Abstract
A Tn402-like class 1 integron was recovered from a prawn-associated bacterium. One of its cassettes included methionine sulfoxide reductase genes, the first example of such genes being captured by an integron. The integron was flanked by direct repeats that resemble miniature inverted-repeat transposable element sequences. Excision of the integron by homologous recombination through these sequences was demonstrated.
Integrons possess a site-specific recombination system that promotes dissemination of mobile gene cassettes (8, 17). About 3% of cells in the general environment contain class 1 integrons, and this integron class is broadly disseminated by lateral gene transfer (1, 6). A subset of class 1 integrons is associated with transposition functions exemplified by transposon Tn402, and they are commonly recovered from clinical environments. The Tn402-like class 1 integrons have a number of characteristic features, including two conserved segments (CS) that flank the site where mobile gene cassettes are inserted (23) (Fig. 1). The ongoing accumulation of resistance genes in Tn402-like integrons makes this integron subtype one of the largest contributors to the spread of multidrug resistance in human pathogens (8, 14).
FIG. 1.
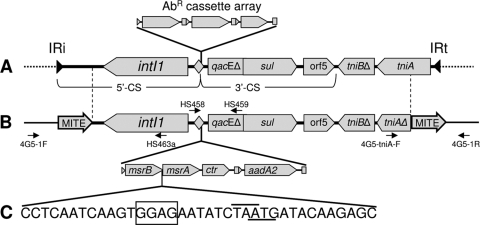
Features of Tn402-like class 1 integrons. (A) Generalized structure of a Tn402-like class 1 integron as commonly recovered from multidrug-resistant pathogens. Black filled horizontal arrows indicate the inverted repeats IRi and IRt, which define the ends of Tn402-like transposons. The gray-shaded diamond represents the attI1 site, at which mobile cassettes are inserted. AbR, antibiotic resistance. A three-cassette array is depicted here, but the type and number of cassettes are variable. The extents of the 5′ CS) and 3′ CS are indicated. (B) The vertical dashed lines define the extent of the sequence of the NFM2 integron common to that of a typical Tn402-like class 1 integron. The open diamond represents the attI1 site with the structure of the two-cassette array in the NFM2 integron indicated immediately below. The small horizontal arrows indicate the relative positions of primers HS458 and HS459 and primers used to assess excision of the integron by homologous recombination between the MITE elements. (C) The sequence of the region at the end of msrB and the start of msrA. Overlined bases are the stop codon for msrB and the underlined bases the start codon of msrA. The boxed sequence is a putative ribosome binding site for msrA. Abbreviations: intI1, class 1 integron integrase gene; qacEΔ1, remnant of quaternary ammonium compound resistance gene; sul, sulfonamide resistance gene; orf5, open reading frame encoding unknown function; tniA and tniB, Tn402-associated transposition genes (“Δ” symbol indicates partial gene deletion). See the text for other abbreviations. The figure is not necessarily drawn to scale.
Tn402-like class 1 integrons are also common in commensal bacteria, including those found in humans, and are spreading back into environmental bacteria (6, 12, 21, 24). Such integrons are then exposed to the highly diverse gene cassette metagenome (11), which includes a variety of potential virulence factors (1, 20). Consequently, environmental Tn402-like class 1 integrons may be a conduit for additional genes encoding functions other than antibiotic resistance to make their way into human pathogens. As part of a project investigating the role of animal commensal bacteria in the spread of resistance genes, we screened bacteria from the digestive tracts of prawns for the presence of class 1 integrons. Here we describe the complete DNA sequence of a novel Tn402-like integron and its surrounds from an Acinetobacter sp. strain isolated from a prawn. Digestive tracts from uncooked prawns were used to separately inoculate 5 ml plate count medium (23) and incubated for 24 h at 25°C. Four prawns, harvested from each of 16 distinct locations from Australia, the South Pacific, and Southeast Asia, were examined. More than 75% of mixed cultures from individual prawns tested positive for class 1 integrons with use of the primers HS915 and HS916 (15). PCR with these primers on pure isolates derived from such mixed cultures revealed that about 3% of 850 colonies were positive for a class 1 integron, a number consistent with findings of other studies (10, 25). We then employed a PCR method that amplified cassette arrays from Tn402-like class 1 integrons (13) using the primers HS458 and HS459 (10) (Fig. 1). Sequencing of one of the HS458/459 amplicons revealed an unusual structure. This unusual integron was recovered from an Acinetobacter sp. strain (here designated strain NFM2) isolated from a prawn (Penaeus plebejus) (19) harvested from ocean waters off the Australian east coast. NFM2 typing was determined by partial 16S rRNA sequencing, with the closest species match being to Acinetobacter johnsonii.
A fosmid library of NFM2 was constructed and an intI1-positive clone completely sequenced using methods previously described (23). A physical map of the relevant region is shown in Fig. 1. The integron possessed features typical of Tn402-derived class 1 integrons, including the presence of a 3′ CS that abutted a truncated tni module. However, the integron was flanked by identical copies of a 439-bp direct repeat, sequence comparisons of which are suggestive of them being miniature inverted repeat transposable elements (MITEs), a family of nonautonomous mobile elements (4). MITEs are small, nonautonomous mobile elements broadly dispersed in prokaryotes that have only recently been described (4). These elements are diverse in their sequence and properties but generally include several of the following characteristics: (i) a target site duplication, (ii) short terminal inverted repeats, (iii) high A/T content, (iv) “TA” motif at each of the termini, and (v) binding sites for host integrative factors and methyltransferases. The MITE-like elements in NFM2 include the first three of these (73% A/T). They also include at least three methyltransferase binding sites, although this domain is short (GANTC), so the significance of their presence is unclear. Many MITEs contain open reading frames, and several produce RNA transcripts, the associated secondary structure of which regulates mRNA (3). The MITEs found in NFM2 do not possess obvious open reading frames. NFM2 MITE transcripts, if produced, would be likely to possess extensive secondary structures; however, we have no information as to the role, if any, in mRNA regulation.
The outer ends of the two MITE direct repeats were in turn flanked by a 5-bp direct repeat (GTTGC). We hypothesize that the MITE sequences captured the class 1 integron in a structure analogous to a composite transposon, and this has subsequently transposed to its present location. The points of insertion of the MITE-like elements are inside the normal boundary of a Tn402-like integron, in that the first 76 bases of the 5′ CS, including IRi, are missing at one end, and IRt and most of tniA are missing at the other end (Fig. 1). In silico analysis by us revealed that portions of the MITE-like sequences in NFM2 are also found in sequences derived from several Acinetobacter sp. clinical isolates derived from soft tissue and bloodstream infections (see reference 16 and accession numbers therein). These MITE-like examples also flank a class 1 integron at the 5′-CS segment end with the same insertion point as seen here. In addition, it has recently been found that a class 1 integron from Enterobacter cloacae is also flanked by MITEs, although these are not related in sequence to those found here (18). In addition, the locations of each of the MITEs at both ends of the respective integrons are different, further suggesting independent capture events.
Transposition of MITEs requires that additional functions be provided in trans (4). The presence of a direct repeat implies that the class 1 integron in NFM2 moved into its identified location by transposition. Indeed, transposition of the Enterobacter cloacae integron mobilization unit has been demonstrated (18). However, attempts to transpose the class 1 integron from NFM2 into a conjugative plasmid in a mating-out assay with an Escherichia coli recipient were unsuccessful (data not shown), suggesting that the required additional transposon functions are not present in this strain. However, the two copies of the MITE-like sequences are identical, and given they are oriented as direct repeats, it seemed highly likely that the integron could be readily excised from the genome by homologous recombination. To test this, PCR primers were designed to detect excision via recombination. The position of these primers is indicated in Fig. 1. Primers 4G5-1F (5′ CCCACACAATAAACGCCG) and 4G5-1R (5′ TGGCGATGGCTCAATGTC) should generate a product of 819 bp, comprising a single MITE-like sequence remaining in the genome if the integron is deleted. Primers HS463a (11) and 4G5-tniA-F (5′TGCGACAAGGTACGGTAGG) should recover a 1,231-bp sequence that includes the junction of the circularized, excised product. A PCR with each of these primer pairs on NFM2 genomic DNA generated PCR products of the predicted lengths (data not shown). DNA sequencing of each PCR product revealed boundaries consistent with excision via homologous recombination from a point within the MITE-like sequences.
The NFM2 integron carried two cassettes, the second of which was an aadA2 gene cassette commonly seen in Tn402-like class 1 integrons. This was consistent with the fact that NFM2 was resistant to streptomycin (25 μg/ml). The first cassette was unusual for a number of reasons. It was 1,874 bases in length and included three genes. These are probably contained in a single transcript and in order are msrB (best match; 78% protein product identity to Xanthobacter autotrophicus Py2 [accession no. CP000781]), msrA (83% protein product identity to Rhodopirellula baltica SH 1 [accession no. BX294154]), and a gene encoding a hypothetical protein (orf255). msrA and msrB encode methionine sulfoxide reductases, involved in the repair of proteins damaged by oxidative stress (7). The A and B forms reduce alternative isomers of methionine sulfoxide (5). The stop codon of msrB overlaps the start codon of msrA by one base (Fig. 1). The presence of a ribosome binding site immediately upstream of msrA suggests that this gene is translated by the process of translational coupling (9). This is the first report of msr-related genes being located in a mobile gene cassette. The attC site associated with this unusual cassette is 71 bases in length and conforms to all the criteria associated with this family of sites, including an overall imperfect repeat structure and two simple sites with the conserved domains 1R, 2R, 2L, and 1L (22). The predicted circular form of the msr cassette would have complementary 1R and 1L sites. We could not identify an obvious promoter in this attC site, so it is likely that the aadA2 gene is expressed from the integron promoter, Pc.
Most Tn402-like class 1 integrons are defective transposons (2), although mobilization by transposition can still occur if the required proteins are provided in trans. The requirement for exogenous components limits mobilization in comparison to autonomous elements. The element found here and that seen in an Enterobacter cloacae isolate (18) represent two independent examples whereby class 1 integrons may be mobilized by families of elements unrelated to Tn402. The fact that one of these is from a clinical isolate and the other from a marine invertebrate further underscores the potential for mobilization of gene cassettes between human pathogens and bacteria found in the general environment. Finally, the NFM2 integron has an unusual cassette in that it includes three genes that may be implicated in a phenotype unrelated to the neutralization of antibiotics. This suggests that Tn402-like class 1 integrons are beginning to recruit new types of functions in some bacteria.
Nucleotide sequence accession numbers.
The integron sequence described here is contained within the fosmid clone 4G5 and is deposited in GenBank under accession number FJ711439. Accession number GQ377756 contains the 16S rRNA sequence from NFM2.
Acknowledgments
This work was supported by the Australian Research Council. N.J.G. was the recipient of a travel bursary from the Quebec Education Ministry.
Footnotes
Published ahead of print on 31 July 2009.
REFERENCES
- 1.Boucher, Y., M. Labbate, J. E. Koenig, and H. W. Stokes. 2007. Integrons: mobilizable platforms that promote genetic diversity in bacteria. Trends Microbiol. 15:301-309. [DOI] [PubMed] [Google Scholar]
- 2.Brown, H. J., H. W. Stokes, and R. M. Hall. 1996. The integrons In0, In2 and In5 are defective transposon derivatives. J. Bacteriol. 178:4429-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Gregorio, E., G. Silvestro, M. Petrillo, M. S. Carlomagno, and P. P. Di Nocera. 2005. Enterobacterial repetitive intergenic consensus sequence repeats in yersiniae: genomic organization and functional properties J. Bacteriol. 187:7945-7954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delihas, N. 2008. Small mobile sequences in bacteria display diverse structure/function motifs. Mol. Microbiol. 67:475-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ezraty, B., L. Aussel, and F. Barras. 2005. Methionine sulfoxide reductases in prokaryotes. Biochim. Biophys. Acta 1703:221-229. [DOI] [PubMed] [Google Scholar]
- 6.Gillings, M., Y. Boucher, M. Labbate, A. Holmes, S. Krishnan, M. Holley, and H. W. Stokes. 2008. The evolution of class 1 integrons and the rise of antibiotic resistance. J. Bacteriol. 190:5095-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grimaud, R., B. Ezraty, J. K. Mitchell, D. Lafitte, C. Briand, P. J. Derrick, and F. Barras. 2001. Repair of oxidized proteins. Identification of a new methionine sulfoxide reductase. J. Biol. Chem. 276:48915-48920. [DOI] [PubMed] [Google Scholar]
- 8.Hall, R. M., C. M. Collis, M. J. Kim, S. R. Partridge, G. D. Recchia, and H. W. Stokes. 1999. Mobile gene cassettes and integrons in evolution. Ann. N. Y. Acad. Sci. 870:68-80. [DOI] [PubMed] [Google Scholar]
- 9.Han, C. G., Y. Shiga, T. Tobe, C. Sasakawa, and E. Ohtsubo. 2001. Structural and functional characterization of IS679 and IS66-family elements. J. Bacteriol. 183:4296-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardwick, S. A., H. W. Stokes, S. Findlay, M. Taylor, and M. R. Gillings. 2008. Quantification of class 1 integron abundance in natural environments using real-time quantitative PCR. FEMS Microbiol. Lett. 278:207-212. [DOI] [PubMed] [Google Scholar]
- 11.Holmes, A. J., M. R. Gillings, B. S. Nield, B. C. Mabbutt, K. M. Nevalainen, and H. W. Stokes. 2003. The gene cassette metagenome is a basic resource for bacterial genome evolution. Environ. Microbiol. 5:383-394. [DOI] [PubMed] [Google Scholar]
- 12.Labbate, M., P. R. Chowdhury, and H. W. Stokes. 2008. A class 1 integron present in a human commensal has a hybrid transposition module compared to Tn402: evidence of interaction with mobile DNA from natural environments. J. Bacteriol. 190:5318-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lévesque, C., L. Piché, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Márquez, C., M. Labbate, C. Raymondo, J. Fernández, A. M. Gestal, M. Holley, G. Borthagaray, and H. W. Stokes. 2008. Urinary tract infections in a South American population: dynamic spread of class 1 integrons and multidrug resistance by homologous and site-specific recombination. J. Clin. Microbiol. 46:3417-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendes, R. E., M. Castanheira, M. A. Toleman, H. S. Sader, R. N. Jones, and T. R. Walsh. 2007. Characterization of an integron carrying blaIMP-1 and a new aminoglycoside resistance gene, aac(6′)-31, and its dissemination among genetically unrelated clinical isolates in a Brazilian hospital. Antimicrob. Agents Chemother. 51:2611-2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Partridge, S. R., G. Tsafnat, E. Coiera, and J. R. Iredell. 2009. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 33:757-784. [DOI] [PubMed] [Google Scholar]
- 18.Poirel, L., A. Carrër, J. D. Pitout, and P. Nordmann. 2009. Integron mobilization unit as a source of mobility of antibiotic resistance genes. Antimicrob. Agents Chemother. 53:2492-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poore, G. C. B. 2004. Marine decapod Crustacea of southern Australia: a guide to identification. CSIRO Publishing, Collingwood, Victoria, Australia.
- 20.Rowe-Magnus, D. A., A. M. Guerout, L. Biskri, P. Bouige, and D. Mazel. 2003. Comparative analysis of superintegrons: engineering extensive genetic diversity in the Vibrionaceae. Genome Res. 13:428-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.San Martín, B., L. Lapierre, J. Cornejo, and S. Bucarey. 2008. Characterization of antibiotic resistance genes linked to class 1 and 2 integrons in strains of Salmonella spp. isolated from swine. Can. J. Microbiol. 54:569-576. [DOI] [PubMed] [Google Scholar]
- 22.Stokes, H. W., D. B. O'Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]
- 23.Stokes, H. W., C. L. Nesbø, M. Holley, M. I. Bahl, M. R. Gillings, and Y. Boucher. 2006. Class 1 integrons potentially predating the association with Tn402-like transposition genes are present in a sediment microbial community. J. Bacteriol. 188:5722-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vinué, L., S. Yáenz, S. Somalo, E. Escudero, M. A. Moreno, F. Ruiz-Larrea, and C. Torres. 2008. Prevalence and diversity of integrons and associated resistance genes in faecal Escherichia coli isolates of healthy humans in Spain. J. Antimicrob. Chemother. 62:934-937. [DOI] [PubMed] [Google Scholar]
- 25.Wright, M. S., C. Baker-Austin, A. H. Lindell, R. Stepanauskas, H. W. Stokes, and J. V. McArthur. 2008. Influence of industrial contamination on mobile genetic elements: class 1 integron abundance and gene cassette structure in aquatic bacterial communities. ISME J. 2:417-428. [DOI] [PubMed] [Google Scholar]