Abstract
DevR activates the transcription of ∼48 genes in response to hypoxia and other stresses and triggers metabolic downshift and dormancy development in Mycobacterium tuberculosis. tgs1 and Rv3131 encode triacylglycerol synthase and a putative nitroreductase, respectively, and both are members of the DevR regulon. This study aimed to understand how a single putative DevR binding site identified previously could sustain powerful induction of divergent tgs1-Rv3131 genes. DNase I footprinting revealed that phosphorylated DevR in fact binds to two sites symmetrically located at −42.5 and −63.5 bp from transcription start points of both genes. DevR first bound to the high-affinity site, P, and cooperatively recruited another DevR molecule to the secondary low-affinity site, S, to activate tgs1-Rv3131 transcription by ∼210- and ∼110-fold, respectively. The presence of a single P site significantly reduced activation of tgs1 expression and abolished Rv3131 activity, reinforcing the requirement of two binding sites for robust expression in both directions. P site inversion abolished tgs1 but not Rv3131 transcription despite DevR occupancy at both sites. The lack of tgs1 expression is most likely due to disruption of its −35 promoter element rather than inversion of the binding site per se. We conclude that (i) an overlap of a DevR binding site and −35 sequence is indispensable for promoter activation, (ii) DevR interaction with two binding sites is obligatory for synergistic activation of tgs1-Rv3131 promoters, and (iii) DevR interaction with binding sites of different affinities offers scope for temporal and differential expression of target genes.
Bacterial persistence is a hallmark of tuberculosis. It is believed that oxygen deprivation and exposure to nitric oxide (NO) or carbon monoxide (CO) trigger a metabolic shift into a state that mimics dormancy in Mycobacterium tuberculosis (6, 9, 22, 29, 30). These signals activate the DevR response regulator (also called DosR), which in turn triggers the expression of ∼48 genes called the DevR regulon (9, 13, 29). These genes are also expressed very early within infected macrophages and dendritic cells (20, 26), in mice (7), and in guinea pigs (21). DevR has also been implicated in the virulence and persistence of M. tuberculosis (10, 12, 15). All these findings underscore the importance of deciphering the activation mechanism(s) of various DevR regulon genes. DevR belongs to the NarL/UhpA subfamily of response regulators (4) and is activated through phosphorylation by either of two sensor kinases, DevS and Rv2027c/DosT (16, 17, 18), in response to hypoxia or exposure to NO or CO (8, 9, 22, 24). As with NarL, phosphorylation is required for DevR binding to specific DNA sequences (1, 11).
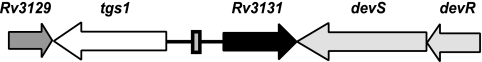
tgs1 is one of the most powerfully induced genes of the DevR regulon (13, 29), and it is divergently transcribed from Rv3131 (Fig. 1). tgs1 encodes a triacylglycerol synthase that synthesizes triacylglycerol, which is proposed as an energy source during dormancy (3). Its disruption has been shown to prevent triacylglycerol accumulation under inducing conditions (23). Rv3131 codes for a putative classical nitroreductase which may be involved in detoxification of nitrogen-containing by-products in the host (14). It was suggested previously that powerfully induced genes had multiple DevR binding sites in their upstream regions (13). While this hypothesis held true for some promoters (1, 2), tgs1 and Rv3131 were powerfully induced under hypoxia apparently from a single DevR binding site only (5, 13). This study aimed to understand the defining features of DevR-dependent promoter activation from a single putative binding site located between divergent tgs1-Rv3131 genes. To our surprise, high-affinity and low-affinity binding sites (Dev box) were detected at −42.5 and at −63.5, respectively, in symmetrical positions with respect to the transcription start points (TSPs) of both genes. Their functional role in divergent transcription was established by examining the effect of mutations and inversion and deletion of binding sites.
FIG. 1.
Schematic representation of tgs1-Rv3131 genes in M. tuberculosis. A putative Dev box in the intergenic region is indicated by a gray rectangle.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
M. tuberculosis and Escherichia coli strains were cultured as described previously (2). All the plasmids used in this study are listed in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant featuresa | Reference |
|---|---|---|
| pFPV27 | E. coli-mycobacterial shuttle plasmid with promoterless gfp; Kmr | 28 |
| pTGS | pFPV27 containing tgs1 promoter (−143 to +45); Hygr | This study |
| p3131 | pFPV27 containing Rv3131 promoter (−150 to +38); Hygr | This study |
| pTGS-P mut | pTGS containing mutated P DevR binding site; Hygr | This study |
| pTGS-S mut | pTGS containing mutated S DevR binding site; Hygr | This study |
| pTGS-PS mut | pTGS containing mutated P and S DevR binding site; Hygr | This study |
| p3131-P mut | p3131 containing mutated P DevR binding sites; Hygr | This study |
| p3131-S mut | p3131 containing mutated S DevR binding site; Hygr | This study |
| p3131-PS mut | p3131 containing mutated P and S DevR binding site; Hygr | This study |
| pTGS P-inv | pTGS containing inverted P DevR binding site; Hygr | This study |
| p3131 P-inv | p3131 containing inverted P DevR binding site; Hygr | This study |
| pTGS-ΔS | pTGS with S DevR binding site deletion; Hygr | This study |
| p3131-ΔS | p3131 with S DevR binding site deletion; Hygr | This study |
Promoter coordinates are with reference to the TSP.
RNA isolation and primer extension.
RNA isolation and TSP mapping were performed as described previously (1). Two separate lots of RNA (40 μg each) were used to confirm the positions of TSPs.
Construction of reporter plasmids and GFP reporter assays.
The tgs1-Rv3131 region containing a 184-bp intergenic sequence was amplified from M. tuberculosis H37Rv DNA using primers Rv3130F and Rv3131R, cloned first in pGEMT-Easy and then into the promoterless green fluorescent protein (GFP) reporter plasmid pFPV27 (28) in both orientations at the EcoRI site as described previously (1), to generate pTGS and p3131. All the cloned inserts were verified by DNA sequencing. Dev box mutants were generated by the mega primer method as described previously (1, 19). Mutations were confirmed by DNA sequencing. The variant promoter plasmids were electroporated into M. tuberculosis H37Rv and also into the ΔdevR mutant strain described earlier (12). The primers used in this study are listed in Table 2.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ → 3′) | Application(s) |
|---|---|---|
| 3130F | TGGCTGCCGGGCCTTTCCCAT | Reporter assay, DNase I footprinting |
| 3131R | CATGGTCAGCGCCTTCCCCGG | Reporter assay, DNase I footprinting |
| 3130TSP | GGCTTCCTGATCGGGAGCCGG | Primer extension |
| 3131TSP | GACTCGTCGGGCATACCCGCC | Primer extension |
| 3130LSM | CGGTGGCAGACCAGGAAGCCTCGCCAGCGG | Mutational analysis (S box) |
| 3131HSM | CGCCAGCGGACGAGCTTTGGCCCTGCCTCA | Mutational analysis (P box) |
| 3130DM | AGCACGAAGCCTGGCCAGCGGACGAGCTTTGGCCC | Mutational analysis (P + S box) |
| ΔLS | CGCGTCGACAGCCGCGGTGGCGGAGGACCTTTGGCCCTGC | Deletion of S box |
| INVERT 3130 | CACGAAGGCTCGCCAGGCAGGGCCAAAGGTCCTCCGCTCAACGGCTCCTC | Inversion of P box |
| LH1 | CGAGTCGACAGAGCACGAAGGCTCGCCAGCGGAGGACCTTTGGCCCTGCGTCGACCGA | Gel shift assay (P + S box) |
| LH2 | TCGGTCGACGCAGGGCCAAAGGTCCTCCGCTGGCGAGCCTTCGTGCTCTGGTCGACTCG | Gel shift assay (P + S box) |
| H1 | CGAGTCGACCGGAGGACCTTTGGCCCTGCGTCGACCGA | Gel shift assay (P box) |
| H2 | TCGGTCGACGCAGGGCCAAAGGTCCTCCGGTCGACTCG | Gel shift assay (P box) |
GFP reporter assays were performed as described previously (1). Briefly, M. tuberculosis cultures were grown under aerobic and standing culture conditions. GFP fluorescence is constant in bacteria harboring the pFPV27 control vector (∼30 relative fluorescence units per optical density unit [RFU/OD at 595 nm]). The percent expression of variant promoters was calculated with respect to the wild-type (WT) promoter. The degree of induction is expressed as the ratio of the number of RFU/OD of standing cultures to that of aerobic shaking cultures at 48 h (without subtracting vector background).
Gel shift assay and DNase I footprinting.
DevR was purified and used in gel shift and DNase I footprinting assays as described previously (1). Single-stranded oligonucleotides corresponding to the primary box (P box) and the primary and secondary box (P + S box) were annealed by incubation at 95°C for 3 min in buffer containing 10 mM Tris-Cl (pH 7.5) and 100 mM NaCl and allowed to cool slowly to 4°C. After electrophoresis, electrophoretic mobility shift assay (EMSA) results were analyzed using Quantity One software (Bio-Rad). The fraction of bound DNA was estimated by subtracting the amount of free DNA from the amount of input DNA.
RESULTS
Presence of two Dev boxes, including a cryptic site in the tgs1-Rv3131 intergenic region.
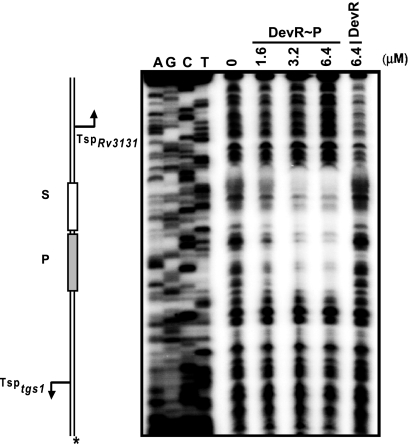
In silico analysis had previously predicted the presence of one putative 18- or 20-bp Dev box in the tgs1-Rv3131 intergenic region (5, 13). However, DNase I footprinting revealed a phosphorylated-DevR-protected ∼40-bp sequence which included the predicted binding site (P box, GCAGGGCCAAAGGTCCTCCG, top strand) and adjacent DNA (Fig. 2). A close examination of this sequence revealed a loosely conserved 20-bp Dev box-like sequence (S box, TGGCGAGCCTTCGTGCTCTG, top strand) adjacent to the P box (Fig. 3A). The sequence of this low-scoring box (0.044) has 10 mismatches with the consensus sequence, and only G5 and C8 are conserved among the nucleotides proposed to be important for specific DNA-DevR interactions. Although the P box has nine mismatches with the consensus box, it had a moderate score of 9.68 (13), since G5, G6, and C8 nucleotides are conserved in it. Both the boxes were occupied simultaneously even at the lowest concentration of phosphorylated DevR tested, suggesting that the interaction with these boxes was highly cooperative.
FIG. 2.
Phosphorylated DevR (DevR∼P) binds to two sites in the tgs1-Rv3131 intergenic region. DNase I footprinting of DevR on tgs1-Rv3131 intergenic DNA (*, Rv3131 coding strand labeled). P and S, Dev boxes. Dideoxy sequencing reactions are shown in lanes A, G, C, and T.
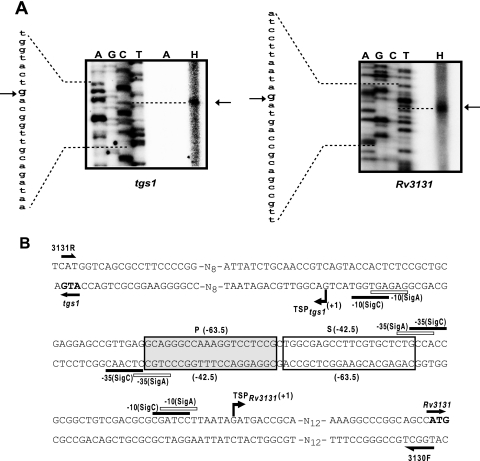
FIG. 3.
TSP mapping. (A) Primer extension analysis with primer Rv3130TSP or Rv3131TSP and M. tuberculosis RNA from aerobic cultures (the section of the gel labeled A) and standing cultures (lane H). Dideoxy sequencing reactions using the same primers are also shown. The position of TSP is indicated by an arrow. (B) Nucleotide sequence of the tgs1-Rv3131 intergenic region. Locations of P and S binding sites are indicated with respect to TSP of Rv3131 (above the sequence) and tgs1 (below the sequence). Primers are indicated by half-headed arrows. The putative −10 and −35 promoter elements similar to SigC and SigA consensus sequences (25, 27) are indicated.
The tgs1-Rv3131 intergenic region has a conserved arrangement of DevR binding sites and adjoining −35 promoter elements.
A single hypoxia-inducible TSP was detected for divergent tgs1 and Rv3131 genes (Fig. 3A). Sequences with partial matches to −10 and −35 elements of SigA and SigC were present upstream of the TSPs (Fig. 3B). Since similar sequences were also identified at other hypoxia-inducible promoters (1), it is likely that these promoters are transcribed by RNA polymerase (RNAP) containing SigA or SigC or both. No TSP was detected under aerobic conditions, which indicates that these promoters are not active in aerobic cultures.
Binding of DevR to P and S sites is essential for full induction of tgs1-Rv3131 transcription.
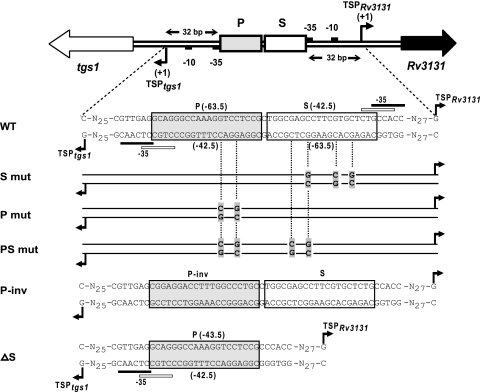
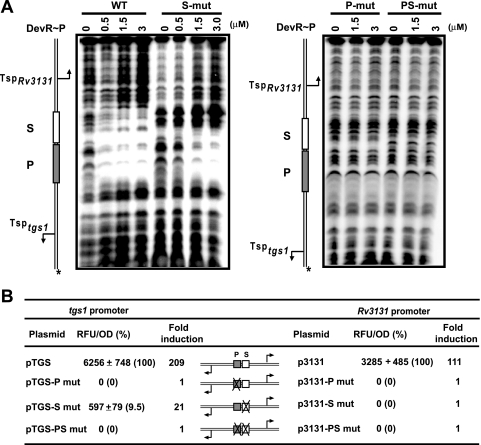
The tgs1-Rv3131 promoter region has an interesting architecture wherein two DevR binding sites are centrally placed and the putative −35 promoter elements of both genes overlap the adjoining Dev boxes (Fig. 3B). In order to decipher the functional role of these boxes, several variant promoter DNA templates were generated (Fig. 4). Mutations at positions G5 and C8 in the cryptic box (S mut) abolished the interaction with it but not with the P box (Fig. 5A). Thus, the S box was established to be a bona fide DevR binding site. Mutation of G5 and C8 nucleotides in one-half of the P box (P mut) abolished the interaction with it and also prevented DevR binding to the S site. This established that DevR first interacts with the P box (primary binding site) and then cooperatively recruits a second DevR molecule to the S box (secondary binding site). When both the boxes were mutated at adjacent half-sites (PS mut), binding was completely abolished. Since mutation of the S site (S mut) also affected DevR occupancy of the P site (Fig. 5A), we believe that DevR bound to the S site may stabilize DevR bound to the P site.
FIG. 4.
Variant promoter constructs used in the study. Map of the tgs1-Rv3131 intergenic region. P and S sites are represented by gray shaded and white rectangular boxes, respectively. Putative −10 and −35 promoter elements are indicated by small black boxes. Various promoter variants used in this study are shown below the map. Nucleotides mutated in P and S boxes are aligned with WT nucleotides. N, WT sequence.
FIG. 5.
Effect of P site and S site mutations on in vitro DNA binding and in vivo transcriptional activation of tgs1 and Rv3131. (A) DNase I footprinting with phosphorylated DevR (DevR∼P) and WT, S mut, P mut, and PS mut DNAs. (B) GFP reporter assay. The GFP fluorescence of M. tuberculosis standing cultures (48 h) is expressed as RFU/OD at 595 nm ± standard deviation.
Next, the role of P and S boxes in tgs1 and Rv3131 promoter activity was assessed by cloning the tgs1-Rv3131 intergenic region upstream of gfp in the promoter probe vector pFPV27. Consistent with the results of TSP mapping, no GFP reporter expression was observed in aerobic M. tuberculosis H37Rv cultures whereas both the genes were highly induced under hypoxia. Interestingly, the tgs1 promoter (near the P site) was twice as active as the Rv3131 promoter (near the S site): expression from pTGS was induced 209-fold, compared to 111-fold induction from p3131 (Fig. 5B). Promoter activity was not detected in the ΔdevR mutant strain (data not shown), and this absence confirmed both promoters to be completely DevR dependent during hypoxia.
When the P box was mutated, tgs1 promoter activity (in pTGS-P mut) was completely abolished, and likewise, when the S box was mutated (p3131-S mut), there was no transcription of Rv3131 (Fig. 5B). Since the −35 promoter elements overlap the adjoining Dev boxes at both promoters, the results suggest that DevR interacts with RNAP and the failure to detect promoter activity is attributed to an inability to recruit RNAP at both sites. Interestingly, mutating the distal S box (pTGS-S mut) reduced pTGS activity by ∼90%, which shows that DevR interaction with P and S sites synergistically activates tgs1 transcription. Inducible promoter activity of p3131 was understandably abolished by mutating the P box (p3131-P mut) because DevR could not bind to the S site (Fig. 5) that abuts this promoter. As expected, mutating both the boxes abolished induction of the divergent genes.
DevR interaction with single and tandem binding sites.
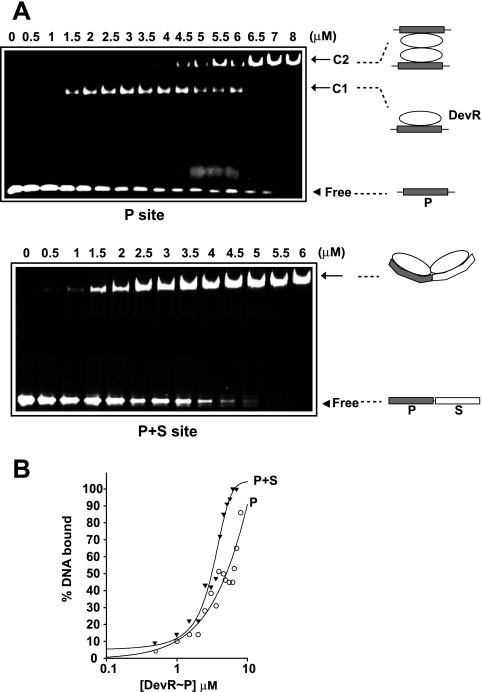
The gel retardation assay was used to further scrutinize DevR interaction with oligonucleotides containing P box or P + S box binding sites. Interestingly, titration of DevR with P site DNA yielded two DNA-protein complexes: a faster-moving complex (C1) at a lower protein concentration was replaced gradually by a DNA-protein complex of low mobility (C2) at a higher protein concentration. In contrast, DevR interaction with P + S box DNA produced a single DNA-protein complex of low mobility even at a low protein concentration (Fig. 6A), suggesting the binding to be highly cooperative (Fig. 6B).
FIG. 6.
EMSA analysis. (A) Double-stranded oligonucleotides having P or P + S box sequences incubated with increasing concentrations of phosphorylated DevR (DevR∼P). Arrow, DNA-protein complex; arrowhead, free DNA. (B) Fraction of bound DNA (from Fig. 6A) plotted against DevR concentration.
P box inversion abolished in vivo transcription activation without affecting DevR binding to DNA.
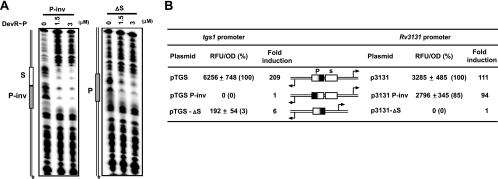
At tgs1 and Rv3131 promoters, since the proximal Dev box, which is essential for transcription activation, overlaps the −35 promoter element, it is possible that DevR could substitute completely for recognition by RNAP. To address this possibility, tgs1 and Rv3131 promoter variants were constructed wherein the P box was inverted (pTGS P-inv and p3131 P-inv [Fig. 4]). Variant promoter DNA (P-inv) bound to DevR at both P and S sites (Fig. 7A). Although four peripheral nucleotides were altered by inversion (CTGC→TCCG [Fig. 4]), the preservation of symmetry in the critical nucleotides, G5, G6, and C8, can explain DevR binding to the altered P box. Interestingly, tgs1 promoter activity was abolished in pTGS P-inv (Fig. 7B), suggesting that −35 promoter sequences may have been disrupted and that the interaction of RNAP with −35 sequences and bound DevR is essential. Importantly, the activity of p3131 P-inv promoter was not significantly affected by P site inversion, which confirmed that Rv3131 and tgs1 were expressed from nonoverlapping promoters. Although the promoters are nonoverlapping, sharing of activating features between P and S sites is clearly demonstrated.
FIG. 7.
Transcription machinery interacts with sequences in Dev box DNA. (A) DNase I footprinting of DevR with phosphorylated DevR (DevR∼P) and P-inv and ΔS promoter variants. (B) GFP reporter assay of WT and tgs1-Rv3131 variant promoter constructs expressed as the number of RFU/OD at 595 nm ± standard deviation.
A single high-affinity Dev box does not support robust divergent gene transcription.
Several DevR-regulated genes are predicted to have a single Dev box in their upstream regions. Therefore to understand the mechanism underlying transcription activation by a single high-affinity box, the P site was placed at an equal distance from both tgs1 and Rv3131 TSPs (Fig. 4, ΔS constructs). In the ΔS construct, DevR bound, albeit weakly, to the P box (Fig. 7A) and tgs1 activity was reduced to a mere 3% relative to WT promoter activity (Fig. 7B). Importantly, juxtaposing the P site with promoter sequences failed to support Rv3131 expression in p3131ΔS, and this is ascribed to deletion/disruption of its own −35 element (within the deleted S box). These results and those reported earlier in this article establish that the presence of a single high-affinity Dev box fails to support robust gene activation.
DISCUSSION
The rationale for carrying out the present study was to understand how powerful induction of tgs1-Rv3131 genes was achievable through one DevR binding site that was predicted computationally. It turned out that there are in fact two binding sites, P and S, for DevR in the tgs1-Rv3131 intergenic region, which provided us with an opportunity to analyze the relative properties and functions of each of these sites in divergent gene expression.
The secondary site, S, escaped detection by in silico approaches (5, 13), as it is a poor match to the proposed consensus binding sequence. A key biological finding of this study was the discovery of DevR interaction with a site that is a poor match to the consensus and the critical role of this binding in regulating tgs1-Rv3131 expression. Surprisingly, the “weak” S site is absolutely essential for Rv3131 promoter induction, and it significantly enhances activation from binding at the P site in the other direction. Such interactions may be relevant to the regulation of other genes of the DevR regulon. A pertinent question for the future is whether a single site or cooperative recruitment of DevR to additional “weak” binding sites is necessary for gene activation.
EMSA using P site oligonucleotide points toward the progressive formation of a DevR dimer-DNA complex (C1) and a DevR tetramer-DNA complex (C2) (Fig. 6). Alternatively, it is conceivable that bound DevR could recruit unbound DevR at higher protein concentrations, in which case the complex would progress from C1 to C2 without a decrease in the amount of free DNA. However, since the progression from C1 to C2 was accompanied by a decrease in free DNA, the second possibility was ruled out. Moreover, visualization of a DevR C-terminal domain tetramer-2 oligonucleotide complex is consistent with the first suggestion (31). The observed cooperativity with the P + S site oligonucleotide was rationalized by DevR-induced DNA bending at the P site, which could facilitate recruitment of DevR to the S site. An important conclusion from these results is that side-by-side placement of binding sites, even of different affinities, highly favors cooperative recruitment of DevR to DNA.
The promoter architecture of tgs1-Rv3131 genes is most interesting, since these genes are induced to different extents despite an equidistant placement of the Dev boxes from their TSPs (∼210-fold and ∼110-fold for the tgs1 and Rv3131 promoters, respectively). The difference is in the correlation with the DevR affinity of the TSP-proximal binding site; higher induction was associated with the presence of a stronger binding site and vice versa. The low-affinity S binding site may permit more frequent dissociation of DevR from DNA and could possibly result in a lower rate of RNAP recruitment at the Rv3131 promoter. In addition, differences in promoter strength and efficiency of the ribosome binding site could also contribute to enhanced tgs1 promoter-derived GFP fluorescence. Many divergently transcribed DevR regulon genes, such as Rv3126c-Rv3127 and Rv2005c-otsB, which were shown to be differentially activated (13), may be similarly regulated by DevR.
The predicted −35 promoter sequences were established to be functionally important by the analysis of variant promoters. First, although DevR bound to an inverted P site and S site in the tgs1 promoter (pTGS P-inv), its transcription was abolished. Since P site inversion did not affect its binding to DevR, we suggest that alteration of the peripheral sequence disrupted the −35 promoter element. The inversion almost entirely removed the putative SigA −35 element but not the SigC −35 element, suggesting the involvement of SigA in tgs1 transcription. Second, when the S box is deleted (p3131-ΔS), Rv3131 promoter activity was abolished, again presumably due to disruption in the −35 sequence. Interestingly, tgs1 promoter activity was consistently lower in the ΔS construct than in the S mut construct (Fig. 5 and 7). The higher promoter activity of S mut could be attributed to an unstable in vivo interaction of DevR with the mutated S site which is absent in the ΔS construct.
In summary, this study provides several new insights into the salient features defining a DevR-dependent promoter. The tgs1 and Rv3131 promoters illustrate a minimum requirement of two binding sites for maximal gene activation. Additionally, an overlap of a DevR binding site and −35 sequences is indispensable for promoter activation. Importantly, at tgs1 and Rv3131 promoters, cooperative recruitment of DevR to P and S sites overcomes the apparent disadvantage of a “weak” S binding site to support powerful expression in both directions. These findings further suggest the possibility that temporal and differential regulation of target genes is achieved via the binding of DevR to sites of different affinities.
Acknowledgments
This work was financially supported by a grant to J.S.T. from the Department of Biotechnology, Government of India.
Santosh Chauhan is grateful to CSIR for a research associateship. We thank Neil Stoker for the generous gift of M. tuberculosis ΔdevR mutant strain. We acknowledge the facilities of the Biotechnology Information Systems, Department of Biotechnology, Government of India.
Footnotes
Published ahead of print on 31 July 2009.
REFERENCES
- 1.Chauhan, S., and J. S. Tyagi. 2008. Cooperative binding of phosphorylated DevR to upstream sites is necessary and sufficient for activation of the Rv3134c-devRS operon in Mycobacterium tuberculosis: implication in the induction of DevR target genes. J. Bacteriol. 190:4301-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chauhan, S., and J. S. Tyagi. 2008. Interaction of DevR with multiple binding sites synergistically activates divergent transcription of narK2-Rv1738 genes in Mycobacterium tuberculosis. J. Bacteriol. 190:5394-5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daniel, J., C. Deb, V. S. Dubey, T. D. Sirakova, B. Abomoelak, H. R. Morbidoni, and P. E. Kolattukudy. 2004. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J. Bacteriol. 186:5017-5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dasgupta, N., V. Kapur, K. K. Singh, T. K. Das, S. Sachdeva, K. Jyothisri, and J. S. Tyagi. 2000. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuber. Lung Dis. 80:141-159. [DOI] [PubMed] [Google Scholar]
- 5.Florczyk, M. A., L. A. McCue, A. Purkayastha, E. Currenti, M. J. Wolin, and K. A. McDonough. 2003. A family of acr-coregulated Mycobacterium tuberculosis genes shares a common DNA motif and requires Rv3133c (dosR or devR) for expression. Infect. Immun. 71:5332-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomez, J. E., and J. D. McKinney. 2004. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis (Edinburgh) 84:29-44. [DOI] [PubMed] [Google Scholar]
- 7.Karakousis, P. C., T. Yoshimatsu, G. Lamichhane, S. C. Woolwine, E. L. Nuermberger, J. Grosset, and W. R. Bishai. 2004. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J. Exp. Med. 200:647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar, A., J. C. Toledo, R. P. Patel, J. R. Lancaster, Jr., and A. J. C. Steyn. 2007. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. USA 104:11568-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar, A., J. S. Deshane, D. K. Crossman, S. Bolisetty, B. S. Yan, I. Kramnik, A. Agarwal, and A. J. Steyn. 2008. Heme oxygenase-1-derived carbon monoxide induces the Mycobacterium tuberculosis dormancy regulon. J. Biol. Chem. 283:18032-18039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malhotra, V., D. Sharma, V. D. Ramanathan, H. Shakila, D. K. Saini, S. Chakravorty, T. K. Das, Q. Li, R. F. Silver, P. R. Narayanan, and J. S. Tyagi. 2004. Disruption of response regulator gene, devR, leads to attenuation in virulence of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 231:237-245. [DOI] [PubMed] [Google Scholar]
- 11.Maris, A. E., M. R. Sawaya, M. Kaczor-Grzeskowiak, M. R. Jarvis, S. M. Bearson, M. L. Kopka, I. Schroder, R. P. Gunsalus, and R. E. Dickerson. 2002. Dimerization allows DNA target site recognition by the NarL response regulator. Nat. Struct. Biol. 9:771-778. [DOI] [PubMed] [Google Scholar]
- 12.Parish, T., D. A. Smith, S. Kendall, N. Casali, G. J. Bancroft, and N. G. Stoker. 2003. Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect. Immun. 71:1134-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park, H. D., K. M. Guinn, M. I. Harrell, R. Liao, M. I. Voskuil, M. Tompa, G. K. Schoolnik, and D. R. Sherman. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48:833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purkayastha, A., L. A. McCue, and K. A. McDonough. 2002. Identification of a Mycobacterium tuberculosis putative classical nitroreductase gene whose expression is coregulated with that of the acr gene within macrophages, in standing versus shaking cultures, and under low oxygen conditions. Infect. Immun. 70:1518-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reed, M. B., S. Gagneux, K. DeRiemer, P. M. Small, and C. E. Barry III. 2007. The W-Beijing lineage of Mycobacterium tuberculosis overproduces triglycerides and has the DosR dormancy regulon constitutively upregulated. J. Bacteriol. 189:2583-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts, D. M., R. P. Liao, G. Wisedchaisri, W. G. Hol, and D. R. Sherman. 2004. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J. Biol. Chem. 279:23082-23087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saini, D. K., V. Malhotra, D. Dey, N. Pant, T. K. Das, and J. S. Tyagi. 2004. DevR-DevS is a bona fide two-component system of Mycobacterium tuberculosis that is hypoxia-responsive in the absence of the DNA-binding domain of DevR. Microbiology 150:865-875. [DOI] [PubMed] [Google Scholar]
- 18.Saini, D. K., V. Malhotra, and J. S. Tyagi. 2004. Cross talk between DevS sensor kinase homologue, Rv2027c, and DevR response regulator of Mycobacterium tuberculosis. FEBS Lett. 565:75-80. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 20.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma, D., A. Bose, H. Shakila, T. K. Das, J. S. Tyagi, and V. D. Ramanathan. 2006. Expression of mycobacterial cell division protein, FtsZ, and dormancy proteins, DevR and Acr, within lung granulomas throughout guinea pig infection. FEMS Immunol. Med. Microbiol. 48:329-336. [DOI] [PubMed] [Google Scholar]
- 22.Shiloh, M. U., P. Manzanillo, and J. S. Cox. 2008. Mycobacterium tuberculosis senses host-derived carbon monoxide during macrophage infection. Cell Host Microbe 3:323-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sirakova, T. D., V. S. Dubey, C. Deb, J. Daniel, T. A. Korotkova, B. Abomoelak, and P. E. Kolattukudy. 2006. Identification of a diacylglycerol acyltransferase gene involved in accumulation of triacylglycerol in Mycobacterium tuberculosis under stress. Microbiology 152:2717-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sousa, E. H., J. R. Tuckerman, G. Gonzalez, and M. A. Gilles-Gonzalez. 2007. DosT and DevS are oxygen-switched kinases in Mycobacterium tuberculosis. Protein Sci. 16:1708-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun, R., P. J. Converse, C. Ko, S. Tyagi, N. E. Morrison, and W. R. Bishai. 2004. Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol. Microbiol. 52:25-38. [DOI] [PubMed] [Google Scholar]
- 26.Tailleux, L., S. J. Waddell, M. Pelizzola, A. Mortellaro, M. Withers, A. Tanne, P. R. Castagnoli, B. Gicquel, N. G. Stoker, P. D. Butcher, M. Foti, and O. Neyrolles. 2008. Probing host pathogen cross-talk by transcriptional profiling of both Mycobacterium tuberculosis and infected human dendritic cells and macrophages. PLoS ONE 3:e1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unniraman, S., M. Chatterji, and V. Nagaraja. 2002. DNA gyrase genes in Mycobacterium tuberculosis: a single operon driven by multiple promoters. J. Bacteriol. 184:5449-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valdivia, R. H., A. E. Hromockyj, D. Monack, L. Ramakrishnan, and S. Falkow. 1996. Applications for green fluorescent protein (GFP) in the study of host-pathogen interactions. Gene 173:47-52. [DOI] [PubMed] [Google Scholar]
- 29.Voskuil, M. I., D. Schnappinger, K. C. Visconti, M. I. Harrell, G. M. Dolganov, D. R. Sherman, and G. K. Schoolnik. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wayne, L. G., and C. D. Sohaskey. 2001. Nonreplicating persistence of mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139-163. [DOI] [PubMed] [Google Scholar]
- 31.Wisedchaisri, G., M. Wu, A. E. Rice, D. M. Roberts, D. R. Sherman, and W. G. Hol. 2005. Structures of Mycobacterium tuberculosis DosR and DosR-DNA complex involved in gene activation during adaptation to hypoxic latency. J. Mol. Biol. 354:630-641. [DOI] [PubMed] [Google Scholar]