Abstract
In addition to di-myo-inositol-1,3′-phosphate (DIP), a compatible solute widespread in hyperthermophiles, the organic solute pool of Thermotoga maritima comprises 2-(O-β-d-mannosyl)-di-myo-inositol-1,3′-phosphate (MDIP) and 2-(O-β-d-mannosyl-1,2-O-β-d-mannosyl)-di-myo-inositol-1,3′-phosphate (MMDIP), two newly identified β-1,2-mannosides. In cells grown under heat stress, MDIP was the major solute, accounting for 43% of the total pool; MMDIP and DIP accumulated to similar levels, each corresponding to 11.5% of the total pool. The synthesis of MDIP involved the transfer of the mannosyl group from GDP-mannose to DIP in a single-step reaction catalyzed by MDIP synthase. This enzyme used MDIP as an acceptor of a second mannose residue, yielding the di-mannosylated compound. Minor amounts of the tri-mannosylated form were also detected. With a genomic approach, putative genes for MDIP synthase were identified in the genome of T. maritima, and the assignment was confirmed by functional expression in Escherichia coli. Genes with significant sequence identity were found only in the genomes of Thermotoga spp., Aquifex aeolicus, and Archaeoglobus profundus. MDIP synthase of T. maritima had maximal activity at 95°C and apparent Km values of 16 mM and 0.7 mM for DIP and GDP-mannose, respectively. The stereochemistry of MDIP was characterized by isotopic labeling and nuclear magnetic resonance (NMR): DIP selectively labeled with carbon 13 at position C1 of the l-inositol moiety was synthesized and used as a substrate for MDIP synthase. This β-1,2-mannosyltransferase is unrelated to known glycosyltransferases, and within the domain Bacteria, it is restricted to members of the two deepest lineages, i.e., the Thermotogales and the Aquificales. To our knowledge, this is the first β-1,2-mannosyltransferase characterized thus far.
Thermotoga maritima was first isolated from hot marine sediments on Vulcano Island, Italy, being able to grow between 55°C and 90°C (14). This strictly anaerobic bacterium ferments a variety of simple and complex carbohydrates to acetate, hydrogen, and CO2 (10). In line with these metabolic traits, a substantial percentage of the genes annotated in the genome of this hyperthermophile are allocated to the metabolism of mono- and polysaccharides (8, 23). Therefore, T. maritima has been pointed out as a source of glycoside hydrolases with potential industrial relevance, namely, in processes of conversion of biomass into biofuels (3, 34).
Like many other hyperthermophiles isolated from marine environments, Thermotoga maritima is slightly halophilic (optimum NaCl concentration of 2.7%, wt/vol) and has developed biochemical strategies to counterbalance the external osmotic pressure. The accumulation of low-molecular-mass organic compounds in the cytoplasm is the most common osmoadaptation mechanism, which enables a rapid response to fluctuations in the salinity of the external medium. Interestingly, the organic solutes encountered in organisms adapted to thrive in hot environments are clearly different from those used by mesophiles, leading to the view that osmolytes of (hyper)thermophiles could play an additional role as protectors of macromolecules and other cellular components against heat damage. This notion is further fuelled by the finding that the total pool of organic solutes of (hyper)thermophiles increases notably not only at supraoptimal salinity but also in response to heat stress conditions (30).
Over the last decade, our team has directed considerable effort to assess the role of osmolytes in the thermo-adaptation strategies of hyperthermophiles. Despite the scarcity of genetic tools for manipulation of marine hyperthermophiles, a number of novel organic solutes were identified and the corresponding biosynthetic pathways characterized at the genetic and biochemical levels (15, 17, 30), providing critical knowledge for engaging in elucidation of the molecular basis of the whole process, from the sensing of stress to the synthesis of specific osmolytes. In this context, we recently reported the characterization of the pathway for synthesis of di-myo-inositol-1,3′-phosphate, the most common solute within hyperthermophiles (5). Additionally, the genes and enzymes involved in the relevant reaction steps were disclosed. The synthesis proceeds via a phosphorylated form of DIP, and the respective synthase is a membrane-associated enzyme that catalyzes the condensation of CDP-inositol with inositol-1-phosphate (26, 27).
The solute pool in members of the order Thermotogales was investigated a few years ago (19). Thermotoga neapolitana responded to heat stress with a strong accumulation of DIP and DIP derivatives. One of the solutes was assigned to a mannosylated form of DIP, at that time designated di-mannosyl-di-myo-inositol phosphate; moreover, the presence of a second DIP derivative was proposed, but its structure remained elusive. Therefore, we set out to fully characterize the solute pool of Thermotoga spp. and to identify the genes and the enzyme(s) involved in the synthesis of the DIP derivatives. Members of the genus Thermotoga accumulated DIP and two mannosylated forms of this compound, herein fully characterized using isotopic labeling, NMR, and mass spectrometry. Moreover, the pathway for the synthesis of these novel solutes was identified, leading to the discovery of a unique β-1,2-mannosyltransferase that catalyzes the transfer of the mannosyl group from GDP-mannose to DIP.
MATERIALS AND METHODS
Abbreviations.
The abbreviations used in this paper are defined as follows: MDIP or mannosyl-di-myo-inositol phosphate, 2-(O-β-d-mannosyl)-di-myo-inositol-1,3′-phosphate; MMDIP or di-mannosyl-di-myo-inositol phosphate, 2-(O-β-d-mannosyl-1,2-O-β-d-mannosyl)-di-myo-inositol-1,3′-phosphate; tri-mannosyl-di-myo-inositol phosphate, 2-(O-β-d-mannosyl-1,2-O-β-d-mannosyl-1,2-O-β-d-mannosyl)-di-myo-inositol-1,3′-phosphate; DIP or di-myo-inositol phosphate, di-myo-inositol-1,3′-phosphate; DIPP, di-myo-inositol-1,3′-phosphate-1′-phosphate; GPI, glycero-phospho-inositol; IPTG, isopropyl β-d-1-thiogalactopyranoside; NMR, nuclear magnetic resonance; HMBC, heteronuclear multiple-bond correlation; HSQC, heteronuclear single-quantum coherence; COSY, homonuclear correlation spectroscopy; PIPES, piperazine-N,N′-bis(2-ethanesulfonic acid); OD600, optical density at 600 nm; MOPS, morpholinepropanesulfonic acid.
Materials.
d,l-Glycerol-3-P, CTP, GDP, GDP-mannose, ADP-glucose, TDP-glucose, UDP-glucose, GDP-glucose, phosphoenolpyruvate, glycerol, myo-inositol, guanidinium chloride, and pyruvate kinase from rabbit muscle were purchased from Sigma-Aldrich (St. Louis, MO). NADH and rabbit muscle lactate dehydrogenase were purchased from Roche Applied Science (Penzberg, Germany). [6-13C]glucose (99% enrichment) was obtained from Omicron Biochemicals, Inc. (South Bend, IN). d-myo-Inositol-1-phosphate was obtained by chemical synthesis (5). l-myo-Inositol-1-phosphate was produced from glucose-6-phosphate and NADH by using recombinant myo-inositol phosphate synthase of Archaeoglobus fulgidus (27). DIPP was obtained as described earlier (5), and di-myo-[1-13C]inositol-1,3′-phosphate was synthesized enzymatically (this work). DIP was supplied by bitop AG (Witten, Germany), and GPI was isolated from Archaeoglobus fulgidus (17).
Strains and culture conditions.
The type strain of Thermotoga maritima (strain 3109T) was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany). Aquifex aeolicus strain VF5 (around 5 g [wet weight]) was kindly provided by M. Tomm and H. Huber, University of Regensburg, Germany. T. maritima was cultivated in a medium containing (per liter) 27 g of NaCl, 0.5 g of yeast extract, 3 g of PIPES, 2.5 g of starch, 2 mg of (NH4)2Ni(SO4)2·7H2O, 1.75 g of MgSO4·7H2O, 0.5 g of MgCl2·6H2O, 0.16 g of KCl, 25 mg of NaBr, 7.5 mg of H3BO3, 3.8 mg of SrCl2·6H2O, 0.025 mg of KI, 0.1 g of CaCl2·2H2O, 0.35 g of K2HPO4, 1 g of NH4Cl, 3 g of cysteine-HCl, and 10 ml of trace mineral solution (2). The final pH was adjusted to 6.5. The medium was degassed with N2 and sterilized by autoclaving. Prior to inoculation, the medium was supplemented with biotin (20 μg/liter), thiosulfate (20 mM), and Na2S (0.05%, wt/vol). The organism was cultured in 2-liter static vessels under anaerobiosis (N2 gas phase); cell growth was assessed by measuring OD600.
To examine the effect of osmotic and heat stresses on the level of intracellular organic solutes, T. maritima was grown under optimal conditions (2.7% NaCl and 80°C), under osmotic stress (4.5% NaCl, 80°C), and under heat stress (2.7% NaCl, 88°C); combined stress conditions (4.5% NaCl, 88°C) were also examined, but growth was not observed.
Extraction, identification, and quantification of intracellular solutes.
Cells were harvested during the late exponential phase of growth by centrifugation (7,000 × g, 10 min, 4°C) and washed twice with an NaCl solution identical in concentration to the growth medium. Cell pellets were suspended in water and disrupted by sonication. Aliquots were removed for determination of total protein content. The remaining cell extract was treated twice with boiling 80% ethanol as described previously (31). Freeze-dried extracts were dissolved in 2H2O and analyzed by NMR (31).
Purification of MDIP.
Cells of T. maritima grown at 88°C with 2.7% NaCl were harvested during the late exponential growth phase (OD600 of 0.3) by centrifugation (7,000 × g, 10 min, 4°C) and washed twice with an NaCl solution identical in concentration to the growth medium. The ethanolic cell extract, obtained as described above, was loaded onto a QAE-Sephadex A-25 column previously equilibrated with 5 mM sodium bicarbonate (pH 9.8), and elution was performed with a linear gradient of the same buffer (5 mM to 1 M). Fractions were analyzed by 31P NMR to monitor the presence of MDIP. Samples containing this compound were desalted using a column of activated Dowex 50W-X8. The pH of the eluted fractions was adjusted to 6 with 1 M KOH prior to lyophilization. The freeze-dried sample was dissolved in 1-propanol-ammonia (1:1.5, vol/vol) and loaded onto a Silica Gel S column; elution was carried out with the same solvent mixture. The fractions enriched in MDIP were applied to a second Silica Gel S column. The fractions containing pure MDIP were pooled, lyophilized, and dissolved in 2H2O for structure determination by NMR. The yield of the purification was only 8% due to the difficulty in removing the contamination with MMDIP.
Preparation of crude cell extracts for detection of enzyme activity.
Thermotoga maritima cells grown at 88°C with 2.7% NaCl were harvested by centrifugation and washed twice with Tris-HCl buffer (20 mM, pH 7.6) containing 2.5% NaCl under anaerobic conditions. The cell pellet was suspended in the same buffer, and cells were disrupted using a French press. Cell debris was removed by centrifugation (30,000 × g, 45 min, 4°C). Low-molecular-mass compounds were removed using a PD-10 column (GE Healthcare Bio-Science AB, Uppsala, Sweden) equilibrated with Tris-HCl (20 mM, pH 7.6) containing 10 mM MgCl2. Protein content was estimated by the Bradford method (6).
Cloning and expression of putative mds genes.
Chromosomal DNA from T. maritima and A. aeolicus was isolated according to the method of Ramakrishnan and Adams (25). The genes of T. maritima (TM_0359) and A. aeolicus (AQ_1141) were amplified by PCR using Pfu DNA polymerase (Fermentas, Burlington, Canada) and cloned in pET23a and pTRC99a, respectively, by following standard protocols (29). The strategies used to clone the genes led to the addition of a methionine residue at the N terminus of the protein from T. maritima and 13 extra residues (Met-His6-Gly-Asp4-Lys) at the N terminus of the A. aeolicus protein. Enterokinase was used to remove the extra peptide, but the yield was very low (around 10%). Escherichia coli BL21(DE3) cells bearing the constructs were grown at 37°C in LB medium with ampicillin (100 μg/ml) to an OD600 of 0.5 and treated with 1 mM IPTG for 4 h. Cells were harvested, suspended in Tris-HCl (20 mM, pH 7.6) containing 10 mM MgCl2, and disrupted using a French press; cell debris was removed by centrifugation (30,000 × g, 4°C, 1 h).
Purification of recombinant T. maritima MDIP synthase.
The cell extract (approximately 40 mg of protein/ml) resulting from the previous step was heated for 20 min at 70°C to precipitate thermo-labile proteins; after centrifugation, the supernatant solution was applied onto a Resource Q column (GE Healthcare Bio-Science AB, Uppsala, Sweden) equilibrated with Tris-HCl buffer (20 mM, pH 7.6). Elution was carried out with a linear gradient (0 to 1 M NaCl). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of fractions eluted between 0.1 and 0.2 M NaCl revealed a strong band with the expected molecular mass of 42 kDa. Moreover, the presence of this band correlated with MDIP synthase activity, as evaluated by 31P NMR for monitoring the formation of MDIP, with DIP and GDP-mannose used as substrates. Fractions containing MDIP synthase were pooled, concentrated using centrifugal filter devices, and dialyzed against Tris-HCl buffer (20 mM, pH 7.6). The solution was applied onto a Mono Q column (GE Healthcare Bio-Science AB, Uppsala, Sweden) equilibrated with Tris-HCl buffer (20 mM, pH 7.6). Fractions containing MDIP synthase eluted between 0.2 and 0.3 M NaCl. This last chromatographic step was repeated, and a fraction with pure MDIP synthase was obtained, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and silver staining.
Purification of recombinant A. aeolicus MDIP synthase.
The cell extracts were treated as described above; after a first chromatographic step (Resource Q), the active fractions were applied onto a HisTrap HP column (GE Healthcare Bio-Science AB, Uppsala, Sweden). The protein eluted with 500 mM imidazole. The active sample was applied to a phenyl-Sepharose column and elution carried out with a linear gradient of ammonium sulfate (1.7 M to 0 M). The activities eluted at 0.5 M of (NH4)2SO4.
Enzyme assays.
MDIP synthase activity was detected in cell extracts of T. maritima and also in cell extracts of E. coli harboring mds genes either from T. maritima or from A. aeolicus. Reaction mixtures (0.4 ml) containing Tris-HCl buffer (20 mM, pH 7.6), 10 mM MgCl2, 4 mM of the substrates, and cell extract (around 2 mg of total protein) were incubated for 1 h at 80°C. The formation of the product, MDIP, was detected by the appearance of a resonance at −0.7 ppm in the 31P NMR spectra. Assays for characterization of MDIP synthase were performed with 0.15 ml of solutions containing 50 mM bis-Tris propane-HCl buffer, pH 7.6, 10 mM MgCl2, 16 mM DIP, and 2.2 mM GDP-mannose; the reaction mixtures were incubated at the specified temperatures in 2-ml glass tubes pressurized with 2 atm of argon and preheated for 2 min. The reaction was initiated by the addition of the enzyme (3 μg) and stopped at different times by immersion in liquid nitrogen. The MDIP synthase activity was measured from the amount of GDP released during incubation with GDP-mannose and DIP. GDP was quantified by monitoring the oxidation of NADH at 340 nm (13). This assay was performed at 30°C with 1-ml reaction mixtures containing Tris-HCl (20 mM, pH 7.6), 10 mM MgCl2, 0.4 mM phosphoenolpyruvate, 0.175 mM NADH, 2.7 U of pyruvate kinase, 4 U of lactate dehydrogenase, and 70 μl of the MDIP synthase assay mixture.
Characterization of T. maritima MDIP synthase.
The temperature profile for the activity was determined to be between 70°C and 105°C. The pH of the buffer as measured at 25°C was 7.6. Using the conversion factor ΔpKa/ΔT °C = −0.015 for bis-Tris propane, we estimated that the working pH value varied between 6.9 and 6.4 in the temperature range studied. Kinetic parameters (Vmax and Km) were determined at 95°C with a calculated pH of 5.7 in reaction mixtures (0.15-ml final volume) containing 5 mM GDP-mannose and 0 to 300 mM DIP or 0 to 3.5 mM GDP-mannose and 98 mM DIP. The reaction was started by the addition of 3 μg of MDIP synthase. Experiments were done in duplicate. Km was determined by fitting the data to Michaelis-Menten equations, using the software program Origin 5.0 Professional (Microcal Software, Inc., MA).
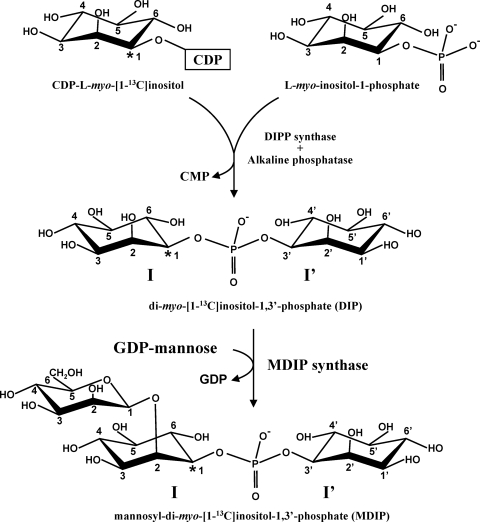
Preparation of di-myo-[1-13C]inositol-1,3′-phosphate and determination of MDIP stereochemistry.
myo-[1-13C]inositol-1-P and CDP-myo-[1-13C]inositol were produced enzymatically. myo-[1-13C]inositol-1-P was produced from d-[6-13C]glucose by coupling hexokinase from Thermoproteus tenax and myo-inositol-1-P synthase from A. fulgidus as described by Rodrigues et al. (27). Subsequently, CDP-myo-[1-13C]inositol was synthesized from CTP and myo-[1-13C]inositol-1-P by using a cell extract of E. coli BL21(DE3) harboring the gene encoding DIPP synthase of Rubrobacter xylanophilus (27). Finally, synthesis of di-myo-[1-13C]inositol-1,3′-phosphate was performed with a reaction mixture containing 4 mM CDP-myo-[1-13C]inositol, 4 mM of d,l-myo-inositol-1-P, 10 mM MgCl2, and 4 mg total protein/ml of a cell extract of A. fulgidus in MOPS buffer (50 mM, pH 8.1). After 30 min of incubation at 80°C, the reaction product, di-myo-[1-13C]inositol-1,3′-phosphate, was purified by anion-exchange chromatography as previously described (5). The final product was 93% enriched in carbon 13 at position 1; the presence of minor amounts of isotopomers without labeling at this position is due to the fact that the inositol moieties of inositol-1-P and CDP-inositol can exchange to a low extent in the reaction catalyzed by DIPP synthase (5). Labeled MDIP was produced from di-myo-[1-13C]inositol-1,3′-phosphate and GDP-mannose supplied to cell extracts of T. maritima or to a partially purified preparation of MDIP synthase. The stereochemistry of MDIP was determined by using 1H NMR, 31P NMR, and 13C NMR.
NMR spectroscopy.
For structure determination, spectra were acquired with a Bruker AVANCE III spectrometer (Bruker, Rheinstetten, Germany) operating at 800.33 MHz for protons. 1H, 13C, and 31P chemical shifts are relative to 4,4-dimethyl-4-silapentane sodium sulfonate, external methanol designated at 49.3 ppm, and external 85% H3PO4, respectively. The homonuclear proton correlation spectra (COSY) as well as the heteronuclear correlation spectra (1H-13C HSQC, 1H-13C HMBC, and 1H-31P HSQC) were acquired at 25°C using standard Bruker pulse programs. The homonuclear correlation spectra were acquired with presaturation of the water signal. In the HSQC sequences, JC,H and JP,H of 145 Hz and 7 Hz were used to calculate the delays for evolution of scalar couplings; in the 1H-13C HMBC spectra, 8 Hz was used for JC,H. For quantification of the organic solutes accumulating in T. maritima, 1H NMR spectra were acquired with a Bruker AVANCE II 500 spectrometer equipped with a broadband inverse probe head. Spectra were acquired with presaturation of the water signal, using a repetition delay of 60 s. Formate was used as an internal concentration standard.
Differential scanning calorimetry.
Measurements were performed with a MicroCal VP-DSC MicroCalorimeter as previously described (12). Samples of MDIP synthase from A. aeolicus (0.35 mg/ml) were prepared by extensive dialysis against 10 mM phosphate buffer, pH 6.2. Guanidinium chloride (1.5 M final concentration) was added before the assay. Scans were run at a heating rate of 1°C/min from 25 to 120°C with an overpressure of approximately 30 lb/in2. Baselines were acquired under the same experimental conditions as for the assays both in the absence and in the presence of guanidinium chloride. The melting temperature was determined using MicroCal software.
RESULTS
Identification of intracellular organic solutes in Thermotoga maritima.
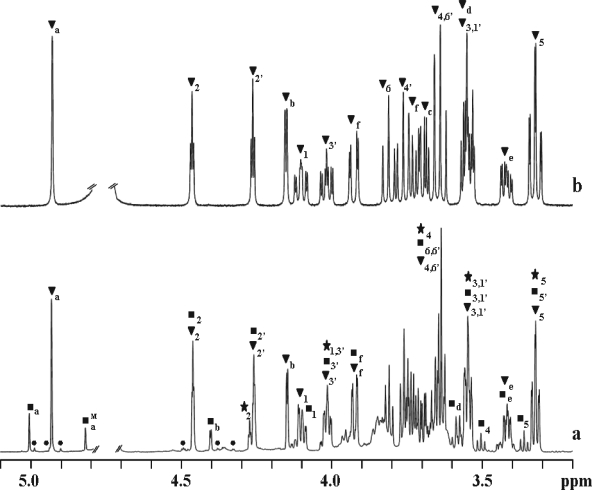
1H, 31P, and 13C NMR spectra of ethanol extracts of T. maritima showed the presence of DIP, several DIP derivatives, and glutamate (α and β forms) (Fig. 1a). The 1H-31P correlation spectra of extracts of cells grown at 80°C showed three signals previously assigned to DIP, to a symmetric mannosylated derivative of DIP, and to an unknown compound, tentatively assigned to a different DIP stereoisomer (see Fig. 3 in reference 19). The enhanced resolution provided by a high-field spectrometer allowed us to conclude that the three sets of resonances corresponded to two compounds only, DIP and an asymmetric mannosylated derivative of DIP, herein firmly identified as MDIP on the basis of an array of spectra (1H COSY, 1H-13C HSQC, 1H-13C HMBC, and 1H-31P HSQC) of the purified MDIP (Fig. 1b). Moreover, the mass spectrum of a partially purified extract of T. maritima revealed two major signals, one with an m/z of 420.9 that perfectly matched the molecular mass of DIP and one with an m/z of 583.1 that was consistent with MDIP. The NMR parameters of MDIP are presented in Table 1. The value of 162 Hz for the 1JC,H coupling constant at position 1 of the mannose residue clearly indicates a β configuration for this sugar moiety (4). The 1H-13C HMBC spectrum placed the glycosidic linkage of the mannosyl residue at position 2 of one inositol group, while the 1H-31P HSQC showed that the phosphate group was linked to positions 1 and 3′ of the inositol moieties. The set of resonances herein assigned to MDIP are also present in spectra of cell extracts of Aquifex pyrophilus (17) and A. aeolicus (data not shown); therefore, we conclude that MDIP is synthesized by these two members of the genus Aquifex.
FIG. 1.
1H NMR spectra of an ethanol extract of Thermotoga maritima grown at 88°C in medium containing 2.7% NaCl (a) and pure 2-(O-β-d-mannosyl)-di-myo-inositol-1,3′-phosphate (b). Resonances due to protons of di-myo-inositol phosphate (DIP) (★), mannosyl-di-myo-inositol phosphate (MDIP) (▾), di-mannosyl-di-myo-inositol phosphate (MMDIP) (▪), and tri-mannosyl-di-myo-inositol phosphate (•) are labeled. Resonances due to protons of the mannose moieties labeled with a, b, c, d, e, and f designate H1, H2, H3, H4, H5, and H6a or H6b, respectively. The label M designates the resonances due to the mannose moiety of MMDIP not directly linked to inositol. The nomenclature used for inositol is in accordance with Rodrigues et al. (27).
FIG. 3.
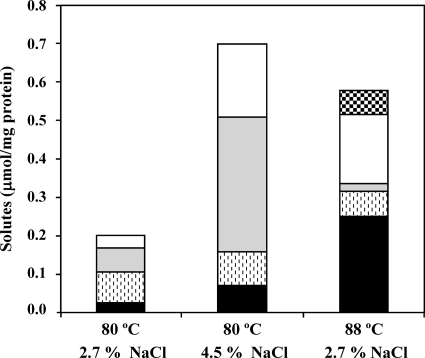
Effect of growth temperature and NaCl concentration on the accumulation of α-glutamate (white), β-glutamate (gray), di-myo-inositol phosphate (stippled), mannosyl-di-myo-inositol phosphate (black), and di-mannosyl-di-myo-inositol phosphate (checkered) in Thermotoga maritima. The optimal growth conditions are 80°C and 2.7% NaCl. Cultures were harvested during the late exponential phase of growth. The data are averages of results from two independent growths under each condition.
TABLE 1.
NMR parameters of MDIP and MMDIPa
| Moiety and position | MDIPb
|
MMDIPb
|
||||
|---|---|---|---|---|---|---|
|
13C NMRc
|
1H NMRd δ (ppm) |
13C NMRc
|
1H NMRd δ (ppm) | |||
| δ (ppm) | 1JC,H (Hz) | δ (ppm) | 1JC,H (Hz) | |||
| Inositol (I) | ||||||
| C1 | 76.3 | 143 | 4.10 | 75.9 | 143 | 4.12 |
| C2 | 79.8 | 153 | 4.46 | 80.5 | 153 | 4.46 |
| C3 | 70.5 | 145 | 3.54 | 70.0 | 143 | 3.57 |
| C4 | 73.0 | 148 | 3.66 | 73.1 | 145 | 3.51 |
| C5 | 74.2 | 144 | 3.32 | 73.7 | 143 | 3.36 |
| C6 | 71.9 | 148 | 3.81 | 72.2 | 146 | 3.67 |
| Inositol (I′) | ||||||
| C1′ | 70.8 | 144 | 3.56 | 76.7 | 144 | 4.02 |
| C2′ | 71.5 | 151 | 4.26 | 71.8 | 151 | 4.26 |
| C3′ | 76.8 | 143 | 4.03 | 70.8 | 143 | 3.55 |
| C4′ | 71.7 | 146 | 3.76 | 72.3 | 145 | 3.64 |
| C5′ | 74.1 | 144 | 3.33 | 74.1 | 143 | 3.32 |
| C6′ | 72.6 | 146 | 3.64 | 71.6 | 147 | 3.76 |
| Mannose | ||||||
| C1 | 100.9 | 162 | 4.93 | 101.1 | 162 | 5.01 |
| C2 | 70.8 | 150 | 4.15 | 78.7 | 150 | 4.41 |
| C3 | 73.1 | 141 | 3.69 | 72.2 | 141 | 3.71 |
| C4 | 67.1 | 145 | 3.55 | 67.2 | 145 | 3.59 |
| C5 | 76.4 | 142 | 3.41 | 76.4 | 142 | 3.44 |
| C6 | 61.3 | 150 | 3.91, 3.73 | 60.9 | ND | 3.92, 3.75 |
| Mannose | ||||||
| C1 | 101.1 | 161 | 4.81 | |||
| C2 | 70.7 | 148 | 4.11 | |||
| C3 | 72.9 | 141 | 3.67 | |||
| C4 | 66.8 | 145 | 3.58 | |||
| C5 | 76.5 | 142 | 3.41 | |||
| C6 | 61.1 | ND | 3.93, 3.74 | |||
The 31P chemical shifts were derived directly from the one-dimensional 31P spectra. 1H and 13C chemical shifts were obtained from the 1H-13C HSQC spectra; the 1JC,H coupling constants were measured for 1H-13C HSQC spectra acquired without carbon decoupling.
Measurements were carried out at 35°C and pH 6.5. The phosphorous resonances of MDIP and MMDIP appear at −0.7 ppm relative to external 85% H3PO4.
Methanol was used as a reference designated at 49.3 ppm. ND, not determined.
4,4-Dimethyl-4-silapentane sodium sulfonate was used as a reference.
In addition to resonances assigned to DIP and MDIP, the proton NMR spectra of extracts of T. maritima grown at 88°C showed an extra set of resonances which comprised two signals at 5.00 and 4.81 ppm, the region typical of the anomeric protons of hexoses (Fig. 1a). Coupling constants 3JH1,H2 at ≤2 Hz and 1JC1,H1 at 161 and 162 Hz were determined and denote mannose residues in the β configuration. A partially purified preparation of the unknown compound was analyzed by 1H, 13C, and 31P NMR. The set of NMR data were consistent with the structure depicted in Fig. 2; the relevant parameters are presented in Table 1. The compound was designated MMDIP. This identification was corroborated by the observation in the mass spectrum of a signal at an m/z of 745.1, corresponding to the expected molecular mass of MMDIP.
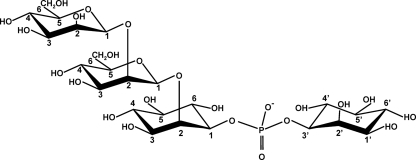
FIG. 2.
Molecular representation of MMDIP, a newly identified organic solute accumulating in cells of Thermotoga maritima grown under heat stress conditions.
The NMR spectra of the ethanolic extract (Fig. 1a) revealed yet another set of very weak resonances which comprised three signals in the region of the anomeric protons of hexoses at 4.98 ppm, 4.92 ppm, and 4.89 ppm. These three signals belong to a vestigial tri-mannosylated derivative of DIP, but the unambiguous assignment of all the signals was precluded by overlapping with the much stronger signals of MMDIP. Nevertheless, it is clear from the 1H-13C HMBC spectra that the three mannosyl residues are linked sequentially through β-(1,2) glycosidic bonds. Therefore, we propose that this minor compound is 2-(O-β-d-mannosyl-1,2-O-β-d-mannosyl-1,2-O-β-d-mannosyl)-di-myo-inositol-1,3′-phosphate, which is in accordance with a peak at an m/z of 907.0 detected in the mass spectrum.
Effect of salt and growth temperature on accumulation of organic solutes in Thermotoga maritima.
Under optimal growth conditions (2.7% NaCl, 80°C), the total amount of organic solutes in T. maritima was around 0.2 μmol·mg of protein−1 and increased up to 0.7 μmol·mg of protein−1 in cells grown under heat or osmotic stress (Fig. 3 and Table 2). The proportion of organic solutes was dependent on the type of stress imposed; for example, the level of glutamate (α and β forms) increased notably in response to osmotic stress, whereas MDIP and MMDIP clearly increased under heat stress. Curiously, the ratio of β-glutamate/α-glutamate was 1.8 in cells grown under osmotic stress but decreased to 0.1 in cells grown at a supraoptimal temperature. When T. maritima was cultivated in medium with 4.5% NaCl (osmotic stress conditions), β- and α-glutamate dominated the solute pool, reaching 77% (molar percentage) of the total organic compounds. In contrast, DIP and DIP derivatives represented 65% of the solute pool of T. maritima grown at 88°C (upshift of 8°C relative to the optimal level), while β- and α-glutamate amounted to the remaining 35%. MDIP was the predominant compound (43% of the total), corresponding to a 10-fold increase in concentration with respect to the optimal temperature conditions. The newly identified DIP derivative, MMDIP, was detected only in cells subjected to heat stress. Interestingly, the level of the parent compound, DIP, was fairly constant in the range of growth conditions examined.
TABLE 2.
Intracellular content of organic solutes in Thermotoga maritima grown at different temperatures and NaCl concentrations
| Growth temp (°C) | NaCl concn (%, wt/vol) | Intracellular concn (μmol/mg protein)a
|
||||
|---|---|---|---|---|---|---|
| α-Glutamate | β-Glutamate | DIP | MDIP | MMDIP | ||
| 80 | 2.7 | 0.03 ± 0.00 (7.1) | 0.06 ± 0.01 (13.8) | 0.08 ± 0.01 (18.0) | 0.02 ± 0.00 (5.5) | |
| 88 | 2.7 | 0.18 ± 0.03 (39.8) | 0.02 ± 0.01 (4.3) | 0.07 ± 0.01 (14.8) | 0.25 ± 0.00 (55.4) | 0.06 ± 0.00 (13.9) |
| 80 | 4.5 | 0.19 ± 0.01 (42.5) | 0.35 ± 0.01 (77.6) | 0.09 ± 0.00 (19.6) | 0.07 ± 0.01 (15.6) | |
Quantification was performed by 1H NMR. The intracellular concentration (mM) based on a cell volume of 4.5 μl/mg protein as determined for Pyrococcus furiosus (20) is given in parentheses.
Identification of the gene(s) for the synthesis of MDIP.
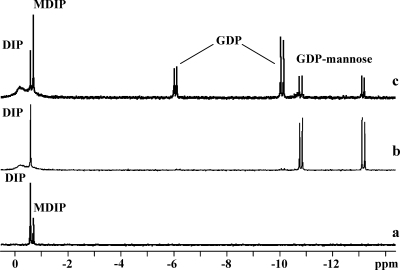
Synthesis of MDIP was investigated by 31P NMR spectroscopy with cell extracts of T. maritima by using GDP-mannose and DIP as the putative sugar donor and acceptor, respectively. In addition to the DIP resonance at −0.57 ppm, a new resonance due to the reaction product, MDIP, appeared at −0.7 ppm (Fig. 4a). This identification was firmly established by spiking with pure MDIP. To identify the gene conferring this activity, we searched Pfam (http://pfam.sanger.ac.uk/) for glycosyltransferases of T. maritima. Fourteen genes whose predicted products belong to family 1 glycosyltransferases were found. Eight of them are annotated, leaving six genes (TM_0392, TM_0359, TM_0619, TM_0628, TM_0744, and TM_1230) as putative candidates for encoding MDIP synthase. A BLAST search in public databases revealed that the gene TM_0359 had homologs (around 35% sequence identity) in the genomes of A. aeolicus (AQ_1141), a hyperthermophilic bacterium that accumulates MDIP (data not shown), and Archaeoglobus profundus (ArcprDRAFT_0086 [http://img.jgi.doe.gov/cgi-bin/geba/main.cgi]). Nearly identical genes (99% identity) were present in the genomes of Thermotoga neapolitana, Thermotoga petrophila, and Thermotoga sp. strain RQ2. No hits were found in members of genera other than Thermotoga, Aquifex, and Archaeoglobus. The genes TM_0359 and AQ_1141 were selected for further analysis. The genes were separately cloned, and cell extracts of E. coli BL21(DE3) harboring pET23a:TM_0359 or pET19b:AQ_1141 were examined for the presence of MDIP synthase activity. The formation of MDIP was proven by 31P NMR analysis (Fig. 4c). We verified that MDIP was not formed in cell extracts of E. coli bearing empty plasmids (pET23a or pET19b) (Fig. 4b). These results showed definitely that TM_0359 and AQ_1141 encode enzymes capable of synthesizing MDIP.
FIG. 4.
31P NMR spectra of the final products resulting from incubation at 80°C of reaction mixtures containing 4 mM GDP-mannose, 4 mM DIP, and 10 mM MgCl2 in 20 mM Tris-HCl buffer, pH 7.6, with cell extract of T. maritima grown under heat stress conditions (a), E. coli cell extract harboring the plasmid pET23a after heat treatment at 70°C for 10 min (b), and E. coli cell extract harboring the construction pET23a:mds after heat treatment at 70°C for 10 min (c). Spectra were run at a temperature of 27°C. The two resonances due to GDP show different intensities due to incomplete relaxation of these signals.
Catalytic properties of T. maritima MDIP synthase.
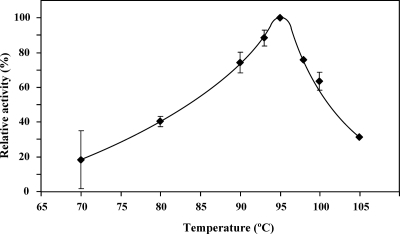
An array of experiments was carried out to evaluate the specificity of MDIP synthase. ADP-glucose, GDP-glucose, UDP-glucose, and TDP-glucose were examined as putative substrates together with DIP, but no reaction product was detected by 31P NMR. GDP-mannose was the only glycosyl donor used by the enzyme. Mannose-activated nucleotides other than GDP-mannose were not examined, due to commercial unavailability. DIP, MDIP, DIPP, GPI, diglycerol phosphate, glycerol, d,l-glycerol-3-P, myo-inositol, l-myo-inositol-1-P, and d-myo-inositol-1-P were tested as putative mannosyl acceptors. DIP was used by the enzyme, resulting in the formation of MDIP and small amounts of MMDIP; it was also verified that the enzyme was able to use MDIP and GDP-mannose to yield MMDIP. Therefore, MDIP synthase catalyzed two reactions: (i) transfer of the mannose group from GDP-mannose to DIP, producing MDIP, and (ii) transfer of the mannose group from GDP-mannose to MDIP, yielding MMDIP. This means that the product of the first reaction is also a substrate of the enzyme. We set out to characterize the kinetic parameters of the enzyme, using GDP-mannose and DIP as substrates. The initial reaction rates were obtained from the quantification of GDP produced using a spectrophotometric method, which has the advantage of a greater sensitivity than that of 1H NMR. MDIP synthase exhibited Michaelis-Menten kinetics; the apparent Km values for DIP and GDP-mannose are presented in Table 3. It should be noted that the spectrophotometric assay assesses the sum of MDIP and MMDIP produced in the two reaction steps; however, if the contribution of the MMDIP-forming reaction is small, the values of the kinetic parameters determined are good approximations for describing the first catalytic reaction. To assess the contribution of the MMDIP-forming reaction, the protocol described to determine the initial rates was followed (see Materials and Methods) with 100 mM DIP and 5 mM GDP-mannose, substrate concentrations intended to be saturating for MDIP synthase; the reaction was allowed to proceed for 30 s at 95°C as in the spectrophotometric assay, and the reaction products MDIP and MMDIP were quantified by NMR. The concentration of MDIP after 30 s was 0.11 mM; MMDIP was not detected (below the sensitivity limit, i.e., 0.03 mM). This shows that the formation of MMDIP is negligible under our assay conditions since DIP is likely to compete with MDIP for the same binding site and the DIP concentration is at least 1,000-fold higher than that of MDIP. Therefore, it is reasonable to assume that the second reaction step does not appreciably affect the values determined for Km and Vmax (Table 3). The kinetic properties of the enzyme for the second reaction (MDIP and GDP-mannose as substrates) were not investigated, due to difficulties in obtaining the gram amounts of pure MDIP required for the assays. However, an estimation of the Km for MDIP was made on the basis of a kinetic model (see Discussion). The presence of Mg2+ was required for maximal activity of MDIP synthase; in the absence of this cation, the activity was only 30% of the maximum value. The activity was 50% lower in the presence of 400 mM NaCl. The temperature for maximal activity was around 95°C (Fig. 5).
TABLE 3.
Kinetic properties of recombinant MDIP synthase of Thermotoga maritima and effect of Mg2+ and Na+ ions
| Parameter | Value for MDIP synthasec |
|---|---|
| Km (mM)a | |
| DIP substrate | 16 ± 4.0 |
| GDP-mannose substrate | 0.7 ± 0.1 |
| Vmax (μmol/min·mg protein) | >60 |
| Mg2+ concn (mM) | |
| 0 | 31b |
| 10 | 100b |
| NaCl concn (mM) | |
| 0 | 100b |
| 400 | 51b |
Km values were determined for MDIP synthase in mixtures containing 5 mM GDP-mannose and 0 to 300 mM DIP or 98 mM DIP and 0 to 5 mM GDP-mannose.
Expressed as percentages of the maximum activity.
All assays were carried out at 95°C.
FIG. 5.
Temperature dependence of the activity of recombinant Thermotoga maritima MDIP synthase. The data are the mean values of results from two independent experiments, except for 98°C and 105°C, which represent single experiments. The experimental conditions are described in Materials and Methods.
Thermodynamic stability of MDIP synthase.
The stability of the recombinant MDIP synthase of A. aeolicus was studied by differential scanning calorimetry. The enzyme from A. aeolicus was used for these studies due to the high production yield. The protein was highly stable, showing a transition temperature of 100°C in the presence of 1.5 M guanidinium chloride. In the absence of denaturants, no transition was observed in the temperature range examined (up to 120°C).
Determination of the stereochemistry of MDIP.
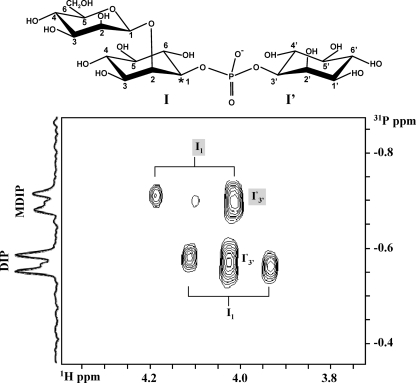
DIP comprises two stereochemically distinct inositol moieties. With the L-stereospecific numbering convention (1, 27), the phosphate bridges between position 1 of the inositol moiety I and position 3′ of the inositol moiety I′ (see the molecular representation of DIP depicted in Fig. 6). The array of NMR data reported above led to the conclusion that the mannosyl group in MDIP is linked at position 2 of one of the inositol moieties, but the identification of this inositol moiety demands specific isotopic labeling. For this purpose, DIP specifically labeled with carbon 13 at position 1 of the inositol moiety I was synthesized and used as a substrate for MDIP synthase (Fig. 6; see also Materials and Methods). The position of the mannosyl residue in MDIP was deduced from the analysis of 1H-31P correlation spectra (Fig. 7). For readers less acquainted with NMR, it is worth pointing out that the NMR resonance of a proton directly bound to a carbon 13 atom (—CH bond) will be split in two signals separated by a large coupling constant (typically greater than 120 Hz). It is also pertinent to note that proton 1 (of inositol I) and proton 3′ (of inositol I′) in DIP resonate at the same frequency since DIP is a symmetric molecule. However, when a mannosyl group is added to one of the inositol moieties of DIP, the symmetry is broken and those protons resonate at 4.10 ppm and 4.03 ppm, the former resonance being assigned to the inositol moiety carrying the mannose group (Table 1). The 1H-31P HSQC spectrum (Fig. 7) reveals a correlation between the phosphorus signal of MDIP (at −0.7 ppm) and the proton signal centered at 4.10 ppm, which is split in two components separated by 143 Hz (≈0.18 ppm); the spectrum also shows a correlation with another proton at a chemical shift of 4.03 ppm whose resonance exhibits no splitting and coincidentally overlaps the component at a lower frequency. These data show that the proton whose signal is split due to direct bonding with carbon 13 resonates at 4.10 ppm, hence belonging to the inositol moiety that carries the mannosyl residue (Table 1). Therefore, the mannosyl residue is linked to inositol I. The stereochemical representation of the MDIP molecule is shown in Fig. 7.
FIG. 6.
Reaction scheme used for the synthesis of mannosyl-di-myo-[1-13C]inositol-1,3′-phosphate. CDP-l-myo-[1-13C]inositol was the labeled precursor for the synthesis of DIP. The latter compound and GDP-mannose were incubated with a cell extract of Thermotoga maritima or with recombinant MDIP synthase to produce MDIP. I and I′ designate the inositol moieties in the l and d configurations, respectively (nomenclature according to Rodrigues et al. [27]). Asterisks designate the positions of labeled carbon atoms.
FIG. 7.
Section of a 1H-31P HSQC correlation spectrum of MDIP produced from incubation of GDP-mannose and DIP with a cell extract of T. maritima. The proton resonance at 4.10 ppm is due to proton 1 of the inositol moiety of MDIP, which is directly linked to mannose; the splitting of this signal, due to scalar coupling with carbon 13, shows that the mannosyl group is definitely linked to the inositol moiety that carries the label, i.e., moiety I (see text for details). Shaded I1 and I′3′ designate proton 1 and proton 3′ of MDIP, respectively; nonshaded I1 and I′3′ designate protons 1 and 3′ of the residual reagent, DIP, respectively.
DISCUSSION
The solute pool of T. maritima is composed exclusively of negatively charged compounds, illustrating the strong preference of hyperthermophilic organisms for charged solutes (30). This bacterium accumulates α- and β-glutamate, DIP, and two novel β-1,2-mannosides, herein identified as mannosyl-di-myo-inositol phosphate and di-mannosyl-di-myo-inositol phosphate. Thus far, we found MDIP and MMDIP only in species of the genera Thermotoga (T. neapolitana and T. maritima) and Aquifex (A. aeolicus and A. pyrophilus). The rareness of these solutes and their close relationship with DIP, the most widespread solute in hyperthermophiles, motivated us to elucidate their biosynthesis.
Herein, we report the characterization of a β-1,2-mannosyltransferase that uses DIP and GDP-mannose for the synthesis of MDIP; the same enzyme catalyzes a subsequent mannosylation reaction that results in the formation of MMDIP. A comparative genomic analysis led to the identification of the gene conferring these activities in the genome of T. maritima. Homologs with high sequence identity are present in other species of the genus Thermotoga (T. neapolitana, T. petrophila, and Thermotoga sp. strain RQ2). Moreover, a gene with 36% identity was found in the genome of A. aeolicus, and we confirmed that, despite the relatively low identity, the gene product exhibited MDIP synthase activity. The enzymes implied in the synthesis of DIP, the substrate for MDIP synthase, also show a relatively low degree of sequence similarity (27). For example, in Thermotoga spp., the relevant activities are conferred by two separate genes, while a fused gene is encountered in A. aeolicus (27); altogether, these findings suggest that the pathways for DIP and MDIP synthesis diverged early during the evolution of these two bacterial genera.
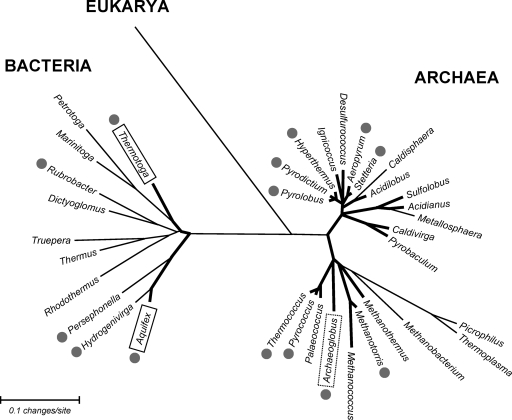
Homologs of MDIP synthase are found only in the genera Thermotoga, Aquifex, and Archaeoglobus, indicating that MDIP synthase is a singular, novel enzyme, thus far restricted to hyperthermophilic organisms. In line with this view, the thermophiles Persephonella marina and Hydrogenivirga sp., members of the order Aquificales, lack a homologous enzyme, although they possess genes for the synthesis of DIP. The available results suggest a correlation between MDIP and hyperthermophily. It is curious, however, that MDIP synthase is absent within the most-extreme hyperthermophiles, the archaeons Pyrolobus fumarii and Pyrodictium occultum, which accumulate DIP as a major solute (Fig. 8).
FIG. 8.
Unrooted phylogenetic tree based on 16S rRNA sequences of thermophilic (thin lines) and hyperthermophilic (thick lines) organisms, highlighting the distribution of DIP (gray circles) and MDIP (boxes) (references 11, 17, and 30 and our unpublished results). The human 18S rRNA sequence was used as an outgroup. The dotted box means that Archaeoglobus profundus possesses the MDIP synthase gene but that the accumulation of MDIP is unknown. The MEGA 4.0 software program (33) was used for sequence alignment and for generation of the phylogenetic tree by use of the neighbor-joining method. Sequences of 16S and 18S rRNA are from the Ribosomal Database Project (http://rdp.cme.msu.edu/hierarchy/hb_intro.jsp).
The MDIP synthase of T. maritima catalyzed not only the synthesis of MDIP but also that of other DIP derivatives containing two- and three-mannosyl groups linked via β-(1,2) glycosidic bonds, i.e., MMDIP and minor amounts of the tri-mannosylated form. All the reactions involve an inverting mechanism in which the α configuration of the substrate, GDP-mannose, was converted into the opposite anomeric configuration. Therefore, MDIP synthase is an inverting β-1,2-mannosyltransferase. The analysis of the amino acid sequence revealed no homology with any of the 91 glycosyltransferase families existing in the Carbohydrate-Active Enzymes database (CAZy) (www.cazy.org [7]) (P. M. Coutinho, CAZy, personal communication), indicating that MDIP synthase is the first member of a new glycosyltransferase family. The number of known β-1,2-mannosyltransferases is extremely low, and the CAZy database contains only one family of β-1,2-mannosyltransferases, comprising the enzymes implicated in the synthesis of glycans of glycoproteins in the cell walls of Pichia pastoris and Candida albicans (21). However, the amino acid sequences of these β-1,2-mannosyltransferases and MDIP synthase are totally unrelated. Therefore, it appears that the MDIP synthase herein disclosed is a novel enzyme, as no counterparts have been found thus far.
The MDIP synthase of T. maritima has a high degree of specificity for GDP-mannose as a glycosyl donor but showed some flexibility in regard to the mannosyl acceptor when DIP, MDIP, and MMDIP were used as substrates. The relatively high Km value (16 mM) for DIP correlates with the accumulation of this solute in the host organism; the intracellular concentration of DIP in T. maritima is 15 to 20 mM, calculated with the assumption that the intracellular volume of Pyrococcus furiosus is a reasonable approximation to that of T. maritima (20). The experimental determination of the Km value of MDIP synthase for MDIP was not possible, due to substrate unavailability. The observation that MMDIP was detected only in cells grown under conditions leading to strong accumulation of MDIP (above 50 mM) suggested that the Km value for MDIP should be considerably higher than that for DIP. Therefore, we deemed it interesting to estimate the Km for MDIP by using a kinetic model to describe the two reaction steps catalyzed by the enzyme (M represents GDP-mannose): DIP + M ↔ MDIP and MDIP + M ↔ MMDIP. The model assumes Michaelis-Menten kinetics for both reactions and the establishment of a steady-state condition in regard to MDIP, i.e., the rate of MDIP formation was considered equal to the rate of MDIP consumption. Hence, the following relationship is derived:
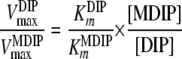 |
(1) |
where 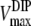 and
and 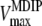 designate the kinetic parameter Vmax for DIP and MDIP, respectively.
designate the kinetic parameter Vmax for DIP and MDIP, respectively.
With the reasonable assumption that the enzyme has equal Vmax values for DIP and MDIP, the above-described relationship becomes
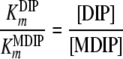 |
(2) |
If the steady-state condition in the cells is a good approximation, a Km value of 60 mM for MDIP is estimated by introducing in equation 2 the DIP and MDIP concentrations determined for cells grown under heat stress (Table 2) and a 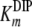 of 16 mM. The magnitude of the
of 16 mM. The magnitude of the 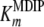 parameter explains the absence of MMDIP under optimal or osmotic stress conditions, when the intracellular concentration of MDIP is far below the estimated value.
parameter explains the absence of MMDIP under optimal or osmotic stress conditions, when the intracellular concentration of MDIP is far below the estimated value.
It is interesting that the level of DIP in T. maritima did not change upon exposure to either heat stress or osmotic stress, in contrast with the observation that the DIP pool increases in response to heat stress in many of the hyperthermophiles examined, like Pyrococcus furiosus, Archaeoglobus fulgidus, A. pyrophilus, and Methanotorris igneus (9, 30). The constancy of the DIP level is probably related to the use of this compound as an intermediate metabolite in the synthesis of MDIP and MMDIP, the solutes whose levels increased notably under heat stress.
The presence of β-1,2-mannosides as components of the solute pool in hyperthermophilic organisms is surprising since the β-1,2-mannoside linkage is very rare in nature. This type of linkage has not been found in mammalian cells and, to our knowledge, has been reported to occur in only a few organisms, mostly pathogens: in glycoproteins of Candida spp., Salmonella enterica serovar Thompson, and Pichia pastoris (17, 21, 32) and in reserve oligosaccharides of Leishmania spp. (24) and of the thermophilic bacterium Truepera radiovitrix (our unpublished results). The absence of β-1,2-mannosides in humans leads to the proposal that β-1,2-mannosyltransferases could be suitable targets for the development of drugs against infectious diseases.
The chemical composition of the new DIP derivatives characterized here leads us to speculate about a connection with the polar heads in the glycolipids of T. maritima. Unfortunately, the information on the glycolipid composition of this bacterium is very limited, and in particular, phosphatidyl-myo-inositol mannosides have not been reported (18). This polar group has been detected in the cell walls of mycobacteria, but the stereochemistry is clearly distinct: the mannose moiety is bound via an α-1,2-linkage to the inositol phosphate group, which has a d configuration instead of the l configuration found in the mannosylated forms of DIP (16, 22, 28). Accordingly, the glycosyltransferases involved in the synthesis of the mycobacterial glycolipids are unrelated to the MDIP synthase of T. maritima.
In conclusion, this work led to the structural characterization of two β-1,2-mannosides accumulating in T. maritima in response to heat stress; these compounds are synthesized by the activity of a novel β-1,2-mannosyltransferase that uses DIP as a mannosyl acceptor and appears to be the first member of a new family of glycosyltransferases. These unusual features fuel interest in the elucidation of the catalytic mechanism. Work is in progress to determine the crystallographic structure of this unique enzyme.
Acknowledgments
This work was funded by Fundação para a Ciência e a Tecnologia (FCT), POCTI Portugal, and FEDER (projects PTDC/BIA-MIC/71146/2006 and PTDC/BIO70806/2006). M.V.R. is supported by a fellowship from FCT (SFRH/BD/25539/2005). The NMR spectrometers at CERMAX are part of the National NMR Network and were acquired with funds from FCT and FEDER.
Mass spectrometry data were obtained from the Mass Spectrometry Laboratory, Analytical Services Unit, Instituto de Tecnologia Química e Biológica. We thank T. Q. Faria for advice on the differential scanning calorimetry experiments and for deducing the kinetic equations.
Footnotes
Published ahead of print on 31 July 2009.
REFERENCES
- 1.Anonymous. 1989. Numbering of atoms in myo-inositol. Recommendations 1988. Biochem. J. 258:1-2. [PMC free article] [PubMed] [Google Scholar]
- 2.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumer-Schuette, S. E., I. Kataeva, J. Westpheling, M. W. Adams, and R. M. Kelly. 2008. Extremely thermophilic microorganisms for biomass conversion: status and prospects. Curr. Opin. Biotechnol. 19:210-217. [DOI] [PubMed] [Google Scholar]
- 4.Bock, K., and C. Pedersen. 1974. A study of 13CH coupling constants in hexopyranoses. J. Chem. Soc. Perkin 2 3:293-297. [Google Scholar]
- 5.Borges, N., L. G. Goncalves, M. V. Rodrigues, F. Siopa, R. Ventura, C. Maycock, P. Lamosa, and H. Santos. 2006. Biosynthetic pathways of inositol and glycerol phosphodiesters used by the hyperthermophile Archaeoglobus fulgidus in stress adaptation. J. Bacteriol. 188:8128-8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Cantarel, B. L., P. M. Coutinho, C. Rancurel, T. Bernard, V. Lombard, and B. Henrissat. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37:D233-D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chhabra, S. R., K. R. Shockley, S. B. Conners, K. L. Scott, R. D. Wolfinger, and R. M. Kelly. 2003. Carbohydrate-induced differential gene expression patterns in the hyperthermophilic bacterium Thermotoga maritima. J. Biol. Chem. 278:7540-7552. [DOI] [PubMed] [Google Scholar]
- 9.Ciulla, R. A., S. Burggraf, K. O. Stetter, and M. F. Roberts. 1994. Occurrence and role of di-myo-inositol-1,1′-phosphate in Methanococcus igneus. Appl. Environ. Microbiol. 60:3660-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conners, S. B., E. F. Mongodin, M. R. Johnson, C. I. Montero, K. E. Nelson, and R. M. Kelly. 2006. Microbial biochemistry, physiology, and biotechnology of hyperthermophilic Thermotoga species. FEMS Microbiol. Rev. 30:872-905. [DOI] [PubMed] [Google Scholar]
- 11.Empadinhas, N., V. Mendes, C. Simoes, M. S. Santos, A. Mingote, P. Lamosa, H. Santos, and M. S. Costa. 2007. Organic solutes in Rubrobacter xylanophilus: the first example of di-myo-inositol-phosphate in a thermophile. Extremophiles 11:667-673. [DOI] [PubMed] [Google Scholar]
- 12.Faria, T. Q., A. Mingote, F. Siopa, R. Ventura, C. Maycock, and H. Santos. 2008. Design of new enzyme stabilizers inspired by glycosides of hyperthermophilic microorganisms. Carbohydr. Res. 343:3025-3033. [DOI] [PubMed] [Google Scholar]
- 13.Gu, X., M. Chen, Q. Wang, M. Zhang, B. Wang, and H. Wang. 2005. Expression and purification of a functionally active recombinant GDP-mannosyltransferase (PimA) from Mycobacterium tuberculosis H37Rv. Protein Expr. Purif. 42:47-53. [DOI] [PubMed] [Google Scholar]
- 14.Huber, R., T. A. Langworthy, H. König, M. Thomm, C. R. Woese, U. B. Sleytr, and K. O. Stetter. 1986. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch. Microbiol. 144:324-333. [Google Scholar]
- 15.Jorge, C. D., P. Lamosa, and H. Santos. 2007. α-D-mannopyranosyl-(1→2)-α-D-glucopyranosyl-(1→2)-glycerate in the thermophilic bacterium Petrotoga miotherma—structure, cellular content and function. FEBS J. 274:3120-3127. [DOI] [PubMed] [Google Scholar]
- 16.Kordulakova, J., M. Gilleron, K. Mikusova, G. Puzo, P. J. Brennan, B. Gicquel, and M. Jackson. 2002. Definition of the first mannosylation step in phosphatidylinositol mannoside synthesis. PimA is essential for growth of mycobacteria. J. Biol. Chem. 277:31335-31344. [DOI] [PubMed] [Google Scholar]
- 17.Lamosa, P., L. G. Goncalves, M. V. Rodrigues, L. O. Martins, N. D. Raven, and H. Santos. 2006. Occurrence of 1-glyceryl-1-myo-inosityl phosphate in hyperthermophiles. Appl. Environ. Microbiol. 72:6169-6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manca, M. C., B. Nicolaus, V. Lanzotti, A. Trincone, A. Gambacorta, J. Peter-Katalinic, H. Egge, R. Huber, and K. O. Stetter. 1992. Glycolipids from Thermotoga maritima, a hyperthermophilic microorganism belonging to Bacteria domain. Biochim. Biophys. Acta 1124:249-252. [DOI] [PubMed] [Google Scholar]
- 19.Martins, L. O., L. S. Carreto, M. S. da Costa, and H. Santos. 1996. New compatible solutes related to di-myo-inositol-phosphate in members of the order Thermotogales. J. Bacteriol. 178:5644-5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martins, L. O., and H. Santos. 1995. Accumulation of mannosylglycerate and di-myo-inositol-phosphate by Pyrococcus furiosus in response to salinity and temperature. Appl. Environ. Microbiol. 61:3299-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mille, C., P. Bobrowicz, P. A. Trinel, H. Li, E. Maes, Y. Guerardel, C. Fradin, M. Martinez-Esparza, R. C. Davidson, G. Janbon, D. Poulain, and S. Wildt. 2008. Identification of a new family of genes involved in β-1,2-mannosylation of glycans in Pichia pastoris and Candida albicans. J. Biol. Chem. 283:9724-9736. [DOI] [PubMed] [Google Scholar]
- 22.Morita, Y. S., J. H. Patterson, H. Billman-Jacobe, and M. J. McConville. 2004. Biosynthesis of mycobacterial phosphatidylinositol mannosides. Biochem. J. 378:589-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelberg, G. G. Sutton, R. D. Fleischmann, J. A. Eisen, O. White, S. L. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed] [Google Scholar]
- 24.Ralton, J. E., T. Naderer, H. L. Piraino, T. A. Bashtannyk, J. M. Callaghan, and M. J. McConville. 2003. Evidence that intracellular β-1-2 mannan is a virulence factor in Leishmania parasites. J. Biol. Chem. 278:40757-40763. [DOI] [PubMed] [Google Scholar]
- 25.Ramakrishnan, V., and M. W. W. Adams. 1995. Preparation of genomic DNA from sulfur-dependent hyperthermophilic Archaea, p. 95-96. In F. T. Robb and A. R. Place (ed.), Archaea: a laboratory manual—thermophiles. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Rodionov, D. A., O. V. Kurnasov, B. Stec, Y. Wang, M. F. Roberts, and A. L. Osterman. 2007. Genomic identification and in vitro reconstitution of a complete biosynthetic pathway for the osmolyte di-myo-inositol-phosphate. Proc. Natl. Acad. Sci. USA 104:4279-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodrigues, M. V., N. Borges, M. Henriques, P. Lamosa, R. Ventura, C. Fernandes, N. Empadinhas, C. Maycock, M. S. da Costa, and H. Santos. 2007. Bifunctional CTP:inositol-1-phosphate cytidylyltransferase/CDP-inositol:inositol-1-phosphate transferase, the key enzyme for di-myo-inositol-phosphate synthesis in several (hyper)thermophiles. J. Bacteriol. 189:5405-5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salman, M., J. T. Lonsdale, G. S. Besra, and P. J. Brennan. 1999. Phosphatidylinositol synthesis in mycobacteria. Biochim. Biophys. Acta 1436:437-450. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Santos, H., P. Lamosa, T. Q. Faria, N. Borges, and C. Neves. 2007. The physiological role, biosynthesis, and mode of action of compatible solutes from (hyper)thermophiles, p. 86-103. In C. Gerday and N. Glansdorff (ed.), Physiology and biochemistry of extremophiles. ASM Press, Washington, DC.
- 31.Santos, H., P. Lamosa, and N. Borges. 2006. Characterization and quantification of compatible solutes in (hyper)thermophilic microorganisms. Methods Microbiol. 35:173-199. [Google Scholar]
- 32.Suzuki, A., N. Shibata, M. Suzuki, F. Saitoh, H. Oyamada, H. Kobayashi, S. Suzuki, and Y. Okawa. 1997. Characterization of β-1,2-mannosyltransferase in Candida guilliermondii and its utilization in the synthesis of novel oligosaccharides. J. Biol. Chem. 272:16822-16828. [DOI] [PubMed] [Google Scholar]
- 33.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 34.VanFossen, A. L., D. L. Lewis, J. D. Nichols, and R. M. Kelly. 2008. Polysaccharide degradation and synthesis by extremely thermophilic anaerobes. Ann. N. Y. Acad. Sci. 1125:322-337. [DOI] [PubMed] [Google Scholar]