Abstract
We report here the identification and characterization of mrdH, a novel chromosomal metal resistance determinant, located in the genomic island 55 of Pseudomonas putida KT2440. It encodes for MrdH, a predicted protein of ∼40 kDa with a chimeric domain organization derived from the RcnA and RND (for resistance-nodulation-cell division) metal efflux proteins. The metal resistance function of mrdH was identified by the ability to confer nickel resistance upon its complementation into rcnA mutant (a nickel- and cobalt-sensitive mutant) of Escherichia coli. However, the disruption of mrdH in P. putida resulted in an increased sensitivity to cadmium and zinc apart from nickel. Expression studies using quantitative reverse transcription-PCR showed the induction of mrdH by cadmium, nickel, zinc, and cobalt. In association with mrdH, we also identified a conserved hypothetical gene mreA whose encoded protein showed significant homology to NreA and NreA-like proteins. Expression of the mreA gene in rcnA mutant of E. coli enhanced its cadmium and nickel resistance. Transcriptional studies showed that both mrdH and mreA underwent parallel changes in gene expression. The mobile genetic elements Tn4652 and IS1246, flanking mrdH and mreA were found to be induced by cadmium, nickel, and zinc, but not by cobalt. This study is the first report of a single-component metal efflux transporter, mrdH, showing chimeric domain organization, a broad substrate spectrum, and a location amid metal-inducible mobile genetic elements.
Bacterial efflux systems for inorganic metal cations and anions play an imperative role in the regulatory network governing metal homeostasis. These efflux systems are also important for the environmental adaptability of the bacteria thriving in metal-rich serpentine environments. Bioinformatic and functional genomic analyses have revealed that the efflux systems belonging to the resistance-nodulation-cell division (RND), cation diffusion facilitator (CDF), and P-type ATPases constitute the majority of the multiple layers of heavy metal resistance in an organism (36). Members of the RND protein family include group of bacterial transport proteins involved in heavy metal resistance, nodulation, and cell division. The best-characterized RND family members include the efflux systems CzcCBA (Cd2+, Zn2+, and Co2+ resistance), CnrCBA (Co2+ and Ni2+ resistance), and NccCBA (Ni2+, Co2+, and Cd2+ resistance) from Cupriavidus metallidurans (Alcaligenes metallidurans or Ralstonia metallidurans) and the CznCBA efflux system (Co2+, Zn2+, and Ni2+ resistance) from Helicobacter pylori (11, 14, 23, 29, 49, 52). CDF family members serve as secondary cation filters in bacteria. The characterized CDF family members CzcD, ZntA, and ZitB were found to have a substrate spectrum consisting of Co2+, Zn2+, Cd2+, and Ni2+(1, 26, 57). P-type ATPase exporters that detoxify heavy metal cations by efflux include Cu-CPx-type ATPases (exporting Cu+ and Ag+) and Zn-CPx-type ATPases (export Zn2+, Cd2+, and Pb2+) (4, 37, 43-45). Other heavy metal export systems include the CHR protein family, NreB, and CnrT-like proteins (36). Recently, a new class of efflux proteins RcnA that confer Ni2+ and Co2+ resistance was identified in Escherichia coli (46).
Simultaneously, analyses of annotated bacterial genomes indicated the increased prevalence of mobile genetic elements (e.g., insertion sequences, integrons, and transposons) in disseminating metal resistance and drug resistance traits. Recent studies provided support for the prominent role of horizontal gene transfer (HGT) in the evolution of metal homeostasis in proteobacteria and also identified putative genomic islands (GIs) containing metal resistance genes in environmental bacteria like C. metallidurans and Pseudomonas putida KT2440 (8, 9, 34, 56). GIs are defined as horizontally acquired genetic elements that share the same structural features of pathogenicity islands (PAIs) and contribute to a microorganism's fitness, metabolic versatility, and adaptability. Most of the novel genes in a prokaryotic genome are more likely to be present in GIs and related horizontally acquired regions (19). Features frequently found to be associated with GIs include the presence of flanking repeats, mobility genes (e.g., integrases and transposases), proximal transfer RNAs (tRNAs), atypical guanine, and cytosine content. More recently, a putative GI was defined as a region containing eight (or more) consecutive open reading frames (ORFs) with dinucleotide bias, or eight consecutive ORFs with dinucleotide bias plus at least one mobility gene in the vicinity (19). The global features of P. putida KT2440 revealed 105 GIs of ≥4,000 bp with atypical GC contents and oligonucleotide signature. Most of these islands enhance the metabolic proficiency of P. putida as a saprophytic omnivore by endowing the organism with determinants of resistance and defense or with constituents and appendages of the cell wall. Major features are the uptake and degradation of organic chemicals, ion transport, and the synthesis and secretion of secondary metabolites (56). Thirteen metal transporters were found to be distributed in GIs of P. putida KT2440 (16, 56).
The genus Pseudomonas encompasses the most diverse group of bacteria thriving on the planet with widespread interactomes and has acquired functions that enable them to exhibit remarkable metabolic versatility and adaptability. The best-studied species include P. aeruginosa PA01, which acts as an opportunistic human pathogen; the soil bacterium P. putida KT2440, which is capable of degrading a wide range of environmental pollutants; the plant pathogen P. syringae DC3000; and the plant growth-promoting P. fluorescens Pf-5 (6, 35, 40, 53). The availability of complete genomic sequences of P. putida KT2440 and P. putida CD2 enabled researchers to study the capacity of these species to endure exposure to heavy metals (7, 20). Functionally well-characterized metal resistance determinants in this genus include the CzrCBA transporter from P. putida CD2 and P. aeruginosa, which confers resistance to Zn2+, Cd2+, and Co2+ (17, 20). Through expression studies this transport system was found to be induced by Zn2+ and Cu2+ (20). Cadmium P-type ATPases (CadA) that confer Cd2+ and Zn2+ resistance were characterized in P. putida 06909, P. fluorescens ATCC 13525, and P. putida CD2 (20, 25, 47). A copper-inducible cin operon encoding an unusual methionine-rich azurin-like protein and a Pre-Q0 reductase was identified in P. putida KT2440 (41). A recent report showed the interplay of the P-type ATPases (CadA1 and CadA2) and RND proteins (CzcCBA1 and CzcCBA2) in mediating the resistance of P. putida KT2440 to divalent heavy metals (27). Except for the studies mentioned above, the diversity of metal resistance determinants in this genus has not been investigated extensively.
In our previous in silico study of Pseudomonas metal transportomes, we carried out data set cataloging of 262 metal ion transporters from metabolically versatile P. putida KT2440, P. fluorescens Pf-5, P. aeruginosa PA01, and P. syringae DC3000 (16). Significantly, 13 metal ion transporters, which were not annotated in their respective genome databases, were identified, and all of the metal transporter genes with GI-associated features were dissected from the metal transportomes of four Pseudomonas species (16). Two of the novel metal resistance determinants identified from P. putida KT2440 (PP_2968) and P. syringae DC3000 (PSPTO_4280) displayed high levels of sequence identity (63%) with the prototypical Ni2+and Co2+ resistance determinant rcnA from E. coli (rcnAEco) (46).
We report here the identification and characterization of PP_2968, as a novel metal resistance determinant mrdH, located in the GI of P. putida KT2440. In the present study, we demonstrate that the mrdH-encoded resistance determinant is a novel metal efflux transporter. In addition, the heavy metal response of the associated cytoplasmic gene mreA and of the flanking mobile genetic elements was also demonstrated.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains, plasmids, and oligonucleotides used in the present study are listed in Table 1 and 2. For the construction and maintenance of plasmids, the E. coli strain DH5α was used. Complementation experiments were performed with strain ARY023, a Ni2+and Co2+ sensitive, rcnA mutant of E. coli (46). Bacteria were routinely grown in Luria-Bertani (LB) or M9 minimal medium (MM; without yeast extract supplementation) (48). Cultures were grown at 37°C (E. coli strains) or 30°C (P. putida KT2440 and its derivatives) on a rotary shaker at 150 rpm. Antibiotics were added to the medium when appropriate at the following concentrations: ampicillin, 100 μg/ml (E. coli) and 500 μg/ml (P. putida); chloramphenicol, 50 μg/ml (P. putida); and kanamycin, 30 μg/ml (P. putida).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or characteristics | Source or reference |
|---|---|---|
| Strains | ||
| P. putida | ||
| KT2440 | Wild type | 35 |
| HMR020 | KT2440 mrdH::ltrB | This study |
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Lab collection |
| JM109 | endA1 recA1 gyrA96 thi-1 hsdR17(rK− mK+) Δel4(ΔmcrA) supE44 relA1 Δ(proAB lac) F′ [proAB+traD36 lacIqlacZΔM15] | 58 |
| ARY023 | F supE thi Δ(lac-proAB) hsd5/F′ proAB+lacIqlacZΔM15 yohM::uidA-Kanr | 46 |
| Plasmids | ||
| pTZ57R/T | Ampr, cloning vector | Fermentas |
| pUC18 | ColE1 replicon; Ampr; 2.7 kb | 58 |
| pUCmrdH | pUC18 containing mrdH gene of P. putida KT2440 | This study |
| pUCrcnAPsy | pUC18 containing the rcnAPsy gene of P. syringae DC3000 | This study |
| pUCmreA | pUC18 containing mreA gene of P. putida KT2440 | This study |
| pJB653 | Ampr; 7.0 kb, derivative of pJB652 in which the PstI fragment containing the Pm promoter was cloned in the opposite orientation | 3 |
| pJBmrdH | pJB653 containing the mrdH gene of P. putida KT2440 | This study |
aAmpr, ampicillin resistance; Kanr, kanamycin resistance.
TABLE 2.
Sequences of the primers used in PCR amplifications
| Primer pair | Sequence (5′-3′)a |
|---|---|
| A1 | CCCGGG ATGTCGAGCTTCGCTGAACTG |
| AAGCTT AGGCCGATCCAGCCATGGA | |
| A2 | CCCGGG ATGCCCGACTTTGCCAGTCTG |
| AAGCTT ACAGACCATTCCAGCCGTG | |
| A3 | CCCGGG ATGAGCGATCACGAACACG |
| AAGCTT TAGAGGTACTTAGTGATTTGCTTGAA | |
| A4 | CCCGGG ATGTCGAGCTTCGCTGAACTG |
| GGATCC AGGCCGATCCAGCCATGGA | |
| A5 | GTAAGCGTTCGGTCGAGAAG |
| CCACCGATGTACGAGGAAAC | |
| A6 | ATGCTGAAGCACAACACCAG |
| TTTAGCAATCGCAACGTCAC | |
| A7 | TGGTCTTGGATAAGCACACG |
| GCCATCAGCACCAAAGTCAT | |
| A8 | GTGTTTGCCTTCACCCATCT |
| CCAGTGGGTTTCGATCAAGT | |
| A9 | GATTCTGTTGCAGCGACTCA |
| GACCAATTCGATGCCTGTCT | |
| A10 | TAACTGGAAGCCAGCCAAAC |
| GAAGTGCTTGCTGTCATGGA |
Restriction sites are indicated by underlining.
DNA manipulations.
The genomic DNA of P. putida KT2440 was isolated with Himedia columns according to the instructions of the manufacturer. Qiagen Mini-Prep and gel purification kits were used for plasmid isolation and for the purification of digested DNA. The enzymes used for DNA manipulation were obtained from Fermentas. PCRs were carried out using Fermentas PCR Master Mix. Plasmids were constructed by using standard recombinant DNA procedures, and E. coli cells were transformed by the routine CaCl2 procedure (48). Competent cells of P. putida KT2440 and the mutant strain HMR020 were prepared by the MgCl2-CaCl2 method, followed by chemical transformation (30).
Complementation of rcnA mutant of E. coli (ARY023 strain) by the hypothetical metal transporters mrdH and rcnAPsy.
The genomic DNA of P. putida KT2440 was used as a template to PCR amplify the 1.1-kbp mrdH gene (PP_2968) with the primer pair A1. The genomic DNA of P. syringae DC3000 was used as a template to amplify the P. syringae rcnA (rcnAPsy) gene (PSPTO_4280) of 879 bp with the primer pair A2. Restriction sites for SmaI and HindIII were incorporated into the primers. The PCR products were cloned into pTZ57R/T (Fermentas), and the constructs were verified by DNA sequencing. Subsequently, these constructs in T/A (pTZ57R/T-mrdH and pTZ57R/T-rcnAPsy) were digested with SmaI and HindIII and then subcloned into pUC18 (ampicillin resistant [Ampr]) at the same restriction sites under the control of the lacZ promoter. The ability of these pUC18 derivatives (pUCmrdH and pUCrcnAPsy) to confer Ni2+and Co2+ resistance was tested by transforming them into ARY023 strain. Cultures of the ARY023 transformants (ARY023, ARY023/pUC18, ARY023/pUCmrdH, and ARY023/pUCrcnAPsy) grown overnight in M9 MM supplemented with 0.4% glucose were diluted 1:100 into fresh M9 medium with the indicated metal ion concentrations (Zn2+, Cd2+, Ni2+, and Co2+). Bacterial growth was monitored as the optical density at 600 nm (OD600) after 16 h of incubation at 37°C. The results are presented as the mean values from three independent experiments.
Expression of the associated cytoplasmic gene mreA in the rcnA mutant of E. coli (ARY023 strain).
The cytoplasmic 288-bp gene, mreA, was PCR amplified from the genomic DNA of P. putida KT2440 with primer pair A3. Restriction sites for SmaI and HindIII were incorporated into the PCR primers. The amplified product was gel purified, cloned into the vector pTZ57R/T, and sequenced. The mreA gene was then subcloned into SmaI- and HindIII-digested pUC18, under the control of the lacZ promoter. The resulting plasmid pUCmreA was then transformed into ARY023 strain of E. coli. Cultures of the ARY023 transformants (ARY023/pUC18 and ARY023/pUCmreA) grown overnight in M9 MM supplemented with 0.4% glucose were diluted 1:100 into fresh M9 medium with various metal ions (Zn2+, Cd2+, Ni2+, and Co2+). Bacterial growth was monitored as the OD600 after 16 h of incubation at 37°C.
Construction of the mrdH gene knockout mutant of P. putida KT2440 (HMR020 strain) using group II introns.
The TargeTron gene knockout system (Sigma-Aldrich) based on the modified group II intron (L1.LtrB intron) was used specifically to disrupt mrdH gene in the chromosome of P. putida KT2440. Group II introns insert themselves via the activity of an RNA-protein complex expressed from the pACD4KC vector (21, 59). To target the intron to mrdH, the group II intron sequence in pACD4K-C vector was modified based on the sequences of predicted insertion sites in the mrdH gene by using Targetron Design (Sigma-Aldrich). The program predicted nine intron insertion sites across the 1,134-bp mrdH gene. The E values (expected number of false positives) of the nine target sites ranged from 0.050 to 0.467. For optimal gene interruption and stable insertion, the insertion site in the antisense strand at position 627 or 628 from the initial ATG with the lowest E value of 0.05 was chosen for intron modification, and primers were designed to retarget the RNA portion of the RNA-protein complex. Subsequently, the retargeted 350-bp PCR gene was ligated into a linearized pACD4K-C (chloramphenicol resistant [Cmr]) under the control of the T7 promoter resulting in pACD4K-mrdHC construct. Since P. putida KT2440 does not express T7 polymerase to drive the expression of the intron component, the pAR1219 vector (Ampr) that expresses T7 polymerase under the control of an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible LacUV5 promoter (10) was used for the functional expression of the retargeted intron. Finally, the retargeted intron containing pACD4K-mrdHC and pAR1219 vectors were cotransformed into competent P. putida KT2440 cells. The transformation mixture was added to LB medium containing ampicillin, chloramphenicol, and glucose (1%). After incubation for 18 h at 30°C, the culture was further diluted into a fresh medium (LB) containing antibiotics and glucose (1%), incubated until the OD600 reached 0.2 at 30°C. These early-log-phase cells were induced with IPTG (0.1 M) for 30 min at 30°C. The cells were centrifuged, resuspended in fresh medium with glucose (1%), and again incubated for 1 h at 30°C. Selection was performed on LB plates with kanamycin (25 μg/ml) that were maintained at room temperature for 3 days. The resulting colonies were then plated onto LB plates containing kanamycin and 1 mM nickel. Colonies unable to grow on final selection plates containing 1 mM Ni2+ were recovered and colony PCRs were performed across the intron-gene (mrdH) junction to verify the correct target site insertion.
Complementation of the HMR020 by expression of mrdH in the broad-range-host vector pJB653.
The mrdH gene was PCR amplified from the genomic DNA of P. putida KT2440 using the primer pair A4. Restriction sites for SmaI and BamHI were incorporated into the primers. The PCR product was cloned into pTZ57R/T, and the construct was verified by DNA sequencing. The mrdH gene was isolated by digesting the pTZ57R/T-mrdHC plasmid with SmaI and BamHI and then subcloned into the SmaI/BamHI-digested pJB653 vector (Ampr), under the control of the Pm promoter, which is induced by m-toluic acid. The resulting plasmid, pJBmrdH, was transformed into HMR020 competent cells by chemical transformation. Part of the transformation reaction was added to LB containing ampicillin and m-toluic acid (0.1 M). After incubation for 1 h at 30°C, the cells were plated onto LB agar containing ampicillin, m-toluic acid (0.1 M), and various concentrations of Ni2+ (0.5 to 3 mM). Nickel was used for the selection of HMR020/pJBmrdH transformants since HMR020 strain has intrinsic ampicillin resistance.
Estimation of metal content.
Overnight cultures of P. putida KT2440 (wild type), HMR020, and HMR020/pJBmrdH were diluted in 50 ml of LB; grown to exponential phase; and then supplemented with 0.5 mM Cd2+, Zn2+, Ni2+, and Co2+. These bacterial suspensions were incubated at 30°C with shaking at 150 rpm for 12 h. The cells were harvested by centrifugation and washed three times with Milli-Q water. The resulting pellet was suspended in Millipore water, and cellular metal was extracted with an acid digestion procedure (55). The Cd2+, Zn2+, Ni2+, and Co2+ concentrations were measured on an Atomic absorption spectrophotometer (GBC). Metal content is expressed as μg of metal/100 mg of dry bacteria.
Isolation of RNA and quantitative real-time PCR.
The cells of P. putida KT2440 were grown at 30°C in conical flasks with 10 ml of LB/M9 MM and shaken at 120 rpm on an orbital shaker until the OD600 reached 0.2 to 0.3 (early log phase). The media were then supplemented with various heavy metals such as Cd2+, Zn2+, Ni2+, and Co2+ and incubated further at 30°C for 40 min. The total RNA was isolated by the Triton X-100 boiling and chloroform extraction method (22). Heavy metal induction and RNA extraction were performed in duplicate using two independent cultures. The quality of RNA was monitored on a 2% agarose gel, followed by spectrophotometric quantification at A260. The material was then digested with RNase-free DNase (Fermentas) to remove the residual DNA. The total RNA (2 μg) was reverse transcribed to cDNA by using Revert Aid H minus Moloney murine leukemia virus reverse transcriptase and random hexamer primers (Fermentas). The target genes selected in the present study were mrdH, mreA, tnpA, tnpC, and IS1246 transposases, and the corresponding primer pairs used were A4, A5, A6, A7, and A8. The expression levels of the target genes were normalized to the housekeeping RPSL gene levels (16S rRNA gene). Real-time PCR was performed on Bio-Rad iCycler machine using Real Master Mix SYBR ROX from 5 PRIME. Both target and normalizer reactions were run in triplicate. The following PCR conditions were used: initial denaturation at 95°C for 60 s, followed by 40 cycles of denaturation at 95°C for 20 s, annealing at 56°C for 20 s, and extension at 68°C for 20 s. The reaction specificity was determined for each reaction by using the melting-curve analysis of the PCR product. To calculate the fold change in gene expression, the 2−ΔΔCT method was used (31).
RESULTS
In silico identification of the novel metal resistance genes mrdH and mreA from P. putida KT2440.
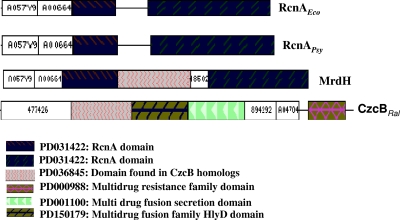
Our previous in silico study identified PP_2968 from P. putida and PSPTO_4280 from P. syringae as RcnA efflux members (nickel/cobalt efflux) exhibiting six transmembrane domains and signature sequences characteristic of the prototypical rcnAEco (16). In the present study we focused on the domain analysis of the proteins encoded by PP_2968 and PSPTO_4280 using ProDom software (release 2005.1/CG171) (50) to identify the compositional differences between them (Fig. 1). The 1,134-bp PP_2968 gene encodes a predicted protein of 41 kDa, composed of an RcnA domain characteristic of RcnA efflux family and a putative domain commonly found in CzcB homologs of the RND efflux family. In contrast, the 879-bp PSPTO_4280 gene from P. syringae DC3000 encodes a predicted protein of 31 kDa, with a protein size and domain composition resembling that of the experimentally characterized RcnAEco (with a molecular mass of 30 kDa and exhibiting only an RcnA domain). Thus, PP_2968 has a significantly different composition due to the presence of an additional domain, which has also been observed in CzcB homologs. The region of PP_2968 encoded protein that comprises the additional domain is located between amino acids 139 and 222, whereas this region is absent in the proteins encoded by PSPTO_4280 and rcnAEco. Due to its novel chimeric domain organization and unidentified substrate spectrum, PP_2968 was annotated as metal resistance determinant mrdH, whereas PSPTO_4280 was annotated as rcnAPsy.
FIG. 1.
Domain organization of putative Pseudomonas metal efflux proteins. ProDom browsing using the MrdH protein sequence as the query gave the domain arrangements of prototypical metal efflux proteins RcnAEco (E. coli) and CzcBRal (Ralstonia species). Their architectures were compared to the output obtained for domain arrangements of putative Pseudomonas metal efflux proteins MrdH (P. putida KT2440) and RcnAPsy (P. syringae DC3000).
We also identified a conserved hypothetical protein, PP_2969, associated with MrdH. This protein composed of 95 amino acids was found to possess a DUF156 domain (PFAM accession no. PF02583). The three protein families found for the DUF156 domain in the SYSTERS database were P151469 (includes hypothetical RcnR proteins), N151470 (includes hypothetical YrkD protein), and P15471 (includes NreA/NreA-like proteins). Among these three families, PP_2969 is homologous to P15471 family members (pairwise identity ranging from 62 to 66%) that include NreA/NreA-like proteins from Sinorhizobium meliloti (accession no. Q92YI5), Legionella pneumophila (accession no. Q93M45), and Achromobacter xylosoxidans (accession no. Q44590) (12, 15). Hence, it was annotated as metal resistance-associated cytoplasmic gene mreA.
Functional complementation of rcnA mutant of E. coli (ARY023 strain) by Pseudomonas metal efflux transporters.
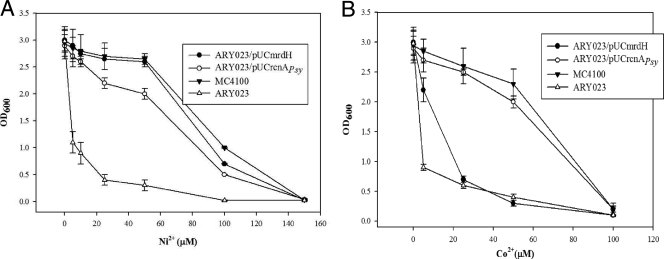
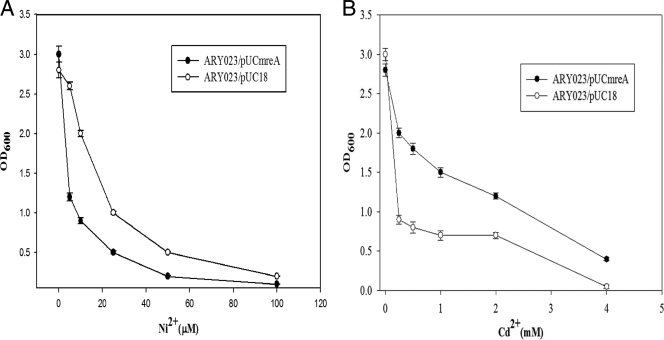
To assess whether the Pseudomonas transporter genes mrdH and rcnAPsy could complement the nickel cobalt-sensitive phenotype of E. coli rcnA mutant (ARY023 strain), these genes were expressed in trans from the multicopy plasmids pUCmrdH and pUCrcnAPsy. Expression of mrdH conferred Ni2+ resistance (fivefold) to the E. coli mutant strain, while Co2+ resistance was least affected. In contrast, the expression of rcnAPsy conferred both Ni2+ and Co2+ resistance (∼3-fold and ∼5-fold, respectively) to the E. coli mutant strain. Compared to ARY023/pUCrcnAPsy, the curve obtained for ARY023/pUCmrdH reflected an increased nickel resistance of the E. coli strain and reached a level similar to that found for the MC4100 (wild-type) strain (Fig. 2). The sensitivity of the ARY023 strain transformed with empty pUC18 vector was similar to that displayed by ARY023 strain alone (data not shown).
FIG. 2.
Effect of nickel and cobalt on growth of ARY023 strains of E. coli. Overnight cultures of ARY023 (open triangle), MC4100 (filled triangle), ARY023/pUCmrdH (filled circle), and ARY023/pUCrcnAPsy (open circle) in M9 MM supplemented with 0.4% glucose were diluted into 1:100 fresh identical medium with the indicated concentrations of Ni2+ (A) and Co2+ (B). Cell growth was monitored as the OD600 after 16 h of incubation at 37°C. Experiments were performed in triplicate, and the values given are averages.
To further support the data obtained in liquid medium, nickel and cobalt viability analysis of the E. coli strains described above was performed on LB plates. Viable counts of ARY023 strain and the complemented strains (ARY023/pUCmrdH and ARY023/pUCrcnAPsy) were measured on LB agar plates after incubation in liquid medium (LB) containing different Ni2+ and Co2+ concentrations. When cells were grown in the absence of metal, viable counts were similar for all of the strains tested. However, with increasing concentrations of Ni2+ and Co2+, the viable counts of the ARY023 strain decreased, indicating its sensitivity to the metals due to the lack of an rcnA efflux system. In contrast, the viable counts of the complemented strains (ARY023/pUCmrdH and ARY023/pUCrcnAPsy) were not affected significantly by increasing concentrations of Ni2+ due to the expression of Ni2+ resistance by mrdH and rcnAPsy genes. However, in the presence of increasing concentrations of Co2+, the viable counts obtained for the mrdH complemented strain (ARY023/pUCmrdH) were relatively less than those obtained for the rcnAPsy complemented strain ARY023/pUCrcnAPsy (see the supplemental material). These data are in accordance with the liquid medium data and show that mrdH confers more Ni2+ resistance than Co2+ resistance.
Thus, the heterologous complementation studies in E. coli provided preliminary evidence for the functional divergence of mrdH from its homologs, rcnAEco and rcnAPsy.
Construction of mrdH mutant strain in P. putida KT2440 with the mrdH targetron.
The targeted disruption of mrdH was accomplished by the cotransformation of pACD4K-mrdC (harboring a retargeted intron) and pAR1219 (to drive the expression of pACD4K-mrdC) into the host strain. Twenty-five colonies were obtained on the final LB selection plates containing kanamycin, and these colonies were further tested for their ability to grow on LB plates containing Ni2+ (1 mM). After 48 h of incubation, the 15 Ni2+-sensitive colonies were further screened for insertion of the targetron and inactivation of mrdH gene by colony PCR. Ten of the fifteen Ni2+-sensitive colonies showed the presence of targetron (in the middle of the mrdH gene) in the PCR analysis, performed using mrdH-specific primer (forward primer) and intron-specific primer (EBS universal) to amplify an 846-bp gene-intron junction (data not shown). Sequence analysis of the 846-bp PCR product from these two colonies indicated that the intron had inserted in the antisense strand at the 627/628 site of the mrdH gene. The stability of the mrdH gene knockout mutant, HMR020, was evaluated after 20 rounds of subculture.
The transporter gene mrdH was cloned into the pJB plasmid under the m-toluic acid inducible Pm promoter and transformed into the HMR020 strain. The HMR020/pJBmrdH transformants were finally selected on a LB agar containing m-toluic acid (0.1 M) and Ni2+ (2 mM) selection plates. The pJB653 vector, without an inserted mrdH gene, was also transformed into HMR020 strain and used as a control in complementation tests.
Disruption of the mrdH gene in P. putida KT2440 conferred sensitivity to cadmium and zinc in addition to nickel and cobalt.
The functionality and substrate range of the mrdH gene was studied by determining the maximum tolerable concentrations (MTCs) of divalent heavy metals for the wild-type, HMR020/pJB653, and HMR020/pJBmrdH strains of P. putida KT2440 on LB plates. Disruption of the mrdH gene affected the resistance to Cd2+, Zn2+, and Ni2+, but Co2+ resistance was only marginally affected (Table 3). The sensitivity of the HMR020/pJB653 strain to the metal ions of interest compared to wild-type and HMR020/pJBmrdH strains was Cd2+ (20-fold) > Zn2+ (4-fold) ≥Ni2+ (4-fold) > Co2+ (2-fold). However, the HMR020/pJB653 strain did not show any sensitivity toward copper, lead, or mercury (Table 3). These data demonstrate the substrate spectrum of mrdH to be Cd2+, Zn2+, Ni2+, and Co2+. In addition, we observed that the HMR020/pJB653 strain had a high level of sensitivity in liquid media to Cd2+ (100-fold) and to a lesser extent to Ni2+ (3-fold) and Zn2+ (3-fold); the levels of HMR020/pJB653 strain sensitivity to Co2+ did not alter significantly from wild-type P. putida KT2440 and HMR020/pJBmrdH strains (data not shown).
TABLE 3.
Determination of MTCs for P. putida KT2440 and derivative strainsa
| Metal | MTC (mM) for P. putida strain:
|
||
|---|---|---|---|
| KT2440 | HMR020/pJB653 | HMR020/pJBmrdH | |
| Cd2+ | 0.2 | 0.01 | 0.2 |
| Ni2+ | 4 | 1 | 4 |
| Zn2+ | 4 | 1 | 4 |
| Co2+ | 0.6 | 0.4 | 0.6 |
| Cu2+ | 2 | 2 | 2 |
| Pb2+ | >1.0 | >1.0 | >1.0 |
| Mn2+ | >1.5 | >1.5 | >1.5 |
P. putida KT2440 (wild type), HMR020/pJB653. and HMR020/pJBmrdH strains were grown till early log phase (OD600 = 0.3) in LB medium at 30°C, diluted 1:100 in fresh medium, and spotted onto LB agar plates containing the indicated concentrations of Cd2+, Zn2+, Ni2+, and Co2+.
mrdH encodes a heavy metal efflux transporter in P. putida KT2440.
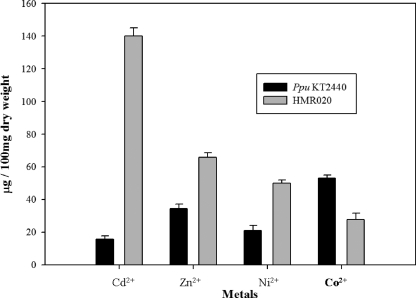
To substantiate the role of mrdH as a metal resistance determinant, we examined the metal accumulation in wild-type P. putida and HMR020 strains of KT2440 (Fig. 3). The HMR020 strain showed a remarkably higher accumulation of Cd2+ (ninefold more) compared to the wild type. Similarly, the accumulation of Ni2+ and Zn2+ was also increased (twofold) in the HMR020 strain. However, the accumulation of Co2+ was twofold higher in the wild-type P. putida than in the HMR020 strain. The preceding results suggests that MrdH can function as an efflux system.
FIG. 3.
Estimation of metal content in P. putida KT2440 and HMR020 strains. Cultures of P. putida KT2440 and HMR020 strains were grown in LB until the exponential phase and then supplemented with 0.5 mM Cd2+, Zn2+, Ni2+, or Co2+. Cells were harvested after 12 h, and the pellet was suspended in Millipore water. Cellular metal was extracted by acid digestion procedure. The concentrations of Cd2+, Zn2+, Ni2+, and Co2+ were measured by atomic absorption spectroscopy.
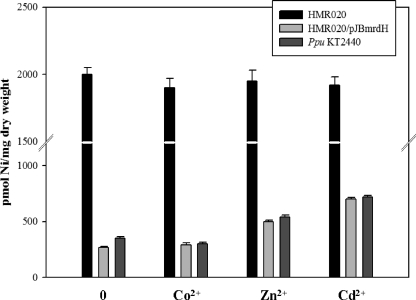
A 63 Ni2+ uptake assay was performed to monitor the in vivo activity of MrdH. If Cd2+, Ni2+, and Zn2+ compete for the same efflux system, then the concentration of Ni2+ in the wild-type strain should be higher when competing metal is present than when it is absent. Our results showed an increased Ni2+ accumulation in wild-type and HMR020/pJBmrdH strains of P. putida in the presence of Cd2+ and Zn2+ (Fig. 4). However, no change was observed for Ni2+ accumulation in the presence or absence of competing metals in the mutant strain, HMR020. These findings also support the proposed role of mrdH as a broad-spectrum metal efflux transporter with a substrate range including Cd2+, Zn2+, and Ni2+.
FIG. 4.
63 Ni uptake analysis. P. putida KT2440, HMR020, and HMR020/pJBmrdH strains were grown in LB medium in the presence of 0.5 mM Ni2+ at 30°C under aerobic conditions until mid-log phase (OD600 = 0.5), harvested, and then washed with buffer consisting of 66 mM KH2PO4-K2HPO4 (pH 6.8) and 0.4% glucose. Cells were resuspended in the same buffer with a fivefold concentration factor. The uptake assay was performed in the presence of 5 μM 63Ni2+ or 5 μM 63Ni2+ and 50 μM Cd2+, Zn2+, or Co2+. Aliquots (100 μl) were filtered after a time period of 45 min (this time point was standardized and kept constant). The intracellular concentration of 63Ni per milligram of bacterial dry weight was determined.
The expression pattern of mreA parallels that of mrdH.
Expression of mreA gene in ARY023 strain of E. coli enhanced its Cd2+ and Ni2+ resistance (Fig. 5), whereas the resistance levels of Zn2+ and Co2+ were not affected.
FIG. 5.
Expression of mreA in E. coli ARY023. Overnight cultures of ARY023/pUC18 (open circle) and ARY023/pUCmreA (filled circle) in M9 MM supplemented with 0.4% glucose were diluted into 1:100 fresh identical medium with the indicated concentrations of metals (Cd2+ and Ni2+). Cell growth was monitored as the OD600 after 16 h of incubation at 37°C.
Expression patterns of mrdH and mreA by Cd2+, Zn2+, and Ni2+ and Co2+ were analyzed in both rich and minimal media by quantitative reverse transcription-PCR (Table 4). The induction of mrdH and mreA genes by different metals was dependent on the growth medium used.
TABLE 4.
Expression analysis of mrdH and mreA in P. putida KT2440
| Metala | Concn (mM) | Mean fold change ± SDb
|
|||
|---|---|---|---|---|---|
|
mrdH
|
mreA
|
||||
| LB | M9 MM | LB | M9 MM | ||
| Cd2+ | 0.02 | 2.2 ± 0.15 | 1.18 ± 0.10 | 3.7 ± 0.10 | 1.40 ± 0.15 |
| 0.2 | 2.5 ± 0.10 | 2.58 ± 0.15 | 4.0 ± 0.15 | 2.27 ± 0.10 | |
| Ni2+ | 0.1 | 1.0 ± 0.20 | 3.89 ± 0.15 | 1.27 ± 0.2 | 2.4 ± 0.20 |
| 1.0 | 2.2 ± 0.21 | 5.5 ± 0.20 | 3.0 ± 0.15 | 4.28 ± 0.30 | |
| Zn2+ | 0.05 | 1.03 ± 0.05 | 1.19 ± 0.15 | 1.03 ± 0.2 | 1.10 ± 0.15 |
| 0.5 | 1.36 ± 0.10 | 7.78 ± 0.25 | 1.36 ± 0.2 | 4.14 ± 0.25 | |
| Co2+ | 0.1 | 1.56 ± 0.15 | 2.29 ± 0.10 | 1.27 ± 0.05 | 2.14 ± 0.15 |
| 1.0 | 2.0 ± 0.05 | 1.00 ± 0.20 | 2.0 ± 0.20 | 1.40 ± 0.20 | |
P. putida KT2440 was grown till early log phase in LB/M9 MM and then induced with the indicated metals for 40 min.
The values indicate the fold change in expression of the mrdH and mreA genes in the presence of various metals.
In LB media, the mrdH gene was induced in the presence of Cd2+ (2.5-fold), Ni2+ (2.2-fold), and Co2+ (2.0-fold). The expression pattern of the mreA gene was similar to that of mrdH and was induced by Cd2+ (4.0-fold), Ni2+ (3.0-fold), and Co2+ (2-fold). In MM, both the mrdH and the mreA genes were induced by Cd2+ (2.58- and 2.27-fold), Zn2+ (7.8- and 4.14-fold), Ni2+ (5.5- and 4.3-fold), and Co2+ (2.29- and 2.14-fold).
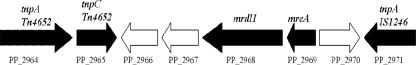
The flanking mobile genetic elements Tn4652 and IS1246 respond to heavy metal stress.
The genetic determinants (mrdH and mreA) involved in the heavy metal resistance of P. putida KT2440 were found to be clustered in a genomic region flanked by Tn4652 and IS1246 elements (Fig. 6). Three other hypothetical proteins, which clustered along with mrdH and mreA, were PP_2966, PP_2967, and PP_2970. Two of these hypothetical genes, PP_2966 and PP_2970, did not have any hits in the database, whereas the third gene, PP_2967, encoded a protein showing homology (69.7% identity) to conserved hypothetical proteins of unknown function from P. fluorescens Pf0-1 (accession no. Pfl_2552) and P. entomophila L48 (accession no. PSEEN3504).
FIG. 6.
Genomic organization of mrdH and mreA in P. putida KT2440. Arrows in the figure indicate the transcriptional orientation of the respective genes. Tn4652 components: tnpA and tnpC (PP_2964 and PP_2965 [transposition function] and PP_2966, PP_2967, and PP_2970 [unknown function]), mrdH (PP_2968 [heavy metal efflux transporter]), mreA (PP_2969 [metal resistance determinant]), and IS1246 tnpA (PP_2971 [transposition function]).
The mobile genetic element Tn4652 belongs to the Tn3 family of transposons and is well characterized in the P. putida strain PaW85. It is a 17-kb derivative of Tn4651 that contains a deletion of its catabolic region, including the xyl gene, likely resulting from the recombination of two copies of IS1246. The other element, IS1246, belongs to the IS5 family and is located within Tn4652 (18, 42).
Expression analysis of the flanking mobile genetic elements showed that their genes were induced by heavy metals, in spite of the fact that they code for transposition functions (Table 5). The tnpA component of the Tn4652 element was induced by Zn2+ (5.3-fold), Ni2+ (5-fold), and Cd2+ (2.5-fold) in LB medium. The same metals caused the induction of tnpA in MM, but a stronger response was observed for Zn2+ (16-fold) than for Ni2+ and Cd2+ (2.5-fold). No induction of the tnpC component was observed with any of the metals in LB, but Ni2+ alone caused the induction of this component in MM (2.5-fold). The other flanking element, IS1246, was induced by Zn2+ (2.5-fold) alone in LB, whereas Zn2+ (8-fold), Ni2+ (4-fold), and Cd2+ (2.6-fold) caused induction in MM.
TABLE 5.
Expression analysis of tnpA (Tn4652), tnpC (Tn4652), and IS1246 in P. putida KT2440
| Metala | Concn (mM) | Mean fold change ± SDb
|
|||||
|---|---|---|---|---|---|---|---|
|
tnpA (Tn4652)
|
tnpC (Tn4652)
|
IS1246
|
|||||
| LB | M9 MM | LB | M9 MM | LB | M9 MM | ||
| Zn2+ | 0.05 | 2.8 ± 0.30 | 8 ± 0.35 | 1 ± 0.2 | 1.10 ± 0.15 | 1.1 ± 0.25 | 8 ± 0.15 |
| 0.5 | 5.3 ± 0.40 | 16 ± 0.40 | 1.4 ± 0.2 | 1.14 ± 0.20 | 2.42 ± 0.2 | 8 ± 0.2 | |
| Ni2+ | 0.1 | 2.46 ± 0.18 | 1.4 ± 0.15 | 1.27 ± 0.10 | 2.3 ± 0.10 | 1.31 ± 0.2 | 2.5 ± 0.1 |
| 1.0 | 4.92 ± 0.10 | 2.5 ± 0.10 | 1.62 ± 0.15 | 2.5 ± 0.25 | 1.50 ± 0.15 | 4 ± 0.2 | |
| Cd2+ | 0.02 | 1.1 ± 0.10 | 1.62 ± 0.10 | 1.0 ± 0.15 | 1.40 ± 0.15 | 1.5 ± 0.1 | 2.2 ± 0.15 |
| 0.2 | 2.51 ± 0.23 | 2.5 ± 0.25 | 1.15 ± 0.12 | 1.27 ± 0.13 | 1.62 ± 0.1 | 2.54 ± 0.10 | |
| Co2+ | 0.1 | 1.51 ± 0.15 | 1.40 ± 0.30 | 1.27 ± 0.15 | 1.14 ± 0.10 | 1.40 ± 0.15 | 1.6 ± 0.15 |
| 1.0 | 1.60 ± 0.25 | 1.75 ± 0.45 | 1.51 ± 0.2 | 1.40 ± 0.2 | 1.60 ± 0.2 | 1.8 ± 0.2 | |
P. putida KT2440 was grown till early log phase in LB/M MM and then induced with the indicated metals for 40 min.
The values indicate the fold change in expression of the tnpA, tnpC, and IS1246 genes in the presence of various metals.
An increased transcriptional response to heavy metals was observed for these flanking elements in MM relative to rich medium. However, Co2+ did not induce the expression of Tn4652 or IS1246 in either of the tested media.
Phylogeny of MrdH and related homologs from environmental bacteria.
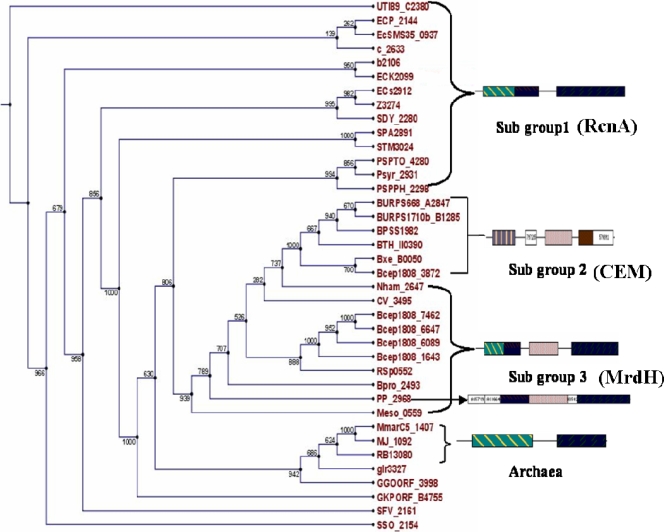
BLAST searching uncovered several homologs of MrdH that are hypothetical membrane transporters from various environmental bacteria, and most of them were annotated as high-affinity Ni2+ transporters or as members of the RcnA efflux family. All of the MrdH homologs exhibited six transmembrane segments and signature sequences characteristically observed for RcnA efflux proteins. However, phylogenetic analysis of the 36 identified MrdH homologs led to their classification into three major subgroups of orthologous genes (Fig. 7). The differences in their core-conserved domains also enabled us to predict distinct domain organizations for the three major subgroups identified. Therefore, we propose division of the MrdH homologs from gram-negative bacteria into three subgroups of efflux proteins: subgroup 1, RcnA proteins (825 bp, 274 amino acids); subgroup 2, cation efflux-related membrane (CEM) proteins (1,270 bp, 422 amino acids); and subgroup 3, MrdH proteins (1,134 bp, 377 amino acids). The RcnA proteins are mainly composed of transmembrane RcnA domain, annotated as PD031422 in the PRODOM database (50). The CEM proteins are composed of three putative domains: the cation efflux transporter system domain (domain identification no. PD001602), the domain found in CzcB homologs (domain PD036845), and the zinc transporter efflux cation system domain (domain PD001369). However, the MrdH proteins (MrdH and related proteins) exhibited chimeric domain organization due to the presence of RcnA domain and the putative domain PD036845, found in CzcB homologs. Intriguingly, all of the MrdH subgroup proteins exhibited a chimeric domain organization and were flanked by mobile genetic elements.
FIG. 7.
Phylogeny of MrdH and related homologs. The neighbor-joining phylogram showed the clustering of MrdH and 35 other homologous proteins into three distinct subgroups. Bootstrap support values are indicated by percentages at the nodes. The accession codes of all of the homologs are indicated in the phylogenetic tree. The domain organizations of the corresponding subgroups are shown on the right side. RcnA proteins constituting subgroup 1 were found to be composed of a cyanobacterial marine plasmid hydrogenase domain (PD129330) and RcnA domains (PD031422 and PD031424). CEM proteins constituting subgroup 2 were predominantly composed of the cation efflux transporter system domain (PD001602), the cation efflux system domain found in CzcB homologs (PD036845), and the zinc transporter efflux cation system domain (PD001369). MrdH proteins constituting subgroup 3 exhibited chimeric domain organization derived from the RcnA domain (PD031422) and the cation efflux system domain found in CzcB homologs (PD036845).
DISCUSSION
This report has demonstrated the presence of a novel metal resistance determinant mrdH, in association with mreA, amid the metal-inducible mobile genetic elements of P. putida KT2440. MrdH was found to contain 377 amino acid residues, with six predicted transmembrane segments, and it contained all of the conserved signature sequences characteristic of an RcnA efflux protein. However, our conserved domain analysis of MrdH showed a chimeric architecture formed by RcnA domain and putative domain found in CzcB homologs. The predicted RcnA domain was found largely in the transmembrane region, whereas the domain derived from CzcB homologs was identified in the cytosolic portion. Such a chimeric domain organization has not been reported for any of the characterized efflux proteins to date. In contrast, RcnAPsy, the putative ortholog of MrdH, is composed entirely of a RcnA domain, as is observed for the prototypical member of the family, RcnAEco (46).
Previously, yohM (rcnA) gene in E. coli possessing RcnA domain conferred increased nickel and cobalt resistance (46). In the czc efflux system (czcABC) of Alcaligenes eutrophus CH34, a component exhibiting the CzcB domain is thought to funnel zinc cations to the CzcA transport protein and facilitate zinc efflux (23). Therefore, the RcnA domain in MrdH could be hypothesized to confer the nickel resistance, whereas the domain derived from CzcB homologs accounts for the observed cadmium and zinc resistance of P. putida KT2440.
A functional status was assigned to the hypothetical membrane transporters mrdH and rcnAPsy by complementation into the ARY023 strain (rcnA) of E. coli. In cross-complementation studies, the mrdH gene was found to confer more Ni2+ resistance than Co2+ resistance, whereas the rcnAPsy gene conferred equal resistance to Ni2+ and Co2+. This result is in agreement with the domain analysis data and strongly suggests that rcnAPsy is an ortholog of rcnAEco, whereas mrdH showed functional divergence from the above homologs. Further, disruption of mrdH in P. putida KT2440 caused increased sensitivity of the strain to Cd2+, Zn2+, and Ni2+ compared to Co2+. To confirm the functionality data of the mrdH transporter derived from the mutant studies, HMR020 strain was complemented with the deleted mrdH transporter (HMR020/pJBmrdH), and the complementation assay confirmed the role of mrdH as a metal resistance determinant. Concomitantly, increased accumulation of Cd2+, Zn2+, and Ni2+ was also observed in the HMR020 strain, indicating the loss of metal efflux activity of the MrdH transporter upon disruption of the corresponding mrdH gene. Transporter studies of MrdH showed that Cd2+ and Zn2+ indeed served as substrates for this transporter and thus competed with 63Ni2+ for metal efflux.
Metal-dependent expression analysis of rcnAEco (by β-glucuronidase assay) showed that the gene was specifically induced by Ni2+/Co2+, and the addition of either Cd2+ or Zn2+ had no effect on the transcription of yohM (46). In the present study, we analyzed the expression of mrdH by real-time PCR and demonstrated the induction of genes by Cd2+, Ni2+, and Co2+ in LB and by all of the four metals in M9 MM. Such growth medium-dependent inducer specificities and expression patterns of the metal transporter genes were reported earlier for the cadA and czcCBA genes of P. putida KT2440 (27).
Most of the previously characterized metal efflux transporters with a broad substrate scope were shown to be members of the tripartite RND family and showed no sequence homology to MrdH. Although MrdH has a high degree of conservation with the archetypal RcnA efflux member (RcnAEco), the presence of an additional domain derived from CzcB homologs may be responsible for conferring Cd2+ or Zn2+ resistance apart from Ni2+ and Co2+ resistance. Thus, mrdH showed a broad substrate profile and compositional differences from the previously reported nickel cobalt efflux protein, rcnAEco. Therefore, we report MrdH as a novel single component metal efflux transporter exhibiting chimeric domain structure and a broad substrate scope.
In the phylogenetic analysis of MrdH homologs, three distinct subgroups comprised of RcnA, CEM, and MrdH proteins were identified in the phylogram with variations in their domain architecture (Fig. 7). Among these three subgroups, MrdH subgroup was predominantly composed of efflux proteins from environmental bacteria such as P. putida KT2440, Burkholderia vietnamiensis, Ralstonia solanacearum, Polaromonas species, Mesorhizobium species, Nitrobacter species, and Chromobacter violaceum that colonize the rhizosphere. Two hits (35 and 37% sequence identities) were also observed from the methanogenic archaea Methanococcus jannaschii DSM2661 (MJ_1092) and Methanococcus maripaludis C5 (Mmarc5_1407), which differed from the other homologs of gram-negative bacteria in possessing five TMDs. The chimeric domain organization of all of the MrdH subgroup proteins and the presence of the mobile genetic elements in the vicinity indicate their acquisition through HGT (except for Nham_2647 and CV_3495). The features described above are apparently absent from members of the RcnA and CEM subgroups. Moreover, the presence of five copies of the mrdH gene in the genome of B. vietnamiensis suggests its pivotal role as a resistance determinant, which might have fostered its evolution and persistence.
We identified, in association with mrdH, a conserved hypothetical cytoplasmic protein encoding gene (PP_2969) that belongs to the DUF156 group of proteins that are widely distributed in bacteria and are speculated to be regulators of gene expression. The reported DUF156 proteins include NreA from Legionella pneumophila (NreABC), NcrB from Serratia marcescens (NcrABC), NirB from Klebsiella oxytoca (NirABCD), and NcrB from Leptospirillum ferriphilum (NcrABCY) (15, 32, 33, 39, 54). Functionally, expression of the NreA gene (L. pneumophila) in E. coli conferred resistance to Ni2+ and Co2+ but not to Cd2+ or Zn2+ (15). In L. ferriphilum the deletion of ncrB gene reduced Ni2+ resistance, suggesting that this gene might assist the associated transporter ncrA in Ni2+ efflux (54). In the present study, expression of mreA endowed rcnA mutant of E. coli with Cd2+ and Ni2+ resistance. However, expression studies of mreA at the transcriptional level demonstrated that transcription was significantly induced by Cd2+, Zn2+, Ni2+, and Co2+ depending on the growth medium used. Moreover, the gene expression of mreA parallels that of mrdH.
The phenomenon of HGT driven by mobile genetic elements, such as plasmids, insertion sequences, integrons, transposons, and phages, has been shown to provide microbes with a wide variety of adaptive traits such as antibiotic resistance, heavy metal resistance, and xenobiotic compound degradation for microbial survival and proliferation (38). Recently, detectable HGT of heavy metal translocating PIB-type ATPases (zntA/cadA/copA-like genes) has been reported in bacterial isolates from pristine, as well as heavy metal-contaminated subsurface environments (8, 9). In fact, most of the novel genes in gammaproteobacteria were found to be distributed in GIs, indicating their acquisition through HGT from distant sources (19). Our previous in silico studies also show HGT to be a source of gene innovation in Pseudomonas metal transportomes (16). In this study, the characterized novel metal resistance determinants mrdH and mreA were found to be located within a GI flanked by mobile genetic elements. Further, the absence of GI-associated features in MrdH homologs from other Pseudomonas (Pmen_2243, PSPTO_4280, PSPPH_2298, and Psyr_2931) and the phylogenetic incongruence observed (Fig. 7) is compelling enough to hypothesize that the mrdH gene in P. putida KT2440 could have been acquired through HGT. Intriguingly, complete sequence of the IncP-9 TOL plasmid pWW0 from P. putida also revealed the presence of two genes (ORF144 and ORF145) encoding putative NreA and RcnA-like metal resistance determinants amid mobile genetic elements (IS1246 and Tn4652 transposase genes) (13). Since they shared 100% identity with the chromosomal metal resistance determinants MrdH and MreA (encoded by PP_2968 and PP_2969) identified here, there is a strong possibility of transposition events in disseminating metal resistance traits from plasmid to chromosome under acute metal stress conditions.
Recent studies have explored the capacity of transposable elements to respond to metal stress by enhancing their transcriptional or transpositional activity. The whole-genome transcriptional profile of a metal-tolerant E. coli strain exhibited an increase in the transcript level of the IS elements, particularly insA, whose expression in E. coli confers tolerance to Zn2+ (5). Transcriptomic analyses of pMOL28 and pMOL30, which encode metal resistance in C. metallidurans, showed metal-mediated upregulation of some transposases (Tn4378 and Tn4380) and truncated IS elements (34). However, not all of the mobile genetic elements flanking metal resistance determinants respond to metal stress. For instance, on the pMOL30 plasmid, an incomplete mer cluster (merRTP) contains genes encoding proteins with significant similarity to Tn4378 and Tn4380, but they were not induced under any of the experimental conditions examined and thus were reported as nonfunctional (34). In Listeria species, Cd2+ resistance genes were found on the Tn5422 transposon, but the flanking elements were not induced by Cd2+ under any of the tested conditions (24). In Pseudomonas spp., mercury resistance determinants were often found to reside on transposons Tn5041, Tn5053, and Tn5056, but there is no functional evidence available for the metal-induced response of the transposable elements in this genus (51). We have demonstrated here a strong induction of the flanking Tn4652 and IS1246 elements by Zn2+ and a moderate induction by Ni2+ and Cd2+. Therefore, Tn4652 and IS1246 in P. putida KT2440 are not remnants of evolutionary rearrangements but are functional genes that have entrapped novel metal resistance genes mrdH and mreA. Since the role of transposons in the coselection of antibiotic and metal resistance has been well documented for Tn21 and Tn21-like transposons, the identification of novel metal inducible mobile genetic elements here provides future research opportunities in metal-antibiotic coselection (2, 28).
Given this body of evidence, we conclude that mrdH, is novel heavy metal efflux transporter exhibiting broad substrate specificity. In association with mrdH, mreA encoding a putative cytoplasmic protein has been identified, with a role in Cd2+/Ni2+ resistance and an expression pattern parallel to that of mrdH. The location of both of these metal-responsive genes amid functional mobile genetic elements implicates their acquisition through HGT. Further investigations are required to understand the molecular mechanism of cation efflux by mrdH, the potential interactions between mrdH and mreA, and the full substrate profile of MrdH subgroup of proteins.
Supplementary Material
Acknowledgments
This manuscript is dedicated to P. Maruthi Mohan, Department of Biochemistry, Osmania University.
We thank Sarah Frank, Zentrum Biochemie und Zentrum Kinderheilkunde, Medizinische Hochschule, Hannover, Germany, for providing P. putida KT2440 and Arun Chaterjee, University of Missouri-Columbia, for providing the genomic DNA of P. syringae DC3000.
This study was financially supported by the Council for Scientific and Industrial Research (9/163/2003 EMR-1) and IFCPAR (3709-1).
Footnotes
Published ahead of print on 31 July 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anton, A., C. Grome, J. ReiMman, T. Pribyl, and D. H. Nies. 1999. CzcD is a heavy metal ion transporter involved in regulation of heavy metal resistance in Ralstonia sp. strain CH34. J. Bacteriol. 181:6876-6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austin, C. B., M. S. Wright, R. Stepanauskas, and J. V. McArthur. 2006. Co-selection of antibiotic and metal resistance. Trends Microbiol. 14:176-182. [DOI] [PubMed] [Google Scholar]
- 3.Blatny, J. M., T. Brautaset, H. C. Winther-Larsen, K. Haugan, and S. Valla. 1997. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl. Environ. Microbiol. 63:370-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borremans, B., J. L. Hobman, A. Provoost, N. L. Brown, and D. Van der Lelie. 2001. Cloning and functional analysis of the pbr lead resistance determinant of Ralstonia metallidurans CH34. J. Bacteriol. 183:5651-5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brocklehurst, K. R., and A. P. Morby. 2000. Metal-ion tolerance in Escherichia coli: analysis of transcriptional profiles by gene-array technology. Microbiology 146:2277-2282. [DOI] [PubMed] [Google Scholar]
- 6.Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, N. Zafar, L. Zhou, J. Liu, Q. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Van Aken, T. V. Feldblyum, M. D'Ascenzo, D. Wen-Ling, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martin, D. J. Schneider, T. Xiaoyan, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100:10181-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canovas, D., I. Cases, and V. de Lorenzo. 2003. Heavy metal tolerance and metal homeostasis in Pseudomonas putida as revealed by complete genome analysis. Environ. Microbiol. 5:1242-1256. [DOI] [PubMed] [Google Scholar]
- 8.Coombs, J. M., and T. Barkay. 2004. Molecular evidence for the evolution of metal homeostasis genes by lateral gene transfer in bacteria from the deep terrestrial subsurface. Appl. Environ. Microbiol. 70:1698-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coombs, J. M., and T. Barkay. 2005. New findings on evolution of metal homeostasis genes: evidence from comparative genome analysis of Bacteria and Archaea. Appl. Environ. Microbiol. 71:7083-7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davanloo, P., A. H. Rosenberg, J. J. Dunn, and F. W. Studier. 1984. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 81:2035-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grass, G., C. Grome, and D. H. Nies. 2000. Regulation of the cnr cobalt/nickel resistance determinant from Ralstonia sp. CH34. J. Bacteriol. 182:1390-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grass, G., B. Fan, B. P. Rosen, K. Lemke, H. G. Schlegel, and C. Rensing. 2001. NreB from Achromobacter xylosoxidans 31A is a nickel-induced transporter conferring nickel resistance. J. Bacteriol. 183:2803-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greated, A., L. Lambertsen, P. A. Williams, and C. M. Thomas. 2002. Complete sequence of the IncP-9 TOL plasmid pWW0 from Pseudomonas putida. Environ. Microbiol. 4:856-871. [DOI] [PubMed] [Google Scholar]
- 14.Grome, C., G. Grass, A. Anton, S. Franke, A. Navarrete Santos, B. Lawley, N. L. Brown, and D. H. Nies. 1999. Transcriptional organization of the czc heavy metal homoeostasis determinant from Alcaligenes eutrophus. J. Bacteriol. 181:2385-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahm, D.-H., Y. Mi-Jung, K. Whae-Min, H. L. Eunjoo, L. Hye-Jung, S. Insop, and K. Hong-Yeoul. 2002. Characterization of the nickel resistance gene from Legionella pneumophila: attenuation of nickel resistance by ppk (polyphosphate kinase) disruption in Escherichia coli. J. Microbiol. Biotechnol. 12:114-120. [Google Scholar]
- 16.Haritha, A., A. Rodrigue, and P. M. Mohan. 2008. A comparative analysis of metal transportomes from metabolically versatile Pseudomonas. BMC Res. Notes 1:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hassan, M. T., D. van der Lelie, D. Springael, U. Romling, N. Ahmed, and M. Mergeay. 1999. Identification of a gene cluster, czr, involved in cadmium and zinc resistance in Pseudomonas aeruginosa. Gene 238:417-425. [DOI] [PubMed] [Google Scholar]
- 18.Hor̃ak, R., and M. Kivisaar. 1998. Expression of the transposase gene tnpA of Tn4652 is positively affected by integration host factor. J. Bacteriol. 180:2822-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsiao, W. W., K. Ung, D. Aeschliman, J. Bryan, B. B. Finlay, and F. S. Brinkman. 2005. Evidence of a large novel gene pool associated with prokaryotic genomic islands. PLoS Genet. 1:e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu, N., and B. Zhao. 2007. Key genes involved in heavy-metal resistance in Pseudomonas putida CD2. FEMS Microbiol. Lett. 267:17-22. [DOI] [PubMed] [Google Scholar]
- 21.Karberg, M., H. Guo, J. Zhong, R. Coon, J. Perutka, and A. M. Lambowitz. 2001. Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat. Biotechnol. 19:1162-1167. [DOI] [PubMed] [Google Scholar]
- 22.Kidon, S., A. Khan Saeed, S. Nawaz Mohamed, and A. Khan Ashraf. 2003. A simple and efficient Triton X-100 boiling and chloroform extraction method of RNA isolation from gram-positive and gram-negative bacteria. FEMS Microbiol. Lett. 229:97-101. [DOI] [PubMed] [Google Scholar]
- 23.Kunito, T., T. Kusano, H. Oyaizu, K. Senoo, S. Kanazawa, and S. Matsumoto. 1996. Cloning and sequence analysis of czc genes in Alcaligenes sp. strain CT14. Biosci. Biotechnol. Biochem. 60:699-704. [DOI] [PubMed] [Google Scholar]
- 24.Lebrun, M., A. Audurier, and P. Cossart. 1994. Plasmid-borne cadmium resistance genes in Listeria monocytogenes are present on Tn5422, a novel transposon closely related to Tn917. J. Bacteriol. 176:3049-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, S. W., E. Glickmann, and D. A. Cooksey. 2001. Chromosomal locus for cadmium resistance in Pseudomonas putida consisting of a cadmium-transporting ATPase and a MerR family response regulator. Appl. Environ. Microbiol. 67:1437-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, S. M., G. Grass, C. J. Haney, B. Fan, B. P. Rosen, A. Anton, D. H. Nies, and C. Rensing. 2002. Functional analysis of the Escherichia coli zinc transporter ZitB. FEMS Microbiol. Lett. 215:273-278. [DOI] [PubMed] [Google Scholar]
- 27.Leedjärv, A., A. Ivask, and M. Virta. 2008. Interplay of different transporters in the mediation of divalent heavy metal resistance in Pseudomonas putida KT2440. J. Bacteriol. 190:2680-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome, Microbiol. Mol. Biol. Rev. 63:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liesegang, H., K. Lemke, R. A. Siddiqui, and H. G. Schlegel. 1993. Characterization of the inducible nickel and cobalt resistance determinant cnr from pMOL28 of Alcaligenes eutrophus CH34. J. Bacteriol. 175:767-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, S. C., D. A. Webster, and B. C. Stark. 1996. An improved method of transformation in pseudomonads. Biotechnol. Techniques 10:683-686. [Google Scholar]
- 31.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 32.Lopez-Maury, L., M. Garcia-Dominguez, F. J. Florencio, and J. C. Reyes. 2002. A two-component signal transduction system involved in nickel sensing in the cyanobacterium Synechocystis sp. PCC 6803. Mol. Microbiol. 43:247-256. [DOI] [PubMed] [Google Scholar]
- 33.Marrero, J., G. Auling, O. Coto, and D. H. Nies. 2007. High-level resistance to cobalt and nickel but probably no transenvelope efflux: metal resistance in the Cuban Serratia marcescens strain C-1. Microbiol. Ecol. 53:123-133. [DOI] [PubMed] [Google Scholar]
- 34.Monchy, S., M. A. Benotmane, P. Janssen, T. Vallaeys, S. Taghavi, D. van der Lelie, and M. Mergeay. 2007. Plasmids pMOL28 and pMOL30 of Cupriavidus metallidurans are specialized in the maximal viable response to heavy metals. J. Bacteriol. 189:7417-7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Hoolmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. C. Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. A. Eisen, K. N. Timmis, A. Dusterhoft, B. Tummler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 36.Nies, D. H. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27:313-339. [DOI] [PubMed] [Google Scholar]
- 37.Nucifora, G., L. Chu, T. K. Misra, and S. Silver. 1989. Cadmium resistance from Staphylococcus aureus plasmid pI258 cadA gene results from a cadmium-efflux ATPase. Proc. Natl. Acad. Sci. USA 86:3544-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 39.Park, J. S., S. J. Lee, H. G. Rhie, and H. S. Lee. 2008. Characterization of a chromosomal nickel resistance determinant from Klebsiella oxytoca CCUG 15788. J. Microbiol. Biotechnol. 18:1040-1043. [PubMed] [Google Scholar]
- 40.Paulsen, I. T., C. M. Press, J. Ravel, D. Y. Kobayashi, G. S. Myers, D. V. Mavrodi, R. T. DeBoy, R. Seshadri, Q. Ren, R. Madupu, R. J. Dodson, A. S. Durkin, L. M. Brinkac, S. C. Daugherty, S. A. Sullivan, M. J. Rosovitz, M. L. Gwinn, L. Zhou, D. J. Schneider, S. W. Cartinhour, W. C. Nelson, J. Weidman, K. Watkins, K. Tran, H. Khouri, E. A. Pierson, L. S. Pierson III, L.S. Thomashow, and J. E. Loper. 2005. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat. Biotechnol. 23:873-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quaranta, D., R. McCarty, V. Bandarian, and C. Rensing. 2007. The copper-inducible cin operon encodes an unusual methionine-rich azurin-like protein and a Pre-Q.0 reductase in Pseudomonas putida KT2440. J. Bacteriol. 181:5361-5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reddy, B. R., L. E. Shaw, J. R. Sayers, and P. A. Williams. 1994. Two identical copies of IS1246, a 1,275 base pair sequence related to other bacterial insertion sequences, enclose the xyl genes of TOL plasmid pWW0. Microbiology 140:2305-2307. [DOI] [PubMed] [Google Scholar]
- 43.Rensing, C., and G. Grass. 2003. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 27:197-213. [DOI] [PubMed] [Google Scholar]
- 44.Rensing, C., M. Ghosh, and B. P. Rosen. 1999. Families of soft metal-ion-transporting ATPases. J. Bacteriol. 181:5891-5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rensing, C., B. Mitra, and B. P. Rosen. 1997. The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc. Natl. Acad. Sci. USA 94:14326-14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodrigue, A., G. Effantin, and M. A. Mandrand-Berthelot. 2005. Identification of rcnA (yohM), a nickel and cobalt resistance gene in Escherichia coli. J. Bacteriol. 187:2912-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rossbach, S., T. L. Wilson, M. L. Kukuk, and H. A. Carty. 2000. Elevated zinc induces siderophore biosynthesis genes and a zntA-like gene in Pseudomonas fluorescens. FEMS Microbiol. Lett. 191:61-70. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis (ed.). 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 49.Schmidt, T., and H. G. Schlegel. 1994. Combined nickel-cobalt-cadmium resistance encoded by the ncc locus of Alcaligenes xylosoxidans 31A. J. Bacteriol. 176:7045-7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Servant, F., C. Bru, S. Carrère, E. Courcelle, J. Gouzy, D. Peyruc, and D. Kahn. 2002. ProDom: automated clustering of homologous domains. Brief. Bioinform. 3:246-251. [DOI] [PubMed] [Google Scholar]
- 51.Sofia, M., M. Leonid, P. Mayya, K. Gennady, M. Svetlana, G. Zhosefine, and N. Vadim. 2005. Present-day mercury resistance transposons are common in bacteria preserved in permafrost grounds since the Upper Pleistocene. Res. Microbiol. 156:994-1004. [DOI] [PubMed] [Google Scholar]
- 52.Stähler, F. N., S. Odenbreit, R. Haas, J. Wilrich, A. H. Van Vliet, J. G. Kusters, M. Kist, and S. Bereswill. 2006. The novel Helicobacter pylori CznABC metal efflux pump is required for cadmium, zinc, and nickel resistance, urease modulation, and gastric colonization. Infect. Immun. 74:3845-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westhrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. S. Wong, Z. Wu, I. T. Paulsen, J. Relzer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 54.Tian, J., N. Wu, J. Li, Y. Liu, J. Guo, B. Yao, and Y. Fan. 2007. Nickel-resistant determinant from Leptospirillum ferriphilum. Appl. Environ. Microbiol. 73:2364-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Venkateswerlu, G., and K. S. Sastry. 1973. Interrelationships in trace element metabolism in metal toxicities in cobalt-resistant strain of Neurospora crassa. Biochem. J. 132:673-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weinel, C., K. E. Nelson, and B. Tummler. 2002. Global features of the Pseudomonas putida KT2440 genome sequence. Environ. Microbiol. 4:809-818. [DOI] [PubMed] [Google Scholar]
- 57.Xiong, A. M., and R. K. Jayaswal. 1998. Molecular characterization of a chromosomal determinant conferring resistance to zinc and cobalt ions in Staphylococcus aureus. J. Bacteriol. 180:4024-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp8 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 59.Yao, J., and A. M. Lambowitz. 2007. Gene targeting in gram-negative bacteria by use of a mobile group II intron (“targetron”) expressed from a broad-host-range vector. Appl. Environ. Microbiol. 73:2735-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.