Abstract
Members of the COG2244 protein family are integral membrane proteins involved in synthesis of a variety of extracellular polymers. In several cases, these proteins have been suggested to move lipid-linked oligomers across the membrane or, in the case of Escherichia coli MviN, to flip the lipid II peptidoglycan precursor. Bacillus subtilis SpoVB was the first member of this family implicated in peptidoglycan synthesis and is required for spore cortex polymerization. Three other COG2244 members with high similarity to SpoVB are encoded within the B. subtilis genome. Mutant strains lacking any or all of these genes (yabM, ykvU, and ytgP) in addition to spoVB are viable and produce apparently normal peptidoglycan, indicating that their function is not essential in B. subtilis. Phenotypic changes associated with loss of two of these genes suggest that they function in peptidoglycan synthesis. Mutants lacking YtgP produce long cells and chains of cells, suggesting a role in cell division. Mutants lacking YabM exhibit sensitivity to moenomycin, an antibiotic that blocks peptidoglycan polymerization by class A penicillin-binding proteins. This result suggests that YabM may function in a previously observed alternate pathway for peptidoglycan strand synthesis.
The Bacillus subtilis spoVB gene was first identified as a locus in which a mutation could produce a block at a late stage of spore development (14, 30). Analysis of this locus revealed that it encoded an apparent integral membrane protein (33), and a detailed analysis of a spoVB null mutant demonstrated a block at a very early step in synthesis of the spore cortex peptidoglycan (PG) (40). The mutant synthesized essentially no cortex and accumulated cytoplasmic PG precursors, the same phenotype found in other mutant strains blocked in functions known to be directly involved in PG polymerization (40). These results suggested that SpoVB plays a direct role in assembly or function of the spore PG synthesis apparatus.
PG synthesis is a highly conserved and complex process that must span the cell membrane (reviewed in reference 38). Soluble nucleotide-linked PG precursors are synthesized in the cytoplasm. N-Acetylmuramic acid with a pentapeptide side chain is then transferred to an undecaprenol lipid carrier to produce lipid I, with subsequent addition of N-acetylglucosamine to produce lipid II, undecaprenyl-pyrophosphoryl-N-acetylmuramic acid (pentapeptide)-N-acetylglucosamine. Lipid II is then flipped across the membrane via an unknown mechanism. Two families of proteins have been postulated to perform this function: the SEDS family of integral membrane proteins, including FtsW, RodA, and SpoVE (13), and, more recently, the COG2244 family (23), which includes SpoVB and the MviN (MurJ) protein of Escherichia coli (35). In both cases, loss of a protein within one of these families has been shown to result in a block in PG synthesis and the accumulation of lipid-linked and/or soluble PG precursors (16, 20, 35, 40).
In the standard model of PG synthesis, flippase activity brings the disaccharide-pentapeptide moieties to the penicillin-binding proteins (PBPs), which polymerize the PG macromolecule on the outer surface of the membrane (39). The class A, high-molecular-weight PBPs possess an N-terminal glycosyl transferase domain that polymerizes the disaccharides into polysaccharide chains (38). These chains are cross-linked via the transpeptidase activity within the penicillin-binding, C-terminal domains of both the class A and the class B PBPs. The N-terminal domains of the class A PBPs and the closely related monofunctional glycosyl transferases found in some species are the only defined PG glycan strand polymerases, and in several species the presence of at least one of these enzymes is essential. However, in B. subtilis (26) and Enterococcus faecalis (3), strains lacking all of these known glycosyl transferases are viable and produce PG walls, indicating the presence of another glycosyl transferase capable of this activity. This alternate glycosyl transferase is distinct in that it is relatively resistant to moenomycin (3, 26), an inhibitor of the class A PBP glycosyl transferase activity (6).
Given the strong and early block in cortex PG polymerization observed to occur in a spoVB mutant (40), we wished to further analyze the potential role of this class of protein. SpoVB is a member of a relatively large family of proteins, COG2244 (23), some of which are involved in polymerization of other polysaccharides in bacteria, archaea, and eukaryotes. Bioinformatic analysis has generally predicted that these proteins span the membrane 12 to 14 times, and in some cases experimental evidence has supported this structure (7, 24). A role generally ascribed to these proteins is the flipping of lipid-linked oligosaccharides, produced on the inner face of a membrane, to the outside, where the oligosaccharides are then further polymerized or transferred to other substrates. Some prominent members of this family include Wzx, which functions in O-antigen synthesis in gram-negative bacteria (41); TuaB, which functions in teichuronic acid synthesis in B. subtilis (36); and Rft1, which functions in protein glycosylation in eukaryotes (12). MviN is essential in some gram-negative species, including Burkholderia pseudomallei, E. coli, and Sinorhizobium meliloti (22, 34), and has been shown to play a role in E. coli PG synthesis (16, 35). A Rhizobium tropici mutation that truncates mviN approximately 50% into the coding sequence was not lethal (29). However, it is not known whether this was the sole mviN homolog in the genome or whether the truncated gene product might be functional.
We have analyzed the phenotypic properties of B. subtilis strains lacking other proteins within the COG2244 family that are most closely related to SpoVB. Results suggest that these proteins also play roles in PG synthesis and that, in one case, this role is in a synthetic system that is relatively moenomycin resistant. We postulate that these proteins function in an alternate pathway for PG synthesis that may involve the flipping of lipid-linked PG oligosaccharides rather than lipid II disaccharides.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains of B. subtilis listed in Table 1 were derivatives of strain 168. Transformation was performed as previously described (2). Transformants were selected and maintained using erythromycin (0.5 mg/ml) plus lincomycin (12.5 mg/ml) (macrolide-lincosamide-streptogramin B [MLS] resistance). Growth and induction of sporulation by nutrient exhaustion were determined by use of 2× SG medium (21) at 37°C with shaking. Antibiotics were omitted in cultures grown for assays. Spore production was determined by heating a culture sample at 80°C for 10 min, followed by serial dilution and plating for viable counts. Spores were purified by repeated water washing (28), and relative heat resistance was determined as previously described (31).
TABLE 1.
Bacillus subtilis strains
| Strain | Genotype | Constructiona | Source |
|---|---|---|---|
| PS832 | Wild type | Setlow lab | |
| DPVB327 | ykvU::MLS | pDPV221→PS832 | This work |
| DPVB335 | yabM::MLS | pDPV220→PS832 | This work |
| DPVB336 | ytgP::MLS | pDPV222→PS832 | This work |
| DPVB450 | ΔykvU | pDPV283→PS832b | This work |
| DPVB488 | ΔyabM | pDPV337→PS832b | This work |
| DPVB489 | ΔytgP | pDPV338→PS832b | This work |
| DPVB520 | ΔyabM ykvU::MLS | DPVB327→DPVB488 | This work |
| DPVB521 | ΔyabM ytgP::MLS | DPVB336→DPVB488 | This work |
| DPVB522 | ΔytgP ykvU::MLS | DPVB327→DPVB489 | This work |
| DPVB523 | ΔytgP yabM::MLS | DPVB335→DPVB489 | This work |
| DPVB532 | ΔyabM ΔytgP ykvU::MLS | DPVB489+DPVB327→DPVB488 | This work |
| DPVB614 | ΔytgP amyE::xylP-ytgP | pDPV411→DPVB489 | This work |
| DPVB615 | ΔyabM amyE::xylP-yabM | pDPV412→DPVB488 | This work |
The designation preceding the arrow indicates the source of donor DNA in a natural transformation of the recipient strain following the arrow.
The indicated transformation was unique and the first in a series of steps in introduction of the in-frame deletion, as described in Materials and Methods.
Construction of mutant strains.
Oligonucleotides used for PCR and plasmid constructions are listed in Table 2. Plasmid insertion mutations with associated lacZ transcriptional fusions were produced by PCR amplification of ∼300-bp internal fragments of the beginning of each coding sequence and ligation into pMUTIN4 (37). These plasmids were transformed into B. subtilis with selection for MLS resistance, and insertion into the proper chromosomal locus was verified by PCR.
TABLE 2.
Bacillus subtilis plasmids and oligonucleotides
| Plasmid | Oligonucleotidea | Sequenceb | Vector |
|---|---|---|---|
| pDPV220 | DLP179 | CGGAATTCCACCATATTAAAAATATCAGC | pMUTIN4 |
| DLP180 | CGGGATCCTCCCGCAGTATAAAGCGAGGC | pMUTIN4 | |
| pDPV221 | DLP175 | CGGAATTCAATCCTAAGTTTCATGAAAGC | pMUTIN4 |
| DLP176 | CGGGATCCGAAATCAGCACAGCCATGTCC | pMUTIN4 | |
| pDPV222 | DLP177 | CGGAATTCAAAGGGGATTATGAGACAAGC | pMUTIN4 |
| DLP178 | CGGGATCCACTCAATAGAAAGATGATGCG | pMUTIN4 | |
| pDPV283 | DLP311 | GCTCTAGAAGTCTGAAAAAGAGTTGC | pBKJ236 |
| DLP312 | CTTCAATTCGCGGGGATCCCACAAATCGATTCATGATG | pBKJ236 | |
| DLP313 | GAATCGATTTGTGGGATCCCCGCGAATTGAAGGATG | pBKJ236 | |
| DLP314 | GAACTGCAGGTGATGATATGATACTCC | pBKJ236 | |
| pDPV337 | DLP307 | GCTCTAGAAGTGCTGATTTGTGATCG | pBKJ236 |
| DLP308 | CCATTTTGTTCGGATCCTATTGAATCGTCCATCC | pBKJ236 | |
| DLP309 | GGACGATTCAATAGGATCCGAACAAAATGGCGGGTAA | pBKJ236 | |
| DLP310 | GAACTGCAGGAAGCCGTCATTTGGTCG | pBKJ236 | |
| pDPV338 | DLP315 | GCTCTAGACTTCCTTCAATTGAATGC | pBKJ236 |
| DLP316 | CTGCATGTCTGCCGGATCCCAGTTTGCTTGACATGTTTC | pBKJ236 | |
| DLP317 | GTCAAGCAAACTGGGATCCGGCAGACATGCAGGCTG | pBKJ236 | |
| DLP318 | GAACTGCAGATCTGTGATTTTAAATGG | pBKJ236 | |
| pDPV411 | DLP468 | GCGCTTAATTAAATGGTGATGAAGAAACATGTC | pSWEET-bgaB |
| DLP469 | GCGCGGATCCTCATCAGCCTGCATGTCTGCC | pSWEET-bgaB | |
| pDPV412 | DLP466 | GCGCTTAATTAAGGGAAGGAGCTTTTGGATGG | pSWEET-bgaB |
| DLP467 | GCGCGGATCCCGACGACTGTAATTTTACCC | pSWEET-bgaB |
The two or four oligonucleotides listed for each plasmid were used to perform PCR as described in Materials and Methods to produce fragments that were cloned into the indicated vector.
Underlined sequences indicate restriction sites used to clone PCR products to the indicated vector. Bold sequences indicate the BamHI sites inserted within in-frame deletion sites.
In-frame deletions were produced by markerless gene replacement as described previously (17, 19), with the following modifications. PCR was used to amplify a fragment containing approximately 500 bp upstream of a coding sequence through the first five codons and a second fragment containing the last five codons plus approximately 500 bp of the downstream flanking region. Primers were designed with tails such that the two PCR products overlapped over the length of the first and last five codons plus two intervening codons produced by the addition of a BamHI site. The two products were mixed and subjected to a second round of PCR using the most-upstream and -downstream primers to produce a product containing the in-frame deletion and with XbaI and PstI restriction sites on the upstream and downstream ends, respectively. The PCR products were digested with XbaI and PstI and were ligated into similarly digested pBKJ236 (17). The plasmids were transformed into B. subtilis with MLS selection at 37°C, resulting in insertion of the plasmid via a single crossover at the chromosomal gene locus. The presence of both the deleted and the wild-type alleles in these strains was verified by PCR using the outermost primers described above. pBKJ223 (17) was transformed into the resulting strains to allow expression of I-SceI, and strains in which the chromosomal plasmid insertion was deleted from the chromosome by a second recombination event were identified by screening for MLS sensitivity. Strains lacking wild-type alleles and carrying the in-frame deletion mutations were verified by PCR and sequencing of the resulting PCR product carrying the deletion. Strain DPVB532 (ΔyabM ΔytgP ykvU::MLS) was constructed by congression of excess chromosomal DNA from DPVB489 (ΔytgP) plus limiting DNA from DPVB327 (ykvU::MLS) into DPVB488 (ΔyabM). The desired strain was identified by selection for MLS resistance followed by PCR screening of transformants for the presence of all three mutations and the absence of wild-type alleles.
Genes were placed at the amyE locus under the control of a xylose-regulated promoter for demonstration of complementation. Either the ytgP coding sequence plus the 18 preceding bp or the yabM coding sequence plus the 14 preceding bp was PCR amplified (Table 2) and inserted between the PacI and BamHI sites in pSWEET-bgaB (5). The resulting plasmids, pDPV411 and pDPV412, respectively, were verified by DNA sequencing of the inserts, linearized with XhoI, and transformed into B. subtilis with selection for chloramphenicol resistance. Recombination into the amyE locus was verified by screening for amylase activity, and maintenance of the deletion mutations at the yabM and ytgP loci was verified by PCR. Due to the low expression levels of the native ytgP and yabM loci, full complementation was observed during growth with very low levels of inducer (0.01% xylose) and partial complementation was observed in the absence of inducer (data not shown).
Assays.
β-Galactosidase was assayed as previously described (28). PG was purified and analyzed by high-pressure liquid chromatography as previously described (4, 26).
Microscopy.
Cultures grown in 2× SG medium at 37°C with shaking were harvested, centrifuged at 5,000 × g for 5 min at 4°C, resuspended in 1/5 volume of cold 0.5 M NaPO4, pH 7.0, and fixed by addition of glutaraldehyde to a final concentration of 1.3%, followed by incubation at 4°C for 12 h. Cells were washed four times with 0.1 M NaPO4, pH 6.7, mounted on polylysine-coated slides, and observed with an Olympus IX81 upright fluorescence microscope. Cell lengths and widths were measured using SlideBook software (Intelligent Imaging Innovations, Inc.). In some cases, the membranes of fixed cells were stained with FM 4-64 (Invitrogen) for easy visualization of septa.
RESULTS
Bioinformatic identification of SpoVB homologues.
A search of the available nucleotide databases for predicted protein sequences with similarity to the 518-amino-acid SpoVB sequence revealed highly similar proteins in all other endospore-forming species and significantly similar proteins in a broad range of species, including both gram-positive and gram-negative bacteria and archaea. These proteins are grouped within the COG2244 family (23), and some have been shown to be involved in the export of polysaccharides from the cell. All are predicted to span the membrane 10 or more times. In B. subtilis, three additional genes encode proteins that are highly similar to SpoVB: yabM, ykvU, and ytgP (Table 3).
TABLE 3.
SpoVB paralogs in B. subtilis
| Protein | Length (amino acids) | Identity/ similarity/alignmenta | Membrane span predictionb | Operon predictionc | Expressionc |
|---|---|---|---|---|---|
| SpoVB | 518 | 12-14 | No | Mother cell, σE | |
| YabM | 532 | 26/48/339 | 12-14 | Probable yabMNO | Weak vegetative |
| YkvU | 445 | 30/52/450 | 10-12 | ykvUV | Mother cell, σE |
| YtgP | 544 | 23/45/421 | 13-14 | No | Vegetative |
Values are percent amino acid identity/percent amino acid similarity/total length of the alignment (amino acids) with B. subtilis SpoVB, determined using BlastP.
Predictions of membrane-spanning helices were produced using five different software packages. The ranges of predictions are shown.
yabM appears to be the first gene in a transcriptional unit, as the well-defined spoVT gene terminates 184 bases upstream of the predicted yabM start codon. There is no obvious transcriptional terminator downstream of yabM, and thus it appears to be in an operon with two downstream genes, yabN and yabO. However, as the end of the yabM coding sequence overlaps the predicted yabN coding sequence by 8 bp, it is possible that yabN begins at an alternate start codon further downstream and that this is not truly an operon. YabN contains domains similar to tetrapyrrole methylases and nucleoside triphosphate pyrophosphohydrolase. YabO contains a domain with similarity to the RNA binding domain of bacterial ribosomal protein S4. No functions have yet been ascribed to yabN or yabO.
YkvU has previously been shown to be expressed within an operon that includes the downstream ykvV (spoIVH, stoA) gene (9, 11, 15). The operon is expressed during sporulation in the mother cell compartment under the control of sigma E. A sigma G-dependent promoter drives expression of only ykvV in the forespore (15). YkvU has been suggested to be involved in spore germination (1), but the mechanism of this effect is not clear. YkvV (StoA) is a thiol-disulfide oxidoreductase postulated to be involved in breaking disulfide bonds in proteins important for cortex synthesis (10). YtgP appears to be monocistronic, as the upstream gene is oriented in the opposite direction and the downstream gene is separated from ytgP by 72 bp and a predicted transcription terminator.
Gene expression.
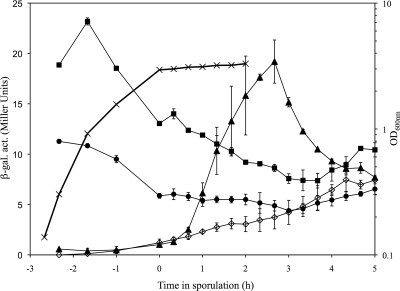
We constructed plasmid insertion mutations in yabM, ykvU, and ytgP such that transcriptional fusions to lacZ were produced, and we assayed expression of each gene throughout growth and sporulation (Fig. 1). Growth rates of the mutant strains in rich medium were comparable to that of the wild type. YabM was expressed at low levels during growth, and expression dropped off to background levels during sporulation. YtgP was expressed somewhat more strongly, but still at low levels, during vegetative growth, with expression decreasing significantly during sporulation. As expected, ykvU was not expressed during vegetative growth but was transcribed between the first and third hours of the sporulation process.
FIG. 1.
Timing of expression of spoVB homologues. Strains were shaken vigorously in 2× SG medium at 37°C, and samples were collected for OD determinations and β-galactosidase assays. Time in sporulation is relative to the entry into stationary phase, which occurs at an OD of ∼3.0 in this medium. Growth rates (×) were similar for all strains, so only a single curve is plotted. β-Galactosidase activity (β-gal. act.) was assayed for PS832 (wild type with no lacZ fusion, ⋄), DPVB335 (yabM-lacZ, •), DPVB327 (ykvU-lacZ, ▴), and DPVB336 (ytgP-lacZ, ▪). Values are averages determined for three independent cultures, with error bars indicating standard deviations.
Topology analysis.
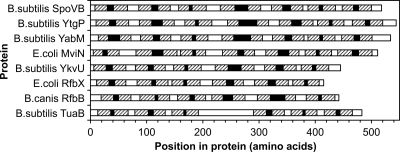
We used five different protein topology prediction programs to analyze the members of this protein family (Fig. 2). These programs all predicted 14 transmembrane helices for SpoVB. They predicted 12 to 14 membrane spans for YabM and YtgP, which are similar in size to SpoVB, but only 10 to 12 membrane spans for YkvU, which is 73 amino acids shorter than SpoVB. We examined the B. subtilis genome sequence to determine if a sequencing error might have led to the prediction of a truncation of ykvU. This does not appear to be the case, as there are stop codons in all three reading frames and the characterized ykvV gene begins 50 bp downstream of the predicted ykvU stop codon.
FIG. 2.
Protein topology predictions. Five different topology prediction programs were used to identify membrane-spanning regions and cytoplasmic versus extracellular domains. The predictions shown are those produced by Phobius (18), which produced the matches most consistent with the majority of the other programs in each instance where differences were apparent. Hatched boxes indicate membrane-spanning helices, white boxes represent cytoplasmic domains, and black boxes indicate extracellular regions. B. canis, Brucella canis.
Growth and morphology of mutant strains.
We constructed two types of null mutations in each gene, the plasmid insertion mutations described above for gene expression analysis, which likely have negative polar effects on expression of downstream genes, and nonpolar in-frame deletion mutations. To determine whether the creation of these mutations required the appearance of second-site suppressor mutations, we determined the transformation frequencies of the plasmid insertion mutations, in the form of chromosomal DNA from the original mutants, into a wild-type background. Each of the mutations could be introduced at high frequencies (between 2 × 104 and 9 × 104 transformants per μg DNA), indistinguishable from the rate of introduction of a known nonlethal mutation (pbpD::Cm in PS1958 [32]). Thus, loss of any of these three individual genes was tolerated without the appearance of suppressor mutations. We also constructed multiple mutants lacking all combinations of the three genes, and these transformations were reproducible with high transformation frequencies (data not shown).
None of the mutant strains, including the insertion or the deletion mutants, exhibited a defect in maximum growth rate in rich medium (Table 4). The triple mutant may have a very slightly, but nearly undetectably, slower growth rate. All of the mutants were examined for their ability to produce heat-resistant spores (Table 4). All of the strains lacking either or both yabM and ykvU were fully competent at producing spores, at a frequency equal to or greater than that for the wild-type strain. All of the strains lacking ytgP produced significantly fewer spores. Eight hours after entry into stationary phase, the ytgP single mutant had produced only 11% of the number of spores produced by the wild type. After 24 h, this had increased to 31%. This was a defect in the number of spores produced rather than in the relative heat resistance of the spores. Spores were harvested after 24 h and were tested for their survival over time at 90°C. Wild-type and ytgP spores exhibited similar D values, losing 1 log of viability in approximately 40 min (data not shown). The ytgP yabM double mutant and the triple mutant produced only about 10% of the number of spores produced by the wild type after 24 h, suggesting that yabM may partially compensate for the loss of ytgP.
TABLE 4.
Mutant phenotypic properties
| Strain | Genotype | Doubling time (min)a | No. of heat-resistant sporesb
|
Avg cell length (μm)c | % Cells in chains ofc:
|
|||
|---|---|---|---|---|---|---|---|---|
| T8 | T24 | 2 | >2 | >8 | ||||
| PS832 | Wild type | 19 ± 1 | 2.5 × 108 | 3.1 × 108 | 4.1 ± 1.1 | 58 | 0 | 0 |
| 3.9 ± 1.0 | 44 | 2 | 0 | |||||
| DPVB488 | ΔyabM | 19 ± 1 | 3.5 × 108 | 5.0 × 108 | 4.6 ± 1.3 | 54 | 0 | 0 |
| 3.9 ± 0.9 | 53 | 0 | 0 | |||||
| DPVB450 | ΔykvU | 20 ± 2 | 1.2 × 108 | 5.3 × 108 | 4.6 ± 1.4 | 47 | 2 | 0 |
| 4.2 ± 2.3 | 39 | 0 | 0 | |||||
| DPVB520 | ΔyabM ΔykvU | 19 ± 1 | 8.3 × 107 | 4.2 × 108 | 4.5 ± 1.2 | 69 | 0 | 0 |
| 3.8 ± 0.9 | 47 | 0 | 0 | |||||
| DPVB489 | ΔytgP | 20 ± 2 | 2.7 × 107* | 9.7 × 107 | 6.4 ± 1.9** | 49 | 42 | 19 |
| 5.7 ± 1.7** | 74 | 17 | 0 | |||||
| DPVB614d | ΔytgP amyE::xylP-ytgP | 20 ± 2 | 2.4 × 108 | ND | 4.5 ± 1.4 | 53 | 2 | 0 |
| DPVB521 | ΔyabM ytgP::MLS | 21 ± 2 | 4.3 × 106* | 3.6 × 107 | 6.5 ± 1.5** | 57 | 39 | 20 |
| 6.3 ± 3.1** | 40 | 52 | 0 | |||||
| DPVB522 | ΔytgP ykvU::MLS | 20 ± 1 | 1.4 × 107* | 9.4 × 107 | 7.3 ± 2.4** | 37 | 45 | 19 |
| 5.7 ± 1.4** | 82 | 9 | 0 | |||||
| DPVB532 | ΔyabM ΔytgP ykvU::MLS | 22 ± 1 | 2.5 × 106* | 2.8 × 107 | 6.9 ± 1.8** | 45 | 45 | 26 |
| 5.6 ± 1.7** | 61 | 22 | 0 | |||||
Values are averages determined for three independent cultures ± standard deviations.
Values are averages determined for three independent cultures. T8, 8 h after entry into stationary phase; T24, 24 h after entry into stationary phase. *, value is significantly different (P ≤ 0.05 using unpaired Student's t test) than that observed for the wild-type strain.
For each strain, the first value is for a mid-log culture (OD of 0.5) and the second value is for a late log culture (OD of 1.5). Average cell lengths are for 200 cells, and standard deviations are given. Cells in chains of >8 are also included in chains of >2. **, value is significantly different (P < 0.0001 using unpaired Student's t test) than that observed for the wild-type strain.
This strain was assayed for ytgP complementation only during mid-log growth with 0.01% xylose.
We examined cells from wild-type and ytgP cultures using transmission electron microscopy to determine the stage of sporulation that was blocked in the mutant. At 2, 4, and 6 h after entry into stationary phase, we did not observe a significant number of cells in the ytgP culture that appeared to be blocked at a particular stage of sporulation. Rather, many of the cells seemed to never enter the sporulation process or to enter very late. Even 6 h after the cessation of significant growth, >50% of the cells showed no sign of asymmetric septation and many were still undergoing symmetric division (data not shown). We believe that decreased spore formation by ytgP strains is due simply to a defect in cell division (see below), leading to slow initiation of sporulation rather than a specific defect in sporulation itself.
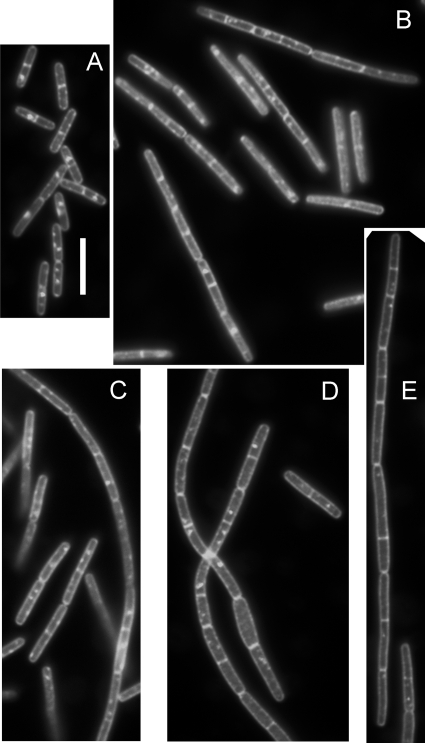
To observe changes in cell morphology, we used phase-contrast and fluorescence microscopy to examine cells at mid-log and late log phases of growth. We photographed multiple fields and then measured approximately 200 cells from each culture and time point. We recorded the numbers of cells present as single cells and in chains of any length, and we measured the length of each cell (Table 4). The yabM and ykvU mutants did not exhibit significant differences from the wild type in cell length or in chain formation. Cells in all strains carrying ytgP mutations tended to be longer and were found in longer chains than those of the wild type (Fig. 3). During mid-log growth, ≥30% of the ytgP cells were in chains of greater than two cells, and many of these chains were longer than eight cells. As the ytgP cultures approached stationary phase, the number of cells in very long chains decreased but did not drop to the levels seen for the wild-type strain. Effects on cell length and chain formation, as well as on spore production, were eliminated by complementation with ytgP in the chromosome at an ectopic locus and under the control of a xylose-regulated promoter (Table 4), indicating that these phenotypic changes are specifically due to the loss of ytgP. The increase in chain formation as well as in cell length suggests that ytgP plays a role in cell division. PG was purified from cells in exponential growth and was digested with muramidase. Muropeptides were separated and quantified by high-pressure liquid chromatography. No reproducible significant difference in PG structure was observed between the wild type and any mutant strain lacking a single gene (data not shown).
FIG. 3.
Cell length increase and chain formation by a ytgP mutant. Cells were grown to mid-log phase, washed, fixed, stained with FM 4-64, and observed for fluorescence of this membrane stain, which appears white in the image. (A) PS832 (wild type); (B and C) DPVB489 (ΔytgP); (D and E) DPVB532 (ΔyabM ΔytgP ykvU::MLS). Bar, 10 μm. All panels are shown at the same magnification.
A quadruple mutant lacking all of the above-mentioned genes in addition to spoVB was constructed (data not shown). As expected from the sporulation-specific expression of spoVB, this strain appeared identical to the triple mutant during growth but produced no detectable heat-resistant spores.
Moenomycin resistance.
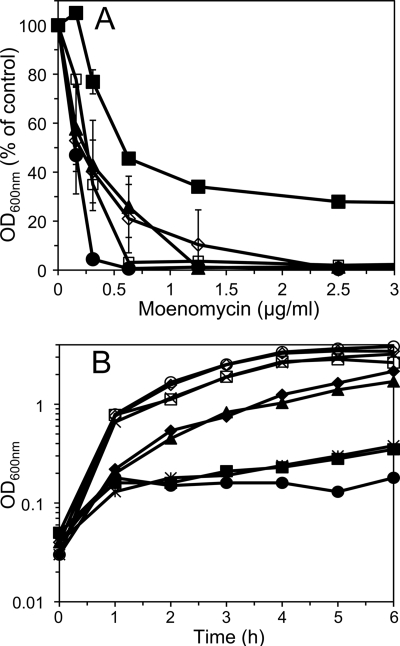
Previous studies demonstrated the presence of two separable B. subtilis PG synthetic systems that can be differentiated based on sensitivity to the antibiotic moenomycin (26), which directly inhibits the glycosyl transferase activity of class A PBPs. The effects of mutations eliminating COG2244 proteins on moenomycin sensitivity were examined. While ykvU had no effect on the B. subtilis minimum inhibitory concentration of moenomycin, loss of yabM reduced the minimum inhibitory concentration approximately fourfold (Fig. 4A). Loss of ytgP had surprising effects. While the ytgP mutant grew similarly to the wild type under optimum aerobic conditions (Fig. 1), ytgP cultures grown in a microtiter plate without vigorous mixing reached a final optical density (OD) of only 25 to 40% of that seen with cultures of the wild type and other mutant strains (data not shown). This was also true of multiple mutants lacking ytgP. On the other hand, loss of ytgP produced a measure of moenomycin resistance. In the presence of moenomycin concentrations up to 10 μg/ml, the ytgP mutant reached final OD values significantly greater than those for other strains and approximately half the OD it reached in the absence of the antibiotic (Fig. 4A and data not shown). However, mutant strains lacking both yabM and ytgP exhibited the moenomycin sensitivity of the yabM strain (Fig. 4A). While the moenomycin sensitivity of the double mutant does not appear as dramatic as that of the yabM single mutant, this is in part due to the low maximum OD achieved by ytgP strains, resulting in a lower percentage drop due to moenomycin. These results indicate that the degree of moenomycin resistance is dependent on YabM.
FIG. 4.
Moenomycin sensitivity of mutant strains. (A) Cultures in exponential growth phase were inoculated into multiwell plates containing medium with various concentrations of moenomycin. One hundred to 200 cells were inoculated into 250 μl of medium. OD values were determined after 16 h at 37°C and are expressed as percentages of that obtained in the absence of moenomycin. Results for PS832 (wild type, ⋄), DPVB488 (ΔyabM, •), DPVB450 (ΔykvU, ▴), DPVB489 (ΔytgP, ▪), and DPVB521 (ΔyabM ytgP::MLS, □) are shown. Values are averages determined for two independent cultures, with error bars indicating standard deviations. Where error bars are not visible, the standard deviations are <2%. (B) Cultures in exponential growth phase were diluted into fresh medium without (open symbols and ×) or with (filled symbols and ✻) moenomycin, and OD was monitored during shaking at 37°C. Results for PS832 (wild type, ⋄ and ♦), DPVB488 (ΔyabM, ○ and •), DPVB521 (ΔyabM ytgP::MLS, □ and ▪), DPVB532 (ΔyabM ΔytgP ykvU::MLS, × and ✻), and DPVB615 (ΔyabM amyE::xylP-yabM, ▴) are shown. Complementation of ΔyabM was observed during growth of DPVB615 in the presence of 0.01% xylose.
The effect of YabM on resistance was also apparent in shaken broth cultures. When exponential-phase cultures were diluted into fresh medium with moenomycin, the wild-type strain exhibited a minor growth inhibition (Fig. 4B). The yabM strain and multiple mutants lacking yabM were severely inhibited for up to 6 h (Fig. 4B). Moenomycin sensitivity of the yabM strain was eliminated by complementation with yabM in the chromosome at an ectopic locus and under the control of a xylose-regulated promoter (Fig. 4B), indicating that this phenotypic change was specifically due to the loss of yabM.
DISCUSSION
Members of the COG2244 protein family play roles in membrane translocation of lipid-linked oligosaccharides in multiple systems. The failure to produce spore cortex PG in a spoVB mutant was the first indication of a role for this type of protein in PG synthesis (40). Demonstrations of PG precursor accumulation in E. coli upon MviN depletion extended this link (16, 35). It was suggested that MviN is the lipid II flippase (35), a role that would be consistent with the spoVB phenotype. However, if these proteins function as lipid II flippases, then in B. subtilis they cannot be the sole source of this activity, as a strain lacking all of these proteins is viable and produces a relatively normal PG structure. The possibility remains that other, more distantly related members of this family in B. subtilis and other species can substitute in the flippase role. Alternatively, there may be multiple classes of proteins that can function as a flippase during PG synthesis, including perhaps the SEDS proteins (13).
Our data do suggest roles for the B. subtilis COG2244 proteins in PG synthesis. Strains lacking YtgP produce slightly longer cells and chains of cells than do wild-type strains, suggesting a role in septum formation. A ytgP strain also exhibits some resistance to moenomycin, an antibiotic that specifically blocks the glycosyl transferase activity involved in PG polymerization by class A PBPs. We suggest that in the absence of YtgP, some important aspect of PG polymerization is shifted toward a pathway that is moenomycin resistant and which potentially involves YabM. On the other hand, strains lacking YabM exhibit increased sensitivity to moenomycin, and this effect dominates over that produced by loss of YtgP. This observation of a role of YabM in moenomycin-resistant PG polymerization suggests a novel model for an alternative method of PG synthesis.
It was observed previously for B. subtilis (26) and E. faecalis (3) that the loss of all class A PBPs did not produce the lethal effect expected, given the accepted model for PG synthesis in which these proteins carried the sole PG glycosyl transferase activities in these species. Rather, these strains grew poorly, but with PG walls, and this growth was now moenomycin resistant, leading to the proposal of an alternative class of moenomycin-insensitive PG glycosyl transferases. We now suggest that YabM and other members of this protein family may function with these alternative glycosyl transferases in an alternate pathway for PG synthesis. By analogy with other systems in which COG2244 proteins function, it is possible that initial glycan polymerization takes place within the cell and that lipid-linked PG oligosaccharides are flipped across the membrane for further polymerization. Depending on the size of the oligosaccharides that are transported, further glycan strand polymerization may or may not be required outside the cytoplasm. The lengths of PG strands produced in the absence of class A PBPs have not been reported. PG cross-link formation via transpeptidation is certainly required outside the cell, and this activity is likely provided by class B PBPs, as some of these proteins become essential in the absence of class A PBPs (unpublished data).
This alternative pathway for PG polymerization is not the major pathway, as growth without the class A PBPs is severely limited, and in some species may be dispensable in the presence of the class A PBP pathway. However, the alternative pathway may play important roles in particular parts of the cell cycle and under certain environmental conditions, as evidenced by the essential nature of MviN in some species (16, 35). One instance where this alternate pathway becomes primary may be during spore formation. While class A PBPs do play a role in synthesis of a minor component of spore PG, the germ cell wall (25), no class A PBP has been demonstrated to be required for production of the major component, the cortex. On the other hand, SpoVB is essential for cortex PG production (40). This may be a situation where the alternative pathway produces all of the PG strands, which are then cross-linked to a very limited degree (27) by the class B PBP SpoVD (8, 40). Further study of the COG2244 family proteins and their roles in PG synthesis will provide new insights into this central bacterial function and may identify novel potential antibiotic targets.
Acknowledgments
This work was supported by grant GM56695 to D.L.P. from the National Institutes of Health.
We thank Allison Fay, Jonathan Dworkin, and John Helmann for communicating results prior to publication.
Footnotes
Published ahead of print on 31 July 2009.
REFERENCES
- 1.Aertsen, A., I. Van Opstal, S. C. Vanmuysen, E. Y. Wuytack, and C. W. Michiels. 2005. Screening for Bacillus subtilis mutants deficient in pressure induced spore germination: identification of ykvU as a novel germination gene. FEMS Microbiol. Lett. 243:385-391. [DOI] [PubMed] [Google Scholar]
- 2.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:74-76. [Google Scholar]
- 3.Arbeloa, A., H. Segal, J. E. Hugonnet, N. Josseaume, L. Dubost, J. P. Brouard, L. Gutmann, D. Mengin-Lecreulx, and M. Arthur. 2004. Role of class A penicillin-binding proteins in PBP5-mediated beta-lactam resistance in Enterococcus faecalis. J. Bacteriol. 186:1221-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atrih, A., G. Bacher, G. Allmaier, M. P. Williamson, and S. J. Foster. 1999. Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J. Bacteriol. 181:3956-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhavsar, A. P., X. Zhao, and E. D. Brown. 2001. Development and characterization of a xylose-dependent system for expression of cloned genes in Bacillus subtilis: conditional complementation of a teichoic acid mutant. Appl. Environ. Microbiol. 67:403-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, L., D. Walker, B. Sun, Y. Hu, S. Walker, and D. Kahne. 2003. Vancomycin analogues active against vanA-resistant strains inhibit bacterial transglycosylase without binding substrate. Proc. Natl. Acad. Sci. USA 100:5658-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunneen, M. M., and P. R. Reeves. 2008. Membrane topology of the Salmonella enterica serovar Typhimurium group B O-antigen translocase Wzx. FEMS Microbiol. Lett. 287:76-84. [DOI] [PubMed] [Google Scholar]
- 8.Daniel, R. A., S. Drake, C. E. Buchanan, R. Scholle, and J. Errington. 1994. The Bacillus subtilis spoVD gene encodes a mother-cell-specific penicillin-binding protein required for spore morphogenesis. J. Mol. Biol. 235:209-220. [DOI] [PubMed] [Google Scholar]
- 9.Eichenberger, P., S. T. Jensen, E. M. Conlon, C. van Ooij, J. Silvaggi, J. E. Gonzalez-Pastor, M. Fujita, S. Ben-Yehuda, P. Stragier, J. S. Liu, and R. Losick. 2003. The sigmaE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 327:945-972. [DOI] [PubMed] [Google Scholar]
- 10.Erlendsson, L. S., M. Moller, and L. Hederstedt. 2004. Bacillus subtilis StoA is a thiol-disulfide oxidoreductase important for spore cortex synthesis. J. Bacteriol. 186:6230-6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feucht, A., L. Evans, and J. Errington. 2003. Identification of sporulation genes by genome-wide analysis of the sigmaE regulon of Bacillus subtilis. Microbiology 149:3023-3034. [DOI] [PubMed] [Google Scholar]
- 12.Helenius, J., and M. Aebi. 2002. Transmembrane movement of dolichol linked carbohydrates during N-glycoprotein biosynthesis in the endoplasmic reticulum. Semin. Cell Dev. Biol. 13:171-178. [DOI] [PubMed] [Google Scholar]
- 13.Henriques, A. O., P. Glaser, P. J. Piggot, and C. P. Moran, Jr. 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28:235-247. [DOI] [PubMed] [Google Scholar]
- 14.Hranueli, D., P. J. Piggot, and J. Mandelstam. 1974. Statistical estimate of the total number of operons specific for Bacillus subtilis sporulation. J. Bacteriol. 199:684-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imamura, D., K. Kobayashi, J. Sekiguchi, N. Ogasawara, M. Takeuchi, and T. Sato. 2004. spoIVH (ykvV), a requisite cortex formation gene, is expressed in both sporulating compartments of Bacillus subtilis. J. Bacteriol. 186:5450-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue, A., Y. Murata, H. Takahashi, N. Tsuji, S. Fujisaki, and J. Kato. 2008. Involvement of an essential gene, mviN, in murein synthesis in Escherichia coli. J. Bacteriol. 190:7298-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janes, B. K., and S. Stibitz. 2006. Routine markerless gene replacement in Bacillus anthracis. Infect. Immun. 74:1949-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kall, L., A. Krogh, and E. L. Sonnhammer. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338:1027-1036. [DOI] [PubMed] [Google Scholar]
- 19.Lambert, E. A., and D. L. Popham. 2008. The Bacillus anthracis SleL (YaaH) protein is an N-acetylglucosaminidase involved in spore cortex depolymerization. J. Bacteriol. 190:7601-7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lara, B., D. Mengin-Lecreulx, J. A. Ayala, and J. van Heijenoort. 2005. Peptidoglycan precursor pools associated with MraY and FtsW deficiencies or antibiotic treatments. FEMS Microbiol. Lett. 250:195-200. [DOI] [PubMed] [Google Scholar]
- 21.Leighton, T. J., and R. H. Doi. 1971. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J. Biol. Chem. 254:3189-3195. [PubMed] [Google Scholar]
- 22.Ling, J. M., R. A. Moore, M. G. Surette, and D. E. Woods. 2006. The mviN homolog in Burkholderia pseudomallei is essential for viability and virulence. Can. J. Microbiol. 52:831-842. [DOI] [PubMed] [Google Scholar]
- 23.Marchler-Bauer, A., J. B. Anderson, M. K. Derbyshire, C. DeWeese-Scott, N. R. Gonzales, M. Gwadz, L. Hao, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, D. Krylov, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, S. Lu, G. H. Marchler, M. Mullokandov, J. S. Song, N. Thanki, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2007. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 35:D237-D240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazur, A., M. Marczak, J. E. Krol, and A. Skorupska. 2005. Topological and transcriptional analysis of pssL gene product: a putative Wzx-like exopolysaccharide translocase in Rhizobium leguminosarum bv. trifolii TA1. Arch. Microbiol. 184:1-10. [DOI] [PubMed] [Google Scholar]
- 25.McPherson, D. C., A. Driks, and D. L. Popham. 2001. Two class A high-molecular-weight penicillin-binding proteins of Bacillus subtilis play redundant roles in sporulation. J. Bacteriol. 183:6046-6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McPherson, D. C., and D. L. Popham. 2003. Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis. J. Bacteriol. 185:1423-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meador-Parton, J., and D. L. Popham. 2000. Structural analysis of Bacillus subtilis spore peptidoglycan during sporulation. J. Bacteriol. 182:4491-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination, and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 29.O'Connell, K. P., S. J. Raffel, B. J. Saville, and J. Handelsman. 1998. Mutants of Rhizobium tropici strain CIAT899 that do not induce chlorosis in plants. Microbiology 144:2607-2617. [DOI] [PubMed] [Google Scholar]
- 30.Piggot, P. J., and J. G. Coote. 1976. Genetic aspects of bacterial endospore formation. Bacteriol. Rev. 40:908-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popham, D. L., B. Illades-Aguiar, and P. Setlow. 1995. The Bacillus subtilis dacB gene, encoding penicillin-binding protein 5*, is part of a three-gene operon required for proper spore cortex synthesis and spore core dehydration. J. Bacteriol. 177:4721-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Popham, D. L., and P. Setlow. 1996. Phenotypes of Bacillus subtilis mutants lacking multiple class A high-molecular-weight penicillin-binding proteins. J. Bacteriol. 178:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Popham, D. L., and P. Stragier. 1991. Cloning, characterization, and expression of the spoVB gene of Bacillus subtilis. J. Bacteriol. 173:7942-7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rudnick, P. A., T. Arcondeguy, C. K. Kennedy, and D. Kahn. 2001. glnD and mviN are genes of an essential operon in Sinorhizobium meliloti. J. Bacteriol. 183:2682-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruiz, N. 2008. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc. Natl. Acad. Sci. USA 105:15553-15557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soldo, B., V. Lazarevic, M. Pagni, and D. Karamata. 1999. Teichuronic acid operon of Bacillus subtilis 168. Mol. Microbiol. 31:795-805. [DOI] [PubMed] [Google Scholar]
- 37.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 38.van Heijenoort, J. 2001. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology 11:25R-36R. [DOI] [PubMed] [Google Scholar]
- 39.van Heijenoort, J. 2007. Lipid intermediates in the biosynthesis of bacterial peptidoglycan. Microbiol. Mol. Biol. Rev. 71:620-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vasudevan, P., A. Weaver, E. D. Reichert, S. D. Linnstaedt, and D. L. Popham. 2007. Spore cortex formation in Bacillus subtilis is regulated by accumulation of peptidoglycan precursors under the control of sigma K. Mol. Microbiol. 65:1582-1594. [DOI] [PubMed] [Google Scholar]
- 41.Whitfield, C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75:39-68. [DOI] [PubMed] [Google Scholar]