Abstract
Assembly of the tubulin-like cytoskeletal protein FtsZ into a ring structure at midcell establishes the location of the nascent division sites in prokaryotes. However, it is not yet known how the assembly and contraction of the Z ring are regulated, especially in cyanobacteria, the environmentally crucial organisms for which only one FtsZ partner protein, ZipN, has been described so far. Here, we characterized SepF and Ftn6, two novel septal proteins, in the spherical-celled strain Synechocystis PCC 6803. Both proteins were found to be indispensable to Synechocystis sp. strain PCC 6803. The depletion of both SepF and Ftn6 resulted in delayed cytokinesis and the generation of giant cells but did not prevent FtsZ polymerization, as shown by the visualization of green fluorescent protein (GFP)-tagged FtsZ polymers. These GFP-tagged Z-ring-like structures often appeared to be abnormal, because these reporter cells respond to the depletion of either SepF or Ftn6 with an increased abundance of total, natural, and GFP-tagged FtsZ proteins. In agreement with their septal localization, we found that both SepF and Ftn6 interact physically with FtsZ. Finally, we showed that SepF, but not Ftn6, stimulates the formation and/or stability of FtsZ polymers in vitro.
Binary fission of a mother cell to form two daughter cells is a widely conserved cell proliferation mechanism. In nearly all bacteria, cell division is initiated by the polymerization into a ring-like structure at midcell of the tubulin homolog GTPase protein FtsZ, which is also found in some archae, as well as in plastids and some mitochondria (for reviews, see references 7, 21, and 33). The Z-ring is subsequently used as a scaffold for recruitment of downstream factors that execute the synthesis of the division septum. The assembly of this complex, also referred to as the divisome, has been thoroughly investigated in studies of the rod-shaped model organisms Escherichia coli and Bacillus subtilis) (for reviews, see references 3, 4, 7, 9, 11, 19, and 21). In E. coli, more than 10 different proteins are required for the progression and completion of cell division. They are designated Fts proteins because their depletion leads to filamentation of the bacteria, and they are recruited to the division site in the following sequential order: FtsZ→FtsA/ZipA/ZapB→FtsK→FtsQ and FtsL/FtsB→FtsW→FtsI and FtsN.
The stability of the FtsZ protofilaments is thought to be important for assembly of the septal Z ring. Four FtsZ-interacting proteins have been shown to promote FtsZ polymerization and/or Z-ring stabilization, namely, ZapA and ZipA (found only in gammaproteobacteria), FtsA (an actin-like protein), and SepF (not found in gammaproteobacteria) (10, 31). Both FtsA and ZipA assemble at the Z-ring early and participate in its anchorage to the inner face of the cytoplasmic membrane of the cell. They also participate in the recruitment of the downstream cytokinetic factor FtsK. Subsequently, the recruitment of FtsQ and the FtsB/FtsL complex allow the progressive assembly of downstream factors (FtsW, FtsI, and FtsN) involved in synthesis of the septal cell wall (7).
By contrast, the negative regulatory proteins MinCDE, DivIVA, EzrA, SulA, and Noc operate in the destabilization and positioning of the Z-ring at midcell (7, 21, 30), sometimes through a direct interaction with FtsZ (SulA, MinC, and ErzA).
Little is known concerning cell division in cyanobacteria, in spite of their crucial importance to the biosphere (5, 27, 34) and their interest for biotechnologists (1, 6, 32). Cyanobacteria are also attractive because many species (such as E. coli and B. subtilis) exhibit a cylindrical morphology with a well-defined middle, whereas many others have a spherical shape (29) and thus possess an infinite number of potential division planes at the point of greatest cell diameter. Furthermore, as the progenitor of the chloroplasts (8), cyanobacteria can be of help for deciphering the stromal chloroplastic division machinery (33). Interestingly, several cell division factors occurring in E. coli and B. subtilis have been shown (FtsZ, MinCDE, and SulA) or proposed (FtsE, FtsI, FtsQ, and FtsW) to be conserved in cyanobacteria (23, 26) and chloroplasts (which lack MinC) (33). In contrast, ftsA, ftsB, zipA, ftsK, ftsL, ftsN, and zapA have not been detected in cyanobacteria.
So far, cyanobacterial cytokinesis has mainly been investigated using the two unicellular species Synechococcus sp. strain PCC 7942 (rod shaped; hereafter S. elongatus) and Synechocystis sp. strain PCC 6803 (spherical-celled; hereafter Synechocystis sp.) and the filamentous strain Anabaena PCC 7120, all of which possess a fully sequenced genome (http://genome.kazusa.or.jp/cyanobase/) that is easily manipulated (16). Both FtsZ and ZipN/Ftn2/Arc6, a protein occurring only in cyanobacteria (ZipN [alternative name, Ftn2]) and plant chloroplasts (Arc6), were found to be crucial for cytokinesis (17, 23, 26) and to physically interact with each other (20, 23). We also reported that the MinCDE system participates in determining the correct positioning of the septal Z ring at midcell (23). In addition, it has recently been shown in studies of Synechococcus sp. that inactivation of both the cdv2 gene (an orthologue of the gene encoding B.subtilis sepF) and the ftn6 gene (present in only some cyanobacteria) promotes filamentation, though their role in cell division has yet to be characterized (16, 26).
In a continuous effort to characterize the divisome machine of Synechocystis sp., we have used a combination of in vivo and in vitro techniques for thorough analysis of the SepF and Ftn6 proteins. We report here that both SepF and Ftn6 are crucial cytokinetic proteins that localize at the division site at midcell and whose depletion leads to the formation of giant cells that remain spherical. In agreement with their septal localization, both SepF and Ftn6 were found to interact physically with FtsZ; also, SepF, but not Ftn6, was found to stimulate the formation and/or stability of FtsZ polymers.
MATERIALS AND METHODS
Bacterial strains, growth, plasmids, and gene transfer procedures.
Synechocystis sp. strain PCC 6803 was grown and transformed at 30°C in BG11 medium (29) enriched with 3.78 mM Na2CO3 under conditions of continuous white light of standard fluence (2,500 lx; 31.25 μE m−2 s−1) as described in reference 2. E. coli strains TOP10 (Invitrogen), CM404, and BL21(DE3), grown at 30°C (strain CM404) or 37°C (other strains) on LB with or without 1% glucose, were used for gene manipulation (strain TOP10), protein production (strain BL21), or conjugative transfer (25) to Synechocystis sp. (CM404) of the plasmids derived from the pSBTac expression vector that replicates at about 10 copies per cell (i.e., one copy per chromosome copy) and expresses the studied genes from the E. coli tac promoter (22). The selective antibiotics and final concentrations were as follows: for E. coli, ampicillin (Ap) at 100 μg ml−1, kanamycin (Km) at 50 μg ml−1, and spectinomycin (Sp) at 100 μg ml−1; and for Synechocystis sp., Km at 50 to 300 μg ml−1, streptomycin (Sm) at 5 μg ml−1, and Sp at 2.5 to 10 μg ml−1.
Gene cloning and DNA manipulations.
All Synechocystis genes, surrounded by their flanking regions (about 300 bp) for homologous recombinations mediating targeted gene replacement in Synechocystis sp. (18), were amplified from wild-type (WT) DNA by PCR, using specific primers. After cloning was carried out with the appropriate plasmids (Table 1), site-directed mutagenesis was performed and disruptions were made through standard PCR-driven overlap extension (12). We used deletion cassettes that carry the antibiotic resistance marker inserted in the same orientation as the gene to be inactivated. The PCR-amplified protein-coding sequences were tagged with six-His through cloning into the E. coli expression vector pET28b(+) (Table 1) before or after fusion with the green fluorescent protein (GFP) sequence performed using overlapping PCR and were cloned into the NheI-NotI sites of pET28b(+). All constructions were verified by PCR and DNA sequencing (BigDye kit; ABI Perking Elmer).
TABLE 1.
Characteristics of the plasmids used in this study
| Plasmid | Relevant feature(s)a | Source or reference |
|---|---|---|
| pGEMT | AT overhang Ampr cloning vector | Promega |
| pUC4K | Source of the Kmr marker gene | Pharmacia |
| psepF | pGEMT with the Synechocystis sepF gene (slr2073) and flanking sequences where part of the SepF CS (from 186 to 426 bp) was replaced by a SmaI site | This study |
| pΔsepF::Kmr | psepF with the Kmr marker inserted into the unique SmaI site | This study |
| pFtn6 | pGEMT with the Synechocystis FTN6 gene (sll1939) and flanking sequences where part of the Ftn6 CS (from 33 to 609 bp) was replaced by a SmaI site | This study |
| pΔftn6::Kmr | pftn6 with the Kmr marker inserted in the unique SmaI site | This study |
| pSBT | Kmr Smr Spr plasmid replicating in Synechocystis sp. and expressing any gene cloned downstream of its E. coli tac promoter | 22 |
| pSM | pSBT derivative lacking the 1.349-kb SmaI segment carrying the Kmr gene; all studied genes were cloned as blunt-end DNA segments downstream of its tac promoter at the unique SmaI site | This study |
| pC-GFP | Source of the enhanced GFP | 23 |
| pSZ-GFP | pSBT producing the Synechocystis FtsZ protein tagged with GFP | 23 |
| pSM-SepF | pSM producing the Synechocystis SepF protein | This study |
| pSM-GFP-SepF | pSM producing the GFP-SepF protein | This study |
| pSM-Ftn6 | pSM producing the Synechocystis Ftn6 protein | This study |
| pSM-GFP-Ftn6 | pSM producing the GFP-Ftn6 protein | This study |
| pCZ | Ampr KmrE. coli plasmid producing the Synechocystis FtsZ protein tagged with a six-His tag at its C terminus | 23 |
| pET28b(+) | KmrE. coli expression vector with T7 and LacO promoters with T7 and six-His tags added and including LacI gene | Novagen |
| p28-SepF | pET28b(+) harboring the SepF CS (with its own stop codon) cloned between the NdeI-BamHI sites | This study |
| p28-Ftn6 | pET28b(+) harboring the Ftn6 CS (with its own stop codon) cloned between the NdeI-BamHI sites | This study |
| p28-GFP-SepF | pET28b(+) harboring the GFP-SepF translational fusion cloned between the NdeI-BamHI sites | This study |
| p28-GFP-Ftn6 | pET28b(+) harboring the GFP-Ftn6 translational fusion cloned between the NdeI-BamHI sites | This study |
CS, protein coding sequence; Δ, deletion.
Light and fluorescence microscopy.
Synechocystis cells (1.25 × 105) from a mid-log-phase culture were placed on microscope slides and immobilized by 5 to 10 min of incubation at room temperature. Images were captured using a Leica DMRXA microscope equipped with a ×100 oil immersion lens, a Ropper Scientific Micromax cooled charge-coupled-device camera, and Metamorph software (Universal Imaging). The final processing of images for presentation was done using Adobe Photoshop.
Fluorescence-activated cell sorter analysis.
Cells from mid-log-phase liquid cultures were harvested, washed twice, and resuspended in phosphate-buffered saline (Sigma-Aldrich) to a final optical density at 580 nm of 0.3 (1.5 × 107 cells ml−1). Then, 2 × 104 cells were analyzed by using a FACSCalibur cytometer (Becton Dickinson) with the following settings: forward scatter (FSC), E01 log; side scatter, 350 V. Results were collected with CellQuest software, version 3.1 (Becton Dickinson). Data were plotted on a two-dimensional graph (x axis, FSC; y axis, number of cells). Then, histograms from WT and mutant strains were superimposed. Experiments were repeated twice.
Protein production and purification.
Recombinant E. coli BL21 cells were grown at 37°C for 2 h (to an optical density at 600 nm of 0.5 to 0.7) prior to induction with IPTG (1 mM isopropyl-β-d-thiogalactopyranoside) for 16 h at 28°C. Cells were harvested by centrifugation for 10 min at 5,000 rpm, washed with lysis buffer (50 mM Tris-HCl [pH 8.0], 300 mM NaCl, 5 mM imidazole), resuspended for 5 min in ice-cold lysis buffer containing 2 mM phenylmethylsulfonyl fluoride, and subsequently incubated with lysozyme (1 mg/ml−1) for 1 h on ice. Cells were disrupted by ultrasonication with three pulses of 30 s each. The lysate was cleared by centrifugation at 12,000 × g for 30 min at 4°C. Six-His-tagged proteins were overexpressed and purified (23) on nickel-nitrilotriacetic acid agarose beads (Invitrogen), washed extensively with lysis buffer containing 25 mM imidazole, and eluted with lysis buffer containing 150 to 300 mM imidazole. Fractions containing the recombinant proteins were pooled and dialyzed against 50 mM Tris-HCl (pH 8.0)-5% (wt/vol) glycerol-300 mM NaCl. The purified proteins were concentrated using Amicon Ultra PL-10 (Millipore) at 4°C, quantified (Bio-Rad protein assay), aliquoted, and stored at −80°C until use. Proteins were estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to be ≥97% pure (data not shown).
In vitro analysis of FtsZ interaction with GFP-tagged proteins.
Analyses of interactions between preassembled FtsZ polymers and the GFP-tagged SepF or Ftn6 proteins were done as described previously (23). The polymers were visualized by phase-contrast microscopy and fluorescence microscopy.
Sedimentation assay.
Analysis of the influence of both SepF and Ftn6 on the polymerization of FtsZ (as determined by the quantity of FtsZ-sedimentable mass) was performed essentially as described previously (31). Purified FtsZ (4.5 μM) was polymerized in assembly buffer (50 mM Tris [pH 7.5], 150 mM NaCl) for 30 min on ice in the presence of various concentrations (0, 2, 3, and 4 μM) of either SepF or Ftn6. Then, 10 mM MgCl2 and 1 mM GTP were added to the reaction mixture, which was then incubated for an additional 20 min at 37°C, and FtsZ polymers were sedimented by centrifugation at 28,000 × g for 30 min at 30°C. The pellets were dissolved in 100 μl of SDS loading buffer and heated at 95°C for 5 min. Samples (20 μl) were subjected to electrophoresis using SDS-PAGE (12%), blotted onto a nitrocellulose membrane, and probed with anti-His tag (MAb 4A12E4; Invitrogen). Secondary M680 antibodies (Odyssey) were used to detect FtsZ and SepF or Ftn6 proteins. Images were obtained using an infrared scanner (Odyssey).
RESULTS
The depletion of both of the indispensable Synechocystis SepF and Ftn6 proteins leads to formation of giant cells.
The Synechocystis genome (14) encodes two proteins, Slr2073 and Sll1939 (CyanoBase; http://genome.kazusa.or.jp/cyanobase), that share significant homology with the Synechoccocus Ftn6 and Cdv2 proteins that are required for formation of cells of normal size (26). Indeed, both of the Synechoccocus mutants lacking Ftn6 or depleted with respect to Cdv2 (an indispensable protein in this organism) exhibited a filamentous phenotype and displayed enlarged cells (17, 26). Also interesting is that whereas Ftn6 was detected in only some cyanobacteria (26), Cdv2 was found to be significantly homologous to the SepF protein of B. subtilis that stimulates FtsZ assembly by binding along its length (10, 31).
To investigate the effect of the presence of SepF and Ftn6 on the morphology and cytokinesis of Synechocystis sp., we constructed ΔsepF::Kmr and Δftn6::Kmr deletion cassettes (Table 1) in which each studied protein-coding sequence was replaced by the transcription-terminator-less Km-resistant (Kmr) marker for selection. After transformation in Synechocystis sp., which harbors about 10 chromosome copies per cell (18), we verified through PCR and DNA sequencing that the Kmr marker had properly replaced the studied gene; then, we assayed whether the segregation of WT and mutant chromosome copies was complete (i.e., whether the gene was dispensable to cell growth) or not (gene indispensable). All transformant clones invariably retained WT chromosome copies, even after several rounds of restreaking in the presence of increasing concentrations of Km to favor the amplification of the mutant chromosome copies (data not shown). These findings indicated that both the sepF and ftn6 genes are essential to the viability of Synechocystis sp. The SepF-depleted mutant (sepF::Kmr/sepF+) appeared to grow slower than both the WT strain and the Ftn6-depleted (ftn6::Kmr/ftn6+) mutant (doubling times about 19 h, 9 h, and 12 h, respectively).
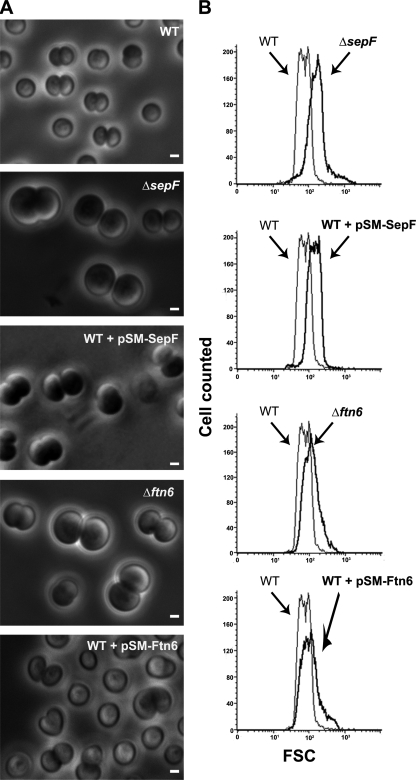
Then, we inspected the cell morphology of these mutants through phase-contrast microscopy (Fig. 1). Both mutants, ΔsepF::Kmr/sepF+ and Δftn6::Kmr/ftn6+, displayed similar aberrant “giant” cell morphologies (45% and 31% of the cell populations, respectively; Fig. 1A), suggesting that cell division had somehow been caused to slow. The Δftn6::Kmr/ftn6+ mutant also displayed an increased amount of “doublet cells” compared to the WT strain (80% versus 60%), again suggesting that cell division had slowed. The global increase in cell size in the mutants compared to the WT strain was also confirmed using flow cytometry (Fig. 1B) by measuring the proportion of the FSC value to cell size (24). The mean FSC values were as follows: for the WT strain, 75.94; for ΔsepF::Kmr/sepF+ (noted as ΔsepF), 183.87; and for Δftn6::Kmr/ftn6+ (noted as Δftn6), 122.64.
FIG. 1.
Influence of the abundance of SepF and Ftn6 proteins on the morphology and size of Synechocystis cells. Phase-contrast image (A) (scale bars, 1 μm) and flow cytometry analysis (B) of WT or mutant cells depleted with respect to (Δ) or producing the natural or tagged versions of the studied proteins from a pSM expression plasmid (Table 1). For each mutant strain, the FSC histogram (bold lines) has been overlaid with that of the WT to better visualize the influence of the mutation on cell size. These experiments were performed twice with two independent clones harboring the same mutation.
To further confirm that both SepF and Ftn6 are important for normal cell size formation, we cloned the corresponding protein-coding sequences in our pSM plasmid (Table 1), a Kms derivative of our previously constructed pSB2T expression vector (22), which harbors the E. coli tac promoter for expression of the studied gene and replicates autonomously at 10 copies per cell, i.e., 1 copy per chromosome copy (22). The resulting pSM-SepF and pSM-Ftn6 plasmids were transferred to Synechocystis sp., where they both modified cell morphology, as expected (Fig. 1). Indeed, both the WT/pSM-SepF and WT/pSM-Ftn6 reporter strains exhibited giant cells (28% and 33%, respectively), likely accounting for their high FSC mean values (123.12 and 119.17, respectively). Interestingly, the WT/pSM-Ftn6 mutant also displayed a total of 33% twisted (nonspherical) cells, a result not observed with the WT/pSM-SepF mutant.
Depletion of SepF and Ftn6 alters both the shape and the positioning of GFP-tagged FtsZ structures.
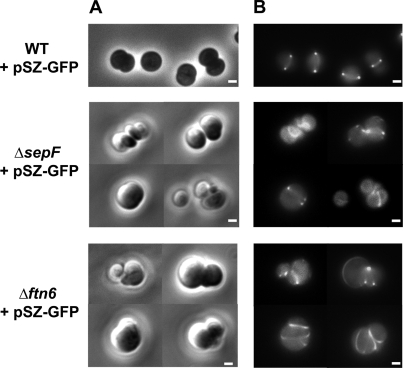
One of the very important features of cytokinetic proteins is their role in assembly and/or positioning of the septal Z ring. For instance, no Z rings are formed at midcell in the absence of both FtsA and ZipA E. coli proteins (28) or after the depletion of the Synechocystis ZipN protein (23). To test the possible influence of the SepF and Ftn6 proteins on FtsZ structures, we introduced into the SepF- and Ftn6-depleted mutants the pSZ-GFP reporter plasmid (Table 1) we had previously used to visualize the septal Z ring at the middle of WT cells (23). In both SepF- and Ftn6-depleted cells, the FtsZ structures appeared to be localized at aberrant (ectopic) positions (Fig. 2) and, consistently, these reporter cells exhibited abnormal morphologies (occurrence of spiraled cells and excrescences) likely resulting from asymmetrical and/or incomplete septation. Collectively, these observations indicate that the depletion of both SepF and Ftn6 resulted in delayed cytokinesis and in the generation of giant cells but did not prevent FtsZ polymerization. That the GFP-tagged Z-ring-like structures appeared to be abnormal in these reporter cells resulted from the fact that they exhibit an increased abundance of FtsZ proteins due to the production of both natural and GFP-tagged FtsZ proteins after the depletion of either SepF or Ftn6.
FIG. 2.
Influence of the abundance of the protein SepF and Ftn6 on the structure and position of FtsZ networks. Phase-contrast images (A) (scale bars, 1 μm) and corresponding fluorescence images (B) (scale bars, 1 μm) of aberrant cells of different strains harboring the pSZ-GFP reporter plasmid (WT strain, ΔsepF::Kmr/sepF+ strain, and Δftn6::Kmr/ftn6+ strain) are shown.
Both GFP-SepF and GFP-Ftn6 proteins are biologically active and localize to the septum at midcell.
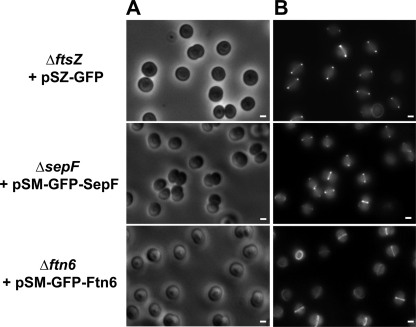
To investigate the subcellular localization of the SepF and Ftn6 proteins, we translationally fused the coding sequence to the C terminus of the GFP and cloned the resulting fusion genes in pSM (Table 1), the Smr Spr derivative of the expression vector we used to visualize the Z ring (23). The pSM-GFP-Ftn6 plasmid was introduced directly in the Ftn6-depleted strain (Δftn6::Kmr/ftn6+), where it restored a normal cell size (Fig. 3). This finding showed that the GFP-Ftn6 reporter protein is functional in that it is able to compensate for the depletion of the natural Ftn6 protein. Very interestingly, the fluorescent GFP-Ftn6 reporter protein exhibited a septal localization at midcell (Fig. 3).
FIG. 3.
Localization of proteins SepF and Ftn6 at the septum. Phase-contrast images (A) (scale bars, 1 μm) and corresponding fluorescence images (B) (scale bars, 1 μm) of indicated strains (ΔftsZ::Kmr + pSZ-GFP, ΔsepF::Kmr + pSM-GFP-SepF, and Δftn6::Kmr + pSM-GFP-Ftn6) are shown.
The pSM-GFP-SepF Smr Spr plasmid could not be directly introduced in the SepF-depleted cells, as demonstrated by the fact that they grew poorly. Alternatively, this plasmid was introduced in WT cells, generating the pSM-GFP-SepF reporter strain, which was subsequently transformed with the sepF deletion cassette ΔsepF::Kmr, yielding the ΔsepF::Kmr/pSM-GFP-SepF reporter strain. These cells exhibited a normal cell morphology, unlike the SepF-depleted ΔsepF::Kmr/sepF+ cells (Fig. 3), showing that the GFP-SepF reporter protein is biologically active in compensating for SepF depletion. In similarity to GFP-Ftn6, the GFP-SepF reporter protein exhibited a septal localization at the midcell (Fig. 3). Collectively, these findings indicate that both Ftn6 and SepF are targeted to the septum, where they might physically interact with FtsZ.
Both GFP-SepF and GFP-Ftn6 proteins can bind to FtsZ polymers preassembled in vitro.
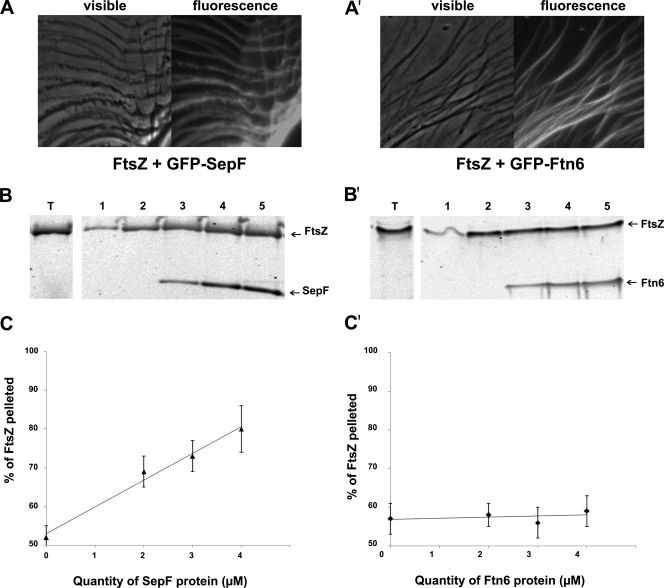
As we found that both the GFP-SepF and GFP-Ftn6 proteins are targeted to the septum at the midcell (Fig. 3), we tested whether they can physically interact with FtsZ. For this purpose, we used our previously published in vitro assay, taking advantage of the self-assembly of FtsZ into polymers, which can be subsequently decorated by a GFP-tagged FtsZ partner protein (23). Hence, preformed FtsZ polymers were incubated with the proteins six-His-GFP-SepF, six-His-GFP-Ftn6, and six-His-GFP (negative control; see reference 23) and subsequently examined using optical and fluorescence microscopy. The FtsZ filaments became highly fluorescent following incubation with six-His-GFP-SepF and six-His-GFP-Ftn6 (Fig. 4) but not after incubation with six-His-GFP (negative control). These findings show that both the SepF protein and the Ftn6 protein are able to interact with FtsZ polymers in vitro.
FIG. 4.
Both SepF and Ftn6 interact with FtsZ, and SepF but not Ftn6 stimulates assembly and/or stability of FtsZ polymers. Phase-contrast images and corresponding fluorescence images showing that GFP-SepF (A) and GFP-Ftn6 (A′) decorate the FtsZ polymer network are presented. Effects of SepF (B and C) and Ftn6 (B′ and C′) on the sedimentable polymeric mass of FtsZ are shown. FtsZ (4.5 μM) was polymerized in the absence or presence of different concentrations of SepF or Ftn6, and the polymers were collected by centrifugation. The protein content in the pellets were visualized by Western blotting using anti-His tag antibodies (B and B′) and then quantified (C and C′) as described in Materials and Methods. Lanes T represent the total amount of FtsZ used for the experiment, which was performed twice. Results show data representing the polymeric mass obtained when FtsZ was incubated without GTP (lanes 1) or with 1 mM GTP (lanes 2 to 5) in the absence (lanes 2) or in the presence of 2 μM (lanes 3), 3 μM (lanes 4), and 4 μM (lanes 5) concentrations of either SepF (B and C) or Ftn6 (B′ and C′).
SepF, but not Ftn6, stimulates assembly and/or stability of FtsZ polymers in vitro.
To test the influence of both SepF and Ftn6 on FtsZ assembly, we performed an in vitro polymerization assay of FtsZ in the presence of increasing concentrations of either SepF or Ftn6. FtsZ polymers were purified by sedimentation, and their quantities were estimated by both Coomassie blue staining (data not shown) and Western blot analysis using antibodies directed against the six-His tag of all these studied proteins (Fig. 4). Interestingly, the amount of FtsZ converted into FtsZ polymers increased in parallel with the concentration of SepF not with that of Ftn6. These findings indicate that SepF, but not Ftn6, stimulates FtsZ polymerization or stabilizes the polymers.
DISCUSSION
As very little is known concerning cell division in the environmentally crucial cyanobacteria, we have thoroughly investigated Ftn6 and SepF, the two cytokinetic proteins, in studies of the widely used unicellular cyanobacterium Synechocystis sp. strain PCC 6803.
SepF has been well characterized in studies of the rod-shaped bacterium B. subtilis, where it was found to be a septal protein and to physically interact with FtsZ (10), thereby promoting assembly and bundling of FtsZ protofilaments (31). In studies of cyanobacteria, analysis of SepF has been initiated using the rod-shaped unicellular strain Synechococcus sp. strain PCC 7942, and it has been found that its depletion generated double-length cells responsible for the filamentous morphology of the corresponding cdv2 mutant (26). As seen with Synechococcus sp., we found SepF to be essential to cell viability in Synechocystis sp. and to participate in the formation of cells of normal size (Fig. 1). In both of these cyanobacterial species, SepF depletion generated giant cells that retained their overall morphology, i.e., cylindrical morphology in Synechococcus sp. (26) and spherical morphology in Synechocystis sp. (Fig. 1). Going deeper into the role of the SepF in cyanobacteria, we found that SepF is targeted to the Synechocystis septum at midcell (Fig. 3), where it likely interacts with FtsZ. Supporting this assumption, we showed that SepF can indeed bind to FtsZ polymers preassembled in vitro and that SepF stimulates assembly and/or stability of FtsZ polymers (Fig. 4), as occurs in B. subtilis. These findings are important, as they show that not only the presence but also the function of a cytokinetic factor can be well conserved in prokaryotes, even in those that differ in the following important aspects: metabolism (photoautotrophy in cyanobacteria versus heterotrophy in B. subtilis), cell envelopes (gram negative in cyanobacteria versus gram positive in B. subtilis), and morphology (spherical in Synechocystis sp. versus cylindrical in B. subtilis).
Ftn6, a cyanobacterium-specific cytokinetic factor, was first identified in the rod-shaped unicellular strain Synechococcus PCC7942. It has been termed Ftn6 because its absence results in longer cells responsible for the filamentous morphology of the ftn6-deletion mutant (17, 26). This observation is reminiscent of what occurs in two intensively studied cylindrical bacteria, E. coli and B. subtilis, for which a wealth of mutants altered in cytokinesis are filamentous (for reviews, see reference 3 and references 4, 7, 9, 11, 19, and 21). The simplest explanation for this phenotype is that the frequency of complete septation decreases while cell growth, i.e., the lateral insertion of the peptidoglycan, continues (13). With respect to spherical cyanobacteria such as Synechocystis sp., for which the consequences for cell morphology of mutations slowing down septation more strongly than cell growth have not been well documented, we believe that such mutations should give rise to giant cells that remain spherical. As is consistent with this hypothesis, we report here that the depletion of either Ftn6 or SepF resulted in the generation of giant cells that remained spherical (Fig. 1). As seen with SepF, Ftn6 was found to be crucial to cell viability in Synechocystis sp. whereas it is dispensable in Synechococcus sp. (17, 26). The reason for this discrepancy between the two cyanobacteria is unclear, but this finding emphasizes the importance of parallel studies of cell division in both spherical organisms (such as Synechocystis sp.) and cylindrical organisms (such as Synechococcus sp.).
Ftn6 was found to be targeted to the septum at the midcell and, consistently, to physically interact with in vitro-preassembled FtsZ polymer, as seen with SepF (Fig. 4). Again, as observed for SepF, the depletion of Ftn6 did not prevent FtsZ polymerization. That the Z ring-like structures often appeared to be abnormal in the Ftn6- and SepF-depleted mutants was likely due to the fact that the cells respond to the depletion of either Ftn6 or SepF with an increased abundance of FtsZ proteins due to the production of both GFP-tagged and untagged FtsZ proteins. However, unlike SepF, Ftn6 was not found to stimulate the assembly and/or stability of FtsZ polymers (Fig. 4).
Although SepF and Ftn6 exhibit similar depletion phenotypes, namely, septal localization and interaction with FtsZ, it is unlikely that they share the same overlapping functions. Indeed, the Ftn6-overproduction mutant displayed a total of 33% twisted (nonspherical) cells, a result not observed in the SepF-overproduction mutant (Fig. 1). In addition, SepF, but not Ftn6, was found to stimulate assembly and/or stabilization of Z-polymers (Fig. 4). Altogether, our data suggest that Ftn6 acts of downstream of SepF, i.e., in the location after the assembly of the Z ring.
In conclusion, both SepF and Ftn6 are new FtsZ-interacting septal proteins in cyanobacteria, for which only one such protein, ZipN, has been characterized so far (23). It will be especially interesting in the future to investigate the likely relations among these three septal proteins.
Acknowledgments
M.M. and C.S. were recipients of Ph.D. and postdoctorate fellowships from the CEA (France).
Footnotes
Published ahead of print on 31 July 2009.
REFERENCES
- 1.Dismukes, G. C., D. Carrieri, N. Bennette, G. M. Ananyev, and M. C. Posewitz. 2008. Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Curr. Opin. Biotechnol. 19:235-240. [DOI] [PubMed] [Google Scholar]
- 2.Domain, F., L. Houot, F. Chauvat, and C. Cassier-Chauvat. 2004. Function and regulation of the cyanobacterial genes lexA, recA and ruvB: LexA is critical to the survival of cells facing inorganic carbon starvation. Mol. Microbiol. 53:65-80. [DOI] [PubMed] [Google Scholar]
- 3.Ebersbach, G., E. Galli, J. Moller-Jensen, J. Lowe, and K. Gerdes. 2008. Novel coiled-coil cell division factor ZapB stimulates Z ring assembly and cell division. Mol. Microbiol. 68:720-735. [DOI] [PubMed] [Google Scholar]
- 4.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferris, M., and B. Palenik. 1998. Niche adaptation in ocean cyanobacteria. Nature 396:226-228. [Google Scholar]
- 6.Ghirardi, M. L., A. Dubini, J. Yu, and P. C. Maness. 2009. Photobiological hydrogen-producing systems. Chem. Soc. Rev. 38:52-61. [DOI] [PubMed] [Google Scholar]
- 7.Goehring, N. W., and J. Beckwith. 2005. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol. 15:R514-R526. [DOI] [PubMed] [Google Scholar]
- 8.Gray, M. W. 1993. Origin and evolution of organelle genomes. Curr. Opin. Genet. Dev. 3:884-890. [DOI] [PubMed] [Google Scholar]
- 9.Haeusser, D. P., and P. A. Levin. 2008. The great divide: coordinating cell cycle events during bacterial growth and division. Curr. Opin. Microbiol. 11:94-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamoen, L. W., J. C. Meile, W. de Jong, P. Noirot, and J. Errington. 2006. SepF, a novel FtsZ-interacting protein required for a late step in cell division. Mol. Microbiol. 59:989-999. [DOI] [PubMed] [Google Scholar]
- 11.Harry, E., L. Monahan, and L. Thompson. 2006. Bacterial cell division: the mechanism and its precision. Int. Rev. Cytol. 253:27-94. [DOI] [PubMed] [Google Scholar]
- 12.Heckman, K. L., and L. R. Pease. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2:924-932. [DOI] [PubMed] [Google Scholar]
- 13.Justice, S. S., D. A. Hunstad, L. Cegelski, and S. J. Hultgren. 2008. Morphological plasticity as a bacterial survival strategy. Nat. Rev. Microbiol. 6:162-168. [DOI] [PubMed] [Google Scholar]
- 14.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:109-136. [DOI] [PubMed] [Google Scholar]
- 15.Reference deleted.
- 16.Koksharova, O. A., and C. P. Wolk. 2002. Genetic tools for cyanobacteria. Appl. Microbiol. Biotechnol. 58:123-137. [DOI] [PubMed] [Google Scholar]
- 17.Koksharova, O. A., and C. P. Wolk. 2002. A novel gene that bears a DnaJ motif influences cyanobacterial cell division. J. Bacteriol. 184:5524-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labarre, J., F. Chauvat, and P. Thuriaux. 1989. Insertional mutagenesis by random cloning of antibiotic resistance genes into the genome of the cyanobacterium Synechocystis strain PCC 6803. J. Bacteriol. 171:3449-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lutkenhaus, J. 2007. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu. Rev. Biochem. 76:539-562. [DOI] [PubMed] [Google Scholar]
- 20.Maple, J., C. Aldridge, and S. G. Moller. 2005. Plastid division is mediated by combinatorial assembly of plastid division proteins. Plant J. 43:811-823. [DOI] [PubMed] [Google Scholar]
- 21.Margolin, W. 2005. FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 6:862-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marraccini, P., S. Bulteau, C. Cassier-Chauvat, P. Mermet-Bouvier, and F. Chauvat. 1993. A conjugative plasmid vector for promoter analysis in several cyanobacteria of the genera Synechococcus and Synechocystis. Plant Mol. Biol. 23:905-909. [DOI] [PubMed] [Google Scholar]
- 23.Mazouni, K., F. Domain, C. Cassier-Chauvat, and F. Chauvat. 2004. Molecular analysis of the key cytokinetic components of cyanobacteria: FtsZ, ZipN and MinCDE. Mol. Microbiol. 52:1145-1158. [DOI] [PubMed] [Google Scholar]
- 24.Meberg, B. M., A. L. Paulson, R. Priyadarshini, and K. D. Young. 2004. Endopeptidase penicillin-binding proteins 4 and 7 play auxiliary roles in determining uniform morphology of Escherichia coli. J. Bacteriol. 186:8326-8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mermet-Bouvier, P., C. Cassier-Chauvat, P. Marraccini, and F. Chauvat. 1993. Transfer and replication of RSF1010-derived plasmids in several cyanobacteria of the genera Synechocystis and Synechococcus. Curr. Microbiol. 26:323-327. [Google Scholar]
- 26.Miyagishima, S. Y., C. P. Wolk, and K. W. Osteryoung. 2005. Identification of cyanobacterial cell division genes by comparative and mutational analyses. Mol. Microbiol. 56:126-143. [DOI] [PubMed] [Google Scholar]
- 27.Partensky, F., W. R. Hess, and D. Vaulot. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63:106-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pichoff, S., and J. Lutkenhaus. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rippka, R., J. Deruelles, J. Waterbury, M. Herdman, and R. Stanier. 1979. Generic assignements, strains histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 30.Singh, J. K., R. D. Makde, V. Kumar, and D. Panda. 2007. A membrane protein, EzrA, regulates assembly dynamics of FtsZ by interacting with the C-terminal tail of FtsZ. Biochemistry 46:11013-11022. [DOI] [PubMed] [Google Scholar]
- 31.Singh, J. K., R. D. Makde, V. Kumar, and D. Panda. 2008. SepF increases the assembly and bundling of FtsZ polymers and stabilizes FtsZ protofilaments by binding along its length. J. Biol. Chem. 283:31116-31124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams, P. G. 2009. Panning for chemical gold: marine bacteria as a source of new therapeutics. Trends Biotechnol. 27:45-52. [DOI] [PubMed] [Google Scholar]
- 33.Yang, Y., J. M. Glynn, B. J. Olson, A. J. Schmitz, and K. W. Osteryoung. 2008. Plastid division: across time and space. Curr. Opin. Plant Biol. 11:577-584. [DOI] [PubMed] [Google Scholar]
- 34.Zehr, J. P., J. B. Waterbury, P. J. Turner, J. P. Montoya, E. Omoregie, G. F. Steward, A. Hansen, and D. M. Karl. 2001. Unicellular cyanobacteria fix N2 in the subtropical North Pacific Ocean. Nature 412:635-638. [DOI] [PubMed] [Google Scholar]