Abstract
Sedimentary hopanes are pentacyclic triterpenoids that serve as biomarker proxies for bacteria and certain bacterial metabolisms, such as oxygenic photosynthesis and aerobic methanotrophy. Their parent molecules, the bacteriohopanepolyols (BHPs), have been hypothesized to be the bacterial equivalent of sterols. However, the actual function of BHPs in bacterial cells is poorly understood. Here, we report the physiological study of a mutant in Rhodopseudomonas palustris TIE-1 that is unable to produce any hopanoids. The deletion of the gene encoding the squalene-hopene cyclase protein (Shc), which cyclizes squalene to the basic hopene structure, resulted in a strain that no longer produced any polycyclic triterpenoids. This strain was able to grow chemoheterotrophically, photoheterotrophically, and photoautotrophically, demonstrating that hopanoids are not required for growth under normal conditions. A severe growth defect, as well as significant morphological damage, was observed when cells were grown under acidic and alkaline conditions. Although minimal changes in shc transcript expression were observed under certain conditions of pH shock, the total amount of hopanoid production was unaffected; however, the abundance of methylated hopanoids significantly increased. This suggests that hopanoids may play an indirect role in pH homeostasis, with certain hopanoid derivatives being of particular importance.
Bacteriohopanepolyols (BHPs) are a class of pentacyclic triterpenoids, generally referred to as hopanoids, that are found in a variety of bacteria. Bacterial hopanoids have been hypothesized to be surrogates of eukaryotic sterols primarily because of similarities in their biosynthesis and structure (Fig. 1 A and B) (36). Sterols have been shown to play an important role in stabilizing eukaryotic cell membranes and regulating membrane fluidity, as well as providing resistance to ethanol and heat shock stress (20, 57, 61). Sterols are produced by all eukaryotes, and the membranes of animals, plants, and fungi have been shown to contain different types of sterols possibly with different physiological roles (20). Usually, these sterols vary in their patterns of A-ring and side chain alkylation and unsaturation (16, 20, 40). There are also a variety of sterol esters and steryl glycosides found in plants and yeast (41, 47). Regardless of which sterols are produced by eukaryotic cells, all eukaryotic organisms require these molecules to survive (40).
FIG. 1.
Biosynthesis of hopanoids in R. palustris TIE-1. (A) Hopanoids produced by R. palustris TIE-1. For 32,33,34,35-bacteriohopanetetrol, R2 is OH; for 35-amino-32,33,34-bacteriohopanetriol, R2 is NH2. (B) Structures of the sterol biosynthesis intermediate lanosterol and of cholesterol, demonstrating the structural similarity to hopanoids. (C) Conversion of squalene to the basic hopene structure by the squalene-hopene cyclase. The Entrez Gene accession number for the squalene-hopene cyclase gene is NC_011004.1.
Hopanoids have been shown to localize to the cytoplasmic and outer membranes of certain bacteria (21, 24, 54), and in vitro experiments have shown that these molecules are efficient in condensing artificial membranes and enhancing the viscosity of liposomes (3, 43). As with eukaryotic sterols, there is also structural variation in hopanoids, where some are methylated at the C-2 or C-3 position and some are unsaturated, and the functionalization of the side chain can differ greatly (59). To date, no genetic, biochemical, physiological, or environmental studies have been done that explain the biological significance of these modifications. A few studies have proposed that hopanoids enhance membrane stability and decrease membrane permeability in some bacteria, including Alicyclobacillus acidocaldarius (43), Zymomonas mobilis (19), Frankia sp. (4), and Streptomyces coelicolor (44). However, these studies did not utilize a genetic approach or in vivo experiments to conclusively elucidate the role of hopanoids in membrane structure and stability. Thus, the extent to which they are sterol surrogates remains an open question.
While understanding the function of hopanoids is interesting from a purely cell biological perspective, there is an additional motivation provided by evolutionary biology. Geochemists and geobiologists frequently utilize these compounds as “molecular fossils” or biomarkers (8). In this context, biomarkers are organic compounds that are preserved in ancient sedimentary rocks and can be linked to specific organisms by analogy with their production in modern organisms (8, 9). The use of these molecular fossils has proven to be a powerful approach to study and reconstruct both ancient and modern microbial ecosystems (8). Sedimentary hopanes comprise one of the most abundant and ubiquitous types of extractable organic compounds in ancient rocks (8) and are unambiguously recognized as the chemical fossils of BHPs (35). Because BHPs with an extended side chain and 35 carbon atoms are known to be produced only by bacteria, extended C31 to C35 sedimentary hopanes are thought to be specific to this domain (34, 49). Furthermore, extended hopanes that are methylated at C-2 have been considered to be indicators of cyanobacteria, whereas C-3 methylated hopanes are thought to be diagnostic for microaerophilic methanotrophs and acetic acid bacteria (56, 64).
To date, no studies have addressed the function of hopanoids in geologically relevant organisms, such as photosynthetic bacteria. The use of hopanoids as biomarkers would be greatly enhanced if they could be attributed to specific bio(geo)chemical processes as opposed to their use solely as taxonomic proxies. Hopanoids are not produced by all bacteria, and their distribution among these groups does not appear to follow a systematic pattern (8, 49). Furthermore, specific sedimentary hopanes have been interpreted as biomarkers for certain metabolisms in modern microbes in the absence of direct experimental evidence linking the two. For example, BHPs methylated at C-2 were known to be produced in abundance by cyanobacteria, and taking this together with their prevalence in ancient marine petroleum source rocks, they were proposed as biomarkers not only for cyanobacteria, but also for oxygenic photosynthesis (49, 56). However, there is no evidence that functionally links 2-methylbacteriohopanepolyols (2-MeBHPs) with oxygenic photosynthesis in modern cyanobacteria. Furthermore, the lack of studies addressing the regulation of the production of hopanoids in modern bacteria limits our ability to constrain these molecules to a specific bacterial group. An example is the recent discovery of significant 2-MeBHP production by the anoxygenic phototroph Rhodopseudomonas palustris, which had been previously tested and was thought not to produce methylated hopanoids (31, 48). This demonstrated not only that organisms other than cyanobacteria are able to make 2-MeBHPs, but also that the production of these molecules depends on the growth conditions (48).
Given these ambiguities, there are inadequate data to fully interpret the sedimentary hopane record. To begin to understand the physiological functions of hopanoids in photosynthetic bacteria and to compare these functions to those of sterols in eukaryotes, we generated and characterized a hopanoid deletion mutant of the genetically tractable anoxygenic phototroph R. palustris TIE-1.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains used in this study are listed in Table 1. Escherichia coli strains were grown in lysogeny broth (LB) at 37°C. E. coli strain BW29427 was supplemented with 60 μM diaminopimelic acid. For aerobic chemoheterotrophic growth, R. palustris strains were grown in unbuffered YP medium (0.3% yeast extract, 0.3% peptone) at 30°C in the dark with shaking at 250 rpm. Buffered YP medium was prepared by adding 100 mM MES (4-morpholineethanosulfonic acid) for pH 4.5, 5, and 6; 100 mM MOPS (4-morpholionepropanesulfonic acid) for pH 7; or 100 mM bicine [N,N-bis(2-hydroyxethylglycine)] for pH 8 and pH 9 and adjusting the medium to the desired pH with concentrated HCl or NaOH prior to autoclaving it. For anaerobic phototrophic growth, R. palustris strains were grown in anaerobic bicarbonate-buffered freshwater (FW) medium (22). Unless otherwise noted, FW medium was supplemented from sterile anoxic stock solutions with 2 mM sodium acetate for photoheterotrophic growth and 2 mM sodium thiosulfate for photoautotrophic growth. All anaerobic cultures were flushed and pressurized to 5 lb/in2 with N2/CO2 (80%/20%) and incubated at 30°C at 50 W/m2 without shaking. For anaerobic photoheterotrophic growth at acidic and alkaline pH, FW medium was prepared without bicarbonate in the medium or CO2 in the headspace to prevent fluctuations in the pH. Anoxic acidic medium was buffered with 50 mM MES, and alkaline medium was buffered with 50 mM HEPES [4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid], and the headspace was flushed with N2 rather than N2/CO2. This medium was supplemented with 10 mM acetate, and the pH was adjusted both before and after autoclaving. For growth on solid medium, LB or YP was solidified with 1.5% agar and supplemented, if necessary, with gentamicin at 20 μg/ml (E. coli) or 800 μg/ml (R. palustris), kanamycin at 50 μg/ml (E. coli) or 400 μg/ml (R. palustris), spectinomycin at 100 μg/ml, or bile salts at 1.5%.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype, markers, or characteristicsa | Source |
|---|---|---|
| Strains | ||
| E. coli S17-1 | thi pro hdsR hdsM+recA; chromosomal insertion of RP4-2 (Tc::Mu Km::Tn7) | 53 |
| E. coli BW29427 | thrB1004 pro thi rpsL hsdS lacZΔM15 RP4-1360 Δ(araBAD)567 ΔdapA1341::[erm pir (wt)] | W. W. Metcalf |
| R. palustris TIE-1 | Isolated from Woods Hole, MA | 22 |
| R. palustris Δshc | R. palustris TIE-1 Δshc | This study |
| R. palustris Δshc+pPVW8 | R. palustris TIE-1 Δshc transformed to Kmr with pPVW8 | This study |
| Plasmids | ||
| pJQ200SK | Mobilizable suicide vector; sacB Gmr | 46 |
| pBBR1MCS-2 | 5.1-kb broad-host-range plasmid; KmrlacZ | 25 |
| pCR8/GW/TOPO | 2.8-kb TA cloning vector; Spr | Invitrogen, Carlsbad, CA |
| pPVW3 | 1-kb shc upstream region PCR amplified with primers PW13 and PW14 and cloned into pJQ200SK at NotI and SpeI | This study |
| pPVW4 | 1-kb shc downstream region PCR amplified with primers PW15 and PW16 and cloned into pJQ200SK at SpeI and XmaI | This study |
| pPVW6 | NotI- and SpeI-cut shc upstream fragment from pPVW3 subcloned into pPVW4 | This study |
| pPVW8 | KpnI- and SpeI-cut 4-kb shc complementation fragment from pPVW23 subcloned into pBBR1MCS-2 | This study |
| pPVW23 | 4-kb shc complementation fragment PCR amplified with primers PW25 and PW26 and cloned into pCR8/GW/TOPO | This study |
Gm, gentamicin; Km, kanamycin; Sp, spectinomycin.
For growth curves and doubling-time calculations, exponential-growth-phase cells were inoculated in triplicate into the appropriate medium with a 1% inoculum and monitored for growth by following the increase in absorbance at 600 nm over time. Absorbance at 600 nm versus time was graphed on a log scale, and the slope of the curve was used to determine the growth rate constant, k. The doubling time, g, was calculated from the following equation: g = ln(2)/k.
DNA methods, plasmid construction, and transformation.
All plasmid constructions and primers used in this study are described in Table 1 and Table 2. A QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA) was used for isolation of plasmid DNA from E. coli. Genomic DNA from R. palustris strains was isolated using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA). DNA sequences of all cloning intermediates were confirmed by sequencing at the Biopolymers Laboratory in the Massachusetts Institute of Technology Center for Cancer Research. E. coli strains were transformed by electroporation using an Electroporator 2510 (Eppendorf, Hamburg, Germany) as recommended by the supplier. Plasmids were mobilized from E. coli S17-1 or BW29427 into R. palustris by conjugation on YP agar plates that were incubated overnight at 30°C (23, 39).
TABLE 2.
Primers used in this study
| Primer | Sequencea | Restriction sites |
|---|---|---|
| PW13 | GGCGCGCCGCGGCCGCGCGCTGGCGGCTGAAGCTCG | AscI/NotI |
| PW14 | GGCGCGCCACTAGTTTTATCTGCTGTGTCTCCGT | AscI/SpeI |
| PW15 | GGCGCGCCACTAGTTTCTGGGGGCAGTGGACGAC | AscI/SpeI |
| PW16 | GGCGCGCCCCCGGGTCGTCGCGCCGAAGGAGTTC | AscI/XmaI |
| PW21 | TTGGGGCGAAATCTGTAAAA | |
| PW22 | TAATTCGTCGGTCAGTGCAA | |
| PW25 | GAATTCGGTACCGCGATTACCTCGGCTTGAT | EcoRI/KpnI |
| PW26 | GAATTCACTAGTGCGACCATCTGTTCGAAGG | EcoRI/SpeI |
| PW136 | TAATACGACTCACTATAGGGAGAGCGCTGCTGAATTATCGTC | |
| PW137 | CGACCAGCAAGAGATCAATG | |
| PW140 | GTCGGCATCGACGAACTATT | |
| PW141 | ATCCGAACAGCTTGAACAGC |
Restriction sites are underlined; the T7 promoter is shown in boldface.
Construction of an R. palustris shc deletion strain.
The shc deletion strain was constructed in R. palustris TIE-1 as previously described (22, 23). Briefly, a deletion plasmid construct, pPVW6, was made by fusing 1 kb of the shc upstream region with 1 kb of the downstream region in the suicide vector pJQ200SK (46). pPVW6 was integrated onto the R. palustris TIE-1 chromosome through homologous recombination with either the shc upstream or downstream region. Because pJQ200SK contains a gentamicin resistance marker, as well as the sacB gene, the resulting R. palustris strain was both gentamicin resistant and sucrose sensitive (46). To remove the plasmid from the chromosome and generate the Δshc mutant, one gentamicin-resistant colony was grown in YP broth without any antibiotic selection for 2 days. Because loss of pPVW6 from the chromosome restored sucrose resistance, serial dilutions were plated on YP agar supplemented with 10% sucrose. Several sucrose-resistant colonies were verified as deletion mutants by PCR (data not shown), and one Δshc colony was picked for further characterization. A more detailed explanation of the method used to generate this mutant is presented in the supplemental material.
Complementation of the Δshc strain.
The shc gene plus 1 kb upstream and downstream of the gene was PCR amplified with primers PW25 and PW26 using the Failsafe PCR System. The 4-kb PCR fragment was cloned into the cloning vector pCR8.1 using the pCR8/GW/TOPO TA cloning kit (Invitrogen, Carlsbad, CA) and electroporated into E. coli DH10B. The complementation fragment was subcloned from pCR8.1 into pBBR1MCS-2, a cloning vector shown to self-replicate in R. palustris TIE-1 (23, 27). The resulting plasmid, pPVW8, was transformed into the E. coli mating strain BW29427 and mated into the Δshc mutant strain on YP agar plates supplemented with diaminopimelic acid (23). Transformants were selected on YP agar containing 400 μg/ml kanamycin. Kanamycin-resistant colonies were screened for the presence of pPVW8 by PCR. All growth conditions included kanamycin at 400 μg/ml to maintain the plasmid in this strain.
Electron microscopy of R. palustris strains.
R. palustris cultures were grown chemoheterotrophically as described above. Mid-exponential- and late-stationary-phase cultures were harvested by centrifugation (5,000 × g for 10 min) and washed twice in 50 mM HEPES buffer (pH 6.8). The cells were enrobed in 2% (wt/vol) Noble agar and placed in 2% glutaraldehyde for 2 h. The agar blocks were then washed twice in HEPES buffer and fixed in 2% osmium tetraoxide (OsO4) for 2 h, followed by 2% (wt/vol) uranyl acetate staining for 2 h. The blocks were then dehydrated through a graded ethanol series (25%, 50%, 75%, 95%, and three times at 100%) for 15 min in each solution. The blocks were suspended in a 50/50 ethanol/LR White resin, a polyhydroxy-aromatic acrylic resin, solution for 30 min, followed by 100% LR White for 1 h. Samples were then embedded in gelatin capsules filled with fresh LR White resin, which was allowed to polymerize at 60°C for 1 h. The capsules were thin sectioned on a Reichert-Jung Ultracut E ultramicrotome, and ultrathin sections were mounted on Formvar carbon-coated copper grids. To improve contrast, the grids containing thin sections were poststained in 2% (wt/vol) uranyl acetate. Electron microscopy was performed on a Jeol JEM-1200EXII transmission electron microscope.
qRT-PCR.
For exponential-phase versus stationary-phase assays, R. palustris TIE-1 was grown aerobically in 10 ml of unbuffered YP medium. All 10 ml of cells was transferred in mid-exponential phase (17 h after inoculation; optical density at 600 nm [OD600], 0.1 to 0.2) or in late stationary phase (72 h after inoculation; OD600, 0.3 to 0.4) into 2 volumes of RNAprotect Bacteria Reagent (Qiagen, Valencia, CA), incubated for 5 min at room temperature, and centrifuged for 20 min at 5,000 × g. RNA was extracted from the cell pellet using the RNeasy Mini Kit (Qiagen, Valencia, CA). Genomic DNA contamination was removed from each RNA sample with Turbo DNA-free DNase (Ambion, Foster City, CA). cDNA was generated from 100 ng of the extracted RNA with the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). The cDNA (2 μl) was then used as a template for quantitative PCR using the iTaq SYBR green Supermix with Rox (Bio-Rad, Hercules, CA) and primers PW140 and PW141, which are specific for the shc gene. All samples were assayed in triplicate, and the cycle time was determined automatically by the Real Time 7500 PCR software (Applied Biosystems, Foster City, CA). Primers (Integrated DNA Technologies, Coralville, IA) for quantitative reverse transcriptase (qRT) PCR were designed using Primer3 software (50) and are listed in Table 2.
For pH shock assays, R. palustris TIE-1 was grown either chemoheterotrophically in 100 ml of YP medium buffered at pH 7 with 100 mM MOPS or photoheterotrophically in 10 ml FW medium supplemented with 10 mM acetate (N2 headspace) and buffered at pH 6.5 with 50 mM MES. For chemoheterotrophic assays, 50 ml of cells was removed in late stationary phase (72 to 96 h; OD600, 0.3 to 0.4) and split into five tubes (10 ml/tube). The cells were centrifuged at 5,000 × g for 20 min at 4°C. The pellets were resuspended in 5 ml of sterile YP medium buffered with 100 mM MES (pH 5 and 6), 100 mM MOPS (pH 7), or 100 mM bicine (pH 8 and 9) and incubated for 30 min at 30°C in the dark with shaking. For photoheterotrophic assays, all treatments were done under anoxic conditions. Eight milliliters of stationary-phase cells (48 h; OD600, 1.3 to 1.5) was split into four tubes (2 ml/tube) and centrifuged at 10,000 × g for 1 min at room temperature. The cells were resuspended in 2 ml of sterile anoxic FW medium buffered with 50 mM MES (pH 6.5) or 50 mM HEPES (pH 7.5, 8.0, or 8.5) and incubated for 30 min at 30°C in the light without shaking. After the pH shock incubation, both chemoheterotrophic and photoheterotrophic cells were immediately transferred to twice the cell volume of RNAprotect Bacteria Reagent (Qiagen, Valencia, CA), incubated for 5 min at room temperature, and centrifuged for 20 min at 5,000 × g. RNA extraction and qRT-PCR were performed as described above.
To generate a standard curve of the shc amplicon (12), an 847-bp fragment of the shc gene with the T7 promoter inserted at the 5′ end was amplified with primers PW136 and PW137. The PCR product was gel purified using the Qiagen Gel Extraction Kit (Qiagen, Valencia, CA), and 100 ng of the PCR product was used in the Megascript T7 Kit (Ambion, Foster City, CA) to generate an in vitro-transcribed sense RNA transcript. The transcription reaction mixture was treated with Turbo DNA-free DNase to remove any DNA template and further purified with the Megaclear Kit (Ambion, Foster City, CA) to remove unincorporated nucleoside triphosphates, enzymes, and buffer components. The concentration of each transcript was determined by averaging triplicate measurements of its concentration on a NanoDrop 1000 (Thermo Scientific, Waltham, MA). Assuming the average mass of a ribonucleotide was 340 Da, the concentration of each transcript was converted to copy numbers/μl, diluted to 2 × 1010 copies/μl, and stored at −80°C. The standard curve was generated by performing two independent 10-fold serial dilutions of the RNA standard and using 5 μl of each dilution in the reverse transcription reaction. Two microliters of the cDNA was used as a template for quantitative PCR using the shc-specific primers PW140 and PW141. A standard curve was generated by plotting cycle time values against the log of the initial copy number. The copy number of shc transcripts in each RNA sample was calculated after real-time amplification from the linear regression of the standard curve.
Total hopanoid quantification in R. palustris TIE-1.
For comparison of hopanoid abundances in exponential versus stationary phase, R. palustris TIE-1 was grown under chemoheterotrophic and photoheterotrophic conditions. Cultures (1 liter) were harvested in log phase (17 h after inoculation; OD600, 0.1 to 0.2) or stationary phase (72 to 96 h after inoculation; OD600, 0.3 to 0.4 for chemoheterotrophic growth and 0.9 to 1.0 for photoheterotrophic growth) by centrifugation at 5,000 × g for 20 min at 4°C. The cells were washed once in 10 ml of unbuffered YP medium, and the cell pellets were frozen and lyophilized overnight.
For hopanoid quantification after pH shock, R. palustris TIE-1 was grown aerobically in YP medium (1 liter) buffered at pH 7 with 100 mM MOPS to late stationary phase (72 to 96 h; OD600, 0.3 to 0.4); 900 ml of culture was removed and split into three centrifuge bottles (300 ml/bottle). The cells were centrifuged at 5,000 × g for 20 min at 4°C. The pellets were resuspended in 150 ml sterile YP medium buffered at pH 5 (100 mM MES), pH 7 (100 mM MOPS), or pH 9 (100 mM bicine) and incubated overnight with shaking at 30°C in the dark. The cells were pelleted at 5,000 × g for 20 min at 4°C and washed once with 10 ml of the appropriate YP medium (at pH 5, 7, or 9). The cell pellets were frozen and lyophilized overnight.
GC-MS analysis.
For initial hopanoid analysis of the Δshc mutant (Fig. 2), cells were grown chemoheterotrophically in 250 ml YP to late stationary phase (5 days). The cells were harvested by centrifugation at 5,000 × g for 20 min at 4°C. The cell pellets were frozen and lyophilized overnight in a VirTis K-series freeze dryer (VirTis, Gardiner, NY). Lipids were extracted by sonicating the cells in Teflon centrifuge tubes (VWR, Bridgeport, NJ) for 15 min at room temperature in 10 ml of 10:5:4 methanol (MeOH)/dichloromethane (DCM)/water (5). Samples were centrifuged for 10 min at 3,000 × g, and the supernatant was transferred to a new tube. The cell pellets were sonicated again in 10 ml of MeOH/DCM/water and centrifuged, and the supernatant was combined with the first extraction. The samples were separated into two phases by adding 10 ml each of DCM and water and centrifuged for 10 min at 3,000 × g, and the organic phase was transferred to a new vial. To the remaining aqueous phase, 10 ml each of DCM and water was added again and centrifuged, and the organic phase was again removed and combined with the previous extraction. This was repeated two more times for a total of four extractions. The total lipid extract was evaporated under N2 and derivatized to acetate esters for gas chromatography/mass spectrometry (GC-MS) analysis by incubation in 100 μl of 1:1 acetic anhydride/pyridine for 1 h at 70°C. To suppress the decomposition of unstable compounds, the derivatized samples were injected directly into acetic anhydride/pyridine. For total hopanoid quantification in R. palustris TIE-1 (see Table 4), dry cells were weighed and then extracted by ultrasonication in a mixture of 10:5:4 MeOH/DCM/water (5). This total lipid extract was dried at room temperature under N2, weighed, and redissolved in DCM. A 0.5-mg aliquot of the total lipid extract was taken for GC-MS analysis, and an internal standard (7 μg epiandrostanol) was added. This mixture was acetylated by heating it with acetic anhydride/pyridine at 80°C for 15 min.
FIG. 2.
GC-MS chromatograms of total lipid extracts from TIE-1 strains. (A) R. palustris TIE-1. (B) R. palustris Δshc. (C) R. palustris Δshc plus pPVW8. The numbered compounds are as follows: I, hopene (desmethyl and 2-methyl); II, tetrahymanol (desmethyl, 2-methyl, and 20-methyl); III, triglycerides; IV, unidentified BHP; V, bacteriohopanetetrol (desmethyl and 2-methyl); VI, aminobacteriohopanetriol; VII, squalene. Retention times for each compound are listed in Table S1 in the supplemental material. At this point, we have not identified compound IV, and it remains unclear whether this is a true hopanoid product of R. palustris TIE-1 or a degradation product of the high-temperature hopanoid analysis method.
TABLE 4.
Hopanoid production and methylation at C-2 by R. palustris TIE-1
| Growth conditiona | Total hopanoidsb
|
Bacteriohopanetetrolb
|
||
|---|---|---|---|---|
| μg/mg dry biomass | % 2-Me | μg/mg dry biomass | % 2-Me | |
| Chemoheterotrophic | ||||
| Exponential | 25.3 ± 5.3 | 6.3 ± 0.9 | 3.4 ± 0.7 | 0.1 ± 0.1 |
| Stationary | 23.1 ± 7.1 | 20.0 ± 0.7 | 3.0 ± 1.4 | 3.4 ± 0.6 |
| Photoheterotrophic | ||||
| Exponential | 36.7 ± 1.6 | 47.1 ± 1.5 | 10.2 ± 0.6 | 9.8 ± 0.5 |
| Stationary | 34.2 ± 3.1 | 65.1 ± 3.2 | 8.2 ± 1.3 | 24.6 ± 5.9 |
| pH shock | ||||
| pH 5 | 20.8 ± 2.6 | 50.7 ± 1.2 | 1.8 ± 0.5 | 6.1 ± 3.3 |
| pH 7 | 27.3 ± 4.5 | 47.3 ± 2.8 | 3.3 ± 0.5 | 5.9 ± 0.9 |
| pH 9 | 27.7 ± 0.9 | 54.4 ± 1.8 | 2.6 ± 0.4 | 13.3 ± 0.8 |
See Materials and Methods for detailed growth and assay conditions.
Each value represents the average and standard error of three biological replicates grown under identical conditions. 2-Me, 2-methyl.
The lipid extracts were analyzed by GC-MS, and we employed a novel protocol for high-temperature GC-MS analysis that allowed functionalized BHPs to be directly analyzed. For the initial analysis of the Δshc mutant (Fig. 2), lipid extracts were separated on a Hewlett-Packard 6890 series gas chromatograph equipped with a DB1-HT column (15 m by 0.25 mm [inside diameter] by 0.1-μm film thickness) with helium as the carrier at a constant flow of 1 ml/minute and programmed as follows: 80°C for 2 min, ramped 10°C /minute to 360°C, and held 20 min. One microliter of the sample was injected into a Gerstel programmable temperature vaporization injector operated in splitless mode at 80°C and programmed to 360°C at 720°C/minute, where it was held for the duration of the run. The gas chromatograph was coupled to a 5975 series MSD (mass selective detector) with the source at 200°C and operated at 70 eV in EI (electron ionization) mode scanning from 50 to 850 Da in 0.5 s.
For total hopanoid quantification of R. palustris TIE-1 (see Table 4), lipid extracts were separated on a Thermo Scientific TraceGC. Samples were injected into a programmable temperature vaporization injector operated in splitless mode at 60°C with a 2-mm (inside diameter) deactivated glass liner and programmed to 325°C for the duration of the run. A DB-XLB column (30 m by 0.25 mm by 0.10-mm film thickness) was employed and was temperature programmed from 100 to 350°C, holding 15 min at the maximum temperature. The GC was coupled to a Thermo DSQ mass spectrometer with the source at 220°C and operated at 70 eV in EI mode scanning from 50 to 750 Da in 0.3 s. Using this program, bacteriohopanetetrol acetate eluted in approximately 38 min, while the desmethyl and 2-methyl homologs remained chromatographically resolved. We did not observe the bacteriohopaneaminotriol using this method and column, although its presence was detected using the DB-1HT GC-MS method described above and has been previously observed by liquid chromatography-MS analyses (48).
Compounds were identified by comparison of retention times and mass spectra to those of authentic compounds (tetrahymanol from Trimyema sp., diplopterol from Methylococcus capsulatus, and bacteriohopanepentol from M. capsulatus) and published mass spectra (hopenes [55] and tetrahymanol [60]). Retention times and molecular ions are listed in Table S1 in the supplemental material. Methylation at C-2 was confirmed for bacteriohopanetetrol by comparison of mass spectra and retention times with those from Phormidium luridum (48). For all other compounds, C-2 methylation was inferred from the shift of fragments with m/z 191 to 205 and m/z 369 to 383 and from the relative retention time compared to the desmethyl homolog (∼0.15 min earlier). Concentrations and 2-methyl/desmethyl ratios were calculated by comparing total ion chromatogram peak areas between the analytes and the internal standard and assuming identical response factors. This assumption was required because no pure standards are available for these compounds.
RESULTS
A squalene-hopene cyclase deletion mutant in R. palustris TIE-1 does not produce hopanoids.
To better understand the physiological role of hopanoids in photosynthetic bacteria, we constructed a deletion mutant in R. palustris TIE-1 that no longer produced any hopanoids. A key step in the biosynthesis of hopanoids is the cyclization of squalene to the basic hopene structure, diploptene (Fig. 1C) (26). This reaction is catalyzed by the squalene-hopene cyclase, which is encoded by the shc gene (42). Utilizing the genomic data available from the R. palustris TIE-1 genome, we identified and made an in-frame deletion of the shc gene in this organism. The resulting Δshc mutant accumulated a significant amount of squalene and did not produce any hopanoids, nor did it produce the triterpenoid tetrahymanol (Fig. 2). As can be seen in Fig. 1A, tetrahymanol differs from the hopanoids in having a six-membered E ring and no functionalized side chain. Previous in vitro characterization of the Shc protein from R. palustris demonstrated that the enzyme was able to convert squalene to diploptene but not to tetrahymanol (26). For that reason, it was presumed that another cyclase was involved in tetrahymanol biosynthesis. However, the deletion of shc in R. palustris showed that the Shc protein is indeed required to form this pentacyclic triterpenoid in vivo.
To verify that the deletion of shc was responsible for the loss of hopanoid production, a copy of the TIE-1 shc gene was introduced into the Δshc mutant on a self-replicating plasmid. Although the complemented (Δshc+pPVW8) strain still accumulated squalene, the strain was now able to produce all of the known R. palustris hopanoids (Fig. 1A and 2). Therefore, the lack of hopanoid production in the Δshc strain was due only to the deletion of shc.
Lack of hopanoids results in increased membrane permeability.
Because hopanoids have been shown to localize to the inner and outer membranes of some bacteria (21, 24, 54), we hypothesized that a lack of hopanoids might lead to membrane damage in R. palustris. A strong indicator of outer membrane damage and permeability in gram-negative bacteria is sensitivity to bile salts and certain antibiotics (2, 51). As seen in Fig. 3, growth of the Δshc mutant was inhibited in the presence of bile salts, while growth of the wild type and complemented strains was not. The mutant strain is also sensitive to the antibiotics erythromycin and rifampin (rifampicin), both inhibitors of protein synthesis (51). The Δshc strain had a 7-mm zone of inhibition with erythromycin, while both TIE-1 and the complemented strain were resistant to the antibiotic (data not shown). The wild-type strain did display some sensitivity to rifampin, with a zone of inhibition of 5 mm. However, the zone of inhibition in the deletion strain increased to 10 mm, indicating that the lack of hopanoids increased sensitivity (data not shown). Together, these data demonstrate that hopanoids may be important in decreasing membrane permeability and, like sterols in eukaryotes, may play a role in maintaining membrane integrity.
FIG. 3.
Δshc cells are more permeable to bile salts. The Δshc strain is unable to grow in the presence of bile salts (1.5%), while both the wild type and the Δshc complemented strain (Δshc+pPVW8) are resistant.
Hopanoids are important in the stationary phase of chemoheterotrophic growth.
The Δshc mutant was able to grow on all growth substrates tested. Although photoautotrophic and photoheterotrophic growth were slightly slower in the Δshc mutant than in the wild-type strain (Table 3), the deletion strain was still able to achieve the same final OD. Thus, hopanoids are not required for photosynthesis in R. palustris.
TABLE 3.
Doubling times of the R. palustris Δshc strain under various growth conditions
| Growth conditiona | Doubling time (h)b
|
|
|---|---|---|
| R. palustris TIE-1 | R. palustris Δshc | |
| Chemoheterotrophic | 3.8 ± 0.1 | 4.1 ± 0.1 |
| Photoheterotrophic | 6.1 ± 0.2 | 7.6 ± 0.7 |
| Photoautotrophic | 8.0 ± 0.2 | 9.0 ± 0.3 |
See Materials and Methods for detailed growth conditions.
Growth rates were measured by monitoring the OD600 during growth, and the doubling times were calculated from three replicate cultures. Each value represents the average and standard deviation of three biological replicates grown under identical conditions.
The growth rate during aerobic chemoheterotrophic growth was the same in the deletion mutant as in the wild-type strain (Table 3). However, a significant drop in OD was observed when cells entered stationary phase during chemoheterotrophic growth (Fig. 4A). Transmission electron microscopy images of both the wild type and deletion mutant illustrated that the Δshc strain was severely damaged in stationary phase but not in exponential phase (Fig. 4C and D). This morphological defect in stationary phase may explain the observed drop in OD.
FIG. 4.
Δshc chemoheterotrophic growth and morphology. (A) Chemoheterotrophic growth of TIE-1 and the Δshc strain in unbuffered medium (A) or buffered medium (B) at pH 7. Each time point represents the average of three replicate cultures (the error bars represent standard deviations and may not be visible beneath the data point markers). Each growth curve was repeated at least three times, and representative growth curves are shown. The circled points in panel A indicate when cultures were harvested for transmission electron microscopy. (C) Transmission electron microscopy of chemoheterotrophically grown TIE-1 and Δshc cells harvested during mid-exponential growth (17 h after inoculation). (D) Transmission electron microscopy of chemoheterotrophically grown TIE-1 and Δshc cells harvested during stationary phase (72 h after inoculation). The arrows indicate the formation of the inner cytoplasmic membrane (lamellar membrane) as anoxic phototrophic growth was induced.
The Δshc mutant is sensitive to alkaline conditions.
Several studies with E. coli have demonstrated that the pH of rich media, such as LB or YP, increases in stationary phase (11). We reasoned that the drop in OD and the morphological damage seen in stationary phase for the Δshc mutant might be due to an increase in pH. We measured the pH of the growth medium at the beginning and end of chemoheterotrophic growth and found that the pH had increased from 7.2 to 8.2 for both TIE-1 and Δshc (Fig. 4A). The growth experiment was repeated in YP medium buffered with 100 mM MOPS at pH 7. In the buffered medium, the pH remained constant throughout the growth curve and the drop in OD observed in unbuffered medium no longer occurred (Fig. 4B). This indicated that the growth defect of the Δshc mutant in stationary phase was related to the increase in pH.
To determine if the drop in OD observed in the Δshc mutant was specific to stationary-phase cells and alkaline conditions, both strains were grown in YP medium buffered at pH 4.5, 5, 6, 8, or 9. Because the medium was buffered, the pH should have remained constant throughout growth and the cells would have experienced either acidic or alkaline stress. Neither the wild type nor the mutant was able to grow at pH 9 (Fig. 5) or at pH 4.5 (see Fig. S1 in the supplemental material). Both strains were able to grow at pH 5, 6, and 8, but the Δshc mutant had a growth defect at pH 5 and pH 8 (Fig. 5). Taken together, these data indicate that the wild type may be sensitive to both acidic and alkaline stress and that the decreased membrane permeability of the Δshc mutant may aggravate this sensitivity. Interestingly, the growth of the Δshc mutant was similar to that of the wild type during exponential phase under each pH tested. It was primarily during stationary phase that the behavior of the strains deviated from one another, particularly at pH 8. Hopanoids thus seem to be most relevant in protecting stationary-phase cells from pH stress.
FIG. 5.
pH effect on Δshc chemoheterotrophic growth. Shown are chemoheterotrophic growth curves of the TIE-1 and Δshc strains in YP medium buffered at pH 5 (A), pH 6 (B), pH 8 (C), and pH 9 (D). Each time point represents the average of three replicate cultures (the error bars represent standard deviations and may not be visible beneath the data point markers). Each growth curve was repeated at least three times, and representative growth curves are shown.
pH stress inhibits phototrophic growth in the Δshc mutant.
The high cell density of stationary-phase cultures often results in oxygen-limiting conditions (45). Given that oxygen limitation stimulates the transition from aerobic chemoheterotrophic growth to anaerobic phototrophic growth in R. palustris (7), it seemed likely that chemoheterotrophically grown TIE-1 cells in stationary phase were switching from oxic growth to anoxic growth. This was confirmed by the presence of photosynthetic lamellar membranes, whose development is stimulated by oxygen deprivation (7), in stationary-phase TIE-1 cells but not in exponential-phase cells (Fig. 4C and D). Therefore, we hypothesized that the acidic and alkaline growth defect observed primarily in stationary-phase Δshc cells may have been related to phototrophic growth. To test this, we grew Δshc and TIE-1 cells photoheterotrophically in 10 mM acetate buffered with 50 mM MES at pH 4.5, 5.0, 5.5, and 6.0 and buffered with 50 mM HEPES at pH 6.5, 7.5, 8, and 8.5.
Both the wild type and the Δshc strain were unable to grow at pH 5.0 or lower (see Fig. S2 in the supplemental material), but the wild-type strain was able to grow to a maximum OD600 of 1.5 when grown at pH 5.5 or higher (Fig. 6). Wild-type cells grown at pH 6.0, pH 8.0, and pH 8.5 exhibited an extensive lag phase compared to growth at pH 6.5 or 7.5, indicating that wild-type cells grown photoheterotrophically, like cells grown chemoheterotrophically, were also sensitive to both acidic and alkaline conditions. While the Δshc strain had growth characteristics similar to those of the wild type at pH 6.5 and 7.5, the mutant strain had a much longer lag phase at pH 6.0 and pH 8.0 and did not grow at pH 5.5 or pH 8.5 (Fig. 6). Therefore, while hopanoids are not required for photosynthetic growth at neutral pH, these data indicate that they are required for photosynthetic growth at acidic or alkaline pH.
FIG. 6.
pH effect on Δshc photoheterotrophic growth. Shown is photoheterotrophic growth of TIE-1 and Δshc strains in FW plus HEPES or FW plus MES medium supplemented with 10 mM sodium acetate under an N2 headspace buffered at pH 5.5 (A), pH 6.0 (B), pH 6.5 (C), pH 7.5 (D), pH 8.0 (E), and pH 8.5 (F). Each time point represents the average of three replicate cultures, and the error bars represent standard errors. Each growth curve was repeated at least three times, and representative growth curves are shown.
Transcription of the shc gene is minimally affected by pH shock.
Given that hopanoids play a role in protecting TIE-1 cells against exposure to high and low pH, we sought to determine whether this role was regulated in response to pH. If hopanoids were specifically required for coping with alkaline stress in stationary phase, we hypothesized that transcription of the shc gene would be upregulated during stationary phase in unbuffered medium. To test this, TIE-1 cells were grown chemoheterotrophically in unbuffered YP medium and RNA was isolated from samples taken during exponential phase (17 h; pH ∼7) and stationary phase (72 h; pH ∼7.8). qRT-PCR was used to measure shc transcript levels in stationary-phase cells relative to log-phase cells, with a change in transcript levels greater than 2-fold or less than 0.5-fold considered significant. Although the Δshc mutant exhibited a severe growth phenotype in stationary phase, transcription of the shc gene did not increase in stationary phase but rather remained constant, with an average change in expression of 0.7- ± 0.2-fold (data not shown).
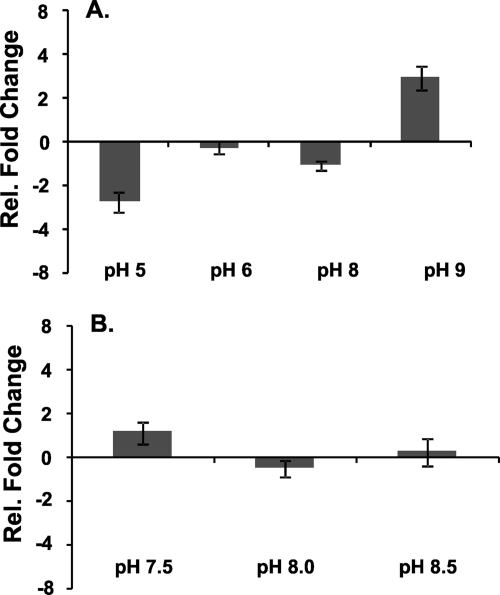
To determine if pH stress would increase the expression of shc, transcript levels of shc were also measured after TIE-1 cells had been shocked with acidic or alkaline medium. TIE-1 cells were grown chemoheterotrophically (buffered at pH 7) or photoheterotrophically (buffered at pH 6.5) to stationary phase. The cells were then shocked for 30 min in the same medium buffered at acidic, neutral, or alkaline pH. qRT-PCR was used to measure shc transcript levels under each shock condition relative to the neutral-pH incubation. As shown in Fig. 7A, we observed slight shifts in shc expression when chemoheterotrophically grown cells were shocked at pH 5 and 9. However, we did not observe any significant changes in shc expression under any pH shock condition for photoheterotrophically grown cells (Fig. 7B).
FIG. 7.
pH effect on shc expression. qRT-PCR was used to measure the expression levels of the shc gene after pH shock. (A) Chemoheterotrophically grown cells (pH 7) were incubated in YP medium buffered at pH 5, 6, 7, 8, or 9 for 30 min. (B) Photoheterotrophically grown cells (pH 6.5) were incubated in FW medium buffered at pH 6.5, 7.5, 8.0, or 8.5 for 30 min. The transcript levels of shc were determined for each sample as described in Materials and Methods. Each data bar represents the change in transcript levels at the indicated pH relative to the pH 7 sample (A) or the pH 6.5 sample (B). The change was graphed on a log2 scale, and a change greater than 2 or less than 0.5 was considered significant. Each data point represents the average of biological triplicates, and the error bars represent standard errors.
Total hopanoid production is not regulated by growth phase or pH.
There are several possible reasons why increases in shc transcript levels during stationary phase or pH shock might not be physiologically important. First, the shc gene may be regulated posttranscriptionally, which qRT-PCR cannot detect. Alternatively, the shc gene may be constitutively expressed, which would prevent significant changes in transcript levels from one condition to another. If this were correct, then we would expect to see similar amounts of hopanoids produced under all conditions. To test this, the total hopanoid content was quantified in chemoheterotrophically and photoheterotrophically grown TIE-1 cells harvested during log phase and stationary phase, as well as cells incubated overnight under acidic or alkaline conditions.
Consistent with the qRT-PCR data, similar amounts of hopanoids were produced during log phase and stationary phase of chemoheterotrophic and photoheterotrophic growth (Table 4). We observed a significant increase in methylation at the C-2 position during stationary phase under both growth conditions (Table 4, 2-Me). A similar result was seen when chemoheterotrophically grown cells were transferred to alkaline pH. The amounts of hopanoids produced were the same under each pH; however, methylation at C-2 increased when the cells were subjected to alkaline conditions. In particular, methylation of one subset of hopanoids, the bacteriohopanetetrols, increased more than twofold at pH 9 compared to incubation at pH 7. These data, along with the qRT-PCR data, indicate that shc is constitutively expressed. Furthermore, these data suggest that the modification of hopanoids, such as methylation at C-2, changes in response to growth conditions.
DISCUSSION
Although hopanoids have been hypothesized to be sterol analogs in bacteria, the true physiological roles of these molecules have received little direct attention. A few studies have proposed that hopanoids are necessary for maintaining membrane integrity, as well as coping with external stresses, such as ethanol tolerance in Z. mobilis (19), oxygen diffusion in Frankia sp. (4), and prevention of water diffusion into Streptomyces spores (44). In vivo genetic studies with Bacillus subtilis have shown that sporulenes, polycyclic terpenoids similar to hopanoids, may be important in alleviating oxidative stress in spores (6). However, no in vivo genetic studies have been used to address the function of hopanoids in any strain, much less photosynthetic bacteria, which are of special interest to geobiologists. Given the ubiquity and abundance of hopanes in the rock record and their role as organic proxies for ancient bacterial life and metabolism, particularly oxygenic photosynthesis, it is important to understand how these molecules are used by photosynthetic bacteria. Therefore, we set out to determine the physiological role of hopanoids in the genetically tractable anoxygenic phototroph R. palustris TIE-1.
Deletion of the squalene-hopene cyclase gene in R. palustris TIE-1 resulted in a strain that no longer produced any hopanoids. Physiological studies of the Δshc mutant revealed that hopanoids are not required for growth in R. palustris TIE-1, as the mutant had growth characteristics similar to those of the wild type under standard chemoheterotrophic, photoautotrophic, and photoheterotrophic conditions. This demonstrates that hopanoids are not directly involved in anoxygenic photosynthesis and suggests that they may not be appropriate biomarkers for photosynthesis more generally.
The sensitivity of the Δshc mutant to acidic and alkaline conditions was unexpected, given that R. palustris TIE-1 was isolated from an iron-rich mat located in a marsh with a pH range of 6 to 7 (22). Because the majority of R. palustris strains have been isolated from freshwater marsh sediments, it seems unlikely that hopanoid production evolved in these organisms as a mechanism to overcome pH stress (32, 33). This is consistent with our finding that hopanoid production is not regulated by acidic or alkaline conditions. Total hopanoid production remained constant when TIE-1 cells were exposed to high pH, either during stationary-phase growth or when they were shocked for a short time. If hopanoid production is not directly affected by pH stress, then what role might these molecules play in helping TIE-1 survive under acidic or alkaline conditions?
Studies of E. coli have demonstrated that acidic and alkaline exposure induces heat shock and SOS cellular responses, showing that extreme pH can be stressful for bacteria (13, 29, 52, 58, 63). The main effect of exposure to high or low pH is an immediate shift in the cytoplasmic pH, which the cell must readjust to maintain the optimal function and integrity of cytoplasmic proteins (37, 62). Several studies have revealed that cells are able to buffer their cytoplasmic pH immediately through the use of sodium/proton antiporters, symporters, and efflux pumps that transport protons in and out of the cytoplasm and, as a result, adjust the pH to more favorable conditions (10, 15, 28, 38).
Because cations and protons play important roles in alleviating acidic and alkaline stress, it is plausible that some bacterial cells have developed secondary mechanisms to prevent the accidental loss of protons and other cations. In particular, alkaliphilic Bacillus spp. have been shown to produce secondary wall polymers that associate with the peptidoglycan layer and presumably are able to bind cations and enhance their availability for pH homeostasis (1, 14). These bacteria also have high levels of squalene in their membranes, and it has been proposed that the high levels of this lipid might lower the permeability of protons across the lipid bilayer (18).
Interestingly, this same role has been put forward for both hopanoids in bacteria and sterols in eukaryotes (17). It has been proposed that both sterols and hopanoids may be able to pack the hydrophobic centers of lipid bilayers, preventing the loss of protons as charged water (17). Therefore, hopanoids may have an indirect but critical function in extreme-pH tolerance by preventing the unintentional loss of cations and protons across the inner and outer membranes. If this is true, then we might expect the lack of hopanoids to result in leaky membranes, making antiporters and overall pH homeostasis less efficient. The transmission electron microscopy images of the Δshc mutant (Fig. 4C and D), together with its sensitivity to bile salts and specific antibiotics, imply that the membranes of the Δshc mutant are damaged. Because the wild type is already somewhat sensitive to acidic and alkaline conditions, the increased membrane permeability resulting from the lack of hopanoids in the Δshc mutant likely exacerbates this sensitivity. It remains unclear why this would affect phototrophically grown cells more than chemoheterotrophically grown cells. Because phototrophic growth induces the production of lamellar membranes for photosynthesis (7), under these growth conditions, more membranes are present. It is conceivable that more protons and cations might leak in the hopanoid mutant during phototrophic growth simply because there is a larger membrane area. Under these conditions, antiporters might be even less efficient, and the cell would be unable to neutralize its cytoplasm, which would result in impaired growth.
The constitutive expression of shc shows that hopanoids are produced under all growth conditions. It is interesting that while the total amount of hopanoids produced remains constant, the percentage of hopanoids methylated at C-2 increases significantly as cells shift from exponential to stationary phase, as well as in response to pH shock (Table 4). This suggests that it may be how hopanoids are modified rather than their total abundance that is important when bacteria encounter environmental stress. This agrees with what has been proposed for other hopanoid-producing bacteria. In A. acidocaldarius, a thermophilic acidophile, it was found that increasing temperature and decreasing pH did not alter the total amount of hopanoids produced, but production was shifted toward hopanoids with extended side chains (43). In Frankia sp., hopanoids were localized primarily to the nitrogenase vesicle; however, nitrogen-depleted conditions did not result in increased hopanoid production. It was speculated that nitrogen limitation causes hopanoids to congregate in the nitrogenase vesicle (4, 30).
In summary, our data suggest that hopanoids, like sterols, may be important in maintaining membrane integrity and for modulating membrane permeability. However, we do not yet know whether these effects are direct or indirect. It is possible that the lack of hopanoids altered the membrane architecture and/or the membrane protein content of the Δshc mutant and that the phenotypes we observed resulted from these effects. Furthermore, our study did not distinguish which cyclic triterpenoids might be important for membrane permeability. Because R. palustris produces a variety of hopanoid molecules, as well as tetrahymanol, more work is needed to address how methylation and side chain modifications of the core hopanoid structure affect the functions of these molecules in R. palustris and other hopanoid-producing bacteria. These and other aspects of the cell biology of hopanoids will be the subjects of our future research.
Supplementary Material
Acknowledgments
This work was supported by grants from the NASA Exobiology Program to A.L.S., D.K.N., and R.E.S. and an NSF Postdoctoral Minority Fellowship to P.V.W. D.K.N. is an Investigator of the Howard Hughes Medical Institute.
We thank David Doughty and Alex Poulain for comments on the manuscript and members of the Newman laboratory for helpful discussions.
Footnotes
Published ahead of print on 10 July 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aono, R., and M. Ohtani. 1990. Loss of alkalophily in cell wall component defective mutants derived from alkalophilic Bacillus C-125. Isolation and partial characterization of the mutants. Biochem. J. 266:933-936. [PMC free article] [PubMed] [Google Scholar]
- 2.Begley, M., C. G. Gahan, and C. Hill. 2005. The interaction between bacteria and bile. FEMS Microbiol. Rev. 29:625-651. [DOI] [PubMed] [Google Scholar]
- 3.Benz, R., D. Hallmann, K. Poralla, and H. Eibl. 1983. Interaction of hopanoids with phosphatidylcholines containing oleic and omega cyclohexyldodecanoic acid in lipid bilayer membranes. Chem. Phys. Lipids 34:7-24. [DOI] [PubMed] [Google Scholar]
- 4.Berry, A. M., O. T. Harriott, R. A. Moreau, S. F. Osman, D. R. Benson, and A. D. Jones. 1993. Hopanoid lipids compose the Frankia vesicle envelope, presumptive barrier of oxygen diffusion to nitrogenase. Proc. Natl. Acad. Sci. USA 90:6091-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bligh, E. G., and W. J. Dyer. 1959. A rapid method for total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 6.Bosak, T., R. M. Losick, and A. Pearson. 2008. A polycyclic terpenoid that alleviates oxidative stress. Proc. Natl. Acad. Sci. USA 105:6725-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braatsch, S., J. R. Bernstein, F. Lessner, J. Morgan, J. C. Liao, C. S. Harwood, and J. T. Beatty. 2006. Rhodopseudomonas palustris CGA009 has two functional ppsR genes, each of which encodes a repressor of photosynthesis gene expression. Biochemistry 45:14441-14451. [DOI] [PubMed] [Google Scholar]
- 8.Brocks, J. J., and A. Pearson. 2005. Building the biomarker tree of life, p. 233-258. In J. E. Banfield, J. Cervini-Silva, and K. H. Nealson (ed.), Molecular geomicrobiology. Reviews in mineralogy and geochemistry vol. 59. Mineralogical Society of America, Chantilly, VA. [Google Scholar]
- 9.Brocks, J. J., and R. E. Summons. 2004. Sedimentary hydrocarbons, biomarkers for early life, p. 63-115. In W. H. Schlesinger (ed.), Treatise on geochemistry, vol. 8. Elsevier-Pergamon, Oxford, United Kingdom. [Google Scholar]
- 10.Cotter, P. A., V. Chepuri, R. B. Gennis, and R. P. Gunsalus. 1990. Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in Escherichia coli is regulated by oxygen, pH, and the fnr gene product. J. Bacteriol. 172:6333-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrell, M. J., and S. E. Finkel. 2003. The growth advantage in stationary-phase phenotype conferred by rpoS mutations is dependent on the pH and nutrient environment. J. Bacteriol. 185:7044-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fey, A., S. Eichler, S. Flavier, R. Christen, M. G. Hofle, and C. A. Guzman. 2004. Establishment of a real-time PCR-based approach for accurate quantification of bacterial RNA targets in water, using Salmonella as a model organism. Appl. Environ. Microbiol. 70:3618-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster, J. W. 2000. Microbial responses to acid stress, p. 99-115. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, DC.
- 14.Gilmour, R., P. Messner, A. A. Guffanti, R. Kent, A. Scheberl, N. Kendrick, and T. A. Krulwich. 2000. Two-dimensional gel electrophoresis analyses of pH-dependent protein expression in facultatively alkaliphilic Bacillus pseudofirmus OF4 lead to characterization of an S-layer protein with a role in alkaliphily. J. Bacteriol. 182:5969-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldberg, E. B., T. Arbel, J. Chen, R. Karpel, G. A. Mackie, S. Schuldiner, and E. Padan. 1987. Characterization of a Na+/H+ antiporter gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 84:2615-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grunwald, C. 1974. Sterol molecular modifications influencing membrane permeability. Plant Physiol. 54:624-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haines, T. H. 2001. Do sterols reduce proton and sodium leaks through lipid bilayers? Prog. Lipid Res. 40:299-324. [DOI] [PubMed] [Google Scholar]
- 18.Hauss, T., S. Dante, N. A. Dencher, and T. H. Haines. 2002. Squalane is in the midplane of the lipid bilayer: implications for its function as a proton permeability barrier. Biochim. Biophys. Acta 1556:149-154. [DOI] [PubMed] [Google Scholar]
- 19.Horbach, S., B. Neuss, and H. Sahm. 1991. Effect of azasqualene on hopanoid biosynthesis and ethanol tolerance of Zymomonas mobilis. FEMS Microbiol. Lett. 79:347-350. [Google Scholar]
- 20.Hossack, J. A., and A. H. Rose. 1976. Fragility of plasma membranes in Saccharomyces cerevisiae enriched with different sterols. J. Bacteriol. 127:67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jahnke, L. L., H. Stan-Lotter, K. Kato, and L. I. Hochstein. 1992. Presence of methyl sterol and bacteriohopanepolyol in an outer-membrane preparation from Methylococcus capsulatus Bath. J. Gen. Microbiol. 138:1759-1766. [DOI] [PubMed] [Google Scholar]
- 22.Jiao, Y., A. Kappler, L. R. Croal, and D. K. Newman. 2005. Isolation and characterization of a genetically tractable photoautotrophic Fe(II)-oxidizing bacterium, Rhodopseudomonas palustris strain TIE-1. Appl. Environ. Microbiol. 71:4487-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiao, Y., and D. K. Newman. 2007. The pio operon is essential for phototrophic Fe(II) oxidation in Rhodopseudomonas palustris TIE-1. J. Bacteriol. 189:1765-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jurgens, U. J., P. Simonin, and M. Rohmer. 1992. Localization and distribution of hopanoids in membrane systems of the cyanobacterium Synechocystis PCC 6714. FEMS Microbiol. Lett. 71:285-288. [DOI] [PubMed] [Google Scholar]
- 25.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 26.Kleemann, G., R. Kellner, and K. Poralla. 1994. Purification and properties of the squalene-hopene cyclase from Rhodopseudomonas palustris, a purple non-sulfur bacterium producing hopanoids and tetrahymanol. Biochim. Biophys. Acta 1210:317-320. [DOI] [PubMed] [Google Scholar]
- 27.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 28.Lewinson, O., E. Padan, and E. Bibi. 2004. Alkalitolerance: a biological function for a multidrug transporter in pH homeostasis. Proc. Natl. Acad. Sci. USA 101:14073-14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maurer, L. M., E. Yohannes, S. S. Bondurant, M. Radmacher, and J. L. Slonczewski. 2005. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J. Bacteriol. 187:304-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nalin, R., S. R. Putra, A. M. Domenach, M. Rohmer, F. Gourbiere, and A. M. Berry. 2000. High hopanoid/total lipids ratio in Frankia mycelia is not related to the nitrogen status. Microbiology 146:3013-3019. [DOI] [PubMed] [Google Scholar]
- 31.Neunlist, S., P. Bisseret, and M. Rohmer. 1988. The hopanoids of the purple non-sulfur bacteria Rhodopseudomonas palustris and Rhodopseudomonas acidophila and the absolute configuration of bacteriohopanetetrol. Eur. J. Biochem. 171:245-252. [DOI] [PubMed] [Google Scholar]
- 32.Oda, Y., F. W. Larimer, P. S. Chain, S. Malfatti, M. V. Shin, L. M. Vergez, L. Hauser, M. L. Land, S. Braatsch, J. T. Beatty, D. A. Pelletier, A. L. Schaefer, and C. S. Harwood. 2008. Multiple genome sequences reveal adaptations of a phototrophic bacterium to sediment microenvironments. Proc. Natl. Acad. Sci. USA 105:18543-18548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oda, Y., W. Wanders, L. A. Huisman, W. G. Meijer, J. C. Gottschal, and L. J. Forney. 2002. Genotypic and phenotypic diversity within species of purple nonsulfur bacteria isolated from aquatic sediments. Appl. Environ. Microbiol. 68:3467-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ourisson, G., and P. Albrecht. 1992. Hopanoids. 1. Geohopanoids—the most abundant natural products on Earth. Acc. Chem. Res. 25:398-402. [Google Scholar]
- 35.Ourisson, G., P. Albrecht, and M. Rohmer. 1979. Hopanoids—palaeochemistry and biochemistry of a group of natural products. Pure Appl. Chem. 51:709-729. [Google Scholar]
- 36.Ourisson, G., M. Rohmer, and K. Poralla. 1987. Prokaryotic hopanoids and other polyterpenoid sterol surrogates. Annu. Rev. Microbiol. 41:301-333. [DOI] [PubMed] [Google Scholar]
- 37.Padan, E., E. Bibi, M. Ito, and T. A. Krulwich. 2005. Alkaline pH homeostasis in bacteria: new insights. Biochim. Biophys. Acta 1717:67-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Padan, E., N. Maisler, D. Taglicht, R. Karpel, and S. Schuldiner. 1989. Deletion of ant in Escherichia coli reveals its function in adaptation to high salinity and an alternative Na+/H+ antiporter system(s). J. Biol. Chem. 264:20297-20302. [PubMed] [Google Scholar]
- 39.Parales, R. E., and C. S. Harwood. 1993. Regulation of the pcaIJ genes for aromatic acid degradation in Pseudomonas putida. J. Bacteriol. 175:5829-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parks, L. W., and W. M. Casey. 1995. Physiological implications of sterol biosynthesis in yeast. Annu. Rev. Microbiol. 49:95-116. [DOI] [PubMed] [Google Scholar]
- 41.Peng, L., Y. Kawagoe, P. Hogan, and D. Delmer. 2002. Sitosterol-beta-glucoside as primer for cellulose synthesis in plants. Science 295:147-150. [DOI] [PubMed] [Google Scholar]
- 42.Perzl, M., I. G. Reipen, S. Schmitz, K. Poralla, H. Sahm, G. A. Sprenger, and E. L. Kannenberg. 1998. Cloning of conserved genes from Zymomonas mobilis and Bradyrhizobium japonicum that function in the biosynthesis of hopanoid lipids. Biochim. Biophys. Acta 1393:108-118. [DOI] [PubMed] [Google Scholar]
- 43.Poralla, K., T. Hartner, and E. Kannenberg. 1984. Effect of temperature and pH on the hopanoid content of Bacillus acidocaldarius. FEMS Microbiol. Lett. 23:253-256. [Google Scholar]
- 44.Poralla, K., G. Muth, and T. Hartner. 2000. Hopanoids are formed during transition from substrate to aerial hyphae in Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 189:93-95. [DOI] [PubMed] [Google Scholar]
- 45.Price-Whelan, A., L. E. Dietrich, and D. K. Newman. 2007. Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J. Bacteriol. 189:6372-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 47.Rajakumari, S., K. Grillitsch, and G. Daum. 2008. Synthesis and turnover of non-polar lipids in yeast. Prog. Lipid Res. 47:157-171. [DOI] [PubMed] [Google Scholar]
- 48.Rashby, S. E., A. L. Sessions, R. E. Summons, and D. K. Newman. 2007. Biosynthesis of 2-methylbacteriohopanepolyols by an anoxygenic phototroph. Proc. Natl. Acad. Sci. USA 104:15099-15104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohmer, M., P. Bouviernave, and G. Ourisson. 1984. Distribution of hopanoid triterpenes in prokaryotes. J. Gen. Microbiol. 130:1137-1150. [Google Scholar]
- 50.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 51.Ruiz, N., B. Falcone, D. Kahne, and T. J. Silhavy. 2005. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121:307-317. [DOI] [PubMed] [Google Scholar]
- 52.Schuldiner, S., V. Agmon, J. Brandsma, A. Cohen, E. Friedman, and E. Padan. 1986. Induction of SOS functions by alkaline intracellular pH in Escherichia coli. J. Bacteriol. 168:936-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 54.Simonin, P., U. J. Jurgens, and M. Rohmer. 1996. Bacterial triterpenoids of the hopane series from the prochlorophyte Prochlorothrix hollandica and their intracellular localization. Eur. J. Biochem. 241:865-871. [DOI] [PubMed] [Google Scholar]
- 55.Summons, R. E., and L. L. Jahnke. 1992. Hopenes and hopanes methylated in ring-A: correlation of the hopanoids of extant methylotrophic bacteria with their fossil analogues, p. 189-200. In J. M. Moldowan, P. Albrecht, and R. P. Philip (ed.), Biomarkers in sediments and petroleum. Prentice Hall, Englewood Cliffs, NJ.
- 56.Summons, R. E., L. L. Jahnke, J. M. Hope, and G. A. Logan. 1999. 2-Methylhopanoids as biomarkers for cyanobacterial oxygenic photosynthesis. Nature 400:554-557. [DOI] [PubMed] [Google Scholar]
- 57.Swan, T. M., and K. Watson. 1998. Stress tolerance in a yeast sterol auxotroph: role of ergosterol, heat shock proteins and trehalose. FEMS Microbiol. Lett. 169:191-197. [DOI] [PubMed] [Google Scholar]
- 58.Taglicht, D., E. Padan, A. B. Oppenheim, and S. Schuldiner. 1987. An alkaline shift induces the heat shock response in Escherichia coli. J. Bacteriol. 169:885-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Talbot, H. M., R. E. Summons, L. L. Jahnke, C. S. Cockell, M. Rohmer, and P. Farrimond. 2008. Cyanobacterial bacteriohopanepolyol signatures from cultures and natural environmental settings. Org. Geochem. 39:232-263. [Google Scholar]
- 60.Ten Haven, H. L., M. Rohmer, J. Rullkotter, and P. Bisseret. 1989. Tetrahymanol, the most likely precursor of gammacerane, occurs ubiquitously in marine sediments. Geochim. Cosmochim. Acta 53:3073-3079. [Google Scholar]
- 61.Thomas, D. S., J. A. Hossack, and A. H. Rose. 1978. Plasma-membrane lipid composition and ethanol tolerance in Saccharomyces cerevisiae. Arch. Microbiol. 117:239-245. [DOI] [PubMed] [Google Scholar]
- 62.Wilks, J. C., and J. L. Slonczewski. 2007. pH of the cytoplasm and periplasm of Escherichia coli: rapid measurement by green fluorescent protein fluorimetry. J. Bacteriol. 189:5601-5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zilberstein, D., V. Agmon, S. Schuldiner, and E. Padan. 1984. Escherichia coli intracellular pH, membrane potential, and cell growth. J. Bacteriol. 158:246-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zundel, M., and M. Rohmer. 1985. Prokaryotic triterpenoids 1. 3 β-Methylhopanoids from Acetobacter species and Methylococcus capsulatus. Eur. J. Biochem. 150:23-27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.