Abstract
Latent tuberculosis represents a high-risk burden for one-third of the world population. Previous analysis of murine tuberculosis identified a novel transcriptional regulator encoded by Rv0348 that could control the establishment of persistent tuberculosis. Disruption of the Rv0348 gene from the genome of the virulent H37Rv strain of Mycobacterium tuberculosis revealed a global impact on the transcriptional profiles of 163 genes, including induction of the mammalian cell entry (mce1) operon and the repression of a significant number of genes involved in hypoxia and starvation responses. Nonetheless, gel shift assays did not reveal direct binding between Rv0348 and a set of regulated promoters, suggesting an indirect regulatory role. However, when expressed in Mycobacterium smegmatis, the Rv0348 transcripts were significantly responsive to different levels of hypoxia and the encoded protein was shown to regulate genes involved in hypoxia [e.g., Rv3130c (tgs1)] and intracellular survival (e.g., mce1), among other genes. Interestingly, the colonization level of the ΔmosR mutant strain was significantly lower than that of the wild-type strain of M. tuberculosis, suggesting its attenuation in the murine model of tuberculosis. Taken together, our analyses indicated that the Rv0348 gene encodes a novel transcriptional factor that regulates several operons involved in mycobacterial survival, especially during hypoxia; hence, we propose that Rv0348 be renamed mosR for regulator of mycobacterial operons of survival.
Tuberculosis remains one of the deadliest diseases worldwide, with 1.8 million deaths each year from infection with Mycobacterium tuberculosis (9). One-third of world's population is already infected with M. tuberculosis, contributing to the high death rate related to the disease. Individuals suffering from latent tuberculosis could act as a reservoir of future infections (21), especially in sub-Saharan Africa and the Indian subcontinent (15, 52). Clinically, human tuberculosis can be divided into three phases: an active, replicative phase; a latent, persistent phase; and a reactivation phase, usually observed in patients with declining immune competence. Examining the persistent stage of tuberculosis is vital to understanding the mechanisms of mycobacterial survival during host immunity. At this stage, M. tuberculosis bacilli are living under several stress conditions invoked by the host immune system, including oxidative, hypoxic, and nutritional stressors (46). Earlier analysis of M. tuberculosis persistence in mice identified several unique genes induced during infection, including a novel transcriptional regulator encoded by the Rv0348 gene (44). The focus of this report is to identify genes under the control of the Rv0348 gene and to characterize its role in the establishment of chronic tuberculosis.
Several in vivo and in vitro models have been developed to mimic the stress conditions encountered by the mycobacterial bacilli inside granulomas, a common pathological feature of chronic tuberculosis (36, 48). Using DNA microarrays, a group of 48 genes constituting what is now known as the “dormancy” regulon was identified in M. tuberculosis cultures exposed to nitric oxide (47), nutrient starvation (3), or low-oxygen tension (32), suggesting an adaptive mechanism activated by mycobacteria to survive the host microenvironments. Such adaptive mechanisms were further confirmed when the M. tuberculosis transcriptome was profiled following macrophage infections (36). Although the in vitro models offered a better understanding of the stress-responsive genes in M. tuberculosis, they did not elucidate survival strategies during infection. Recently, a knockout of the dormancy regulon did not impact M. tuberculosis survival in mice (32), suggesting a minor role for the dosR regulon (discovered through in vitro modeling) in M. tuberculosis survival strategies in mice. Using in vivo microarray analysis, the transcriptional profiles of M. tuberculosis were determined in murine tuberculosis, identifying a unique transcriptional profile of M. tuberculosis during progression from the active to the chronic stage of tuberculosis (42, 44). Furthermore, the in vivo microarray analysis showed that the number of bacilli remained stable during chronic tuberculosis while several metabolic pathways and transcriptional regulators were activated (44). Recently, the dynamic change in M. tuberculosis bacillus levels during chronic infection in mice was confirmed (13).
Among the potential transcriptional regulators that showed high induction during chronic tuberculosis was the Rv0348 gene (44). Transcripts for Rv0348 were upregulated ∼200-fold after 60 and 140 days of M. tuberculosis infection in mice, and the Rv0348 protein was shown to bind to its own promoter (44). In the present work, we generated an M. tuberculosis mutant with an inactivated Rv0348 gene to examine the role of the gene in modulating the M. tuberculosis transcriptional profile. Interestingly, the transcriptional profiles of the isogenic mutant relative to its parental strain, H37Rv, indicated the regulation of several gene groups organized into operons and regulons, including those involved in mammalian cell entry (mce1), hypoxia (tgs1), and starvation. The ability of the Rv0348 protein to affect the transcription of these genes was further analyzed using a lacZ reporter assay and the Wayne model of hypoxia in Mycobacterium smegmatis. Finally, mouse infection with the ΔRv0348 strain provided evidence that the gene could participate in M. tuberculosis virulence. Because of the broad regulatory role of Rv0348 in mycobacterial survival strategies, we propose to rename Rv0348 mosR for regulator of mycobacterial operons of survival.
MATERIALS AND METHODS
Strains, media, and plasmids.
Escherichia coli DH5α and HB101 bacteria were used as host cells for cloning purposes in all experiments presented here. M. tuberculosis H37Rv and M. smegmatis mc2155 strains were grown in 7H9 Middlebrook liquid medium (BD Biosciences, Rockville, MD) and on 7H10 Middlebrook plates supplemented with albumin dextrose catalase and antibiotics when needed (25 μg/ml kanamycin or 50 μg/ml hygromycin). The protocols for DNA manipulations employed throughout this study, including PCR, cloning, DNA ligations, and electroporation, were performed as described previously (33) or according to the manufacturers' recommendations. A list of plasmids and constructs used in this study is presented in Table 1. The procedures for cloning, overexpression, and purification of M. tuberculosis Rv0348 in E. coli were detailed previously (44). Total RNA samples were extracted from mycobacterial cultures grown to an optical density at 600 nm (OD600) of 0.5 or 1.5 using Trizol (Invitrogen, Carlsbad, CA) as described previously by our group (42, 44). Extracted mycobacterial total RNA samples were treated with DNase I (Ambion, Austin, TX) until no DNA was detected using PCR primers for the 16S rRNA gene. The primers used in this study are listed in the supplemental material.
TABLE 1.
Vectors and constructs used in this study
| Plasmid | Characteristics | Reference |
|---|---|---|
| pYUB854 | Cosmid for disruption construct | 2 |
| pMV361 | Integrative mycobacterial shuttle vector; Kanr | 39 |
| M. smegmatis::pML21 | Recombinant M. smegmatis harboring the whole MosR operon under the control of its own promoter, in addition to hsp60; Kanr | This work |
| pML23 | pMV361 harboring the MosR gene; Kanr | This work |
| pCV77 | Replicative shuttle vector with promoterless LacZ transcriptional fusion; Kanr | MedImmune |
| pML24 | pCV77 with hygromycin cassette in opposite direction to LacZ.; Kanr Hygr | This work |
| pML25 | pML24 harboring the promoter region of the Rv0347 operon; Kanr Hygr | This work |
| pML26 | pML24 harboring the promoter region of Rv0167 (mce1 operon); Kanr Hygr | This work |
| pML27 | pML24 harboring the promoter region of Rv0700 (rpsJ); Kanr Hygr | This work |
| pML28 | pML24 harboring the promoter region of Rv3130c (tgs1); Kanr Hygr | This work |
| pML29 | pML24 harboring the promoter region of hsp60; Kanr Hygr | This work |
Generation of Rv0348 (mosR) knockout and complemented strains.
A specialized transduction protocol was adopted with a few modifications to inactivate the Rv0348 gene using the virulent strain of M. tuberculosis H37Rv (2). Approximately 800-bp fragments flanking the Rv0348 open reading frame (specifically, flanking base number 269) were amplified using standard PCR protocols. The amplicons were cloned into a pGEM-T vector (Promega, Madison, WI) and the sequences were verified before ligation into the pYUB845 vector (2), using SpeI and HindIII for the left arm and XbaI and Acc65I for the right arm, to form the allelic-exchange substrate. Construction of specialized transducing mycobacteriophages and transduction protocols were performed as described by Bardarov et.al. (2). Following 6 weeks of incubation at 37°C, hygromycin-resistant colonies were selected for further analysis. PCR and Southern blot analyses were used to verify the mutant genotypes as described previously by members of our laboratory (43, 53).
For complementation experiments, the coding sequence of the entire Rv0348 operon (2.3 kb) or the coding sequence of the Rv0348 gene alone (∼654 bp) was amplified by PCR. The amplicons were cloned into the pGEM-T vector and subsequently verified by DNA sequencing. The vectors were double digested by EcoRI and HindIII restriction enzymes, followed by ligation of gel-purified inserts into pMV361 to give rise to pML21 (OpRv0348) and pML23 (Rv0348) shuttle vectors for the expression of the whole operon or the Rv0348 gene, respectively. Both plasmids (pML21 and pML23) were independently electroporated into electrocompetent M. smegmatis and M. tuberculosis H37Rv cells (50). Transformants were selected and subsequently analyzed by PCR to verify integration of the delivered sequences into the M. tuberculosis genome. Expression of the Rv0348 protein was further examined using immunoblotting as described above.
Stress treatments of M. tuberculosis cultures.
Cultures of M. tuberculosis H37Rv, H37RvΔRv0348, a complemented strain, ΔRv0348::Rv0348, or H37Rv M.tb:rv0348 were grown to early log phase (OD600 = 0.5), and their colony counts were determined by plating them on Middlebrook 7H10 agar plates in order to calculate the viable cells at the beginning of the experiment. Aliquots (10 ml) were subjected to 0.05% sodium dodecyl sulfate (SDS) treatment (Sigma) for 4 h at 37°C or to heat shock at 45°C for 24 h in a slowly shaking incubator. To test static growth conditions, 50-ml aliquots of M. tuberculosis constructs were allowed to grow for 2 and 6 months without shaking in closed Falcon tubes. At the designated times, culture aliquots were plated and counted on Middlebrook 7H10 agar. Other aliquots were used for RNA isolation to assess the expression of Rv0348 under the examined stress conditions using quantitative real-time reverse transcriptase PCR (qRT-PCR) as detailed previously (41).
Transcriptional analysis.
Before DNA microarray hybridizations, double-stranded cDNA was synthesized from 10 μg of total RNA using the SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen), as directed by the manufacturer, in the presence of 250 ng genome-directed primers (41, 49). The double-stranded cDNA was cleaned up and labeled following the NimbleGen gene expression analysis protocol (NimbleGen Systems, Inc., Madison, WI) and hybridized to NimbelGen-manufactured microarrays following a protocol we established earlier (49). In this microarray, each of the 3,989 open reading frames in the genome of M. tuberculosis strain H37Rv was represented by 19 probes of 60-mer oligonucleotides. Further, the whole genome was represented five times on each chip (i.e., five technical replicates per chip) for a total of 95 probes/gene. All hybridizations (3 μg of double-stranded cDNA/chip) were performed using NimbleGen hybridization buffer and commercial hybridization chambers (TeleChem International, Inc., Sunnyvale, CA) overnight at 42°C. Following hybridization, washing steps were performed using NimbleGen washes I, II, and III as recommended by the manufacturer. The slides were scanned using an Axon GenePix 4000B scanner (Molecular Devices Corporation, Sunnyvale, CA), and fluorescence intensity levels were extracted using NimbleScan (NimbleGen) and normalized to a mean value of 1,000. Determination of significantly changed genes was performed using a flexible empirical Bayes model, specifically, the LNN model in the EBArrays package (19) employing an R language (http://www.bioconductor.org/). A cutoff of 0.50 for the probability of differential expression of >0.5 was used to determine significantly changed genes. Statistical enrichment of gene groups within the microarray genes versus other transcriptomes was calculated using a standard hypergeometric distribution function in Microsoft Excel.
qRT-PCR.
For qRT-PCR, cDNA was synthesized from 1 μg of total RNA using SuperScript III (Invitrogen), as directed by the manufacturer, in the presence of SYBR green and 250 ng of mycobacterial genome-directed primers (41, 49). SYBR green qRT-PCR was done using gene-specific primers (see the supplemental material) at a concentration of 200 nM. The thermocycling conditions were 95°C for 3 min and 40 cycles of 95°C for 15 s and 60°C for 30 s. qRT-PCRs were performed in triplicate, the threshold cycle values were normalized to the levels of 16S rRNA transcripts, and the changes were calculated by the 2−ΔΔCT method (49).
EMSA.
The MosR/Rv0348 protein was purified as detailed previously (44). For the electrophoresis mobility shift assay (EMSA), probes were generated using standard PCR amplification protocols and primers designed by Primer3 (http://frodo.wi.mit.edu/primer3/) by providing the upstream probable regulatory sequences of selected genes. All EMSA reactions were resolved on 6% native polyacrylamide gels as detailed previously (44).
Antibody generation and immunoblotting.
To generate antibodies against purified Rv0348, two adult male New Zealand White rabbits were inoculated with 125 μg of the recombinant fusion protein in Freund's incomplete adjuvant (Sigma, St. Louis, MO) using our protocol approved by the Institutional Animal Care and Use Committee (IACUC). The rabbits were housed individually in cages at 15 to 18°C and given antibiotic-free food and water ad libitum. Each immunization was administered subcutaneously (12.5 μg), intradermally (37.5 μg), intramuscularly (50 μg), and intraperitoneally (25 μg) in accordance with the manufacturer's suggestions. Injections of the antigen-adjuvant mixture were administered every 3 weeks for a total of three immunizations. Antibody titers for seroconverted rabbits were measured by enzyme-linked immunosorbent assay and immunoblotting using recombinant purification tag-specific antibodies.
For immunoblotting, mycobacterial cultures were harvested and lysed by boiling them in phosphate-buffered saline (PBS). The total crude extracts were centrifuged, and equal amounts of soluble lysates or insoluble pellets were separated on 12% SDS-polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane (Hybond-P; Amersham Biosciences). The membranes were saturated with 5% dried milk, and rabbit polyclonal antibody was used as the primary antibody at a dilution of 1/5,000 for 2 h. Horseradish peroxidase conjugated to goat anti-rabbit immunoglobulin G (Pierce Thermo Scientific, Rockford, IL) was used as a secondary antibody at 1/30,000. The membranes were developed with a chemiluminescence kit according to the manufacturer's protocol (Pierce).
Lipid extraction and MALDI-TOF analysis.
Lipids were extracted from mycobacterial cultures, first with a mixture of chloroform and methanol (1:2 [vol/vol]) for 24 h at room temperature and then twice with CHCl3/CH3OH (2:1 [vol/vol]) for 2 days. The different organic phases were pooled and evaporated to dryness. The crude lipid extracts were dissolved in chloroform and analyzed by thin-layer chromatography on Durasil 25 silica gel-precoated plates (Macherey-Nagel) using various types of eluents (28). Additionally, lipid samples were subjected to alkaline hydrolysis with 1 M sodium methanolate for 1 h at 37°C. The resulting mixture was neutralized with glacial acetic acid and dried under nitrogen. The lipids were then extracted twice with diethyl ether and washed with water prior to analysis by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry as detailed previously (5, 20). Detection of lipid samples was performed in reflectron mode on a 4700 Proteomics Analyzer (Voyager DE-STR; Applied Biosystems, Framingham, MA) equipped with an Nd:YAG laser (355-nm wavelength) operating in pulses of 500 ps at a frequency of 200 Hz. A total of 2,500 shots were accumulated in positive-ion mode, and mass spectrometry data were acquired using the instrument default calibration.
Construction of lacZ vectors and β-galactosidase assays.
The DNA fragments corresponding to the putative promoter regions of the Rv3130c, Rv0167, Rv0700, Rv0347, and hsp60 genes were cloned by PCR using gene-specific primers (see the supplemental material). The different promoters were cloned into the pML24 shuttle vector (a derivative of the pCV77 vector, in which a hygromycin resistance cassette was cloned into a SpeI site). M. smegmatis was first electroporated with pML21, and positive clones were selected and verified by PCR and Western blotting to ensure the expression of the Rv0348 protein. Recombinant M. smegmatis was electroporated with a different shuttle vector (a pML24 derivative) and incubated on selective LB plates supplemented with 5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside (X-Gal). All recombinant M. smegmatis cells developed blue color on the plates, except the negative control (pML24). Assessment of β-galactosidase activities in different constructs was performed in sonicated extracts of M. smegmatis strains using a β-galactosidase assay kit (Stratagene, Cedar Creek, TX) according to the manufacturer's protocol. Experiments were carried out in triplicate and repeated twice for independent cultures with different amounts of soluble-fraction proteins. β-Galactosidase was expressed as Miller units per mg of soluble lysate.
Growth of M. smegmatis::pML21 under anaerobic conditions.
The study of mosR expression under hypoxic conditions in M. smegmatis::pML21was performed using the Wayne model of hypoxia in M. tuberculosis (51), which has proven equally useful in studies of M. smegmatis (24). Briefly, a single colony of M. smegmatis::pML21 harboring the mosR operon was grown with shaking at 37°C to an OD600 of 1.0 in Dubos Tween Albumin medium (BD Biosciences) supplemented with kanamycin (30 μg/ml). This culture was used to inoculate six 30-ml screw-cap tubes containing stir bars to an OD600 of 0.1 in Dubos medium containing methylene blue (1.5 μg/ml) to serve as an indicator of oxygen levels. Three tubes with loose caps and a headspace ratio of 1.5 were used as aerobic controls and were stirred at 200 rpm. The remaining three tubes with tightened, parafilm-sealed caps and a headspace ratio of 0.5 were used for the anaerobic cultures and were stirred at 120 rpm. The colors of the tubes were monitored, and aerobic/anaerobic tubes were taken for analysis when the methylene blue first showed signs of fading (day 1) and after the anaerobic cultures became completely colorless (days 6 and 7). An aliquot (100 μl) of each sample was plated for colony counts, while the rest was used for total RNA extraction and qRT-PCR as described above. The whole experiment was repeated three times.
For some experiments, M. smegmatis and M. smegmatis::pML21, both harboring pML28, were similarly grown using the Wayne model of hypoxia (24) to compare the effect of MosR on the expression of Rv3130c (tgs1) under both aerobic and anaerobic conditions. M. smegmatis::pML28 cultures were collected based on the fading of the methylene blue (1.5 μg/ml) color. In one experiment, samples were collected at 0, 1, 7, and 8 days postinoculation, and in a second experiment, samples were collected at 0, 1, 13, and 14 days. In all cases, these times represented the time points at which the cultures had yet to fade, were first observed to fade, were nearly colorless, and were completely colorless, respectively. β-Galactosidase activity was assessed at these time points as described above.
Animal infections.
Three groups (n = 40) of 5-week-old BALB/c mice were infected with M. tuberculosis strains and housed in a biosafety level 3 environment using our protocol approved by the University of Wisconsin IACUC. Cultures of M. tuberculosis H37Rv, ΔRv0348, and ΔRv0348::Rv0348 strains were grown to mid-log phase (OD600 = 1). A total of 10-ml cultures adjusted to an OD600 of 0.3 were suspended in sterile PBS and used for aerosolization using a Glass-Col (Terra-Haute, IN) aerosol chamber to generate an infectious dose of 200 to 400 CFU per mouse (44). Two mice were sacrificed at 4 to 6 h postinfection to enumerate the infectious dose by plating the bacteria on 7H10 Middlebrook plates in the presence of hygromycin and/or kanamycin for the mutant and complemented strains, respectively. Mice (n = 5) were sacrificed at different times postinfection to remove the lungs for plating on Middlebrook 7H10 agar for colony counting. Portions of the livers, spleens, and lungs were fixed in 10% neutral buffered formalin for at least 2 h before being sectioned and stained with hematoxylin and eosin and Ziehl-Neelsen stain (44).
Statistical analysis.
Student's t test implemented in Microsoft Excel was used to assess the significance of differences among samples at a P level of <0.05 for the reporter assays.
RESULTS AND DISCUSSION
Generation of M. tuberculosis strains with various copies of Rv0348 (mosR).
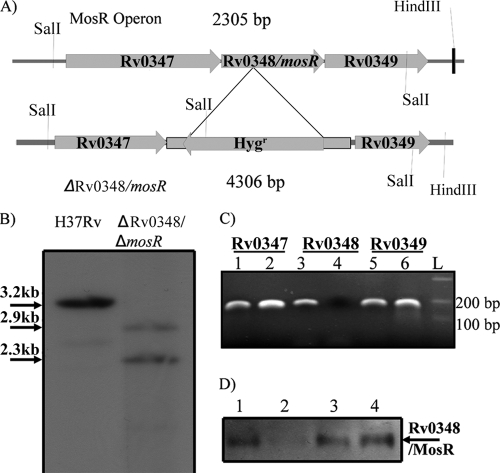
Our strategy to construct the ΔmosR mutant included the insertion of a hygromycin cassette within the coding sequence of the mosR gene using a specialized transduction-based protocol (2). Attempts to delete the whole gene with 200-bp flanking sequences failed to yield any mutants. Earlier transposon mutagenesis indicated that the Rv0347 gene flanking the mosR sequence was essential (34), explaining the failure to delete the whole mosR gene where flanking sequences were disrupted. However, following specialized transduction of the insertion constructs (introducing the Hygr gene sequence at 269 bp after the start of mosR) to the M. tuberculosis H37Rv strain, several transductants were obtained. PCR, sequencing, and Southern blot analyses (Fig. 1A and B) of several transductants verified the desired genotype (ΔmosR) in all transductants, and one transducant was chosen for the rest of the analyses.
FIG. 1.
Deletion, complementation and overexpression of Rv0348. (A) Organization of the Rv0348 operon and our strategy for gene disruption. See Materials and Methods for details. (B) Southern blot analysis of SalI-digested genomic DNA of the H37Rv wild type and the ΔmosR mutant. (C) PCR analysis of cDNA synthesized from RNA samples purified from H37Rv (lanes 1, 3, and 5) or the ΔmosR mutant (lanes 2, 4, and 6). (D) Western blot analysis of different M. tuberculosis strains using polyclonal antibodies raised in rabbits against the MBP-MosR protein. Pellets from M. tuberculosis H37Rv (lane 1), the ΔmosR mutant (lane 2), the ΔmosR::mosR complemented strain (lane 3), and the H37Rv::mosR overexpression strain (lane 4) were subjected to immunoblotting. The blot for soluble fractions was negative (data not shown).
To examine the polarity of the generated mutant, PCR analysis of RNA samples isolated from the ΔmosR strain showed the presence of transcripts of the genes upstream and downstream of the mosR gene, indicating that the ΔmosR mutant is nonpolar (Fig. 1C). The selected ΔmosR mutant was further used to construct the complementation strain, in which the coding sequence of mosR is expressed in trans using pMV361-mosR to yield the ΔmosR::mosR construct. Attempts to introduce the whole operon into M. tuberculosis were unsuccessful due to several genomic rearrangements (data not shown). Furthermore, we examined the level of expression of the Rv0348/MosR protein in M. tuberculosis H37Rv compared to other constructs (Fig. 1D). As expected, MosR was detectable when mycobacterial pellets, not culture filtrate, were analyzed using Western blotting, suggesting intracellular expression of the MosR protein. Growth curves for all strains (wild-type H37Rv, its isogenic mutant ΔmosR, the complemented ΔmosR::mosR strain, and the overexpression strain M. tuberculosis H37Rv:mosR) showed no measurable difference during in vitro growth in Middlebrook 7H9 broth (see the supplemental material).
mosR expression under various stress conditions.
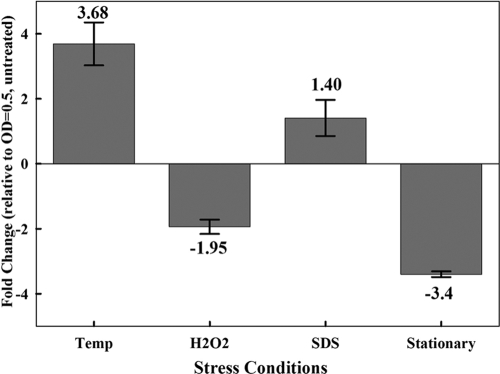
Previously, transcripts constituting the mosR operon were activated during starvation (3) and under extended anaerobic conditions (32). To examine mosR expression under other stresses, cultures of the wild-type strain H37Rv and its isogenic ΔmosR mutant were exposed to stress conditions that are thought to be activated during intracellular survival of M. tuberculosis (36, 46, 47). Following exposure to variable stressors, both the colony counts and the transcriptional profiles of mosR transcripts (for the H37Rv strain only) were assayed. Interestingly, qRT-PCR showed that the transcripts of mosR remained unchanged (<2-fold change) when M. tuberculosis cultures were exposed to SDS (0.05%). However, the transcriptional profile of mosR indicated its significant induction at 45°C and its significant repression following exposure to high levels of H2O2 or during the stationary phase of growth (Fig. 2). On the other hand, no difference in colony counts was found when mycobacterial cultures (H37Rv and ΔmosR strains) were exposed to any of the examined stressors, including culture under static conditions for 2 to 6 months at 37°C (see Fig. S1 in the supplemental material). Such disparity between CFU counts and qRT-PCR suggests that mosR might not directly impact mycobacterial survival during exposure to in vitro stressors but might have a role in regulating the expression of other genes under stress conditions, such as high temperature or the stationary phase of growth, in addition to starvation, as shown previously (3, 32).
FIG. 2.
Transcriptional profile of mosR in M. tuberculosis H37Rv under various stressors. Shown is qRT-PCR of mosR transcripts under various stress conditions, such as high temperature (45°C) (Temp), H2O2 (10 mM), and SDS (0.05%) treatments, as well as transition to stationary phase (OD600 = 1.5). The change was calculated relative to transcripts in untreated cultures of M. tuberculosis strain H37Rv (OD600 = 0.5). The error bars represent standard deviations from the means.
Global changes in the M. tuberculosis transcriptome triggered by mosR.
Previously, it was suggested that MosR could play a role in the transcriptional regulation of M. tuberculosis during transition to the chronic stage of murine tuberculosis (44). To identify genes under the control of MosR, cultures of both H37Rv and its isogenic ΔmosR mutant were grown in vitro for DNA microarray analysis. A high-sensitivity oligonucleotide microarray platform was used in this experiment (7, 37). Replicate microarray hybridizations were performed for at least two biological samples of both the wild-type and mutant strains and showed a high correlation level (r = 0.9). Using a standard protocol for Bayesian statistics (18, 19), we identified significantly regulated genes with a probability of differential expression of >0.5 and more than a ±2-fold change between H37Rv and ΔmosR cultures. Using these criteria, a significant change in a set of 163 genes was identified between the transcriptomes for H37Rv and ΔmosR (mosR regulon). A partial list of significantly changed genes organized into operons (as determined by an operon prediction algorithm [29]) is shown in Table 2. Induced genes in the H37Rv transcriptome compared to the ΔmosR mutant (n = 98 genes) are suggested to be under the positive control of mosR, while repressed genes (n = 65) are suggested to be under its negative control. A representative sample of genes that showed transcriptional changes by DNA microarray analysis was verified by qRT-PCR. In all of the examined genes (n = 10), there was agreement of the transcriptional change (either induction or repression) between DNA microarray and qRT-PCR analyses (see Fig. S2 in the supplemental material).
TABLE 2.
Mycobacterial operons under positive and negative control of MosR
| Regulation | Gene ID | Operon namea | Putative function |
|---|---|---|---|
| Positive | Rv0167-Rv0177 | mce1 | Mammalian cell entry operon |
| Rv0684-Rv0685 | fusA-tuf | Elongation factor | |
| Rv0700-Rv0710 | rpsJ-rpsQ | 30S ribosomal protein S10 | |
| Rv0718-Rv0723 | rpsH-rplO | 30S ribosomal protein S10 | |
| Rv1184c-Rv1185c | Rv1184c-Rv1185c | Conserved hypothetical protein, acyl-coenzyme A (CoA) synthase | |
| Rv1613-Rv1614 | trpA-ltg | Tryptophan synthase α chain; prolipoprotein diacylglyceryl transferase | |
| Rv2391, Rv2392 | nirA-cysH | Probable nitrite reductase/sulfite reductase | |
| Rv2948c-Rv2950c | fadD22-fadD29 | Acyl-CoA synthase | |
| Rv3148-Rv3154 | nuoD-nuoJ | NADH dehydrogenase chain D-J | |
| Rv3460cb | rpsM-rpsJ | 30S ribosomal protein S13-L36 | |
| Rv3824c-Rv3825c | papA1-pks2 | PKS-associated protein; unknown function; polyketide synthase | |
| Rv3921c-Rv3924c | rnpA, rpmH | Unknown membrane protein | |
| Negative | Rv0823c-Rv0824c | desA1 | Transcriptional regulator; ntrB (NifR3/Smm1 family) |
| Rv1622c, Rv1623c | cydB, appC | Cytochrome d ubiquinol oxidase subunit II | |
| Rv2031cb | hspX | 14-kDa antigen; heat shock protein Hsp20 family | |
| Rv2629-Rv2630 | Rv2629-Rv2630 | Hypothetical protein | |
| Rv3048cb | nrdG | Ribonucleoside diphosphate small subunit | |
| Rv3053cb | nrdH | Glutaredoxin electron transport component of NrdEF | |
| Rv3139-Rv3140 | fadE24, fadE23 | Acyl-CoA dehydrogenase |
Operon predications are based on an earlier analysis (29).
Single gene of a larger operon or regulon.
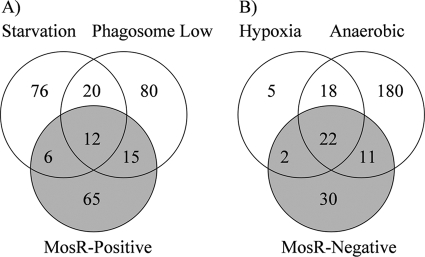
Genes involved in survival during stationary and persistent phases (rpoB) (16) of growth, as well as those regulating transcription (e.g., rho and rpmE) (TubercuList database) were among the genes under the positive control of MosR. The positively regulated operons (Table 2) included the mce1 operon (Rv0167 to Rv0177) (8), suggesting a role for mosR in regulating the virulence of M. tuberculosis (45). Other genes induced in the presence of mosR included a tryptophan biosynthesis gene (trpA), a translation apparatus operon (fusA-tuf), and the ribosomal biosynthesis operon (Rv0700 to Rv0723) (Table 2). In E. coli, the expression of the tryptophan operon is regulated by inhibition of ribosomal binding sites (11). It is worthy of mention here that functional orthologues of the trp operon regulatory genes are induced by mosR (e.g., the 50S ribosomal operon and the rho transcription termination gene), suggesting the ability of MosR to exert its regulatory role(s) through transcriptional inhibition. Further comparative analysis of the starvation-induced transcriptome reported previously (3) identified a set of 18 genes that are positively regulated by MosR (Fig. 3A). Interestingly, among the positively mosR-regulated genes is a group of 29 genes that were repressed during macrophage infection (36). This profile suggests the ability of M. tuberculosis to modulate the levels of gene transcripts to survive the macrophage environment using a mosR-dependent mechanism.
FIG. 3.
Comparative analysis of the transcriptome of M. tuberculosis exposed to various conditions. (A) Venn diagram representing the number of positively mosR-regulated genes compared to genes induced under nutrient starvation (3) and those repressed in the phagosome environment (36). (B) Venn diagram representing the number of negatively mosR-regulated genes compared to genes induced under hypoxia (27) and anaerobic conditions (25).
In contrast, negatively mosR-regulated genes included a significant number of phagosome-activated genes (n = 33) (36). Notable among this group is Rv3130c (tgs1), which encodes triglyceride synthase, a protein that is involved in triglyceride synthesis in M. tuberculosis (38). Interestingly, a set of the negatively mosR-regulated genes (n = 24) were among the 47 genes (Fig. 3B) responsive to hypoxia (27) or the 48 genes responsive to reactive nitrogen intermediates and gradual adaptation to low levels of oxygen (26, 47). Additionally, a set of 33 genes that were activated during anaerobic growth of M. tuberculosis (25) was also found among the negatively mosR-regulated genes in this study (Fig. 3B). Previously, a significant level of overlap existed between the hypoxia and reactive nitrogen intermediate regulons, and they were shown to be under the two-component regulator dosR (27), including the acr gene. The acr gene was previously confirmed to contribute to M. tuberculosis survival in macrophages (54), and hence, its inclusion under the negative control of mosR suggests a potential role for mosR in downregulating genes involved in hypoxia in stages when they are not needed. Finally, hypergeometric distribution analysis of the MosR-dependent genes and each of the compared transcriptomes indicated significant association between the mosR-induced transcriptome and starvation, phagosome survival, hypoxia, and anaerobic conditions (P < 0.001) (see the supplemental material). Overall, the analyses presented show the broad potential regulatory roles exerted by mosR in M. tuberculosis survival strategies. Therefore, we suggest that the Rv0348 gene be renamed mosR for regulator of mycobacterial operons of survival.
MosR binding to new promoter regions.
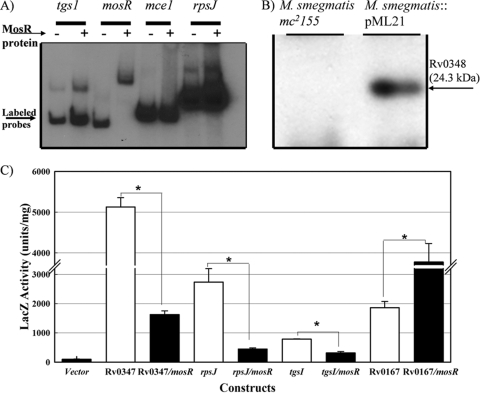
Previously, we showed that mosR (also called Rv0348) encodes a transcriptional regulator that binds to its own promoter (44). To examine the ability of MosR to regulate other genes, we performed EMSAs using the predicted regulatory sequences of 10 genes that were shown by using DNA microarrays to be members of the mosR regulon. Surprisingly, the presence of MosR did not impact the migration pattern of DNA fragments representing any of the 10 putative promoters (see Fig. S3 in the supplemental material), suggesting an indirect regulatory function for MosR. Only when the positive control was used (the upstream region of the mosR operon) was a retardation of DNA migration noticeable (Fig. 4A). It is possible that other regulatory elements or transcriptional cofactors are needed to induce the binding ability of MosR.
FIG. 4.
Transcriptional activity of the mosR operon. (A) EMSA of MosR with different promoter regions of significantly regulated genes as detected by DNA microarrays. EMSA was performed as described previously (44). Gene names are listed above the lanes. (B) Western blot analysis of the recombinant strain of M. smegmatis mc2155 expressing MosR protein. (C) β-Galactosidase activities of constructs for Rv3130c (tgs1), Rv0347, Rv0700, and Rv0167 promoters fused to the coding sequence of lacZ in the presence/absence of the mosR operon using M. smegmatis cells as host cells. The asterisks denote significant changes in a Student t test (P < 0.001). The error bars indicate standard deviations.
lacZ fusion assay for MosR regulatory functions.
Because of the lack of direct binding of MosR to the putative promoter regions of any of the examined genes of the mosR regulon, we employed the lacZ reporter gene to examine the regulatory role of MosR. For this purpose, the generated M. smegmatis::mosR (M. smegmatis::pML21) construct was used to examine the transcriptional regulation of a selected list of genes in the mosR regulon. Previously, similar construct systems were employed (6, 14, 22) as surrogates for lacZ assays in M. tuberculosis. BLAST analysis indicated that the mosR operon is absent from the genome of M. smegmatis, allowing us to express MosR and assess its function(s) in the rapidly growing M. smegmatis. The expression of MosR was verified in several colonies of the recombinant strain of M. smegmatis::mosR by using Western blot analysis (Fig. 4B). To examine the regulatory function of mosR, a verified clone of the M. smegmatis::mosR strain was electroporated with derivatives of the pML24 plasmid (Table 1), in which the putative promoter regions of several genes were cloned upstream of a promoterless reporter gene (lacZ). As expected, screening of transformants showed that all constructs formed blue colonies on plates supplemented with X-Gal except when a promoterless lacZ vector was used for transformation (see Fig. S4 in the supplemental material). Quantitative analysis of the β-galactosidase activity of each construct in the presence/absence of the mosR operon showed significant differences among constructs depending on the presence of the mosR operon (Fig. 4C).
In all examined promoters, a significant change in the expression level of LacZ was found between constructs in which the mosR operon was present and those without it. Both the repression of the Rv0347 (the promoter for mosR) and Rv3130c promoters and the induction of Rv0167 (the promoter for mce1) were in agreement with negative and positive regulation by MosR, respectively, as indicated by DNA microarrays. However, in the case of Rv0700 (the promoter for the ribosomal-protein operon), the lacZ assay indicated its repression despite evidence that it is under the positive control of MosR, suggesting the presence of other regulatory mechanisms besides MosR that control Rv0700 in M. smegmatis. In general, the lacZ reporter assay was able to show differential regulation for the mce1, Rv3130c, and Rv0700 genes in M. smegmatis expressing MosR, despite the inability of MosR to bind to their putative promoter regions, suggesting the need for other transcriptional factors to allow the mosR regulatory role(s).
A role for mosR in M. tuberculosis hypoxic response.
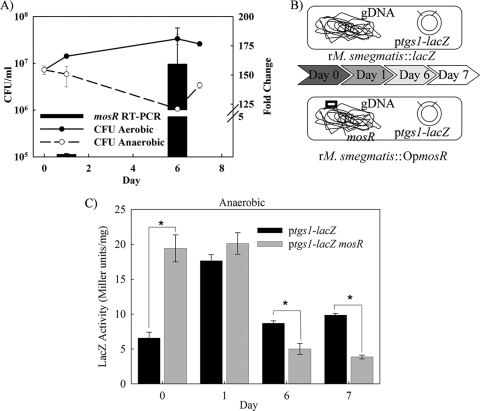
Using the Wayne model of hypoxia in M. tuberculosis, transcripts of the mosR operon were modestly induced under anaerobic conditions (25), suggesting its involvement in hypoxic responses. Since a large number of the dormancy regulon genes were suggested to be controlled by mosR, it is possible that mosR could be involved in mycobacterial hypoxic responses. To test this hypothesis, we adopted an in vitro model of hypoxia in which the influences of hypoxia and anaerobic conditions on the mosR operon could be studied in a recombinant strain of M. smegmatis (M. smegmatis::pML21). Using a modified version of the Wayne model of hypoxia (24), we found that aerobic cultures of M. smegmatis::pML21 grew to a higher density than the anaerobic cultures, as expected (Fig. 5A). Interestingly, transcripts of mosR were significantly upregulated in the cultures grown under hypoxic conditions (at day 1), with a more profound induction when the cultures reached anaerobic phase by day 6 of incubation. This dramatic increase in mosR transcripts strongly supports the hypothesis that mosR participates in M. tuberculosis anaerobic responses. Currently, experiments are under way to examine the survival of the ΔmosR mutant under anaerobic conditions.
FIG. 5.
A regulatory role for mosR under anaerobic conditions. (A) The survival curve of M. smegmatis::pML21 under aerobic and anaerobic conditions (left scale) and the change in mosR transcripts for hypoxic versus aerobic cultures as measured by qRT-PCR (right scale). The error bars indicate standard deviations. (B) Experimental design for the regulation of the tgs1 gene under the control of mosR. Cultures of recombinant M. smegmatis (rM. smegmatis) harboring the mosR operon (OpmosR) were grown under anaerobic conditions using the Wayne model of hypoxia. Samples were collected at different times following incubation based on fading of the methylene blue color and transition from aerobic (dark blue) to anaerobic (colorless) stages. gDNA, genomic DNA. (C) β-Galactosidase activities of anaerobic cultures of recombinant M. smegmatis harboring pML28 with or without the mosR operon. All activities were normalized to the total protein content in each sample. The asterisks denote significant changes in a Student t test (P < 0.001).
Despite the induction of mosR under anaerobic stress, we did not know whether such induction could help in regulating other members of the dormancy regulon. To address this hypothesis, we adopted the Wayne model to analyze the regulation of Rv3130c (tgs1), a member of the dormancy/hypoxia regulon (25). Interestingly, β-galactosidase activities showed a significant repression of the tgs1 promoter in the late microaerobic and anaerobic cultures depending on the presence of the mosR operon (Fig. 5B and C). On the other hand, downregulation of tgs1 was not observed in control, paired samples grown under aerobic conditions (see Fig. S5 in the supplemental material). This analysis was further confirmed on the RNA level using real-time PCR (data not shown). These results are consistent with the observed upregulation of mosR transcription under anaerobic relative to aerobic conditions in M. smegmatis::pML21. Overall, the lacZ reporter assays supported a regulatory role for MosR as a transcriptional factor, especially in an anaerobic microenvironment.
Murine infection with a mosR mutant.
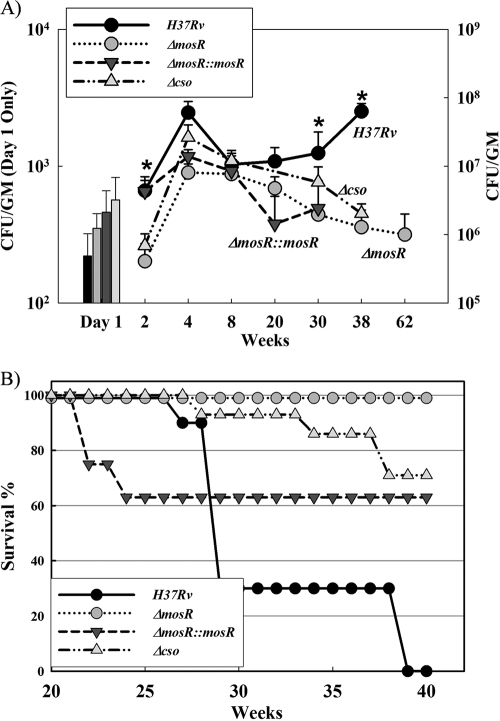
To examine the role of mosR in M. tuberculosis survival during infection, we used the murine model of tuberculosis following aerosol infection with H37Rv, ΔmosR, and mosR-complemented (ΔmosR::mosR) strains. The complemented strain was constructed by amplifying the coding sequence of the Rv0348 gene (0.6 kb) and cloning it into a pMV361-derived plasmid (4). Three groups (n = 40/group) of BALB/c mice were infected with M. tuberculosis strains using the Glas-Col (Terra-Haute, IN) inhalation system to generate an infectious dose of 200 to 400 CFU per mouse (44), according to our protocol approved by the University of Wisconsin IACUC. Mice (n = 5) were sacrificed at different times postinfection to remove the lungs, livers, and spleens for plating on Middlebrook 7H10 agar for colony counting and histopathological staining (44).
Culturing lung tissues at 1 day postinfection confirmed that all groups of mice received comparable infectious doses of each strain (220 CFU/mouse for H37Rv, 350 CFU/mouse for the ΔmosR strain, and 460 CFU/mouse for the ΔmosR::mosR strain) (Fig. 6A). As shown in Fig. 6B, all mice infected with the ΔmosR mutant survived the infection up to 40 weeks postinfection (the end of experiment) with a median survival time of >40 weeks, while mice infected with the wild-type strain, H37Rv, started to die at 27 weeks postinfection, with a median survival time of 29 weeks. In fact, an additional group of mice infected with the ΔmosR strain survived up to 62 weeks postinfection, when they were sacrificed for colony counting. Additionally, the mycobacterial count for the ΔmosR-infected group was significantly different from those of other groups at 2, 30, and 38 weeks postinfection. At these times, the bacterial loads of the ΔmosR strain were significantly lower than those of the wild-type strain (Fig. 6A). The difference in CFU counts was mostly observed following 30 weeks postinfection, suggesting a critical role for mosR in the chronic phase of tuberculosis, as well as a minor role during the early phase, when the difference was observable only at 2 weeks postinfection.
FIG. 6.
Colonization of M. tuberculosis with various copies of mosR. (A) Lung colonization levels following aerosol infection with H37Rv, ΔmosR, ΔmosR::mosR, and Δcso strains. The asterisks denote significant differences in these times (Student's t test). The error bars indicate standard deviations. GM, gram. (B) Survival curves of mouse groups (n = 10) following aerosol infection with various strains of M. tuberculosis. The Δcso mutant was included as an example of a high inducer of PDIM.
The initial colony counts of the complemented ΔmosR::mosR strain at 2 weeks postinfection were similar to those of the wild-type H37Rv strain. Also, 40% of mice infected with the ΔmosR::mosR strain died within the first 24 weeks postinfection, probably because of the higher dose of infection with the complemented strain relative to other strains, in addition to the partial restoration of virulence. However, the ΔmosR::mosR strain colonization level did not persist until the end of infection, suggesting only partial complementation. PCR analysis of the colonies retrieved from mouse lungs infected with the ΔmosR::mosR strain at all times indicated the stability of the complementation construct, even after 30 weeks postinfection in mice (data not shown). Failure of complementation after 8 weeks of infection could be explained by the strength of the mosR operon promoter in comparison to the constitutive but lower expression under the control of the hsp60 promoter used in the complementation construct. Deficiency of in vivo complementation was encountered for several other genes when similar complementation vectors and strategies were used (17, 31).
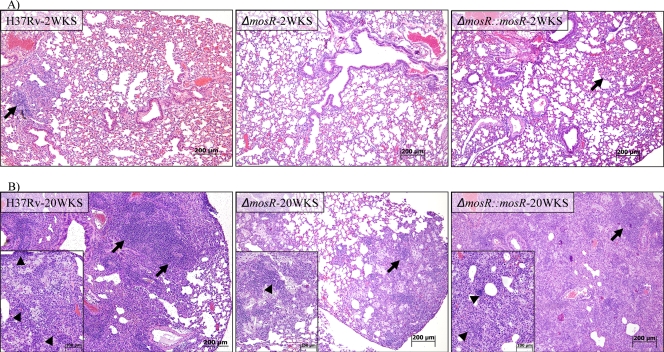
Histopathological analysis of infected tissues showed typical, progressive granulomatous lesions in the lungs of mice infected with the wild type and, to a lesser extent, the complemented strain (Fig. 7). Notably, inflammatory lesions were visible in 100% of the lung sections of mice infected with the ΔmosR::mosR and H37Rv strains, starting at 20 weeks postinfection and continuing, while lesions were observed in only 50% of the lung sections of mice infected with the ΔmosR mutant even at 62 weeks postinfection (see Fig. S6 in the supplemental material). Bacterial loads and survival curves, in addition to histopathological analyses, suggested the attenuation of the ΔmosR mutant, especially during chronic tuberculosis. Nonetheless, the inability to restore the colonization levels associated with the complemented strain to the wild-type level warranted further investigation. To confirm the ability of pMV361-mosR to complement the mosR regulatory function, we employed qRT-PCR to examine the restoration of transcripts identified by DNA microarrays to be under the control of mosR. In all 10 randomly selected genes, transcripts in the complemented strain were restored to the wild-type level (±1), confirming the ability of the pMV361 construct to complement mosR activities (see Fig. S7 in the supplemental material).
FIG. 7.
Lesions associated with infection with M. tuberculosis with different copies of mosR. Shown is a histological analysis of sections of mouse lungs at 2 and 20 weeks (WKS) postinfection with the ΔmosR, ΔmosR::mosR, and H37Rv strains. Note the levels of accumulation of inflammatory cells (arrows) in hematoxylin- and eosin-stained sections. Aggregates of macrophages and lymphocytes (arrowheads) are displayed inside insets at higher magnification. Bars, 100 μm or 200 μm.
Analysis of the lipid profiles of M. tuberculosis strains.
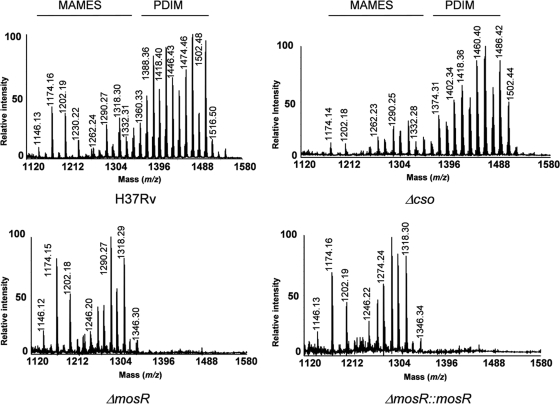
In earlier reports (17, 23), the failure of complementation was suspected to include the spontaneous loss of phthiocerol dimycocerosates (PDIM), key components of the mycobacterial cell wall associated with virulence (10). To examine lipids present in the strains used for animal infection, we profiled the lipid content of each strain used for mouse infection. Lipids extracted from the wild-type H37Rv, its isogenic ΔmosR mutant, and the complemented ΔmosR::mosR strain were first analyzed by thin-layer chromatography (28). Serendipitously, we analyzed an additional cso operon mutant (Δcso) that was shown to be involved in copper homeostasis in M. tuberculosis (22). This analysis did not reveal any obvious differences between the various classes of the major lipids produced by the four strains, including trehalose dimycolates, triacyl glycerol, and phospholipids. The species-specific PDIM, as well as quantitatively minor substances, were hardly visible by this technique. To visualize these compounds, the major and ubiquitous ester-linked lipids were hydrolyzed using alkaline methanolysis. This hydrolysis cleaves nonbranched fatty acyl-containing compounds, e.g., phospholipids and triacyl glycerol, but not mycolate- and multimethyl branched fatty acyl-containing products, such as trehalose mycolates and PDIM. The resulting alkali-treated lipids were analyzed by MALDI-TOF as detailed previously (5, 20).
Analysis of the mass spectra showed a series of pseudomolecular ion (M + Na)+ peaks at m/z 1,146, 1,174, and 1,202, corresponding to C76, C78, and C80 α-mycolic acid methyl esters (MAMEs), respectively (Fig. 8). The other series of peaks, observed at higher mass values between m/z 1,246 and 1,346, corresponded to the C82 to C89 oxygenated methoxy- and keto-MAMEs (20). No differences were observed between the MAME profiles of the three strains examined. In sharp contrast, the MALDI mass spectra of the wild-type and Δcso strains showed an additional envelope of pseudomolecular ion peaks at m/z 1,376, 1,390, 1,418, 1,432, 1,460, 1,488, and 1,502; these corresponded to compounds with 91 to 100 carbon atoms. The last series of peaks, observed only in the mass spectra of the wild-type and Δcso strains and absent from other strains (ΔmosR and ΔmosR::mosR), were assignable to lipid compounds belonging to the PDIM family (5, 28). These data were further confirmed by thin-layer chromatography analysis of the alkaline methanolysis products (see Fig. S8 in the supplemental material). Generally, the lipid profiles of the examined strains suggested a loss of PDIM that could be responsible for the attenuation phenotype observed with the ΔmosR mutation. However, this possibility is unlikely based on the analysis of the colonization and survival data of mice infected with the Δcso strain (Fig. 6), with a high level of PDIM production (Fig. 8). We also reasoned that the loss of PDIM in the ΔmosR::mosR strain could be related to spontaneous loss of PDIM, as suggested previously (12, 23), or to the inability of the pMV361 vector to restore genes involved in PDIM production. Previously, the fadD22-fadD29 gene cluster was shown to be involved in the synthesis of PDIM (28), which could explain the absence of PDIM in the ΔmosR, but not the ΔmosR::mosR, strain. To examine this possibility, we used qRT-PCR to analyze transcripts for the three-gene operon (fadD22-Rv2949c-fadD29). The qRT-PCR analysis confirmed the downregulation of this operon in both the ΔmosR and ΔmosR::mosR strains compared to the wild-type strain (see Fig. S7B in the supplemental material), confirming the partial complementation in the ΔmosR::mosR strain, as well as the regulatory role played by mosR on the fadD22 gene cluster, as detected earlier using DNA microarrays (Table 2).
FIG. 8.
MALDI-TOF mass spectra of alkali-treated lipids from M. tuberculosis H37Rv, ΔmosR, ΔmosR::mosR, and Δcso strains. The values indicate the masses of the major pseudomolecular ions [M + Na]+ of MAMEs and PDIM as indicated above the spectra. Lipid samples were dissolved in chloroform and applied to the target plate. 2,5-dihydroxy benzoic acid was used as a matrix (10 mg/ml in CHCl3/CH3OH; 1:1 [vol/vol]).
Interestingly, the earlier analysis of the role of PDIM in M. tuberculosis virulence (10) was based only on lungs during early tuberculosis (3 weeks postinfection) following intravenous injection of a high dose of M. tuberculosis, a model that we did not employ here because of the superiority of the aerosol model. Moreover, levels of a PDIM-deficient strain of M. tuberculosis were not reduced in spleens of infected mice, and complementation analysis for the PDIM phenotype was not shown in the earlier report (10). Later analysis of the role of PDIM in cellular infection revealed no significant effect on macrophage transcriptional responses measured by DNA microarrays (35). On the contrary, our model continued for up to 40 weeks postinfection with no difference in colonization and survival data for strains with low (e.g., ΔmosR) or high (Δcso) levels of PDIM. Additionally, levels of the ΔmosR strain were significantly lower in spleens (in addition to lungs) when samples collected at 8 and 20 weeks postinfection were cultured (data not shown). Notably, the H37Rv strain is not a strong producer of PDIM (1, 17), with the presence of minor changes in their production depending on the culture growth phase (see Fig. S9 in the supplemental material). Given the impact of mosR on the transcriptional regulation of several gene operons, we expect its contribution to M. tuberculosis virulence to be more complex and dependent on several gene groups.
Conclusions.
In summary, we have shown that mosR encodes a transcriptional regulator with both inducer and repressor activities that could be involved in M. tuberculosis virulence. Based on the analyses presented, we generated a model that delineates possible pathways that involve MosR to explain its role in establishing chronic tuberculosis. In this model (see Fig. S10 in the supplemental material), MosR can bind to its own promoter in order to maintain a low level of expression, especially during log-phase culture or in vitro growth in general. Under certain stressors (e.g., a high O− level), the expression of MosR is even lower, which relieves its repression of other genes, such as hypoxia- and phagosome-responsive genes. However, under other stressors (e.g., high temperature or in vivo growth), mosR is induced (44), most likely through the activities of other transcriptional regulators (e.g., SigC) that share a transcriptional binding site (30) upstream of the mosR operon. Such binding could prevent the binding of MosR to its promoter, and hence, its own expression would be induced, which in turn could activate genes involved in starvation and invasion in the mosR regulon. The role of SigC in mosR operon regulation requires more investigation, as a ΔsigC mutant created in M. tuberculosis CDC 1551 did not show significant differential expression patterns of mosR operon genes during different growth phases (40). Under all of these scenarios (relief of repression of hypoxic genes or induction of starvation genes), the general outcome of the induction of the mosR regulon is the fitness of M. tuberculosis to persist in various host microenvironments. Despite the conclusions that can be drawn from the proposed model, the exact function of the mosR operon remains elusive, and further studies are warranted. Specifically, it will be interesting to delineate the mechanism(s) employed by mosR to regulate other gene groups. Additionally, testing mosR mutants in other animal models (e.g., guinea pigs or immunodeficient mice) will further help to dissect the role of mosR in the immunopathogenesis of M. tuberculosis. This knowledge base could improve our understanding of the mechanisms employed by M. tuberculosis to survive in the lung microenvironments.
Supplementary Material
Acknowledgments
We acknowledge the technical assistance of Emma Hsu. Also, we are indebted to Matyas Sandor for reading the manuscript.
This work was partially supported by NIH-R21AI066235 and Animal Formula Funds awarded to A.M.T. Portions of this work were also funded by the USDA Agricultural Research Service.
Footnotes
Published ahead of print on 31 July 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Andreu, N., and I. Gibert. 2008. Cell population heterogeneity in Mycobacterium tuberculosis H37Rv. Tuberculosis 88:553-559. [DOI] [PubMed] [Google Scholar]
- 2.Bardarov, S., M. S. Pavelka, V. Sambandamurthy, M. Larsen, J. Tufariello, J. Chan, G. Hatfull, and W. R. Jacobs. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007-3017. [DOI] [PubMed] [Google Scholar]
- 3.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 4.Braunstein, M., S. S. Bardarov, and W. R. Jacobs. 2002. Genetic methods for deciphering virulence determinants of Mycobacterium tuberculosis. Methods Enzymol. 358:67-99. [DOI] [PubMed] [Google Scholar]
- 5.Camacho, L. R., P. Constant, C. Raynaud, M. A. Laneelle, J. A. Triccas, B. Gicquel, M. Daffe, and C. Guilhot. 2001. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis: evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 276:19845-19854. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, D. R., K. E. Chapman, K. J. Waldron, S. Tottey, S. Kendall, G. Cavallaro, C. Andreini, J. Hinds, N. G. Stoker, N. J. Robinson, and J. S. Cavet. 2007. Mycobacterial cells have dual nickel-cobalt sensors: sequence relationships and metal sites of metal-responsive repressors are not congruent. J. Biol. Chem. 282:32298-32310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerrina, F., F. Blattner, W. Huang, Y. Hue, R. Green, S. Singh-Gasson, and M. Sussman. 2002. Biological lithography: development of a maskless microarray synthesizer for DNA chips. Microelectron 2:33-40. [Google Scholar]
- 8.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornby, and K. Jagels. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-538. [DOI] [PubMed] [Google Scholar]
- 9.Corbett, E. L., C. J. Watt, N. Walker, D. Maher, B. G. Williams, M. C. Raviglione, and C. Dye. 2003. The growing burden of tuberculosis—global trends and interactions with the HIV epidemic. Arch. Intern. Med. 163:1009-1021. [DOI] [PubMed] [Google Scholar]
- 10.Cox, J. S., B. Chen, M. McNeil, and W. R. Jacobs. 1999. Complex lipids determine tissue specific replication of Mycobacterium tuberculosis in mice. Nature 402:79-83. [DOI] [PubMed] [Google Scholar]
- 11.Cruz-Vera, L. R., and C. Yanofsky. 2008. Conserved residues Asp16 and Pro24 of TnaC-tRNAPro participate in tryptophan induction of tna operon expression. J. Bacteriol. 190:4791-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domenech, P., M. B. Reed, C. S. Dowd, C. Manca, G. Kaplan, and C. E. Barry III. 2004. The Role of MmpL8 in sulfatide biogenesis and virulence of Mycobacterium tuberculosis. J. Biol. Chem. 279:21257-21265. [DOI] [PubMed] [Google Scholar]
- 13.Gill, W. P., N. S. Harik, M. R. Whiddon, R. P. Liao, J. E. Mittler, and D. R. Sherman. 1 February 2009. A replication clock for Mycobacterium tuberculosis. Nat. Med. doi: 10.1038/nm.1915. [DOI] [PMC free article] [PubMed]
- 14.Hahn, M. Y., S. Raman, M. Anaya, and R. N. Husson. 2005. The Mycobacterium tuberculosis extracytoplasmic-function sigma factor SigL regulates polyketide synthases and secreted or membrane proteins and is required for virulence. J. Bacteriol. 187:7062-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harries, A. D. 1997. Tuberculosis in Africa: clinical presentation and management. Pharmacol. Ther. 73:1-50. [DOI] [PubMed] [Google Scholar]
- 16.Hu, Y. M., J. A. Mangan, J. Dhillon, K. M. Sole, D. A. Mitchison, P. D. Butcher, and A. R. M. Coates. 2000. Detection of mRNA transcripts and active transcription in persistent Mycobacterium tuberculosis induced by exposure to rifampin or pyrazinamide. J. Bacteriol. 182:6358-6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kana, B. D., B. G. Gordhan, K. J. Downing, N. Sung, G. Vostroktunova, E. E. Machowski, L. Tsenova, M. Young, A. S. Kaprelyants, G. Kaplan, and V. Mizrahi. 2008. The resuscitation-promoting factors of Mycobacterium tuberculosis are required for virulence and resuscitation from dormancy but are collectively dispensable for growth in vitro. Mol. Microbiol. 67:672-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kendziorski, C. M., R. A. Irizarry, K. S. Chen, J. D. Haag, and M. N. Gould. 2005. On the utility of pooling biological samples in microarray experiments. Proc. Natl. Acad. Sci. USA 102:4252-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kendziorski, C. M., M. A. Newtone, H. Lan, and M. N. Goululd. 2003. On parameteric empirical Bayes methods for comparing multiple groups using replicated gene expression profiles. Stat. Med. 22:3899-3914. [DOI] [PubMed] [Google Scholar]
- 20.Laval, F., M. A. Laneelle, C. Deon, B. Monsarrat, and M. Daffe. 2001. Accurate molecular mass determination of mycolic acids by MALDI-TOF mass spectrometry. Anal. Chem. 73:4537-4544. [DOI] [PubMed] [Google Scholar]
- 21.Lillebaek, T., A. Dirksen, I. Baess, B. Strunge, V. O. Thomsen, and A. B. Andersen. 2002. Molecular evidence of endogenous reactivation of Mycobacterium tuberculosis after 33 years of latent infection. J. Infect. Dis. 185:401-404. [DOI] [PubMed] [Google Scholar]
- 22.Liu, T., A. Ramesh, Z. Ma, S. K. Ward, L. Zhang, G. N. George, A. M. Talaat, J. C. Sacchettini, and D. P. Giedroc. 2007. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat. Chem. Biol. 3:60-68. [DOI] [PubMed] [Google Scholar]
- 23.Manjunatha, U. H., H. Boshoff, C. S. Dowd, L. Zhang, T. J. Albert, J. E. Norton, L. Daniels, T. Dick, S. S. Pang, and C. E. Barry. 2006. Identification of a nitroimidazo-oxazine-specific protein involved in PA-824 resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 103:431-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayuri, G. Bagchi, T. K. Das, and J. S. Tyagi. 2002. Molecular analysis of the dormancy response in Mycobacterium smegmatis: expression analysis of genes encoding the DevR-DevS two-component system, Rv313c and chaperone alpha-crystallin homologues. FEMS Microbiol. Lett. 211:231-237. [DOI] [PubMed] [Google Scholar]
- 25.Muttucumaru, D. G. N., G. Roberts, J. Hinds, R. A. Stabler, and T. Parish. 2004. Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis 84:239-246. [DOI] [PubMed] [Google Scholar]
- 26.Ohno, H., G. F. Zhu, V. P. Mohan, D. Chu, S. Kohno, W. R. Jacobs, and J. Chan. 2003. The effects of reactive nitrogen intermediates on gene expression in Mycobacterium tuberculosis. Cell Microbiol. 5:637-648. [DOI] [PubMed] [Google Scholar]
- 27.Park, H. D., K. M. Guinn, M. I. Harrell, R. Liao, M. I. Voskuil, M. Tompa, G. K. Schoolnik, and D. R. Sherman. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48:833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez, E., P. Constant, F. Laval, A. Lemassu, M. A. Laneelle, M. Daffe, and C. Guilhot. 2004. Molecular dissection of the role of two methyltransferases in the biosynthesis of phenolglycolipids and phthiocerol dimycoserosate in the Mycobacterium tuberculosis complex. J. Biol. Chem. 279:42584-42592. [DOI] [PubMed] [Google Scholar]
- 29.Roback, P., J. Beard, D. Baumann, C. Gille, K. Henry, S. Krohn, H. Wiste, M. I. Voskuil, C. Rainville, and R. Rutherford. 2007. A predicted operon map for Mycobacterium tuberculosis. Nucelic Acids Res. 35:5085-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodrigue, S., J. Brodeur, P. E. Jacques, A. L. Gervais, R. Brzezinski, and L. Gaudreau. 2007. Identification of Mycobacterial σ factor binding sites by chromatin immunoprecipitation assays. J. Bacteriol. 189:1505-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russell-Goldman, E., J. Xu, X. Wang, J. Chan, and J. M. Tufariello. 2008. A Mycobacterium tuberculosis Rpf double-knockout strain exhibits profound defects in reactivation from chronic tuberculosis and innate immunity phenotypes. Infect. Immun. 76:4269-4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rustad, T. R., M. I. Harrell, R. Liao, and D. R. Sherman. 2008. The enduring hypoxic response of Mycobacterium tuberculosis. PLoS ONE 3:e1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 35.Scandurra, G. M., R. B. H. Williams, J. A. Triccas, R. Pinto, B. Gicquel, B. Slobedman, A. Cunningham, and W. J. Britton. 2007. Effect of phthiocerol dimycocerosate deficiency on the transcriptional response of human macrophages to Mycobacterium tuberculosis. Microbes Infect. 9:87-95. [DOI] [PubMed] [Google Scholar]
- 36.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh-Gasson, S., R. D. Green, Y. J. Yue, C. Nelson, F. Blattner, M. R. Sussman, and F. Cerrina. 1999. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol. 17:974-978. [DOI] [PubMed] [Google Scholar]
- 38.Sirakova, T. D., V. S. Dubey, C. Deb, J. Daniel, T. A. Korotkova, B. Abomoelak, and P. E. Kolattukudy. 2006. Identification of a diacylglycerol acyltransferase gene involved in accumulation of triacylglycerol in Mycobacterium tuberculosis under stress. Microbiology 152:2717-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stover, C. K., F. De la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, S. B. Snapper, R. G. Barletta, W. R. Jacobs, Jr., and B. R. Bloom. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 40.Sun, R. G., P. J. Converse, C. Ko, S. Tyagi, N. E. Morrison, and W. R. Bishai. 2004. Mycobacterium tuberculosis ECF sigma factor SigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol. Microbiol. 52:25-38. [DOI] [PubMed] [Google Scholar]
- 41.Talaat, A. M., P. Hunter, and S. A. Johnston. 2000. Genome-directed primers for selective labeling of bacterial transcripts for DNA microarray analysis. Nat. Biotechnol. 18:679-682. [DOI] [PubMed] [Google Scholar]
- 42.Talaat, A. M., R. Lyons, S. T. Howard, and S. A. Johnston. 2004. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc. Natl. Acad. Sci. USA 101:4602-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Talaat, A. M., and M. Trucksis. 2000. Transformation and transposition of the genome of Mycobacterium marinum. Am. J. Vet. Res. 61:125-128. [DOI] [PubMed] [Google Scholar]
- 44.Talaat, A. M., S. K. Ward, C.-W. Wu, E. Rondon, C. Tavano, J. P. Bannantine, R. Lyons, and S. A. Johnston. 2007. Mycobacterial bacilli are metabolically active during chronic tuberculosis in murine lungs: insights from genome-wide transcriptional profiling. J. Bacteriol. 189:4265-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uchida, Y., N. Casali, A. White, L. Morici, L. V. Kendall, and L. W. Riley. 2007. Accelerated immunopathological response of mice infected with Mycobacterium tuberculosis disrupted in the mce1 operon negative transcriptional regulator. Cell Microbiol. 9:1275-1283. [DOI] [PubMed] [Google Scholar]
- 46.Voskuil, M. I. 2004. Mycobacterium tuberculosis gene expression during environmental conditions associated with latency. Tuberculosis 84:138-143. [DOI] [PubMed] [Google Scholar]
- 47.Voskuil, M. I., D. Schnappinger, K. C. Visconti, M. I. Harrell, G. M. Dolganov, D. R. Sherman, and G. K. Schoolnik. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voskuil, M. I., K. C. Visconti, and G. K. Schoolnik. 2004. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis 84:218-227. [DOI] [PubMed] [Google Scholar]
- 49.Ward, S. K., E. A. Hoye, and A. M. Talaat. 2008. The global responses of Mycobacterium tuberculosis to physiological levels of copper. J. Bacteriol. 190:2939-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wards, B. J., and D. M. Collins. 1996. Electroporation at elevated temperatures substantially improves transformation efficiency of slow-growing mycobacteria. FEMS Microbiol. Lett. 145:101-105. [DOI] [PubMed] [Google Scholar]
- 51.Wayne, L. G., and L. G. Hayes. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.WHO. 2009. Global tuberculosis control: epidemiology, strategy, financing: WHO report 2009. World Health Organization Press, Geneva, Switzerland. http://www.globalhealthreporting.org/tb.asp.
- 53.Wu, C. W., S. K. Schmoller, S. J. Shin, and A. M. Talaat. 2007. Defining the stressome of Mycobacterium avium subspecies paratuberculosis in vitro and in naturally infected cows. J. Bacteriol. 189:7877-7886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan, Y., D. D. Crane, R. M. Simpson, Y. Q. Zhu, M. J. Hickey, D. R. Sherman, and C. E. Barry. 1998. The 16-kDa alpha-crystallin (acr) protein of Mycobacterium tuberculosis is required for growth in macrophages. Proc. Natl. Acad. Sci. USA 95:9578-9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.