Abstract
Streptomyces griseus mutants exhibiting deficient glucose repression of β-galactosidase activity on lactose-containing minimal medium supplemented with a high concentration of glucose were isolated. One of these mutants had a 12-bp deletion in cebR, which encodes a LacI/GalR family regulator. Disruption of cebR in the wild-type strain caused the same phenotype as the mutant, indicating that CebR is required for glucose repression of β-galactosidase activity. Recombinant CebR protein bound to a 14-bp inverted-repeat sequence (designated the CebR box) present in the promoter regions of cebR and the putative cellobiose utilization operon, cebEFG-bglC. The DNA-binding activity of CebR was impaired by cellooligosaccharides, including cellobiose, cellotriose, cellotetraose, cellopentaose, and cellohexaose. In agreement with this observation, transcription from the cebE and cebR promoters was greatly enhanced by the addition of cellobiose to the medium. Seven other genes containing one or two CebR boxes in their upstream regions were found in the S. griseus genome. Five of these genes encode putative secreted proteins: two cellulases, a cellulose-binding protein, a pectate lyase, and a protein of unknown function. These five genes and cebEFG-bglC were transcribed at levels 4 to 130 times higher in the ΔcebR mutant than in the wild-type strain, as determined by quantitative reverse transcription-PCR. These findings indicate that CebR is a master regulator of cellulose/cellooligosaccharide catabolism. Unexpectedly, the ΔcebR mutant formed very few aerial hyphae on lactose-containing medium, demonstrating a link between carbon source utilization and morphological development.
The gram-positive filamentous bacteria of the genus Streptomyces are representative soil microorganisms. Although recent metagenomic studies revealed the surprising genetic diversity of soil microorganisms, including bacteria, archaea, and fungi (10, 18), Streptomyces spp. have been considered to comprise the major fraction of soil decomposers (26). Accounting for its niche, studies of complete Streptomyces genome sequences have revealed a huge number of genes encoding enzymes for the utilization of divergent nutrient sources, such as cellulose, chitin, xylan, and their hydrolysis products. For example, the model organism Streptomyces coelicolor A3(2) encodes 172 sets of secreted proteins that probably function in the metabolism of complex soil substrates and 81 ATP-binding cassette (ABC) transporters that might be used in the uptake of saccharides, oligopeptides, and nucleosides (5, 6).
Another characteristic of Streptomyces is that these prokaryotic cells undergo complex morphological development resembling that of eukaryotic filamentous fungi. The typical stages of the streptomycete life cycle on solid medium include germination from spores, substrate growth, formation of multinucleoid aerial hyphae, and sporulation by septation of aerial hyphae. Studies of the bld and whi series of morphological differentiation mutants, which are arrested at the stages of substrate growth and formation of aerial hyphae, respectively, have made extraordinary contributions to our knowledge of Streptomyces morphological development (8). The checkpoint mechanisms for these complicated developmental events have been investigated and elucidated in S. coelicolor A3(2) and Streptomyces griseus (9). Interestingly, most of the bld mutants also lack the capability for catabolite control or “carbon catabolite repression” (31). However, the molecular mechanism that links morphogenesis to the regulation of carbon source utilization, including carbon catabolite repression, is largely unknown.
In our studies of carbon catabolite repression in S. griseus, we have observed that S. griseus colonies turn blue when grown on lactose-containing minimal medium with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). The blue color indicates the production of β-galactosidase, which hydrolyzes the β-galactoside bond of X-Gal to yield a blue pigment. However, the colonies remain white when glucose is present at a high concentration, indicating that glucose represses β-galactosidase production, while the mechanism of induction of β-galactosidase activity by lactose is unknown in Streptomyces.
Here, we isolated several S. griseus mutants that formed blue colonies even in the presence of high concentrations of glucose, indicating an apparent escape from glucose catabolite repression of β-galactoside activity. Analysis of one of the mutants, GRD1, revealed that a mutation in the cebR gene encoding a LacI/GalR family transcriptional regulator was responsible for the phenotype. The purpose of this study was to characterize the gene product, CebR, and investigate the involvement of CebR in catabolite repression. CebR was found to be a master regulator of cellulose/cellooligosaccharide catabolism in S. griseus, indicating that CebR is indirectly involved in glucose catabolite repression. Although molecular mechanisms for the apparent escape from catabolite repression in a strain with cebR deleted (ΔcebR) remained to be elucidated, we found that the ΔcebR strain formed very few aerial hyphae on lactose-containing medium. Thus, the ΔcebR mutant is the first conditional bald mutant that almost fails to form aerial mycelia in the presence of lactose rather than glucose. This result reemphasizes a possible link between morphogenesis and the regulation of carbon source utilization, including carbon catabolite repression.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The wild-type strain S. griseus IFO13350 (equivalent to NBRC102592) was obtained from the Institution of Fermentation (Osaka, Japan) (29). The S. griseus mutant GRD1 was derived from the wild-type strain IFO13350 by UV-induced mutagenesis. Escherichia coli strains JM109 and JM110 (Takara Biochemicals) and BL21(DE3)Gold (Stratagene) and plasmids pUC19 and pET-26b(+) (Invitrogen) were used for DNA manipulation in E. coli.
S. griseus was routinely cultured at 30°C in YMP medium or standard minimal medium (SMM) supplemented with 5 μg/ml thiostrepton or neomycin when necessary. YMP is a nutrient-rich medium (pH 7.2) consisting of 0.2% yeast extract, 0.14% Ehrlich's fish extract (Kyokuto), 0.75% meat extract powder (Kyokuto), 0.4% Bacto peptone, 0.5% NaCl, and 0.2% MgSO4·7H2O. SMM (pH 7.2) consisted of 0.9% glucose, 0.9% l-asparagine, 0.2% (NH4)2SO4, 0.24% Tris, 0.1% NaCl, 0.05% K2SO4, 0.02% MgSO4·7H2O, 0.01% CaCl2, 1% trace element solution (14), and 2.5 mM KH2PO4. To examine the morphological development of S. griseus strains, glucose or another carbon source was added to the media at various concentrations. R5 medium (14) was used for the regeneration of protoplasts. Inorganic salt (IS) medium (0.07% K2HPO4, 0.03% KH2PO4, 0.05% MgSO4·7H2O, 0.001% FeSO4·7H2O, 0.03% NH4NO3, and appropriate sugar [27]) was used for screening for S. griseus mutants defective in glucose catabolite repression. The media and growth conditions for E. coli were described by Maniatis et al. (24). Ampicillin (50 μg/ml) or kanamycin (20 μg/ml) was used when necessary. The Streptomyces plasmid pIJ922 (22), which contained the thiostrepton resistance gene and had an estimated copy number of one per genome, was used for construction of a chromosomal DNA library of S. griseus.
General recombinant DNA studies.
Restriction enzymes, T4 DNA ligase, PrimeStar HS DNA polymerase, ExTaq DNA polymerase, and other DNA-modifying enzymes were purchased from Takara Biochemicals. [γ-32P]ATP (220 TBq/mmol) for 5′-end labeling with T4 polynucleotide kinase was purchased from Perkin-Elmer Inc. DNA was manipulated in Streptomyces (14, 20) and in E. coli (3, 24) as described previously. When DNA fragments were amplified by PCR, the absence of PCR errors was confirmed by nucleotide sequencing on a Beckman CEQ 8000 DNA analysis system. All primers used in this study are shown in Table S1 in the supplemental material.
UV mutagenesis.
Spores of S. griseus IFO13350 were irradiated with long-wavelength UV light to yield a survival ratio of approximately 0.5% and spread on IS agar medium (IS-GLX) containing 1% glucose, 0.5% lactose, and 0.006% X-Gal. After incubation for 7 days at 28°C, blue colonies were picked for further analyses.
Shotgun cloning.
S. griseus IFO13350 chromosomal DNA was partially digested with Sau3AI. The resultant 6- to 10-kb DNA fragments were ligated into BamHI-digested pIJ922, and the ligation mixture was introduced by protoplast transformation into the S. griseus mutant strain GRD1. Thiostrepton (5 μg/ml)-resistant transformants were selected on R5 regeneration medium and then inoculated onto IS-GLX agar medium. Transformants that formed white colonies on the medium were isolated and further analyzed.
Gene disruption and complementation.
A 7.3-kb EcoRI-BglII fragment of pGRL2 (Fig. 1A), which contained the full-length shotgun-cloned S. griseus sequence and part of the pIJ922 sequence, was subcloned into pUC19 between the EcoRI and BamHI sites to form pUC-GRL2. A 2.3-kb NruI fragment containing a 3′ portion of cebG, all of bglC, and a 5′ portion of cebR was excised from pUC-GRL2 and introduced into the HincII site of pUC19 to yield pDCEBR-1. A 0.6-kb fragment containing a 3′ portion of cebR and a 5′ portion of SGR4740 was amplified by PCR with the primers DR-for (containing EcoRI and NcoI sites) and DR-rev (containing a HindIII site), digested with EcoRI and HindIII, and subcloned between the EcoRI-HindIII sites of pUC18 to yield pDCEBR-2. The 0.5-kb NcoI-XbaI fragment of pDCEBR-2 and the 2.9-kb XbaI-EcoRI fragment of pUC-GRL2, which contains the 3′ portion of SGR4740, ornA, the 5′ portion of adpA, and a 0.6-kb fragment derived from pIJ922, were subcloned together into pDCEBR-1 between the NcoI and EcoRI sites by three-fragment ligation to yield pDCEBR. On pDCEBR, a 2.3-kb fragment upstream from cebR and a 2.3-kb fragment downstream from cebR are directly connected. The 3.4-kb kanamycin resistance (Kmr) determinant obtained by digestion of Tn5 (4) with HindIII was cloned into the HindIII site of pUC19. A 1.3-kb fragment containing the Kmr gene (aphII) was excised from the plasmid by digestion with BamHI (on the multiple-cloning site of pUC19) and SmaI (downstream from aphII) and subcloned between the BamHI-SmaI sites of pUC19 to yield pUC-APH. The 1.3-kb HindIII fragment containing the Kmr gene (aphII) was excised from pUC-APH and introduced into the HindIII site of pDCEBR, resulting in pDCEBR-aph. On pDCEBR-aph, aphII is located next to a 4.6-kb recombinant fragment containing both upstream and downstream sequences from cebR. This plasmid was amplified in E. coli JM110, isolated, alkali denatured, and introduced by protoplast transformation into S. griseus IFO13350, and neomycin (5 μg/ml)-resistant colonies by a single crossover were isolated. After one of the neomycin-resistant transformants had been cultured in the absence of neomycin several times, neomycin-sensitive colonies by a second crossover were isolated as candidates for true cebR disruptants. The disruption was confirmed by PCR and sequencing of the amplified DNA fragment.
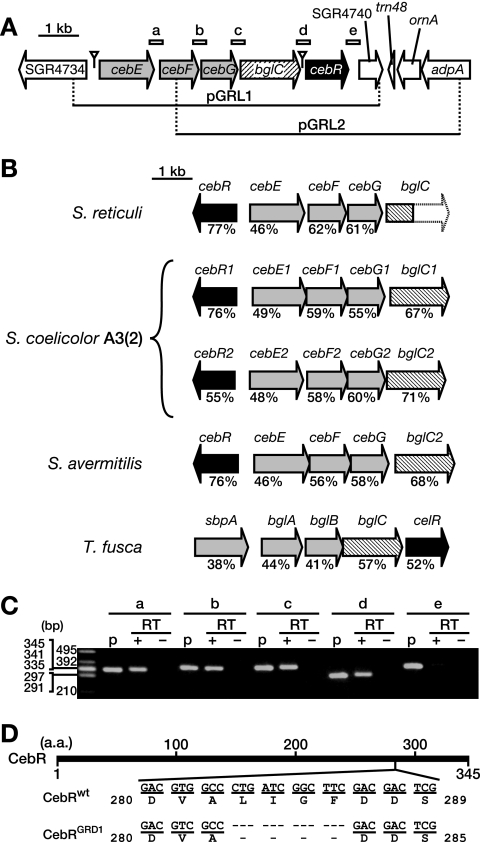
FIG. 1.
Cloning and sequence analysis of the ceb operon in S. griseus. (A) Gene organization of the insert DNA fragments (indicated as horizontal lines) of pGRL1 and pGRL2. The ORFs and their directions on the cloned fragments are indicated by arrows. Light-gray, hatched, and black arrows indicate the genes encoding ABC transporter subunits, a β-glucosidase, and a LacI/GalR family transcriptional regulator, respectively. The open triangles indicate complete CebR boxes (Table 1). The regions amplified by RT-PCR in panel C (described below) are indicated by short bars labeled a, b, c, d, and e. (B) Gene organization of the ceb homologs in actinomycetes. The values below the arrows indicate amino acid identity with the corresponding genes in S. griseus. The genome of S. coelicolor A3(2) includes two ceb gene clusters. The entire sequence of bglC of S. reticuli has not yet been determined. (C) RT-PCR analysis of the transcriptional units in the ceb gene cluster. Amplification of five intergenic regions, a (between cebE and cebF), b (cebF and cebG), c (cebG and bglC), d (bglC and cebR), and e (cebR and SGR4740) (Fig. 1A), was attempted. To confirm amplification of the DNA fragments, pGRL1 was used as a PCR template (lanes p). No amplification occurred when reverse transcriptase was omitted from the reaction mixture (lanes RT−), indicating the absence of DNA in the mRNA samples. (D) Site of the cebR mutation in the GRD1 mutant. The bold horizontal line represents the CebR protein. The nucleotide and deduced amino acid (a.a.) sequences of cebR of the wild-type strain (CebRwt) and the GRD1 mutant (CebRGRD1) are shown.
Peptidoglycan degradation activity assay.
To examine the peptidoglycan degradation activities of S. griseus strains, we supplemented YMP solid medium with dead cells of Micrococcus lysodeikticus ATCC 4698 (Sigma) at a concentration of 0.25%. S. griseus cells were grown on the medium at 28°C for 36 h, and peptidoglycan degradation activity was detected by the observation of clear halos caused by degradation of the M. lysodeikticus cell walls.
Production and purification of CebR.
The cebR sequence was amplified by PCR with the primers PR-for (containing BamHI and NdeI sites) and PR-rev (containing EcoRI and XhoI sites). The cebR coding sequence was excised as a BamHI-EcoRI fragment and subcloned into BamHI/EcoRI-digested pUC19. The NdeI-XhoI fragment was excised from this plasmid and subcloned into NdeI/XhoI-digested pET-26b(+) to create pET-CEBR. E. coli BL21(DE3)Gold cells harboring pET-CEBR were cultured at 16°C for 24 h in the presence of 0.3 mM isopropyl-β-d-thiogalactopyranoside. CebR with a C-terminal His tag (CebR-H) was purified from the soluble fraction with an Ni-nitrilotriacetic acid spin column (Qiagen) according to the manufacturer's instructions. Protein concentrations were measured using a Bio-Rad protein assay kit with bovine serum albumin as a standard.
The molecular mass of the recombinant protein CebR-H was determined by Superdex 200 gel filtration chromatography on an AKTA FPLC Purifier (GE Healthcare) in a mobile phase consisting of 50 mM Tris-HCl (pH 8.0), 200 mM NaCl, and 10% glycerol at a flow rate of 0.2 ml/min.
EMSA.
DNA fragments were amplified by PCR using appropriate primers, 5′ 32P labeled, and used as probes in electrophoretic mobility shift assays (EMSAs). For the binding assay, 0.5 nM of 32P-labeled probe DNA was incubated with CebR-H at room temperature for 15 min in a buffer containing 20 mM HEPES (pH 7.6), 1 mM EDTA, 10 mM (NH4)2SO4, 1 mM dithiothreitol, 30 mM KCl, 0.2% (vol/vol) Tween-20, and 10% (vol/vol) glycerol in a total volume of 20 μl. The resulting DNA/CebR-H complexes and free DNA were resolved on nondenaturing 6% polyacrylamide gels (mono/bis ratio, 79:1) in a running buffer containing 45 mM Tris-borate and 1 mM EDTA. The dissociation constant was calculated as the concentration of CebR-H that caused 50% of the amount of the probe DNA to form a complex with the protein.
S1 nuclease mapping.
Spores (2 × 106 CFU) were inoculated into 200 ml of liquid SMM and incubated at 30°C for 50 h with reciprocal shaking (120 rpm). Cells were collected by centrifugation, and total RNA was prepared from the cells using Isogen (Nippon Gene). S1 nuclease mapping was performed as described by Bibb et al. (7) and Kelemen et al. (19). Hybridization probes were prepared by PCR with a pair of 32P-labeled and nonlabeled primers. The PCR primers used for high-resolution S1 nuclease mapping were HR-for and HR-rev* for cebR and HLE-for and HE-rev* for cebE. The asterisks indicate 5′ labeling performed using T4 polynucleotide kinase with [γ-32P]ATP before PCR. The primers for low-resolution S1 mapping were LR-for and LR-rev* for cebR and HLE-for and LE-rev* for cebE. The hrdB (SGR1701) gene encoding a principal sigma factor of RNA polymerase was used as a control for the purity and amount of RNA, as described previously (51). Protected fragments were analyzed on 6%-polyacrylamide DNA-sequencing gels by the method of Maxam and Gilbert (25).
RT-PCR.
Total RNA was used for a standard reverse transcription (RT) reaction using the ThermoScript RT-PCR system (Invitrogen) with random hexamers according to the manufacturer's instructions. The resulting cDNA fragments, a to e, were amplified by PCR with the primer pairs rA-for/A-rev, rB-for/rB-rev, rC-for/rC-rev, rD-for/rD-rev, and rE-for/rE-rev, respectively (Fig. 1A). The amplified PCR fragments were analyzed by agarose gel electrophoresis.
Quantitative RT-PCR.
A pair of primers for each gene was designed using the SYBR green method. The housekeeping gene hrdB was used as an internal standard because it is presumably transcribed consistently throughout growth. The cDNA was synthesized using the ThermoScript RT-PCR system (Invitrogen) with random hexamers according to the manufacturer's instructions. All reactions were performed in the Takara SYBR Premix ExTaq II reaction mixture using a SmartCycler II real-time PCR system (Cepheid) with an initial denaturation at 95°C for 10 s and 50 cycles of 5 s at 95°C (denaturing) and 30 s at 60°C (annealing and extension). All reactions were performed in triplicate, and the data were normalized to the average of the internal standard.
RESULTS
Isolation of S. griseus GRD (for glucose catabolite repression-deficient) mutants by UV mutagenesis.
During our study of carbon catabolite repression in S. griseus, we noticed that β-galactosidase production was repressed by glucose. To find novel genes involved in carbon catabolite repression, we attempted to isolate mutants with deficiencies of glucose catabolite repression such that β-galactosidase activity was not repressed in the presence of a high concentration of glucose. More than 10,000 colonies formed from UV-mutated spores were screened on IS-GLX medium (1% glucose, 0.5% lactose, and 0.006% X-Gal), and 14 blue colonies that appeared to produce β-galactosidase even in the presence of a high concentration of glucose, were obtained. The wild-type S. griseus strain formed no blue colonies on this IS-GLX medium.
The morphological development of these 14 strains was tested on solid YMP medium containing a high concentration of glucose. Of the 14 mutant strains, 4 were distinct from the wild-type strain; while the wild-type strain exhibited vegetative arrest on 2% glucose-containing medium, the 4 mutant strains formed aerial mycelia even on 3% glucose-containing medium (data not shown). These mutants were designated GRD1, -2, -3, and -4.
Previous studies showed that mutations in the glucose kinase gene glkA cause a defect in glucose catabolite repression in Streptomyces (1, 2, 13, 16, 21, 35, 47). Therefore, we next determined the nucleotide sequence of the glkA locus of each mutant. The GRD2 mutant was found to harbor a point mutation at position +238 of glkA (taking the first nucleotide of the start codon as position +1), resulting in an Asp80→His mutation. However, no mutations were found in the glkA loci of the other three mutants. Because the GRD1 mutant exhibited the most extreme glucose catabolite repression-deficient phenotype (data not shown), we chose it for further analysis.
Cloning and nucleotide sequencing of DNA fragments complementing the glucose repression deficiency in the S. griseus mutant GRD1.
A chromosomal DNA library of wild-type S. griseus was constructed using a low-copy-number plasmid, pIJ922, as the cloning vector. This library was introduced into mutant GRD1 by protoplast transformation. The β-galactosidase activities of approximately 5,000 transformants were examined on IS-GLX medium, and two white colonies with apparently repressed β-galactosidase activities were isolated. Like the wild-type strain, the two transformants exhibited vegetative arrest on medium containing 2% glucose (data not shown). Two plasmids (designated pGRL1 and pGRL2) were individually extracted from these transformants, and their insert DNA fragments were identified by end sequencing and found to overlap (Fig. 1A).
Plasmid pGRL1 was found to contain five complete open reading frames (ORFs), SGR4735 to SGR4739. On the basis of sequence homology and synteny, these five ORFs were predicted to compose a gene cluster for cellobiose/cellotriose utilization; similar gene clusters in Streptomyces reticuli and Thermobifida fusca (Fig. 1B) have been characterized and identified as cellobiose/cellotriose (37) and cellobiose (41) utilization systems, respectively. Therefore, these genes were designated cebE (SGR4735), cebF (SGR4736), cebG (SGR4737), bglC (SGR4738), and cebR (SGR4739). The cebE, cebF, and cebG genes appeared to encode subunits of a sugar ABC transporter, since CebE was a homolog of sugar-binding proteins and CebF and CebG were homologs of membrane transport proteins. BglC, an apparent β-glucosidase, probably hydrolyzed cellobiose (or cellotriose) imported through the ABC transporter consisting of CebE, CebF, CebG, and an ATP-binding protein encoded by a gene not linked to the ceb operon. CebR, homologous to LacI/GalR regulators, was probably a transcriptional repressor for the gene cluster. S. coelicolor A3(2) has two gene clusters homologous to the ceb gene cluster (5), and Streptomyces avermitilis has one such cluster (17), as shown in Fig. 1B. Plasmid pGRL2 was found to contain two complete ORFs, SGR4740, encoding a chitin-binding protein, and ornA, encoding an oligoribonuclease (30), in addition to cebG, bglC, and cebR.
In S. reticuli, cebE, cebF, cebG, and bglC are cotranscribed (37). Therefore, the possible cotranscription of ORFs SGR4735 (cebE) through SGR4740 was analyzed by RT-PCR. The intervening regions between cebE and cebF (Fig. 1A, a, and C, a), cebF and cebG (b), cebG and bglC (c), and bglC and cebR (d) were amplified by RT-PCR (Fig. 1C), indicating that these five genes (cebE through cebR) were cotranscribed as an operon. In contrast, no amplification of the intervening region between cebR and SGR4740 was detected (Fig. 1C, e), indicating that SGR4740 was not cotranscribed with its upstream gene cebR. As described below, cebR appeared to harbor its own promoter in the intergenic region between bglC and cebR. Thus, cebR seems to be transcribed from not only the cebE promoter, but also the cebR promoter.
Identification of the GRD1 mutation.
As described above, three complete ORFs, cebG, bglC, and cebR, were common to pGRL1 and pGRL2. Therefore, we thought that the GRD1 mutant chromosome might harbor mutations in the coding or regulatory regions of cebG, bglC, and cebR. Nucleotide sequencing of the region covering these genes revealed a 12-bp deletion in the cebR coding region (positions +847 to +858, taking the first nucleotide of the start codon as +1), causing a deletion of amino acid residues Leu283 through Phe286 of CebR (Fig. 1D).
The glucose repression-defective phenotype of a cebR-disrupted strain.
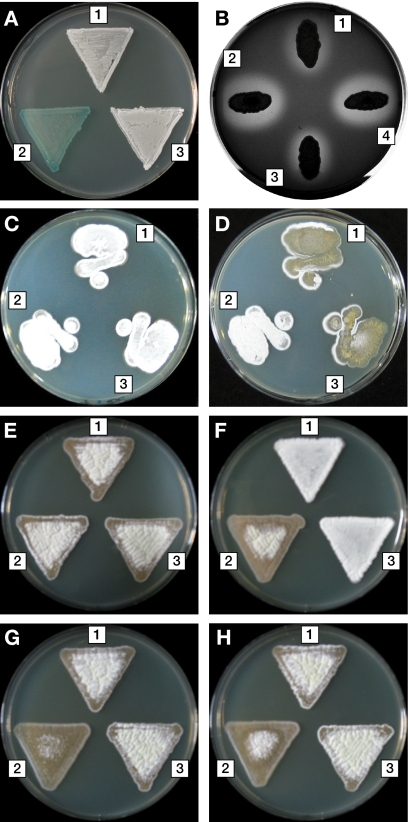
To confirm that the chromosome DNA mutation found in GRD1 was responsible for its glucose repression-defective phenotype, we disrupted the chromosomal cebR gene of S. griseus IFO13350 by deleting approximately 70% of the cebR coding sequence. This ΔcebR mutant showed the same phenotype as GRD1, i.e., defective glucose repression of β-galactosidase activity (Fig. 2A) and morphological development on 2% glucose-containing medium (Fig. 2C and D). Moreover, we found that deletion of cebR caused a defect in glucose repression of peptidoglycan degradation activity (Fig. 2B). When carried on the low-copy-number plasmid pIJ922, cebR was sufficient to rescue the glucose repression-deficient phenotype of the ΔcebR mutant (Fig. 2A to D). On the other hand, a plasmid containing cebR with the GRD1-type 12-bp deletion (pIJ922-cebRGRD1) failed to complement glucose repression of peptidoglycan degradation activity in the ΔcebR mutant, confirming that the 12-bp deletion was sufficient to abolish the function of CebR (Fig. 2B).
FIG. 2.
Glucose catabolite repression-defective phenotypes of the S. griseus ΔcebR mutant. (A) β-Galactosidase activity on IS-GLX agar medium. (B) Halo formation by peptidoglycan degradation activity on YMP medium containing 1% glucose. (C to H) Morphology on YMP agar medium supplemented with no carbohydrates (C), 2.0% glucose (D), 1.5% glucose (E), 1.5% lactose (F), 1.0% glucose plus 0.5% lactose (G), and 1.0% glucose plus 0.5% lactose and 100 mM TES (pH 7.5) (H). 1, the wild-type strain harboring pIJ922; 2, the ΔcebR mutant harboring pIJ922; 3, the ΔcebR mutant harboring pIJ922-cebRwt; and 4, the ΔcebR mutant harboring pIJ922-cebRGRD1.
Binding of CebR to a CebR box located upstream of cebE.
Two CebR homologs in S. reticuli and T. fusca have been characterized; CebR of S. reticuli is a regulator for cellobiose/cellotriose utilization genes and the avicelase gene cel1, and CelR of T. fusca is a regulator for six cellulase genes. Both proteins have been shown to bind to the same 14-bp inverted-repeat sequence, TGGGAGCGCTCCCA, located in the promoter regions of their target genes (37, 40). We found the same inverted-repeat sequence (hereafter designated the “CebR box”) in the ceb gene cluster of S. griseus; the CebR box was present in the promoter regions of cebE and cebR (Fig. 1A and Table 1).
TABLE 1.
CebR boxes found in the genome of S. griseus
| Gene ID | Deduced function | Sequence of CebR boxa | Positionb | Location | Transcription level (ΔcebR/wt)c |
|---|---|---|---|---|---|
| SGR199 | Secreted cellulose-binding protein | TGGGAGCGCTCCCA, CGGGAGCGCTCCCC | −324.5, −519.5 | TL | 39 |
| SGR217 | Secreted pectate lyase | TGGGAGCGCTCCCG | −103.5 | TL | 4.2 |
| SGR1971 | Secreted protein of unknown function | TGGGAGCGCTCCCC | −298.5 | TL | 5.5 |
| SGR2445 | Secreted cellulase | TGGGAGCGCTCCCA | −23.5 | TL | 15 |
| SGR3391 | Acetyltransferase | CGGGAGCGCTCCCG | −25.5 | TL | 1.6 |
| SGR4735 | cebE; cellobiose ABC transporter solute-binding protein | TGGGAGCGCTCCCA | +13.5 | TC | 10 |
| SGR4738 | bglC; β-glucosidase | AGGGAGCGCTCCCA | −8.5 | TL | 5.8 |
| SGR4739 | cebR; LacI/GalR family transcriptional regulator | TGGGAGCGCTCCCA | −21.5d | TC | -e |
| SGR6927f | Hypothetical protein | TGGGAGCGCTCCCA, TTGGAGCGCTCCCA | −296.5, −209.5 | TL | 2.6 |
| SGR6928f | Secreted cellulase | TGGGAGCGCTCCAA, TGGGAGCGCTCCCA | −199.5, −112.5 | TL | 130 |
Nucleotides that match the consensus CebR box sequence are underlined.
Nucleotide position of the center of the CebR box relative to the transcriptional start site (TC) or the translational start site (TL).
Average value from three quantitative-RT-PCR experiments. wt, wild type.
From the transcriptional start site of cebRp1 (Fig. 5E).
Not detected because the cebR coding region was deleted in the ΔcebR mutant.
SGR6927 and SGR6928 are divergent. Two CebR boxes exist in the intergenic region between SGR6927 and SGR6928.
To examine binding of the S. griseus CebR protein to the CebR box, we first expressed cebR in E. coli and purified recombinant CebR (Fig. 3A). The recombinant CebR having the sequence CebR-Leu-Glu-His6 was designated CebR-H. The subunit structure of CebR-H was determined using gel filtration chromatography under nondenaturing conditions. A single peak representing CebR-H eluted at 81 kDa, as determined on the basis of its elution volume, Ve. Because CebR-H had a calculated molecular mass of 38 kDa, we concluded that it formed a homodimer in solution. We confirmed that the Ve did not vary in the presence of the CebR effector molecule cellobiose (10 mM) (see below), indicating that the binding of the effector molecule does not affect the subunit structure of CebR-H.
FIG. 3.
Binding of CebR-H to the cebE promoter region. (A) CebR-H used in this study was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Lane 1, crude extract of E. coli BL21(DE3)Gold cells harboring pET-CebR; lane 2, purified CebR-H; lane 3, molecular mass standards (phosphorylase b [94 kDa], bovine serum albumin [67 kDa], ovalbumin [43 kDa], carbonic anhydrase [30 kDa], and soybean trypsin inhibitor [20 kDa]). (B) EMSA using CebR-H and a DNA fragment containing the cebE promoter region. The CebR-H homodimer was diluted stepwise by 75% from 57 nM (lane 2) to 1.4 nM (lane 15). Lane 1 shows a negative control without CebR-H. The positions of free probe (solid arrowhead) and DNA-CebR-H complexes (open arrowhead) are shown. (C) Plot of percentages of CebR-H-bound probe versus log[CebR-H (nM)] from 18 nM (lane 6) to 3.2 nM (lane 12). The apparent Kd value for CebR-H is indicated by a dashed line.
Finally, we confirmed binding of CebR-H to the CebR box using EMSAs. The recombinant protein bound to a 120-bp DNA fragment of the cebE promoter region containing a CebR box (Fig. 3B). The apparent Kd (dissociation constant) value was 8.6 nM (Fig. 3C), indicating that the DNA/CebR-H complex was quite stable.
Effects of cellooligosaccharides on DNA binding by CebR.
Regulators of the LacI/GalR family generally bind low-molecular-weight effectors that abolish their DNA-binding activities (50). The major effector of CebR of S. reticuli is cellopentaose, a cellooligosaccharide comprising five glucose units joined with β(1→4) linkages (38). On the other hand, the effector of T. fusca CelR is cellobiose rather than cellopentaose (40).
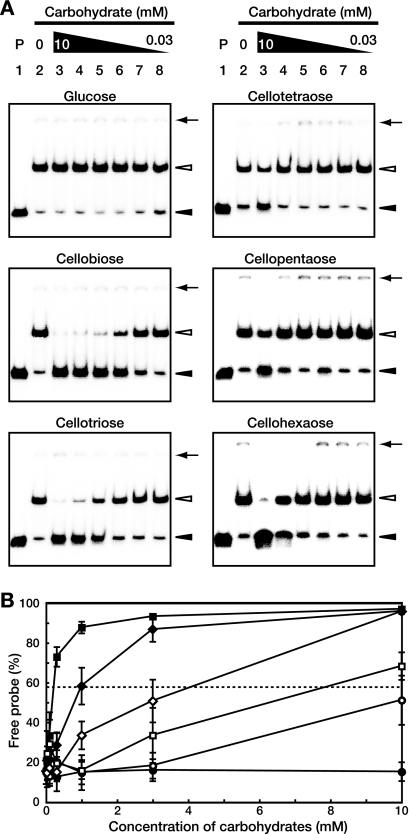
To investigate the possible effectors of S. griseus CebR, we used EMSAs to examine the binding of cellooligosaccharides consisting of two to six glucose units, i.e., cellobiose, cellotriose, cellotetraose, cellopentaose, and cellohexaose. The DNA-binding activity of CebR-H, unlike that of its S. reticuli and T. fusca CebR homologs, was impaired by all cellooligosaccharides tested (Fig. 4A). For each cellooligosaccharide, the apparent Kd was calculated as the concentration that caused 50% dissociation of the protein-DNA complex. These values were 0.22 mM for cellobiose, 0.99 mM for cellotriose, 4.1 mM for cellohexaose, 7.9 mM for cellopentaose, and approximately 12 mM for cellotetraose (Fig. 4B). Monosaccharides and other oligosaccharides, such as glucose (Fig. 4), glucose-6-phosphate, fructose-6-phosphate, maltose, lactose, or gentiobiose (data not shown), had no effect on DNA binding by CebR at concentrations of at least 20 mM. Although cellobiose was bound about 50 times more effectively than cellotetraose, the apparent Kd value of cellotetraose for CebR-H (12 mM) was nonetheless remarkable compared to those of CebR homologs from other species. For example, the DNA-binding activity of T. fusca CelR was not affected by 50 mM cellotriose (40), and the DNA-binding activity of S. reticuli CebR was not affected by 10 mM cellotriose or cellopentaose (38).
FIG. 4.
Inhibition of the DNA-binding activity of CebR by cellooligosaccharides. (A) EMSAs using CebR-H and a DNA fragment containing the cebE promoter region in the presence of the indicated carbohydrates. Lane 1, a negative control without CebR-H; lanes 2 to 8, CebR-H homodimer (at 15 nM, so that approximately 85% of the probe is shifted in the absence of carbohydrates) with carbohydrate at 0 mM (lane 2; control), 10 mM (lane 3), 3 mM (lane 4), 1 mM (lane 5), 0.3 mM (lane 6), 0.1 mM (lane 7), and 0.03 mM (lane 8). The positions of free probes (solid arrowheads), DNA-CebR-H complexes (open arrowheads), and the wells (solid arrows) are shown. (B) Plot of percentages of free probe versus concentrations of each carbohydrate: glucose (•), cellobiose (▪), cellotriose (♦), cellotetraose (○), cellopentaose (□), and cellohexaose (⋄). The values shown are averages and standard deviations from triplicate assays. A dashed line indicates 50% dissociation of the protein-DNA complex for calculation of the apparent Kd value for each cellooligosaccharide.
Transcriptional analysis of cebE and cebR by S1 nuclease mapping.
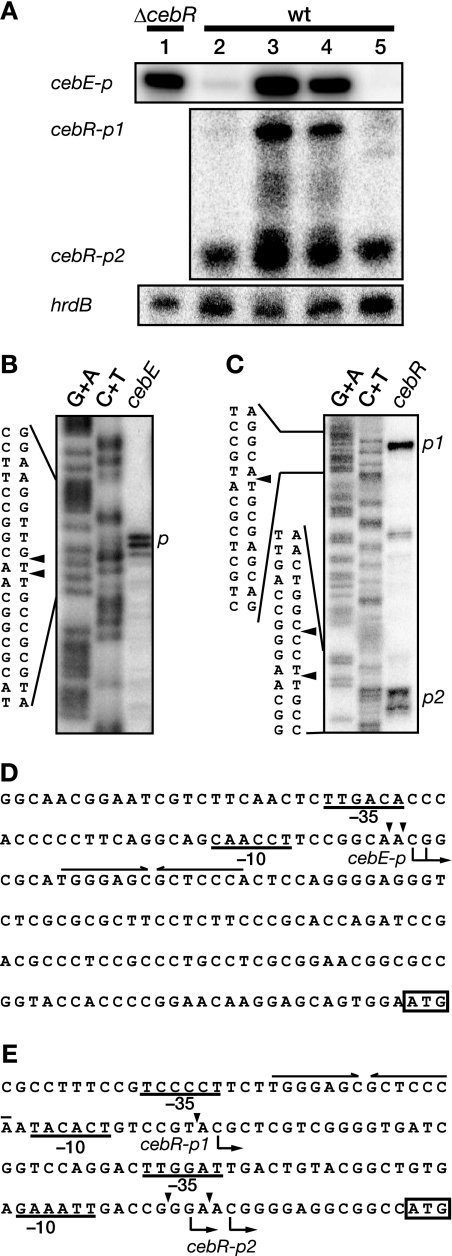
As described above, cellobiose was the most effective inhibitor of the in vitro DNA-binding activity of CebR. Therefore, we used low-resolution S1 nuclease mapping to determine whether cellobiose in the medium affected transcription from the CebR box-containing cebE and cebR promoters. RNAs were prepared from the wild-type strain grown in liquid SMM containing (i) 1% glucose, (ii) 1% cellobiose, (iii) 1% glucose plus 1% cellobiose, and (iv) 1% glycerol. The hrdB gene, which is transcribed throughout growth, was used as an internal control for the integrity and amount of mRNA. As shown in Fig. 5A, the cebE transcript was detected at a high level in the wild-type strain grown in cellobiose-containing medium (lane 3), whereas it was barely detectable in the wild-type strain grown in glucose- or glycerol-containing medium (lanes 2 and 5). Furthermore, we also analyzed transcription of cebE in the ΔcebR mutant grown in liquid SMM containing 1% glucose. As we expected, the cebE transcript was clearly detected in the ΔcebR mutant even in the absence of cellobiose (lane 1).
FIG. 5.
Transcriptional analysis of cebE and cebR. (A) Transcription of cebE and cebR in S. griseus was examined by low-resolution S1 nuclease mapping. Total RNA was prepared from the ΔcebR mutant (lane 1) or the wild-type strain (lanes 2 to 5) grown in liquid SMM containing 1% glucose (lanes 1 and 2), 1% cellobiose (lane 3), 1% glucose plus 1% cellobiose (lane 4), and 1% glycerol (lane 5) at 30°C for 50 h. (B and C) High-resolution S1 nuclease mapping of cebE (B) and cebR (C). S1-protected fragments were analyzed in parallel with the sequence ladders (lanes G+A and C+T) (24). The arrowheads indicate the positions of S1 nuclease-protected fragments. The 5′ terminus of the mRNA was assigned to the positions shown in panels D and E because fragments generated by the chemical sequencing reactions migrated 1.5 nucleotides further than the corresponding fragments generated by S1 nuclease digestion of the DNA/RNA hybrids (one-half residue from the presence of the 3′-terminal phosphate group and one residue from the elimination of the 3′-terminal nucleo tide). (D and E) Nucleotide sequences of the upstream regions of cebE (D) and cebR (E). Transcriptional start points are shown by arrows below the sequences. The probable −35 and −10 sequences are underlined. A CebR box is indicated with a pair of convergent arrows above the sequence. Probable translational start codons are boxed.
Using high-resolution S1 nuclease mapping, we determined the transcriptional start sites of cebE to be the C and G located 128 and 127 nucleotides, respectively, upstream from the translational start point of cebE (Fig. 5B). The −35 and −10 sequences for the cebE promoter (cebEp) appeared to be TTGACA and CAACCT, respectively (Fig. 5D), although the putative −10 sequence differed somewhat from the consensus −10 sequence (TAGRRT, where R = A or G) for promoters of housekeeping genes (43). Thus, the CebR box was located immediately downstream of the two adjacent transcriptional start sites of cebE. Together, these results indicated that CebR acted as a repressor for cebE and that cellobiose released the transcriptional repression of cebE by CebR. A considerable amount of cebE transcript was detected in medium containing cellobiose and glucose (lane 4), suggesting that transcription of cebE was not subject to glucose catabolite repression.
In contrast to cebE, cebR was transcribed from two different promoters (Fig. 5A). Transcription from the upstream promoter (cebRp1) was detected only in the presence of cellobiose, like transcription from cebEp (Fig. 5A). However, transcription from the downstream promoter (cebRp2) was detected regardless of the carbon source in the medium (Fig. 5A). Although cellobiose induced a low level of transcription from cebRp2, this result suggested that cebRp2 was constitutively active at a basal level.
The transcriptional start sites of cebR were determined by high-resolution S1 nuclease mapping (Fig. 5C). Transcription from cebRp1 was found to begin at the C located 78 nucleotides upstream from the translational start point of cebR. The −35 and −10 sequences for cebRp1 appeared to be TCCCCT and TACACT (Fig. 5E), respectively, although the −35 sequence differed somewhat from the consensus −35 sequence (TTGACR) for promoters of housekeeping genes (43). Thus, the CebR box was located only in cebRp1, consistent with the observation that cebRp1 was regulated by CebR.
Transcription from cebRp2 was found to begin at the G and C located 16 and 13 nucleotides, respectively, upstream from the translational start point of cebR (Fig. 5C and E). Its −35 and −10 sequences for cebRp2 appeared to be TTGGAT and GAAATT (Fig. 5E). Thus, the CebR box was located far from cebRp2, in agreement with the observation that cebRp2 was essentially independent of CebR.
Regulation of the other CebR box-containing genes by CebR.
When we searched the S. griseus genome (29) for the 14-bp CebR box sequence (TGGGAGCGCTCCCA), allowing up to two mismatches, we identified 11 sequences (Table 1) in addition to those located in the cebE and cebR upstream regions. These CebR boxes were generally located in the upstream regions of genes encoding secreted proteins for cellulose/cellooligosaccharide catabolism (Table 1). Two CebR boxes were found in the upstream region of SGR199, and two additional CebR boxes were found in the intergenic region between the divergent genes SGR6927 and SGR6928. Using EMSAs, we confirmed that CebR-H bound to the nine DNA fragments containing one or two CebR boxes (Fig. 6). CebR-H appeared to have similar binding affinities for these DNA fragments, although two shifted bands were detected for the probes for SGR199 and SGR6927/SGR6928, both of which had two CebR boxes.
FIG. 6.
Binding of CebR-H to DNA fragments containing CebR boxes, as determined by EMSAs. DNA fragments of the indicated upstream gene regions were used as probes. Fragments containing two CebR boxes are marked with asterisks. The CebR-H dimer was used at 0, 10, 20, and 40 nM. The positions of free probes (solid arrowheads), DNA-CebR-H complexes (open arrowheads), and the wells (solid arrows) are shown.
We next compared the transcription of these CebR box-containing genes in the wild-type and ΔcebR strains using quantitative RT-PCR. The strains were grown on IS agar medium supplemented with 1% glucose as the sole carbon source. Although the expression ratio (ΔcebR/wild-type) varied from 1.6 to 130, all of these genes were upregulated in the ΔcebR mutant compared with the wild-type strain (Table 1). We concluded that at least five genes (SGR199, SGR217, SGR1971, SGR2445, and SGR6928), all of which were clearly upregulated in the ΔcebR mutant (by at least fourfold), were CebR target genes, as well as cebEFG-bglC and cebR.
Carbon source-dependent morphological defects of the ΔcebR mutant.
We observed consistently incomplete morphological differentiation for the ΔcebR mutant on IS-GLX medium. To confirm this observation, the ΔcebR mutant was inoculated onto YMP solid medium, which has been routinely used for culture of S. griseus in our laboratory, supplemented with various carbon sources. The mutant was unable to complete morphological differentiation on YMP solid medium supplemented with 1.5% lactose as a carbon source (Fig. 2F). The morphological defect was enhanced for YMP medium supplemented with 1.0% glucose and 0.5% lactose (YMP-GL) (Fig. 2G), whereas the ΔcebR mutant was indistinguishable from the wild-type strain when the carbon source was 1.5% glucose (Fig. 2E) or 1.5% maltose, 1.5% galactose, 1.5% glycerol, or 1.0% glucose plus 0.5% galactose (data not shown).
In previous studies, the morphological differentiation of some S. coelicolor A3(2) mutants was inhibited by acidification of the medium, and the addition of appropriate buffers to the medium allowed these mutants to develop normally (44, 48, 49). Therefore, we added 50 mM HEPES or TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid] to the YMP-GL solid medium to maintain the pH at ∼7.5. The morphological differentiation of the ΔcebR mutant did not improve, even on solid YMP-GL media containing HEPES (data not shown) and TES (Fig. 2H), indicating that its morphological defect on lactose-containing medium was not caused by acidification of the medium.
DISCUSSION
In this study, we identified cebR as a gene required for glucose catabolite repression of β-galactosidase activity in S. griseus. A ΔcebR mutant was deficient in glucose catabolite repression; it exhibited β-galactosidase and peptidoglycan degradation activities in the presence of a high concentration of glucose, and it exhibited morphological development on medium containing 3% glucose. However, cebR is not directly involved in glucose catabolite repression; rather, we found that CebR acts as a master regulator of cellulose/cellooligosaccharide catabolism. Identification of genes encoding a β-galactosidase(s) and an enzyme(s) for peptidoglycan degradation is essential for elucidation of the molecular mechanism for catabolite repression in S. griseus. Disruption of some candidate genes on the S. griseus chromosome is in progress in our laboratory.
Concerning the β-galactosidase activity observed in the ΔcebR mutant in the presence of glucose at a high concentration, a β-glucosidase, BglC, which is ectopically overproduced in the mutant, may have some activity toward β-galactoside in addition to β-glucoside, because BglC of T. fusca, which shows 57% identity to S. griseus BglC in their amino acid sequences, can hydrolyze a β-galactosidic bond of o-nitrophenyl-β-d-galactopyranoside (41). If this is the case, the release from the catabolite repression of β-galactosidase activity in the ΔcebR mutant must be ascribed to a kind of “artifact.” However, because catabolite repression of not only β-galactosidase activity, but also peptidoglycan degradation activity and morphological development, was abolished in the ΔcebR mutant, disruption of cebR undoubtedly caused a defect in catabolite repression through some unknown mechanisms. Again, identification of genes that encode a β-galactosidase(s) and an enzyme(s) for peptidoglycan degradation is important.
Unexpectedly, the ΔcebR mutant produced very few aerial hyphae on medium containing 1.0% glucose and 0.5% lactose, although the IS-GLX medium that was used for the isolation of glucose catabolite repression-deficient mutants also contained 1.0% glucose and 0.5% lactose. Why is aerial mycelium production in the ΔcebR mutant so poor in medium containing lactose? We assume that the lack of CebR in the presence of lactose triggers a drastic disturbance in the sugar-sensing and utilization systems, which might result in vegetative arrest. Deficient morphological development caused by a disturbance of the chitin/chitooligosaccharide utilization system has been reported in S. coelicolor A3(2). In S. coelicolor A3(2), the transcriptional repressor DasR, a member of the GntR family (HutC subfamily), acts as a global regulator of chitin/chitooligosaccharide catabolism and regulates the phosphotransferase system components specific to the uptake of N-acetylglucosamine (a monomer of chitin), the ABC transporter system for the import of chitobiose, and several enzymes for chitin/chitooligosaccharide utilization (32, 33, 36). Glucosamine 6-phosphate, a central molecule in N-acetylglucosamine metabolism, is an effector of DasR that abolishes its DNA-binding activity (32). Disruption of dasR results in a bald phenotype on glucose-containing media, not only in S. coelicolor A3(2), but also in S. griseus (32, 39). Rigali et al. (32) have suggested that the significant deregulation of gene expression in the dasR mutant is a major reason for its vegetative arrest. Additionally, they have proposed that, since the polysaccharides cellulose and chitin are the most abundant on Earth, both compounds are therefore potentially crucial sources of carbon (and chitin also of nitrogen) and can constitute an important marker for nutrient availability in soil-dwelling microorganisms, implying their possible involvement in developmental control. DasR is apparently involved in the sensing of chitin and chitooligosaccharides. Here, we propose that CebR is involved in the sensing of cellulose and cellooligosaccharides in S. griseus. It is very interesting that disruption of either dasR or cebR results in vegetative arrest in medium containing a sugar that is not apparently related to the metabolites controlled by the regulator; the dasR and cebR mutants showed a bald phenotype on media containing glucose and lactose, respectively. We speculate that ectopic production of a sugar-utilizing system, in the absence of its substrate and in the presence of some other sugar, should dramatically disturb the balance in glucose catabolite repression and/or morphological development. Further studies are required to examine this speculation.
We showed that CebR binds to CebR boxes in S. griseus to repress the transcription of its target genes. In addition to the cellobiose/cellotriose uptake ABC-transporter genes cebEFG-bglC and cebR itself, at least five genes encoding secreted proteins (two cellulases, a cellulose-binding protein, a pectate lyase, and a protein with unknown function) were identified as target genes of CebR. The binding of CebR to the short intergenic region between cebG and bglC (Table 1) may inhibit elongation of transcription from cebEFG to bglC, which would enable strict regulation of the cebEFG-bglC operon by CebR. In contrast, we cannot exclude the possibility that bglC additionally has its own promoter and the promoter is repressed by the binding of CebR to the intergenic region between cebG and bglC. Because only two cellulases, both of which have been shown to be regulated by CebR in this study, are annotated in the S. griseus genome and because CebEFG, which seems to be the only transporter for cellobiose (cellooligosaccharide), is also regulated by CebR, we conclude that CebR is a master regulator of cellulose/cellooligosaccharide catabolism in S. griseus.
Despite the high frequency of CebR boxes in the T. fusca genome (e.g., in the upstream regions of six cellulase-encoding genes), Lykidis et al. (23) reported that CelR appears not to be a global regulator in T. fusca; celR transcription was diminished when T. fusca was grown on glucose- or xylan-containing medium. In comparing the transcriptional control of cebR in S. griseus with that of celR in T. fusca, it is noteworthy that cebR is transcribed constitutively from cebRp2 and in a cellobiose-dependent manner from cebRp1 in S. griseus. The constitutive transcription of cebR from cebRp2 is important for the repression of transcription of the target genes in the absence of cellulose/cellooligosaccharide. On the other hand, the cellobiose-dependent transcription from cebRp1 may contribute to strict control of CebR at the intracellular level, allowing a quick and adequate response to extracellular cellulose/cellooligosaccharide. A similar regulation (self-repression) of sugar utilization by a LacI/GalR family repressor has been reported for GlyR3, a regulator of cellulose/hemicellulose catabolism in Clostridium thermocellum (28).
We found that the DNA-binding activity of CebR was impaired by all cellooligosaccharides tested (cellobiose, cellotriose, cellotetraose, cellopentaose, and cellohexaose). This result was unexpected, because relatively strict ligand specificities have been reported for two CebR homologs. The DNA-binding activity of CelR of T. fusca is impaired by cellobiose (apparent Kd, 0.5 mM), but only minimally by cellotriose, sophorose, or xylobiose at 50 mM and not at all by other tested mono- or disaccharides at up to 100 mM (40). The DNA-binding activity of CebR of S. reticuli is abolished by cellopentaose (apparent Kd, 0.5 mM), only slightly impaired by cellobiose at 10 mM, and not at all affected by glucose, cellotriose, or cellotetraose at up to 10 mM (38). As an effector, S. griseus CebR accepts neither lactose, which differs from cellobiose only in the stereochemistry at the C-4 position of the nonreducing end of the disaccharide, nor maltose, an α-1,4-linked glucose dimer. These results suggest that S. griseus CebR recognizes only the terminal cellobiose structure of cellooligosaccharides, rather than their entire structures.
The morphological development of Streptomyces spp. involves several DNA-binding proteins belonging to large families of bacterial regulatory proteins. A representative example is WhiH, a regulatory protein of the GntR family in S. coelicolor A3(2). WhiH is specifically needed for the orderly multiple-sporulation septation of aerial hyphae (34). Many DNA-binding proteins have recently been identified as regulators of morphological development in S. coelicolor A3(2): a TetR family regulator (XdhR) (12), an AsnC family regulator (BkdR) (42), two IclR family regulators (SamR [45] and SsgR [46]), and three GntR family regulators (DevA [15], DasR [32], and Agl3R [11]). In particular, DasR and Agl3R are regulators of carbon source utilization. In this study, we have identified the first LacI/GalR family regulator involved in morphological development in Streptomyces. As described by Weickert and Adhya (50), proteins belonging to the LacI/GalR family appear to be regulators of primary metabolism, including the utilization of sugars and nucleotides. Further analyses of the integral relationships between morphological development, nutrient utilization regulation, and primary metabolism may reveal the nutrient signals required for morphological development in Streptomyces.
Supplementary Material
Acknowledgments
We thank Takeshi Fujii for technical advice concerning the β-galactosidase activity plate assay.
K. Marushima was supported by the Japan Society for the Promotion of Science. This research was supported by a Grant-in-Aid for Scientific Research on Priority Area “Applied Genomics” from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Published ahead of print on 31 July 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Angell, S., C. G. Lewis, M. J. Buttner, and M. J. Bibb. 1994. Glucose repression in Streptomyces coelicolor A3(2): a likely regulatory role for glucose kinase. Mol. Gen. Genet. 244:135-143. [DOI] [PubMed] [Google Scholar]
- 2.Angell, S., E. Schwarz, and M. J. Bibb. 1992. The glucose kinase gene of Streptomyces coelicolor A3(2): its nucleotide sequence, transcriptional analysis and role in glucose repression. Mol. Microbiol. 6:2833-2844. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingstone, D. O. Moore, J. S. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 4.Beck, E., G. Ludwig, E. A. Auerswald, B. Reiss, and H. Schaller. 1982. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene 19:327-336. [DOI] [PubMed] [Google Scholar]
- 5.Bentley, S. D., K. F. Chater, A.-M. Cerdeño-Tárraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C.-H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M.-A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 6.Bertram, R., M. Schlicht, K. Mahr, H. Nothaft, M. H. Saier, Jr., and F. Titgemeyer. 2004. In silico and transcriptional analysis of carbohydrate uptake systems of Streptomyces coelicolor A3(2). J. Bacteriol. 186:1362-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bibb, M. J., G. R. Janssen, and J. M. Ward. 1986. Cloning and analysis of the promoter region of the erythromycin-resistance gene (ermE) of Streptomyces erythraeus. Gene 41:E357-E368. [DOI] [PubMed] [Google Scholar]
- 8.Chater, K. F., and G. Chandra. 2006. The evolution of development in Streptomyces analysed by genome comparisons. FEMS Microbiol. Rev. 30:651-672. [DOI] [PubMed] [Google Scholar]
- 9.Chater, K. F., and S. Horinouchi. 2003. Signalling early developmental events in two highly diverged Streptomyces species. Mol. Microbiol. 48:9-15. [DOI] [PubMed] [Google Scholar]
- 10.Fierer, N., M. Breitbart, J. Nulton, P. Salamon, C. Lozupone, R. Jones, M. Robeson, R. A. Edwards, B. Felts, S. Rayhawk, R. Knight, F. Rohwer, and R. B. Jackson. 2007. Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl. Environ. Microbiol. 73:7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hillerich, B., and J. Westpheling. 2006. A new GntR family transcriptional regulator in Streptomyces coelicolor is required for morphogenesis and antibiotic production and controls transcription of an ABC transporter in response to carbon source. J. Bacteriol. 188:7477-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hillerich, B., and J. Westpheling. 2008. A new TetR family transcriptional regulator required for morphogenesis in Streptomyces coelicolor. J. Bacteriol. 190:61-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodgson, D. A. 1982. Glucose repression of carbon source uptake and metabolism in Streptomyces coelicolor A3(2) and its perturbation in mutants resistant to 2-deoxyglucose. J. Gen. Microbiol. 128:2417-2430. [Google Scholar]
- 14.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces: a laboratory manual. The John Innes Foundation, Norwich, United Kingdom.
- 15.Hoskisson, P. A., S. Rigali, K. Fowler, K. C. Findlay, and M. J. Buttner. 2006. DevA, a GntR-like transcriptional regulator required for development in Streptomyces coelicolor. J. Bacteriol. 188:5014-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikeda, H., E. T. Seno, C. J. Bruton, and K. F. Chater. 1984. Genetic mapping, cloning and physiological aspects of the glucose kinase gene of Streptomyces coelicolor. Mol. Gen. Genet. 196:501-507. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Ômura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 18.Janssen, P. H. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelemen, G. H., P. Brian, K. Flärdh, L. Chamberlin, K. F. Chater, and M. J. Buttner. 1998. Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3(2). J. Bacteriol. 180:2515-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 21.Kwakman, J. H., and P. W. Postma. 1994. Glucose kinase has a regulatory role in carbon catabolite repression in Streptomyces coelicolor. J. Bacteriol. 176:2694-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lydiate, D. J., F. Malpartida, and D. A. Hopwood. 1985. The Streptomyces plasmid SCP2*: its functional analysis and development into useful cloning vectors. Gene 35:223-235. [DOI] [PubMed] [Google Scholar]
- 23.Lykidis, A., K. Mavromatis, N. Ivanova, I. Anderson, M. Land, G. DiBartolo, M. Martinez, A. Lapidus, S. Lucas, A. Copeland, P. Richardson, D. B. Wilson, and N. Kyrpides. 2007. Genome sequence and analysis of the soil cellulolytic actinomycete Thermobifida fusca YX. J. Bacteriol. 189:2477-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press Cold Spring Harbor, NY.
- 25.Maxam, A. M., and W. Gilbert. 1980. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65:499-560. [DOI] [PubMed] [Google Scholar]
- 26.McCarthy, A. J., and S. T. Williams. 1992. Actinomycetes as agents of biodegradation in the environment. Gene 115:189-192. [DOI] [PubMed] [Google Scholar]
- 27.Miyashita, K., T. Fujii, and Y. Sawada. 1991. Molecular cloning and characterization of chitinase genes from Streptomyces lividans 66. J. Gen. Microbiol. 137:2065-2072. [Google Scholar]
- 28.Newcomb, M., C.-Y. Chen, and J. H. D. Wu. 2007. Induction of the celC operon of Clostridium thermocellum by laminaribiose. Proc. Natl. Acad. Sci. USA 104:3747-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohnishi, Y., J. Ishikawa, H. Hara, H. Suzuki, M. Ikenoya, H. Ikeda, A. Yamashita, M. Hattori, and S. Horinouchi. 2008. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO13350. J. Bacteriol. 190:4050-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohnishi, Y., Y. Nishiyama, R. Sato, S. Kameyama, and S. Horinouchi. 2000. An oligoribonuclease gene in Streptomyces griseus. J. Bacteriol. 182:4647-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pope, M. K., B. D. Green, and J. Westpheling. 1996. The bld mutants of Streptomyces coelicolor are defective in the regulation of carbon utilization, morphogenesis and cell-cell signalling. Mol. Microbiol. 19:747-756. [DOI] [PubMed] [Google Scholar]
- 32.Rigali, S., H. Nothaft, E. E. Noens, M. Schlicht, S. Colson, M. Müller, B. Joris, H. K. Koerten, D. A. Hopwood, F. Titgemeyer, and G. P. van Wezel. 2006. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol. Microbiol. 61:1237-1251. [DOI] [PubMed] [Google Scholar]
- 33.Rigali, S., M. Schlicht, P. Hoskisson, H. Nothaft, M. Merzbacher, B. Joris, and F. Titgemeyer. 2004. Extending the classification of bacterial transcription factors beyond the helix-turn-helix motif as an alternative approach to discover new cis/trans relationships. Nucleic Acids Res. 32:3418-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryding, N. J., G. H. Kelemen, C. A. Whatling, K. Flärdh, M. J. Buttner, and K. F. Chater. 1998. A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol. 29:343-357. [DOI] [PubMed] [Google Scholar]
- 35.Saito, A., T. Fujii, T. Yoneyama, and K. Miyashita. 1998. glkA is involved in glucose repression of chitinase production in Streptomyces lividans. J. Bacteriol. 180:2911-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saito, A., T. Shinya, K. Miyamoto, T. Yokoyama, H. Kaku, E. Minami, N. Shibuya, H. Tsujibo, Y. Nagata, A. Ando, T. Fujii, and K. Miyashita. 2007. The dasABC gene cluster, adjacent to dasR, encodes a novel ABC transporter for the uptake of N,N′-diacetylchitobiose in Streptomyces coelicolor A3(2). Appl. Environ. Microbiol. 73:3000-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlösser, A., J. Jantos, K. Hackmann, and H. Schrempf. 1999. Characterization of the binding protein-dependent cellobiose and cellotriose transport system of the cellulose degrader Streptomyces reticuli. Appl. Environ. Microbiol. 65:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlösser, A., T. Aldekamp, and H. Schrempf. 2000. Binding characteristics of CebR, the regulator of the ceb operon required for cellobiose/cellotriose uptake in Streptomyces reticuli. FEMS Microbiol. Lett. 190:127-132. [DOI] [PubMed] [Google Scholar]
- 39.Seo, J. W., Y. Ohnishi, A. Hirata, and S. Horinouchi. 2002. ATP-binding cassette transport system involved in regulation of morphological differentiation in response to glucose in Streptomyces griseus. J. Bacteriol. 184:91-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spiridonov, N. A., and D. B. Wilson. 1999. Characterization and cloning of CelR, a transcriptional regulator of cellulase genes from Thermomonospora fusca. J. Biol. Chem. 274:13127-13132. [DOI] [PubMed] [Google Scholar]
- 41.Spiridonov, N. A., and D. B. Wilson. 2001. Cloning and biochemical characterization of BglC, a β-glucosidase from the cellulolytic actinomycete Thermobifida fusca. Curr. Microbiol. 42:295-301. [DOI] [PubMed] [Google Scholar]
- 42.Sprusansky, O., K. Stirrett, D. Skinner, C. Denoya, and J. Westpheling. 2005. The bkdR gene of Streptomyces coelicolor is required for morphogenesis and antibiotic production and encodes a transcriptional regulator of a branched-chain amino acid dehydrogenase complex. J. Bacteriol. 187:664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strohl, W. R. 1992. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 20:961-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Süsstrunk, U., J. Pidoux, S. Taubert, A. Ullmann, and C. J. Thompson. 1998. Pleiotropic effects of cAMP on germination, antibiotic biosynthesis and morphological development in Streptomyces coelicolor. Mol. Microbiol. 30:33-46. [DOI] [PubMed] [Google Scholar]
- 45.Tan, H. R., Y. Q. Tian, H. H. Yang, G. Liu, and L. P. Nie. 2002. A novel Streptomyces gene, samR, with different effects on differentiation of Streptomyces ansochromogenes and Streptomyces coelicolor. Arch. Microbiol. 177:274-278. [DOI] [PubMed] [Google Scholar]
- 46.Traag, B. A., G. H. Kelemen, and G. P. van Wezel. 2004. Transcription of the sporulation gene ssgA is activated by the IclR-type regulator SsgR in a whi-independent manner in Streptomyces coelicolor A3(2). Mol. Microbiol. 53:985-1000. [DOI] [PubMed] [Google Scholar]
- 47.van Wezel, G. P., M. König, K. Mahr, H. Nothaft, A. W. Thomae, M. Bibb, and F. Titgemeyer. 2007. A new piece of an old jigsaw: glucose kinase is activated posttranslationally in a glucose transport-dependent manner in Streptomyces coelicolor A3(2). J. Mol. Microbiol. Biotechnol. 12:67-74. [DOI] [PubMed] [Google Scholar]
- 48.Viollier, P. H., K. T. Nguyen, W. Minas, M. Folcher, G. E. Dale, and C. J. Thompson. 2001. Roles of aconitase in growth, metabolism, and morphological differentiation of Streptomyces coelicolor. J. Bacteriol. 183:3193-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viollier, P. H., W. Minas, G. E. Dale, M. Folcher, and C. J. Thompson. 2001. Role of acid metabolism in Streptomyces coelicolor morphological differentiation and antibiotic biosynthesis. J. Bacteriol. 183:3184-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weickert, M. J., and S. Adhya. 1992. A family of bacterial regulators homologous to Gal and Lac repressors. J. Biol. Chem. 267:15869-15874. [PubMed] [Google Scholar]
- 51.Yamazaki, H., Y. Ohnishi, and S. Horinouchi. 2000. An A-factor-dependent extracytoplasmic function sigma factor (σAdsA) that is essential for morphological development in Streptomyces griseus. J. Bacteriol. 182:4596-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.