Abstract
Porphyromonas gingivalis is a major periodontal pathogen that has the pathogenic proteinases Arg-specific gingipain and Lys-specific gingipain. We previously found that a cell surface component on P. gingivalis is able to induce Toll-like receptor 2 (TLR2)- and TLR4-independent signaling in 7.19 cells and that this component can be degraded by gingipains. In this study, we purified this component from the P. gingivalis gingipain-null mutant KDP136 and obtained two candidate proteins. Matrix-assisted laser desorption ionization-time-of-flight mass spectrometry analysis showed that the proteins, with molecular masses of 123 and 43 kDa, were encoded by PGN_0748 and PGN_0728 (pgm6), respectively, in the P. gingivalis ATCC 33277 genome sequence. The PGN_0748-encoded protein, which we refer to as gingipain-sensitive ligand A (GslA), reacted with antiserum that could effectively inhibit the activity of KDP136 to induce NF-κB activation in 7.19 cells, but Pgm6 did not. To further determine what protein is responsible for the NF-κB activation, we constructed gslA, pgm6, and pgm6 pgm7 deletion mutants from KDP136. When 7.19 cells were exposed to those mutants, the gslA deletion mutant did not induce NF-κB activation, whereas the pgm6 and pgm6 pgm7 deletion mutants did. Furthermore, NF-κB activation in 7.19 cells induced by KDP136 was partially inhibited by antiserum against a recombinant protein expressed from the 5′-terminal third of gslA. These results indicate that GslA is one of the factors that induce NF-κB activation in 7.19 cells. Interestingly, the gslA gene was present in four of seven P. gingivalis strains tested. This restricted distribution might be associated with the virulence potential of each strain.
Porphyromonas gingivalis is an anaerobic gram-negative bacterium that is frequently isolated from advanced periodontal lesions (25). The number of P. gingivalis cells is closely associated with the depth of periodontal pockets and is significantly reduced after treatment (7). Thus, this organism is thought to play an important role in the development and progression of periodontitis.
P. gingivalis has two major cysteine proteinases, Arg-specific gingipain and Lys-specific gingipain. These proteinases have been reported to cleave various host immune effector molecules, such as immunoglobulin G (IgG) and IgM (22); several cytokines and cytokine receptors (1, 2, 10, 11, 17); and a pattern recognition receptor, CD14 (21, 23). These modifications of host immune regulatory molecules enable P. gingivalis to escape from the host immune system. This activity of gingipain seems to play an important role in the colonization of P. gingivalis in the oral cavity.
Besides degradation of the host molecules, we previously found that gingipains could degrade a ligand expressed on the P. gingivalis cell surface (8). A CHO cell-derived nuclear factor (NF)-κB reporter cell line, 7.19, was stimulated with wild-type (ATCC 33277) and gingipain-deficient P. gingivalis (KDP136) bacterial cells. Since bacterial cells possess a number of ligands for Toll-like receptor 2 (TLR2) and TLR4, 7.19 cells, which lack both TLR2- and TLR4-signaling pathways, enable analysis of TLR2- and TLR4-independent signaling (18). Interestingly, 7.19 cells were activated by gingipain-null mutant KDP136 but not by its parental strain ATCC 33277, suggesting that the ligand of P. gingivalis was degraded by gingipains in the wild-type bacterial cells. In fact, the ability of KDP136 to induce activation of NF-κB in 7.19 cells was diminished after treatment of the bacterial cells with gingipains. In a previous study (8), we partially purified components with the ability to activate NF-κB in 7.19 cells from KDP136. The activities of the components were also diminished by treatment with gingipains.
The aim of the present study was to purify and identify the gingipain-sensitive ligand from gingipain-deficient P. gingivalis cells. We tried further purification and obtained two proteins encoded by protein-coding sequence (CDS) PGN_0748 and CDS PGN_0728 (pgm6) in the P. gingivalis ATCC 33277 genome sequence (14). We then constructed CDS mutants from KDP136 and determined which protein is responsible for the activity that induces NF-κB activation in 7.19 cells.
MATERIALS AND METHODS
Reagents.
Phosphate-buffered saline (PBS), Ham's F-12 medium, penicillin-streptomycin, and trypsin-EDTA were obtained from Life Technologies (Rockville, MD). Fetal bovine serum was obtained from JRH Biosciences (Lenexa, KS). Hygromycin B was obtained from Calbiochem (San Diego, CA). DEAE Sepharose CL-6B and Superose 6 gel filtration columns were obtained from GE Healthcare (Buckinghamshire, United Kingdom). Anti-CD25 monoclonal antibody conjugated with fluorescein isothiocyanate (FITC) was obtained from Becton Dickinson (Bedford, MA). IgG1-FITC isotype control from murine myeloma was obtained from Sigma-Aldrich (St. Louis, MO). The DNA primers used in this study are summarized in Table 1.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′-3′)a |
|---|---|
| 0748UPFOR | GGTATTCATTTGTCCTTGCAGAGAC |
| 0748UPREV | CAGGGGATCCGTTAGACATCAACCACACCCGC |
| 0748DOFOR | TAACGGATCCCCTGACTCCTTCAAAGGCCTG |
| 0748DOREV | GAATACGACTCTAGCCCCATGC |
| 0728UPFOR | GCGGCCGCATCATGGGATGATCTACAGGGT |
| 0728UPREV | GGATCCTCTCAAATATCCCCACAAATAAAT |
| 0728DOFOR | GGATCCAATTCTGTATGTCATTTTATATTA |
| 0728DOREV | TAATTCCATGGGTAGGTGTTGGCTACCAAC |
| 0729DOFOR | GGATCCAGTTTTACTTTTCTAAGTGTATTT |
| 0729DOREV | GAAGTCGCCAAAGGTGTCCTTTGCAGAACG |
| CEPFOR | GGATCCGACGTCAAAAGAGTTAAGGAAAGTGAAGC |
| CEPREV | GGATCCGACGTCTTTCAAGTCACCGATAGTG |
| GSLAFOR1 | CATATGCCTGATCAAGAAAATAAGGAAAAGGC |
| GSLAREV1 | GAATTCGCCTCTTCATACAAGAGTTTGACCC |
| GSLAFOR2 | CATATGGATAAAAAGAAAAAAGAGGAGGGTACAACG |
| GSLAREV2 | GAATTCAGATAGGCCCTGAGGTCATCACA |
| GSLAFOR3 | CATATGTCTGTATTTGAGCTAAATATAAGTGGCAG |
| GSLAREV3 | GAATTCTCCTTTTTAGGATTAGGCTCCTTCCC |
All primers are from this study. Restriction sites incorporated into oligonucleotides for subcloning are underlined.
Cell lines and flow cytometric analysis.
The engineering of the CHO/CD14 reporter cell line has been previously described in detail (3). The 7.19 cell line is a CHO/CD14 cell line-derived mutant that has a point mutation at position 284 of the coding region of MD-2 that results in conversion of the codon for cysteine to tyrosine (18). Therefore, this cell line is defective in both TLR4- and TLR2-signaling pathways but can express a reporter molecule, CD25, on the cell surface through NF-κB activation induced by interleukin-1β (8). This cell line was grown as adherent monolayers in Ham's F-12 medium supplemented with 10% fetal bovine serum and hygromycin B (400 U/ml) at 37°C in a 5% CO2 atmosphere.
For flow cytometric analysis, 7.19 cells were plated in 24-well tissue culture dishes at a density of 2.0 × 105 cells per well. After incubation for 20 h, a confluent monolayer of 7.19 cells was stimulated with freeze-dried bacteria (200 μg/ml). Following incubation for 20 h, the cells were treated with 30 mM EDTA for 1 min, and detached cells were assessed by flow cytometry for the presence of surface CD25 as described previously (8).
Bacterial strains and culture conditions.
P. gingivalis ATCC 33277 and its isogenic mutants used in this study are shown in Table 2. The cells were grown anaerobically (10% CO2, 10% H2, 80% N2) at 37°C in enriched brain-heart infusion medium supplemented with vitamin K1 (1 μg/ml) and hemin (5 μg/ml). For maintenance of KDP136, erythromycin (10 μg/ml) was added to the medium. For maintenance of the other antibiotic-resistant strains of P. gingivalis, erythromycin (10 μg/ml) and ampicillin (10 μg/ml) were added to the medium. For reporter cell stimulation experiments, the microorganisms were harvested by centrifugation, washed three times with distilled water, and freeze-dried.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| P. gingivalis strains | ||
| ATCC 33277 | Wild type | ATCC |
| W83 | Wild type | Gift from M. J. Duncana |
| TDC60 | Wild type | Gift from K. Ishiharab |
| TDC117 | Wild type | |
| TDC275 | Wild type | |
| SU63 | Wild type | Gift from M. Yonedac |
| GAI-7802 | Wild type | Gift from E. Hoshinod |
| KDP136 | kgp-2::cat rgpA2::[ermFermAM] rgpB2::tetQ | 19 |
| KDP377 | kgp-2::cat rgpA2::[ermFermAM] rgpB2::tetQ Δ[gslA]::cepA | This study |
| KDP378 | kgp-2::cat rgpA2::[ermFermAM] rgpB2::tetQ Δ[pgm6]::cepA | This study |
| KDP379 | kgp-2::cat rgpA2::[ermFermAM] rgpB2::tetQ Δ[λσθβ]pgm6pgm7]::cepA | This study |
| E. coli strains | ||
| XL-1 Blue | General-purpose host strain for recombinant protein production | Stratagene |
| BL21(DE3) | Host strain for production of recombinant proteins | 20 |
| Plasmids | ||
| pCR4Blunt-TOPO | Apr Kmr; plasmid vector for cloning | Invitrogen |
| pBluescript ΙΙ SK(+) | Apr; plasmid vector for cloning | Stratagene |
| pET22b | Apr; plasmid vector for cloning | Novagenf |
| pCS22 | Apr; contains the cepA DNA cassette at the AatII site of pCS14 | Gift from E. C. Reynoldse |
| pKD1001 | Apr Kmr; contains 0.6-kb upstream and 0.9-kb downstream gslA DNA fragments in pCR4BluntII-TOPO | This study |
| pKD1002 | Apr Kmr; contains the cepA DNA cassette at the BamHI site of pKD1001 | This study |
| pKD1003 | Apr; contains 0.8-kb upstream and 0.8-kb downstream pgm6 DNA fragments in pBluescript ΙΙ SK(+) | This study |
| pKD1004 | Apr; contains 0.8-kb upstream and 0.8-kb downstream pgm7 DNA fragments in pBluescript ΙΙ SK(+) | This study |
| pKD1005 | Apr; contains the cepA DNA cassette at the BamHI site of pKD1003 | This study |
| pKD1006 | Apr; contains the cepA DNA cassette at the BamHI site of pKD1004 | This study |
| pKD1007 | Apr; contains the 5′-terminal third of gslA in pET22b | This study |
Department of Molecular Genetics, The Forsyth Institute, Boston, MA.
Department of Microbiology, Tokyo Dental College, Chiba, Japan.
Department of General Dentistry, Fukuoka Dental College, Fukuoka, Japan.
Department of Oral Health Science, Graduate School of Medical and Dental Sciences, Niigata University, Niigata, Japan.
Cooperative Research Centre for Oral Health Science, School of Dental Science, University of Melbourne, Victoria, Australia.
San Diego, CA.
Purification of the cell surface component from KDP136.
The cell surface component of KDP136 was partially purified as described previously (8). Then, further purification was carried out with gel filtration chromatography (1.0-cm by 30-cm, 24-ml-column-volume Superose 6 gel filtration column) at a flow rate of 0.4 ml/min with 50 mM potassium phosphate buffer (pH 7.4) containing 0.05% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} and 0.5 M NaCl. Proteins in each fraction were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the activity of each fraction that induces NF-κB activation was determined using 7.19 reporter cells.
Protein analysis by MS.
Proteins separated by SDS-PAGE were analyzed by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS). After in-gel tryptic digestion, peptides were extracted, concentrated, and analyzed using a Voyage-DE STR BioSpectrometry work station (Applied Biosystems, Foster City, CA). The identities of the proteins were deduced from MS peaks via the MS-Fit peptide mass fingerprinting methods in Mascot software (http://www.matrixscience.com/).
Polyclonal antibodies.
To obtain antiserum against the ligand of P. gingivalis which can activate 7.19 cells, BALB/c mice were immunized with 30 to 250 μg of the partially purified components from KDP136 in conjunction with Freund's incomplete adjuvant. The immunization was performed at 2-week intervals, with blood being collected from the tail vein every two or three times of injection. The serum was tested for inhibitory activity to induce NF-κB activation in 7.19 cells stimulated with KDP136.
To obtain a polyclonal antibody against the PGN_0748-encoded protein, which was designated the gingipain-sensitive ligand A (GslA), rabbits were immunized with 250 μg of the recombinant protein in conjunction with Freund's incomplete adjuvant. The immunization was performed at 2-week intervals, with blood being collected every two or three times of injection. The antibody levels in the serum were measured by enzyme-linked immunosorbent assay.
Following elevation of the titer of serum antibody, animals were boosted with another immunization, which resulted in antisera. Animal care and experimental procedures were conducted in accordance with the Guidelines for Animal Experimentation of Nagasaki University and with approval of the Institutional Animal Care and Use Committee. Rabbit polyclonal antibody was purified from antiserum using protein A-conjugated Sepharose 4B (GE Healthcare).
Subcellular fractionation of P. gingivalis.
P. gingivalis at mid-log phase was subjected to the following fractionation, as described previously (12). Briefly, the cells were harvested by centrifugation at 10,000 × g for 20 min and washed twice with 10 mM HEPES buffer (pH 7.4), suspended with 10 mM HEPES buffer (pH 7.4) that included 1 mM TLCK (Nα-p-tosyl-l-lysine chloromethyl ketone) and 1 mM leupeptin, and disrupted in a French pressure cell at 100 MPa. Unbroken cells and large debris were removed by centrifugation at 1,000 × g for 10 min, and the supernatants (whole-cell lysates) were subjected to ultracentrifugation at 100,000 × g for 1 h. The precipitates were suspended in 10 mM HEPES buffer (pH 7.4) supplemented with Triton X-100 at a final concentration of 1% and mixed gently at room temperature (RT) for 30 min. The solution was subjected to ultracentrifugation at 100,000 × g for 1 h to yield the bacterial outer membrane fraction as the precipitate.
Construction of P. gingivalis mutants.
To construct a gslA deletion mutant, two DNA fragments corresponding to the upstream and downstream regions of gslA were PCR amplified from the chromosomal DNA of P. gingivalis ATCC 33277 with two primer pairs, 0748UPFOR/0748UPREV and 0748DOFOR/0748DOREV. The two DNA fragments were annealed and subjected to PCR amplification with the primers 0748UPFOR and 0748DOREV. The resulting amplified fragment, in which gslA was replaced with a BamHI site, was ligated to the pCR4Blunt-TOPO vector (Invitrogen, Carlsbad, CA), resulting in pKD1001. The ampicillin resistance gene cepA was amplified from pCS22 with two primers, CEPFOR and CEPREV, and inserted into the BamHI site of pKD1001, resulting in a gslA deletion cassette, pKD1002. KDP136 was transformed to be ampicillin resistant by electroporation with linearized pKD1002 plasmid DNA to yield a gslA deletion mutant, KDP377 (kgp-2::cat rgpA2::[ermF ermAM] rgpB2::tetQ gslA::cepA).
To construct pgm6 and pgm6 PGN_0729 (pgm7) deletion mutants, a DNA fragments corresponding to an upstream region of pgm6 was PCR amplified with the primers 0728UPFOR and 0728UPREV. A downstream fragment of pgm6 was PCR amplified with the primers 0728DOFOR and 0728DOREV. A downstream fragment of pgm7 was PCR amplified with the primers 0729DOFOR and 0729DOREV. Each resulting amplified fragment was ligated to the pCR4Blunt-TOPO vector. The vector containing the upstream fragment of pgm6 was digested with NotI and BamHI, and the other vectors were digested with BamHI and EcoRI. The NotI-BamHI fragment, BamHI-EcoRI fragment, and pBluescript SK(−) vector (Stratagene, La Jolla, CA) digested with NotI and EcoRI were ligated, resulting in pKD1003 (target, pgm6) and pKD1004 (target, pgm6 pgm7). The amplified cepA gene was inserted into the BamHI site of pKD1003 or pKD1004, resulting in a pgm6 deletion cassette, pKD1005, or a pgm6 pgm7 deletion cassette, pKD1006. Each cepA-containing plasmid was subjected to transformation of KDP136 to yield a pgm6 deletion mutant, KDP378, or a pgm6 pgm7 deletion mutant, KDP379.
Recombinant protein.
Recombinant GslA protein was prepared as described previously (15). The DNA fragment coding for the 5′-terminal 510 amino acids was amplified from gslA of P. gingivalis ATCC 33277 with two primers, GSLAFOR1 and GSLAREV1. The resulting fragments were then inserted into the NdeI-EcoRI site of plasmid pET22b to yield pKD1007. E. coli BL21(DE3) cells harboring the resulting plasmids were grown in LB broth. Then, isopropyl β-d-thiogalactoside was added to the culture at 0.1 μM, followed by incubation for 2 h, to overproduce the recombinant protein. The induced recombinant protein was purified using a Ni-nitrilotriacetic acid purification system (Invitrogen).
Western blot analyses.
The samples were dissolved in Laemmli sample buffer and separated by SDS-PAGE. The gels were stained with Coomassie brilliant blue (CBB) or transblotted onto Immun-Blot polyvinylidene difluoride membranes (Bio-Rad Laboratories, Hercules, CA). Each membrane was blocked with 5% skim milk in PBS for 2 h at RT and reacted with the mouse polyclonal antibody against the Triton X-114-extracted bacterial cell surface components or the rabbit polyclonal antibody against the recombinant protein from the 5′-terminal third of gslA in 5% skim milk in PBS for 2 h at RT. Each membrane was then washed with PBS containing 0.05% Tween 20 and subjected to immunodetection using peroxidase-conjugated goat anti-mouse Ig or peroxidase-conjugated swine anti-rabbit Ig and an ECL Plus detection system (GE Healthcare).
Southern hybridization analyses.
A DNA probe for the upstream region of gslA was prepared with PCR amplification from ATCC 33277 chromosomal DNA using the primer pair 0748UPFOR and 0748UPREV. A DNA probe for cepA was prepared with PCR amplification from pCS22 plasmid DNA using CEPFOR and CEPREV. DNA probes a, b, and c for internal regions of gslA were prepared with PCR amplification from ATCC 33277 chromosomal DNA with the three primer pairs GSLAFOR1 and GSLAREV1, GSLAFOR2 and GSLAREV2, and GSLAFOR3 and GSLAREV3, respectively. These probes were labeled with the Alkphos Direct system for chemiluminescence (GE Healthcare). Chromosomal DNA was digested with BamHI or PstI and electrophoresed in an 0.8% agarose gel. Southern blotting was performed using a Hybond-N+ membrane (GE Healthcare) and developed with CDP-Star detection reagent (GE Healthcare).
RESULTS
Purification and identification of the protein from KDP136 that induces NF-κB activation in 7.19 reporter cells.
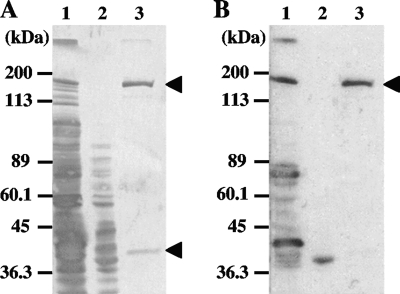
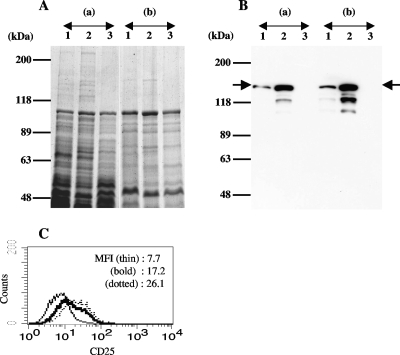
The bacterial cell surface components of P. gingivalis KDP136 extracted with Triton X-114 were subjected to ion-exchange chromatography as described previously (8). They were further purified by gel filtration chromatography, and the resulting fractions were analyzed for NF-κB activation-inducing activity in 7.19 cells. The fractions with high activity were subjected to SDS-PAGE, and a 43-kDa protein and a 123-kDa protein were detected (Fig. 1A). The 123-kDa protein was found in the Triton X-114-extracted bacterial cell surface components of KDP136 but not in those of P. gingivalis ATCC 33277.
FIG. 1.
Results of SDS-PAGE and Western blot analyses of bacterial components. Cell surface components of P. gingivalis were extracted with Triton X-114 (lanes 1, gingipain-null mutant KDP136; lanes 2, wild-type ATCC 33277). Then, the cell surface components of KDP136 were purified by ion-exchange chromatography and gel filtration chromatography as described in Materials and Methods (lanes 3). The samples were loaded onto 15% SDS-PAGE gels, and the gels were stained with CBB (A) or subjected to Western blot analysis using mouse antiserum that could effectively inhibit NF-κB activation in 7.19 cells stimulated with KDP136 (B).
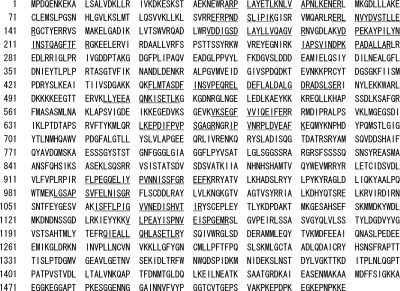
MALDI-TOF MS analysis revealed the 123-kDa and the 43-kDa proteins as proteins encoded by PGN_0748 and PGN_0728 (pgm6), respectively. The PGN_0748 protein consists of 1,530 amino acids, and the peptides of the 123-kDa protein obtained by MALDI-TOF MS analysis were matched to the amino acid sequence in the region from A38 to R1219 of PGN_0748, indicating that the 123-kDa protein was encoded by at least the 5′-terminal three-quarters of the PGN_0748 gene without the putative signal peptide-encoding region (Fig. 2).
FIG. 2.
Results of MALDI-TOF MS analysis of the 123-kDa protein. The predicted peptides that matched to GslA are shown with underlines.
Western blot analyses were performed using mouse antiserum that was obtained by immunization with the Triton X-114-extracted bacterial cell surface components and could effectively inhibit NF-κB activation in 7.19 cells stimulated with KDP136. The antiserum reacted to the 123-kDa protein but not to the 43-kDa protein (Fig. 1B). The antiserum also reacted to a protein with a molecular mass of 40 kDa, shown in Fig. 1B, lane 2 (ATCC 33277). Since the 40-kDa protein seems not to be present in the cell surface components extracted from KDP136, the protein may be a protein product degraded from the 123-kDa protein. These results suggested that the 123-kDa protein, which we refer to as gingipain-sensitive ligand A (GslA), was a principle candidate for the ligand that induces NF-κB activation in 7.19 cells.
Responses of 7.19 cells to gslA, pgm6, and pgm6 pgm7 deletion mutant cells.
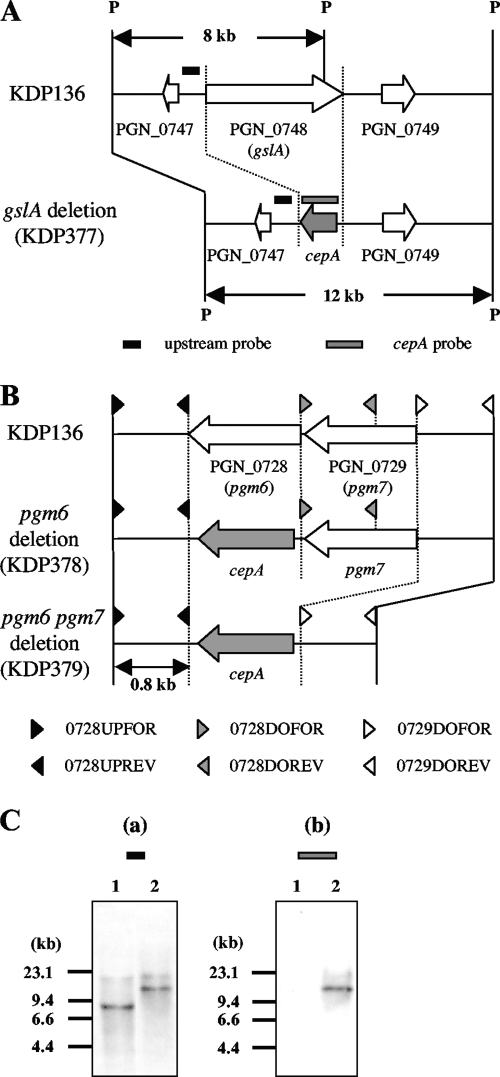
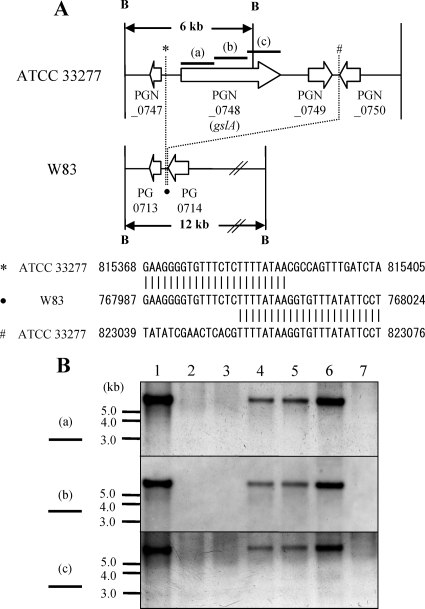
To determine which protein is responsible for the induction of NF-κB activation in 7.19 cells, the GslA-encoding gene (gslA) and pgm6 and pgm6 pgm7 deletion mutants (KDP377, KDP378, and KDP379, respectively) were constructed from the gingipain-null mutant KDP136 (rgpA rgpB kgp). Since proteins encoded by pgm6 and pgm7 form stable heterotrimers (13), we constructed the pgm6 pgm7 mutant (KDP379) to exclude the possibility that the function of those heterotrimers was replaced by the homotrimer of Pgm7. Each target gene was replaced by cepA, an ampicillin resistance gene. The designs for construction of the mutants are shown in Fig. 3A and B. Correct construction of the mutants was revealed by Southern blot hybridization and PCR analyses (Fig. 3C and data not shown).
FIG. 3.
(A and B) Construction of gslA, pgm6, and pgm6 pgm7 deletion mutants. gslA (A) and pgm6 alone or pgm6 pgm7 (B) were replaced with cepA genes as described in Materials and Methods. Arrows show CDSs. PGN numbers are CDS numbers of P. gingivalis ATCC 33277. Hybridization probes for the upstream region of gslA (black box) and for cepA (gray box) are indicated (A). P, restriction enzyme-recognized site for PstI. (C) Chromosomal DNA from gingipain-null mutant KDP136 and its gslA deletion mutant KDP377 was digested with PstI and subjected to Southern hybridization using a probe specific for the upstream region of gslA (a) and a probe specific for cepA (b). Lanes: 1, KDP136 (kgp rgpA rgpB); 2, KDP377 (kgp rgpA rgpB gslA::cepA).
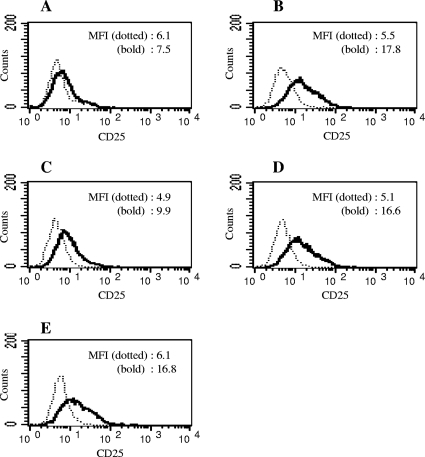
7.19 cells were stimulated with wild-type or mutant P. gingivalis cells and subjected to flow cytometric analysis. Freeze-dried KDP136 cells induced a considerable level of NF-κB activation in 7.19 cells, whereas ATCC 33277 induced only a marginal level of activation (Fig. 4A and B). Cells of strain KDP377 showed a much lower level of NF-κB activation than did cells of strain KDP136 (Fig. 4C). Cells of strains KDP378 and KDP379 induced the same level of CD25 expression as was induced by cells of strain KDP136 (Fig. 4D and E). These results indicated that GslA was essential for the activation of NF-κB in 7.19 cells.
FIG. 4.
NF-κB activation in 7.19 cells exposed to wild-type or mutant P. gingivalis. 7.19 cells were stimulated with 200 μg/ml of freeze-dried wild-type P. gingivalis ATCC 33277 (A), gingipain-null mutant KDP136 (kgp rgpA rgpB) (B), gslA deletion mutant KDP377 (kgp rgpA rgpB gslA::cepA) (C), pgm6 deletion mutant KDP378 (kgp rgpA rgpB pgm6::cepA) (D), and pgm6 pgm7 deletion mutant KDP379 (kgp rgpA rgpB Δ[pgm6 pgm7]::cepA) (E). Following 20 h of incubation, the cells were stained with FITC-labeled anti-CD25 monoclonal antibody (bold line) or isotype-matched control monoclonal antibody (dotted line) and subjected to flow cytometric analysis for NF-κB-driven CD25 expression. Representative results of one of three experiments performed are shown. MFI, mean fluorescence intensity.
Western blot and inhibition assay with polyclonal antibody against recombinant GslA protein.
A polyclonal antibody against the recombinant protein encoded by the 5′-terminal third of gslA was raised, and Western blot analysis was performed using this antibody. The antibody reacted to the 123-kDa protein in the whole-cell lysates of ATCC 33277 and KDP136, but there were no protein bands in the lysate of KDP377. In addition, the lysate of KDP136 contained a much larger amount of the 123-kDa protein than did the lysate of ATCC 33277. Similar results were obtained when outer membrane fractions were used. These results confirmed the identity of the 123-kDa protein and gslA product (Fig. 5A and B). Next, an experiment was performed to determine whether this antibody could inhibit the activation of NF-κB in 7.19 cells induced by freeze-dried KDP136. The activation of NF-κB in 7.19 cells was partially inhibited by this antibody compared to the level of activation induced in the presence of the IgG fraction purified from the serum of a preimmunized rabbit (Fig. 5C). Together with the results of the genetic experiments, these results indicated that GslA was responsible for the activation of NF-κB in 7.19 cells.
FIG. 5.
Inhibition of KDP136-induced NF-κB activation in 7.19 cells by an antibody against the recombinant GslA protein. Whole-cell lysates and outer membrane fractions of P. gingivalis were subjected to 7% SDS-PAGE and stained with CBB (A) or reacted with the polyclonal antibody against the recombinant protein encoded by the 5′-terminal third of gslA (B). (a) Whole-cell lysates. (b) Outer membrane fraction. Lanes: 1, wild-type P. gingivalis ATCC 33277; 2, gingipain-null mutant KDP136 (kgp rgpA rgpB); 3, gslA deletion mutant KDP377 (kgp rgpA rgpB gslA::cepA). Arrows indicate the 123-kDa protein. (C) Following 20 h of incubation, 7.19 cells were stained with FITC-labeled anti-CD25 monoclonal antibody and subjected to flow cytometric analysis for NF-κB-driven CD25 expression. 7.19 cells remained unstimulated (thin line) or were stimulated with 200 μg/ml of freeze-dried KDP136 bacterial cells in the presence of 100 μg/ml of the polyclonal antibody used for Western blot analysis (bold line) or the IgG fraction from serum of a preimmunized rabbit (dotted line). Representative results of one of three experiments performed are shown. MFI, mean fluorescence intensity.
Distribution of gslA among various strains of P. gingivalis.
Interestingly, the gslA gene is not found in the genome of P. gingivalis W83 (16). A DNA region (7.7 kb) in the ATCC 33277 genome that contains gslA between direct repeats (TTTTATAA) was deleted in the W83 genome (Fig. 6A). To investigate the presence of gslA in other P. gingivalis strains, Southern blot analysis was performed. Three probes for gslA hybridized to the genomic DNA from P. gingivalis strains ATCC 33277, TDC117, TDC275, and SU63 but not to genomic DNA from strains W83, TDC60, and GAI-7802 (Fig. 6B).
FIG. 6.
Distribution of gslA among various strains of P. gingivalis. Hybridization probes a, b, and c for internal regions of gslA, described in Materials and Methods, are indicated in both panels. (A) A DNA region (7.7 kb) in the ATCC 33277 genome that contains gslA between direct repeats (TTTTATAA) was deleted in the W83 genome. B, restriction enzyme-recognized site for BamHI; *, #, and •, detailed DNA sequences of indicated regions are shown below. Arrows show CDSs. PGN numbers are CDS numbers from P. gingivalis ATCC 33277. PG numbers are those of P. gingivalis W83. (B) Chromosomal DNA from ATCC 33277 and other strains of P. gingivalis was digested with BamHI and subjected to Southern hybridization using the probes mentioned above. Lanes: 1, P. gingivalis ATCC 33277; 2, W83; 3, TDC60; 4, TDC117; 5, TDC275; 6, SU63; 7, GAI-7802.
DISCUSSION
We purified cell surface components of KDP136 and found two proteins with molecular masses of 123 and 43 kDa in the fraction with high ability to induce NF-κB activation in 7.19 cells. In Western blot analysis, mouse antiserum that could inhibit the activation of NF-κB in 7.19 cells reacted to the 123-kDa protein but not to the 43-kDa protein. When 7.19 cells were stimulated with freeze-dried P. gingivalis, a gslA deletion mutant lacking the 123-kDa protein did not induce NF-κB activation in 7.19 cells, whereas a pgm6 or pgm6 pgm7 deletion mutant lacking the 43-kDa protein induced NF-κB activation. These results all indicate that the 123-kDa protein encoded by gslA is a crucial factor for inducing NF-κB activation in 7.19 cells.
The polyclonal antibody against the 123-kDa protein partially inhibited the activation of NF-κB in 7.19 cells induced by freeze-dried KDP136, indicating that the 123-kDa protein was involved in the induction of NF-κB activation in 7.19 cells. The partial inhibition might be due to the existence of another gingipain-sensitive ligand that could induce NF-κB activation in 7.19 cells. In accordance with this hypothesis, stimulation with the gslA deletion mutant induced a slightly higher level of NF-κB activation in 7.19 cells than that induced by stimulation with the wild-type strain.
The lysate of the gingipain-null mutant KDP136 contained a much larger amount of the 123-kDa protein than did the lysate of the wild-type strain ATCC 33277. In addition, the Triton X-114-extracted cell surface protein fraction of ATCC 33277 had no 123-kDa protein, whereas that of KDP136 did have the 123-kDa protein. It is likely that the 123-kDa protein was degraded by surface-associated gingipains after transportation of the protein to the cell surface in the wild-type P. gingivalis. The degradation of the 123-kDa protein on the cell surface of wild-type P. gingivalis is consistent with the findings by other investigators that gingipains cleave and process the products encoded by the fimA, mfa1, rgpA, rgpB, kgp, hagA, and ragA genes, which are located on the bacterial cell surface (5, 12, 24). Masuda et al. reported that the expression of mRNA for gingipains is modulated by environmental stress (9). The expression and activities of gingipains gradually decreased with the increase of the growth rate of P. gingivalis. The activities of gingipains in the culture fluids of P. gingivalis under aerated conditions were approximately eightfold lower than those in anaerobic conditions. In those conditions, the 123-kDa protein may remain intact on the cell surface and induce NF-κB activation even in wild-type P. gingivalis cells.
The physiological function of the 123-kDa protein encoded by gslA is unknown, and no homologue to GslA was found by BLASTP analysis. Interestingly, gslA was found in four of seven strains of P. gingivalis. Although strain W83 has been found to be more virulent than strain ATCC 33277 in a mouse subcutaneous chamber model, rats challenged with strain ATCC 33277 had more periodontal bone loss than those challenged with other strains (4, 6). Although further studies are needed to determine whether the presence of gslA is influential in the pathogenesis of P. gingivalis, the restricted distribution of gslA might be associated with the virulence potential of each strain. The common features of the four strains possessing gslA could be a clue for finding the pathogenic function of GslA.
The 123-kDa protein could induce NF-κB activation in 7.19 cells in a TLR2- and TLR4-independent manner. Although the activity to induce NF-κB activation was analyzed using only 7.19 cells, it is important to determine whether and how this ligand stimulates host cells derived from human periodontal tissue. Supposing that GslA is able to induce NF-κB activation in periodontal tissue, it would lead to the secretion of proinflammatory cytokines and chemokines that promote the acceleration of inflammatory responses in periodontal tissue. Regulation of gingipain expression by environmental conditions may affect this process.
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (grant 20592431 to A.Y., 20791617 to M.K., 20592142 to M.N., and 20249073 to K.N.).
Editor: J. B. Bliska
Footnotes
Published ahead of print on 10 August 2009.
REFERENCES
- 1.Banbula, A., M. Bugno, A. Kuster, P. C. Heinrich, J. Travis, and J. Potempa. 1999. Rapid and efficient inactivation of IL-6 gingipains, lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Biochem. Biophys. Res. Commun. 261:598-602. [DOI] [PubMed] [Google Scholar]
- 2.Calkins, C. C., K. Platt, J. Potempa, and J. Travis. 1998. Inactivation of tumor necrosis factor-α by proteinases (gingipains) from the periodontal pathogen, Porphyromonas gingivalis. Implications of immune evasion. J. Biol. Chem. 273:6611-6614. [DOI] [PubMed] [Google Scholar]
- 3.Delude, R. L., A. Yoshimura, R. R. Ingalls, and D. T. Golenbock. 1998. Construction of a lipopolysaccharide reporter cell line and its use in identifying mutants defective in endotoxin, but not TNF-α, signal transduction. J. Immunol. 161:3001-3009. [PubMed] [Google Scholar]
- 4.Genco, C. A., C. W. Cutler, D. Kapczynski, K. Maloney, and R. R. Arnold. 1991. A novel mouse model to study the virulence of and host response to Porphyromonas (Bacteroides) gingivalis. Infect. Immun. 59:1255-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kadowaki, T., K. Nakayama, F. Yoshimura, K. Okamoto, N. Abe, and K. Yamamoto. 1998. Arg-gingipain acts as a major processing enzyme for various cell surface proteins in Porphyromonas gingivalis. J. Biol. Chem. 273:29072-29076. [DOI] [PubMed] [Google Scholar]
- 6.Katz, J., D. C. Ward, and S. M. Michalek. 1996. Effect of host responses on the pathogenicity of strains of Porphyromonas gingivalis. Oral Microbiol. Immunol. 11:309-318. [DOI] [PubMed] [Google Scholar]
- 7.Kawada, M., A. Yoshida, N. Suzuki, Y. Nakano, T. Saito, T. Oho, and T. Koga. 2004. Prevalence of Porphyromonas gingivalis in relation to periodontal status assessed by real-time PCR. Oral Microbiol. Immunol. 19:289-292. [DOI] [PubMed] [Google Scholar]
- 8.Kishimoto, M., A. Yoshimura, M. Naito, K. Okamoto, K. Yamamoto, D. T. Golenbock, Y. Hara, and K. Nakayama. 2006. Gingipains inactivate a cell surface ligand on Porphyromonas gingivalis that induces TLR2- and TLR4-independent signaling. Microbiol. Immunol. 50:315-325. [DOI] [PubMed] [Google Scholar]
- 9.Masuda, T., Y. Murakami, T. Noguchi, and F. Yoshimura. 2006. Effects of various growth conditions in a chemostat on expression of virulence factors in Porphyromonas gingivalis. Appl. Environ. Microbiol. 72:3458-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mezyk-Kopec, R., M. Bzowska, J. Potempa, M. Bzowska, N. Jura, A. Sroka, R. A. Black, and J. Bereta. 2005. Inactivation of membrane tumor necrosis factor alpha by gingipains from Porphyromonas gingivalis. Infect. Immun. 73:1506-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikolajczyk-Pawlinska, J., J. Travis, and J. Potempa. 1998. Modulation of interleukin-8 activity by gingipains from Porphyromonas gingivalis: implications for pathogenicity of periodontal disease. FEBS Lett. 440:282-286. [DOI] [PubMed] [Google Scholar]
- 12.Murakami, Y., M. Imai, H. Nakamura, and F. Yoshimura. 2002. Separation of the outer membrane and identification of major outer membrane proteins from Porphyromonas gingivalis. Eur. J. Oral Sci. 110:157-162. [DOI] [PubMed] [Google Scholar]
- 13.Nagano, K., E. K. Read, Y. Murakami, T. Masuda, T. Noguchi, and F. Yoshimura. 2005. Trimeric structure of major outer membrane proteins homologous to OmpA in Porphyromonas gingivalis. J. Bacteriol. 187:902-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naito, M., H. Hirakawa, A. Yamashita, N. Ohara, M. Shoji, H. Yukitake, K. Nakayama, H. Toh, F. Yoshimura, S. Kuhara, M. Hattori, T. Hayashi, and K. Nakayama. 2008. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res. 15:215-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakayama, K., D. B. Ratnayake, T. Tsukuba, T. Kadowaki, K. Yamamoto, and S. Fujimura. 1998. Haemoglobin receptor protein is intragenically encoded by the cysteine proteinase-encoding genes and the haemagglutinin-encoding gene of Porphyromonas gingivalis. Mol. Microbiol. 27:51-61. [DOI] [PubMed] [Google Scholar]
- 16.Nelson, K. E., R. D. Fleischmann, R. T. DeBoy, I. T. Paulsen, D. E. Fouts, J. A. Eisen, S. C. Daugherty, R. J. Dodson, A. S. Durkin, M. Gwinn, D. H. Haft, J. F. Kolonay, W. C. Nelson, T. Mason, L. Tallon, J. Gray, D. Granger, H. Tettelin, H. Dong, J. L. Galvin, M. J. Duncan, F. E. Dewhirst, and C. M. Fraser. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 185:5591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oleksy, A., A. Banbula, M. Bugno, J. Travis, and J. Potempa. 2002. Proteolysis of interleukin-6 receptor (IL-6R) by Porphyromonas gingivalis cysteine proteinases (gingipains) inhibits interleukin-6-mediated cell activation. Microb. Pathog. 32:173-181. [DOI] [PubMed] [Google Scholar]
- 18.Schromm, A. B., E. Lien, P. Henneke, J. C. Chow, A. Yoshimura, H. Heine, E. Latz, B. G. Monks, D. A. Schwartz, K. Miyake, and D. T. Golenbock. 2001. Molecular genetic analysis of an endotoxin nonresponder mutant cell line: a point mutation in a conserved region of MD-2 abolishes endotoxin-induced signaling. J. Exp. Med. 194:79-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi, Y., D. B. Ratnayake, K. Okamoto, N. Abe, K. Yamamoto, and K. Nakayama. 1999. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J. Biol. Chem. 274:17955-17960. [DOI] [PubMed] [Google Scholar]
- 20.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 21.Sugawara, S., E. Nemoto, H. Tada, K. Miyake, T. Imamura, and H. Takada. 2000. Proteolysis of human monocyte CD14 by cysteine proteinases (gingipains) from Porphyromonas gingivalis leading to lipopolysaccharide hyporesponsiveness. J. Immunol. 165:411-418. [DOI] [PubMed] [Google Scholar]
- 22.Sundqvist, G., J. Carlsson, B. Herrmann, and A. Tarnvik. 1985. Degradation of human immunoglobulins G and M and complement factors C3 and C5 by black-pigmented Bacteroides. J. Med. Microbiol. 19:85-94. [DOI] [PubMed] [Google Scholar]
- 23.Tada, H., S. Sugawara, E. Nemoto, N. Takahashi, T. Imamura, J. Potempa, J. Travis, H. Shimauchi, and H. Takada. 2002. Proteolysis of CD14 on human gingival fibroblasts by arginine-specific cysteine proteinases from Porphyromonas gingivalis leading to down-regulation of lipopolysaccharide-induced interleukin-8 production. Infect. Immun. 70:3304-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veith, P. D., G. H. Talbo, N. Slakeski, S. G. Dashper, C. Moore, R. A. Paolini, and E. C. Reynolds. 2002. Major outer membrane proteins and proteolytic processing of RgpA and Kgp of Porphyromonas gingivalis W50. Biochem. J. 363:105-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang, H. W., Y. F. Huang, and M. Y. Chou. 2004. Occurrence of Porphyromonas gingivalis and Tannerella forsythensis in periodontally diseased and healthy subjects. J. Periodontol. 75:1077-1083. [DOI] [PubMed] [Google Scholar]