five different sites. We also investigated the phylogenetic diversity of 1,340 “Bacteroidales” 16S rRNA gene sequences derived from swine feces, swine waste lagoons, swine manure pits, and waters adjacent to swine operations. Most swine fecal samples were positive for the host-specific Prevotella-based PCR assays (80 to 87%), while fewer were positive with the methanogen-targeted PCR assay (53%). Similarly, the Prevotella markers were detected more frequently than the methanogen-targeted assay markers in waters historically impacted with swine fecal contamination. However, the PF163 PCR assay cross-reacted with 23% of nontarget fecal DNA extracts, although Bayesian statistics suggested that it yielded the highest probability of detecting pig fecal contamination in a given water sample. Phylogenetic analyses revealed previously unknown swine-associated clades comprised of clones from geographically diverse swine sources and from water samples adjacent to swine operations that are not targeted by the Prevotella assays. While deeper sequencing coverage might be necessary to better understand the molecular diversity of fecal Bacteroidales species, results of sequence analyses supported the presence of swine fecal pollution in the studied watersheds. Overall, due to nontarget cross amplification and poor geographic stability of currently available host-specific PCR assays, development of additional assays is necessary to accurately detect sources of swine fecal pollution.
The size of swine farming operations has increased significantly during the last few decades as a result of the high demand for pork products. In fact, pork is now considered the most popular meat worldwide (15). In the United States, the number of large confined swine animal units increased by 3 orders of magnitude from 1982 to 1997 (18), making the swine industry among the top three producers of domesticated animal feces. A direct consequence of this trend is the increase in swine fecal waste, which in turn has raised environmental concerns. When introduced to water, swine fecal waste can present a risk to human health because this waste can harbor a variety of human pathogens (5, 13, 15, 21, 36). The diversity and relatively high frequency of human pathogens in swine feces make swine important reservoirs of zoonotic pathogens. Moreover, the marked increase in the number of large operations has resulted in increased manure production and application in small geographic areas, creating an imbalance between the assimilative capacity of manure-treated farmland and the amount of manure nutrients produced on each farm. This imbalance is evidenced by the 20% increase (from 1982 to 1997) in nitrogen and phosphorus produced in swine operations, thus potentially contributing to the detrimental eutrophication of aquatic ecosystems (18). Swine manure spills and leaks are commonplace in the top hog production states, such as Iowa and North Carolina, due to failure or overflow of manure storage, uncontrolled runoff from open feedlots, improper manure application on cropland, deliberate pumping of manure onto the ground, and intentional breaches in storage lagoons (28, 37).
Recently, swine-associated PCR-based methods targeting members of the “Bacteroidales” order (i.e., Prevotella species) and methanogen populations (12, 29, 35) have been proposed to discriminate swine fecal pollution events from other potential fecal contributions (i.e., human, bovine, and wildlife) to environmental waters. Nevertheless, the value of these assays in reliably detecting fecal pollution sources in watershed-based studies has not been thoroughly investigated. The main goals of this study were to determine host specificity, frequency of detection, and detection limits of currently available swine-associated PCR-based, microbial source tracking assays. To achieve these objectives, assays were tested against swine and nontarget fecal samples, samples from swine manure pits and swine waste lagoons, and water samples presumed to be impacted by swine fecal sources. Furthermore, we investigated the phylogenetic diversity of Bacteroidales 16S rRNA gene sequences derived from some of the aforementioned samples to resolve the level of specificity, relative abundance, and environmental occurrence of Bacteroidales-specific 16S rRNA gene sequences.
MATERIALS AND METHODS
Sample collection.
Fecal (n = 215), manure pit (n = 4), and waste lagoon (n = 3) samples were collected from different sites in Illinois, Nebraska, Ohio, Texas, Delaware, and West Virginia (see Table S1 in the supplemental material). Selection of source material was based on the goal of including as many different animal types as possible to check for host specificity, with emphasis on hosts considered to be important sources of fecal pollution in the United States. Approximately 1.0 to 2.0 g of the fecal material was placed into individual sterile vials and processed as previously described (9, 20). One liter of manure pit and lagoon liquid was collected in autoclaved bottles and transported on ice to the laboratory. To ease filtration, manure pit and lagoon samples were first centrifuged at 8,000 × g for 10 min at 4°C and the supernatant was then filtered onto 0.45-μm polycarbonate filters. DNA extractions were performed for both pellets and filters immediately after the centrifugation and filtration process.
Water samples were collected from multiple sites within two Texas watersheds and three sites in Illinois known to be impacted by fecal pollution sources (see Fig. S1 in the supplemental material). Specifically, water samples from Texas were collected (n = 5) from the Red River basin (TX) in section 207A of Buck Creek (Collingsworth, TX), which is currently on the impaired waters 303(d) list for exceeding fecal bacterial concentrations. Water samples (n = 5) were also collected from Lake Granbury (segment 1205) in the Brazos River basin (TX), which serves as a critical water supply in north Texas and provides water for more than 250,000 customers. Texas water samples (100 ml) were collected, placed on ice for transportation to the laboratory, and filtered within 6 hours of sample collection. Samples were filtered through 0.22-μm-pore-size filters as previously described (22). DNA was extracted using the QIAamp DNA minikit (Qiagen, Valencia, CA) and stored at −80°C until further analyses.
Duplicate water samples were also collected from monitoring wells located on three commercial swine operations in Illinois and surface waters adjacent to these operations, herein described as sites A, C, and E (see Fig. S1 in the supplemental material). Specifically, samples were collected from 16 wells from site A, six wells from site C, and 12 wells from site E. Site A is a 4,000-pig finishing operation that uses a two-stage waste handling system in which a concrete settling basin collects solids prior to the supernatant liquid passively moving into an unlined lagoon. The aim of the two-stage waste handling system is to reduce fecal loading into the lagoon. Site C is a farrowing and nursery operation that houses up to 2,500 sows and that utilizes a single-stage 6-m-deep unlined lagoon to directly collect both feces and urine. Site E is a 2,300-hog finishing facility that uses a concrete-lined pit system for manure storage. Well installation and groundwater sample collection have been previously described (20). Surface water samples were also collected from a groundwater field seep at site C and streams north and south of site A and south of site C.
Groundwater samples (250 ml) from the sites in Illinois were collected in sterile plastic bottles and transported to the laboratory on ice. Samples were centrifuged at 17,700 × g for 20 min at 4°C, and the supernatants were discarded. The pellets were then washed three times with 0.1 volume of phosphate-buffered saline (120 mM NaH2PO4 [pH 8.0], 0.85% NaCl) before extraction of total DNA (20). Three surface water samples (500 ml) were also collected from a stream located less than 100 m from a swine operation housing approximately 200 animals in Loudonville, OH. Samples were stored in ice coolers and transported to the laboratory within 4 hours of collection. Each sample was centrifuged at 3,600 × g for 20 min at 4°C, after which the supernatants were filtered onto 0.22-μm polycarbonate membranes (22). Filters and pellets were stored at −20°C until further processing.
DNA extraction.
Fecal DNA was extracted with the FastDNA spin kit (MP Biomedicals, Inc., Solon, OH) according to the manufacturer's instructions by using 250 μl of each fecal slurry. For Ohio water samples, DNA was extracted directly from whole filters and pellets using the FastDNA spin kit. For Illinois water samples, DNA was extracted from pellets also using the FastDNA spin kit. Total DNA from corresponding filters and pellets was eluted in 100 μl of 10 mM Tris and combined in a sterile tube. DNA was then quantified using a NanoDrop ND-1000 UV spectrophotometer (NanoDrop Technologies, Wilmington, DE). To test for the presence of extraneous DNA contamination introduced during laboratory procedures, extraction blanks (n = 8) were included in the PCR assays. DNA extracts were stored at −20°C until further processing.
PCR assays and limits of detection.
Four different PCR assays, including three assays reported to be swine specific, were tested using DNA extracts from fecal sources and water samples as templates. Two of the swine-associated PCR assays, PF163 and PigBac1, target the 16S rRNA gene of members of the Bacteroidales order, specifically Prevotella spp. (12, 29), while P23-2 targets the methyl coenzyme M reductase (mcrA) gene of methanogenic bacteria (35) (Table 1). Additionally, a general Bacteroidales 16S rRNA gene PCR assay (Bac32) (2) was used to detect potential PCR inhibition and the overall presence of Bacteroidales fecal anaerobic bacteria in water samples and to assess Bacteroidales phylogenetic diversity in different fecal sources and environmental samples impacted by fecal pollution via sequencing studies.
TABLE 1.
Description of primers tested in this study
| Primer name | Primer sequence | Target | Reference |
|---|---|---|---|
| P23-2f | 5′-TCTGCGACACCGGTAGCCATTGA-3′ | mcrA gene of methanogens | 35 |
| P23-2r | 5′-ATACACTGGCGACATTCTTGAGGATTAC-3′ | ||
| Bac32F | 5′-AACGCTAGCTACAGGCTT-3′ | 16S rRNA gene of Bacteroidales | 2 |
| Bac708R | 5′-CAATCGGAGTTCTTCGTG-3′ | ||
| PigBac1f | 5′-CGGGTTGTAAACTGCTTTTATGAAG-3′ | 16S rRNA gene of Prevotella-related group | 29 |
| PigBac1r | 5′-CGCTCCCTTTAAACCCAATAAA-3′ | ||
| PF163 | 5′-GCGGATTAATACCGTATGA-3′ | 16S rRNA gene of Prevotella-related group | 12 |
| Bac708R | 5′-CAATCGGAGTTCTTCGTG-3′ |
Reactions for the general Bacteroidales assay were conducted using the previously described protocol (2). PCR conditions for the PF163 swine-associated assay have not been previously described and were determined as follows: 94°C for 5 min, followed by 35 cycles of 94°C for 20 s, 58°C for 20 s, and 72°C for 30 s, and a final extension step consisting of 72°C for 5 min. The other swine-associated assays, PigBac1 and P23-2, were used as described elsewhere (29, 35). Fecal and water DNA template concentrations used in the PCR assays were adjusted based on published detection limits. Specifically, 0.2 and 1 ng were used for Bac32, 1 and 10 ng were used for the PF163 and PigBac1 assays, and 50 ng was used for the P23-2 reactions, as suggested by the authors who originally designed these assays. Multiple template concentrations were used, as the commonly found levels of the targeted populations for each assay are different. Final PCR solutions (25-μl total volume) contained 2.5 μl of Takara Ex Taq 10× buffer (20 mM Mg2+), 2 μl of deoxynucleoside triphosphate mixture (2.5 mM each), 1 μl of 25% acetamide, 17.5 μl of UltraPure water, 12.5 pmol of each forward and reverse primer, and 0.625 units of Ex Taq DNA polymerase (Takara Mirus Bio., Madison, WI). Reactions were conducted on a DNA Engine 2 Tetrad thermal cycler (Bio-Rad Laboratories, Inc., Hercules, CA). Amplification products were visualized using 1% agarose gels and GelSTAR nucleic acid stain (Cambrex BioScience, East Rutherford, NJ). PCR inhibition was tested in water DNA extracts by using 8F and 787R general bacterial 16S rRNA gene-targeted primer sets, as described by Buchholz-Cleven et al. (4).
The performance of each swine-associated assay was determined in PCR mixtures containing known concentrations of fecal and water DNA extracts. Using this approach, it was possible to determine the detection limits of an assay against environmental extracts (22). PCR assays were performed using templates consisting of serial fecal DNA dilutions (1 × 10−8 to 1 × 10−16 g DNA) of composite fecal samples of swine from different age groups, lagoons, manure pits, and selected water samples yielding positive PCR results.
Cloning and sequencing analyses.
General Bacteroidales (Bac32F/Bac708R) PCR products were used in cloning experiments to qualitatively assess the molecular diversity of Bacteroidales species in different hosts. Sequencing and data analyses were performed as previously described (22). Briefly, PCR products were purified using the QIAquick PCR purification kit according to the manufacturer's instructions (Qiagen, Valencia, CA). Representative PCR products derived from swine feces (feral and domesticated), manure pits, lagoons, and water adjacent to swine farms in Illinois and Ohio were cloned into the pCR4.1 Topo vector as described by the manufacturer (Invitrogen, Carlsbad, CA). Individual Escherichia coli clones were subcultured into 300 μl of Luria broth containing 50 μg/ml ampicillin and screened for inserts using M13 PCR. Clones were submitted to the Children's Hospital DNA Core Facility (Cincinnati, OH) for sequencing using Big Dye sequencing chemistry (Applied Biosystems, Foster City, CA), M13 forward and reverse primers, and an Applied Biosystems Prism 3730XL DNA analyzer. Sequences were manually verified and cleaned using Sequencher 4.7 software (Gene Codes, Ann Arbor, MI). Chimeric sequences detected using Bellerophon (17) were not included in further analyses. Nonchimeric sequences were submitted to Greengenes for alignment using the Nearest Alignment Space Termination algorithm (10, 11). Sequences were also submitted to homology search algorithms to assess sequence similarity to the Greengenes database (1, 10). The distance matrix and phylogenetic tree were generated using ARB software (26). Trees were inferred from 650 sequence positions using neighbor-joining (using a Kimura correction) and maximum parsimony (using the Phylip DNAPARS tool) (26). To statistically evaluate branching confidence, bootstrap values were obtained from a consensus of 100 parsimonious trees using MEGA software (http://www.megasoftware.net). Werenella sp. 16S rRNA gene sequence (accession number AJ234059) was used as the outgroup, while cultured Bacteroidales species were included in the analyses as points of reference.
Statistical analyses and molecular diversity estimates.
The ability of each marker to accurately detect swine feces within a given water sample was determined using Bayes' theorem as described by Kildare et al. with minor modifications (19). Briefly, the posterior probability or P(S|T) that a given pig-specific PCR assay generated a true-positive signal in a water sample was estimated using the following formula:
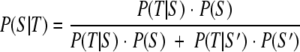 |
where P(T|S) is the proportion of positive signals in swine fecal samples, P(T|S′) is the proportion of positive signals in nonswine fecal samples (false positives in fecal samples), P(S) is the prior probability of swine fecal contamination in a water sample, and P(S′) is the probability that a given water sample is not contaminated with pig feces. P(S|T) was calculated over a range of possible prior probabilities, as in all cases the prior probability of swine fecal contamination was unknown. The posterior probability that each swine-associated marker generated a false-negative result in a water sample was calculated using the same Bayesian framework as that described above, with the exception that P(T|S) is the proportion of false-negative signals in swine fecal samples and P(T|S′) is the proportion of true-negative signals in nonswine fecal samples. Additionally, to understand if the combination of any two assays increased the confidence that swine contamination was present in a water sample, the posterior probability from one assay (e.g., PigBac1) was used as the prior probability of the second assay (e.g., P23-2). A similar approach was used to determine the confidence of using more than two markers, although in this case the posterior probability of two combined assays (e.g., PigBac1) was used as the new prior probability of the third assay (e.g., P23-2).
Molecular diversity analysis and assemblage comparison of clone libraries were performed using DOTUR and SONS software, respectively (31, 32). Specifically, DOTUR was used to place sequences into operational taxonomic units (OTUs); to compute Chao 1 indices, abundance-based coverage estimators, and Shannon and Simpson diversity indices; and to perform rarefaction analyses. These diversity estimates were calculated for lagoon, manure pit, feces, and water sequences to determine if the current level of sequencing performed in this study saturated Bacteroidales diversity and to confirm the level of inclusiveness of the Bacteroidales-based assays. SONS was used to characterize community structure overlap among clone libraries derived from lagoons, manure pits, feces, and water adjacent to swine farms.
Nucleotide sequence accession numbers.
Representative sequences generated in this study have been deposited in the GenBank database under accession numbers FJ596647 to FJ596751.
RESULTS AND DISCUSSION
Detection limits and performance of PCR assays for fecal sources.
With the exception of fecal samples collected from 8-week-old piglets, results from the detection limit experiments indicated that the general Bacteroidales assay had lower detection limits than did the swine-associated markers in fecal and environmental samples (Table 2). This was expected because host-specific populations represent a small group within general bacterial classes (2, 22). The abundance of fecal populations targeted by the PF163 assay changed with the host age, suggesting that physiological and dietary changes can have an impact on the dynamics of these bacterial populations. In contrast, the abundance of the populations targeted by the PigBac1 assay did not change over time. Interestingly, the PF163 and PigBac1 assays target different Prevotella clades. Each Prevotella subgroup might occupy different niches in the swine gut and therefore be involved in different interactions with the host. Because it is not well understood how 16S rRNA gene sequence similarity can predict the level of convergence of genes relevant to survival in the gut environment (25), it is possible that these populations coexist by sharing limited overlapping niches, particularly niches involved in polysaccharide utilization (3), as has been suggested for Bacteroides fragilis and Bacteroides thetaiotaomicron in the human gut (38). The detection limits for the mcrA-based assay were similar for all fecal samples, regardless of age, suggesting no major changes in the abundance of swine-associated methanogen populations. This is in contrast with the general belief that methanogen densities increase with age in pigs and humans (27).
TABLE 2.
Environmental limits of detection for general Bacteroidales and swine-associated PCR assays for DNA extracted from fecal and water samples
| Sample type | Detection limit (g of DNA extract)
|
|||
|---|---|---|---|---|
| General Bacteroidales (Bac32) | Swine-associated Prevotella (PF163) | Swine-associated methanogens (P23-2) | Swine-associated Prevotella (PigBac1) | |
| Pig fecal (8 wk)a | 1 × 10−15 | 1 × 10−15b | 1 × 10−8 | 1 × 10−12c |
| Pig fecal (6 mo) | 1 × 10−15 | 1 × 10−13 | 1 × 10−8 | 1 × 10−12 |
| Pig fecal (3 to 5 yr) | 1 × 10−15b | 1 × 10−12 | 1 × 10−8 | 1 × 10−12 |
| Lagoon (site A) | 1 × 10−12 | 1 × 10−9 | 5 × 10−7d | >1 × 10−8e |
| Lagoon (site C) | 1 × 10−14 | 1 × 10−9 | 1 × 10−8 | >1 × 10−8e |
| Manure pit (site E) | 1 × 10−12 | 1 × 10−8 | >1 × 10−7d | >1 × 10−8e |
| Water (site A9) | 1 × 10−10 | 1 × 10−9 | >1 × 10−7d | >1 × 10−8e |
| Water (site C, south stream) | 1 × 10−9 | 1 × 10−9 | 1 × 10−7 | >1 × 10−8e |
| Water (site E8) | 1 × 10−14 | 1 × 10−8 | >1 × 10−7d | >1 × 10−8e |
| Water (site C2) | 1 × 10−12 | 1 × 10−8 | 5 × 10−7 | >1 × 10−8e |
| Water (Lake Granbury, site 18015) | 1 × 10−10 | 1 × 10−9 | >1 × 10−7d | 1 × 10−9 |
| Water (Buck Creek, site 10A) | 1 × 10−11 | 1 × 10−9 | >1 × 10−7d | >1 × 10−8e |
For pig fecal DNA detection limits, four fecal samples from each age class (i.e., 8 weeks, 6 months, and 3 to 5 years) were pooled.
Detection limit for the duplicate sample was 1 × 10−14 grams of DNA.
Detection limit for the duplicate sample was 1 × 10−13 grams of DNA.
Detection limit was more than 1 × 10−7 grams of DNA extract.
Detection limit was more than 1 × 10−8 grams of DNA extract.
In most cases, the detection limits for all assays were lower in swine feces than in lagoons and manure pit samples, suggesting the poor survival of these fecal anaerobic bacterial populations during waste management practices. The PF163 assay had the lowest detection limits for most fecal, lagoon, and manure samples. The PigBac1 assay was the least sensitive for both lagoon and manure samples. Specifically, when swine fecal DNA extracts were used as PCR templates, the methanogen assay (P23-2) was at least 4 orders of magnitude less sensitive than each of the other two host-specific assays. This is not surprising, as methanogens have been reported to be 3 to 4 orders of magnitude less prevalent than the total number of anaerobic bacteria in pigs and humans (34). However, when the lagoon samples were tested, the difference between the methanogen assay and the PF163 assay was only 1 to 1.5 orders of magnitude. Overall, these results may suggest differences in survival rates between different host-specific bacterial groups in manure pits and lagoons. As these are the most likely swine pollution sources, these results have practical implications when selecting specific assays for source tracking studies. For example, differences in detection limits among pig-associated assays may suggest that pig-associated populations prevalent in lagoons are different than those prevalent in manure pits. Thus, targeting these different manure pit and lagoon populations may enhance the usefulness of these PCR-based assays as risk assessment tools and in estimating fecal load rates for total maximum daily loads. Our data are consistent with differential distribution among host populations and differential survivorship with respect to the fate and transport of the marker targets as noted by others (23).
With the exception of raccoon feces, of which only a third of the individual samples were positive, most fecal sources produced positive signals with the general fecal Bacteroidales assay (Table 3 ). Differences were noted, however, in host distribution and geographical stability for each of the swine-associated assays. For example, PigBac1 and PF163 assays were positive for at least 80% of swine fecal samples, while the methanogen-based PCR assay (i.e., P23-2) yielded positive results in only 53% of swine fecal samples. Specifically, the methanogen assay performed poorly in fecal samples derived from Ohio and Texas, perhaps due to differences in animal husbandry practices used at these locations (Table 3; see also Table S1 in the supplemental material). Moreover, while all swine manure pit and lagoon DNA extracts (n = 6) yielded positive signals with the PF163 assay, only one sample was positive with either the PigBac1 or the P23-2 assay. Altogether, these data demonstrate that PF163 was the most frequently detected of the targets tested in this study.
TABLE 3.
Specificity of general Bacteroidales and swine-associated PCR markers using fecal DNA extracts
| Fecal type (origin) | % Positive PCR resultsa (no. of positive PCR results/total no. of source type samples tested)
|
|||
|---|---|---|---|---|
| General Bacteroidales (Bac32) | Pig Prevotella (PF163) | Pig methanogen (P23-2) | Pig Prevotella (PigBac1) | |
| Pig feces (DE) | 100 (9/9) | 44 (4/9) | 78 (7/9) | 44 (4/9) |
| Pig feces (OH) | 100 (52/52) | 98 (51/52) | 42 (22/52) | 81 (42/52) |
| Pig feces (TX) | 100 (7/7) | 100 (7/7) | 29 (2/7) | 100 (7/7) |
| Pig feces (TX) | 100 (9/9) | 89 (8/9) | 22 (2/9) | 100 (9/9) |
| Pig feces (WV) | 90 (18/20) | 70 (14/20) | 90 (18/20) | 90 (18/20) |
| Cattle (WV) | 100 (20/20) | 40 (9/20) | 5 (1/20) | 10 (2/20) |
| Pig manure pits (OH) | 100 (3/3) | 100 (3/3) | 0 (0/3) | 33 (1/3) |
| Pig manure pit (IL) | 100 (1/1) | 100 (1/1) | 0 (0/1) | 0 (0/1) |
| Pig lagoons (IL) | 100 (2/2) | 100 (2/2) | 0 (0/2) | 0 (0/2) |
| Human feces (WV) | 100 (10/10) | 30 (3/10) | 30 (3/10) | 60 (6/10) |
| Chicken (DE) | 88 (7/8) | 50 (4/8) | 38 (3/8) | 63 (5/8) |
| Raccoon (NE) | 34 (23/68) | 4 (3/68) | 1 (1/68) | 29 (20/68) |
| Horse (WV) | 100 (12/12) | 67 (8/12) | 0 (0/12) | 50 (6/12) |
| Cattle lagoon (OH) | 100 (1/1) | 0 (0/1) | 0 (0/1) | 0 (0/1) |
Percent positive PCR results using a given marker on a given source type. Diagnostic specificity, the number of nonpig fecal source samples that produce negative PCR results divided by the total number of nonpig fecal source samples tested (n = 119), was 0.77, 0.93, and 0.67 for pig Prevotella (PF163), pig methanogen (P23-2), and pig Prevotella (PigBac1), respectively. Diagnostic sensitivity, the number of pig source samples testing positive divided by the total number of pig source samples tested (n = 103), was 0.87, 0.49, and 0.79 for pig Prevotella (PF163), pig methanogen (P23-2), and pig Prevotella (PigBac1), respectively.
While PF163 and PigBac1-like populations were present in most pig fecal samples, the host specificity tests showed higher false-positive rates for these assays than for the methanogen-based assay. For example, false-positive signals were obtained for several nontarget hosts, including horse, human, and chicken fecal DNA extracts (Table 3). Gourmelon et al. (14) reported cross-reactivity for the PF163 assay, particularly when using chicken fecal DNA templates. These data indicate that these host-specific Prevotella populations are also present in the feces of animals other than swine, and therefore, assays based on these specific populations are prone to introducing false-positive signals when environmental samples are analyzed.
PCR assay performance with water samples adjacent to three swine farms in Illinois.
Water samples collected in Illinois were positive at a relatively high frequency (i.e., 40 to 78%) by the general Bacteroidales assay (Table 4; see also Table S2 in the supplemental material), suggesting the presence of fecal contamination. Considering that all Illinois sampling stations were adjacent to swine farms and no other domesticated animal practices are known to occur near these sampling stations, it is assumed that swine are the primary source of fecal contamination in these sites. Indeed, water samples tested positive for the swine-associated assays at many of the sites, although at a relatively low frequency. For example, at site A the general Bacteroidales marker produced positive signals in 40 to 45% of the different water DNA extracts tested, while the host-specific assays produced positive PCR signals for only 5 to 20% of the samples. Of each of the swine-associated markers, the methanogen-targeted P23-2 assay yielded the lowest frequency of detection, with a positive PCR result in only 0 to 22% of the water sample DNA extracts from the three Illinois sampling locations. This may be explained by the lower abundance of methanogens than of Bacteroidales isolates found in the pig gut (6) and by the fact that managed swine fecal waste might select for different methanogenic populations (35). Interestingly, all three markers produced the highest proportion of positive pig-associated signals at site C (22 to 56%), compared to site A (0 to 20%) and site E (0 to 38%), which may be explained by the different waste handling strategies employed at each farm. The two-stage waste handling system used at site A may have resulted in reduced fecal loading into the shallow (1.5-m) lagoon, supporting our finding of lower proportions of general and pig-associated signals at site A. Additionally, few positive pig signals were observed at site E, which may be explained by the use of a concrete-lined manure pit, limiting the direct flow of waste into groundwater at this site. In contrast, site C had the highest frequency of fecal and pig-associated signals, which may be a result of the single-stage, deep (6-m) waste management system. It should also be noted that there were no consistent spatial relationships associated with the positive swine-associated PCR results and proximity to waste storage.
TABLE 4.
Proportion of positive PCR results using general Bacteroidales and swine-associated markers on water DNA extracts
| Water sampling site (n) | No. of positive PCR results/total no. of PCR results at DNA template concn
|
|||||
|---|---|---|---|---|---|---|
| General Bacteroidales (Bac32)
|
Swine-associated Prevotella (PF163)
|
Swine-associated methanogens (P23-2)
|
Swine-associated Prevotella (PigBac1)
|
|||
| 0.2 ng | 1 ng | 1 ng | 10 ng | 50 ng | 10 ng | |
| Illinoisa | ||||||
| Site A | ||||||
| Wells (16) | 6/16 | 7/16 | 1/16 | 3/16 | 1/16 | 2/16 |
| Lagoon (1) | 1/1 | 0/1 | 0/1 | 1/1 | 0/1 | 1/1 |
| North stream (1) | 1/1 | 1/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| North tile (1) | 1/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Site C | ||||||
| Wells (6) | 3/6 | 1/6 | 2/6 | 2/6 | 2/6 | 1/6 |
| Lagoon (1) | 1/1 | 1/1 | 1/1 | 1/1 | 0/1 | 1/1 |
| Seep from field (1) | 1/1 | 1/1 | 0/1 | 1/1 | 0/1 | 0/1 |
| South stream (1) | 1/1 | 1/1 | 1/1 | 1/1 | 0/1 | 0/1 |
| Site E | ||||||
| Wells (11) | 5/11 | 6/11 | 4/11 | 3/11 | 0/11 | 0/11 |
| Manure pit (n = 1) | 1/1 | 1/1 | 0/1 | 1/1 | 0/1 | 1/1 |
| Texas | ||||||
| Lake Granbury (10) | 10/10 | NDb | ND | 6/10 | 2/10 | 10/10 |
| Buck Creek (10) | 10/10 | ND | ND | 6/10 | 0/10 | 0/10 |
With use of the general bacterial 16S rRNA gene-targeted PCR assay (8F/787R), 37/40 environmental DNA extracts from Illinois yielded positive PCR results.
ND, not determined.
PCR assay performance with Texas surface water samples.
A high frequency of positive signals was obtained for the general Bacteroidales marker and for PF163 in water samples from Lake Granbury and Buck Creek (Table 4; see also Table S2 in the supplemental material). Interestingly, while the PigBac1 assay was positive for all the samples from Lake Granbury, none of the samples tested from Buck Creek yielded positive PCR signals. In addition, the methanogen-targeted marker produced a positive PCR signal in only one of the water samples from either Lake Granbury or Buck Creek. PF163 was also the only swine-associated marker detected in three water samples collected close to a swine farming operation in Ohio (data not shown). These results suggest that PF163 might be more frequently detected in different environmental settings than are the other two swine-associated assays tested in this study.
Probabilities of swine-associated PCR detection at sites using Bayesian statistics.
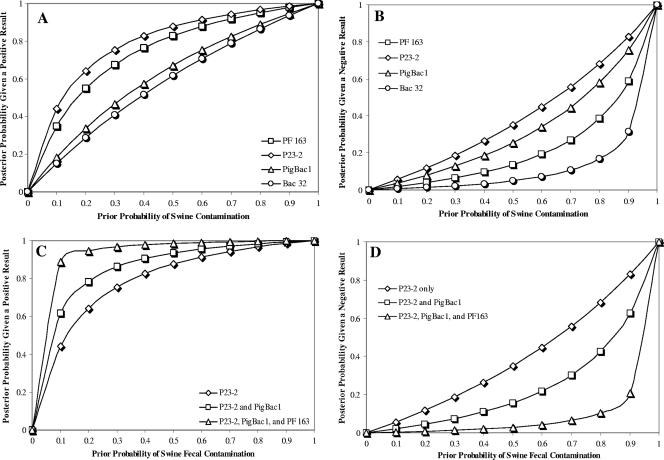
Because all of the swine PCR assays showed some level of cross-reactivity with nontarget fecal sources, the probability of detecting feces originating from swine-associated sources within a given water sample was estimated using Bayesian statistics (19). When the confidence of each assay was tested for water samples using a range of prior probabilities, a positive result from either the P23-2 or the PF163 assay always yielded higher confidence of detecting a true-positive pig fecal signal than did PigBac1 (Fig. 1A). The probability of pig fecal contamination given a negative PCR result indicated that, of the three pig-specific markers, PF163 yielded the lowest probability of false negatives (Fig. 1B). Altogether, these data indicate that the PF163 assay yielded the highest probability of yielding true-positive and negative PCR results in environmental waters, compared to the other pig-specific markers tested in this study.
FIG. 1.
Probability of swine fecal contamination using a Bayesian statistical model. (A) Posterior probability of contamination given a positive PCR result using each of the four primer sets tested in this study over a range of prior probabilities. (B) Posterior probability of contamination given a negative PCR result using each of the four primer sets tested in this study over a range of prior probabilities. (C) Posterior probability of contamination given a positive PCR result using the P23-2 primer set alone or in combination with PigBac1 and all three host-specific assays together. (D) Posterior probability of contamination given a negative PCR result using the P23-2 primer set alone or in combination with PigBac1 and all three host-specific assays together.
The Bayesian analysis also reveals important limitations regarding the utility of some of the currently available assays. Specifically, while a combination of assays could increase the accuracy of detecting swine fecal pollution in environmental waters (Fig. 1C and D), the results from this study suggest that the currently available assays have limited value as environmental monitoring tools. For example, only 40% of the water samples tested in this study were positive using any of the three swine-associated assays. This number is lower than expected considering the close proximity of the sites to swine operations and the high occurrence of the general Bacteroidales marker (75% of the samples). While PCR inhibition is one possible explanation for the low occurrence of the swine-associated markers, there were only two occurrences where a swine-associated marker produced a PCR product and the general Bacteroidales marker did not. Additionally, all but three water DNA extracts produced a positive PCR result when using the general bacterial 16S rRNA gene-targeted assay (8F/787R), suggesting that PCR inhibition was not impacting PCR results associated with these environmental samples. Assuming that most Bacteroidales clades are of fecal origin, the results suggest that the targeted host-specific populations survive poorly under conditions associated with waste management and transport into environmental waters. Bayesian statistics indicated that a positive PCR result from the three host-specific markers used in this study would result in greater than 90% confidence that a given water sample is indeed contaminated with swine feces [at P(S) > 0.2]. However, only eight of all water samples tested (n = 43) were positive for two or more of the swine-associated assays. Moreover, only two samples were positive for all four swine-associated assays. Thus, to improve statistical confidence that a water sample is indeed contaminated with swine feces, better assays are needed, particularly assays capable of detecting multiple groups of environmentally relevant swine-associated fecal bacteria.
Phylogenetic analysis and population diversity of fecal and environmental Bacteroidales clones.
Because several studies have suggested that Bacteroidales 16S rRNA gene sequences are promising targets for method development (2, 12, 22, 29), clone libraries were developed from feces and fecally impacted waters to assess the level of diversity, host specificity, and membership of Bacteroidales associated with swine fecal sources and waters impacted with swine fecal pollution. Clone libraries were also developed to understand why the Bacteroidales-based assays did not perform as well as expected in environmental scenarios. By unveiling the diversity of the current pig-associated targets, it was possible to provide information on the level of diversity relevant to microbial source tracking assay development.
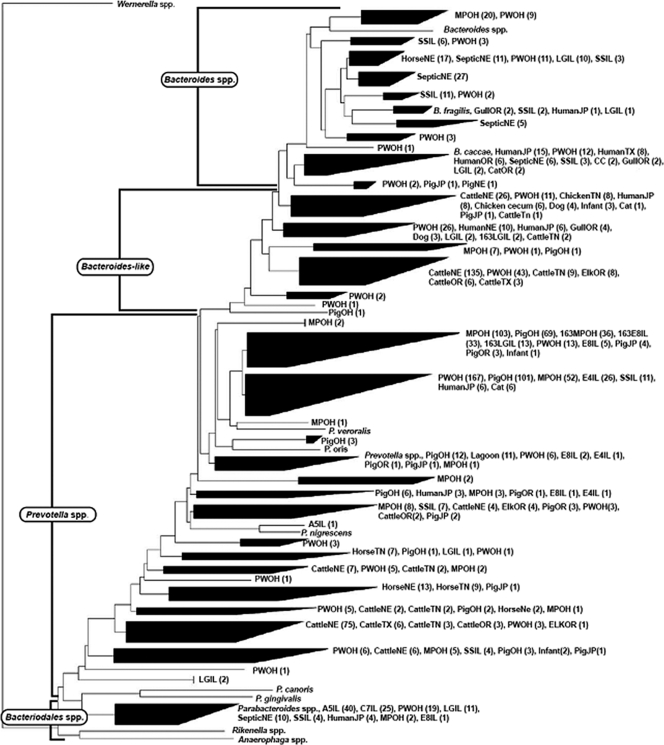
A total of 1,502 partial 16S rRNA gene Bacteroidales sequences derived from nine different animal fecal sources, three waste storage facilities (i.e., manure pits, lagoons, and septic tanks), and six sampling sites adjacent to swine operations were analyzed (Table 5). The final phylogenetic analysis included sequences from cattle (n = 294), pig (n = 219), human (n = 67), horse (n = 48), chicken (n = 16), elk (n = 13), cat (n = 9), seagull (n = 8), and human infant (n = 6) feces, as well as from swine manure pit (n = 245), human septic (n = 59), swine lagoon (n = 55), and surface water (n = 360) samples adjacent to a swine operation in Ohio and surface water (n = 51) and groundwater (n = 135) adjacent to a swine operation in Illinois (Fig. 2). A total of 245 anomalous sequences were excluded in the analysis (10). More than half of all unique clone sequences (i.e., 56%) exhibited low sequence similarity (<97%) to Bacteroidales 16S rRNA gene sequences, indicating that the phylogenetic diversity of Bacteroidales-like sequences may be currently underrepresented in the publicly available databases. Interestingly, fewer than 2% of all swine fecal, manure pit, and lagoon sequences derived from geographically diverse locations were associated with Bacteroides spp. As the latter sequences clustered with clones retrieved from other nonswine sources, these populations could be considered cosmopolitan. Similarly, Hong et al. (16) concluded that Bacteroides spp. were present at a relatively low abundance in swine feces, even after targeting 14 different Bacteroides species. The results in this study further indicate that Bacteroides spp. may account for only a limited number of PCR-amplified Bacteroidales 16S rRNA genes from the swine fecal wastes (24) and therefore are not ideal sequences for the development of inclusive host-specific assays.
TABLE 5.
Similarity-based OTUs and richness estimates for general Bacteroidales swine source and environmental samples
| Sample source | No. of sequences | No. of OTUsa | Richness estimateb
|
Diversity index
|
|||
|---|---|---|---|---|---|---|---|
| Chao 1 (95% CIc) | ACE (95% CI) | Bootstrap | Shannon-Weaver (95% CI) | Simpson | |||
| Swine lagoon | 43 | 10 | 15 (11-42) | 19 (12-49) | 12 | 1.75 (1.52-1.97) | 0.17 |
| Swine manure pit | 209 | 59 | 122 (85-210) | 131 (92-215) | 74 | 3.18 (2.98-3.37) | 0.079 |
| Water adjacent to swine farms | 582 | 86 | 125 (103-173) | 138 (113-188) | 103 | 2.79 (2.63-2.95) | 0.19 |
| Swine feces | 199 | 55 | 86 (68-132) | 104 (77-164) | 68 | 2.84 (2.59-3.08) | 0.17 |
OTUs have been defined as each of the sequences that are at most 3% distant from the most similar sequence in a given OTU.
The Chao 1 estimator and abundance-based coverage estimator (ACE) are nonparametric methods used to estimate richness by adding a correction factor to the observed number of species (7, 8). Bootstrap is a nonparametric estimate of species richness as described by Smith and van Belle (33).
CI, confidence interval.
FIG. 2.
Phylogenetic tree of 1,502 Bacteroidales 16S rRNA gene sequences derived from different animal hosts and from water, lagoon, and manure samples, based on a neighbor-joining algorithm. Numbers in parentheses indicate the number of sequences associated in each clade for a given host. Sequences for cultured Bacteroidales members were added to the analyses as reference points. MPOH, manure pit sample from Ohio; PigOH, pig fecal sample from Ohio; PWOH, water sample impacted with pig feces from Ohio; SSIL, sample from a stream south of a site in Illinois; LGIL, lagoon sample from Illinois; TN, OR, NE, JP, and TX, samples taken from Tennessee, Oregon, Nebraska, Japan, and Texas, respectively; CC, chicken cecum; 163, clones generated using PF163 and Bac708 primers; A5, E4, C7, and E8, samples taken from groundwater wells from Illinois sites A, C, and E.
In contrast, nearly all (i.e., 98%) swine-associated sequences clustered with members related to Prevotella species, suggesting that this group exists in higher abundance within swine fecal waste. Analysis of Prevotella-like sequences associated with swine fecal pollution sources revealed interesting diversity patterns. For example, more than 80% of swine manure pit and swine fecal Bacteroidales 16S rRNA gene sequences belong to the same Prevotella clades. More importantly, half of the sequences retrieved from the water samples examined in this study (i.e., 250/495) clustered within the aforementioned Prevotella clades, supporting the contribution of swine fecal sources in these waters. However, compared to all the water clones from this study, the PF163 primer did not match with the vast majority of the environmental water sequences (i.e., 474 out of 495), suggesting that this assay can underestimate swine pollution in the environment. It should be noted that the PF163 primer matched 24% and 47% of fecal and manure pit sequences, respectively, and that depending on the site, 5 to 56% of the water samples tested were positive for the PF163 assay. Hence, not only does the PF163 assay target a subset of host-specific populations of apparently low abundance, but these populations might not survive well in aquatic environments. Furthermore, a lower number of sequences from fecal sources (i.e., 13/245 manure pit sequences and 5/219 fecal sequences) and water samples (0/495) matched the PigBac1 primer, which may explain the lower frequency of detection of this marker in manure pit and water samples. Altogether, these data suggest that multiple swine-associated assays should be sought to maximize the coverage of these environmentally relevant host-specific populations.
More than 80% (i.e., 44 of 55) of the swine lagoon clones were associated with either Bacteroides or Parabacteroides species and not with Prevotella-like sequences, which dominated the feces and manure pit clone libraries. Interestingly, all sequences derived from site A and site C, which used lagoon-type waste management, clustered with 11 lagoon sequences, theoretically supporting swine contamination within these wells. Nevertheless, the majority (i.e., 42 of 55) of other swine lagoon sequences could be considered cosmopolitan, as they also cluster with clones derived from human septic tanks, human feces, and horse feces. While more sequencing of isolates from swine lagoons is needed, these data suggest that many of the numerically dominant Bacteroidales populations derived from swine lagoons can withstand the conditions found in this type of fecal waste storage.
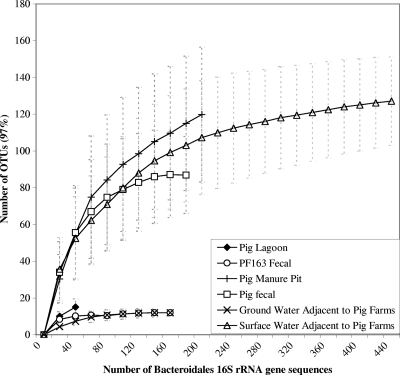
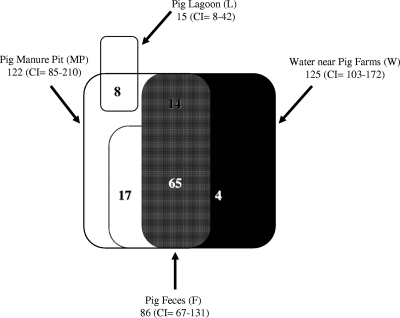
Rarefaction of the Chao 1 richness estimator indicated that Bacteroidales assemblages derived from surface water samples from Ohio showed that deeper sequencing coverage might not reveal significant novel diversity, as the curve was approaching a horizontal asymptote at approximately 138 OTUs (Fig. 3). Richness estimators and rarefaction analysis also indicated that in the current sequencing effort, swine feces, manure pits, and surface water contained more diverse Bacteroidales assemblages than did sequences derived from groundwater and clones developed using PF163 PCR products from feces (Table 5; Fig. 3). When pairwise comparisons were inferred between pig feces, pig manure pits, pig lagoons, and water adjacent to swine operations, it was found that Bacteroidales sequences derived from pig fecal samples were a subset of water and fresh manure pit samples using OTU0.03 memberships with the SONS program (Fig. 4). This may suggest that manure pits may be a better surrogate for fecal sources, as these samples represent a mixture of several individual pigs and are more representative of fecal waste entering the environment. Using community fingerprint data, Ziemer et al. (39) showed that Bacteroides-Prevotella species present within the fecal and manure pit samples are different. More interestingly, 65 OTUs were shared among water, fecal, and manure pit sequences, indicating that several source fecal and manure pit Bacteroidales populations are indeed found in water samples believed to be contaminated with swine pollution. Sequences representing these OTUs clustered together in host-specific Prevotella phylogenetic clades (Fig. 2). These populations may be promising pig fecal source tracking targets as they are also relatively high in abundance in environmental samples. This finding supports our conclusion that several swine-specific assays will be required to track swine fecal sources in environmental samples due to the diversity of populations that show host specificity. By choosing assays that target multiple groups, the limitations associated with using a single marker (16, 30) can be circumvented, including limitations for assays developed using newly discovered Bacteroidales host-specific clades.
FIG. 3.
Rarefaction of Chao 1 richness estimators using sampling without replacement for swine source and environmental Bacteroidales assemblages. Sequences within an OTU are at most 3% distant from the most similar sequence in that given OTU.
FIG. 4.
Venn diagram comparing community structures of Bacteroidales populations derived from pig feces, pig manure pits, pig lagoons, and water adjacent to swine operations. Numbers indicate the OTUs shared at 97% similarity (OTU0.03) by each of the overlapping communities. Pairwise comparisons were inferred between all four communities to generate a shared Chao estimate for any two overlapping communities. To better define the number of OTUs in overlap regions, shared Chao estimates were generated using SONS where one community (e.g., water) was compared to the other three communities (e.g., lagoon, manure pit, and fecal) pooled as one community. This was repeated for each community to generate a Venn diagram of all studied Bacteroidales assemblages. Shared Chao indices were as follows: MP-W, 79; MP-F, 82; W-F, 70; MP-W/F, 95; W-MP/F, 84; F-MP/W, 87; MP/F, 15 (MP, manure pit; W, water; F, fecal). The Chao 1 richness estimate of all four libraries pooled together was 263 (confidence interval [CI] = 221 to 337).
In this study, we evaluated the utility of currently available swine-targeted PCR assays within multiple environmental scenarios. Overall, the assays targeting Prevotella populations were found to more frequently detect swine fecal pollution than did the methanogen-based assay in the environmental samples analyzed in this study. Both Prevotella-targeted assays cross-reacted with nontarget fecal sources, questioning their potential value as stand-alone assays in complex multiuse watersheds. However, the application of Bayes' theorem shows how a probability model can inform users about the utility of host-specific markers within watershed-based studies. Sequence analysis of the general Bacteroidales clones demonstrated the presence of novel Prevotella clades that are not accounted for when using the currently available pig-specific assays. Additionally, phylogenetic analyses discriminated between endemic and cosmopolitan Bacteroidales populations. By comparing membership compositions and structures of fecal and environmental microbial communities, it was possible to demonstrate the presence of swine-associated populations, which could also be found in swine-impacted waters. While additional tests are necessary to examine host specificity and universal occurrence of the latter populations in environmental waters, the findings in this study suggest that these sequences are promising targets for swine fecal source tracking assay development. Understanding the molecular diversity of both fecal and environmental microbial populations clearly provides an additional method for confirming specific fecal pollution sources in environmental waters and will likely unveil novel targets for future method development.
Supplementary Material
Acknowledgments
This research was supported in part by an Augmentation Award to J.W.S.D. from the National Center for Computational Toxicology of the U.S. EPA, Office of Research and Development.
We are grateful to Jingrang Lu and Donald Stoeckel for sharing their fecal sample collections. We thank the Brazos River Authority and Phyllis Dyer for water sample collection. We also thank Elizabeth Casarez, Joy Truesdale, and Nicholas Garcia for technical assistance.
The U.S. Environmental Protection Agency, through its Office of Research and Development, funded and managed, or partially funded and collaborated in, the research described herein. It has been subjected to the Agency's peer and administrative review and has been approved for external publication. Any opinions expressed are those of the author(s) and do not necessarily reflect the views of the Agency; therefore, no official endorsement should be inferred. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Published ahead of print on 24 July 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bernhard, A. E., and K. G. Field. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjursell, M. K., E. C. Martens, and J. I. Gordon. 2006. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J. Biol. Chem. 281:36269-36279. [DOI] [PubMed] [Google Scholar]
- 4.Buchholz-Cleven, B. E., B. Rattunde, and K. L. Straub. 1997. Screening for genetic diversity of isolates of anaerobic Fe(II)-oxidizing bacteria using DGGE and whole-cell hybridization. Syst. Appl. Microbiol. 20:301-309. [Google Scholar]
- 5.Buckholt, M. A., J. H. Lee, and S. Tzipori. 2002. Prevalence of Enterocytozoon bieneusi in swine: an 18-month survey at a slaughterhouse in Massachusetts. Appl. Environ. Microbiol. 68:2595-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butine, T. J., and J. A. Leedle. 1989. Enumeration of selected anaerobic bacterial groups in cecal and colonic contents of growing-finishing pigs. Appl. Environ. Microbiol. 55:1112-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 8.Chao, A., and S. M. Lee. 1992. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 87:210-217. [Google Scholar]
- 9.Chee-Sanford, J. C., R. I. Aminov, I. J. Krapac, N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl. Environ. Microbiol. 67:1494-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeSantis, T. Z., P. Hugenholtz, K. Keller, E. L. Brodie, N. Larsen, Y. M. Piceno, R. Phan, and G. L. Andersen. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:394-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dick, L. K., A. E. Bernhard, T. J. Brodeur, J. W. Santo Domingo, J. M. Simpson, S. P. Walters, and K. G. Field. 2005. Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification. Appl. Environ. Microbiol. 71:3184-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fratamico, P. M., L. K. Bagi, E. J. Bush, and B. T. Solow. 2004. Prevalence and characterization of Shiga toxin-producing Escherichia coli in swine feces recovered in the National Animal Health Monitoring System's Swine 2000 study. Appl. Environ. Microbiol. 70:7173-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gourmelon, M., M. P. Caprais, R. Segura, C. Le Mennec, S. Lozach, J. Y. Piriou, and A. Rince. 2007. Evaluation of two library-independent microbial source tracking methods to identify sources of fecal contamination in French estuaries Appl. Environ. Microbiol. 73:4857-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan, T. Y., and R. A. Holley. 2003. Pathogen survival in swine manure environments and transmission of human enteric illness—a review. J. Environ. Qual. 32:383-392. [DOI] [PubMed] [Google Scholar]
- 16.Hong, P.-Y., J.-H. Wu, and W.-T. Liu. 2008. Relative abundance of Bacteroides spp. in stools and wastewaters as determined by hierarchical oligonucleotide primer extension. Appl. Environ. Microbiol. 74:2882-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 18.Kellog, R. L., C. H. Lander, D. C. Moffitt, and N. Gollehon. 2000. Manure nutrients relative to the capacity of cropland and pastureland to assimilate nutrients: spatial and temporal trends for the United States. U.S. Department of Agriculture, Washington, DC.
- 19.Kildare, B. J., C. M. Leutenegger, B. S. McSwain, D. G. Bambic, V. B. Rajal, and S. Wuertz. 2007. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: a Bayesian approach. Water Res. 41:3701-3715. [DOI] [PubMed] [Google Scholar]
- 20.Koike, S., I. G. Krapac, H. D. Oliver, A. C. Yannarell, J. C. Chee-Sanford, R. I. Aminov, and R. I. Mackie. 2007. Monitoring and source tracking of tetracycline resistance genes in lagoons and groundwater adjacent to swine production facilities over a 3-year period. Appl. Environ. Microbiol. 73:4813-4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krapac, I. G., W. S. Dey, W. R. Roy, C. A. Smyth, E. Storment, S. L. Sargent, and J. D. Steele. 2002. Impacts of swine manure pits on groundwater quality. Environ. Pollut. 120:475-492. [DOI] [PubMed] [Google Scholar]
- 22.Lamendella, R., J. W. Santo Domingo, D. B. Oerther, J. R. Vogel, and D. M. Stoeckel. 2007. Assessment of fecal pollution sources in a small northern-plains watershed using PCR and phylogenetic analyses of Bacteroidetes 16S rRNA gene. FEMS Microbiol. Ecol. 59:651-660. [DOI] [PubMed] [Google Scholar]
- 23.Leach, M. D., S. L. Broschat, and D. R. Call. 2008. A discrete, stochastic model and correction method for bacterial source tracking. Environ. Sci. Technol. 42:524-529. [DOI] [PubMed] [Google Scholar]
- 24.Leser, T. D., J. Z. Amenuvor, T. K. Jensen, R. H. Lindecrona, M. Boye, and K. Moller. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lozupone, C. A., M. Hamady, B. L. Cantarel, P. M. Coutinho, B. Henrissat, J. I. Gordon, and R. Knight. 2008. The convergence of carbohydrate active gene repertoires in human gut microbes. Proc. Natl. Acad. Sci. USA 105:15076-15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Heier, I. Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, L. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, R. Hermann, A. Jost, T. König, R. Liss, M. Lüßmann, B. May, B. Nonhoff, S. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maczulak, A. E., M. J. Wolin, and T. L. Miller. 1989. Increase in colonic methanogens and total anaerobes in aging rats. Appl. Environ. Microbiol. 55:2468-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merkel, M. 2004. Threatening Iowa's future: Iowa's failure to implement and enforce the Clean Water Act for livestock operations. Environmental Integrity Project, Washington, DC. http://www.environmentalintegrity.org/pub194.cfm.
- 29.Okabe, S., N. Okayama, O. Savichtcheva, and T. Ito. 2007. Quantification of host-specific Bacteroides-Prevotella 16S rRNA genetic markers for assessment of fecal pollution in freshwater. Appl. Microbiol. Biotechnol. 74:890-901. [DOI] [PubMed] [Google Scholar]
- 30.Santo Domingo, J. W., D. G. Bambic, T. A. Edge, and S. Wuertz. 2007. Quo vadis source tracking? Towards a strategic framework for environmental monitoring of fecal pollution. Water Res. 41:3539-3552. [DOI] [PubMed] [Google Scholar]
- 31.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schloss, P. D., and J. Handelsman. 2006. Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures. Appl. Environ. Microbiol. 72:6773-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, E. P., and G. van Belle. 1984. Nonparametric estimation of species richness. Biometrics 40:119-129. [Google Scholar]
- 34.Sorlini, C., T. Brusa, G. Ranalli, and A. Ferrari. 1988. Quantitative determination of methanogenic bacteria in the feces of different mammals. Curr. Microbiol. 17:33-36. [Google Scholar]
- 35.Ufnar, J. A., D. F. Ufnar, S. Y. Wang, and R. D. Ellender. 2007. Development of a swine-associated fecal pollution marker based on host differences in methanogen mcrA genes. Appl. Environ. Microbiol. 73:5209-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wegener, H. C., and D. L. Baggesen. 1996. Investigation of an outbreak of human salmonellosis caused by Salmonella enterica ssp. enterica serovar Infantis by use of pulsed field gel electrophoresis. Int. J. Food Microbiol. 32:125-131. [DOI] [PubMed] [Google Scholar]
- 37.Wing, S., S. Freedman, and L. Band. 2002. The potential impact of flooding on confined animal feeding operations in eastern North Carolina. Environ. Health Perspect. 110:387-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu, J., M. A. Mahowald, R. E. Ley, C. A. Lozupone, M. Hamady, E. C. Martens, B. Henrissat, P. M. Coutinho, P. Minx, P. Latreille, H. Cordum, A. Van Brunt, K. Kim, R. S. Fulton, L. A. Fulton, S. W. Clifton, R. K. Wilson, R. D. Knight, and J. I. Gordon. 2007. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 5:e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ziemer, C. J., M. A. Cotta, and T. R. Whitehead. 2004. Application of group specific amplified rDNA restriction analysis to characterize swine fecal and manure storage pit samples. Anaerobe 10:217-227. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.