Abstract
The toxin MazF in Escherichia coli cleaves single-stranded RNAs specifically at ACA sequences. MazF overexpression virtually eliminates all cellular mRNAs to completely block protein synthesis. However, protein synthesis can continue on an mRNA that is devoid of ACA triplets. The finding that ribosomal RNAs remain intact in the face of complete translation arrest suggested a purpose for such preservation. We therefore examined the sequences of all transcribed RNAs to determine if there was any statistically significant bias against ACA. While ACA motifs are absent from tmRNA, 4.5S RNA, and seven of the eight 5S rRNAs, statistical analysis revealed that only for tmRNA was the absence nonrandom. The introduction of single-strand ACAs makes tmRNA highly susceptible to MazF cleavage. Furthermore, analysis of tmRNA sequences from 442 bacteria showed that the discrimination against ACA in tmRNAs was seen mostly in enterobacteria. We propose that the unusual bias against ACA in tmRNA may have coevolved with the acquisition of MazF.
Toxin-antitoxin (TA) modules, originally found in plasmids, are believed to specifically prevent the growth of dividing cells that do not inherit the plasmid (13, 20). This type of selection process was given the term “postsegregational killing.” New findings demonstrate that the prevalence of these genes on plasmids is due to their ability to inhibit the replication of TA-negative plasmids after within-host competition (7, 8).
Surprisingly, many species of bacteria and archaea contain homologues of these TA pairs in their genomes, often containing more than one copy (35, 51). Current research is revealing how diverse the targets of these toxins are, from topoisomerase to mRNAs (21, 36, 37, 44). Whatever its target, the defining characteristic of a toxin is its ability to slow the growth of the organism in which the toxin is overexpressed or in which its cognate antitoxin is degraded. Considering the apparent loss of fitness during toxin expression, the functional importance of the many TA modules found throughout the bacterial and archaeal kingdoms, often in the same organism, is still in dispute. Theories involving quorum sensing (28) and programmed cell death have been proposed, where the death of one population may benefit the survival of the species (32). Others have proposed that rather than being irreversible, toxin-induced growth inhibition may be the cell's last resort to temporarily conserve its metabolic stores under extreme environmental conditions (5, 6, 11, 12). The radically different theory put forth by Saavedra De Bast et al. (38) argues that postsegregational killing was the selective pressure for the fixation of TA systems in the genome and that these genes may no longer have any biological role once the plasmid-borne TA modules evolve enough that they cannot be neutralized. Those authors showed that the possession of a cognate antitoxin in its genome does indeed allow the bacterium to survive the toxic effects of losing a TA-containing plasmid. However, that work did not address the fact that most antitoxins in genomes are found together with their toxins, the presence of which would be unnecessary, according to their theory.
One of the best-studied toxins, MazF, is a sequence-specific endoribonuclease. This toxin cleaves single-strand ACA sequences and, when overexpressed, rapidly blocks protein synthesis (53). However, we demonstrated that translation itself is unaffected during MazF induction by overexpressing a protein whose mRNA is ACA-less (41). As both soluble and membrane-bound proteins can be expressed upon MazF induction (41), we inferred that the noncoding RNAs involved in protein synthesis and translocation must also be protected from MazF cleavage. This finding prompted us to analyze all RNAs involved in translation for the presence of ACAs in their secondary structures.
In bacteria, the repertoire of RNAs involved in protein synthesis includes not only rRNAs, tRNAs, and mRNAs but also 4.5S and tmRNA. 4.5S RNA, in a complex with the Ffh protein, plays a pivotal role in cotranslational protein targeting and translocation (31). The ssrA gene encodes another small RNA (tmRNA) that is required for normal growth because it mediates the release of stalled ribosomes from defective mRNAs (48). All eubacterial genomes examined so far contain the ssrA gene (1).
Here we examined the occurrence of every possible nucleotide triplet in the RNAs of Escherichia coli K-12. By ranking ACA among the 64 triplets possible, we found the frequency of ACAs to be generally low in the RNAs involved in translation. However, statistical analysis showed that only tmRNA had a significantly low number of ACAs in its single-strand region. Furthermore, examination of over 400 tmRNA sequences from different bacteria revealed a significant bias against single-strand ACAs most often in enterobacteria. As MazF cleavage of ACAs in tmRNA was biochemically confirmed, we propose that tmRNA function may be critical for the MazF-related stress response and that the unusual bias against ACA in tmRNA may have coevolved with the acquisition of a sequence-specific endoribonuclease.
MATERIALS AND METHODS
Sequence analysis.
A short program was written in Perl to count the frequencies of all 64 possible triplets in a given sequence.
Probability of random ACA accumulation.
tmRNA sequences and predicted secondary structures were obtained from an online database (http://www.indiana.edu/∼tmrna/). Incomplete sequences and sequences from unculturable bacteria were excluded from any analyses. As a result, a total of 442 tmRNAs were examined, including 12 plastid tmRNAs and two virus tmRNAs. E. coli K-12 tRNA sequences and predicted secondary structures were obtained from the Genomic tmRNA Database (http://lowelab.ucsc.edu/GtRNAdb/Esch_coli_K12/). The secondary structures of 5S, 16S, and 23S RNAs of E. coli were obtained from the University of Texas at Austin Comparative Web Site and Project online database (http://www.rna.ccbb.utexas.edu/). The sequences of 4.5S, 5S, 16S, 23S, and all other noncoding RNA genes from E. coli K-12 were obtained from GenBank. The secondary structure of 4.5S RNA was obtained from the SRPDB (http://rnp.uthct.edu/rnp/SRPDB/SRPDB.html).
Since ACA sequences in secondary structures are protected from MazF (52), a program was written in Perl to compare the probabilities of ACA triplets occurring randomly in the single-strand versus the double-strand regions of each structural RNA. In order to fairly compare two RNAs with different numbers of individual single-strand segments, we calculated one probability for all the individual single-strand regions and one probability for all the individual double-strand regions for each RNA. To do this, we first calculated the total number of A's and the total number of C's for all single-strand regions and likewise for double-strand regions. The probability, p, of ACA appearing anywhere in a single-strand segment is calculated as follows: p = (percentage of A)2(percentage of C). To determine the expected number, E, of ACA in single-strand regions, we multiplied the total number of triplets able to be counted in all single-strand regions, L, by the percentage of ACA: E = p × L. If K is the actual number of ACAs in single-strand regions, then the probability, P, of having K or fewer ACAs in single-strand regions is calculated as follows:
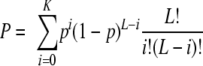 |
As the seven 23S genes of E. coli contain a nontrivial number of ACAs at the junction of single- and double-strand regions, the following rule was used to account for these “junction” ACAs. If two bases were single stranded, then the triplet was counted as single stranded; if two bases were double stranded, then the ACA triplet was counted as double stranded. Otherwise, the number of ACAs in a given RNA would appear artificially low.
As mRNA and the other noncoding small RNAs were analyzed for general comparison purposes, the random likelihood of ACAs for the whole RNA was deemed sufficient to indicate if there was a genome-wide bias against ACAs. More importantly, the secondary structures of most mRNAs are unknown and most likely unstable.
Strains and plasmids used.
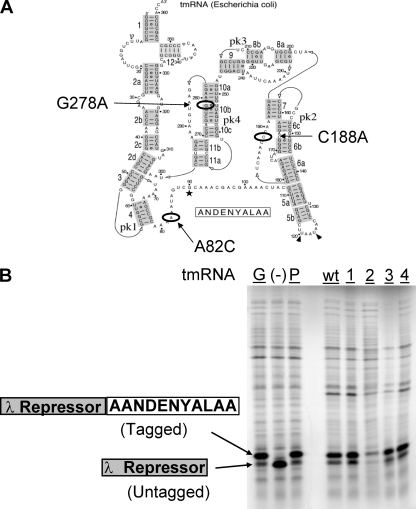
tmRNA was deleted from strain BW25113 by homologous recombination using a method described previously (9). The kanamycin cassette was inserted between the following sequences: 5′-TCTGGTATACTTACCTTTAC-3′ and 5′-GTCCCGCGAAGTCCGAAGAG-3′. The corresponding deletion was made in strain BL21(DE3) by P1 transduction using a lysate from BW25113ssrA. The kanamycin cassette was then removed by the expression of the FLP recombinase from pCP20 (9) and confirmed by PCR. For the expression of tmRNA from a plasmid, approximately 1.3 kb of the ssrA locus was amplified from strain CP78 (10, 45) (gift from Barbara Wright) and cloned into pCR2.1-TOPO (Invitrogen) using primers SB1 (5′-CCCGTCACGAATACTTTATC-3′) and SB2 (5′-CGGCATCTTTAGGTTTGGCG-3′). An XmaI site was introduced downstream of the terminator sequence of ssrA by site-directed PCR mutagenesis in order to insert a 1.2-kb kanamycin cassette (including its own promoter) to produce pSSWB1. PCR mutagenesis was then performed sequentially to create a triple-ACA mutant, pSSWB2 (see Fig. 5A). The final 2.5-kb fragments of the wild type and the triple mutant were subcloned into the EcoRI site of pBR322 to produce pSSWB3 and pSSWB4, respectively. mazF expression was driven by a T7 promoter and induced by the Lac operator sequence in pACYCDuet (Novagen). pPW500 contains a truncated version of the λ repressor gene that lacks a stop codon but has a strong transcription terminator (25).
FIG. 5.
Mutant tmRNAs have tagging function. (A) Diagram of the three ACA sites introduced into E. coli K-12 tmRNA (adapted from reference 57 with permission of the publisher). Two ACA mutations were introduced into single-strand regions, while the third was introduced into a double-strand region. (B) Radiolabeled lambda repressor is tagged by genomic tmRNA (G), wild-type (wt) tmRNA plasmid (P), each of the single tmRNA mutant plasmids (lanes 1, 2, and 3), and the tmRNA triple mutant plasmid (lane 4). The lambda repressor is not tagged (and hence smaller) in cells lacking tmRNA (−).
Primer extension.
Total RNA was extracted using the hot-phenol method (39) and quantitated by UV spectrophotometry. The following oligonucleotides were synthesized to detect potential cleavage sites at A82C, C188A, and G278A, respectively: OligoSS1 (5′-CCTGCTTAGAGCCCTCTCTC-3′), OligoSS2 (5′-CAGGCTAGTTTGTTAGTGGC-3′), and OligoDS (5′-CGGACGCGGGTTCAACTCCC-3′). They were 5′ labeled by T4 polynucleotide kinase using [γ-32P]ATP at 37°C. Primer extensions were carried out using 2.5 μg of total RNA with Moloney murine leukemia virus reverse transcriptase (Superscript II; Invitrogen) at 44°C, according to the manufacturer's protocol. Sequencing reactions were performed using the Sequenase kit from US Biologicals, with pSSWB4 as the template and [α-32P]CTP to internally label the amplicons. Primer extension and sequencing reaction mixtures were run on a 6% polyacrylamide sequencing gel, dried for 2 h at 80°C, and then exposed to a PhosphorImager apparatus (Molecular Dynamics) overnight at room temperature.
Test of ssrA tagging function.
Cells were transformed with pCR2.1 containing either wild-type or mutant ssrA and pPW500 (25). These transformants were then grown in Luria-Bertani broth with ampicillin (100 μg/ml) and kanamycin (50 μg/ml) at 37°C to an optical density at 600 nm of 0.4. Lambda repressor expression was induced with 1 mM isopropyl-β-d-thiogalactoside (IPTG). After 30 min of induction at 37°C, the cells were metabolically labeled with 75 μCi/ml of [35S]methionine for 1 min and then harvested by centrifugation for 1 min at 16,000 × g. The cells were then lysed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer and run on a 12% polyacrylamide gel. The gel was then dried and exposed to a PhosphorImager apparatus.
RESULTS
ACA sequence analysis in RNAs may explain the preservation of translation. (i) Multiple factors may preserve tRNA function during MazF induction.
In the 87 tRNAs of E. coli K-12 (obtained from an online tRNA database [http://lowelab.ucsc.edu/GtRNAdb/Esch_coli_K12/]), ACA was found to be the least frequent triplet out of the possible 64 triplets. Of the 19 tRNAs that do contain ACA sequences, 16 are most likely protected from cleavage due to their ACAs being found in a secondary structure or due to ACA methylation (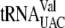 [2]) (Fig. 1A shows the 10 tRNA genes with unique ACA sites). Indirect experimental evidence that these ACAs are protected was obtained by examining the gene sequences of EnvZb, HR91, and human eotaxin. These proteins were well expressed upon MazF induction (40, 41) when their ACAs were removed by mutagenesis. The codon usage of these genes would determine which tRNAs were functional for protein expression. The codons that specifically require
[2]) (Fig. 1A shows the 10 tRNA genes with unique ACA sites). Indirect experimental evidence that these ACAs are protected was obtained by examining the gene sequences of EnvZb, HR91, and human eotaxin. These proteins were well expressed upon MazF induction (40, 41) when their ACAs were removed by mutagenesis. The codon usage of these genes would determine which tRNAs were functional for protein expression. The codons that specifically require 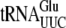 ,
, 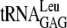 ,
, 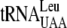 , and
, and 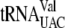 for translation are all found in the EnvZb gene sequence. The codons that bind
for translation are all found in the EnvZb gene sequence. The codons that bind 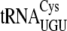 and
and 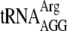 are found in eotaxin and HR91, respectively. All of these tRNAs have an ACA in a double-strand region or an ACA that is methylated (
are found in eotaxin and HR91, respectively. All of these tRNAs have an ACA in a double-strand region or an ACA that is methylated (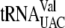 [2]). The presence of their cognate codons in these genes strongly suggests that these tRNAs are preserved during MazF induction. The three tRNAs which have unmodified ACAs in their single-strand regions (tyrU, valV, and valW) have redundant functions with other tRNAs (30) so that their cleavage during MazF induction would not necessarily inhibit translation. As many ACA-less genes were well expressed upon MazF induction (40, 41), these factors probably contribute to the preservation of overall tRNA function during MazF induction.
[2]). The presence of their cognate codons in these genes strongly suggests that these tRNAs are preserved during MazF induction. The three tRNAs which have unmodified ACAs in their single-strand regions (tyrU, valV, and valW) have redundant functions with other tRNAs (30) so that their cleavage during MazF induction would not necessarily inhibit translation. As many ACA-less genes were well expressed upon MazF induction (40, 41), these factors probably contribute to the preservation of overall tRNA function during MazF induction.
FIG. 1.
Location of all ACA triplets in tRNA and 5S rRNA sequences of E. coli K-12. (A) Nucleotide sequences (Seq) of any tRNAs containing ACAs are shown (for a total of 10 unique ACA-containing tRNAs), with the predicted single-strand and double-strand (Str) regions indicated by dots and arrowheads, respectively. ACA triplets are underlined, and the methylation site of 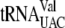 is marked with an arrow. (B) The nucleotide sequence of the 5S rRNA gene, rrfF, is similarly shown.
is marked with an arrow. (B) The nucleotide sequence of the 5S rRNA gene, rrfF, is similarly shown.
(ii) Ribosomal RNAs are preserved from MazF induction.
Although 16S and 23S rRNAs contain 18 and 32 ACA triplets, respectively, these rRNAs are highly resistant to MazF cleavage because of their association with ribosomal proteins and their extensive secondary structures (53) (see Fig. S1 in the supplemental material). However, MazF can effectively digest naked 16S and 23S rRNAs (53).
(iii) 5S RNAs have one ACA, while 4.5S and tmRNA have none.
The ribosomal 5S RNAs are encoded by eight independent genes. Seven of these eight 5S genes lack the ACA triplet. One remaining 5S rRNA contains one ACA in a double-strand region (Fig. 1B), suggesting that 5S rRNA is completely protected from MazF cleavage.
4.5S RNA encoded by the ffs gene has an essential role in the targeting of secretory and membrane proteins to the cytoplasmic membrane in bacteria during translation (26). E. coli 4.5S RNA does not contain any ACA triplets (Table 1), which may explain how membrane proteins can be expressed during MazF induction (41).
TABLE 1.
Numbers of ACA triplets in RNAs involved in protein synthesis in E. coli K-12
| RNA | Length (nucleotides)a | Avg no. of ACAsb | No. of ACA-less RNAs/total no. of genes | Probability of single-strand ACAc | Probability of double-strand ACAc |
|---|---|---|---|---|---|
| tmRNA | 363 | 0 | 1/1 | 0.02 | 0.51 |
| 4.5S RNA | 114 | 0 | 1/1 | 0.73 | 0.70 |
| tRNA | 78.5 | 0.30 | 68/87 | 0.74 | 0.87 |
| 5S rRNA | 120 | 0.13 | 7/8 | 0.49 | 0.83 |
| 16S rRNA | 1,542 | 18.4 | 0/7 | 0.50 | 1.00 |
| 23S rRNA | 2,904 | 32.0 | 0/7 | 0.51 | 1.00 |
| mRNA | 952 | 11.1 | 51/4,243 | 0.21d | NA |
For RNAs with more than one gene, we indicate the average length.
Total number of ACA triplets found in all genes of each category divided by number of genes in each category.
The random probability of ACA was calculated as described in Materials and Methods. NA, not applicable.
The random probability of ACA was calculated for the whole RNA and averaged by the total number of mRNAs.
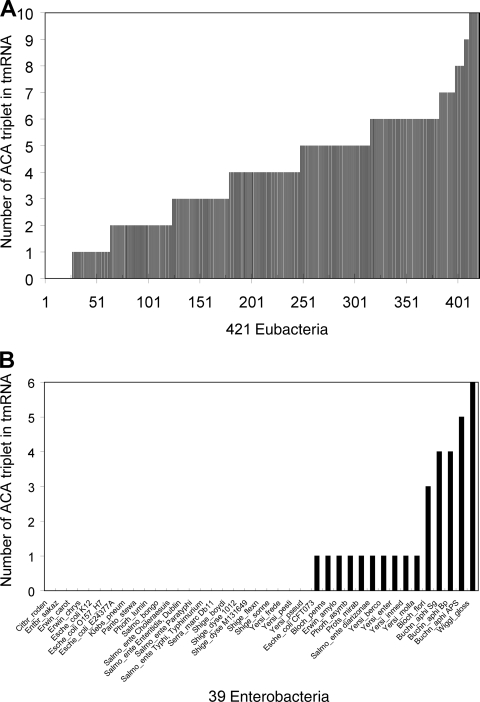
tmRNA was also analyzed, as it functions to rescue ribosomes stalled on damaged mRNAs (48), such as those produced during MazF induction. tmRNA of E. coli K-12 is 363 nucleotides long but does not contain any ACAs. The complete absence of ACAs in this RNA led to the examination of tmRNAs from other bacterial species. The sequences of 421 tmRNAs were retrieved from an online database (http://www.indiana.edu/∼tmrna/). At the time of analysis, this database contained a total of 609 eubacterial tmRNA sequences from which incomplete sequences or those from unculturable bacteria were excluded. As shown in Fig. 2A, most of the tmRNAs contain ACA triplets. However, 26 out of 421 tmRNAs did not contain any ACA triplets. Almost all of these (24 of 26 tmRNAs) were from the Enterobacteriaceae, such as Escherichia, Salmonella, Shigella, Photorhabdus, Enterobacter, Serratia, Pantoea, Klebsiella, Citrobacter, Yersinia, and Erwinia species. The other two ACA-less tmRNAs were encoded by Carboxydothermus hydrogenoformans (extremely thermophilic anaerobic gram-positive organism) and Moorella thermoacetica (thermophilic gram-positive organism).
FIG. 2.
Distribution of ACA frequencies in tmRNAs of 421 species of bacteria (A) and in tmRNAs of 39 species of enterobacteria (B) (http://www.indiana.edu/∼tmrna/). Citbr_roden, Citrobacter rodentium; Entbr_sakaz, Enterobacter sakazakii; Erwin_carot, Erwinia carotovora; Erwin_chrys, Erwinia chrysanthemi; Klebs_pneum, Klebsiellia pneumonaie; Panto_stewa, Pantoea stewartii; Phorh_lumin, Photorhabdus luminescens; Salmo_bongo, Salmonella bongori; Salmo_ente, Salmonella enterica; Serra_marc, Serratia marcescens; Shige_boydi, Shigella boydii; Shige_dyse, Shigella dysenteriae; Shige_flexn, Shigella flexneri; Shige_sonne, Shigella sonnei; Yersi_frede, Yersinia frederiksenii; Yersi_pesti, Yersinia pestis; Yersi_pseud, Yersinia pseudotuberculosis; Bloch_penns, Candidatus Blochmannia pennsylvanicus; Erwin_amylo, Erwinia amylovora; Phorh_asymb, Photorhabdus asymbiotica; Prots_mirab, Proteus mirabilis; Yersi_berco, Yersinia bercovieri; Yersi_enter, Yersinia enterocolitica; Yersi_inmed, Yersinia intermedia; Yersi_molla, Yersenia mollaretii; Bloch_flori, “Candidatus Blochmannia floridanus”; Buchn_aphi, Buchnera aphidicola; Wiggl_gloss, Wigglesworthia glossinidia.
(iv) Almost all mRNAs contain ACAs.
As the overexpression of MazF effectively cleaves cellular mRNAs at ACA sequences (53), we examined the usage of ACA triplets in all of the mRNAs in E. coli K-12. While roughly 1% (51 out of 4,243 mRNAs) do not contain any ACA triplets in their coding sequences (Table 1), ACA triplets are found quite frequently (17th rare triplet among 64) in the other 99%. Their abundance correlates well with the halt in protein synthesis caused by MazF. The probabilities for the random occurrence of ACA in all mRNAs were averaged (see Materials and Methods) to determine if there was a general bias against ACAs in E. coli K-12. The mRNAs do not have a general bias against ACA.
The absence of ACA in single-strand regions of tmRNA is statistically significant.
To determine the statistical significance of the number of ACAs in the RNAs involved in translation, we calculated the probability of ACAs occurring by chance given the AC content and length of each RNA (Table 1). Single-strand ACA probabilities were calculated separately from double-strand ACA probabilities, as the absence of ACAs in single-strand regions would be more biologically significant than their absence in double-strand regions. In addition, the AC content for double-strand regions did generally tend to be lower than that in single-strand regions due to GC and GU base pairing. Due to this bias, the absence of ACA in single-strand regions of tmRNA was statistically significant, whereas the probabilities for single-strand ACAs in the other RNAs were not too far from the expected number of ACAs, given the AC content and length of these RNAs. On the other hand, the absence of ACA in double-strand regions of tmRNA was not that far from the expected number of ACAs based on AC content and length.
Bias against ACAs in tmRNAs of enterobacteria is not random.
We next assessed the likelihood that the absence of ACAs in tmRNAs of enterobacteria occurred by chance. To do this, the same statistical analysis for single-strand ACA triplets and double-strand ACA triplets was performed on all complete tmRNA sequences available based on their AC content and sequence length. The secondary structures that were used were derived by comparative sequence analysis (47). Histograms of values for the two different regions are shown in Fig. 3. When the probabilities for enterobacteria were plotted separately from other bacteria, their single-strand distribution was significantly different from their double-strand distribution with respect to the population as a whole. Enterobacteria also make up a disproportionately large number of bacteria with a low probability of random ACAs in their single-strand regions. These results strongly suggest a selective pressure against single-strand ACAs in tmRNA sequences from enterobacteria.
FIG. 3.
Distribution of probabilities for the random occurrence of single-strand and double-strand ACAs in tmRNAs from 442 bacteria. The probability was calculated as described in Materials and Methods. Enterobacteria were plotted separately from other bacteria.
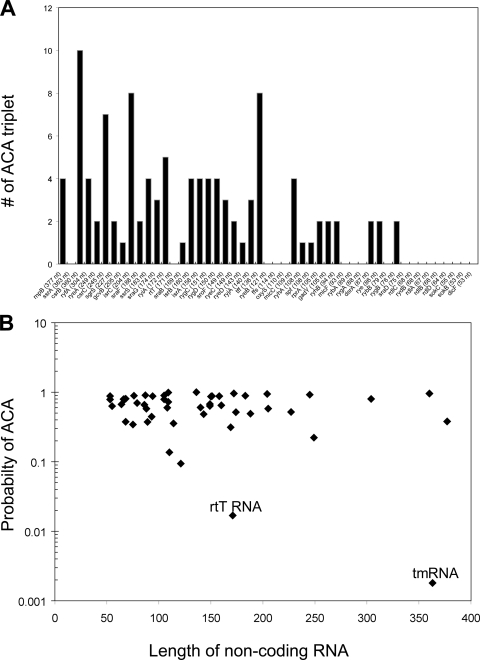
ACA bias in other noncoding RNAs.
Bacteria transcribe numerous small noncoding RNAs (15) besides tmRNA and 4.5S RNA. The function of most of these RNAs is still unknown. In this study we examined 49 noncoding RNAs of E. coli K-12 to determine if there is any unique feature in the ACA content of these RNAs. As shown in Fig. 4A, many noncoding small RNAs contain ACA triplets. For instance, we found it significant that 6S RNA, involved in transcriptional regulation through a direct interaction with RNA polymerase (42), contains three ACA triplets in its 183 nucleotides. Two of these ACA triplets are located at single-strand regions (3, 43), exposing this RNA to potential MazF cleavage. Other RNAs, such as rtT, ryeB, and oxyS, were found to lack ACA sequences. However, none of these RNAs had a statistical bias against ACAs except rtT (P = 0.0169) and ssrA (P = 0.0018) (Fig. 4B). Interestingly, rtT RNA is associated with the stringent response (3), while mazEF gene expression is known to be induced during the stringent response (6).
FIG. 4.
(A) Frequency of ACA sequences in 49 noncoding, small RNAs in E. coli K-12 (not including rRNAs and tRNAs). Numbers in parentheses are the lengths of each RNA molecule in nucleotides (nt). (B) Probability of ACA sequences in noncoding RNAs plotted by sequence length. The probability was calculated as described in Materials and Methods.
Correlation of MazF homologues and the absence of ACAs in tmRNAs.
While most enterobacteria lack ACAs in their tmRNAs, a few exceptions have been observed for obligate-symbiotic species such as Buchnera aphidicola, Wigglesworthia glossinidia, and “Candidatus Blochmannia floridanus” (14), which contain one to six ACA triplets (Fig. 2B). However, these symbiotic bacteria do not contain the mazF gene or any of its homologues in their genomes (35).
Notably, when tmRNAs contain ACA sequences in mazF-containing bacteria, the ACA triplets are located only in double-strand regions (some Yersinia species, Erwinia amylovora, and Salmonella enterica subsp. diarizonae). Interestingly, while both Proteus vulgaris and E. coli CFT073 have one ACA sequence in their tmRNAs at single-strand regions, neither one has the mazF gene. Only one species, Photorhabdus asymbiotica, has both a MazF homologue (http://www.sanger.ac.uk/Projects/P_asymbiotica/) and an unprotected ACA in its tmRNA. Although this bacterium is classified as being a member of the Enterobacteriaceae, its biochemical characteristics are quite different from those of most other enterobacteria (46). Its distinct physiology might require a different response to stress than MazF-induced growth regulation.
There remain a number of enterobacteria that have neither ACA triplets in their tmRNAs nor MazF. These enterobacteria, which may have originally possessed MazF and subsequently lost ACA sequences in their tmRNAs, may have recently lost MazF due to environmental changes. As the mazE-mazF locus is an area of unusual variability (35), a high rate of recombination may have facilitated such adaptiveness. Overall, the enterobacteria examined have an unusually low number of ACA sequences in their tmRNAs.
MazF effectively cleaves tmRNA containing single-stranded ACAs.
In order to assess the biochemical consequence of ACA triplets in tmRNA during MazF induction, three ACA triplets were introduced into tmRNA cloned into a plasmid. Two ACAs (A82C and C188A) were introduced into single-strand regions, and one ACA (G278A) was introduced into a double-strand region (Fig. 5A). As there are different structural intermediates during the folding (49) and function (19, 50) of tmRNA, we wanted to confirm that ACAs in the single-strand region of the structure proposed previously by Zwieb et al. (57) are more accessible to MazF cleavage than are double-strand ACAs.
To ensure that these mutations did not interfere with the function of tmRNA, each of the single-mutant tmRNAs and the triple mutant were tested for their ability to tag a truncated λ repressor whose mRNA did not contain a stop codon (25). All of these mutants were able to tag the peptide, as shown by the increase in size of the radiolabeled repressor compared to the size of the untagged repressor (Fig. 5B). The translation of the proteolysis tag of tmRNA would also provide the ribosome with a stop codon for dissociation. With the exception of the C188A single mutant, the tagging was comparable to wild-type levels.
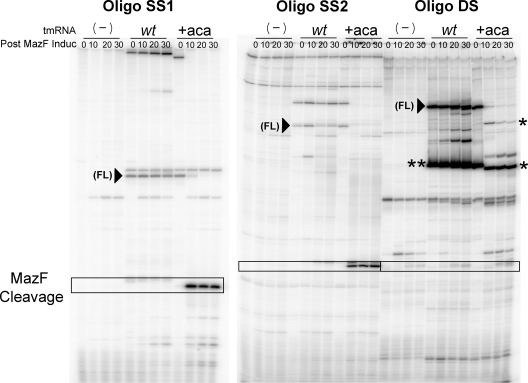
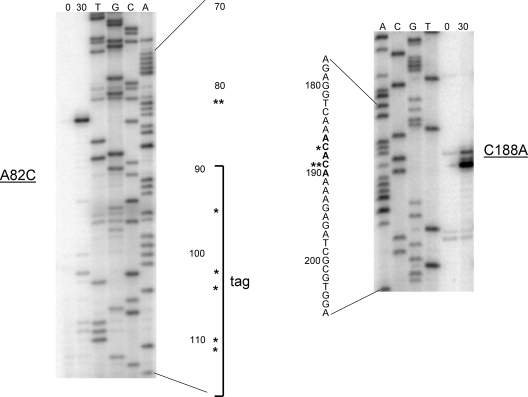
Primer extension analysis was then performed to determine whether the ACA sequences were accessible to MazF. tmRNA deletion strains were cotransformed with pACYCDuet-MazF and a low-copy plasmid containing no, wild-type, or triple-mutant tmRNA. After MazF was induced, total RNA was extracted at different time points. Primer extension analysis with each RNA sample was performed in triplicate using radiolabeled oligonucleotides detecting each of the three potential cutting sites. Only the ACAs in the single-strand region were cleaved, while the double-strand region was not cut at all (Fig. 6). As the oligonucleotides to detect cleavage at C188A (single-strand ACA) and at G278A (double-strand ACA) are both 73 bp away from their respective ACA sites, if there was cleavage in both samples, the cleavage products should migrate the same distance on the gel. However, at the position where the C188A cleavage product is migrating, there is no corresponding G278A band. Sequencing reactions for the triple-mutant tmRNA were run alongside the primer extension reactions to determine the exact single-strand cutting sites (Fig. 7). In both cases, the major bands appearing after the induction of MazF are at an ACA sequence.
FIG. 6.
Induction of MazF cleaves tmRNA at single-strand ACA sites. Primer extensions were run on total RNA from strains containing pBR322 alone (−), wild-type tmRNA (wt), or triple mutant tmRNA (+aca) over a time course of 30 min. (Left) Cleavage at ACA81 (boxed) in the triple mutant but not in the wild type or vector alone. (Right) Cleavage at ACA188 in the triple mutant only but no cleavage at ACA278 with any of the vectors. Full-length (FL) primer extension products are marked with triangles. Single asterisks indicate primer extension products ending at single-strand ACA sites. The double asterisk indicates incomplete primer extension, ending at regions of stable secondary structure.
FIG. 7.
MazF cleavage occurs at ACA sites. Sequencing reactions for tmRNA were run next to primer extension reactions using total RNA harvested 0 and 30 min after MazF induction. (Left) Cleavage at A82C. (Right) Cleavage at C188A. Part of the proteolysis tag-encoding region of tmRNA is shown in brackets. Single asterisks indicate minor cleavage bands, while double asterisks indicate major cleavage bands.
DISCUSSION
The widespread presence of such self-destructive toxin proteins is a paradox in a unicellular organism. Even if these genes had been fixed by a previous selective pressure (38), there is surely a fitness cost to maintaining so many harmful genes. One may also wonder, if these genes truly are relics of past evolutionary events, why are their open reading frames still intact? The same question might be asked of “nonessential genes,” which are so classified when there is no phenotype associated with their deletion under laboratory conditions. A recent study by Hillenmeyer et al. (17) directly addressed the usefulness or accuracy of the notion of “nonessential genes” by demonstrating that every gene in Saccharomyces cerevisiae (genome size of 12.5 Mbp) contributed to a phenotype under different environmental stress conditions.
While science has yet to arrive at the true function of these toxins, we propose that sequence analysis may be a reasonable tool to determine genetic interactions for a highly specific and potent endoribonuclease. Our observation that translation itself is uninhibited during this halt in growth (40, 41) led to the sequence analysis of rRNAs and then, later, all RNAs to look for any unusual ACA distribution. While multiple mechanisms may preserve these RNAs involved in translation, a statistical survey of tmRNA sequences from known bacteria revealed that the absence of ACAs in enterobacteria is unlikely to have occurred by chance. These two observations have led us to wonder whether it is the preservation of translation or the preservation of tmRNA function that is advantageous to the cell during recovery from MazF induction. There have already been preliminary studies of the importance of tmRNA after toxin induction (4, 6), but the specific function of tmRNA that ameliorates toxicity is unknown and has not yet been explored.
The function of tmRNA has been proposed to rescue stalled ribosomes due to the ability of tmRNA to transfer protein synthesis to its own coding region (25). However, rather than rescuing protein synthesis, there are data from many reports suggesting that tmRNA may alter gene expression by targeting specific substrates for tagging and proteolysis. For instance, in the alphaproteobacterium Caulobacter crescentus, many proteins were found to be normally tagged by tmRNA, some of which included genes involved in regulating cell division (18). As the deletion of tmRNA causes cell cycle defects (24), the function of this tagging may be to alter the half-life of these proteins. tmRNA may also alter gene expression by targeting mRNAs lacking a stop codon for faster degradation. This was shown by introducing silent mutations into the tag-encoding portion of tmRNA and observing an increase in the half-life of the target mRNA (29). Whether tmRNA influences the stability of specific intact mRNA for purposes of gene regulation remains to be determined.
In E. coli, very few natural substrates have been confirmed, and a deletion of tmRNA produces only a mild growth defect under optimal laboratory conditions (33). However, in two other enterobacteria, the deletion of tmRNA greatly reduces the pathogenicity of the bacteria (22, 34), sometimes at the level of transcription of virulence factors. Similarly, tmRNA may provide E. coli a significant growth advantage only under specific environmental conditions, such as those stress conditions which induce MazF (6, 16, 27). Furthermore, the role of tmRNA under these stress conditions may be something other than recycling ribosomes.
As E. coli K-12 contains another sequence-specific endoribonuclease, ChpBK, we checked the small RNAs for bias against its cleavage site, which are single-strand RNAs containing ACY sequences (Y is U, A, or G) (54). We found that 5S rRNA, tRNAs, 4.5S RNA, and tmRNA all contain potential ChpBK cleavage sites (ACU and ACG). However, as it was previously shown that MazF cleavage activity is much more efficient than that of ChpBK (54), it is possible that sequence bias may not be necessary to protect these RNAs from ChpBK activity.
During our analysis of overall triplet use in RNAs, we did notice that, like ACA, UAU was often missing in tmRNAs. In E. coli K-12, no UAU sequence is found in tmRNA, 4.5S rRNA, or 5S rRNA, while UAU is the fourth-least-frequent triplet in its 87 tRNAs. If the absence of this UAU were found to be statistically significant, it might be interesting to determine if introducing a UAU triplet into a reporter sequence would promote the cleavage of the RNA by an unidentified endoribonuclease.
UAU was also found to be missing frequently in the tmRNAs of members of the Mycobacteriaceae. The family of mycobacteria, which includes the human pathogen Mycobacterium tuberculosis, is made up of obligate aerobes. While a number of MazF homologues in M. tuberculosis were recently characterized (56), it remains to be seen whether the absence of UAU triplets protects the tmRNA from MazF cleavage in this bacterium.
We did not find any trends in triplet sequences in tmRNAs from other bacteria. However, there are toxins recognizing 4 or 5 nucleotides, such as the plasmid-encoded toxin Kid/PemK (23) and the recently characterized MazF-mt3 and MazF-mt7 (55), which may also have exerted a sequence pressure on RNAs. Proving a nonrandom absence of a 5-nucleotide sequence in a small RNA, though, is difficult, even if there were a real selective pressure, due to the greater number of unique 5-nucleotide sequences possible. There is also the possibility that an increase in number, rather than the absence, of cleavage sites in some RNAs may also have conferred a survival advantage. Examining such sequence bias and proving their biochemical significance may help shed light on new functions of TA modules.
While there is still the possibility that other sequence constraints limited the appearance of ACAs in tmRNA, the specificity of MazF and the absence of its cutting site, not just in E. coli but in most enterobacteria, are difficult to ignore. Our future work will explore the functional relationship between MazF and tmRNA and determine what kind of fitness advantage tmRNA may provide a given population.
Supplementary Material
Acknowledgments
Part of this work was funded by a research grant from Takara Bio., Inc.
We are grateful to Oleg Mirochnitchenko for technical support and to Vikas Nanda, Marilyn Kozak, Michael Travisano, Liliana Falzon, Yoshihiro Yamaguchi, and Raphael Margueron for valuable comments and suggestions on the manuscript.
Footnotes
Published ahead of print on 24 July 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Andersen, E. S., M. A. Rosenblad, N. Larsen, J. C. Westergaard, J. Burks, I. K. Wower, J. Wower, J. Gorodkin, T. Samuelsson, and C. Zwieb. 2006. The tmRDB and SRPDB resources. Nucleic Acids Res. 34:D163-D168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bjork, G. R. 1996. Stable RNA modification, vol. 2. ASM Press, Washington, DC.
- 3.Bosl, M., and H. Kersten. 1991. A novel RNA product of the tyrT operon of Escherichia coli. Nucleic Acids Res. 19:5863-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen, S. K., and K. Gerdes. 2003. RelE toxins from bacteria and archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol. Microbiol. 48:1389-1400. [DOI] [PubMed] [Google Scholar]
- 5.Christensen, S. K., M. Mikkelsen, K. Pedersen, and K. Gerdes. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. USA 98:14328-14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen, S. K., K. Pedersen, F. G. Hansen, and K. Gerdes. 2003. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332:809-819. [DOI] [PubMed] [Google Scholar]
- 7.Cooper, T. F., and J. A. Heinemann. 2000. Postsegregational killing does not increase plasmid stability but acts to mediate the exclusion of competing plasmids. Proc. Natl. Acad. Sci. USA 97:12643-12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper, T. F., and J. A. Heinemann. 2005. Selection for plasmid postsegregational killing depends on multiple infection: evidence for the selection of more virulent parasites through parasite-level competition. Proc. Biol. Sci. 272:403-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiil, N., and J. D. Friesen. 1968. Isolation of “relaxed” mutants of Escherichia coli. J. Bacteriol. 95:729-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerdes, K. 2000. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J. Bacteriol. 182:561-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerdes, K., S. K. Christensen, and A. Lobner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371-382. [DOI] [PubMed] [Google Scholar]
- 13.Gerdes, K., P. B. Rasmussen, and S. Molin. 1986. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA 83:3116-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gil, R., F. J. Silva, E. Zientz, F. Delmotte, F. Gonzalez-Candelas, A. Latorre, C. Rausell, J. Kamerbeek, J. Gadau, B. Holldobler, R. C. van Ham, R. Gross, and A. Moya. 2003. The genome sequence of Blochmannia floridanus: comparative analysis of reduced genomes. Proc. Natl. Acad. Sci. USA 100:9388-9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottesman, S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58:303-328. [DOI] [PubMed] [Google Scholar]
- 16.Hazan, R., and H. Engelberg-Kulka. 2004. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol. Genet. Genomics 272:227-234. [DOI] [PubMed] [Google Scholar]
- 17.Hillenmeyer, M. E., E. Fung, J. Wildenhain, S. E. Pierce, S. Hoon, W. Lee, M. Proctor, R. P. St. Onge, M. Tyers, D. Koller, R. B. Altman, R. W. Davis, C. Nislow, and G. Giaever. 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320:362-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong, S. J., F. H. Lessner, E. M. Mahen, and K. C. Keiler. 2007. Proteomic identification of tmRNA substrates. Proc. Natl. Acad. Sci. USA 104:17128-17133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanova, N., M. Lindell, M. Pavlov, L. H. Schiavone, E. G. Wagner, and M. Ehrenberg. 2007. Structure probing of tmRNA in distinct stages of trans-translation. RNA 13:713-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaffé, A., T. Ogura, and S. Hiraga. 1985. Effects of the ccd function of the F plasmid on bacterial growth. J. Bacteriol. 163:841-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, Y., J. Pogliano, D. R. Helinski, and I. Konieczny. 2002. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 44:971-979. [DOI] [PubMed] [Google Scholar]
- 22.Julio, S. M., D. M. Heithoff, and M. J. Mahan. 2000. ssrA (tmRNA) plays a role in Salmonella enterica serovar Typhimurium pathogenesis. J. Bacteriol. 182:1558-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamphuis, M. B., A. M. Bonvin, M. C. Monti, M. Lemonnier, A. Munoz-Gomez, R. H. van den Heuvel, R. Diaz-Orejas, and R. Boelens. 2006. Model for RNA binding and the catalytic site of the RNase Kid of the bacterial parD toxin-antitoxin system. J. Mol. Biol. 357:115-126. [DOI] [PubMed] [Google Scholar]
- 24.Keiler, K. C., and L. Shapiro. 2003. tmRNA is required for correct timing of DNA replication in Caulobacter crescentus. J. Bacteriol. 185:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keiler, K. C., P. R. Waller, and R. T. Sauer. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990-993. [DOI] [PubMed] [Google Scholar]
- 26.Koch, H. G., M. Moser, and M. Muller. 2003. Signal recognition particle-dependent protein targeting, universal to all kingdoms of life. Rev. Physiol. Biochem. Pharmacol. 146:55-94. [DOI] [PubMed] [Google Scholar]
- 27.Kolodkin-Gal, I., and H. Engelberg-Kulka. 2006. Induction of Escherichia coli chromosomal mazEF by stressful conditions causes an irreversible loss of viability. J. Bacteriol. 188:3420-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolodkin-Gal, I., R. Hazan, A. Gaathon, S. Carmeli, and H. Engelberg-Kulka. 2007. A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science 318:652-655. [DOI] [PubMed] [Google Scholar]
- 29.Mehta, P., J. Richards, and A. W. Karzai. 2006. tmRNA determinants required for facilitating nonstop mRNA decay. RNA 12:2187-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitra, S. K., F. Lustig, B. Åkesson, and U. Lagerkvist. 1977. Codon-anticodon recognition in the valine codon family. J. Biol. Chem. 252:471-478. [PubMed] [Google Scholar]
- 31.Nagai, K., C. Oubridge, A. Kuglstatter, E. Menichelli, C. Isel, and L. Jovine. 2003. Structure, function and evolution of the signal recognition particle. EMBO J. 22:3479-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nariya, H., and M. Inouye. 2008. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell 132:55-66. [DOI] [PubMed] [Google Scholar]
- 33.Oh, B. K., and D. Apirion. 1991. 10Sa RNA, a small stable RNA of Escherichia coli, is functional. Mol. Gen. Genet. 229:52-56. [DOI] [PubMed] [Google Scholar]
- 34.Okan, N. A., J. B. Bliska, and A. W. Karzai. 2006. A role for the SmpB-SsrA system in Yersinia pseudotuberculosis pathogenesis. PLoS Pathog. 2:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pandey, D. P., and K. Gerdes. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33:966-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pedersen, K., A. V. Zavialov, M. Y. Pavlov, J. Elf, K. Gerdes, and M. Ehrenberg. 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112:131-140. [DOI] [PubMed] [Google Scholar]
- 37.Ruiz-Echevarria, M. J., G. Gimenez-Gallego, R. Sabariegos-Jareno, and R. Diaz-Orejas. 1995. Kid, a small protein of the parD stability system of plasmid R1, is an inhibitor of DNA replication acting at the initiation of DNA synthesis. J. Mol. Biol. 247:568-577. [DOI] [PubMed] [Google Scholar]
- 38.Saavedra De Bast, M., N. Mine, and L. Van Melderen. 2008. Chromosomal toxin-antitoxin systems may act as antiaddiction modules. J. Bacteriol. 190:4603-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarmientos, P., J. E. Sylvester, S. Contente, and M. Cashel. 1983. Differential stringent control of the tandem E. coli ribosomal RNA promoters from the rrnA operon expressed in vivo in multicopy plasmids. Cell 32:1337-1346. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki, M., R. Roy, H. Zheng, N. Woychik, and M. Inouye. 2006. Bacterial bioreactors for high yield production of recombinant protein. J. Biol. Chem. 281:37559-37565. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki, M., J. Zhang, M. Liu, N. A. Woychik, and M. Inouye. 2005. Single protein production in living cells facilitated by an mRNA interferase. Mol. Cell 18:253-261. [DOI] [PubMed] [Google Scholar]
- 42.Trotochaud, A. E., and K. M. Wassarman. 2006. 6S RNA regulation of pspF transcription leads to altered cell survival at high pH. J. Bacteriol. 188:3936-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trotochaud, A. E., and K. M. Wassarman. 2005. A highly conserved 6S RNA structure is required for regulation of transcription. Nat. Struct. Mol. Biol. 12:313-319. [DOI] [PubMed] [Google Scholar]
- 44.Van Melderen, L. 2002. Molecular interactions of the CcdB poison with its bacterial target, the DNA gyrase. Int. J. Med. Microbiol. 291:537-544. [DOI] [PubMed] [Google Scholar]
- 45.Watson, R. J., J. Parker, N. P. Fiil, J. G. Flaks, and J. D. Friesen. 1975. New chromosomal location for structural genes of ribosomal proteins. Proc. Natl. Acad. Sci. USA 72:2765-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weissfeld, A. S., R. J. Halliday, D. E. Simmons, E. A. Trevino, P. H. Vance, C. M. O'Hara, E. G. Sowers, R. Kern, R. D. Koy, K. Hodde, M. Bing, C. Lo, J. Gerrard, R. Vohra, and J. Harper. 2005. Photorhabdus asymbiotica, a pathogen emerging on two continents that proves that there is no substitute for a well-trained clinical microbiologist. J. Clin. Microbiol. 43:4152-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams, K. P., and D. P. Bartel. 1996. Phylogenetic analysis of tmRNA secondary structure. RNA 2:1306-1310. [PMC free article] [PubMed] [Google Scholar]
- 48.Withey, J. H., and D. I. Friedman. 2002. The biological roles of trans-translation. Curr. Opin. Microbiol. 5:154-159. [DOI] [PubMed] [Google Scholar]
- 49.Wong, T. N., T. R. Sosnick, and T. Pan. 2007. Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. Proc. Natl. Acad. Sci. USA 104:17995-18000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wower, I. K., C. Zwieb, and J. Wower. 2005. Transfer-messenger RNA unfolds as it transits the ribosome. RNA 11:668-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamaguchi, Y., and M. Inouye. 2009. mRNA interferases, sequence-specific endoribonucleases from the toxin-antitoxin systems. Prog. Mol. Biol. Transl. Sci. 85:467-500. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, Y., J. Zhang, H. Hara, I. Kato, and M. Inouye. 2005. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 280:3143-3150. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, Y., J. Zhang, K. P. Hoeflich, M. Ikura, G. Qing, and M. Inouye. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12:913-923. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, Y., L. Zhu, J. Zhang, and M. Inouye. 2005. Characterization of ChpBK, an mRNA interferase from Escherichia coli. J. Biol. Chem. 280:26080-26088. [DOI] [PubMed] [Google Scholar]
- 55.Zhu, L., S. Phadtare, H. Nariya, M. Ouyang, R. N. Husson, and M. Inouye. 2008. The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol. Microbiol. 69:559-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu, L., Y. Zhang, J. S. Teh, J. Zhang, N. Connell, H. Rubin, and M. Inouye. 2006. Characterization of mRNA interferases from Mycobacterium tuberculosis. J. Biol. Chem. 281:18638-18643. [DOI] [PubMed] [Google Scholar]
- 57.Zwieb, C., I. Wower, and J. Wower. 1999. Comparative sequence analysis of tmRNA. Nucleic Acids Res. 27:2063-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.