Abstract
Recent work has highlighted a number of compounds that target bacterial virulence by affecting gene regulation. In this work, we show that small-molecule inhibitors affect the expression of the type III secretion system (T3SS) of Escherichia coli O157:H7 in liquid culture and when this bacterium is attached to bovine epithelial cells. Inhibition of T3SS expression resulted in a reduction in the capacity of the bacteria to form attaching and effacing lesions. Our results show that there is marked variation in the abilities of four structurally related compounds to inhibit the T3SS of a panel of isolates. Using transcriptomics, we performed a comprehensive analysis of the conserved and inhibitor-specific transcriptional responses to these four compounds. These analyses of gene expression show that numerous virulence genes, located on horizontally acquired DNA elements, are affected by the compounds, but the number of genes significantly affected varied markedly for the different compounds. Overall, we highlight the importance of assessing the effect of such “antivirulence” agents on a range of isolates and discuss the possible mechanisms which may lead to the coordinate downregulation of horizontally acquired virulence genes.
Outbreaks of enterohemorrhagic Escherichia coli (EHEC) infection occur sporadically in both the developed world and the developing world, causing diarrheal disease that can progress to hemorrhagic colitis and hemolytic-uremic syndrome (22). E. coli O157:H7 is the dominant serotype in North America, parts of Europe, and Japan, and its pathogenicity has been studied extensively at the molecular level. A number of key virulence factors are known to be required for disease progression, including the Shiga toxins and the type III secretion (T3S) system (T3SS) required for characteristic attaching and effacing (A/E) lesion formation (13). The T3SS is encoded in a locus of enterocyte effacement (LEE) pathogenicity island containing 41 genes in five main operons (18). Deletion of the LEE4 operon prevents A/E lesion formation and colonization of the terminal rectum, the principle colonization site in cattle (24). Thus, vaccines that target the EHEC T3SS are being developed for use in cattle (28). Furthermore, given the central role of the T3SS in bovine colonization, it is highly likely that inhibition of a functional T3SS could prevent colonization of humans, thereby limiting disease.
As the prevalence of antibiotic-resistant strains increases, compounds that target virulence determinants of pathogenic bacteria have become an increasingly attractive alternative to bactericidal antibiotics. The major advantage of these compounds is their significantly less positive selection for resistant mutants as the nonpathogenic normal flora is not affected by treatment (14). A class of such “virulence blockers” that inhibit T3SS in Yersinia spp. has been described, and these compounds have been shown to be broadly effective against a number of pathogenic bacteria that utilize T3SS, including Chlamydia spp., Salmonella enterica serovar Typhimurium, and enteropathogenic E. coli (EPEC). The mechanism of inhibition in each organism is not completely understood. However, it is clear that transcription of T3S genes in these pathogens is reduced. Whether this is due to feedback from the T3S machinery or due to direct effects on transcription factors remains to be determined. Treatment of bacteria with micromolar concentrations of inhibitors is sufficient to reduce Salmonella serovar Typhimurium invasion of cultured epithelial cells and induction of enteritis in vivo (11), to arrest maturation of Chlamydia sp. inclusions (36), and to reduce proliferation of Yersinia pseudotuberculosis (25). Further experiments have shown that the antichlamydial activity of this family of inhibitors can be reversed by addition of iron, suggesting that the activity of these compounds may be linked to iron availability in the cell, although the mechanisms behind this reversal are unknown. In Vibrio cholerae a “virulence blocker” that reduces expression of cholera toxin and the toxin-coregulated pilus has been described. This inhibitor has been shown to act by posttranslationally inactivating the AraC family regulator ToxT, suggesting that regulators of virulence genes may be an important target for this class of inhibitors (35). A recent study has also shown that a structurally distinct compound, LED209, can prevent phosphorylation of the E. coli O157 QseC membrane protein, a sensor of host adrenergic molecules (30). The inhibition of QseC function leads to impaired expression of virulence factors and affects colonization of tissue culture cells by downregulation of Ler, the master regulator of the T3SS of E. coli O157 (30). Of particular importance to this study is a previous report that described screening of virulence blockers in EPEC (9). This previous report showed that addition of a compound with structural similarity to the compounds described in this work transcriptionally downregulated the major operons of the EPEC LEE. However, the extent to which other genes were affected was not examined. Here we characterize the effects of four structurally related salicylidene acylhydrazide virulence blockers on global gene expression in E. coli O157:H7. Our analyses show that transcriptional repression of the T3SS is accompanied by repression of virulence genes encoded on mobile genetic elements, demonstrating that there are global effects on virulence gene regulation. We also show that these inhibitors effectively reduce formation of actin pedestals in vitro.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Unless otherwise stated, all experiments were performed using E. coli O157:H7 strain TUV-930, a Shiga toxin-negative derivative of strain EDL933 (3). To examine the effects of inhibitors on a range of strains and phage types (PTs), six additional E. coli O157 strains were used: ZAP 3 (PT 2), ZAP 11 (PT 21/28), ZAP 12 (PT 21/28), ZAP 229, (PT 2), ZAP 231 (PT 4), and ZAP 243 (PT 8). The green fluorescent protein (GFP) reporter plasmids pAJR71 to pAJR75 used in this study have been described previously (32, 33).
Inhibitors and media.
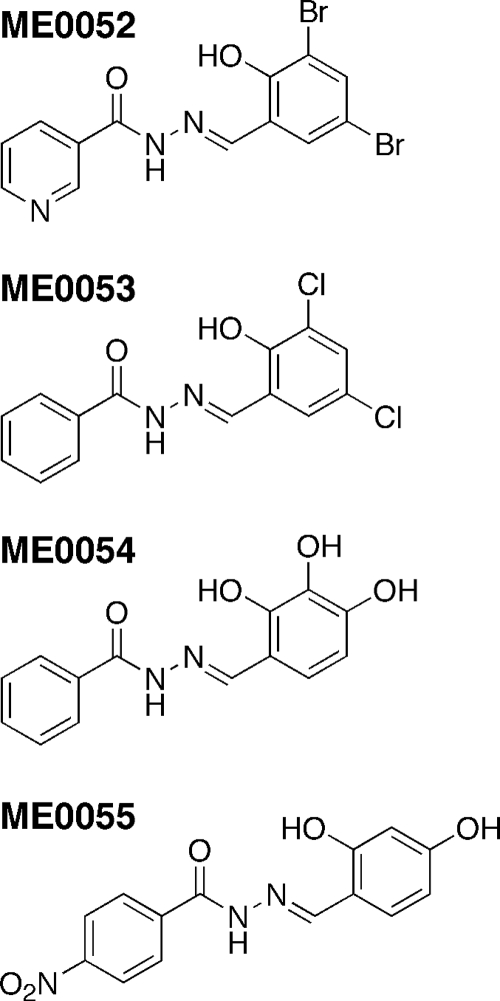
Compounds ME0052 to ME0055 (Fig. 1), which were selected on the basis of their ability to inhibit the T3SS of related pathogens (2, 25, 36), were prepared and supplied by Mikael Elofsson. These compounds have been described previously using different names, as follows: ME0052, INP0010 and compound 8; ME0053, INP0403 and compound 11; ME0054, INP0401 and compound 10; and ME0055, INP0031 and compound 17 (25). Stock solutions were prepared in dimethyl sulfoxide (DMSO), aliquoted, and stored at −20°C in the dark. The final DMSO concentration added to the bacteria was less than 2% in all experiments. Minimal essential medium with HEPES modification (MEM-HEPES) (Sigma M7278) was used in this study to promote expression of the LEE pathogenicity island. This medium was supplemented with 250 nM Fe(NO3)2 and glucose at a final concentration of 0.2%. Antibiotics were included when they were required at the following concentrations: 12.5 μg·ml−1 chloramphenicol and 25 μg·ml−1 kanamycin. To examine the effects of higher concentrations of iron on the effects of the inhibitors, 200 μM FeSO4 was added to the growth medium where indicated below.
FIG. 1.
Structures of ME0052, ME0053, ME0054, and ME0055. The compounds have been previously described using different names, as follows: ME0052, INP0010 and compound 8; ME0053, INP0403 and compound 11; ME0054, INP0401 and compound 10; and ME0055, INP0031 and compound 17.
Protein secretion and Western blot analyses.
Bacteria were cultured overnight in LB and diluted to obtain an optical density at 600 nm (OD600) of ≤0.05 in prewarmed MEM-HEPES supplemented with 250 nM Fe(NO3)2 and glucose (final concentration, 0.2%). When required, the specific growth rate was reduced by addition of 0.1 M or 0.2 M NaCl to this medium. Cultures were grown at 37°C to an OD600 of 0.8, and then the secreted proteins were extracted by trichloroacetic acid precipitation as described previously (34). For protein localization experiments, whole-cell fractions were prepared by centrifugation (20 min, 4,000 × g), washed twice in 20 ml phosphate-buffered saline (PBS), sonicated, and resuspended in 400 μl protein A buffer (34). Proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Western blotting for Tir, EspD, and GroEL was carried out as described previously (34). The Western blots were quantified using Scion image software, which allowed comparisons of different samples.
Transcriptomics.
Transcriptional profiling of cultures treated with the inhibitors was carried out essentially as described previously (33). In brief, overnight cultures of E. coli TUV-930 were grown in MEM-HEPES (Sigma) supplemented with 250 nM Fe(NO3)2 and glucose (final concentration, 0.2%) and then diluted to obtain an OD600 of 0.1 in the same medium. An inhibitor was added to test cultures at a final concentration of 20 μM, and an equal volume of DMSO was added to reference cultures. At an OD600 of 0.8, 15-ml cultures were stabilized using an equal volume of RNAprotect (Qiagen), and RNA was extracted using a Qiagen RNeasy mini kit. Contaminating DNA was removed by DNase I treatment (Ambion). The total RNA was assessed to determine its quality and was quantified using an Aglient 2100 bioanalyzer. Synthesis of cDNA and labeling of total RNA were performed using an Amersham CyScribe postlabeling kit according to the manufacturer's instructions. The cDNA was hybridized to 70-mer spotted oligonucleotide arrays containing open reading frames from E. coli K-12, E. coli Sakai VT2, and E. coli EDL933 (University of Birmingham, United Kingdom) using a Genomic Solutions GeneTac hybridization machine. Hybridized slides were scanned using a Genepix 4000A scanner and GenePix 7.0 software (Axon Instruments, Union City, CA). Data were analyzed using Genespring GX 7.3.1 (Agilent).
Regulon analysis for global regulators.
In order to determine if posttranslational modification of regulators contributed to regulation by ME0052 to ME0055, the regulons for nucleoid structuring protein (H-NS), LexA, Fur, and PchA were defined using previously described data and analyzed using Genespring 7.3.1. The H-NS, LexA, and Fur regulons have not been elucidated for E. coli O157 but have been described for E. coli K-12, and they provide an indicator of the activity in the E. coli common backbone. The H-NS regulon has been described previously by Oshima et al. (26), and the LexA regulon has been described for E. coli K-12 using transcriptomics and chromatin immunoprecipitation studies (39). For both of these regulons, E. coli K-12 gene designations were converted to O157 “z” numbers using Genespring 7.3.1 and by BLAST analysis. The Fur regulon has been described by Gama-Castro et al. (8) and was acquired from RegulonDB (http://regulondb.ccg.unam.mx/), and the gene designations were similarly converted to “z” numbers using Genespring 7.3.1 and BLAST. The PchA and Ler regulons were recently defined by transcriptomics and chromatin immunoprecipitation studies for E. coli O157 strain Sakai, a sequenced O157 outbreak strain from Japan. The Sakai PchA and Ler regulons were converted to EDL933 gene designations using the nucmer genomic alignment tool at coliBASE (http://xbase.bham.ac.uk/colibase/) and by BLAST analysis. Analyses of overlaps between these regulons and genes affected by inhibitor treatment were performed using Genespring 7.3.1.
Determination of LEE expression using GFP reporter fusions.
Expression of the LEE was determined by using a series of plasmid-based promoter::GFP reporter fusions described previously (pAJR71 to pAJR75) (34). To measure population fluorescence, triplicate 100-μl aliquots of transformed bacteria were dispensed into 96-well black microtiter plates and analyzed using a BMG Fluostar plate reader at 37°C. The OD600 of cultures were used to monitor growth. Fluorescence was plotted against optical density using Microsoft Excel software, and the line of best fit was obtained. The promoterless plasmid pAJR70 (34) was employed to correct for the background fluorescence of the strain and medium. Comparisons between treated cultures and cultures treated with DMSO alone were made using an OD600 of 0.8 to obtain the best comparison with the data obtained by transcriptomics.
Fluorescence microscopy analysis of LEE1 and flagellum expression.
Embryonic bovine cells (German Collection of Microorganisms and Cell Cultures no. ACC192) were prepared and cultured as described previously (32). TUV-930(pAJR71) (LEE1::GFP) was grown in MEM-HEPES to an OD600 0.6 at 37°C with 20 μM ME0055 or the same volume of DMSO. Bacteria were added to a multichamber slide and incubated at 37°C. Samples were fixed at intervals by removing portions of the culture and adding 4% paraformaldehyde (PFA). Flagellum expression was detected as described previously (10). The adherent bacteria were stained with rabbit anti-O157 and anti-H7 polyclonal antiserum (diluted 1:500 in PBS; Mast Diagnostics) at room temperature for 30 min, washed, and then incubated with tetramethyl rhodamine isocyanate (TRITC)-labeled secondary antibody (anti-rabbit antibody diluted 1:1,000; Sigma). The time points analyzed were 60, 120, 180, and 300 min after addition of bacteria. To measure single-cell expression by fluorescence microscopy, a z stack of 20 images with a spacing of 0.1 μm was captured with a Zeiss Axioimager M1 fluorescence microscope equipped with a Hamamatsu ORCA AG camera using Volocity software (Improvision). These images were used to create a composite image that reduced the spatial effects of bacteria in different focal planes. GFP intensity was determined using Volocity software with a threshold of 2,000 relative fluorescence units as the cutoff point to separate “GFP-positive” and “GFP-negative” bacteria.
Detection and quantification of A/E lesions.
To detect host cell actin, cells were permeabilized using 0.2% Triton X-100 and then treated for 20 min at room temperature with TRITC-phalloidin (5 μg·ml−1; Sigma), washed twice with PBS, and mounted in fluorescent mounting medium (Dako). Improvision Volocity software (Quantification) was used to examine three independent replicates with sampling of 15 separate fields per replicate. Bacteria and areas of concentrated actin were identified using size and the fluorophore as selection parameters. The percentage of total bacteria in each field associated with condensed actin was then determined.
Array data accession number.
The MIAME-compliant array data have been deposited in the Gene Expression Omnibus (GEO) database at NCBI under GEO accession number GSE10319.
RESULTS
Effects of salicylidene acylhydrazide inhibitors on the growth rate.
In our initial work we aimed to determine the effects of the compounds studied on the bacterial growth rate. We were aware that marked changes in the growth rate result in changes in global gene expression, making any specific effects of the compounds hard to determine. Bacteria were cultured in MEM-HEPES containing a range of concentrations (10 to 50 μM) of the four compounds. At both 10 and 20 μM we observed only minor changes in the growth rate (Table 1) for all four compounds. Addition of a higher concentration (50 μM) of the compounds had more marked effects on the specific growth rate (Table 1). We therefore focused on the 20 μM concentration to minimize any effects of an altered growth rate on gene expression.
TABLE 1.
Growth rates of strain TUV-930 with and without inhibitors
| Compound | Concn (μM) | Specific growth rate (h−1, mean ± SD) | % Growth inhibition |
|---|---|---|---|
| None | 0.54 ± 0.01 | ||
| ME0052 | 10 | 0.49 ± 0.01 | 10 |
| ME0052 | 20 | 0.44 ± 0.01 | 12 |
| ME0052 | 50 | 0.37 ± 0.02 | 46 |
| ME0053 | 10 | 0.49 ± 0.02 | 10 |
| ME0053 | 20 | 0.44 ± 0.01 | 12 |
| ME0053 | 50 | 0.06 ± 0.01 | 900 |
| ME0054 | 10 | 0.54 ± 0.03 | 0 |
| ME0054 | 20 | 0.50 ± 0.02 | 8 |
| ME0054 | 50 | 0.49 ± 0.01 | 10 |
| ME0055 | 10 | 0.54 ± 0.02 | 0 |
| ME0055 | 20 | 0.50 ± 0.02 | 8 |
| ME0055 | 50 | 0.44 ± 0.03 | 12 |
Effects of salicylidene acylhydrazide inhibitors on secretion of T3SS effector proteins.
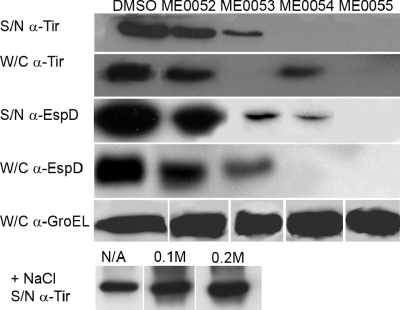
In order to characterize the effects of the four inhibitors on the secretion of effector proteins by the T3SS, total secreted proteins were prepared from strain TUV-930 cultured in MEM-HEPES containing an inhibitor at a concentration of 20 μM or the equivalent volume of DMSO. The secreted protein profiles were analyzed by colloidal blue staining of the total secreted proteins produced by the bacteria, as well as by Western blotting to quantify both the whole-cell expression and secretion of the translocated intimin receptor (Tir). Addition of the compounds at a concentration of 20 μM resulted in inhibition of Tir expression by all four compounds; ME0053 and ME0055 were the most effective inhibitors, reducing the Tir expression in the whole-cell fraction by >90% (Fig. 2). Analysis of the secreted fractions showed that there was no detectable Tir when the bacteria were treated with ME0055, which correlates with the major reduction in whole-cell expression with this compound. Treatment with ME0053 resulted in a reduced but detectable signal, implying that, while whole-cell expression of Tir was reduced, Tir secretion was not strongly inhibited. ME0052 and ME0054 were less effective, reducing Tir expression by 39% and 54%, respectively. Treatment with ME0054 resulted in strong inhibition of Tir secretion, whereas treatment with ME0052 did not appear to reduce the secretion of Tir and the secreted fraction correlated closely with the whole-cell fraction.
FIG. 2.
ME0052, ME0053, ME0054, and ME0055 inhibit expression and secretion of the effector proteins Tir and EspD from E. coli O157:H7. Secreted (S/N) and whole-cell (W/C) fractions were prepared from bacteria cultured in MEM-HEPES. Samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by Western blotting. The antibodies used for the blots are indicated on the left (α-Tir, anti-Tir antibody; α-EspD, anti-EspD antibody; α-GroEL, anti-GroEL antibody). To examine the effect of a decreased growth rate on Tir secretion, E. coli O157:H7 was cultured in the absence (N/A) or presence of NaCl, and secreted Tir was detected by Western blotting. The concentrations of NaCl added are indicated above the blots.
The same samples were also tested to determine the levels of the translocator protein EspD, which is transcribed from a different operon in the LEE. Addition of ME0054 or ME0055 resulted in strong inhibition of whole-cell EspD expression (>95% reduction) and corresponding low (ME0054) or undetectable (ME0055) levels of EspD in the supernatant fraction (Fig. 2). ME0053 blocked whole-cell expression less effectively, although the level of secreted EspD was markedly reduced by this inhibitor. ME0052 was the least effective inhibitor, and both whole-cell expression and the secreted levels showed reductions similar to those observed for Tir.
These data were confirmed by colloidal blue visualization of the total secreted proteins, which showed that there was a concentration-dependent inhibitory effect on the overall secretion profile that varied with the compound; ME0055 was the most effective inhibitor, followed by ME0054, ME0053, and ME0052 (data not shown). As a control, intracellular GroEL levels were also assessed by Western analysis (Fig. 2). The results showed that the four compounds did not affect general protein expression or stability, and less than 15% variation between the samples was observed.
To examine if the observed inhibition of translocator and effector protein expression and secretion was due to the slight reduction in the bacterial growth rate caused by addition of the compounds, we tested the relationship between the growth rate and the secretion phenotype. To reduce the bacterial growth rate independent of the salicylidene acylhydrazide inhibitors, NaCl was added to increase the osmolarity of the growth medium. Compared to the data for bacteria cultured in MEM-HEPES alone, addition of NaCl resulted in an increase in Tir protein secretion (Fig. 2). Additional of 0.1 M NaCl reduced the bacterial growth rate by 18% (data not shown) and increased the level of secreted Tir by 34% (Fig. 2). Addition of 0.2 M NaCl reduced the growth rate by 44%, which led to a 61% increase in the level of Tir secretion. These data suggest that the decrease in the growth rate after addition of the salicylidene acylhydrazide inhibitors is unlikely to account for the inhibition of T3S.
To examine if the compounds acted with a range of strains, six other E. coli O157 isolates were tested with all four compounds (Table 2). These isolates included three isolates from human outbreaks (ZAP 229, ZAP 231, and ZAP 243) and three isolates obtained from bovine samples (ZAP3, ZAP 11, and ZAP 12). These strains have four different PTs (PTs 2, 4, 8, and 21/28), all commonly associated with disease outbreaks in the United Kingdom. For these assays, variations in the extent of inhibition of secretion were determined by blotting for secreted Tir (Table 2). While addition of ME0052 was largely ineffective with TUV-930, it resulted in inhibition of Tir secretion in the six strains mentioned above (Table 2). Addition of ME0053 resulted in inhibition of TUV-930 and the six other strains, but the extent of inhibition varied considerably. ME0054 was the most effective inhibitor for the entire strain collection. In comparison, ME0055, which was highly effective with four strains, was far less inhibitory when it was tested with ZAP229, ZAP231, and ZAP243 and reduced Tir secretion by only 43 to 60% (Table 2).
TABLE 2.
Relative inhibition of T3S by salicylidene acylhydrazide compounds in different E. coli O157 strains
| Strain | Source | PT | % Inhibition witha:
|
|||
|---|---|---|---|---|---|---|
| ME0052 | ME0053 | ME0054 | ME0055 | |||
| TUV-930 | Human | NTb | 23 | 68 | 98 | 99 |
| ZAP3 | Bovine | 2 | 100 | 100 | 100 | 100 |
| ZAP11 | Bovine | 21/28 | 100 | 100 | 100 | 100 |
| ZAP12 | Bovine | 21/28 | 100 | 100 | 100 | 100 |
| ZAP229 | Human | 2 | 80 | 59 | 97 | 60 |
| ZAP231 | Human | 4 | 79 | 75 | 95 | 43 |
| ZAP243 | Human | 8 | 86 | 94 | 91 | 44 |
The amount of secreted Tir effector protein was determined by Western analysis and compared to the amount for the strain cultured in DMSO alone to determine the percentage of inhibition. Therefore, 100% inhibition was complete inhibition of Tir secretion.
NT, not typed.
Analysis of transcriptional changes resulting from inhibitor addition.
In an effort to understand the molecular mechanism by which the inhibitory compounds function, we carried out transcriptional profiling experiments. Strain TUV-930 was grown to an OD600 of 0.8 in MEM-HEPES in the presence or absence of an inhibitor at a concentration of 20 μM, and RNA was harvested and labeled for microarray analysis. Table 3 shows the cumulative results for genes with >2-fold changes with all four inhibitors. The data show that all four inhibitors reduce the expression of all genes in the LEE (Fig. 3). In addition to repressing the LEE, all of the inhibitors tested also repressed transcription of the non-LEE effectors nleA, espJ, and espN and pO157 open reading frames L7027 and L7037. The inhibitors predominantly repressed gene expression, and they varied in the total number of genes that they affected. For example, using a cutoff of a >3-fold change in transcription, ME0052 significantly (P ≤ 0.05) affected the transcription of 181 genes, whereas ME0055 significantly affected the transcription of 24 genes (Tables 3 and 4; see Table S1 in the supplemental material).
TABLE 3.
Genes with >2-fold changes with all inhibitor treatments (P < 0.05; Benjamini-Hochberg false discovery rate) in three of four experimentsa
| Systematic name | Common name | OI no. | Effector | Mobile element | Presence of Ler and PchA | ME0052
|
ME0053
|
ME0054
|
ME0055
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ratio | P value | Ratio | P value | Ratio | P value | Ratio | P value | ||||||
| L7027 | L7027 | pO157 | 0.361 | 3.08E−03 | 0.483 | 1.62E−01 | 0.328 | 5.89E−02 | 0.337 | 8.44E−02 | |||
| L7031 | L7031 | pO157 | Regulator | 0.141 | 1.27E−03 | 0.464 | 3.68E−02 | 0.151 | 1.05E−01 | 0.32 | 6.75E−02 | ||
| Z0955 | Z0955 | 36 | CP-933K | PchA + Ler | 0.083 | 2.13E−04 | 0.214 | 4.11E−02 | 0.096 | 9.60E−02 | 0.155 | 6.75E−02 | |
| Z1823 | Z1823 | 50 | CP-933N | PchA | 0.277 | 3.70E−05 | 0.349 | 8.09E−02 | 0.298 | 4.37E−02 | 0.218 | 2.86E−05 | |
| Z1824 | Z1824 | 50 | espN | CP-933N | PchA | 0.147 | 5.33E−04 | 0.214 | 1.42E−03 | 0.17 | 7.79E−03 | 0.435 | 3.50E−04 |
| Z2478b | pspD | 3.648 | 6.53E−05 | 2.338 | 6.46E−02 | 2.45 | 1.03E−01 | 3.567 | 5.04E−03 | ||||
| Z2479b | pspC | 3.9 | 5.05E−05 | 2.444 | 5.19E−02 | 2.512 | 9.44E−02 | 3.97 | 7.69E−03 | ||||
| Z3071 | Z3071 | 79 | espJ | CP-933U | PchA + Ler | 0.152 | 3.49E−04 | 0.236 | 9.20E−03 | 0.168 | 3.01E−03 | 0.478 | 2.45E−01 |
| Z3572 | argT | 0.494 | 1.35E−03 | 0.492 | 1.00E−01 | 0.387 | 1.48E−02 | 0.262 | 6.02E−03 | ||||
| Z5100 | espF | 148 | espF1 | CP-933L | Ler | 0.126 | 8.48E−03 | 0.452 | 4.74E−04 | 0.145 | 1.70E−01 | 0.268 | 8.42E−02 |
| Z5102 | Z5102 | 148 | CP-933L | Ler | 0.062 | 4.34E−03 | 0.357 | 2.87E−05 | 0.103 | 1.92E−01 | 0.125 | 5.12E−02 | |
| Z5103 | escF | 148 | CP-933L | Ler | 0.044 | 1.85E−03 | 0.306 | 3.23E−05 | 0.087 | 1.54E−01 | 0.162 | 7.38E−02 | |
| Z5104 | Z5104 | 148 | CP-933L | Ler | 0.045 | 1.80E−03 | 0.305 | 5.35E−04 | 0.085 | 1.48E−01 | 0.138 | 5.03E−02 | |
| Z5105 | espB | 148 | espB | CP-933L | Ler | 0.037 | 1.01E−03 | 0.216 | 3.90E−02 | 0.071 | 1.16E−01 | 0.122 | 7.56E−02 |
| Z5106 | espD | 148 | CP-933L | Ler | 0.033 | 3.62E−03 | 0.261 | 5.87E−02 | 0.072 | 1.17E−01 | 0.061 | 4.71E−02 | |
| Z5107 | espA | 148 | CP-933L | Ler | 0.045 | 3.04E−03 | 0.437 | 2.11E−01 | 0.074 | 1.23E−01 | 0.037 | 2.42E−02 | |
| Z5108 | sepL | 148 | CP-933L | Ler | 0.083 | 3.97E−05 | 0.252 | 1.21E−03 | 0.065 | 1.01E−01 | 0.238 | 5.33E−02 | |
| Z5109 | escD | 148 | CP-933L | PchA + Ler | 0.107 | 5.47E−04 | 0.435 | 3.59E−05 | 0.105 | 1.59E−01 | 0.237 | 3.92E−02 | |
| Z5110 | eae | 148 | CP-933L | PchA + Ler | 0.040 | 2.24E−03 | 0.211 | 4.34E−05 | 0.061 | 9.04E−02 | 0.132 | 3.86E−02 | |
| Z5111 | Z5111 | 148 | CP-933L | PchA + Ler | 0.056 | 7.15E−03 | 0.329 | 1.64E−02 | 0.084 | 1.48E−01 | 0.081 | 3.12E−02 | |
| Z5112 | tir | 148 | tir | CP-933L | Ler | 0.079 | 2.12E−03 | 0.344 | 1.40E−05 | 0.103 | 1.24E−01 | 0.156 | 5.16E−02 |
| Z5113 | Z5113 | 148 | CP-933L | PchA + Ler | 0.072 | 3.23E−04 | 0.287 | 1.06E−03 | 0.082 | 1.26E−01 | 0.155 | 3.80E−02 | |
| Z5114 | Z5114 | 148 | CP-933L | PchA + Ler | 0.057 | 5.71E−03 | 0.312 | 1.13E−03 | 0.075 | 1.24E−01 | 0.191 | 3.64E−02 | |
| Z5115 | Z5115 | 148 | espH | CP-933L | PchA + Ler | 0.071 | 2.57E−04 | 0.235 | 2.21E−03 | 0.065 | 6.92E−02 | 0.224 | 6.52E−02 |
| Z5116 | sepQ | 148 | CP-933L | PchA + Ler | 0.033 | 9.11E−03 | 0.156 | 3.88E−04 | 0.047 | 6.05E−02 | 0.204 | 1.28E−02 | |
| Z5119 | escN | 148 | CP-933L | PchA + Ler | 0.134 | 7.62E−03 | 0.388 | 1.47E−04 | 0.123 | 1.32E−01 | 0.333 | 1.32E−02 | |
| Z5120 | escV | 148 | CP-933L | PchA + Ler | 0.088 | 1.01E−02 | 0.285 | 5.95E−04 | 0.114 | 1.06E−01 | 0.398 | 1.91E−02 | |
| Z5121 | Z5121 | 148 | CP-933L | PchA + Ler | 0.064 | 1.08E−02 | 0.273 | 2.96E−03 | 0.092 | 1.27E−01 | 0.328 | 8.38E−03 | |
| Z5123 | Z5123 | 148 | CP-933L | PchA + Ler | 0.139 | 5.37E−03 | 0.361 | 1.69E−02 | 0.118 | 1.16E−01 | 0.176 | 3.22E−02 | |
| Z5124 | escJ | 148 | CP-933L | PchA + Ler | 0.081 | 4.69E−03 | 0.272 | 5.05E−04 | 0.111 | 7.00E−02 | 0.259 | 4.04E−02 | |
| Z5125 | Z5125 | 148 | CP-933L | PchA + Ler | 0.047 | 1.05E−02 | 0.233 | 1.79E−02 | 0.085 | 1.01E−01 | 0.27 | 9.86E−03 | |
| Z5126 | escC | 148 | CP-933L | PchA + Ler | 0.112 | 6.86E−03 | 0.307 | 7.42E−03 | 0.087 | 1.04E−01 | 0.173 | 2.91E−02 | |
| Z5127 | cesD | 148 | CP-933L | PchA + Ler | 0.075 | 6.20E−03 | 0.237 | 2.02E−02 | 0.062 | 8.64E−02 | 0.178 | 1.73E−02 | |
| Z5128 | Z5128 | 148 | CP-933L | PchA + Ler | 0.04 | 5.09E−03 | 0.177 | 5.26E−03 | 0.056 | 7.90E−02 | 0.179 | 5.51E−02 | |
| Z5129 | Z5129 | 148 | CP-933L | PchA + Ler | 0.083 | 1.06E−03 | 0.244 | 8.03E−04 | 0.076 | 8.62E−02 | 0.204 | 2.14E−02 | |
| Z5131 | Z5131 | 148 | CP-933L | PchA + Ler | 0.035 | 3.05E−03 | 0.256 | 1.10E−02 | 0.076 | 1.09E−01 | 0.397 | 3.56E−03 | |
| Z5132 | escU | 148 | CP-933L | PchA + Ler | 0.025 | 1.07E−02 | 0.14 | 4.94E−03 | 0.044 | 2.57E−02 | 0.463 | 7.15E−02 | |
| Z5133 | escT | 148 | CP-933L | PchA + Ler | 0.043 | 8.33E−07 | 0.178 | 1.54E−02 | 0.088 | 4.77E−02 | 0.3 | 1.82E−03 | |
| Z5134 | escS | 148 | CP-933L | PchA + Ler | 0.088 | 1.57E−03 | 0.18 | 1.67E−02 | 0.085 | 7.07E−02 | 0.374 | 4.24E−02 | |
| Z5135 | escR | 148 | CP-933L | PchA + Ler | 0.117 | 2.84E−04 | 0.179 | 2.74E−02 | 0.086 | 4.95E−02 | 0.304 | 4.41E−04 | |
| Z5137 | Z5137 | 148 | CP-933L | PchA + Ler | 0.14 | 1.12E−02 | 0.216 | 2.29E−02 | 0.091 | 2.44E−02 | 0.148 | 1.09E−05 | |
| Z5139 | Z5139 | 148 | CP-933L | PchA + Ler | 0.233 | 2.46E−03 | 0.295 | 4.66E−02 | 0.143 | 4.93E−03 | 0.173 | 3.92E−05 | |
| Z5142 | Z5142 | 148 | espG | CP-933L | PchA + Ler | 0.089 | 9.46E−04 | 0.221 | 1.72E−03 | 0.092 | 3.57E−02 | 0.298 | 7.90E−02 |
| Z5953b | yjiY | 12.27 | 2.22E−06 | 4.792 | 2.84E−03 | 4.997 | 2.60E−02 | 2.653 | 1.66E−01 | ||||
| Z6024 | Z6024 | 71 | nleA | CP-933P | PchA + Ler | 0.077 | 1.49E−03 | 0.196 | 3.13E−03 | 0.075 | 8.08E−02 | 0.38 | 6.11E−02 |
| Z4668 | rpsM | 0.673 | 0.015 | 0.926 | 0.159 | 1.103 | 0.219 | 0.965 | 0.585 | ||||
| Z3013b | fliC | 4.305 | 0.061 | 33.830 | 0.020 | 5.064 | 0.111 | 0.311 | 0.340 | ||||
Transcription of rpsM and transcription of fliC were included as controls.
Gene upregulated by inhibitor treatment.
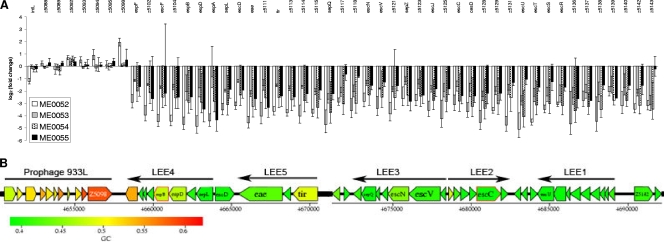
FIG. 3.
(A) Effects of ME0052, ME0053, ME0054, and ME0055 on transcription of the LEE. The changes (log2) in the transcription of treated cultures compared with cultures treated with DMSO alone are shown for all of the genes in the LEE (OI-148A). The strong repressive effects of the compounds on LEE1 to LEE5 can be contrasted to the effects on the prophage 933L genes indicated on the left. (B) Reference diagram of the LEE from strain EDL933, showing the five polycistronic operons (LEE1 to LEE5). The diagram was produced using Colibase (http://xbase.bham.ac.uk/colibase/).
TABLE 4.
Total numbers of genes with >2-fold changes (>3-fold changes for inhibitors ME0052 and ME0054) in transcription for the four inhibitors and numbers of genes in the OI and a cryptic prophage (CP-933)
| Inhibitor or genome | Total no. of genes | OI genes
|
CP-933 genes
|
||
|---|---|---|---|---|---|
| No. of genes | Pa | No. of genes | Pa | ||
| ME0052 | 181 | 118 | 7.2e−32 | 94 | 1.3e−52 |
| ME0053 | 72 | 46 | 8.2e−13 | 41 | 1.5e−25 |
| ME0054 | 23 | 18 | 2.2e−08 | 18 | 1.2e−19 |
| ME0055 | 36 | 7 | 0.22 | 7 | 0.41 |
| Genomeb | 5,481 | 1,449 | 0.98 | 727 | 0.98 |
P values were calculated using a chi-square test with Yates correction. Expected values were generated from the total numbers of OI or CP-933 genes in the genome as described by Perna et al. (27) and are expressed as relative proportions of the total number of genes with a twofold change. The P values represent the significance of the bias toward OI or CP-933 genes.
Total numbers of genes in E. coli O157 EDL933 (annotation of Perna et al. [27]).
Repression of cryptic prophage genes was observed for all inhibitors, and 52% (ME0052), 57% (ME0053), 78% (ME0054), and 19% (ME0055) of the affected genes were on a cryptic prophage (Table 4). Genes with >2-fold or >3-fold changes for each inhibitor were found to be significantly biased toward laterally acquired O-island (OI) DNA (27) for compounds ME0052, ME0053, and ME0054, as determined using a chi-square test (P < 0.0001) (Table 4). The level of significance for ME0055 fell short of this level, probably due to the small subset of genes affected by this compound.
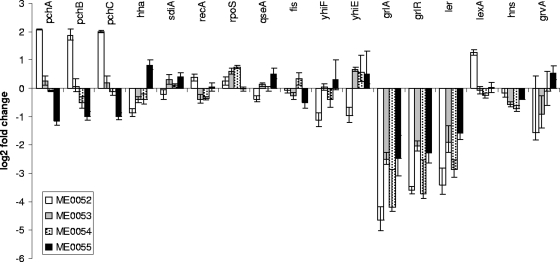
The finding that the transcriptional changes induced by the salicylidene acylhydrazide compounds are biased toward a subset of OI genes suggested that a global regulator of horizontally acquired DNA was affected by these inhibitors. We therefore examined the array data to determine if any of the established regulators of the LEE and other horizontally acquired genes was affected by the compounds. Seventeen previously characterized regulators (for a review, see reference 19) were analyzed to determine if addition of the compounds resulted in transcriptional changes in their expression. For several of these regulators, the analysis was expanded to examine the genes known to be controlled by them. Figure 4 shows the effects of all four compounds on the regulators. It is clear that the majority of the genes (hha, sdiA, recA, rpoS, qseA, fis, yhiF, yhiE, lexA, and hns) showed no marked changes in transcription. To examine if the compounds could affect the regulators posttranscriptionally, the regulons of H-NS, LexA, and RecA were examined.
FIG. 4.
Effects of ME0052, ME0053, ME0054, and ME0055 on the transcription of known regulators of the LEE. The changes (log2) in transcription of treated cultures compared with cultures treated with DMSO alone are shown for the 17 regulators that were characterized.
The histone-like H-NS protein is a pleiotropic regulator of gene expression that plays an important role in transcriptional inactivation of horizontally acquired DNA (7, 23). We used the E. coli K-12 H-NS regulon described by Oshima et al. (26) to look for transcriptional changes in the E. coli K-12 “backbone” (genes that are common to both K-12 and O157) that would be indicative of changes in H-NS regulation. We found that 10 of the 63 genes affected by deletion of H-NS were significantly (P ≤ 0.05) up- or downregulated at least twofold; however, the majority of the regulon genes (53 genes) were not affected by addition of an inhibitor (see Fig. S1A and B in the supplemental material). These data suggest that the compounds do not function by affecting global H-NS regulation.
LexA and RecA mediate the SOS response to DNA damage in E. coli K-12. The SOS response is known to be a key regulator of lambdoid bacteriophage transcription as RecA stimulates autocleavage of the phage repressor, cI, in response to DNA damage, triggering a cascade that leads to transcription of the Q antiterminator transcript and the lytic cycle. LexA has also been shown to regulate the LEE in EPEC, placing both the LEE and potentially phage-regulated transcripts under the control of the SOS response (21). However, transcripts in the LexA regulon were not significantly repressed in inhibitor-treated cultures. It therefore seems unlikely that inhibition of LexA derepression is the mechanism of transcriptional silencing of the T3SS (see Fig. S1C in the supplemental material).
Whereas the majority of the regulators showed no changes in expression when the inhibitors were added, the LEE regulator genes ler (encoding Ler), grlA, and grlR all showed consistent downregulation upon treatment with all four compounds. The finding that ler and grlAR are downregulated coordinately is consistent with previous reports (6, 12). The pchABC genes also showed some changes, particularly after addition of ME0055, the most potent inhibitor of Tir secretion. The T3S regulators Ler and PchA have been shown to bind and positively activate transcription of a number of genes in OIs (1). A comparison of the genes affected by inhibitor treatment and the Ler-PchA regulon defined by Abe and coworkers (1) indicates that there is significant overlap between these regulons (see Table S1 in the supplemental material). Fifty-two of 100 genes repressed by ME0052, 39 of 60 genes repressed by ME0053, 11 of 21 genes repressed by ME0054, and 6 of 19 genes repressed by ME0055 are bound or regulated by Ler or PchA. PchA binds and positively regulates a number of secreted effectors encoded on horizontally acquired elements, such as a cryptic prophage. Table S1 in the supplemental material shows that a number of the non-LEE-encoded effectors are repressed by inhibitor treatment, indicating that in addition to repressing transcription of the T3SS machinery, translocated effectors not encoded by the LEE are also downregulated.
Iron starvation has been proposed to play a key role in the virulence-attenuating properties of the T3S inhibitors described here in Chlamydia pneumoniae (36). Related compounds have been shown to sequester iron, and addition of a high concentration (200 μM) of iron sulfate or iron chloride to inhibitor-treated cultures can reverse the inhibitory effects of these compounds. Our data support this finding since addition of 200 μM FeSO4 reversed the effect of ME0053 in TUV-930 (data not shown). In order to assess the effects of the inhibitors on iron availability in the cell, we used the RegulonDB database definition of the Fur regulon in E. coli K-12 (8; http://regulondb.ccg.unam.mx/). None of 16 genes known to be Fur regulated were significantly affected by addition of an inhibitor (see Fig. S1D in the supplemental material). Similarly, when the GO ontology “iron transport” subset of 29 genes (GO6826) was used, no genes involved in iron transport were affected by all four inhibitors, indicating that intracellular iron supplies are not affected at the concentration of inhibitor used in this study (see Fig. S1E in the supplemental material). It should be noted that the growth medium contained 250 nM Fe(NO3)2, which might have masked any effects on iron availability. Finally, to confirm that an inhibitor at a concentration of 20 μM did not modulate gene expression by perturbing the bacterial growth rate, we analyzed both the rpoS (Z4049) and groEL genes (Z5748) and found that they were not significantly affected by addition of the four compounds (data not shown).
While the majority of transcriptional changes conserved across all four inhibitors were downregulated, pspDC and yjiY were significantly upregulated by inhibitor treatment (P ≤ 0.05). pspBC is part of the phage shock protein response, and analysis of the transcriptional data indicated that the rest of the operon, including pspAB and pspE, was induced, albeit to a lesser degree. The phage shock protein response has previously been reported to be induced by loss of the proton motive force, notably through insertion of the filamentous phage pIV secretin. This response is also essential for membrane insertion of the YscC secretin of the Yersinia T3SS (5). yjiY was also consistently induced in inhibitor-treated cultures; however, there has been little characterization of this protein apart from its induction under stress conditions, such as growth in urine and pH stress (17, 37).
Reporter gene analysis.
For independent verification of the array data, we used a series of GFP reporter fusions to analyze the effects of the inhibitors on the promoters of the LEE and the rpsM gene as a control. Previous work has shown that the majority of LEE genes are transcribed by promoters that encompass five polycistronic operons designated LEE1 to LEE5 (20). We therefore used LEE1 to LEE5 plasmid-based GFP reporter fusions (34) to verify the transcriptome data and show inhibition of LEE transcription. The concentration of each compound (20 μM) was same as that used for transcriptional profiling so that the data could be directly compared. Expression of LEE1 to LEE5 was repressed in the presence of the inhibitors, consistent with the microarray results (Table 5). The control fusion, consisting of the rpsM promoter which drives expression of the small rRNA protein, was not affected by addition of the inhibitors, which is also consistent with the microarray findings (Table 4).
TABLE 5.
Analysis of promoter-GFP constructs for cultures in MEM-HEPES in presence and absence of the small-molecule inhibitors
| Plasmid | Promoter | Ratiosa
|
|||
|---|---|---|---|---|---|
| ME0052 | ME0053 | ME0054 | ME0055 | ||
| pAJR71 | LEE1 | 0.23 | 0.25 | 0.25 | 0.38 |
| pAJR72 | LEE2 | 0.38 | 0.55 | 0.21 | 0.120 |
| pAJR73 | LEE3 | 0.21 | 0.41 | 0.16 | 0.328 |
| pAJR74 | LEE4 | 0.21 | 0.34 | 0.17 | 0.17 |
| pAJR75 | LEE5 | 0.19 | 0.45 | 0.19 | 0.25 |
| pAJR145 | rpsM | 0.98 | 0.99 | 1.1 | 0.98 |
The ratios were calculated by dividing the fluorescence value for the strain cultured in the presence of the inhibitor by the fluorescence value for the strain cultured in the absence of the inhibitor. All values were corrected for the background by subtracting the fluorescence value for the strain transformed with pAJR70 (promoterless gfp) obtained at the same optical density.
Effect on A/E lesion formation.
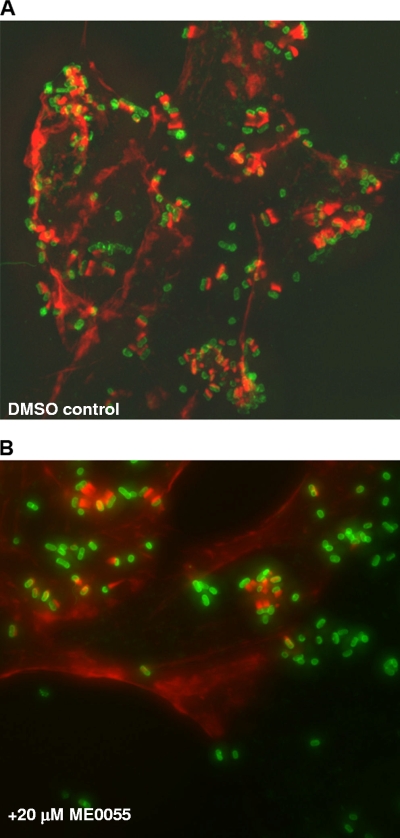
The Western analyses demonstrated that compound ME0055 was the most effective of the four compounds for reducing expression and, therefore, secretion of the key virulence-associated effector protein Tir. However, all the data presented above were obtained with in vitro assays, the majority of which used tissue culture medium to stimulate expression of the LEE. This raised the question of whether the inhibitor would affect the ability of E. coli O157 to bind host cells and form A/E lesions. Using a bovine epithelial cell line (EBL), we analyzed the ability of TUV-930 to form A/E lesions. Following a 6-h infection period, we observed that bacteria cultured in the presence of 20 μM ME0055 were still capable of condensing host cell actin and forming distinctive pedestals (Fig. 5B). However, it was evident that the proportion of bacteria forming A/E lesions was reduced compared to the proportion in the culture treated with DMSO alone (Fig. 5A). In order to quantify any differences, we used Volocity Quantification (Improvision) to identify bacteria associated with areas of dense actin in the three-dimensional images. This was achieved by using size and fluorescence intensity thresholds to identify bacteria, condensed actin, and then areas of clear colocalization. This automated approach allowed us to collate the data from 15 individual fields collected across triplicate repeats of the experiment. These analyses indicated that after 6 h, 28% ± 3% of bacteria treated with ME0055 were associated with A/E lesions (Fig. 5b). This value was significantly less (P < 0.001) than the value for the culture treated with DMSO alone, in which 69% ± 16% of the bacteria formed A/E lesions (Fig. 5A).
FIG. 5.
Addition of ME0055 affects formation of A/E lesions on EBL bovine epithelial cells. Bacteria were cultured in the presence of DMSO (A) or 20 μM ME0055 (B) and added to the EBL cell line. After 6 h, the cells were fixed using 4% PFA and stained to reveal host cell actin (red) (TRITC-phalloidin) and bacteria (green) (anti-O157 and Alexifluor 488-conjugated secondary antibodies). Composite, overlay images obtained using 15 z stack sections are shown.
Effect on LEE1 gene expression and flagellum production during contact with bovine epithelial cells.
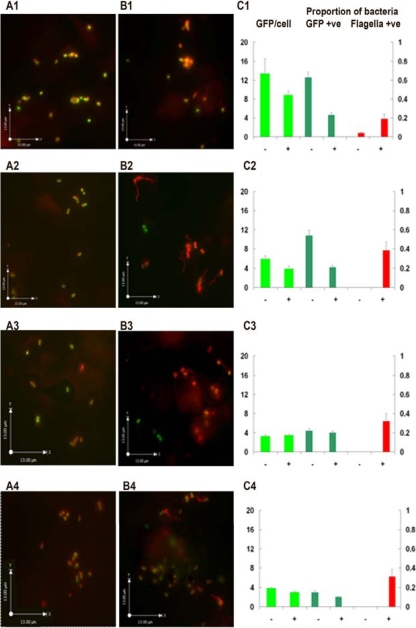
The reduction in the ability of TUV-930 to form A/E lesions implied that addition of compound ME0055 resulted in suppression of LEE expression even during contact with host epithelial cells. To examine this result further, we performed cell binding assays using TUV-930 transformed with a LEE1::GFP reporter plasmid (pAJR71) that allowed measurement of ler expression during contact with epithelial cells. Additionally, our observation that the inhibitors affected grlA and grlR expression implied that flagellum production may be increased as these regulators have been shown to cross-regulate the LEE and flagella (12). Furthermore, recent work has shown that during the cell infection process, E. coli O157 undergoes a rapid temporal shift from expression of H7 flagella to production of the T3SS (15). We therefore asked if the inhibitory compounds affected this transition. To examine this possibility, immunofluorescence antibody staining was used to examine any effects on the expression of H7 flagella. After addition of the bacteria to the EBL cell line, the expression of LEE1::GFP was determined at fixed time points by addition of 4% PFA. Figure 6 shows that 60 min after addition of TUV-930 to the EBL cell line there was a significant difference in LEE1::GFP expression when ME0055-treated cells were compared with the culture treated with DMSO alone. Both the proportion of bacteria expressing GFP and the GFP intensity in bacteria which were “GFP positive” were found to be significantly different. Over the time course, the GFP levels decreased in the bacteria treated with DMSO alone. The ME0055-treated cells exhibited a similar pattern in that the GFP levels decreased throughout the time course, but both the proportion of GFP-positive bacteria and the GFP intensity of these bacteria were observed to be consistently reduced compared to the results for the culture treated with DMSO alone. Staining for H7 revealed that addition of ME0055 led to an increase in flagellum expression compared to the results for the bacteria treated with DMSO alone. Over the time course, the level of H7 expression increased from 20% of the population at 60 min to 40% at 120 min and was still high (30%) 5 h after addition of the bacteria to the eukaryotic cell line. These data indicate that addition of ME0055 suppresses LEE1 expression, even during host cell contact, and leads to an increase in H7 expression. Clearly, both these observations correlate well with the reduction in A/E lesion formation.
FIG. 6.
Effect of ler gene expression during contact with EBL bovine epithelial cells in DMSO-treated (A1 to A4) or ME0055-treated (B1 to B4) bacteria. Bacteria transformed with the LEE1::GFP reporter plasmid (pAJR71) were added to EBL cells, and the samples were fixed using 4% PFA and visualized using anti-O157:H7 antibodies and Alexifluor 555-conjugated secondary antibodies. The time points analyzed were 60 min (A1 and B1), 120 min (A2 and B2), 180 min (A3 and B3), and 300 min (A4 and B4) after bacterial addition. Composite, overlay images obtained using 20 0.1-μm z stack sections are shown. (C) Quantification of LEE1::GFP expression during bacterial contact with host cells. Minus and plus signs indicate samples that contained DMSO and samples that contained ME0055, respectively. The graphs show the amounts of GFP per bacterial cell (expressed in relative fluorescence units) for the time points described above. The proportion of bacteria that showed GFP expression greater than a threshold level (GPF +ve) and the proportion of bacteria that were found to express flagella (Flagella +ve) are also indicated (red bars).
DISCUSSION
The central role of T3SSs in the pathogenesis of many bacterial species provides the impetus to develop strategies that interfere with their function. To do this, “virulence blockers” have been described that selectively target the expression or function of T3SSs in a number of bacterial species, including Y. pseudotuberculosis, Salmonella serovar Typhimurium, C. pneumoniae, and EPEC. There is marked conservation of elements of the T3SS between EPEC and EHEC, yet the transcriptional control of these systems is varied and many regulators of T3S are encoded by horizontally acquired DNA in these polylysogenic pathogens (31, 40, 41). The variation in T3S regulation observed for isolates of EHEC O157 has also been suggested to play an important role in the epidemiology of this pathogen (4, 16). As salicylidene acylhydrazide inhibitors of T3S repress transcription of the T3SS, a detailed investigation of the effects of these compounds on EHEC O157 is warranted. In this study we attempted to address two questions. First, is the T3S inhibition that has been demonstrated in other pathogens observed in EHEC O157, and is there serotype- and strain-specific variation in the responses? Second, are there conserved and/or inhibitor-specific transcriptional responses that may help us understand the mode of action?
In agreement with the previous studies examining repression of bacterial T3SS, we observed that all four compounds studied were capable of inhibiting secretion of virulence proteins from E. coli O157:H7. Addition of higher concentrations of the compounds had marked effects on the growth rate and were not used in subsequent experiments. At a concentration of 20 μM, inhibition of Tir and EspD was observed without major effects on the GroEL level or the growth rate. Further, the relative transcription levels of the rpoS (Z4049) and groEL genes (Z5748) were not found to be significantly (P ≤ 0.05) affected by addition of any of the four compounds. This indicated that the transcriptional changes observed could not be attributed simply to changes in the bacterial growth rate.
The repression of T3S in a diverse range of pathogens suggests that there is a common, conserved target. The most conserved region of the T3SS is the basal apparatus proteins, and this region has recently been proposed to be the target of related inhibitors in Shigella flexneri (38). We noted that the results presented in the study of Veenendaal et al. do not contradict our findings as these authors showed that treatment with an inhibitor decreased the amount of the basal apparatus in the membrane, suggesting that inhibition occurs at a very early stage of assembly and insertion or at the level of transcription. We anticipate that if the inhibitors bind proteins of the T3S complex, all related strains with conserved basal apparatus proteins would likely be affected by the compounds to similar extents, if not the same extent. In the present study we found significant variation in the level of inhibition of T3S between strains. We propose that this reflects inhibition through a more variable route, such as the repertoire of LEE regulators in any one strain. Indeed, we previously reported that the EHEC O157:H7 Sakai strain differed from the EHEC O26:H- strain 193 by the presence of the ETT2-encoded negative regulators of LEE Z3720 and Z3734 (41), and variability in expression of the pchABCX genes has recently been described (40). We therefore favor a model of inhibition involving transcriptional repression of T3S that would be consistent with the strain variation and the global transcriptional changes that we observed.
To examine the effect of the inhibitors on gene regulation, we examined the transcriptional profiles of cells treated with all four compounds. The results presented here indicate that ME0052 to ME0055 represses transcription of the LEE- and non-LEE-encoded virulence factors. Surprisingly, ME0052, which was the least effective compound for limiting Tir secretion, had the greatest effect on global gene transcription. Conversely, ME0055 showed much greater repression of protein secretion but more limited global repression of transcription. Perhaps critically, transcription of espA, encoding the T3 translocation filament, was strongly repressed by ME0055, and this may indicate that repression of the translocon can have a more significant overall effect on protein secretion.
Related salicylidene acylhydrazide compounds have been shown to repress expression of the T3SS in C. pneumoniae, a phenotype that can be reversed by addition of high concentrations of iron. These findings are supported by our results which show that addition of 200 μM FeSO4 can reverse the effect of compound ME0053 in TUV-930. In order to assess the effects of these compounds on iron availability in the cell, we analyzed the transcriptome to look for changes that would indicate iron starvation. The ferric uptake regulator (Fur) is a key regulator of iron uptake under iron starvation conditions, repressing expression of iron uptake pathways and virulence determinants such as Shiga toxin under iron-replete conditions. We did not observe marked changes in the Fur regulon or in genes associated with iron transport in inhibitor-treated cells (see Fig. S2D and E in the supplemental material), indicating that they were not iron starved.
Repressed transcripts were biased toward OI genes, particularly those in a cryptic prophage (Table 4), and many of these genes encode secreted effectors of the T3SS. One explanation for the bias toward repression of the OI genes is that one or more key regulators of horizontally acquired genes are affected by addition of the compounds. On this basis we examined the expression of regulators known to play a role in coordinate expression of virulence genes, including H-NS, LexA, Fur, PchA, and Ler. There was little indication in the transcriptional data that H-NS and LexA regulons were significantly affected by inhibitor treatment (see Fig. S1C in the supplemental material). In contrast, PchA and Ler were significantly repressed by inhibitor treatment.
Recent transcriptional profiling and chromatin immunoprecipitation experiments performed by Abe and coworkers (1) have shown that both Ler and PchA can globally regulate transcription of the LEE and a significant number of non-LEE-encoded effectors. For many of these recently identified effectors this is the first indication of a regulatory mechanism that coordinates transcription with the LEE. Transcriptional profiling of inhibitor-treated cells indicated that repression of transcription is not confined to the LEE but extends to pathogenicity island-encoded virulence determinants, such as the non-LEE-encoded effectors, elements of the type II secretion pathway, and hemolysins encoded ion pO157. Comparison of the Ler-PchA regulon with genes conserved across all four inhibitor treatments showed that 6 of the 12 non-LEE genes repressed by all inhibitor treatments are bound or regulated by PchA and/or Ler, and 30 of 166 genes are significantly repressed by at least one inhibitor treatment. There are PchA homologues in many pathogenic and nonpathogenic bacteria, including Yersinia (YpsIP31758_0776), Salmonella (NP_700416.1), Shigella (NP_599072.1), EPEC (PerC), uropathogenic E. coli (NP_754054.1 and NP_755073.1), and nonpathogenic E. coli (yfdN) (29), and they may contribute to expression of horizontally acquired elements in these bacteria. Our results suggest that modulating the activity of the PchA regulon may contribute to the broad antivirulence effects of the inhibitors in EHEC and, potentially, in other pathogenic bacteria. Clearly, inhibition of Pch function through changes in its expression or activity is one possible mechanism to explain how numerous OI genes are coordinately downregulated by addition of the inhibitors. Simultaneous repression of a number of virulence determinants could contribute to the effectiveness of these inhibitors to limit colonization of cattle and disease in humans.
In conclusion, we have shown that salicylidene acylhydrazide inhibitors selectively inhibit the transcription of the T3SS and other OI genes of E. coli O157. Our data suggest that in E. coli O157 these compounds act through regulators rather than directly at the level of the T3SS apparatus. We have also found inhibitor- and strain-specific effects that should be taken into consideration when the mode of action and therapeutic potential of such molecules are determined. Our ongoing studies are using a combination of transposon mutagenesis screening and affinity purification to identify possible protein targets.
Supplementary Material
Acknowledgments
This research was supported by funds from Tenovus Scotland to A.J.R. and by a Medical Research Scotland grant (reference no. 223 ORG G 0709) to A.J.R. and D.W. D.L.G. and J.J.T. were supported by grants VF0304 and VT0102 from the Department for Environment and Rural Affairs (DEFRA). M.P.S. and A.L. gratefully acknowledge support provided by the Biotechnology & Biological Sciences Research Council (grant BB/D010632/1). Infection assays were performed by A.M. at Easter Bush.
We are grateful to Tim Mitchell and his pathogenesis group at the University of Glasgow for providing helpful ideas and support.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 27 July 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Abe, H., A. Miyahara, T. Oshima, K. Tashiro, Y. Ogura, S. Kuhara, N. Ogasawara, T. Hayashi, and T. Tobe. 2008. Global regulation by horizontally transferred regulators establishes the pathogenicity of Escherichia coli. DNA Res. 15:25-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, L., A. Gylfe, C. Sundin, S. Muschiol, M. Elofsson, P. Nordstrom, B. Henriques-Normark, R. Lugert, A. Waldenstrom, H. Wolf-Watz, and S. Bergstrom. 2007. Small molecule inhibitors of type III secretion in Yersinia block the Chlamydia pneumoniae infection cycle. FEBS Lett. 581:587-595. [DOI] [PubMed] [Google Scholar]
- 3.Campellone, K. G., and J. M. Leong. 2003. Tails of two Tirs: actin pedestal formation by enteropathogenic E. coli and enterohemorrhagic E. coli O157:H7. Curr. Opin. Microbiol. 6:82-90. [DOI] [PubMed] [Google Scholar]
- 4.Chase-Topping, M., D. Gally, C. Low, L. Matthews, and M. Woolhouse. 2008. Super-shedding and the link between human infection and livestock carriage of Escherichia coli O157. Nat. Rev. Microbiol. 6:904-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darwin, A. J. 2005. The phage-shock-protein response. Mol. Microbiol. 57:621-628. [DOI] [PubMed] [Google Scholar]
- 6.Deng, W. Y., J. L. Puente, S. Gruenheid, Y. Li, B. A. Vallance, A. Vazquez, J. Barba, J. A. Ibarra, P. O'Donnell, P. Metalnikov, K. Ashman, S. Lee, D. Goode, T. Pawson, and B. B. Finlay. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. USA 101:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorman, C. J. 2004. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2:391-400. [DOI] [PubMed] [Google Scholar]
- 8.Gama-Castro, S., V. Jimenez-Jacinto, M. Peralta-Gil, A. Santos-Zavaleta, M. I. Penaloza-Spinola, B. Contreras-Moreira, J. Segura-Salazar, L. Muniz-Rascado, I. Martinez-Flores, H. Salgado, C. Bonavides-Martinez, C. Abreu-Goodger, C. Rodriguez-Penagos, J. Miranda-Rios, E. Morett, E. Merino, A. M. Huerta, L. Trevino-Quintanilla, and J. Collado-Vides. 2008. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 36:D120-D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gauthier, A., M. L. Robertson, M. Lowden, J. A. Ibarra, J. L. Puente, and B. B. Finlay. 2005. Transcriptional inhibitor of virulence factors in enteropathogenic Escherichia coli. Antimicrob. Agents Chemother. 49:4101-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giron, J. A., A. G. Torres, E. Freer, and J. B. Kaper. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44:361-379. [DOI] [PubMed] [Google Scholar]
- 11.Hudson, D. L., A. N. Layton, T. R. Field, A. J. Bowen, H. Wolf-Watz, M. Elofsson, M. P. Stevens, and E. E. Galyov. 2007. Inhibition of type III secretion in Salmonella enterica serovar Typhimurium by small-molecule inhibitors. Antimicrob. Agents Chemother. 51:2631-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iyoda, S., N. Koizumi, H. Satou, Y. Lu, T. Saitoh, M. Ohnishi, and H. Watanabe. 2006. The GrlR-GrlA regulatory system coordinately controls the expression of flagellar and LEE-encoded type III protein secretion systems in enterohemorrhagic Escherichia coli. J. Bacteriol. 188:5682-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keyser, P., M. Elofsson, S. Rosell, and H. Wolf-Watz. 2008. Virulence blockers as alternatives to antibiotics: type III secretion inhibitors against Gram-negative bacteria. J. Internal Med. 264:17-29. [DOI] [PubMed] [Google Scholar]
- 15.Mahajan, A., C. G. Currie, S. Mackie, J. Tree, S. McAteer, I. McKendrick, T. N. McNeilly, A. Roe, R. M. La Ragione, M. J. Woodward, D. L. Gally, and D. G. Smith. 2009. An investigation of the expression and adhesin function of H7 flagella in the interaction of Escherichia coli O157:H7 with bovine intestinal epithelium. Cell. Microbiol. 11:121-137. [DOI] [PubMed] [Google Scholar]
- 16.Matthews, L., J. C. Low, D. L. Gally, M. C. Pearce, D. J. Mellor, J. A. Heesterbeek, M. Chase-Topping, S. W. Naylor, D. J. Shaw, S. W. Reid, G. J. Gunn, and M. E. Woolhouse. 2006. Heterogeneous shedding of Escherichia coli O157 in cattle and its implications for control. Proc. Natl. Acad. Sci. USA 103:547-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maurer, L. M., E. Yohannes, S. S. Bondurant, M. Radmacher, and J. L. Slonczewski. 2005. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J. Bacteriol. 187:304-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mellies, J. L., A. M. S. Barron, and A. M. Carmona. 2007. Enteropathogenic and enterohemorrhagic Escherichia coli virulence gene regulation. Infect. Immun. 75:4199-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 21.Mellies, J. L., K. R. Haack, and D. C. Galligan. 2007. SOS regulation of the type III secretion system of enteropathogenic Escherichia coli. J. Bacteriol. 189:2863-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Navarre, W. W., M. McClelland, S. J. Libby, and F. C. Fang. 2007. Silencing of xenogeneic DNA by H-NS—facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 21:1456-1471. [DOI] [PubMed] [Google Scholar]
- 24.Naylor, S. W., A. J. Roe, P. Nart, K. Spears, D. G. E. Smith, J. C. Low, and D. L. Gally. 2005. Escherichia coli O157:H7 forms attaching and effacing lesions at the terminal rectum of cattle and colonization requires the LEE4 operon. Microbiology 151:2773-2781. [DOI] [PubMed] [Google Scholar]
- 25.Nordfelth, R., A. M. Kauppi, H. A. Norberg, H. Wolf-Watz, and M. Elofsson. 2005. Small-molecule inhibitors specifically targeting type III secretion. Infect. Immun. 73:3104-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oshima, T., S. Ishikawa, K. Kurokawa, H. Aiba, and N. Ogasawara. 2006. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 13:141-153. [DOI] [PubMed] [Google Scholar]
- 27.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 28.Peterson, R. E., T. J. Klopfenstein, R. A. Moxley, G. E. Erickson, S. Hinkley, D. Rogan, and D. R. Smith. 2007. Efficacy of dose regimen and observation of herd immunity from a vaccine against Escherichia coli O157:H7 for feedlot cattle. J. Food Prot. 70:2561-2567. [DOI] [PubMed] [Google Scholar]
- 29.Porter, M. E., P. Mitchell, A. Free, D. G. E. Smith, and D. L. Gally. 2005. The LEE1 promoters from both enteropathogenic and enterohemorrhagic Escherichia coli can be activated by PerC-like proteins from either organism. J. Bacteriol. 187:458-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasko, D. A., C. G. Moreira, R. Li de, N. C. Reading, J. M. Ritchie, M. K. Waldor, N. Williams, R. Taussig, S. Wei, M. Roth, D. T. Hughes, J. F. Huntley, M. W. Fina, J. R. Falck, and V. Sperandio. 2008. Targeting QseC signaling and virulence for antibiotic development. Science 321:1078-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roe, A. J., and D. L. Gally. 2000. Enteropathogenic and enterohaemorrhagic Escherichia coli and diarrhoea. Curr. Opin. Infect. Dis. 13:511-517. [DOI] [PubMed] [Google Scholar]
- 32.Roe, A. J., S. W. Naylor, K. J. Spears, H. M. Yull, T. A. Dransfield, M. Oxford, I. J. McKendrick, M. Porter, M. J. Woodward, D. G. Smith, and D. L. Gally. 2004. Co-ordinate single-cell expression of LEE4- and LEE5-encoded proteins of Escherichia coli O157:H7. Mol. Microbiol. 54:337-352. [DOI] [PubMed] [Google Scholar]
- 33.Roe, A. J., L. Tysall, T. Dransfield, D. Wang, D. Fraser-Pitt, A. Mahajan, C. Constandinou, N. Inglis, A. Downing, R. Talbot, D. G. E. Smith, and D. L. Gally. 2007. Analysis of the expression, regulation and export of NleA-E in Escherichia coli O157:H7. Microbiology 153:1350-1360. [DOI] [PubMed] [Google Scholar]
- 34.Roe, A. J., H. Yull, S. W. Naylor, M. J. Woodward, D. G. E. Smith, and D. L. Gally. 2003. Heterogeneous surface expression of EspA translocon filaments by Escherichia coli O157:H7 is controlled at the posttranscriptional level. Infect. Immun. 71:5900-5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shakhnovich, E. A., D. T. Hung, E. Pierson, K. Lee, and J. J. Mekalanos. 2007. Virstatin inhibits dimerization of the transcriptional activator ToxT. Proc. Natl. Acad. Sci. USA 104:2372-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slepenkin, A., P. A. Enquist, U. Hagglund, L. M. de la Maza, M. Elofsson, and E. M. Peterson. 2007. Reversal of the antichlamydial activity of putative type III secretion inhibitors by iron. Infect. Immun. 75:3478-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snyder, J. A., B. J. Haugen, E. L. Buckles, C. V. Lockatell, D. E. Johnson, M. S. Donnenberg, R. A. Welch, and H. L. Mobley. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 72:6373-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Veenendaal, A. K., C. Sundin, and A. J. Blocker. 2009. Small-molecule type III secretion system inhibitors block assembly of the Shigella type III secreton. J. Bacteriol. 191:563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wade, J. T., N. B. Reppas, G. M. Church, and K. Struhl. 2005. Genomic analysis of LexA binding reveals the permissive nature of the Escherichia coli genome and identifies unconventional target sites. Genes Dev. 19:2619-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, Z., J. Kim, C. Zhang, M. Zhang, J. Nietfeldt, C. M. Southward, M. G. Surette, S. D. Kachman, and A. K. Benson. 2009. Genomic instability in regions adjacent to a highly conserved pch prophage in Escherichia coli O157:H7 generates diversity in expression patterns of the LEE pathogenicity island. J. Bacteriol. 191:3553-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, L. H., R. R. Chaudhuri, C. Constantinidou, J. L. Hobman, M. D. Patel, A. C. Jones, D. Sarti, A. J. Roe, I. Vlisidou, R. K. Shaw, F. Falciani, M. P. Stevens, D. L. Gally, S. Knutton, G. Frankel, C. W. Penn, and M. J. Pallen. 2004. Regulators encoded in the Escherichia coli type III secretion system 2 gene cluster influence expression of genes within the locus for enterocyte effacement in enterohemorrhagic E. coli O157:H7. Infect. Immun. 72:7282-7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.