Abstract
Moraxella catarrhalis is a causative agent of otitis media in children and lower respiratory tract infections in adults suffering from chronic obstructive pulmonary disease (COPD). This strict human pathogen continues to be a significant cause of disease in this broad spectrum of patients because there is no available vaccine. Although numerous putative vaccine antigens have been described, little is known about the human immune response to M. catarrhalis infection in vivo. Human serum antibodies are directed at a number of surface proteins, and lipooligosaccharides (LOS) and detoxified LOS may be an effective immunogen in mice. In this study, we used a specific LOS-based enzyme-linked immunosorbent assay (ELISA), containing the three major M. catarrhalis serotypes together with a complete series of truncated LOS mutants, to detect the development of new antibodies to specific regions of the oligosaccharide molecule. We compared serum samples from COPD patients who had recently cleared an M. catarrhalis infection to serum samples collected prior to their infection. Variability in the antibody response to LOS was observed, as some patients developed serotype-specific antibodies, others developed antibodies to the LOS of each serotype, others developed broadly cross-reactive antibodies, and some did not develop new antibodies. These newly developed human antibodies are directed at both side chains and core structures in the LOS molecule. This LOS-based ELISA can be used to dissect the human antibody response to both internal and external carbohydrate epitopes, thus providing a better understanding of the humoral immune response to M. catarrhalis LOS epitopes developed during natural infection.
Chronic obstructive pulmonary disease (COPD) is currently the fourth leading cause of death in the United States and Europe. The morbidity and mortality associated with exacerbations of COPD together with the associated health care-related costs of $32 billion reported in the United States in 2002 demonstrate that there is a need for a better understanding of the etiology and pathogenesis of these events (6, 36, 43). Bacteria have been isolated in large numbers from the lower respiratory tract during exacerbations, and up to 50% of COPD exacerbations are due to a bacterial agent, primarily nontypeable Haemophilus influenzae, Moraxella catarrhalis, and Streptococcus pneumoniae (31). M. catarrhalis accounts for up to 10% of these infections in adults, and this strictly human pathogen is currently among the three most prominent causes of otitis media in children (13, 15, 28). Some of the primary reasons why M. catarrhalis continues to cause disease can be attributed to the fact that greater than 90% of the clinical isolates express beta-lactamase, there is a high frequency of recurrent disease observed for children that have recovered from infection, and there is a lack of a vaccine (13, 27, 42, 47). Thus, the identification of potential drug targets and vaccine antigens is clearly a priority.
One of the major problems hindering the identification of putative vaccine antigens involves the fact that M. catarrhalis is a strictly human pathogen, and the human immune response to this bacterium remains poorly understood. Previous studies investigated the production of new antibodies against different bacterial pathogens in patients suffering from COPD and lower respiratory tract infections. These patients exhibited increased antibody responses to bacterial outer membrane proteins (OMP) and surface-exposed lipooligosaccharides (LOS) after clearing the bacterial strain following an exacerbation (4, 29, 38, 51). Human serum immunoglobulin G (IgG), sputum IgA, or salivary IgA antibodies against M. catarrhalis surface proteins such as UspA1, UspA2, Hag, TbpB, CopB, OMP CD, OMP E, and OMP G1b have been developed (1, 3, 25, 28, 29). In addition, new antibodies to LOS have also been detected in some COPD patients (3, 28, 29).
The LOS structure of M. catarrhalis has been well studied. There are three major serotypes, serotypes A, B, and C, previously defined by polyclonal antisera and structural analyses (8-10, 23, 46). The LOS glycosyltransferase (lgt) genes that encode the enzymes required to transfer carbohydrate residues to the M. catarrhalis LOS molecule were previously identified and characterized (11, 33, 41, 49). In addition, the lgt present in a given strain of M. catarrhalis can be used to identify the specific LOS serotype of that isolate using our previously described multiplex PCR method (12). Serotypes A and B are the predominant glycoforms expressed by most clinical isolates analyzed to date (12, 46). In recently reported animal studies, other researchers suggested that detoxified M. catarrhalis LOS has potential as a vaccine antigen in a mouse pulmonary clearance model (16, 19, 52, 53). While these data are both valuable and interesting, it is sometimes difficult to link observations of animals to those of humans (34).
Currently, we have constructed a panel of defined LOS mutants that are defective in the expression of each specific glycosyltransferase gene identified in all three major M. catarrhalis LOS serotypes. These truncations are a comprehensive set of mutations with various oligosaccharide (OS) chain lengths representing most, if not all, possible LOS epitopes (11, 33, 41, 49). Purified LOS samples from these mutants were used in enzyme-linked immunosorbent assays (ELISAs) to assess the development of new human antibodies to all LOS epitopes developed following an M. catarrhalis infection. ELISAs were previously employed to determine levels of antibody to Neisseria meningitidis lipopolysaccharide (LPS), including inner core mutations, in patients following disease (35). Thus, this LOS-based ELISA system with the full set of mutations has the potential to determine the regions of the LOS molecule that elicit new antibodies in both children and adults following infection, providing a unique opportunity to analyze the human immune response to these major surface glycolipids in the native host.
MATERIALS AND METHODS
COPD study clinic.
All COPD bacterial isolates and serum samples were obtained from patients enrolled in a prospective COPD study clinic at the Buffalo Veterans Affairs Medical Center. This study was previously described (42). Patients are seen on a monthly basis and when an exacerbation of COPD is suspected. Clinical evaluations were performed, and sputum and serum samples were obtained.
Bacterial strains and culture conditions.
The bacterial strains used in this study are described in Table 1. COPD strains were isolated and identified from patient sputum samples and the LOS serotype was determined as previously described (12, 29, 42). M. catarrhalis strains were cultured on brain heart infusion agar plates at 35.5°C in 5% CO2 or shaking aerobically in brain heart infusion broth at 37°C. Escherichia coli XL1-Blue cells, cultured in Luria-Bertani (LB) medium at 35.5°C in 5% CO2, were used as the host for plasmid DNA manipulations. The following antibiotics were supplemented as necessary: 50 μg/ml kanamycin and 100 μg/ml ampicillin.
TABLE 1.
Bacterial strains and plasmids
| Strain or Plasmid | Description | Source and/or reference(s) |
|---|---|---|
| Strains | ||
| E. coli | ||
| XL1-Blue | Host strain used for cloning | Stratagene |
| M. catarrhalis | ||
| 25238 | LOS serotype A, wild type | American Type Culture Collection; 9 |
| 25238::lgt1 | lgt1 kanamycin-resistant isogenic mutant in strain 25238 | This study |
| 25238::lgt2A | lgt2A kanamycin-resistant isogenic mutant in strain 25238 | This study |
| 25238::lgt3 | lgt3 kanamycin-resistant isogenic mutant in strain 25238 | This study |
| 25238::lgt4 | lgt4 kanamycin-resistant isogenic mutant in strain 25238 | This study |
| 25238::lgt5 | lgt5 kanamycin-resistant isogenic mutant in strain 25238 | This study |
| 25238::lgt6 | lgt6 kanamycin-resistant isogenic mutant in strain 25238 | 41 |
| 7169 | LOS serotype B, wild type, otitis media isolate | 11 |
| 7169::lgt2B/C | lgt2B/C kanamycin-resistant isogenic mutant in strain 7169 | 11 |
| 7169::lgt5 | lgt5 kanamycin-resistant isogenic mutant in strain 7169 | This study |
| 26391 | LOS serotype C, wild type | Culture Collection, University of Göteborg, Göteborg, Sweden; 17 |
| 26391::lgt2B/C | lgt2B/C kanamycin-resistant isogenic mutant in strain 26391 | This study |
| 26391::lgt4 | lgt4 kanamycin-resistant isogenic mutant in strain 26391 | This study |
| 26391::lgt5 | lgt5 kanamycin-resistant isogenic mutant in strain 26391 | This study |
| 3P34B1 | LOS serotype A, COPD clinical isolate | Timothy Murphy |
| 6P29B1 | LOS serotype A, COPD clinical isolate | Timothy Murphy; 3, 39 |
| 7P33B1 | LOS serotype A, COPD clinical isolate | Timothy Murphy; 3 |
| 7P94B1 | LOS serotype C, COPD clinical isolate | Timothy Murphy; 12, 28, 34 |
| 12P22B1 | LOS serotype A, COPD clinical isolate | Timothy Murphy; 12 |
| 13P6B1 | LOS serotype B, COPD clinical isolate | Timothy Murphy |
| 18P7B1 | LOS serotype B, COPD clinical isolate | Timothy Murphy |
| 19P54B1 | LOS serotype A, COPD clinical isolate | Timothy Murphy |
| 39P10B1 | LOS serotype B, COPD clinical isolate | Timothy Murphy; 3, 34 |
| 51P9B1 | LOS serotype A, COPD clinical isolate | Timothy Murphy; 3, 12 |
| 87P15B1 | LOS serotype A, COPD clinical isolate | Timothy Murphy; 29 |
| Plasmids | ||
| pGEM-T Easy | Commercial vector used for TA cloning and mutagenesis | Promega |
| pUC18K | Contains aphA-3, nonpolar kanamycin resistance cassette | 26 |
Serum samples.
Preacquisition serum samples were obtained 4 to 8 weeks prior to the first acquisition of an M. catarrhalis strain (indicated by the patient number followed by the clinic visit number, i.e., patient 6, homologous strain 6P29B1, had an exacerbation on clinic visit 29) based on an analysis of strains by pulsed-field gel electrophoresis. Postclearance serum samples were obtained 4 to 8 weeks after the M. catarrhalis strain was cleared from the respiratory tract based on monthly sputum cultures as previously described (29). M. catarrhalis strains and the corresponding serum samples were obtained from patients who developed new serum IgG against their homologous infecting strain as described previously (30).
Nucleic acid techniques.
Standard molecular biological techniques were used as previously described (40). Plasmids were isolated using QIAprep Spin miniprep kits (Qiagen, Chatsworth, CA). Chromosomal DNA was obtained from M. catarrhalis using standard methods, and DNA was purified using Qiagen purification kits. PCR was performed using Platinum Taq high-fidelity DNA polymerase (Invitrogen, Carlsbad, CA). Fast-Link DNA ligase (Epicentre, Madison, WI) and all enzymes obtained from New England Biolabs (Beverly, MA) were used according to company protocols. PCR amplicons were ligated into the TA cloning vector pGEM-T Easy (Promega, Madison, WI) for further modifications. DNA nucleotide sequences were obtained for all cloning and mutant constructs via automated DNA sequencing at the RPCI Biopolymer Facility (Roswell Park Cancer Institute, Buffalo, NY), and all sequences were analyzed using Sequencher, version 4.5 (Gene Codes Corporation, Ann Arbor, MI), and MacVector, version 7.2 (Accelrys, San Diego, CA), software. RNA was isolated from cells at the mid-log phase of growth using an RNeasy minikit (Qiagen) followed by RQ1 RNase-free DNase treatment (Promega).
Construction of M. catarrhalis isogenic mutants.
The PCR primers used to sequence, clone, and mutagenize the lgt genes are listed in Table 2. The primers were designed based on the sequence homologies between M. catarrhalis patent number WO0078968 (patent located at the NCBI database under Incyte Genomics accession number AX067456) and the reported sequence of other known lgt genes (11, 12, 49). An inverse PCR strategy was used to construct isogenic mutants as previously described (11, 14, 21, 22). This resulted in an internal deletion of the cloned lgt gene with engineered restriction sites in each individual gene that allowed for the directed insertion of a nonpolar kanamycin resistance cassette (Table 1) (26). The resulting insertionally inactivated lgt-Kan mutant constructs were amplified by PCR, and purified linear amplicons were introduced into M. catarrhalis cells by natural transformation (14, 22). Kanamycin-resistant transformants were selected, and the inactivated gene was amplified from chromosomal DNA and sequenced to ensure the proper integration of the mutant construct. Reverse transcriptase PCR analysis was completed using a OneStep reverse transcriptase PCR kit (Qiagen) with primers shown in Table 2 to ensure that the surrounding genes are still transcribed in the disrupted mutants.
TABLE 2.
Nucleotide sequences of oligonucleotide primers used for sequencing, PCR-based cloning, mutagenesis procedures, and transcriptional analysis in this study
| Primer | Primer sequence (5′-3′)a | Gene specificity |
|---|---|---|
| 913F | TGGGCATTGTATCAAGGGC | Sequencing and cloning of lgt4 |
| 914R | ATTGGGTGGGTAGATTCTGTG | Sequencing and cloning of lgt4 |
| 915F | CTCGAGAGCACCCAAAGCAAATCAGAGATG | lgt4 mutagenesis |
| 916R | AGATCTGCCCAATACAGCCCATTATGAAC | lgt4 mutagenesis |
| 508F | AAGAAGTGGGGCTTTTGTCAGAG | Mutagenesis and cloning of lgt2B/C mutant in 26391 |
| 509R | GAGAGTATGTCATTCGTGGCGAC | Mutagenesis and cloning of lgt2B/C mutant in 26391 |
| 1102R | GTCATCAGCATAAAAACTGC | Sequencing and cloning of lgt2A from 25238 |
| 1103F | ATAGCGATTCGTTGGC | Sequencing and cloning of lgt2A from 25238 |
| 1104F | AGATCTCGGTGAGTCTTTGGTTGTTGA | Mutagenesis of lgt2A from 25238 |
| 1105R | CTCGAGTCAGGTGCTAAGCGGCTACTA | Mutagenesis of lgt2A from 25238 |
| 1086F | TTCATTGCCGATACCACC | Sequencing and cloning of lgt1 from 25238; lgt1 transcription |
| 1098R | AAACCGCATAACTGTCTGAC | Sequencing and cloning of lgt1 from 25238 |
| 1099R | AGATCTCACTGTTACTGTGCTCTGTGC | Mutagenesis of lgt1 from 25238 |
| 1100F | CTCGAGATCTTTCCTGTGTTGTGGCA | Mutagenesis of lgt1 from 25238 |
| 1264F | ATTTTGGGGCAGGCAAG | Sequencing and cloning of lgt5 |
| 1265R | TGGATTGGTGATACACTCGG | Sequencing and cloning of lgt5 |
| 1297R | CTCGAGCTGCCCAAAGAAAGTGAGT | lgt5 mutagenesis |
| 1298F | AGATCTCAGAAAATAACGCATAACTGCC | lgt5 mutagenesis |
| 1087R | TTACTCCGCCAACCATTCC | lgt1 transcription |
| 649F | ATCCTGCTCCAACTGACTTTC | lgt2A transcription |
| 650R | GGTAACAGAACGCTCAACCC | lgt2A transcription |
| 671F | GTGTCATCTTCTGATACTAAATGGG | lgt2B/C transcription |
| 672R | AAACTTTTCATACAGGCACTGC | lgt2B/C transcription |
BglII restriction sites are in italics, and XhoI restriction sites are underlined.
Preparation of M. catarrhalis LOS for ES-MS.
For mass spectrometric analyses, plate-grown bacterial cells were treated with proteinase K followed by successive treatments with DNase and RNase to release the LOS, which was O-deacylated in situ with anhydrous hydrazine (20). This microanalytical procedure proved particularly convenient for the rapid profiling of LOS glycoforms since single-plate growth provided sufficient material for subsequent analyses by electrospray mass spectrometry (ES-MS). For linkage analysis, cells from 6 liters of stationary-phase bacteria were harvested and treated with 1% phenol (final concentration). The cell pellets obtained by centrifugation were successively washed once with ethanol, twice with acetone, and twice with petroleum ether (35 to 60°C) (23). The LOS was extracted from the dried cell pellet by the hot-phenol-water extraction procedure (48).
Glycoform profiling and linkage analyses.
Lyophilized O-deacylated LOS samples were dissolved in 1 M ammonium acetate solution and analyzed directly on a Crystal model 310 capillary electrophoresis instrument (ATI Unicam, Boston, MA) coupled to a 4000 Qtrap mass spectrometer (Applied Biosystems/MDS Sciex, Canada). Separations were obtained on 90-cm-length bare fused silica using 30 mM aqueous morpholine acetate (pH 9) containing 5% methanol. A voltage of −5 kV was applied at the injection end of the capillary, and spectra were obtained in the negative-ion mode. Calculations of molecular mass based on proposed compositions were done by using average mass units as follows: hexose (Hex), 162.14; heptose (Hep), 192.17; N-acetylhexosamine (HexNAc), 203.19; 2-keto-3-deoxyoctulosonic acid (KDO), 220.20; phosphoethanolamine (PEA), 123.05; lipid A-OH, 1,020.
Core OS obtained from LOS by mild acid hydrolysis was used for linkage analyses as previously described (23). The core OS sample was methylated with iodomethane in dimethyl sulfoxide containing an excess of NaOH (7). Methylated core OS was hydrolyzed with 2 M trifluoroacetic acid at 100°C for 3 h, and the liberated glycoses were reduced with NaBD4 and acetylated by acetic anhydride (Ac2O) at 100°C for 1.5 h (23). Partially methylated alditol acetates were separated by gas-liquid chromatography and identified by electron impact mass spectrometry on a Varian Saturn 2000 apparatus fitted with a DB-17 fused-silica capillary column (0.25 mm by 25 m), utilizing an ionization potential of 70 eV and a temperature program consisting of 180°C followed by an increase to 260°C at 3.5°C/min.
Preparation of M. catarrhalis LOS for ELISA.
LOS was extracted from M. catarrhalis strains by a modified whole-cell proteinase K procedure. Briefly, plate-grown organisms were washed with sterile phosphate-buffered saline and boiled in lysing buffer (2% sodium dodecyl sulfate, 10% glycerol, and 4% β-mercaptoethanol in 1 M Tris [pH 6.8]) for 10 min, followed by a 6-h incubation at 60°C with proteinase K at a final concentration of 0.4 mg/ml. The proteinase K-treated sample was precipitated with 3 M sodium acetate and 100% ethyl alcohol, dried, weighed, and resuspended in ELISA coating buffer (Kirkegaard & Perry Laboratories, Inc. [KPL], Gaithersburg, MD) to a final concentration of 5 to 10 mg/ml. The LOS was resolved by urea sodium dodecyl sulfate-18% polyacrylamide gel electrophoresis using a bilayer stacker and visualized by silver staining as previously described to confirm LOS glycoforms (18, 45).
ELISA.
Immunon 1B 96-well plates (Thermo Electron Corporation, Milford, MA) were coated overnight at 4°C, with gentle agitation, using approximately 20 to 30 μg of LOS per well suspended in coating buffer from the Protein Detector ELISA kit (KPL). The wells were blocked for 15 min at room temperature with KPL bovine serum albumin blocking solution. The wells were incubated with 80-μl serum samples diluted 1:500 in phosphate-buffered saline for 1 h at room temperature. The plates were washed five times with KPL washing solution. The plates were incubated for 1 h at room temperature with KPL horseradish peroxidase-conjugated goat anti-human IgG diluted 1:1,000 in KPL bovine serum albumin blocking solution to detect any IgG serum attached to the LOS-coated wells. The plates were then washed five additional times with KPL wash solution and developed with KPL ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)] peroxide substrate for up to 1.5 h. The absorbance at 405 nm was determined using a SPECTRAmax Plus plate reader (Molecular Devices Corporation, Sunnyvale, CA). At least three independent assays were performed with samples run in triplicate for each ELISA. The preacquisition and postclearance means were calculated, and the background was subtracted for each replicate. According to the Kolmogorov-Smirnov test of normality, the majority of data can be assumed to be normally distributed (P value of >0.05). Therefore, a one-sided paired Student's t test was used to analyze all ELISA data.
Nucleotide sequence accession numbers.
The sequences of the cluster of genes reported in this paper were deposited in the GenBank database and assigned accession numbers AY789049, EF032481, and EF032482.
RESULTS AND DISCUSSION
Construction and structural analysis of defined LOS mutants.
All serotypes of M. catarrhalis contain the genes lgt1, lgt2, lgt3, lgt5, and lgt6. Lgt2 is present in two distinct alleles: lgt2A is expressed in serotype A, and lgt2B/C is expressed in serotypes B and C. Serotypes A and C also contain lgt4, which encodes an α-(1-2)-N-acetylglucosaminyltransferase, which accounts for their difference from serotype B. In order to complete the panel of LOS mutants defective in individual Lgt-encoding genes, a single lgt4 mutant was constructed. As this was the first description of such a construct, ES-MS analysis of LOS-OH from the lgt4 mutants (Table 3) in both the serotype A and serotype C backgrounds was performed. These studies revealed that Hex7 and Hex5 glycoform populations lack the HexNAc substitution, confirming that Lgt4 functions as an α-(1-2)-N-acetylglucosaminyltransferase (33). Thus, M. catarrhalis LOS mutants in every defined glycosyltransferase were constructed, either in this study or previously, as listed in Table 3 (11, 41).
TABLE 3.
Negative-ion ES-MS data and proposed compositions of O-deacylated LPS from M. catarrhalis wild-type and mutant strains
| Strain | Observed ion (m/z)
|
Molecular mass (Da)a
|
Relative intensity (%) | Proposed composition | |||
|---|---|---|---|---|---|---|---|
| (M − 2H)2− | (M − 3H)3− | (M − 4H)4− | Observed | Calculated | |||
| 25238b | 973.2 | 730.0 | 2,923.3 | 2,921.5 | 44.1 | PEA + GlcNAc + Hex7 + KDO2 + lipid A-OH | |
| 932.0 | 698.9 | 2,799.3 | 2,798.4 | 42.4 | GlcNAc + Hex7 + KDO2 + lipid A-OH | ||
| 891.3 | 2,676.9 | 2,675.4 | 13.8 | GlcNAc + Hex7 + KDO2 + lipid A-OH-(PEA) | |||
| 25238 lgt1 | 811.1 | 608.0 | 2,436.2 | 2,435.0 | 31.0 | PEA + GlcNAc + Hex4 + KDO2 + lipid A-OH | |
| 769.7 | 577.2 | 2,312.5 | 2,311.6 | 14.7 | GlcNAc + Hex4 + KDO2 + lipid A-OH | ||
| 743.1 | 557.4 | 2,233.0 | 2,232.0 | 39.7 | PEA + Hex4 + KDO2 + lipid A-OH | ||
| 702.4 | 2,110.2 | 2,109.0 | 14.7 | Hex4 + KDO2 + lipid A-OH | |||
| 25238 lgt2A | 1,297.5 | 865.2 | 648.8 | 2,598.3 | 2,597.2 | 64.5 | PEA + GlcNAc + Hex5 + KDO2 + lipid A-OH |
| 1,237.0 | 824.0 | 617.9 | 2,474.5 | 2,474.1 | 35.5 | GlcNAc + Hex5 + KDO2 + lipid A-OH | |
| 25238 lgt3 | 871.9 | 581.0 | 1,745.9 | 1,745.6 | 55.1 | PEA + Hex1 + KDO2 + lipid A-OH | |
| 540.6 | 1,624.8 | 1,622.5 | 44.9 | Hex1 + KDO2 + lipid A-OH | |||
| 25238 lgt4 | 905.3 | 678.2 | 2,717.9 | 2,718.5 | 5.2 | PEA + Hex7 + KDO2 + lipid A-OH | |
| 1,196.2 | 797.2 | 597.9 | 2,394.9 | 2,394.2 | 67.5 | PEA + Hex5 + KDO2 + lipid A-OH | |
| 1,134.8 | 756.2 | 567.1 | 2,271.9 | 2,271.1 | 27.3 | Hex5 + KDO2 + lipid A-OH | |
| 25238 lgt5 | 1,379.3 | 2,760.6 | 2,759.3 | 35.3 | PEA + GlcNAc + Hex6 + KDO2 + lipid A-OH | ||
| 1,317.8 | 2,637.6 | 2,636.3 | 64.7 | GlcNAc + Hex6 + KDO2 + lipid A-OH | |||
| 25238 lgt6b | 790.8 | 1,583.6 | 1,583.4 | 0.7 | PEA + KDO2 + lipid A-OH | ||
| 729.3 | 1,460.6 | 1,460.3 | 1.0 | KDO2 + lipid A-OH | |||
| 7169b | 1,601.7 | 1,067.6 | 3,205.6 | 3,204.8 | 0.4 | PEA + Hex10 + KDO2 + lipid A-OH | |
| 1,539.8 | 1,026.2 | 3,081.6 | 3,081.7 | 0.5 | Hex10 + KDO2 + lipid A-OH | ||
| 1,520.8 | 1,013.7 | 759.1 | 3,043.9 | 3,042.6 | 1.0 | PEA + Hex9 + KDO2 + lipid A-OH | |
| 1,459.0 | 972.8 | 2,920.7 | 2,919.6 | 1.0 | Hex9 + KDO2 + lipid A-OH | ||
| 1,439.7 | 959.7 | 2,881.9 | 2,880.5 | 1.0 | PEA + Hex8 + KDO2 + lipid A-OH | ||
| 1,378.4 | 918.4 | 2,758.8 | 2,757.4 | 0.9 | Hex8 + KDO2 + lipid A-OH | ||
| 7169 lgt2c | PEA + Hex6 + KDO2 + lipid A-OH | ||||||
| Hex6 + KDO2 + lipid A-OH | |||||||
| 7169 lgt5 | 1,439.4 | 959.4 | 719.4 | 2,881.1 | 2,880.6 | 30.0 | PEA + Hex8 + KDO2 + lipid A-OH |
| 1,377.6 | 918.3 | 688.8 | 2,759.3 | 2,757.6 | 50.0 | Hex8 + KDO2 + lipid A-OH | |
| 1,316.1 | 877.5 | 657.9 | 2,635.6 | 2,634.5 | 20.0 | Hex8 + KDO2 + lipid A-OH(-PEA) | |
| 26391b | 1,081.1 | 810.6 | 3,246.5 | 3,245.8 | 20.8 | PEA + GlcNAc + Hex9 + KDO2 + lipid A-OH | |
| 1,027.3 | 770.4 | 3,085.3 | 3,083.6 | 41.7 | PEA + GlcNAc + Hex8 + KDO2 + lipid A-OH | ||
| 986.6 | 739.5 | 2,962.4 | 2,960.6 | 37.5 | GlcNAc + Hex8 + KDO2 + lipid A-OH | ||
| 26391 lgt2B/C | 865.3 | 648.8 | 2,599.1 | 2,597.2 | 64.0 | PEA + GlcNAc + Hex5 + KDO2 + lipid A-OH | |
| 824.0 | 618.0 | 2,475.5 | 2,474.1 | 36.0 | GlcNAc + Hex5 + KDO2 + lipid A-OH | ||
| 26391 lgt4 | 905.2 | 678.9 | 2,719.1 | 27,18.5 | 27.2 | PEA + Hex7 + KDO2 + lipid A-OH | |
| 864.5 | 648.3 | 2,596.9 | 2,595.4 | 29.3 | Hex7 + KDO2 + lipid A-OH | ||
| 797.3 | 597.8 | 2,395.1 | 2,394.2 | 29.3 | PEA + Hex5 + KDO2 + lipid A-OH | ||
| 756.3 | 567.4 | 2,272.8 | 2,271.1 | 14.1 | Hex5 + KDO2 + lipid A-OH | ||
| 26391 lgt5 | 973.3 | 729.8 | 2,923.1 | 2,921.5 | 54.2 | PEA + GlcNAc + Hex7 + KDO2 + lipid A-OH | |
| 932.0 | 699.0 | 2,799.5 | 2,798.4 | 45.8 | GlcNAc + Hex7 + KDO2 + lipid A-OH | ||
Average mass units were used for calculations of molecular mass based on the proposed composition as follows: Hex, 162.15; GlcNAc, 203.19; KDO, 220.18; PEA, 123.05. O-deacylated lipid A (lipid A-OH) is (GlcN)2 + (3-OH-C12)2 + PPEA + P (m/z 1,020). In some cases, an extra PEA molecule is elaborated on the lipid A-OH moiety.
These negative-ion ES-MS data were previously reported (41).
This mutation was previously characterized by matrix-assisted laser desorption ionization-time of flight mass spectrometry (11).
Human antibody response to OS epitopes.
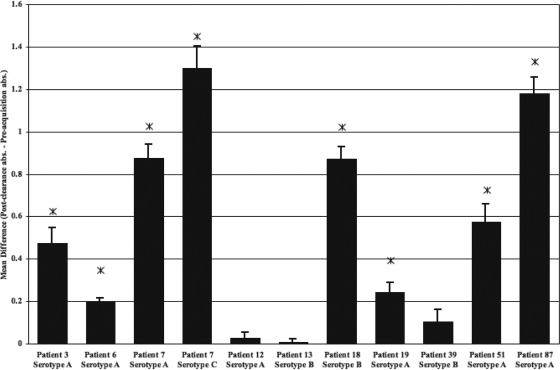
Multiplex PCR was used to determine the LOS serotype of each clinical isolate from the COPD patients analyzed in these studies (12). Initially, preacquisition and postclearance sera from each patient were assayed for the presence of new IgG antibodies that were reactive to full-length LOS of the homologous strain. Preacquisition and postclearance serum samples were assayed in parallel in the same ELISA with LOS purified from the homologous strain. The mean difference in absorbance was determined from at least three independent assays. A one-sided paired t test was used to determine whether a significant increase in the antibody level was observed from the preacquisition sample to the postclearance sample. Figure 1 shows the results for the 10 patient sera infected with 11 different M. catarrhalis strains. These data demonstrated that 7 of the 10 patients developed a statistically significant level of new antibodies to the homologous LOS following the acquisition and clearance of a strain of M. catarrhalis, although the levels of reactivity were quite variable. For example, although patients 6 and 19 had a significant rise in antibody titers to the homologous LOS in postclearance sera, the level of reactivity was well below that detected for sera from patients 3, 7, 18, 51, and 87. In addition, patients 12, 13, and 39 did not develop any significant level of new antibodies to their homologous LOS.
FIG. 1.
Identification of newly developed serum IgG antibodies to the LOS of the homologous M. catarrhalis isolate. ELISAs were used to test the level of serum IgG antibody (abs) in response to LOS obtained from the exacerbating strain of bacterium. COPD patients are indicated in addition to the LOS serotype of the exacerbating strain of M. catarrhalis isolated from that patient. Serum samples obtained preacquisition and postclearance of the infecting M. catarrhalis strain were incubated in a 96-well plate coated with 20 to 30 μg of the homologous LOS per well. The bars represent the mean differences in absorbance for data from at least three independent ELISAs of serum samples taken after clearance of the exacerbating strain to preacquisition of that strain, with the error bars representing the standard errors of the means. Asterisks indicate a P value of ≤0.05 as determined by a one-sided paired t test.
While the development of new antibodies to the full-length LOS is not surprising, there is currently no data describing specific M. catarrhalis OS epitopes that stimulate antibodies in humans. In order to identify the regions of the LOS molecule to which the newly developed anti-LOS antibodies were directed, we extended our ELISAs to include a comprehensive set of truncated LOS isolated from defined M. catarrhalis mutants (11, 33, 41, 49). These mutants (structures described in Table 4) were compared to the full-length LOS of all three major serotypes. Mutations in lgt3 and lgt6 yield the same LOS structures when constructed in all M. catarrhalis LOS serotypes; therefore, only one of these mutations was included in the panel. The serotype A and C mutations in lgt2 also yielded the same carbohydrate structure, but this mutant was included in the panel to serve as an internal control. The LOS of the lgt1 mutation is similar among all three serotypes except for the occasional addition of an N-acetylglucosamine residue.
TABLE 4.
LOS truncation structuresa
| Strain | LOS structure
|
||
|---|---|---|---|
| R1 | R2 | R3 | |
| 25238 (serotype A) | αGal-(1-4)-βGal-(1-4)-αGlc-(1-2)-βGlc-(1-6)-αGlc-(1-5)- | αGlcNAc-(1-2)-βGlc-(1-4)- | βGlc-(1-3)- |
| 25238 lgt1 | βGlc-(1-6)-αGlc-(1-5)- | αGlcNAc-(1-2)-βGlc-(1-4)- | βGlc-(1-3)- |
| βGlc-(1-4)- | |||
| 25238 lgt2 | αGlc-(1-2)-βGlc-(1-6)-αGlc-(1-5)- | αGlcNAc-(1-2)-βGlc-(1-4)- | βGlc-(1-3)- |
| 25238 lgt3 | αGlc-(1-5)- | H- | H- |
| 25238 lgt4 | αGal-(1-4)-βGal-(1-4)-αGlc-(1-2)-βGlc-(1-6)-αGlc-(1-5)- | βGlc-(1-4)- | βGlc-(1-3)- |
| αGlc-(1-2)-βGlc-(1-6)-αGlc-(1-5)- | |||
| 25238 lgt5 | βGal-(1-4)-αGlc-(1-2)-βGlc-(1-6)-αGlc-(1-5)- | αGlcNAc-(1-2)-βGlc-(1-4)- | βGlc-(1-3)- |
| 25238 lgt6 | H- | H- | H- |
| 7169 (serotype B) | αGal-(1-4)-βGal-(1-4)-αGlc-(1-2)-βGlc-(1-6)-αGlc-(1-5)- | αGal-(1-4)-βGal-(1-4)-αGlc-(1-2)-βGlc-(1-4)- | βGlc-(1-3)- |
| βGal-(1-4)-αGlc-(1-2)-βGlc-(1-6)-αGlc-(1-5)- | βGal-(1-4)-αGlc-(1-2)-βGlc-(1-4)- | ||
| 7169 lgt2 | αGlc-(1-2)-βGlc-(1-6)-αGlc-(1-5)- | αGlc-(1-2)-βGlc-(1-4)- | βGlc-(1-3)- |
| 7169 lgt5 | βGal-(1-4)-αGlc-(1-2)-βGlc-(1-6)-αGlc-(1-5)- | βGal-(1-4)-αGlc-(1-2)-βGlc-(1-4)- | βGlc-(1-3)- |
| 26391 (serotype C) | αGal-(1-4)-βGal-(1-4)-αGlc-(1-2)-βGlc-(1-6)-αGlc-(1-5)- | αGal-(1-4)-βGal-(1-4)-αGlcNAc-(1-2)-βGlc-(1-4)- | βGlc-(1-3)- |
| βGal-(1-4)-αGlcNAc-(1-2)-βGlc-(1-4)- | |||
| 26391 lgt2 | αGlc-(1-2)-βGlc-(1-6)-αGlc-(1-5)- | αGlcNAc-(1-2)-βGlc-(1-4)- | βGlc-(1-3)- |
| 26391 lgt4 | αGal-(1-4)-βGal-(1-4)-αGlc-(1-2)-βGlc-(1-6)-αGlc-(1-5)- | βGlc-(1-4)- | βGlc-(1-3)- |
| αGlc-(1-2)-βGlc-(1-6)-αGlc-(1-5)- | |||
| 26391 lgt5 | βGal-(1-4)-αGlc-(1-2)-βGlc-(1-6)-αGlc-(1-5)- | βGal-(1-4)-αGlcNAc-(1-2)-βGlc-(1-4)- | βGlc-(1-3)- |
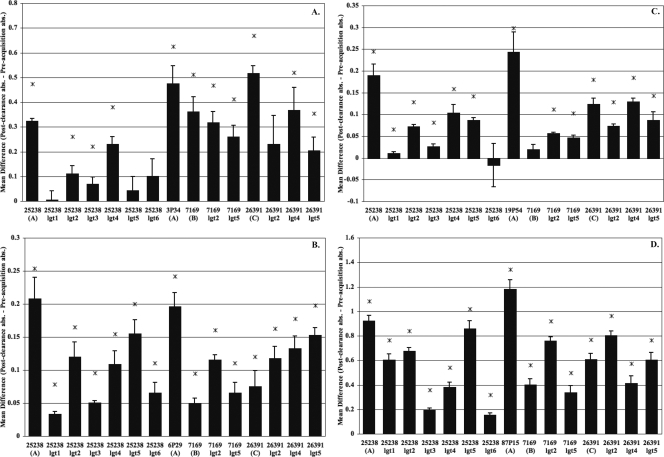
Although it is evident that the responses were diverse from patient to patient, some general observations were apparent. For example, Fig. 2 demonstrates that patients 3, 6, 19, and 87 all exhibited a significant rise in antibodies to the LOS of their infecting strains while also developing new antibodies directed toward multiple LOS epitopes in all three serotypes. The increase in the amount of antibody binding of the postclearance serum compared to that of the preacquisition serum from each patient is significant for almost all of the antigens tested, as determined by a one-sided paired t test. Although the responses vary in levels, the data suggest a possible pattern of reactivity among these patients. For example, these patients all developed a robust IgG response to the full-length LOS of their homologous strain. In addition, patient 3 (Fig. 2a), infected with a serotype A strain (3P34B1), also developed a significant antibody response that was equivalent to or greater than that for the prototypical serotype A, B, and C LOS. While patient 6 (Fig. 2b), infected with a serotype A strain (6P29B1), also developed a significant antibody response to all three major LOS serotypes, there appeared to be more antibodies that were reactive to the homologous LOS and the prototypical serotype A LOS (25238) than to the LOS of the strains representing serotype B (7169) and serotype C (26391). This pattern of reactivity was also evident for patient 19 (Fig. 2c) and patient 87 (Fig. 2d). These two individuals were also infected with a serotype A strain, suggesting a more directed serotype-specific response.
FIG. 2.
COPD patients who developed serotype-specific and externally directed antibody responses to different LOS epitopes. Shown are data from an evaluation of COPD patient IgG serum levels against various truncated LOS molecules. The OS used as antigens are listed along the x axis. Patient 3 (A) was infected with 3P34B1 (serotype A), patient 6 (B) was infected with 6P29B1 (serotype A), patient 19 (C) was infected with 19P54B1 (serotype A), and patient 87 (D) was infected with 87P15B1 (serotype A). ELISAs were used to test the level of COPD serum IgG antibody (abs) in response to truncated LOS obtained from a panel of glycosyltransferase mutants and full-length wild-type structures representing serotype A (25238), serotype B (7169), and serotype C (26391). Serum samples obtained preacquisition and postclearance of the infecting M. catarrhalis strain were incubated in a 96-well plate coated with 20 to 30 μg of LOS per well. The bars represent the mean differences in absorbance for at least three independent ELISAs of serum samples taken postclearance of the exacerbating strain to preacquisition of that strain, with the error bars representing the standard errors of the means. Asterisks indicate a P value of ≤0.05 as determined by a one-sided paired t test.
Figure 2 also shows that this group of patients developed more antibodies directed to the terminal regions of M. catarrhalis LOS, as all of these individuals exhibited diminished reactivity to the LOS assembled by the lgt1, lgt3, and lgt6 mutants, which represent truncations in the inner core region of the LOS molecule (Table 4). Taken together, these results suggest that this group of patients appears to have developed more antibodies directed toward terminal LOS epitopes following the clearance of M. catarrhalis from the respiratory tract. Therefore, we conclude that the R1 (1-6-linked) and R2 (1-4-linked) chains of the LOS molecule may represent two possible targets of human antibodies following the clearance of M. catarrhalis from the respiratory tract in adults with COPD.
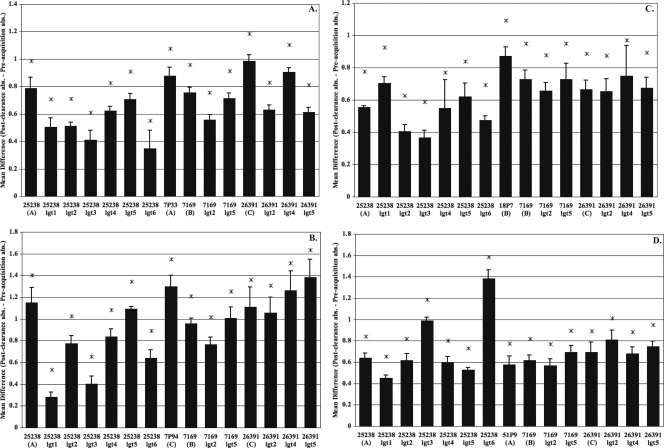
Figure 3 shows the results for three other patients that developed a more broadly cross-reactive antibody response, in contrast to the patient responses shown in Fig. 2. Patient 7, infected with a serotype A strain (7P33B1) (Fig. 3a) and subsequently infected with a serotype C strain (7P94B1) (Fig. 3b); patient 18, (Fig. 3c) infected with a serotype B strain (18P7B1); and patient 51 (Fig. 3d), infected with a serotype A strain (51P9B1), all developed a significant level of new IgG antibodies to every carbohydrate epitope tested in these studies. The reactivity elicited in these patients to even the most truncated LOS glycoforms (lgt1, lgt3, and lgt6) suggests that antibodies in the postclearance sera of these patients are directed to core structures such as lipid A-KDO2 (lgt6 mutant LOS) and lipid A-KDO2-(1-5)-d-glucose (lgt3 mutant LOS), as these regions are shared by all of the LOS mutants included in this ELISA. Thus, we conclude that the core structures of the LOS molecule represent another target of human antibodies to LOS following the clearance of M. catarrhalis from the respiratory tract in adults with COPD.
FIG. 3.
COPD patients who developed more broadly cross-reactive and internally directed antibody responses to different LOS epitopes. Shown are data from an evaluation of levels of IgG against various truncated LOS molecules in sera from COPD patients. The OS used as antigens are listed along the x axis. Patient 7 (A) was infected with 7P33B1 (serotype A), patient 7 (B) was infected with 7P94B1 (serotype C), patient 18 (C) was infected with 18P7B1 (serotype B), and patient 51 (D) was infected with 51P9B1 (serotype A). ELISAs were used to test the level of IgG antibody in sera of COPD patients in response to truncated LOS obtained from a panel of glycosyltransferase mutants and full-length wild-type structures representing serotype A (25238), serotype B (7169), and serotype C (26391). Serum samples obtained preacquisition and postclearance of the infecting M. catarrhalis strain were incubated in a 96-well plate coated with 20 to 30 μg of LOS per well. The bars represent the mean differences in absorbance for at least three independent ELISAs of serum samples taken postclearance of the exacerbating strain to preacquisition of that strain, with the error bars representing the standard errors of the means. Asterisks indicate a P value of ≤0.05 as determined by a one-sided paired t test.
Finally, sera from patients 12, 13, and 39 were examined by using the LOS-based ELISA described in this study. Just as there was a lack of a significant increase in the level of antibody to their homologous strains that developed (Fig. 1), there was no significant development of serum antibodies to the panel of truncation mutant LOS (data not shown).
One factor that must be considered in the interpretation of our data is the previous exposure to M. catarrhalis throughout childhood and adulthood experienced by all adults. Similar antigens or epitopes may have been presented in these patients through past infections or colonization with other gram-negative organisms that express common LOS and LPS epitopes (5). Haemophilus influenzae occupies the same ecological niche in the respiratory tract as M. catarrhalis; therefore, these patients have been exposed to these LOS moieties prior to infection with M. catarrhalis. While some of the patients investigated likely had detectable levels of antibodies to M. catarrhalis LOS prior to infection, these antibodies do not present a confounding issue because we have compared the preacquisition sera to the matched postclearance sera from each individual patient. This approach effectively eliminates baseline antibodies to any region of the LOS molecule and serves as the appropriate control.
Most of the structural variability in M. catarrhalis LOS is present in the terminal regions of the main LOS branch and in the side chains. However, compared with LOS and LPS of other pathogens, M. catarrhalis LOS does not exhibit a high degree of structural diversity. Even though the LOS serotypes of M. catarrhalis display distinct carbohydrate structures, there are shared epitopes (46). Table 4 shows that the carbohydrate structures of all three LOS serotypes contain the common OS α-d-Galp-(1-4)-β-d-Galp-(1-4)-α-d-Glcp. This epitope is also present in other gram-negative mucosal pathogens such as H. influenzae, Neisseria meningitidis, and Neisseria gonorrhoeae and is found in glycolipids of human mucous membrane epithelial cells (37, 38). It is also important that this common LOS contains the α-d-Galp-(1-4)-β-d-Galp moiety, which is involved in resistance to the bactericidal activity of human serum in one strain of M. catarrhalis (54). A recent investigation found that the loss of this same epitope decreased the recognition of bactericidal antibodies produced in mice against an M. catarrhalis conjugate vaccine (53). Our data suggest that some patients develop more antibodies to the terminal regions of M. catarrhalis LOS following infection; thus, the α-d-Galp-(1-4)-β-d-Galp-(1-4)-α-d-Glcp moiety could represent a possible target of the immune response, although more studies are needed to confirm this observation.
Previous reports suggested that the lack of a serotype-specific antibody response in patients indicated that M. catarrhalis LOS is weakly immunogenic in humans (37, 38, 46). However, a more recent study detected the presence of anti-LOS antibodies in sera of COPD patients infected with M. catarrhalis (29). The current study demonstrates that a group of patients with COPD develop new antibodies to M. catarrhalis LOS in their postclearance sera following an infection. Perhaps more importantly, the ELISA system that we have developed clearly has the potential to discern antibody reactivity to specific regions of the M. catarrhalis LOS molecule regardless of serotype.
One limitation of the present study is the relatively small number of patient samples analyzed, precluding definitive conclusions regarding the importance of antibody reactivity to specific regions of the LOS molecule and also the potential protective effect of these antibodies. The latter issue is very difficult to address, especially as it pertains to M. catarrhalis infections, and it is important that the new antibodies developed in these patients' sera represent a polyclonal response with the potential to cross-react with other bacterial epitopes. In order to determine if any of the human antibodies directed toward specific carbohydrate epitopes are functional, these antibodies would have to be isolated from each serum sample and then evaluated, which is extremely difficult, if not impossible. In addition, M. catarrhalis is a strictly human pathogen, and there is currently no good animal model that can effectively replicate natural infection. Furthermore, there have been data reported previously that suggested that antibodies elicited in animals may be directed at different epitopes than antibodies that are developed in humans following M. catarrhalis infections (2).
Perhaps one of the most important issues that must be considered when attempting to analyze the antibody response elicited in humans is the effect of the pathogen. Recent studies suggested that some pathogens express immunodominant epitopes that induce highly restrictive antibody reactivity, thus limiting the overall effectiveness of the immune response. This appears to result from the fact that the human immune system is geared to respond to these immunodominant epitopes, a phenomenon referred to as deceptive imprinting (44). While this has been more carefully studied for viral pathogens, there have been numerous studies published describing immunodominant epitopes expressed by bacterial pathogens such as H. influenzae (24, 32, 50). Novotny and Bakaletz previously described an immunodominant epitope expressed by nontypeable H. influenzae that acts as a decoy, eliciting antibodies that do not protect the host (32). Thus, the development of new antibodies to bacterial epitopes in humans, while potentially beneficial to the host, could also be nonprotective in certain situations. For this reason, as a first step, it is critical to identify the specific M. catarrhalis epitopes that elicit new antibodies in humans, which is a very important aspect of the work that we have presented in this study. Although more studies using a more extensive cohort of patients including children and adults are clearly needed, this is the first LOS-based ELISA system that can define the M. catarrhalis LOS epitopes that elicit new antibodies following natural infection in the native host. The identification of these antibodies lays the foundation for discerning the M. catarrhalis LOS epitopes that warrant further analysis to determine if these LOS regions confer a protective humoral immune response in humans.
Acknowledgments
This research was supported by Public Health Service research grants AI46422 (A.A.C.), DC005837 (A.A.C.), and AI28304 (T.F.M.) and the Department of Veterans Affairs. K.J.E. was also supported as a graduate student fellow by NIH training grant AI07614.
We thank Perry Fleming for bacterial growth and Jacek Stupak for assistance in recording capillary electrophoresis-ES-MS data.
Editor: S. R. Blanke
Footnotes
Published ahead of print on 3 August 2009.
REFERENCES
- 1.Adlowitz, D. G., C. Kirkham, S. Sethi, and T. F. Murphy. 2006. Human serum and mucosal antibody responses to outer membrane protein G1b of Moraxella catarrhalis in chronic obstructive pulmonary disease. FEMS Immunol. Med. Microbiol. 46:139-146. [DOI] [PubMed] [Google Scholar]
- 2.Ala'Aldeen, D. A., P. Stevenson, E. Griffiths, A. R. Gorringe, L. I. Irons, A. Robinson, S. Hyde, and S. P. Borriello. 1994. Immune responses in humans and animals to meningococcal transferrin-binding proteins: implications for vaccine design. Infect. Immun. 62:2984-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakri, F., A. L. Brauer, S. Sethi, and T. F. Murphy. 2002. Systemic and mucosal antibody response to Moraxella catarrhalis after exacerbations of chronic obstructive pulmonary disease. J. Infect. Dis. 185:632-640. [DOI] [PubMed] [Google Scholar]
- 4.Bogaert, D., P. van der Valk, R. Ramdin, M. Sluijter, E. Monninkhof, R. Hendrix, R. de Groot, and P. W. Hermans. 2004. Host-pathogen interaction during pneumococcal infection in patients with chronic obstructive pulmonary disease. Infect. Immun. 72:818-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buttenschoen, K., M. Kornmann, D. Berger, G. Leder, H. G. Beger, and C. Vasilescu. 2008. Endotoxemia and endotoxin tolerance in patients with ARDS. Langenbeck's Arch. Surg. 393:473-478. [DOI] [PubMed] [Google Scholar]
- 6.Celli, B. R., W. MacNee, and ATS/ERS Task Force. 2004. Standards for the diagnosis and treatment of patients with COPD: a summary of the ATS/ERS position paper. Eur. Respir. J. 23:932-946. [DOI] [PubMed] [Google Scholar]
- 7.Ciucanu, I., and F. Kerek. 1984. A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131:209-217. [Google Scholar]
- 8.Edebrink, P., P. E. Jansson, M. M. Rahman, G. Widmalm, T. Holme, and M. Rahman. 1995. Structural studies of the O-antigen oligosaccharides from two strains of Moraxella catarrhalis serotype C. Carbohydr. Res. 266:237-261. [DOI] [PubMed] [Google Scholar]
- 9.Edebrink, P., P. E. Jansson, M. M. Rahman, G. Widmalm, T. Holme, M. Rahman, and A. Weintraub. 1994. Structural studies of the O-polysaccharide from the lipopolysaccharide of Moraxella (Branhamella) catarrhalis serotype A (strain ATCC 25238). Carbohydr. Res. 257:269-284. [DOI] [PubMed] [Google Scholar]
- 10.Edebrink, P., P. E. Jansson, G. Widmalm, T. Holme, and M. Rahman. 1996. The structures of oligosaccharides isolated from the lipopolysaccharide of Moraxella catarrhalis serotype B, strain CCUG 3292. Carbohydr. Res. 295:127-146. [DOI] [PubMed] [Google Scholar]
- 11.Edwards, K. J., S. Allen, B. W. Gibson, and A. A. Campagnari. 2005. Characterization of a cluster of three glycosyltransferase enzymes essential for Moraxella catarrhalis lipooligosaccharide assembly. J. Bacteriol. 187:2939-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards, K. J., J. M. Schwingel, A. K. Datta, and A. A. Campagnari. 2005. Multiplex PCR assay that identifies the major lipooligosaccharide serotype expressed by Moraxella catarrhalis clinical isolates. J. Clin. Microbiol. 43:6139-6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enright, M. C., and H. McKenzie. 1997. Moraxella (Branhamella) catarrhalis—clinical and molecular aspects of a rediscovered pathogen. J. Med. Microbiol. 46:360-371. [DOI] [PubMed] [Google Scholar]
- 14.Furano, K., and A. A. Campagnari. 2003. Inactivation of the Moraxella catarrhalis 7169 ferric uptake regulator increases susceptibility to the bactericidal activity of normal human sera. Infect. Immun. 71:1843-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holme, T., M. Rahman, P. E. Jansson, and G. Widmalm. 1999. The lipopolysaccharide of Moraxella catarrhalis: structural relationships and antigenic properties. Eur. J. Biochem. 265:524-529. [DOI] [PubMed] [Google Scholar]
- 16.Hu, W. G., J. Chen, J. F. Battey, and X. X. Gu. 2000. Enhancement of clearance of bacteria from murine lungs by immunization with detoxified lipooligosaccharide from Moraxella catarrhalis conjugated to proteins. Infect. Immun. 68:4980-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, W. G., J. Chen, J. C. McMichael, and X. X. Gu. 2001. Functional characteristics of a protective monoclonal antibody against serotype A and C lipooligosaccharides from Moraxella catarrhalis. Infect. Immun. 69:1358-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inzana, T. J., and M. A. Apicella. 1999. Use of a bilayer stacking gel to improve resolution of lipopolysaccharides and lipooligosaccharides in polyacrylamide gels. Electrophoresis 20:462-465. [DOI] [PubMed] [Google Scholar]
- 19.Jiao, X., T. Hirano, Y. Hou, and X. X. Gu. 2002. Specific immune responses and enhancement of murine pulmonary clearance of Moraxella catarrhalis by intranasal immunization with a detoxified lipooligosaccharide conjugate vaccine. Infect. Immun. 70:5982-5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, J., P. Thibault, A. Martin, J. C. Richards, W. W. Wakarchuk, and W. van der Wilp. 1998. Development of an on-line preconcentration method for the analysis of pathogenic lipopolysaccharides using capillary electrophoresis-electrospray mass spectrometry. Application to small colony isolates. J. Chromatogr. A 817:325-336. [DOI] [PubMed] [Google Scholar]
- 21.Ling, M. M., and B. H. Robinson. 1997. Approaches to DNA mutagenesis: an overview. Anal. Biochem. 254:157-178. [DOI] [PubMed] [Google Scholar]
- 22.Luke, N. R., S. Allen, B. W. Gibson, and A. A. Campagnari. 2003. Identification of a 3-deoxy-d-manno-octulosonic acid biosynthetic operon in Moraxella catarrhalis and analysis of a KdsA-deficient isogenic mutant. Infect. Immun. 71:6426-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masoud, H., M. B. Perry, J.-R. Brisson, D. Uhrin, and J. C. Richards. 1994. Structural elucidation of the backbone oligosaccharide for the lipopolysaccharide of Moraxella catarrhalis serotype A. Can. J. Chem. 72:1466-1477. [Google Scholar]
- 24.McMahon, M., T. F. Murphy, J. Kyd, and Y. Thanavala. 2005. Role of an immunodominant T cell epitope of the P6 protein of nontypeable Haemophilus influenzae in murine protective immunity. Vaccine 23:3590-3596. [DOI] [PubMed] [Google Scholar]
- 25.Meier, P. S., N. Heiniger, R. Troller, and C. Aebi. 2003. Salivary antibodies directed against outer membrane proteins of Moraxella catarrhalis in healthy adults. Infect. Immun. 71:6793-6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy, T. F. 1996. Branhamella catarrhalis: epidemiology, surface antigenic structure, and immune response. Microbiol. Rev. 60:267-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy, T. F., A. L. Brauer, C. Aebi, and S. Sethi. 2005. Antigenic specificity of the mucosal antibody response to Moraxella catarrhalis in chronic obstructive pulmonary disease. Infect. Immun. 73:8161-8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy, T. F., A. L. Brauer, C. Aebi, and S. Sethi. 2005. Identification of surface antigens of Moraxella catarrhalis as targets of human serum antibody responses in chronic obstructive pulmonary disease. Infect. Immun. 73:3471-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy, T. F., A. L. Brauer, B. J. Grant, and S. Sethi. 2005. Moraxella catarrhalis in chronic obstructive pulmonary disease: burden of disease and immune response. Am. J. Respir. Crit. Care Med. 172:195-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy, T. F., S. Sethi, and M. S. Niederman. 2000. The role of bacteria in exacerbations of COPD. A constructive view. Chest 118:204-209. [DOI] [PubMed] [Google Scholar]
- 32.Novotny, L. A., and L. O. Bakaletz. 2003. The fourth surface-exposed region of the outer membrane protein P5-homologous adhesin of nontypable Haemophilus influenzae is an immunodominant but nonprotective decoying epitope. J. Immunol. 171:1978-1983. [DOI] [PubMed] [Google Scholar]
- 33.Peak, I. R., I. D. Grice, I. Faglin, Z. Klipic, P. M. Collins, L. van Schendel, P. G. Hitchen, H. R. Morris, A. Dell, and J. C. Wilson. 2007. Towards understanding the functional role of the glycosyltransferases involved in the biosynthesis of Moraxella catarrhalis lipooligosaccharide. FEBS J. 274:2024-2037. [DOI] [PubMed] [Google Scholar]
- 34.Plamondon, P., N. R. Luke, and A. A. Campagnari. 2007. Identification of a novel two-partner secretion locus in Moraxella catarrhalis. Infect. Immun. 75:2929-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plested, J. S., M. A. Gidney, P. A. Coull, H. G. Griffiths, M. A. Herbert, A. G. Bird, J. C. Richards, and E. R. Moxon. 2000. Enzyme linked immunosorbent assay (ELISA) for the detection of serum antibodies to the inner core lipopolysaccharide of Neisseria meningitidis group B. J. Immunol. Methods 237:73-84. [DOI] [PubMed] [Google Scholar]
- 36.Rabe, K. F., S. Hurd, A. Anzueto, P. J. Barnes, S. A. Buist, P. Calverley, Y. Fukuchi, C. Jenkins, R. Rodriguez-Roisin, C. van Weel, and J. Zielinski. 2007. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease—GOLD executive summary. Am. J. Respir. Crit. Care Med. 176:532-555. [DOI] [PubMed] [Google Scholar]
- 37.Rahman, M., and T. Holme. 1996. Antibody response in rabbits to serotype-specific determinants in lipopolysaccharides from Moraxella catarrhalis. J. Med. Microbiol. 44:348-354. [DOI] [PubMed] [Google Scholar]
- 38.Rahman, M., T. Holme, I. Jonsson, and A. Krook. 1995. Lack of serotype-specific antibody response to lipopolysaccharide antigens of Moraxella catarrhalis during lower respiratory tract infection. Eur. J. Clin. Microbiol. Infect. Dis. 14:297-304. [DOI] [PubMed] [Google Scholar]
- 39.Ruckdeschel, E. A., C. Kirkham, A. J. Lesse, Z. Hu, and T. F. Murphy. 2008. Mining the Moraxella catarrhalis genome: identification of potential vaccine antigens expressed during human infection. Infect. Immun. 76:1599-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Schwingel, J. M., F. St. Michael, A. D. Cox, H. Masoud, J. C. Richards, and A. A. Campagnari. 2008. A unique glycosyltransferase involved in the initial assembly of Moraxella catarrhalis lipooligosaccharides. Glycobiology 18:447-455. [DOI] [PubMed] [Google Scholar]
- 42.Sethi, S., N. Evans, B. J. Grant, and T. F. Murphy. 2002. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N. Engl. J. Med. 347:465-471. [DOI] [PubMed] [Google Scholar]
- 43.Sethi, S., and T. F. Murphy. 2001. Bacterial infection in chronic obstructive pulmonary disease in 2000: a state-of-the-art review. Clin. Microbiol. Rev. 14:336-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tobin, G. J., J. D. Trujillo, R. V. Bushnell, G. Lin, A. R. Chaudhuri, J. Long, J. Barrera, L. Pena, M. J. Grubman, and P. L. Nara. 2008. Deceptive imprinting and immune refocusing in vaccine design. Vaccine 26:6189-6199. [DOI] [PubMed] [Google Scholar]
- 45.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 46.Vaneechoutte, M., G. Verschraegen, G. Claeys, and A. M. Van Den Abeele. 1990. Serological typing of Branhamella catarrhalis strains on the basis of lipopolysaccharide antigens. J. Clin. Microbiol. 28:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verduin, C. M., C. Hol, A. Fleer, H. van Dijk, and A. van Belkum. 2002. Moraxella catarrhalis: from emerging to established pathogen. Clin. Microbiol. Rev. 15:125-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Westphal, O., and K. Jann. 1965. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of procedure, p. 83-91. In R. Whistler (ed.), Methods in carbohydrate chemistry, vol. 5. Academic Press, New York, NY. [Google Scholar]
- 49.Wilson, J. C., P. M. Collins, Z. Klipic, I. D. Grice, and I. R. Peak. 2006. Identification of a novel glycosyltransferase involved in LOS biosynthesis of Moraxella catarrhalis. Carbohydr. Res. 341:2600-2606. [DOI] [PubMed] [Google Scholar]
- 50.Yi, K., and T. F. Murphy. 1997. Importance of an immunodominant surface-exposed loop on outer membrane protein P2 of nontypeable Haemophilus influenzae. Infect. Immun. 65:150-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yi, K., S. Sethi, and T. F. Murphy. 1997. Human immune response to nontypeable Haemophilus influenzae in chronic bronchitis. J. Infect. Dis. 176:1247-1252. [DOI] [PubMed] [Google Scholar]
- 52.Yu, S., and X. X. Gu. 2005. Synthesis and characterization of lipooligosaccharide-based conjugate vaccines for serotype B Moraxella catarrhalis. Infect. Immun. 73:2790-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu, S., H. Xie, A. Datta, N. Naidu, and X.-X. Gu. 2008. Galactose residues on the lipooligosaccharide of Moraxella catarrhalis 26404 form the epitope recognized by the bactericidal antiserum from conjugate vaccination. Infect. Immun. 76:4251-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zaleski, A., N. K. Scheffler, P. Densen, F. K. Lee, A. A. Campagnari, B. W. Gibson, and M. A. Apicella. 2000. Lipooligosaccharide Pk (Galα1-4Galβ1-4Glc) epitope of Moraxella catarrhalis is a factor in resistance to bactericidal activity mediated by normal human serum. Infect. Immun. 68:5261-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]



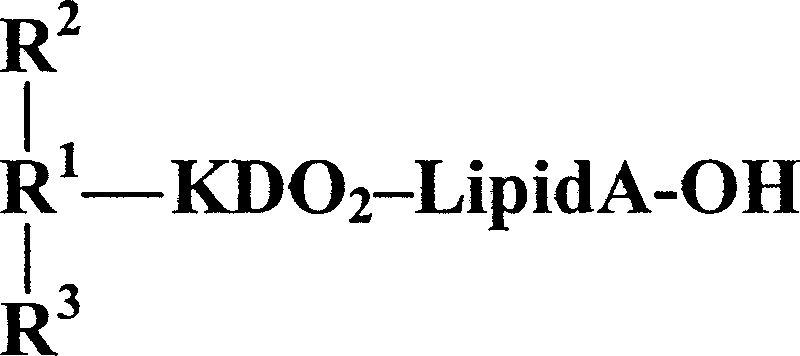 .
.