Abstract
Translesion synthesis is a DNA damage tolerance mechanism by which damaged DNA in a cell can be replicated by specialized DNA polymerases without being repaired. The Escherichia coli umuDC gene products, UmuC and the cleaved form of UmuD, UmuD′, comprise a specialized, potentially mutagenic translesion DNA polymerase, polymerase V (UmuD′2C). The full-length UmuD protein, together with UmuC, plays a role in a primitive DNA damage checkpoint by decreasing the rate of DNA synthesis. It has been proposed that the checkpoint is manifested as a cold-sensitive phenotype that is observed when the umuDC gene products are overexpressed. Elevated levels of the beta processivity clamp along with elevated levels of the umuDC gene products, UmuD′C, exacerbate the cold-sensitive phenotype. We used this observation as the basis for genetic selection to identify two alleles of umuD′ and seven alleles of umuC that do not exacerbate the cold-sensitive phenotype when they are present in cells with elevated levels of the beta clamp. The variants were characterized to determine their abilities to confer the umuD′C-specific phenotype UV-induced mutagenesis. The umuD variants were assayed to determine their proficiencies in UmuD cleavage, and one variant (G129S) rendered UmuD noncleaveable. We found at least two UmuC residues, T243 and L389, that may further define the beta binding region on UmuC. We also identified UmuC S31, which is predicted to bind to the template nucleotide, as a residue that is important for UV-induced mutagenesis.
DNA damage arises from numerous sources, including exogenous and endogenous sources, as well as spontaneously (14). DNA damage must be repaired or it can lead to mutagenesis and genomic instability (14). All cells possess several overlapping pathways to repair DNA damage and maintain genomic integrity. In response to DNA damage, the Escherichia coli SOS response regulates the expression of at least 57 genes involved in DNA repair, DNA damage tolerance, and cell division (10, 14, 41).
Among the genes induced as part of the SOS response are the E. coli Y family DNA polymerase genes dinB (polymerase IV [pol IV]) and umuDC (pol V, UmuD′2C). The Y family DNA polymerases are characterized by a specialized ability to replicate DNA templates that contain lesions that typically block highly accurate, replicative DNA polymerases, a process termed translesion synthesis (14, 39). They accomplish this by virtue of their relatively open active sites, their lack of an intrinsic proofreading domain, and their reduced protein contacts with the lesion-containing template (14). The Y family DNA polymerases also replicate undamaged DNA, albeit with lower fidelity than replicative DNA polymerases (14). Y family polymerases adopt a right-hand fold reminiscent of replicative DNA polymerases, although the Y family polymerases are typically found in the “open” state. Y family polymerases also have a “little finger” domain that provides additional binding contact with the DNA (14, 54).
The umuDC genes encode the Y family DNA polymerase UmuD′2C, which has the striking ability to copy DNA that contains thymine-thymine cyclobutane pyrimidine dimers or abasic sites (37, 51, 52). This activity comes at a potentially mutagenic cost, as Y family DNA polymerases copy undamaged DNA with lower fidelity than replicative DNA polymerases (14). As Y family polymerase activity is potentially mutagenic, it is elaborately regulated in order to employ translesion synthesis only when necessary.
Expression of the umuDC genes is transcriptionally regulated as part of the SOS response (14, 39). UmuC activity is regulated by the accessory protein UmuD. UmuD is a homodimeric protein composed of a C-terminal globular domain and N-terminal arms (5, 12, 35, 42, 47). After SOS induction, UmuD initially persists in the full-length dimeric form, UmuD2. The presence of UmuD results in mainly accurate DNA replication and repair (18, 30, 50). Between 20 and 40 min after SOS induction, interaction of UmuD2 with RecA—single-stranded DNA (ssDNA) nucleoprotein filaments stimulates the latent autocatalytic ability of UmuD2 to cleave its N-terminal arms between residues C24 and G25 to form UmuD′2. The UmuD′2 form is required for UmuC to be active as a translesion DNA polymerase (37, 51).
In addition to the translesion DNA synthesis activity of UmuD′2C, the umuDC gene products also play a role in a primitive DNA damage checkpoint. Elevated levels of umuD and umuC gene products specifically decrease the rate of DNA synthesis (27). The lag in the resumption of DNA synthesis allows time for accurate repair processes, such as nucleotide excision repair, to take place (30, 33) before the cell engages the potentially mutagenic process of translesion synthesis. Elevated levels of the umuD and umuC gene products cause a cold-sensitive growth phenotype that has been interpreted to be an exaggeration of the checkpoint function (50). Variants of umuC that genetically separate its roles in a primitive DNA damage checkpoint and UV-induced mutagenesis have been discovered. The umuC125(A29V) allele is proficient for UV-induced mutagenesis but does not confer the cold-sensitive phenotype (26), while the umuC104(D101N) allele is deficient for UV-induced mutagenesis but confers a cold-sensitive growth phenotype (50).
Interactions with the β clamp confer high processivity and efficiency on replicative DNA polymerases (22) and increase the processivity of Y family polymerases to various extents (16, 25, 53). The presence of the β clamp is also thought to play a key role in managing polymerases at the replication fork (15, 17). UmuD and UmuD′ interact with the β clamp physically, as well as genetically (5, 42, 46, 48, 49). Deletion of the N-terminal 9 residues of UmuD has little effect on its ability to cross-link to the β clamp, while deletion of the N-terminal 19 residues results in a dramatic decrease in, but not a complete loss of, cross-linking efficiency (48). Thus, the β clamp interacts with both the N-terminal arms and the C-terminal globular domain of UmuD, as well as with the globular domain of UmuD′.
There are also two identified regions of UmuC that interact with the β clamp. UmuC possesses a canonical β clamp binding motif (357QLNLF361) that has been shown to interact with β and to be critically important for both the umuC-mediated cold-sensitive phenotype and the UV-induced mutagenesis function (2, 3, 11). A second β interaction site of UmuC has been identified, 313LTP315, by analogy to an observed interaction in the cocrystal structure of the DinB little finger domain with the β clamp (7) that is important for mediating the cold-sensitive phenotype but is dispensable for UV-induced mutagenesis (3).
Elevated levels of UmuD and UmuC cause host cells to exhibit cold sensitivity for growth (27). Intriguingly, cooverexpression of the β processivity clamp with umuDC exacerbates umuDC-mediated cold sensitivity. This finding was exploited to uncover eight new β clamp alleles that did not exacerbate the cold-sensitive phenotype when they were cooverexpressed with the umuDC gene products, suggesting that these mutations in the β clamp impair its interactions with the umuDC gene products (46). Further characterization of these β clamp alleles uncovered several variants that are critically important for pol V-dependent translesion synthesis (20, 45).
In this work, we used a similar strategy to find new variants of UmuD′ and UmuC that disrupt the interactions of these proteins with the β clamp and thus identify β binding sites. We identified nine alleles of the umuDC genes that suppress the cold-sensitive phenotype observed with elevated levels of the β clamp, four of which resulted in two different amino acid changes. The variants obtained included those with 2 deduced amino acid substitutions in UmuD′ and variants with 11 deduced amino acid substitutions in UmuC. Furthermore, several of the umuC alleles obtained in this selection experiment genetically separate the roles of umuC in UV-induced mutagenesis and in the cold-sensitive growth phenotype.
MATERIALS AND METHODS
Bacteriological techniques.
The E. coli strains and plasmids used in this study are listed in Table 1. The low-copy-number plasmids pGY9739 and pGY9738 carry the umuDC operon and a synthetic umuD′C operon, respectively, under control of a promoter that varies from the normal promoter by a single base substitution that leads to increased expression (44). Strains were routinely grown in Luria-Bertani broth at 37°C unless otherwise noted, and the medium was supplemented with the following antibiotics when necessary: spectinomycin (60 μg/ml) and/or ampicillin (100 μg/ml). Competent cells were prepared using the CaCl2 method (38). Site-directed mutants were constructed using QuikChange (Stratagene). Mutagenic primer sequences are available on request. Quantitative transformation assays were performed as described previously (4). UV mutagenesis and survival assays were performed as described previously (4, 5).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant genotype and/or characteristics | Source or reference |
|---|---|---|
| Strains | ||
| AB1157 | argE3 | Laboratory stock |
| GW8017 | AB1157 ΔumuDC | 19 |
| Plasmids | ||
| pGY9738 | o1C umuD′C; pSC101 derived | 43 |
| pGY9739 | o1C umuDC; pSC101 derived | 43 |
| pGB2 | Vector; pSC101 derived | 9 |
| pJRC210 | pBR22; overproduces β | Charles McHenry |
Genetic assay for selection of umuD′C alleles.
Strains harboring the pGY9739 plasmid have a cold-sensitive phenotype, exhibited as reduced growth at 30°C compared to growth at 37°C or 42°C, which is thought to be due to a hyperactive DNA damage checkpoint that likely results from interactions of UmuD with the replicative DNA polymerase (27, 49, 50). Elevated levels of the β clamp exacerbate the cold-sensitive phenotype observed in cells that overexpress the umuDC gene products (46). The analogous plasmid pGY9738, which carries a synthetic operon for umuD′C, does not normally cause the cold-sensitive phenotype, except when it is combined with elevated levels of the β clamp (46, 49). We used this fact to screen for alleles of umuD′C that do not result in the cold-sensitive phenotype in the presence of elevated levels of the β clamp. Plasmid pGY9738 (Table 1), which expresses umuD′C, was mutagenized by treatment with hydroxylamine (HA) as described previously (29). AB1157 cells harboring a plasmid expressing the β clamp, pJRC210, and HA-treated pGY9738 that could grow at 30°C (i.e., the cold-sensitive phenotype was suppressed) were selected as follows. Chemically competent AB1157 cells bearing pJRC210 were transformed with HA-treated pGY9738. To minimize the isolation of siblings, transformation reaction mixtures were aliquoted prior to recovery. Following a 60-min recovery, an aliquot of the transformation mixture was plated for growth at a permissive temperature, 42°C, to permit calculation of the transformation efficiency. The remainder of each transformation mixture was plated for growth at 30°C to select for alleles of umuD′C that allowed growth at the nonpermissive temperature. To confirm that growth at 30°C was due to the plasmid umuD′C alleles, the selected plasmid variants were isolated and retransformed into AB1157(pJRC210). Equal volumes were plated for growth at 30°C and for growth at 42°C, and plates were scored after overnight incubation. In our hands pJRC210, the plasmid used to express the β clamp, was somewhat unstable. At extremely elevated expression levels, when induced with isopropyl-β-d-thiogalactopyranoside (IPTG), the β clamp confers lethality in E. coli strains with elevated levels of umuD′C at 30°C (46). We used this observation to confirm that pJRC210 expressed elevated levels of the β clamp and to eliminate any “hits” that might be due to changes in pJRC210. IPTG was used only in this validation step, not in the selection or quantitative transformation assays; that is, we took advantage of the observation mentioned above to confirm that there was β clamp overexpression-dependent lethality in all of the selected clones at 30°C. Verified plasmids were then sequenced at the Tufts University Core Facility (Boston, MA) using the following three primers for complete coverage of the umuD′C operon: UmuDseq5′ (5′-GCCTGAATCAGTATTGATCTGCTGGC), UmuCseqFor (5′-GGTGATCCACGTCGTTAAGGCGA), and UmuC3′seq (5′-CGCTAATCCATTCGGCGCTCCTGC). Sequencing results were compared to the sequence of the parental plasmid using Lasergene.
Protein expression and purification and UmuD cleavage assay.
UmuD expression was carried out as described previously (4). Cells were harvested by centrifugation at 8,000 × g for 10 min at 4°C. Cell pellets were collected and stored at −80°C overnight before they were lysed. Frozen cells were thawed on ice and suspended in 25 ml of lysis buffer (50 mM HEPES [pH 7.5], 100 mM NaCl, 10% glycerol, 1 mM dithiothreitol, 10 μg/ml phenylmethanesulfonyl fluoride, 0.5 protease inhibitor cocktail tablet [Roche Diagnostics]). Cell lysis was performed by sonication at 50% output with alternating 15-s bursts and 3-min rest periods. Cell lysate was treated with lysozyme (300 μg/ml; Sigma) and DNase I (1 μg/ml; Roche) on ice for 45 min, followed by a 10-min freeze-thaw cycle using −80°C and 37°C. The cell debris was then pelleted by centrifugation at 12,000 × g and 4°C for 1 h, and the supernatant was collected and filtered through a 0.45-μm filter before purification.
The supernatant was diluted 1:1 with Qa buffer (20 mM HEPES [pH 7.5], 0.1 mM EDTA [pH 8.0], 100 mM NaCl, 1 mM dithiothreitol, 10 μg/ml phenylmethanesulfonyl fluoride, 0.5 protease inhibitor cocktail tablet per liter of buffer). The sample was then loaded onto a 5-ml HiTrap Fast Flow DEAE column (GE Healthcare) preequilibrated with Qa buffer. Wild-type UmuD and the G129S variant were eluted with a gradient of 0 to 50% Qb buffer (same as Qa buffer except that the NaCl concentration was 1 M) over 20 column volumes at a flow rate of 2.5 ml/min. Fractions were collected at the beginning of the gradient, after the Qb buffer concentration reached 20%. Aliquots (10 μl) of each fraction were analyzed by 14% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), and protein was detected by staining with Coomassie blue.
Fractions containing UmuD were pooled, filtered, and loaded onto a Hi-Trap Fast Flow phenyl-Sepharose column (GE Healthcare) preequilibrated with PSA (Qa buffer containing 1 M ammonium sulfate). Wild-type UmuD was eluted with a gradient of 0 to 100% Qa over 20 column volumes at a flow rate of 4 ml/min. UmuD(G129S) was eluted with a gradient of 0 to 100% Qa over 25 column volumes at a flow rate of 2 ml/min. In both cases, fractions were collected at the end of the gradient after the Qa concentration reached 70%. Fractions were analyzed by 14% SDS-PAGE, and protein was detected by staining with Coomassie blue.
Fractions chosen for further purification were concentrated to 1 to 3% of the volume of an approximately using spin concentrators with a 5-kDa-cutoff membrane (VivaSpin; GE Healthcare). A sample was then filtered and injected onto a 200-ml Superdex 75 gel filtration column (GE Healthcare), where it was isocratically eluted with Qa buffer at a flow rate of 1.5 ml/min. The purity of wild-type UmuD fractions was analyzed by SDS-PAGE. Pure UmuD fractions were combined and concentrated to obtain a concentration of about 500 μM. UmuD(G129S) was purified further using a DEAE column as described above. Fractions of UmuD(G129S) for which the yield of protein was low were concentrated and analyzed by immunoblotting as described below. The UmuD concentration was determined by the Bradford assay. The UmuD cleavage assay was carried out as described previously using LG buffer with 0.25 μM UmuD and approximately 0.2 μM UmuD(G129S) (4).
Immunoblotting.
Western blots were used to determine levels of UmuD, UmuD′, and UmuC expression from low-copy-number plasmids. The antibodies used do not detect UmuD, UmuD′, or UmuC in non-UV-induced cells under most conditions; therefore, we UV irradiated cells harboring plasmids expressing the variants of interest (3, 4). To detect UV-induced expression, ca. 2.5 × 1010 cells of an exponentially growing culture at an optical density at 600 nm of 0.2 to 0.3 were harvested, washed in 0.85% saline, and UV irradiated (25 J/m2) with a USHIO 15-W G15T8 UV lamp, essentially as described previously (4). Irradiated cells were then transferred to Luria-Bertani medium supplemented with spectinomycin and grown at 37°C for 2 to 3 h. After the cells were harvested by centrifugation, they were resuspended in 50 μl 0.85% saline and 50 μl loading buffer (25 mM Tris-HCl, 5% glycerol, 1% SDS, 25 mM β-mercaptoethanol, 0.05% bromophenol blue). Cells were then lysed by boiling them for 15 min, and 15-μl aliquots were loaded on 4 to 20% SDS-polyacrylamide gradient gels (Pierce). Electrophoresed proteins were transferred to a polyvinylidene difluoride membrane (Millipore) in 10 mM CAPS (N-cyclohexyl-3-aminopropanesulfonic acid) (pH 10) (adjusted with NaOH), 10% methanol. After blocking with 5% milk in TBS-Tween (20 mM Tris-HCl [pH 7.6], 137 mM NaCl, 0.1% Tween 20), the membranes were probed with anti-UmuC (3) or anti-UmuD/D′ (5) in 2.5% milk in TBS-Tween, which was followed by several washes in TBS-Tween, and subsequently probed with DyLight 649 goat anti-rabbit antibody (Pierce). Bound antibodies were detected with a Storm 860 phosphorimager using excitation at 635 nm. Alternatively, blots were probed with horseradish peroxidase-conjugated goat anti-rabbit antibody, developed with SuperSignal chemiluminescence reagent (Pierce), and imaged using X-ray film.
Peptide array blots.
A peptide array containing UmuC peptide sequences representing the entire protein was prepared with an Abimed peptide arrayer (MIT Biopolymers Core Facility). Each peptide consisted of 12 residues offset by two residues from the preceding peptide. The arrays were probed overnight with 50 nM β protein, which was detected with antibodies against β clamp (gifts from Charles McHenry, University of Colorado Health Sciences Center). The blocking, washing, and development procedures used have been described previously (31).
RESULTS
Overexpression of the β processivity clamp exacerbates the umuDC-mediated cold-sensitive phenotype: basis for selection.
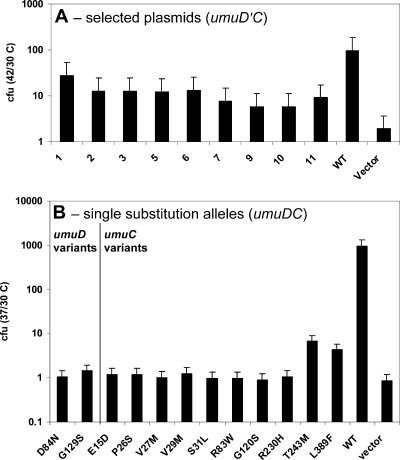
Overexpression of the umuDC gene products leads to a cold-sensitive phenotype, which has been hypothesized to be a manifestation of a hyperactive DNA damage checkpoint that results from a decrease in the rate of DNA replication after a cell experiences DNA damage (27, 50). Elevated levels of the β processivity clamp exacerbate the umuDC- and umuD′C-mediated cold-sensitive phenotype in E. coli, likely due to specific protein-protein interactions (46, 50). This observation has been used to uncover novel variants of the β clamp that do not interact with the umuD′C gene products (46). We sought to perform a complementary experiment in the hope of identifying variants that perturb the interaction of the umuD′C gene products with the β clamp. We screened for variants of umuD′C that do not exhibit the cold-sensitive phenotype in the presence of elevated levels of the β clamp, expecting that the variants that were selected would include variants that altered the ability of the umuD′C gene products to interact with the β clamp. We transformed cells that express elevated levels of the β clamp with a collection of mutagenized plasmids and selected, confirmed, and sequenced transformants that grew at a nonpermissive temperature, 30°C (Fig. 1A and Table 2). We identified nine alleles of umuD′C that suppress the cold-sensitive phenotype in the presence of elevated levels of the β clamp (Table 2).
FIG. 1.
Novel alleles of umuD′C suppress the cold-sensitive growth phenotype. The ratio of the number of CFU at the permissive temperature to the number of CFU at the nonpermissive temperature is plotted for (A) each allele in the presence of elevated levels of the β clamp (pJRC210) or (B) each single amino acid change in the absence of elevated levels of the β clamp. (A) The selected alleles suppress cold sensitivity due to elevated levels of umuD′C (pGY9738) and the β clamp (pJRC210). The wild-type plasmid is pGY9738. (B) All but two of the individual point mutations in umuDC suppress umuDC-mediated cold sensitivity in AB1157. The UmuC(T243M) and UmuC(L389F) variants confer a partial cold-sensitive phenotype compared to wild-type UmuC. The wild-type plasmid is pGY9739. In both panels A and B, the empty vector is pGB2. WT, wild type; cfu 42/30 C, 42°C/30°C ratio; cfu 37/30 C, 37°C/30°C ratio.
TABLE 2.
Nucleotide sequence analysis of umuD′C alleles containing missense mutations
| Plasmid | Nucleotide substitutiona | Deduced amino acid substitutionb |
|---|---|---|
| pGYHA-1 | G732A (TGC→TAC) | UmuC (Cys105Tyr) |
| G841A (CAG→CAA) | Silent | |
| pGYHA-2 | G384A (GGT→AGT) | UmuD′ (Gly129Ser) |
| G841A (CAG→CAA) | Silent | |
| pGYHA-3 | C1583T (CTC→TTC) | UmuC (Leu389Phe) |
| pGYHA-5 | C1146T (ACG→ATG) | UmuC (Thr243Met) |
| pGYHA-6 | G463C (GAG→GAC) | UmuC (Glu15Asp) |
| C665T (CGG→TGG) | UmuC (Arg83Trp) | |
| pGYHA-7 | C494T (CCG→TCG) | UmuC (Pro26Ser) |
| C510T (TCG→TTG) | UmuC (Ser31Leu) | |
| pGYHA-9 | G497A (GTG→ATG) | UmuC (Val27Met) |
| G503A (GTG→ATG) | UmuC (Val29Met) | |
| pGYHA-10 | G249A (GAT→AAT) | UmuD′ (Asp84Asn) |
| pGYHA-11 | G778A (GGC→AGC) | UmuC (Gly120Ser) |
| G1107A (CGC→CAC) | UmuC (Arg230His) |
For nucleotide numbering the full-length first codon of UmuD (ATG) was considered position 1.
Amino acid numbering begins with position 1 for each protein (UmuD or UmuC).
We also wanted to determine whether these variants were able to suppress the cold-sensitive phenotype caused by elevated levels of the umuDC gene products, independent of elevated levels of the β clamp. As noted above, elevated levels of the umuD′C gene products cause the cold-sensitive phenotype only in the presence of the β clamp (46), and such a system was used for the selection described above, but elevated levels of the umuDC gene products confer cold sensitivity on their own. Thus, we reconstructed each of the mutations obtained in the selection experiment discussed above in the context of a plasmid (pGY9739) expressing full-length umuDC, which confers a cold-sensitive phenotype on strains harboring this plasmid. Several of the plasmids identified in the screen contained two mutations (Table 2), and these mutations were constructed individually. These constructs having mutations in the full-length umuDC genes were then assayed for the ability to cause the cold-sensitive phenotype using a quantitative transformation assay (Fig. 1B). We found that nearly all of the single variants of either UmuD or UmuC suppressed the cold-sensitive phenotype in the absence of elevated levels of the β clamp, suggesting that they may suppress the cold-sensitive phenotype through a β clamp-independent mechanism. However, strains harboring plasmids expressing either the UmuC(L389F) or UmuC(T243M) variant exhibited an intermediate cold-sensitive phenotype for growth in the absence of elevated levels of the β clamp (Fig. 1B). This observation suggests that UmuC residues 389 and 243 are specifically involved in interactions with the β clamp that result in exacerbation of the cold-sensitive phenotype. Notably, no transformants were obtained with either AB1157 or its ΔumuDC derivative GW8017 when pGY9739(umuDC)-C105Y was used at either temperature, whereas strains with pGY9738(umuD′C)-C105Y (pGYHA-1) exhibited a much milder detrimental effect on growth. This suggests that in the context of umuDC, the umuC(C105Y) variant is lethal, whereas in the context of the shorter umuD′C construct, the C105Y substitution does not affect viability.
Levels of steady-state expression of selected UmuDC variants.
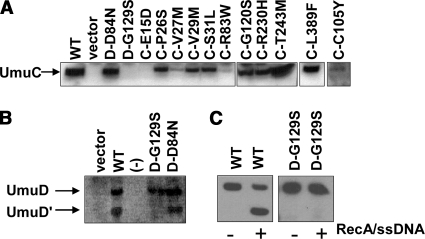
In principle, the cold-sensitive phenotype could be suppressed due to specific mutations that disrupt interaction of the umuD′C or umuDC gene products either with the β clamp or with other cellular components important for the phenotype (46, 50). However, the cold-sensitive phenotype could also be suppressed by mutations, such as a promoter mutation, that decrease the expression of umuD′C or umuDC or by mutations that decrease the stability of UmuD′, UmuD, or UmuC. In order to identify alleles that possibly suppressed cold sensitivity because of the latter processes, we determined the expression levels of the selected variants in UV-irradiated cells by immunoblotting with antibodies specific to UmuD/D′ or UmuC (Fig. 2). Several of the selected UmuC variants that suppress the cold-sensitive phenotype result in wild-type or nearly wild-type levels of UmuC expression (Fig. 2A). A few of the selected variants result in reduced steady-state levels of UmuC; in particular, UmuC variants E15D, V27M, R83W, and C105Y result in reduced steady-state levels (Fig. 2A). In addition, there is a substantially reduced amount of UmuC with the plasmid that harbors the UmuD(G129S) variant (Fig. 2A).
FIG. 2.
Steady-state levels of UmuC (A) and UmuD (B) variants as determined by immunoblotting. (A) UmuC was expressed from the pGY9739 (umuDC) plasmids in all cases except UmuC(C105Y), which was expressed from pGY9738 (umuD′C). (B) In vivo cleavage efficiency of UmuD variants expressed from the pGY9739 plasmids. Cells were harvested 3 h after UV irradiation. (C) Purified wild-type UmuD and UmuD(G129S) were assayed for proficiency for cleavage in vitro with a RecA-ssDNA nucleoprotein filament, the presence of which is indicated by a plus sign and the absence of which is indicated by a minus sign. We did not detect cleavage of UmuD(G129S). Because of the low yield of purified UmuD(G129S), the results were detected by immunoblotting. WT, wild type; D, UmuD; C, UmuC.
The levels of UmuD expressed from plasmids expressing the UmuD(D84N) or UmuD(G129S) variant appear to be normal (Fig. 2B). This experiment also measured UmuD cleavage, as indicated by the amount of UmuD′ present (Fig. 2B). The UmuD(D84N) variant appears to have normal cleavage activity, but the UmuD(G129S) variant does not show detectable cleavage under the conditions of the assay. It is possible that this variant is cleaved but the resulting UmuD′ protein is rapidly degraded. In order to assay the cleavage proficiency of UmuD(G129S) in vitro, we overexpressed and purified the UmuD(G129S) variant and found that it cannot be cleaved in vitro (Fig. 2C). It has previously been observed that the UmuD(G129D) variant is noncleavable (1, 28), suggesting that UmuD cleavage is exquisitely sensitive to changes at position 129.
Effect of selected umuD′C alleles on UV mutagenesis.
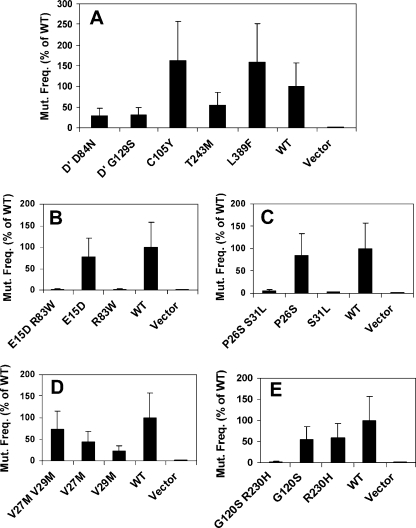
The UmuD′2C function is required for induced mutagenesis upon exposure to UV light (14), as detected by the reversion of an auxotrophic marker, argE3 (4). To assess whether the variants that are predicted to disrupt interactions with the β clamp also have an effect on induced mutagenesis, we determined the mutation frequency after exposure to UV irradiation of each of the selected umuD′C variants, as well as the analogous single variants constructed in the context of the plasmid harboring umuD′C (Fig. 3). In these experiments, we used a synthetic plasmid construct that contains the umuD′ and umuC genes in order to circumvent the complication that UmuD(G129S) prevents UmuD cleavage, which is required for UV-induced mutagenesis. Strains harboring plasmids expressing either of the two umuD′ alleles obtained in this selection experiment, umuD′(D84N) and umuD′(G129S), each exhibit an approximately threefold reduction in UV-induced mutagenesis.
FIG. 3.
UV-induced mutation frequency of selected alleles in plasmid pGY9738 (umuD′C) in strain GW8017 (ΔumuDC). The wild-type plasmid was pGY9738, and the empty vector was pGB2. (A) Mutation frequency conferred by each single mutation in the umuD′C genes. (B to E) Mutation frequencies conferred by the double mutations found in the selection experiment together with the corresponding single mutations. WT, wild type; Mut. Freq., mutation frequency; D′, UmuD′.
It has previously been shown that there are at least two classes of UmuC variants that disrupt binding to the β clamp: variants that essentially abolish UV-induced mutagenesis and umuDC-mediated cold sensitivity (2, 3) and variants that have no effect on UV-induced mutagenesis but abolish umuDC-mediated cold sensitivity (3). Here we identified a third category of UmuC variants that disrupt binding to the β clamp: UmuC(T243M) and UmuC(L389F) each have only modest effects on UV-induced mutagenesis (±2-fold) (Fig. 3) and on the umuDC-mediated cold sensitivity seen in the absence of the β clamp (Fig. 1), but they disrupt the cold-sensitive phenotype observed when both umuD′C and the β clamp are present at elevated levels (Fig. 1).
Several of the variants obtained in this selection experiment confer severely reduced UV-induced mutagenesis (Fig. 3). Based on a homology model of UmuC (3), we predict that many of changes in these variants are either near the active site or on the surface of the protein (Fig. 4). Also notable is the apparent lack of correlation between the levels of expression of UmuC (Fig. 2) and UV-induced mutagenesis (Fig. 3). This is especially clear when UmuC(V27M) (reduced expression; UV-induced mutagenesis is 43% of wild-type UV-induced mutagenesis), UmuC(V29M) (nearly wild-type expression levels; UV-induced mutagenesis is 22% of wild-type UV-induced mutagenesis), and UmuC(E15D) (severely reduced expression; UV-induced mutagenesis is 77% of wild-type UV-induced mutagenesis) are compared. Under the conditions used here, we were not be able to detect expression of chromosomally encoded UmuC, yet the activity of this protein in UV-induced mutagenesis was observed under comparable conditions.
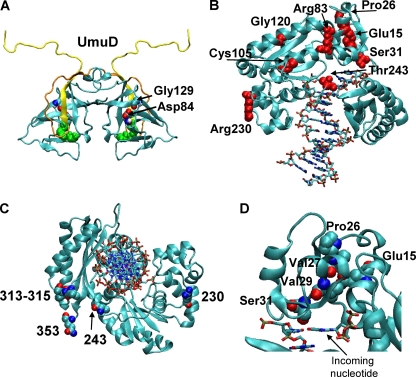
FIG. 4.
Predicted locations of selected residues in homology models of UmuD (A) and UmuC (B, C, and D). (A) Homology model of UmuD (5) with Asp84 and Gly129 highlighted by space-filling rendering, with different colors indicating different kinds of atoms. The Ser60-Lys97 active site dyad is highlighted by green space-filling rendering. The N-terminal 24 amino acids that are removed to form UmuD′ are yellow, while the rest of the arm is orange. (B, C and D) Homology model for residues 1 to 353 of UmuC (3) with the DNA indicated by lines. (B) Eight of the selected residues are shown in red space-filling rendering and labeled explicitly. (C) Residues, shown in space-filling rendering, of UmuC implicated as residues that are important for binding to the β clamp. The model of UmuC with DNA is shown oriented along the DNA helical axis. The canonical β clamp binding motif (residues 357 to 361) is not included in the model; therefore, the C-terminal residue of the model, residue 353, is explicitly indicated in space-filling rendering. Residues 313 to 315 comprise the second β-interaction site. (D) Five of the selected residues, four of which lie along one β strand, are shown in space-filling rendering, with different colors indicating different kinds of atoms. The images were created with VMD (21).
UV sensitivity of selected umuD alleles.
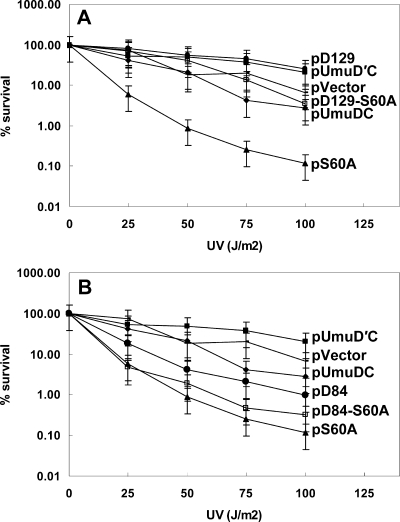
Noncleavable variants of UmuD typically exacerbate the cold-sensitive phenotype that is due to elevated levels of the umuDC gene products (34), as well as cause sensitivity to UV light (1, 33). Given the observation that the UmuD(G129S) variant is noncleavable (Fig. 2B and C) and the observation that this variant protein suppresses the cold-sensitive phenotype (Fig. 1), we examined whether strains containing it were sensitive to UV light. Strains harboring plasmids expressing the G129S variant were not sensitive to UV light, similar to strains expressing UmuD′ (Fig. 5A). In contrast, cells expressing the UmuD(S60A) variant, which is not cleavable because the catalytic serine has been mutated, show marked sensitivity to UV light (Fig. 5A). At least one noncleavable UmuD variant (the T14A L17A F18A triple mutant) that does not confer extreme sensitivity to UV light has been identified (5). We suspected that the noncleavable UmuD(G129S) variant might suppress the sensitivity of the UmuD(S60A) variant to UV light. We constructed a variant with G129S in combination with the UmuD(S60A) active site variant and assayed cells containing the double mutant for resistance to UV light. The sensitivity of cells containing the UmuD double variant, UmuD(S60A G129S), to killing by UV irradiation was essentially the same as that of a strain expressing wild-type UmuD, indicating that the G129S mutation suppresses the UV killing conferred by the S60A mutation (Fig. 5A). Thus, we identified a UmuD mutation in the globular domain of the protein that renders UmuD noncleavable, yet suppresses the UV sensitivity of the noncleavable S60A variant. Mutations in the N-terminal arm (T14A L17A F18A) of UmuD that result in similar behavior were identified previously (5). Strains expressing the UmuD(D84N) variant exhibited normal sensitivity to UV light, as expected for a cleavable UmuD variant, whereas strains expressing UmuD(S60A D84N) exhibited increased sensitivity to killing by UV light that is comparable to that of the S60A variant (Fig. 5B).
FIG. 5.
UV survival of GW8017 strains harboring plasmids expressing UmuD variants (A) G129S and (B) D84N. (A) Noncleavable UmuD(G129S) suppresses UV sensitivity that is characteristic of noncleavable UmuD variants. UmuD(G129S) also suppresses the extreme UV sensitivity of UmuD(S60A). •, pGY9739-umuDC-D129 (pD129); ▪, pGY9738-umuD′C (pUmuD′C); −, pGB2 (pVector); □, pGY9739- umuDC-DS60A, G129S (pD129-S60A); ⧫, pGY9739-umuDC (pUmuDC); ▴, pGY9739-umuDC-DS60A (pS60A). (B) UmuD(D84N) is cleavable, and strains harboring plasmids expressing this variant are resistant to UV light. A strain expressing UmuD(S60A D84N) is sensitive to UV light, which is characteristic of a strain expressing UmuD(S60A). ▪, pGY9738-umuD′C (pUmuD′C); −, pGB2 (pVector); ⧫, pGY9739-umuDC (pUmuDC); •, pGY9739-umuDC-D84 (pD84); □, pGY9739-umuDC-D84, S60A (pD84-S60A); ▴, pGY9739-umuDC-DS60A (pS60A).
Binding defects of selected umuD′C alleles.
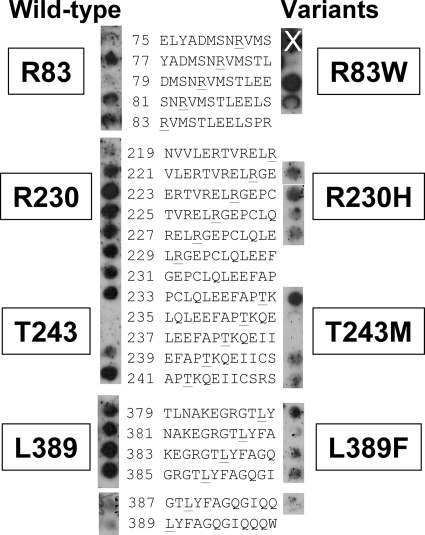
We next investigated whether the UmuC residues implicated by selection as residues that interact with the β clamp could affect direct physical contact with the β clamp. We focused on four residues which, based on our homology model (Fig. 4), seem likely to be exposed or partially exposed on the surface of UmuC: R83, R230, T243, and L389 (Fig. 4 and 6). In order to test the contributions of the selected UmuC residues to the physical interactions between UmuC and the β clamp, we constructed peptide arrays on derivatized nitrocellulose membranes of overlapping 12-mer peptides spanning the entire UmuC protein (13). The peptides represent both the wild-type UmuC sequence and the mutations found in the selection experiment. The membrane was probed with purified β and washed, and the bound protein was detected using anti-β clamp antibodies. Importantly, it appears that the β clamp interacts with peptides that include each of the four amino acids of interest, R83, R230, T243, and L389. Thus, the mutations obtained in the selection experiment identify potential β clamp interaction sites on the surface of UmuC, and peptides from these regions of UmuC interact directly with the β clamp. The R83W mutation seems to cause a slight decrease in binding of the β clamp, but only in one of the three peptides examined. The presence of mutations at residues R230, T243, and L389 decreases but does not completely disrupt binding by the β clamp (Fig. 6).
FIG. 6.
Peptide array blot showing selected UmuC residues and their corresponding mutations, probed with wild-type β protein. Only the UmuC residues predicted to be surface exposed are shown. The blots on the left contained the wild-type UmuC sequences shown, and the blots on the right contained the sequences with the underlined residue mutated, as indicated in Table 2. The “X” indicates a spot that is not meaningful, because the antibody control reacted at this position (data not shown).
Model-based analysis of selected umuD′C alleles.
We mapped the two residues of UmuD identified in the selection experiment on the available structures of UmuD′2 (12, 35) and our homology model of UmuD2 (5). Although the D84N and G129S mutations conferred similar phenotypes in the context of UmuD′, they had remarkably different effects in the context of UmuD. The G129S variant is noncleavable yet confers resistance to UV light, whereas the D84N variant is cleavable and, as is typical for cleavable UmuD variants, also confers resistance to UV light. Both variants suppress the cold-sensitive phenotype and show somewhat reduced UV-induced mutagenesis. Both variants were mapped onto a homology model of UmuD (Fig. 4), which was derived from the crystal structure of LexA (5, 24). In both UmuD and UmuD′, G129 appears to be mostly buried and D84 appears to be partially buried and involved in main chain and side chain hydrogen bonding interactions with S81. Intriguingly, G129 does not appear to be near the active site, involved in interactions with the N-terminal arms, or near the dimer interface, although mutations in this residue might decrease dimer formation (see Discussion) (28, 42).
We used our previously developed homology model of UmuC (3), based on the crystal structure of Dpo4 (23), to analyze the potential roles of the UmuC variants. This model includes only residues 1 to 353, so L389 cannot be mapped in it. Based on this model, it seems likely that only residues R83, R230, and T243 are exposed on the UmuC surface (Fig. 4). R83 is predicted to be in the middle of α-helix D (6) and nearly fully exposed. R83W, the variant obtained in the selection experiment, results in nearly complete loss of UV-induced mutagenesis, loss of the cold-sensitive phenotype, and reduced steady-state protein levels (Table 3). Thus, it seems likely that this variant results in a perturbed protein conformation; however, it is possible that it disrupts other protein-protein interactions necessary for both UV-induced mutagenesis and the cold-sensitive phenotype. R230 is predicted to be at the C-terminal end of α-helix K (6) and nearly fully exposed. The R230H variant is proficient for UV-induced mutagenesis and results in nearly wild-type steady-state protein levels, but it suppresses the cold-sensitive phenotype (Table 3). It also appears to result in a decrease in binding to the β clamp (Fig. 6). T243 is in the middle of a long peptide that spans the width of the DNA and joins the little finger domain to the polymerase domain; it is predicted to be nearly fully exposed. UmuC(T243M), found in this work, results in a modest reduction in UV-induced mutagenesis and unperturbed steady-state protein levels (Table 3). UmuC(T243M) also suppresses the cold-sensitive phenotype that results from elevated levels of both the β clamp and the umuDC gene products, but it only partially suppresses the umuDC-dependent cold-sensitive phenotype (Table 3), suggesting that this residue is an interaction site for the β clamp (Fig. 1). Peptide array mapping also suggests that this residue is an interaction site for the β clamp, and the T243M mutation results in a slight reduction in binding (Fig. 6). Mapping these exposed residues on the UmuC homology model allowed us to more fully define the β clamp interaction sites on UmuC (Fig. 4C).
TABLE 3.
Effects of selected variantsa
| Variant | Cold sensitivity (37°C/30°C ratio)b | SOS mutagenesis (% of wild type)c | Protein levelsd | Cleavage (UmuD)/ β binding (UmuC)e |
|---|---|---|---|---|
| UmuD | ||||
| Wild typef | 950 | 100 | + | + |
| D84N | 1.1 | 30 | + | + |
| G129S | 1.4 | 32 | + | − |
| UmuC | ||||
| Wild typef | 950 | 100 | + | NA |
| E15D | 1.2 | 77 | − | ND |
| P26S | 1.2 | 84 | + | ND |
| V27M | 1.0 | 43 | ± | ND |
| V29M | 1.3 | 22 | + | ND |
| S31L | 1.0 | 2.7 | + | ND |
| R83W | 1.0 | 1.9 | ± | + |
| C105Y | ND | 160 | ± | ND |
| G120S | 0.89 | 55 | + | ND |
| R230H | 1.1 | 59 | + | + |
| T243M | 6.7 | 54 | + | + |
| L389F | 4.3 | 160 | + | + |
| Vector (pGB2) | 0.87 | 1.0 | − | NA |
ND, not determined; NA, not applicable.
See Fig. 1B.
See Fig. 3.
See Fig. 2. +, ±, and −, wild type, intermediate, and undetectable, respectively.
In cold sensitivity and immunoblotting experiments, pGY9739 (umuDC) is the wild-type plasmid; in SOS mutagenesis experiments, pGY9738 (umuD′C) is the wild-type plasmid.
There are now β clamp interaction sites that have been identified, including the canonical β clamp interaction motif (residues 357 to 361) and the second β clamp interaction site (residues 313 to 315), along with R230 and T243, that form a ring around UmuC (Fig. 4C). R83 is unlikely to be part of this ring as it is likely to be quite far from this “back” side of UmuC. The mutations found in the experiment described here would be expected to disrupt β clamp binding to UmuC as they result in nonconservative amino acid changes. The T243M mutation changes a small, polar amino acid to a large, aliphatic amino acid, whereas the R230H mutation replaces a large, basic amino acid with a smaller amino acid. L389F is a conservative substitution, which suggests that position 389 is exquisitely sensitive to the identity of the amino acid.
Several mutations identified in this work are at positions that appear to be buried. It is interesting that while umuD′C(C105Y) has no defect in UV-induced mutagenesis, a plasmid harboring umuDC(C105Y) is apparently toxic, as AB1157 transformants of the plasmid containing this variant could not be obtained (but XL1-Blue transformants were obtained). G120 is buried and predicted to be in the middle of α-helix E (6). The G120S mutation, a mutation from a small nonpolar amino acid to polar amino acid, suppresses the cold-sensitive phenotype, but it does not result in perturbed UmuC protein steady-state expression levels or UV-induced mutagenesis (Table 3).
Closer to the predicted location of the active site, UmuC residue E15 is in the middle of α-helix A (6) and appears to be nearly buried. E15D, a fairly conservative substitution, suppresses the umuDC-dependent cold-sensitive phenotype and results in a dramatic decrease in the steady-state UmuC protein levels. However, the E15D mutation results in little change in UV-induced mutagenesis compared to that of the wild type.
Residues P26, V27, V29, and S31 lie along β strand 2 (6), and S31 is predicted to make a close approach to the incipient base pair (Fig. 4D). Mutations in each of these residues suppress the umuDC-dependent cold-sensitive phenotype. The UV-induced mutagenesis of strains harboring plasmids expressing umuC variants at these positions decreases with increasing proximity to the incipient base pair, suggesting that this β strand plays a role in efficient UV-induced mutagenesis. UmuC residue S31, in particular, is important for UV-induced mutagenesis and has been identified as a “flue” residue that is responsible for plugging the DNA- and lesion-binding channel (8). Moreover, according to our model, the side chain of S31 is predicted to be within approximately 5 Å of the deoxyribose moiety of the template base, which may imply that it has a role in correct positioning of the template to facilitate efficient base pair formation. The S31L mutation, which changes a small, polar amino acid to a larger, aliphatic amino acid, may result in a decrease in UV-induced mutagenesis due to an inability to correctly position the DNA template.
DISCUSSION
UmuC, together with UmuD′2 and other factors, acts in translesion synthesis, particularly in the bypass of abasic sites and adducts resulting from UV light (16, 36, 37, 40, 51). UmuC, together with intact UmuD2, plays a role in a primitive DNA damage checkpoint, specifically decreasing the rate of DNA synthesis (27, 33, 50). The cold-sensitive growth phenotype that is observed upon umuDC overexpression is thought to be an exaggeration of this checkpoint (50). A similar effect on the reduction of the rate of pol III-dependent DNA synthesis has also been observed for DNA pol IV (17). It has been found that pol IV can displace pol III from the β clamp, suggesting that pol IV also acts in a primitive DNA damage checkpoint, a function that depends on its interactions with the β clamp (17).
Modestly elevated levels of umuD′C products, when combined with elevated levels of the β clamp, also confer a cold-sensitive growth phenotype (34, 46). This observation was previously exploited to select for variants of the β clamp that do not confer the cold-sensitive growth phenotype when they are present at elevated levels together with umuD′C (46). In this study, we carried out a similar selection experiment, but instead we selected for alleles of umuD′C that suppressed the cold-sensitive growth phenotype when they were present with elevated levels of the β clamp. In this selection experiment we found nine alleles of umuD′C that suppress the cold-sensitive growth phenotype; these alleles were two alleles of umuD′ and seven alleles of umuC, four of which result in two amino acid substitutions. A minority of the amino acid substitutions that we obtained in the selection experiment are implicated in direct interactions with the β clamp. The other amino acid substitutions were likely obtained because of their ability to disrupt interactions with other proteins or with DNA. Although the precise mechanism that results in the cold-sensitive phenotype is unknown, multiple protein-protein interactions are likely to regulate it, including interactions between UmuD or UmuD′ and UmuC (46, 49, 50).
This paper presents three major findings that result from characterization of the selected alleles. First, we defined additional sites where there are likely interactions between UmuC and the β clamp, specifically UmuC residues R230, T243, and L389. Unlike mutation of other known UmuC residues that interact with the β clamp (2, 3), mutation of these residues has modest effects on umuDC-mediated cold sensitivity and on UV-induced mutagenesis. However, each of the variants has a specific defect in conferring cold sensitivity in strains expressing elevated levels of both the umuD′C variant and the β clamp. This suggests that the phenotype that was the basis for selection is exquisitely sensitive to changes in interactions between the umuD′C gene products and the β clamp.
Second, we found four mutations (P26S, V27M, V29M, and S31L) in amino acids that lie along a β strand in UmuC that is predicted to lead directly to the UmuC active site. The proficiency of strains with these variants for UV-induced mutagenesis directly correlates with the distance of the variants from the active site. A detailed molecular modeling analysis of UmuC bound to DNA containing a benzo-[a]-pyrene lesion suggests that UmuC has a smaller channel (“chimney”) than DinB between the fingers and little finger with which to accommodate bulky lesions (8). Furthermore, this analysis suggested that a trio of residues, S31, N32 and N33, form a cap, or “flue” of the chimney, that is considered closed in the case of UmuC and contributes to the smaller gap of UmuC (8). We provide experimental evidence that UmuC residue S31 is important for UV-induced mutagenesis and presumably for translesion synthesis.
Finally, we obtained the G129S variant of UmuD′; the G129 residue is in the C-terminal globular domain of the protein and appears to be partially buried. UmuD(G129S) is likely only the second example (5) of a noncleavable UmuD variant that confers proficiency for UV-induced mutagenesis and suppresses the extreme UV sensitivity of the noncleavable UmuD(S60A) variant. Mutations in residue G129 have been found using other approaches; in particular, a screen carried out to identify variants of umuD that were defective in UV-induced mutagenesis found the UmuD(G129D) variant among the 10 unique missense alleles obtained (1). UmuD(G129D) was found to confer dramatically reduced UV-induced mutagenesis and to be defective for autocleavage, as well as to be moderately defective for homodimer formation as determined by cross-linking analysis (1, 32). A similar screen designed to find variants of umuD′ that do not promote SOS-dependent spontaneous mutagenesis uncovered G129D and G129S (28). Using a yeast two-hybrid assay, it was determined that UmuD′(G129S) is severely defective in dimerization, while UmuD′(G129D) is more proficient than wild-type UmuD′ in dimer formation (28). This is intriguing as G129 is predicted to be located in the middle of the C-terminal β strand that ends at the dimer interface of UmuD (Fig. 4A) (5, 12), and thus disruptions in the β strand could result in dimerization defects. A defect in dimerization, which would likely affect all UmuD functions, could explain several of the activities that we observed for the G129S variant, including suppression of the cold-sensitive phenotype, the lack of RecA-ssDNA-dependent cleavage, and suppression of UmuD(S60A)-dependent UV sensitivity. However, in our hands strains harboring plasmids expressing umuD′(G129S) together with umuC exhibited a small but significant decrease in UV-induced mutagenesis, a cellular function that is most likely dependent on UmuD′ homodimerization (16, 36, 37, 51).
The β clamp interacts with UmuD via both the N-terminal arm and the C-terminal globular domain of UmuD (48). The two mutations of umuD′ that we uncovered are in the C-terminal globular domain, but they are likely to be at least partially buried (Fig. 4A). In nearly every allele found in this selection experiment, for both umuD′ and umuC, the predicted amino acid change either is a change to a larger amino acid or results in a change in the charge (i.e., a change from a charged amino acid to an uncharged amino acid). This could easily result in a slightly altered protein conformation that would lead to perturbed presentation of an interaction surface of a protein. In the case of UmuD(G129S), it may also result in perturbation of the UmuD dimer interface (28).
In the case of UmuC, two regions are known to be involved in interactions with the β clamp, namely, the canonical β clamp binding motif (residues 357 to 361) and the so-called second β interaction site (residues 313 to 315) (2, 3). We obtained mutations at residues 230 and 243 in our selection experiment; these residues could complete a ring of residues around UmuC that form binding sites for the β clamp (Fig. 4C). This set of β clamp binding residues likely includes L389, which cannot be mapped on the UmuC model because homology between UmuC and other proteins with known structures ends near the canonical β clamp binding motif (residues 357 to 361) of UmuC (3, 23). There is no or extremely weak homology between the C-terminal 70 residues of UmuC and any protein whose structure is known.
Several of the residues of UmuC identified in our selection experiment are predicted to be buried or partially buried. It is unlikely that these residues were found in this experiment due to specific defects in binding to the β clamp. It seems more likely that there are myriad subtle structural defects that result in suppression of the cold-sensitive phenotype. Moreover, we found that apparently extremely low levels of UmuC are still proficient for UV-induced mutagenesis.
Y family DNA polymerases that are specialized for lesion bypass DNA synthesis play additional biological roles in the cell. In particular, elevated levels of the umuDC gene products also confer a cold-sensitive phenotype, which has been inferred to be an exaggeration of a DNA damage checkpoint. In this work, we obtained new alleles of umuC that genetically separate its roles in UV-induced mutagenesis and in a cold-sensitive phenotype, supporting the idea that these roles are distinct roles of the umuDC gene products.
Acknowledgments
This work was supported by a Dreyfus Foundation New Faculty Award to P.J.B. P.J.B. is a Cottrell Scholar of the Research Corporation for Science Advancement. This work was also supported by grant CA21615 from the National Cancer Institute and NIEHS Center grant P30ES02109 from the MIT Center for Environmental Health Sciences to G.C.W., by a postdoctoral fellowship from the Damon Runyon Cancer Research Foundation to P.J.B., and by funds from the MIT UROP program to S.C. G.C.W. is an American Cancer Society Research Professor.
We thank Charles McHenry for plasmid pJRC210. We also thank Susan Cohen, Kathryn Jones, and Sharotka Simon for helpful discussions and for critical reading of the manuscript.
Footnotes
Published ahead of print on 24 July 2009.
REFERENCES
- 1.Battista, J. R., T. Ohta, T. Nohmi, W. Sun, and G. C. Walker. 1990. Dominant negative umuD mutations decreasing RecA-mediated cleavage suggest roles for intact UmuD in modulation of SOS mutagenesis. Proc. Natl. Acad. Sci. USA 87:7190-7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becherel, O. J., R. P. P. Fuchs, and J. Wagner. 2002. Pivotal role of the β-clamp in translesion DNA synthesis and mutagenesis in E. coli cells. DNA Repair 1:703-708. [DOI] [PubMed] [Google Scholar]
- 3.Beuning, P. J., D. Sawicka, D. Barsky, and G. C. Walker. 2006. Two processivity clamp interactions differentially alter the dual activities of UmuC. Mol. Microbiol. 59:460-474. [DOI] [PubMed] [Google Scholar]
- 4.Beuning, P. J., S. M. Simon, V. G. Godoy, D. F. Jarosz, and G. C. Walker. 2006. Characterization of Escherichia coli translesion synthesis polymerases and their accessory factors. Methods Enzymol. 408:318-340. [DOI] [PubMed] [Google Scholar]
- 5.Beuning, P. J., S. M. Simon, A. Zemla, D. Barsky, and G. C. Walker. 2006. A non-cleavable UmuD variant that acts as a UmuD′ mimic. J. Biol. Chem. 281:9633-9640. [DOI] [PubMed] [Google Scholar]
- 6.Boudsocq, F., H. Ling, W. Yang, and R. Woodgate. 2002. Structure-based interpretation of missense mutations in Y-family DNA polymerases and their implications for polymerase function and lesion bypass. DNA Repair 1:343-358. [DOI] [PubMed] [Google Scholar]
- 7.Bunting, K. A., S. M. Roe, and L. H. Pearl. 2003. Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the β-clamp. EMBO J. 22:5883-5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandani, S., and E. L. Loechler. 2009. Y-family DNA polymerases may use two different dNTP shapes for insertion: a hypothesis and its implications. J. Mol. Graph. Model. 27:759-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Churchward, G., D. Belin, and Y. Nagamine. 1984. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 31:165-171. [DOI] [PubMed] [Google Scholar]
- 10.Courcelle, J., A. Khodursky, B. Peter, P. O. Brown, and P. C. Hanawalt. 2001. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158:41-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalrymple, B. P., K. Kongsuwan, G. Wijffels, N. E. Dixon, and P. A. Jennings. 2001. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc. Natl. Acad. Sci. USA 98:11627-11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferentz, A. E., T. Opperman, G. C. Walker, and G. Wagner. 1997. Dimerization of the UmuD′ protein in solution and its implications for regulation of SOS mutagenesis. Nat. Struct. Biol. 4:979-983. [DOI] [PubMed] [Google Scholar]
- 13.Frank, R. 2002. The SPOT-synthesis technique: synthetic peptide arrays on membrane supports—principles and applications. J. Immunol. Methods 267:13-26. [DOI] [PubMed] [Google Scholar]
- 14.Friedberg, E. C., G. C. Walker, W. Siede, R. D. Wood, R. A. Schultz, and T. Ellenberger. 2006. DNA repair and mutagenesis, 2nd ed. ASM Press, Washington, DC.
- 15.Fujii, S., and R. P. Fuchs. 2004. Defining the position of the switches between replicative and bypass DNA polymerases. EMBO J. 23:4342-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujii, S., V. Gasser, and R. P. Fuchs. 2004. The biochemical requirements of DNA polymerase V-mediated translesion synthesis revisited. J. Mol. Biol. 341:405-417. [DOI] [PubMed] [Google Scholar]
- 17.Furukohri, A., M. F. Goodman, and H. Maki. 2008. A dynamic polymerase exchange with Escherichia coli DNA polymerase IV replacing DNA polymerase III on the sliding clamp. J. Biol. Chem. 283:11260-11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godoy, V. G., D. F. Jarosz, S. M. Simon, A. Abyzov, V. Ilyin, and G. C. Walker. 2007. UmuD and RecA directly modulate the mutagenic potential of the Y family DNA polymerase DinB. Mol. Cell 28:1058-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guzzo, A., M. H. Lee, K. Oda, and G. C. Walker. 1996. Analysis of the region between amino acids 30 and 42 of intact UmuD by a monocysteine approach. J. Bacteriol. 178:7295-7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heltzel, J. M. H., S. K. Scouten Ponticelli, L. H. Sanders, J. M. Duzen, V. Cody, J. Pace, E. H. Snell, and M. D. Sutton. 2009. Sliding clamp-DNA interactions are required for viability and contribute to DNA polymerase management in Escherichia coli. J. Mol. Biol. 387:74-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD—visual molecular dynamics. J. Mol. Graph. 14:33-38. [DOI] [PubMed] [Google Scholar]
- 22.Kornberg, A., and T. A. Baker. 1992. DNA replication, 2nd ed. W.H. Freeman & Company, New York, NY.
- 23.Ling, H., F. Boudsocq, R. Woodgate, and W. Yang. 2001. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell 107:91-102. [DOI] [PubMed] [Google Scholar]
- 24.Luo, Y., R. A. Pfuetzner, S. Mosimann, M. Paetzel, E. A. Frey, M. Cherney, B. Kim, J. W. Little, and N. C. J. Strynadka. 2001. Crystal structure of LexA: a conformational switch for regulation of self-cleavage. Cell 106:585-594. [DOI] [PubMed] [Google Scholar]
- 25.Maor-Shoshani, A., and Z. Livneh. 2002. Analysis of the stimulation of DNA polymerase V of Escherichia coli by processivity proteins. Biochemistry 41:14438-14446. [DOI] [PubMed] [Google Scholar]
- 26.Marsh, L., T. Nohmi, S. Hinton, and G. C. Walker. 1991. New mutations in cloned Escherichia coli umuDC genes: novel phenotypes of strains carrying a umuC125 plasmid. Mutat. Res. 250:183-197. [DOI] [PubMed] [Google Scholar]
- 27.Marsh, L., and G. C. Walker. 1985. Cold sensitivity induced by overproduction of UmuDC in Escherichia coli. J. Bacteriol. 162:155-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLenigan, M., T. S. Peat, E. G. Frank, J. P. McDonald, M. Gonzalez, A. S. Levine, W. A. Hendrickson, and R. Woodgate. 1998. Novel Escherichia coli umuD′ mutants: structure-function insights into SOS mutagenesis. J. Bacteriol. 180:4658-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Murli, S., T. Opperman, B. T. Smith, and G. C. Walker. 2000. A role for the umuDC gene products of Escherichia coli in increasing resistance to DNA damage in stationary phase by inhibiting the transition to exponential growth. J. Bacteriol. 182:1127-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niebuhr, K., and J. Wehland. 1997. Screening of antibody epitopes and regions of protein-protein interaction site using SPOT peptides, p. 797-800. In I. Lefkovits (ed.), Immunology methods manual: the comprehensive sourcebook of techniques. Academic Press, San Diego, CA.
- 32.Ohta, T., M. D. Sutton, A. Guzzo, S. Cole, A. E. Ferentz, and G. C. Walker. 1999. Mutations affecting the ability of the Escherichia coli UmuD′ protein to participate in SOS mutagenesis. J. Bacteriol. 181:177-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Opperman, T., S. Murli, B. T. Smith, and G. C. Walker. 1999. A model for a umuDC-dependent prokaryotic DNA damage checkpoint. Proc. Natl. Acad. Sci. USA 96:9218-9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Opperman, T., S. Murli, and G. C. Walker. 1996. The genetic requirements for UmuDC-mediated cold sensitivity are distinct from those for SOS mutagenesis. J. Bacteriol. 178:4400-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peat, T. S., E. G. Frank, J. P. McDonald, A. S. Levine, R. Woodgate, and W. A. Hendrickson. 1996. Structure of the UmuD′ protein and its regulation in response to DNA damage. Nature 380:727-730. [DOI] [PubMed] [Google Scholar]
- 36.Pham, P., S. Rangarajan, R. Woodgate, and M. F. Goodman. 2001. Roles of DNA polymerases V and II in SOS-induced error-prone and error-free repair in Escherichia coli. Proc. Natl. Acad. Sci. USA 98:8350-8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reuven, N. B., G. Arad, A. Maor-Shoshani, and Z. Livneh. 1999. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA, and SSB and is specialized for translesion replication. J. Biol. Chem. 274:31763-31766. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Schlacher, K., and M. F. Goodman. 2007. Lessons from 50 years of SOS DNA-damage-induced mutagenesis. Nat. Rev. Mol. Cell Biol. 8:587-594. [DOI] [PubMed] [Google Scholar]
- 40.Schlacher, K., K. Leslie, C. Wyman, R. Woodgate, M. Cox, and M. Goodman. 2005. DNA polymerase V and RecA protein, a minimal mutasome. Mol. Cell 17:561-572. [DOI] [PubMed] [Google Scholar]
- 41.Simmons, L. A., J. J. Foti, S. E. Cohen, and G. C. Walker. 2008. The SOS regulatory network. In A. Bock, R. Curtiss III, J. B. Kaper, P. D. Karp, F. C. Neidhardt, T. Nystrom, J. M. Slauch, C. L. Squires, and D. Ussary (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org.
- 42.Simon, S. M., F. J. Sousa, R. Mohana-Borges, and G. C. Walker. 2008. Regulation of Escherichia coli SOS mutagenesis by dimeric intrinsically disordered umuD gene products. Proc. Natl. Acad. Sci. USA 105:1152-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sommer, S., F. Boudsocq, R. Devoret, and A. Bailone. 1998. Specific RecA amino acid changes affect RecA-UmuD′C interaction. Mol. Microbiol. 28:281-291. [DOI] [PubMed] [Google Scholar]
- 44.Sommer, S., J. Knezevic, A. Bailone, and R. Devoret. 1993. Induction of only one SOS operon, umuDC, is required for SOS mutagenesis in Escherichia coli. Mol. Gen. Genet. 239:137-144. [DOI] [PubMed] [Google Scholar]
- 45.Sutton, M. D., J. M. Duzen, and R. W. Maul. 2005. Mutant forms of the Escherichia coli beta sliding clamp that distinguish between its roles in replication and DNA polymerase V-dependent translesion DNA synthesis. Mol. Microbiol. 55:1751-1766. [DOI] [PubMed] [Google Scholar]
- 46.Sutton, M. D., M. F. Farrow, B. M. Burton, and G. C. Walker. 2001. Genetic interactions between the Escherichia coli umuDC gene products and the β processivity clamp of the replicative DNA polymerase. J. Bacteriol. 183:2897-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutton, M. D., A. Guzzo, I. Narumi, M. Costanzo, C. Altenbach, A. E. Ferentz, W. L. Hubbell, and G. C. Walker. 2002. A model for the structure of the Escherichia coli SOS-regulated UmuD2 protein. DNA Repair 1:77-93. [DOI] [PubMed] [Google Scholar]
- 48.Sutton, M. D., I. Narumi, and G. C. Walker. 2002. Posttranslational modification of the umuD-encoded subunit of Escherichia coli DNA polymerase V regulates its interactions with the beta processivity clamp. Proc. Natl. Acad. Sci. USA 99:5307-5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sutton, M. D., T. Opperman, and G. C. Walker. 1999. The Escherichia coli SOS mutagenesis proteins UmuD and UmuD′ interact physically with the replicative DNA polymerase. Proc. Natl. Acad. Sci. USA 96:12373-12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sutton, M. D., and G. C. Walker. 2001. umuDC-Mediated cold sensitivity is a manifestation of functions of the UmuD2C complex involved in a DNA damage checkpoint control. J. Bacteriol. 183:1215-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang, M., I. Bruck, R. Eritja, J. Turner, E. G. Frank, R. Woodgate, M. O'Donnell, and M. F. Goodman. 1998. Biochemical basis of SOS-induced mutagenesis in Escherichia coli: reconstitution of in vitro lesion bypass dependent on the UmuD′2C mutagenic complex and RecA protein. Proc. Natl. Acad. Sci. USA 95:9755-9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang, M., P. Pham, X. Shen, J. S. Taylor, M. O'Donnell, R. Woodgate, and M. F. Goodman. 2000. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature 404:1014-1018. [DOI] [PubMed] [Google Scholar]
- 53.Wagner, J., S. Fujii, P. Gruz, T. Nohmi, and R. P. Fuchs. 2000. The β clamp targets DNA polymerase IV to DNA and strongly increases its processivity. EMBO Rep. 1:484-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang, W. 2003. Damage repair DNA polymerases Y. Curr. Opin. Struct. Biol. 13:23-30. [DOI] [PubMed] [Google Scholar]