Abstract
We investigated the role of the 58-kDa FTT0918 protein in the iron metabolism of Francisella tularensis. The phenotypes of SCHU S4, a prototypic strain of F. tularensis subsp. tularensis, and the ΔFTT0918 and ΔfslA isogenic mutants were analyzed. The gene product missing in the ΔfslA mutant is responsible for synthesis of a siderophore. When grown in broth with various iron concentrations, the two deletion mutants generally reached lower maximal densities than SCHU S4. The ΔFTT0918 mutant, but not the ΔfslA mutant, upregulated the genes of the F. tularensis siderophore locus (fsl) operon even at high iron concentrations. A chrome azurol sulfonate plate assay confirmed siderophore production by all strains except the ΔfslA strain. In a cross-feeding experiment using medium devoid of free iron, SCHU S4 promoted growth of the ΔfslA strain but not of the ΔFTT0918 strain. The sensitivity of SCHU S4 and the ΔFTT0918 and ΔfslA strains to streptonigrin demonstrated that the ΔFTT0918 strain contained a smaller free intracellular iron pool and that the ΔfslA strain contained a larger one than SCHU S4. In contrast to the marked attenuation of the ΔFTT0918 strain, the ΔfslA strain was as virulent as SCHU S4 in a mouse model. Altogether, the data demonstrate that the FTT0918 protein is required for F. tularensis to utilize iron bound to siderophores and that it likely has a role also in siderophore-independent iron acquisition. We suggest that the FTT0918 protein be designated Fe utilization protein A, FupA.
Francisella tularensis is a facultative intracellular bacterium of many mammalian species capable of causing a zoonotic disease, tularemia (28, 29). Strains of F. tularensis subsp. tularensis (type A) and subsp. holarctica (type B) are the causative agents of tularemia in humans. In mice as few as 10 CFU of type A strains cause a lethal infection, and this dose is also sufficient to establish tularemia in humans (7). The high virulence of the type A strain SCHU S4 was in part shown to depend on the expression of a 58-kDa protein, annotated as the FTT0918 protein, since a mutant lacking expression of the protein was markedly attenuated (35). It was further shown that immunization with a ΔFTT0918 strain elicited protection in mice against a subsequent challenge of a fully virulent type A isolate. Thus, the ΔFTT0918 strain is a promising vaccine candidate. Development of a new vaccine is of definite interest since the existing vaccine, the F. tularensis live vaccine strain (LVS), is not licensed. LVS is a spontaneous type B mutant (6). Interestingly, in LVS, as well as in some other spontaneous F. tularensis mutants, parts of the genes FTT0918 and the other member of the putative operon, FTT0919, are deleted (33). A recent study demonstrated that the deletion of FTT0918 makes a very important contribution to the attenuation of LVS (25).
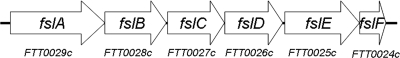
FTT0918 shows no similarity to any genes in other bacterial species but shows high similarity to fslE, a gene located in the F. tularensis siderophore locus (fsl operon), which is involved in synthesis and uptake of an F. tularensis-specific siderophore (11, 32). The operon contains six genes, fslA to fslF (FTT0029 to FTT0024), and is depicted in Fig. 1. FslA has been shown to be responsible for synthesis of the siderophore, and recently it has been suggested that FslE is responsible for the uptake of the siderophore in Francisella novicida and in SCHU S4 (16, 23). FslB, FslC, and FslD all display sequence similarities to proteins in other bacteria predicted to have a role in iron uptake or synthesis. The siderophore contributes to the growth of F. tularensis in vitro because it is necessary for the uptake of iron from media, but its role in the virulence of the pathogen is unknown (11, 32).
FIG. 1.
The fsl operon of Francisella tularensis SCHU S4.
Siderophores chelate ferric iron with high affinity and often participate in bacterial iron acquisition. Iron is pivotal to bacterial growth, as it functions as a cofactor or as a prosthetic group for enzymes essential for many cellular functions and metabolic pathways, such as electron transport, glycolysis, DNA synthesis, and defense against oxidative stress (26). Although iron is abundant both in the environment and in mammalian hosts, it is not readily accessible to bacteria. In nature, iron is oxidized and forms insoluble ferric salts and minerals. Mammals limit the availability of free iron by sequestering it in the ferric form to high-affinity chelators, such as transferrin, ferritin, lactoferrin, and heme, and thereby generate a natural biochemical barrier that a potential invader has to overcome (36). In view of this and the pivotal role of iron in the survival of bacteria, it is not surprising that factors related to iron uptake in bacterial pathogens are often critically related to their virulence.
Besides siderophores, bacteria can express receptors for the direct interaction and sequestration of iron from transferrin, ferritin, and heme-containing proteins or produce hemolysins to sequester iron from hemoglobin of erythrocytes (26, 30). Some bacteria, like Legionella pneumophila, reside in an acidified phagosome (31) where ferrous iron is released from transferrin, which may then be taken up by the bacteria by diffusion through porins into the periplasmic space, with the subsequent uptake into the bacterial cytoplasm by the ferrous iron transport (feo) system (4, 36). Although F. tularensis has a predominant cytoplasmic location, it also possesses a putative feo operon, but, besides the fsl operon, no other iron uptake mechanisms have been described. Since F. tularensis survives in many niches both in nature and in mammals, it can be postulated that iron acquisition is essential for its rapid replication and that it has evolved multiple such mechanisms.
Since the FTT0918 protein is an important virulence factor of F. tularensis and, in addition, a ΔFTT0918 strain is a promising vaccine candidate, we sought to determine why the protein plays such an essential role. Based on its sequence similarity to FslE, we asked if the FTT0918 protein was involved in the iron metabolism of F. tularensis. To this end, we characterized the phenotypes of the ΔFTT0918, ΔfslA, and SCHU S4 strains with regard to growth in iron-limiting media, expression of the fsl operon, and production and utilization of siderophores.
MATERIALS AND METHODS
Bacterial strains.
The F. tularensis subsp. tularensis strain SCHU S4 was obtained from the Francisella Strain Collection of the Swedish Defense Research Agency, Umeå, Sweden.
All bacteriological work was carried out in biosafety level 3 facilities certified by the Swedish Work Environment Authority.
Generation of mutant strains.
The flanking upstream and downstream regions of fslA (FTT0029; 928 and 1,025 bp) and FTT0918 (each ∼1,500 bp) were amplified by PCR using the Phusion high-fidelity DNA polymerase (Finnzyme, Espoo, Finland). One primer in each pair contained complementary sequences to generate “sticky” sequences for a second round of PCR that resulted in one fragment incorporating the flanking regions. The ΔfslA fragment was ligated to an EcoRV-digested pGEM-T Easy vector and the ΔFTT0918 fragment to an XbaI/SalI-digested pGEM-T Easy vector (Promega, Madison, WI), and the vectors were cloned into the Escherichia coli TOP10 strain (Invitrogen, Carlsbad, CA), recloned to NotI-digested pDMK2 (accession no. FJ824848), and introduced to E. coli S17-1 λpir by electroporation. The resulting plasmids, pDMK2-ΔfslA and pDMK2-ΔFTT0918, were conjugated to SCHU S4 as described previously (12).
Clones with pDMK2-ΔfslA or pDMK2-ΔFTT0918 integrated in the SCHU chromosome by recombination were selected on plates containing kanamycin and polymyxin B, and integration was verified by PCR. Clones were then subjected to sucrose selection (12). This procedure selected for a second crossover event in which the integrated plasmid, carrying sacB, was excised from the chromosome. Kanamycin-sensitive clones were examined by PCR to verify the deletion of the expected fragment and confirmed by sequencing. The deletions were in frame, thereby not affecting transcription downstream. Nucleotides 67 to 1893 of the 1,920 bp of the fslA gene and 449 to 1626 of the 1,674 bp of the FTT0918 gene were deleted.
For complementation in trans, the intact FTT0918 gene lacking the codon for the first amino acid, so that translation was initiated from the second GTG, was amplified by PCR and cloned to pKK289Km, resulting in plasmid pKK289KmFTT0918. pKK289Km is a derivative of the previously described vector pKK214GFP (1) without the green fluorescent protein gene and encodes kanamycin resistance as the only antibiotic resistance mechanism. The resulting plasmid was then introduced into the ΔFTT0918 strain by cryotransformation, and the resulting strain was designated FUU075.
Preparation of growth media.
F. tularensis was cultured in Chamberlain's defined medium (CDM) (5) or in CDM that had been iron depleted by chelation (C-CDM). C-CDM was produced as described previously (9, 32). Briefly, 1% (wt/vol) Chelex-100 (Bio-Rad, Hercules, CA) was added to CDM, and the mixture was kept in rotation for 24 h at 4°C. The Chelex-100 was removed by filtering the medium through a 0.2-μm Millipore filter (Biosciences, Stockholm, Sweden), and the chelating step was repeated once. The medium was thereafter supplemented with essential cations (0.55 mM MgSO4, 1.4 μM ZnCl2, 0.2 μM CuSO4, 1.0 μM MnCl2, and 5 μM CaCl2). Sterility was ensured by a second filtration. The C-CDM was supplemented with FeSO4 at various concentrations as indicated in the legends for Fig. 3, 4, 5, and 8.
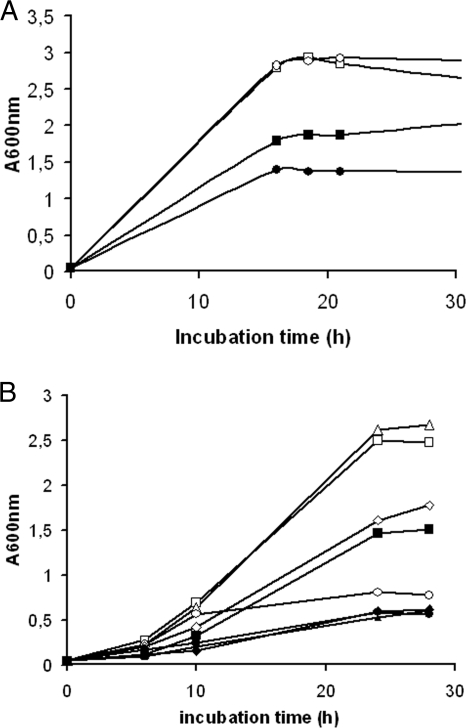
FIG. 3.
Growth of F. tularensis strains in C-CDM supplemented with 0.2 (filled symbols) or 2.0 (open symbols) μg/ml FeSO4. The strains were grown to logarithmic growth phase in CDM (A) or overnight in C-CDM (B) prior to subculturing in C-CDM. SCHU S4 (squares), the ΔfslA strain (triangles), the ΔFTT0918 strain (circles), and FUU075 (diamonds) were grown. Similar growth curves were obtained in at least two additional experiments.
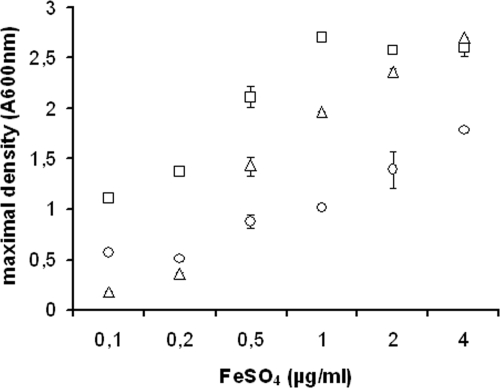
FIG. 4.
Maximal density of SCHU S4 (squares) and the ΔfslA (triangles) and ΔFTT0918 (circles) strains in C-CDM supplemented with the indicated concentrations of FeSO4. Values represent the averages of three observations obtained on separate days, and the error bars are the standard errors of means.
FIG. 5.
Regulation of the fsl operon by SCHU S4 (black bars), the ΔFTT0918 strain (light gray bars), FUU075 (white bars), and the ΔfslA strain (dark gray bars) after 10 h of growth in C-CDM with 2.0 or 0.2 μg/ml FeSO4. Values are expressed as ratios relative to the expression level of SCHU S4 grown in C-CDM with 2.0 μg/ml of FeSO4. The bars represent the averages of six (FUU075 and the ΔfslA strain) or nine (SCHU S4 and the ΔFTT0918 strain) observations obtained from two or three samples from experiments performed on separate occasions. The error bars indicate standard errors of means.
FIG. 8.
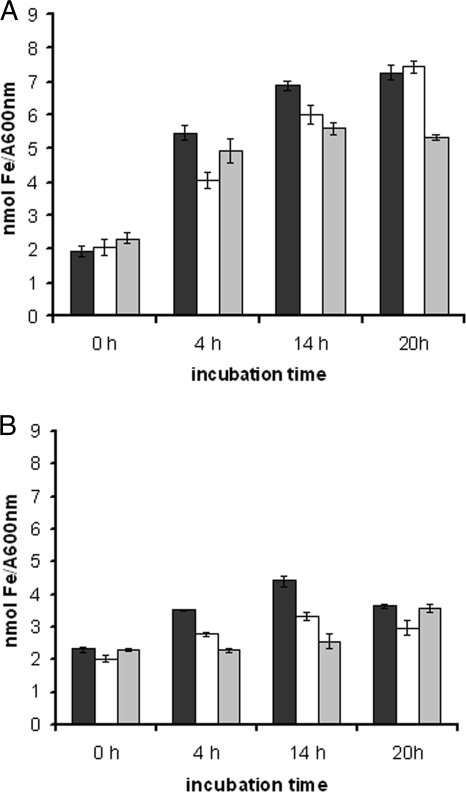
The iron pools of SCHU S4 (black bars) and the ΔFTT0918 (white bars) and ΔfslA (gray bars) strains were measured by the ferrozine assay during growth in 2.0 μg/ml (A) or 0.2 μg/ml FeSO4 (B). The values represent the averages of data from six samples obtained from experiments performed on two separate occasions. The error bars indicate standard errors of means.
To avoid contamination of iron, all glassware was rinsed with 18 mM HCl and thereafter washed twice with ultrapure water. The medium was stored in plastic flasks that prior to their usage had been treated with 1% (vol/vol) HNO3 overnight and washed and rinsed with ultrapure water to remove any contaminating iron.
The chrome azurol sulfonate-C-CDM agar (CAS) plates were prepared essentially as described by Deng et al. (11). Briefly, 40 ml of CAS-Fe(III)-hexadecyltrimethylammonium solution (21) was mixed with 50 ml of a 4% (wt/vol) solution of GC II agar base (BD Diagnostic Systems, MD) and 110 ml of C-CDM. The resulting CAS solution was poured into 20-ml petri dishes. The components of the CAS solution were all purchased from Sigma-Aldrich, Buchs, Switzerland.
The agar plates used to test the strains for streptonigrin sensitivity were composed of equal volumes of 4% GC II agar base (BD Diagnostic Systems, MD) and CDM with no iron supplied.
Growth experiments.
F. tularensis strains grown on GC II agar base supplemented with IsoVitaleX at 37°C in 5% CO2 overnight were suspended in CDM or C-CDM to an optical density (OD) (A600) of 0.2 and incubated overnight at 37°C. The bacteria grown in C-CDM were washed twice in C-CDM to remove the original growth medium and thereafter resuspended in C-CDM supplemented with defined concentrations of FeSO4. Bacteria grown in CDM overnight were diluted in fresh CDM to an OD of 0.2 and then grown to log phase (OD of 0.6 to 0.7). The log phase bacteria were washed two times in C-CDM and thereafter resuspended in C-CDM supplemented with defined concentrations of FeSO4.
Analysis of gene expression.
Bacteria, 3 × 109 CFU, were collected from cultures after 10 to 11 h of incubation, and RNA was extracted using Trizol according to the manufacturer's protocol (Invitrogen). cDNA was synthesized from this RNA, and quantitative real-time PCR (RT-PCR) was used to analyze the cDNA samples. In order to remove contaminating DNA, the RNA samples were DNase treated (DNA-free kit; Ambion, Inc., Austin, TX) in accordance with the protocol supplied by the manufacturer. The RNA was quantified by Nanodrop (Thermo Fisher Scientific, Wilmington, DE). cDNA was synthesized from 1 μg of the extracted RNA using iscript (Bio-Rad) according to the protocol provided by the manufacturer. A negative control was prepared by excluding the enzyme from the cDNA synthesis. In order to degrade any remaining RNA, the cDNA was treated with 2.0 μl of 2.5 M NaOH at 42°C for 10 min, after which the pH was adjusted by the addition of 5 μl of 1 M HCl. The samples were thereafter diluted and stored at −20°C.
RT-PCR was performed with the ABI Prism 7900HT sequence detection system (Applied Biosystems, Foster City, CA) using the SYBR green I PCR kit (Applied Biosystems) as recommended by the manufacturer. Each reaction mixture contained 12.5 μl of the SYBR green mix, 250 nM of forward and reverse primers, and 5 μl of a cDNA, and the total volume was adjusted with water to 25 μl. Forward and reverse primers were obtained from Invitrogen, and their sequences are listed in Table 1. The reactions were performed in a MicroAmp 96-well plate (Applied Biosystems) capped with MicroAmp optical adhesive seal. The reaction mixtures were incubated at 50°C for 2 min and then for 10 min at 95°C, followed by 45 cycles of 15 s at 95°C and 1 min at 60°C. The final cycle consisted of incubation at 95°C for 15 s, 60°C for 15 s, and 95°C for 15 s. The housekeeping gene TUL4 was used to normalize the samples (18). The relative levels of gene expression were calculated using the ΔΔCT method, where CT is the threshold cycle (19). Normalized CT values were used for statistical evaluation of the data.
TABLE 1.
Sequences of primers used in the RT-PCR
| Gene | Primer sequences (5′-3′)a |
|---|---|
| fslA | GACAAGCTACTTCATCCGAGTCTA, |
| TTAGCTGATATTGCTGGTATCCAT | |
| fslB | GCAAAGGTAACTCCAAGTATGAATC, |
| TGCAATATCCACTGTAGCAATTTA | |
| fslC | AATATGACAATGTAGCCCAGCTAA, |
| TCTAGATTTGGATTTGCCTTAACA | |
| fslD | GGCTCCAAGTAGGTGAGCTATTA, |
| GGTGAATCAGCATTTAACACAAAT | |
| fslE | CGCCAAAGAATATCGAATAACTATC, |
| ACCTTCTTAGGAGCATACTCTGGT | |
| FTT0918 | CAGTATTCCAATTGGTCAGATTTC, |
| GCTTCTATAAAGCCACCAAAGAAT |
Forward and reverse primers are indicated on the top and bottom lines, respectively.
CAS plate assay.
The bacteria were cultivated overnight in C-CDM and thereafter washed twice in C-CDM before dilution in C-CDM to a density of 3 × 109 CFU/ml. The suspension was added as a droplet of 2.5 μl to the center of each CAS plate. The plates were incubated at 37°C in 5% CO2, and the size and appearance of the halo formed around the bacterial spot were scored at 24, 48, and 72 h.
Cross-feeding experiment.
The bacteria were cultivated overnight in C-CDM and thereafter washed twice and diluted in the same medium to the final concentration. Fifty microliters of the receiver strain at a density of 3 × 106 CFU/ml was evenly distributed on the CAS plate and dried at 37°C for 2 h. Then, the donor strain at a density of 3 × 109 CFU/ml was added as a droplet of 2.5 μl, containing 7.5 × 106 CFU, to the center of each CAS plate. The plates were incubated at 37°C in 5% CO2, and the size and appearance of the receiver strain and donor strain were recorded after 72 h.
Ferrozine assay.
A ferrozine-based method was used to measure the total amount of intracellular iron (24). Ferrozine forms a complex with Fe2+ that absorbs strongly at 562 nm. A sample volume corresponding to an A600 of 1.0, equal to approximately 3 × 109 bacteria, was collected. The bacteria were washed twice in C-CDM, and the resulting bacterial pellet was lysed with 100 μl of 50 mM NaOH. The solution was mixed thoroughly to ensure complete lysis of the bacteria. One hundred microliters of 10 mM HCl was added to the lysate. To release protein-bound iron, the samples were treated with 100 μl of a freshly prepared solution of 0.7 M HCl and 2.25% (wt/vol) KMnO4 in H2O and incubated for 2 h at 60°C. All chemicals used were from Sigma-Aldrich. Thereafter, the samples were mixed with 100 μl of the iron detection reagent, composed of 6.5 mM ferrozine, 6.5 mM neocuproine, 2.5 M ammonium acetate, and 1.0 M ascorbic acid dissolved in water. The samples were incubated for 30 min, and insoluble particles were removed by centrifugation. Two hundred microliters of the supernatant was transferred to a 96-well plate, and the A562 was determined in a microplate reader. The iron content of the sample was calculated by comparing its absorbance to those of a range of FeCl3 concentrations (0 to 40 μM) in equal volumes that had been prepared similarly to the sample. The correlation coefficients of the standard curves varied between 0.998 and 0.999. The detection limit of the assay was 1 μM Fe. The intrasample variations (i.e., samples from the same culture) were less than 0.3 nmol/3 × 109 bacteria.
Streptonigrin sensitivity.
Before being tested, bacteria were cultured in C-CDM overnight (0 h) and thereafter cultured in C-CDM with 2.0 μg/ml FeSO4 for 14 and 20 h. The bacteria collected from the cultures were washed twice and diluted in C-CDM. One hundred microliters of the bacterial suspension at a density of 3 × 108 CFU/ml was evenly distributed on the agar plate and allowed to dry at room temperature. A sterile disk, 1 cm in diameter (Sigma-Aldrich), soaked with 10 μl of 2-μg/ml streptonigrin (Sigma-Aldrich) was placed in the middle of the plate. The plate was incubated at 37°C in 5% CO2 for 72 h. The diameter of growth inhibition was measured after subtraction of the diameter of the filter disc.
Western blot analysis.
Bacterial lysates in Laemmli sample buffer were separated on a 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel. Each lane was loaded with lysate corresponding to 3 × 107 bacteria. Proteins were then transferred onto a nitrocellulose membrane using a semidry blotter (EBU 4000; C.B.S. Scientific Co., Del Mar, CA). Membranes were then incubated overnight at 4°C with a previously described primary chicken antibody (25) directed against the 58-kDa protein, diluted 1:1,000 (Agrisera, Vännäs, Sweden). Secondary goat anti-chicken horseradish peroxidase-conjugated antibodies were diluted 1:30,000 (Santa Cruz Biotechnology, Santa Cruz, CA) and incubated with the membrane for 1 h. The proteins were visualized using Hyperfilm ECL and the ECL Plus Western blotting detection system (Amersham Biosciences, Uppsala, Sweden).
Mouse infections.
For testing of SCHU S4 and the ΔFTT0918 and ΔfslA strains, specific-pathogen-free, female BALB/c mice were purchased from Charles River Laboratories (St. Constant, Quebec, Canada). Mice were maintained and used in accordance with the recommendations of the Canadian Council on Animal Care Guide to the Care and Use of Experimental Animals in a federally licensed, select agent-approved, small-animal containment level 3 facility (National Research Council, Ottawa, Canada). F. tularensis strains were injected in a volume of 50 μl intradermally. Actual concentrations of inocula were determined by plating 10-fold serial dilutions. Mice were examined daily for signs of infection and were euthanized by CO2 asphyxiation as soon as they displayed signs of irreversible morbidity.
Statistical analysis.
For statistical evaluation, a one-tailed Student t test in the statistical software program SPSS was used.
RESULTS
Characterization of protein expression by mutants and complemented strains.
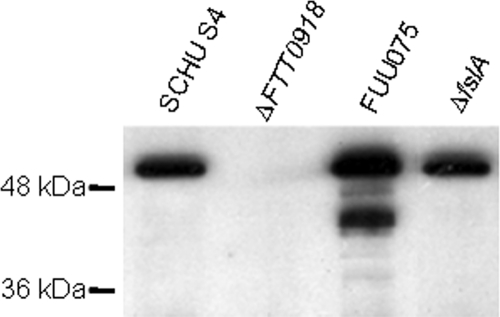
All strains were assayed for protein expression by Western blotting with a polyclonal rabbit antibody directed against the FTT0918 protein. Levels of the protein were similar in SCHU S4 and the ΔfslA mutant (Fig. 2). Thus, the deletion did not affect expression of the FTT0918 protein. A high expression of the protein was observed in FUU075, the ΔFTT0918 strain expressing the FTT0918 protein in trans, indicating that the complementation in the null background was successful and also that the use of the GroE promoter of the plasmid resulted in strong transcription (Fig. 2).
FIG. 2.
Western blot. Bacterial lysates in Laemmli sample buffer were separated on a 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel. Each lane was loaded with lysate corresponding to 3 × 107 bacteria of the indicated strain, and the filter was probed with a polyclonal antiserum directed against the FTT0918 protein.
The influence of iron on the growth of SCHU S4, the ΔFTT0918 strain, FUU075, and the ΔfslA strain.
The ΔFTT0918 strain and SCHU S4 were grown to logarithmic growth phase in CDM, pelleted, and then resuspended in C-CDM with 2.0 μg/ml or 0.2 μg/ml of FeSO4. After 20 h, SCHU S4 reached a significantly greater density in 2.0 than in 0.2 μg/ml of FeSO4 (Fig. 3A). The ΔFTT0918 strain reached a similar density as SCHU S4 in the high concentration, but in the low concentration, the density was about 0.5 A600 unit below that of SCHU S4 (Fig. 3A).
To analyze the ability of the bacteria to grow in media with different iron concentrations after iron starvation, the strains were grown overnight in C-CDM to reduce the bacterial intracellular iron pool. In these experiments, the ΔfslA strain, defective for siderophore production, was included to determine if siderophores contributed to the growth of SCHU S4. After the overnight incubation, SCHU S4, the ΔFTT0918 strain, FUU075, and the ΔfslA strain were resuspended in C-CDM with graded concentrations of 0.1, 0.2, 0.5, 1.0, 2.0, and 4.0 μg/ml of FeSO4. Representative growth curves for 0.2 μg/ml and 2.0 μg/ml are shown in Fig. 3B. The maximal density of SCHU S4 was reached after approximately 20 h of incubation, and it gradually increased when iron concentration was increased up to 1.0 μg/ml (Fig. 4). The ΔFTT0918 strain showed a similar response to increasing concentrations of iron, but for a given iron concentration, the maximal density was at least 1.0 A600 unit below that of SCHU S4 (Fig. 4). Expression of FTT0918 in trans (FUU075) partly restored the growth of this mutant in C-CDM with 2.0 μg/ml but not with 0.2 μg/ml of FeSO4 (Fig. 3B). The ΔfslA strain reached a lower maximal density than SCHU S4 when the iron concentrations were below 2 μg/ml but grew as well at higher concentrations (Fig. 3B and 4).
In summary, iron concentrations below 1.0 μg/ml limited the growth of SCHU S4 and to a greater extent that of the ΔFTT0918 strain, with the ΔfslA strain showing intermediate growth rates. In addition, depletion of the intracellular iron pool prior to cultivation resulted in impaired growth of the ΔFTT0918 strain at iron concentrations that did not affect the growth of SCHU S4. The restricted growth of the ΔfslA strain at iron concentrations below 2.0 μg/ml indicated that siderophore-mediated uptake of iron promoted growth of SCHU S4 at the lower concentrations tested. Henceforth, F. tularensis strains were always cultivated overnight in C-CDM to reduce their intracellular iron pool prior to analysis.
Although the complemented strain, FUU075, demonstrated partly restored growth at the high iron concentration, the impaired phenotype of the ΔFTT0918 strain was not restored at the lower iron concentration, and this showed either that the very strong expression of the FTT0918 protein was detrimental to the function or that the nonphysiological control of the gene when it was expressed in trans resulted in a lack of complementation under these conditions.
Gene expression by SCHU S4, the ΔFTT0918 strain, FUU0075, and the ΔfslA strain during growth in low or high iron concentrations.
Siderophore production is tightly regulated and normally occurs in bacteria only in response to iron deficiency. The expression of the genes of the fsl operon was therefore analyzed by RT-PCR to judge how SCHU S4, the ΔFTT0918 and ΔfslA strains, and FUU075 adapted to various iron concentrations. As expected, all strains grown in C-CDM with 0.2 μg/ml of FeSO4 upregulated (P < 0.01) the genes of the fsl operon relative to SCHU S4 grown in 2.0 μg/ml of FeSO4 (Fig. 5). Additionally, the ΔFTT0918 strain expressed increased levels of most fsl genes (P < 0.05 for fslA to fslD, P = 0.1 for fslE, and P > 0.1 for fslF) when grown in C-CDM with 2.0 μg/ml FeSO4 (Fig. 5). fslF was expressed at increased levels by the ΔfslA strain, in particular, in the low iron concentration. Expression of FTT0918 in trans (FUU075) reduced the expression of the fsl operon, and levels of fslA, fslD, and fslE were similar to those in SCHU S4, whereas fslB and fslD were still upregulated (P < 0.05). As expected, fslA and FTT0918 expression was not detected in the corresponding deletion mutants. FTT0918 was not differentially expressed in any strain under any of the conditions tested. Also expression of FTT0919 was monitored since it is likely that it is expressed in the same operon as FTT0918. There was no difference in its expression between the ΔFTT0918 strain and SCHU S4, and this demonstrates that there is no polar effect of the deletion of FTT0918.
In summary, all strains upregulated the fsl genes when grown in C-CDM with 0.2 μg/ml of iron and the ΔFTT0918 strain also expressed increased levels at the high iron concentration.
Siderophore production by F. tularensis strains.
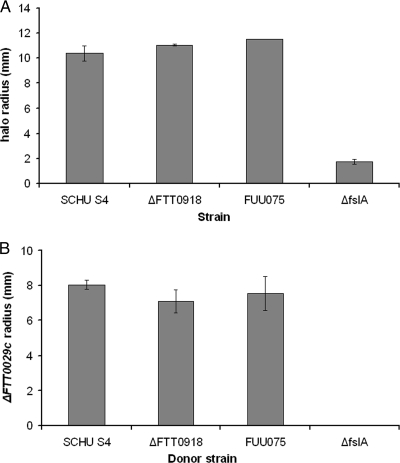
The ability of SCHU S4, the ΔFTT0918 strain, FUU075, and the ΔfslA strain to produce siderophores was determined by the CAS assay. Bacteria, 7.5 × 106 CFU, in 2.5 μl were added to the CAS plate as a droplet. The appearance and size of a halo around the droplet, which are an indication of siderophore production, were measured after 24, 48, and 72 h of incubation. During the first 24 h, the ΔFTT0918 strain grew slower than SCHU S4 but not thereafter. Since the halo formation may be affected by these initial differences in growth rates, the data are presented as increases between days 1 and 3 (Fig. 6A). The halos around the strains increased about 10 mm with the exception of that around the ΔfslA strain, which increased <2 mm, and this was expected as FslA has been demonstrated to produce the siderophore (23). All strains, including the ΔFTT0918 strain, grew after overnight incubation on C-CDM plates, nutritionally identical to CAS plates but with 2.0 μg/ml of FeSO4 added to the agar. Thus, the availability of iron and not the composition of the CAS plate appeared to be the limiting factor for growth of the ΔFTT0918 strain.
FIG. 6.
Siderophore production by the F. tularensis strains as determined by the CAS assay (A) and a cross-feeding experiment (B), with data presented as the increases of the halo between days 1 and 3 (A) and the increases in the growth radius of the ΔfslA receiver strain between days 2 and 3 (B). The bars represent the averages of values obtained from experiments performed on three separate occasions. The error bars indicate standard errors of means.
To further evaluate the ability of the strains to produce siderophores, a cross-feeding experiment was performed, which investigated if a donor strain could promote the growth of a receiver strain under iron-restricted growth conditions. As the ΔfslA strain does not produce siderophores, it was used as the receiver strain, and its growth was measured after 24, 48, and 72 h. Regardless of donor strain, no growth occurred during the first 24 h. As in the CAS assay, the ΔFTT0918 strain grew slower than the other strains, and therefore the data are presented as increases between days 2 and 3. During this period, the ΔfslA strain did not promote any growth whereas an approximately 8-mm zone appeared around the other strains (Fig. 6B).
In conclusion, the CAS assay and the cross-feeding experiment demonstrated that all strains tested except the ΔfslA strain possessed intact siderophore production.
Siderophore utilization by F. tularensis strains.
The capacity of the strains to utilize siderophores was tested by a cross-feeding experiment. SCHU S4 was used as the donor, and the growth of the receiver strains, SCHU S4, the ΔFTT0918 strain, FUU075, and the ΔfslA strain, was assessed after 72 h. The growth of SCHU S4 increased approximately 24 mm, whereas the ΔFTT0918 strain did not grow at all (Fig. 7), not even after 144 h. In contrast, FUU0075, the ΔFTT0918 strain complemented with FTT0918, and the ΔfslA strain showed an increase of approximately 16 mm. Importantly, none of the receiver strains grew on the CAS plate in the absence of a donor strain or with the ΔfslA strain as a donor. This confirms that the receiver strain grew as a response to the siderophore produced by the donor strain.
FIG. 7.
Siderophore utilization tested by a cross-feeding experiment. The data indicate the increases in growth radius of the receiver strain after 72 h. The bars represent the averages of values obtained from experiments performed on three separate occasions. The error bars indicate standard errors of means.
In summary, the failure of SCHU S4 to promote growth of the ΔFTT0918 strain indicated that the latter has an impaired ability to utilize iron bound by the siderophore. This showed that the FTT0918 protein has a direct or indirect role in the siderophore-mediated utilization of iron. FUU075 showed a partial complementation of the defect.
Bacterial iron acquisition in low or high iron concentrations.
To determine if the growth rates of SCHU S4 and the ΔFTT0918 and ΔfslA strains under various iron concentrations were related to their uptake of iron, their intracellular iron pools were determined by the ferrozine assay. After cultivation in C-CDM overnight, the iron pool was approximately 2 nmol/A600 unit (Fig. 8A and B). When these iron-starved bacteria were further cultivated in C-CDM with 2.0 μg/ml of FeSO4, all strains increased their iron pools significantly (Fig. 8A). At 4 and 14 h SCHU S4 contained more iron than the ΔFTT0918 strain (P < 0.001 and < 0.05, respectively), whereas the amounts were similar at 20 h. In contrast, SCHU S4 contained amounts of iron similar to those in the ΔfslA strain at the early time point but more iron at the later time points (P < 0.001). Despite the differences, the data showed that all strains very significantly increased their iron pools over the 20-h period. Complementation of the ΔFTT0918 strain with the FTT0918 protein in trans (FUU075) resulted in iron levels similar to those in SCHU S4 at 4 and 14 h (data not shown).
When cultivated in 0.2 μg/ml of FeSO4, all strains had diminished iron pools compared to cultivation in 2.0 μg/ml FeSO4 (Fig. 8A and B). When iron-starved SCHU S4 was cultivated in the lower iron concentration, there was a 1.5-fold increase at 4 h and approximately a 2-fold increase at 14 h. The ΔFTT0918 strain also increased its iron pool at 4 h but not thereafter and contained significantly less iron than SCHU S4 at all time points examined (P < 0.001) (Fig. 8B). The ΔfslA strain, in contrast to SCHU S4 and the ΔFTT0918 strain, did not increase its iron pool at 4 or 14 h (Fig. 8B). Between 14 h and 20 h, the iron pool of SCHU S4 decreased but increased for the ΔfslA strain, so the iron pools were similar at the latter time point (Fig. 8B).
In summary, in low iron concentrations, the iron uptake was dependent on both the siderophore and the FTT0918 protein since both the ΔfslA and ΔFTT0918 strains contained significantly less iron than SCHU S4 during the first 14 h. In 2.0 μg/ml FeSO4, the uptake was dependent on the presence of the FTT0918 protein at the early stage of growth and on the siderophore at the later stages.
Streptonigrin sensitivity of F. tularensis strains.
Streptonigrin is an antibiotic with an antibacterial effect that is proportional to the size of the free intracellular iron pool (10). The sensitivities of SCHU S4 and the ΔFTT0918 and ΔfslA strains to the antibiotic were tested after culturing in C-CDM overnight (0 h) and in C-CDM with addition of 2.0 μg/ml FeSO4 for 14 and 20 h. The growth inhibition was measured after 72 h. We found that the ΔFTT0918 strain was significantly less sensitive and the ΔfslA strain significantly more sensitive than SCHU S4 (Table 2). In separate tests, FUU075 was tested, and it displayed the same sensitivity as SCHU S4, demonstrating that the defect of the ΔFTT0918 strain could be complemented.
TABLE 2.
Streptonigrin sensitivity
| Strain | Diam of growth inhibition (mm) at:
|
||
|---|---|---|---|
| 0 h | 14 h | 20 h | |
| SCHU S4 | 9.5 ± 0.5 | 9.4 ± 0.4 | 9.2 ± 0.5 |
| ΔFTT0918 mutant | 3.5 ± 1.3a | 3.8 ± 0.4a | 4.3 ± 0.2a |
| ΔfslA mutant | 18.7 ± 5.0b | 17.2 ± 1.5b | 20.8 ± 3.1b |
P < 0.001 compared to SCHU S4.
P < 0.01 compared to SCHU S4.
The data indicated that the ΔFTT0918 strain contained a smaller free intracellular iron pool and the ΔfslA strain contained a larger one than SCHU S4. Moreover, these data and those of the ferrozine test indicated that there were distinct differences between the iron acquisition capacities of the strains and their free intracellular iron pools.
Virulence of mutant F. tularensis strains in mice.
In order to define the contribution of the siderophore to the virulence of F. tularensis, BALB/c mice were infected with SCHU S4 or the ΔfslA strain by the intradermal route. The median survival of mice receiving 10 CFU of the ΔfslA strain was 6 days, and this was the same as that for mice infected with SCHU S4. Thus, in the mouse model siderophores are dispensable for the virulence of SCHU S4. To determine the survival of mice infected with the ΔFTT0918 strain, they were given graded doses of the mutant strain. Mice given a dose of 103 CFU all survived, but no mice survived a dose of 104 CFU (n = 5). Thus, the lack of FTT0918 resulted in an ∼1,000-fold attenuation compared to the virulence of SCHU S4. In contrast, the complemented strain, FUU075, killed all mice (n = 5) at a dose of 10 CFU within 10 days.
DISCUSSION
The study was aimed at characterizing the phenotype of the ΔFTT0918 strain and thereby to understand the molecular mechanisms behind the important contribution of the FTT0918 protein to the virulence of F. tularensis. FTT0918 is highly similar to fslE, and the corresponding genes in F. novicida and in SCHU S4 were recently shown to be part of the fsl operon (11, 20, 32). Interestingly, using the TribeMCL method, a previous study identified that the two genes encoded proteins that belonged to a protein family with five members, also including the FTT0267, FTT0602, and FTT0919 proteins (17). No homologs from other species were identified in the databases; thus, the cluster represents a novel family. However, no significant associations among the proteins were found when a motif search was performed (17), so the functional importance of the other proteins, besides FslE, is enigmatic.
The FslE protein likely serves as a receptor for the F. tularensis siderophore in both F. tularensis and F. novicida (23). This led us to ask if the FTT0918 protein has a role in the iron metabolism of SCHU S4. Our findings suggest that the FTT0918 protein is required for the acquisition of siderophore-bound iron. Several pieces of evidence supported this conclusion since the ΔFTT0918 strain showed impaired growth and iron uptake in broth under iron-limiting conditions and did not grow in the cross-feeding experiment. The specific role of the FTT0918 protein in iron acquisition was corroborated by the finding that FUU075 grew as well SCHU S4 in the cross-feeding experiment, demonstrating that the phenotype could be complemented. This function resembles the recently demonstrated role of the homologous protein FslE, which was suggested to serve as a receptor for siderophore-mediated uptake of iron in SCHU S4 (23). The fslE mutant was defective for growth in broth under iron-limiting conditions, but its virulence was not tested. Using an assay similar to the one described herein, it was demonstrated that the ΔfslE strain secreted siderophores but was unable to utilize iron bound to siderophores (23). The bacterial outer membrane proteins that function as receptors for high-affinity iron chelators, e.g., siderophores or transferrin, belong to a group of ligand-gated porins and are induced in response to iron starvation (8, 14). In line with this, the study by Ramakrishnan et al. demonstrated that fslE was induced under iron starvation (23). A bioinformatic analysis showed that the FTT0918 protein contains a signal peptide, and it was recently demonstrated to be localized in the outer membrane (15). Expression of FTT0918 was, however, not affected by the iron concentration, and thus the FTT0918 protein does not behave like members of this group. Therefore, although the ΔFTT0918 and ΔfslE strains showed the same phenotype in the cross-feeding experiment, i.e., they did not grow with the siderophore as the iron source, their regulatory mechanisms appear to be fundamentally different, and this suggests that the two proteins have distinct functions. Since both mutants appear to be deficient for growth under iron-limiting conditions and the ΔFTT0918 strain shows a dramatic attenuation, there seems to be little or no overlap in their functions.
Even if siderophores are clearly required to enhance the growth of F. tularensis under low iron concentration in broth, we found that siderophore-dependent uptake was not essential in vivo since the ΔfslA strain was not attenuated. This implies that F. tularensis predominantly relies on non-siderophore-dependent iron acquisition in vivo, and the presence of such a system was further supported by our finding of substantial accumulation of iron by the ΔfslA strain during growth in iron-replete medium. Most bacteria have developed multiple iron transport systems, and their functions are often overlapping (34, 37). For instance, Shigella dysenteriae produces two different siderophores and two systems (feo and sit) for uptake of ferrous iron and possesses a heme transport system as well (22). So far, only the fsl operon and the feo genes have been identified in F. tularensis. In view of the high virulence of the ΔfslA strain and the approximately 1,000-fold attenuation of the ΔFTT0918 strain, it appears likely that the FTT0918 protein, in addition to having a role in the utilization of siderophore-bound iron, has a siderophore-independent function that explains its important contribution to the virulence of F. tularensis.
We found additional evidence that supported a siderophore-independent role for the FTT0918 protein since the ΔFTT0918 strain, after iron starvation, did not grow to the same density as SCHU S4 even in high iron concentrations, whereas the ΔfslA strain did not show impaired growth. This growth defect appeared to be unrelated to the iron uptake as determined by the ferrozine assay since iron levels of the ΔFTT0918 strain were at least as high to those in the ΔfslA strain. Despite this finding, the ΔFTT0918 strain showed higher expression of most genes of the fsl operon than did SCHU S4, indicating a relative iron deficiency. In contrast, the ΔfslA strain did not show any differential regulation of the operon despite lower iron levels than those in SCHU S4. To clarify these apparently paradoxical findings, we tested the sensitivity of the strains to streptonigrin, which is correlated to the levels of free intracellular iron (3, 38). We found that the ΔFTT0918 strain was less sensitive and the ΔfslA strain much more sensitive than SCHU S4, and it is therefore possible that the free intracellular iron is a determining factor for growth and the regulation of the fsl operon in the two mutants.
The high sensitivity of the ΔfslA strain to streptonigrin suggests that the siderophore produced by F. tularensis may function as an iron storage molecule. This has also been demonstrated for siderophores produced by fungi and yeasts (13, 27). In contrast to what was found for the ΔfslA strain, inactivation of the corresponding gene (FTN1682) in F. novicida resulted in increased resistance to streptonigrin (10). Thus, in F. tularensis subsp. tularensis the siderophore appears to have a dual function, iron acquisition in iron-deplete medium and iron storage, whereas in F. novicida it has only the latter function.
Intracellular iron has very potent biological effects, and therefore most of it is bound to iron storage molecules. Also mobilization of iron needs to be tightly regulated to avoid detrimental effects, and it is coordinated with subsequent incorporation of ferrous iron into heme and nonheme apoproteins (2). The coordination of the mobilization and metabolism of iron is important since free intracellular iron needs to be carefully regulated to avoid generation of highly toxic hydroxyl radicals through the iron-driven Fenton reaction, but still be sufficient to allow normal metabolism to proceed. In particular, fine-tuning of free iron is delicate for a bacterium like F. tularensis since it will encounter an environment replete with reactive oxygen species intracellularly. A possible role of the FTT0918 protein would be the fine-tuning of iron acquisition and intracellular utilization and thereby the control of free intracellular iron. This could explain the multiple defects of the ΔFTT0918 strain demonstrated herein.
In summary, the results presented demonstrate that the FTT0918 protein plays an important role in iron metabolism of the highly virulent SCHU S4 strain. This occurs under both iron-depleted and -replete conditions and also in the absence or presence of the F. tularensis siderophore. The multiple effects produced by the FTT0918 protein may be an important reason for its major contribution to the virulence of F. tularensis. Based on these findings, we suggest the designation FupA (Fe utilization protein A) for the FTT0918 protein.
Acknowledgments
The work was funded in part by grant AI60689 from the U.S. National Institutes of Health. Grant support was also obtained from the Swedish Medical Research Council and the Medical Faculty, Umeå University, Umeå, Sweden.
The work was performed in part at the Umeå Centre for Microbial Research.
We thank Hua Shen for her skilled help with the animal experiments.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 3 August 2009.
REFERENCES
- 1.Abd, H., T. Johansson, I. Golovliov, G. Sandstrom, and M. Forsman. 2003. Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl. Environ. Microbiol. 69:600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baysse, C., S. Matthijs, M. Schobert, G. Layer, D. Jahn, and P. Cornelis. 2003. Co-ordination of iron acquisition, iron porphyrin chelation and iron-protoporphyrin export via the cytochrome c biogenesis protein CcmC in Pseudomonas fluorescens. Microbiology 149:3543-3552. [DOI] [PubMed] [Google Scholar]
- 3.Bolzan, A. D., and M. S. Bianchi. 2001. Genotoxicity of streptonigrin: a review. Mutat. Res. 488:25-37. [DOI] [PubMed] [Google Scholar]
- 4.Cartron, M. L., S. Maddocks, P. Gillingham, C. J. Craven, and S. C. Andrews. 2006. Feo-transport of ferrous iron into bacteria. Biometals 19:143-157. [DOI] [PubMed] [Google Scholar]
- 5.Chamberlain, R. E. 1965. Evaluation of live tularemia vaccine prepared in a chemically defined medium. Appl. Microbiol. 13:232-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conlan, J. W. 2004. Vaccines against Francisella tularensis—past, present and future. Expert Rev. Vaccines 3:307-314. [DOI] [PubMed] [Google Scholar]
- 7.Conlan, J. W., W. Chen, H. Shen, A. Webb, and R. KuoLee. 2003. Experimental tularemia in mice challenged by aerosol or intradermally with virulent strains of Francisella tularensis: bacteriologic and histopathologic studies. Microb. Pathog. 34:239-248. [DOI] [PubMed] [Google Scholar]
- 8.Cornelissen, C. N. 2003. Transferrin-iron uptake by gram-negative bacteria. Front. Biosci. 8:d836-d847. [DOI] [PubMed] [Google Scholar]
- 9.Cox, C. D. 1994. Deferration of laboratory media and assays for ferric and ferrous ions. Methods Enzymol. 235:315-329. [DOI] [PubMed] [Google Scholar]
- 10.Crosa, L. M., J. H. Crosa, and F. Heffron. 2009. Iron transport in Francisella in the absence of a recognizable TonB protein still requires energy generated by the proton motive force. Biometals 22:337-344. [DOI] [PubMed] [Google Scholar]
- 11.Deng, K., R. J. Blick, W. Liu, and E. J. Hansen. 2006. Identification of Francisella tularensis genes affected by iron limitation. Infect. Immun. 74:4224-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golovliov, I., A. Sjostedt, A. Mokrievich, and V. Pavlov. 2003. A method for allelic replacement in Francisella tularensis. FEMS Microbiol. Lett. 222:273-280. [DOI] [PubMed] [Google Scholar]
- 13.Haas, H. 2003. Molecular genetics of fungal siderophore biosynthesis and uptake: the role of siderophores in iron uptake and storage. Appl. Microbiol. Biotechnol. 62:316-330. [DOI] [PubMed] [Google Scholar]
- 14.Hancock, R. E., and F. S. Brinkman. 2002. Function of pseudomonas porins in uptake and efflux. Annu. Rev. Microbiol. 56:17-38. [DOI] [PubMed] [Google Scholar]
- 15.Huntley, J. F., P. G. Conley, K. E. Hagman, and M. V. Norgard. 2007. Characterization of Francisella tularensis outer membrane proteins. J. Bacteriol. 189:561-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiss, K., W. Liu, J. F. Huntley, M. V. Norgard, and E. J. Hansen. 2008. Characterization of fig operon mutants of Francisella novicida U112. FEMS Microbiol. Lett. 285:270-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsson, P., P. C. Oyston, P. Chain, M. C. Chu, M. Duffield, H. H. Fuxelius, E. Garcia, G. Halltorp, D. Johansson, K. E. Isherwood, P. D. Karp, E. Larsson, Y. Liu, S. Michell, J. Prior, R. Prior, S. Malfatti, A. Sjostedt, K. Svensson, N. Thompson, L. Vergez, J. K. Wagg, B. W. Wren, L. E. Lindler, S. G. Andersson, M. Forsman, and R. W. Titball. 2005. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37:153-159. [DOI] [PubMed] [Google Scholar]
- 18.Lauriano, C. M., J. R. Barker, S. S. Yoon, F. E. Nano, B. P. Arulanandam, D. J. Hassett, and K. E. Klose. 2004. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc. Natl. Acad. Sci. USA. 101:4246-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 20.Milne, T. S., S. L. Michell, H. Diaper, P. Wikstrom, K. Svensson, P. C. Oyston, and R. W. Titball. 2007. A 55 kDa hypothetical membrane protein is an iron-regulated virulence factor of Francisella tularensis subsp. novicida U112. J. Med. Microbiol. 56:1268-1276. [DOI] [PubMed] [Google Scholar]
- 21.Payne, S. M. 1994. Detection, isolation, and characterization of siderophores. Methods Enzymol. 235:329-344. [DOI] [PubMed] [Google Scholar]
- 22.Payne, S. M., E. E. Wyckoff, E. R. Murphy, A. G. Oglesby, M. L. Boulette, and N. M. Davies. 2006. Iron and pathogenesis of Shigella: iron acquisition in the intracellular environment. Biometals 19:173-180. [DOI] [PubMed] [Google Scholar]
- 23.Ramakrishnan, G., A. Meeker, and B. Dragulev. 2008. fslE is necessary for siderophore-mediated iron acquisition in Francisella tularensis Schu S4. J. Bacteriol. 190:5353-5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riemer, J., H. H. Hoepken, H. Czerwinska, S. R. Robinson, and R. Dringen. 2004. Colorimetric ferrozine-based assay for the quantitation of iron in cultured cells. Anal. Biochem. 331:370-375. [DOI] [PubMed] [Google Scholar]
- 25.Salomonsson, E., K. Kuoppa, A. L. Forslund, C. Zingmark, I. Golovliov, A. Sjöstedt, L. Noppa, and A. Forsberg. 2009. Reintroduction of two deleted virulence loci restores full virulence to the live vaccine strain of Francisella tularensis. Infect. Immun. 77:3424-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaible, U. E., and S. H. Kaufmann. 2004. Iron and microbial infection. Nat. Rev. Microbiol. 2:946-953. [DOI] [PubMed] [Google Scholar]
- 27.Schrettl, M., G. Winkelmann, and H. Haas. 2004. Ferrichrome in Schizosaccharomyces pombe—an iron transport and iron storage compound. Biometals 17:647-654. [DOI] [PubMed] [Google Scholar]
- 28.Sjöstedt, A. 2006. Intracellular survival mechanisms of Francisella tularensis, a stealth pathogen. Microbes Infect. 8:561-567. [DOI] [PubMed] [Google Scholar]
- 29.Sjöstedt, A. 2007. Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann. N. Y. Acad. Sci. 1105:1-29. [DOI] [PubMed] [Google Scholar]
- 30.Stojiljkovic, I., and D. Perkins-Balding. 2002. Processing of heme and heme-containing proteins by bacteria. DNA Cell Biol. 21:281-295. [DOI] [PubMed] [Google Scholar]
- 31.Sturgill-Koszycki, S., and M. S. Swanson. 2000. Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J. Exp. Med. 192:1261-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sullivan, J. T., E. F. Jeffery, J. D. Shannon, and G. Ramakrishnan. 2006. Characterization of the siderophore of Francisella tularensis and role of fslA in siderophore production. J. Bacteriol. 188:3785-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svensson, K., P. Larsson, D. Johansson, M. Bystrom, M. Forsman, and A. Johansson. 2005. Evolution of subspecies of Francisella tularensis. J. Bacteriol. 187:3903-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsolis, R. M., A. J. Baumler, F. Heffron, and I. Stojiljkovic. 1996. Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella typhimurium in the mouse. Infect. Immun. 64:4549-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Twine, S., M. Byström, W. Chen, M. Forsman, I. Golovliov, A. Johansson, J. Kelly, H. Lindgren, K. Svensson, C. Zingmark, W. Conlan, and A. Sjöstedt. 2005. A mutant of Francisella tularensis strain SCHU S4 lacking the ability to express a 58-kilodalton protein is attenuated for virulence and is an effective live vaccine. Infect. Immun. 73:8345-8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wandersman, C., and P. Delepelaire. 2004. Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58:611-647. [DOI] [PubMed] [Google Scholar]
- 37.Wyckoff, E. E., A. R. Mey, A. Leimbach, C. F. Fisher, and S. M. Payne. 2006. Characterization of ferric and ferrous iron transport systems in Vibrio cholerae. J. Bacteriol. 188:6515-6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeowell, H. N., and J. R. White. 1982. Iron requirement in the bactericidal mechanism of streptonigrin. Antimicrob. Agents Chemother. 22:961-968. [DOI] [PMC free article] [PubMed] [Google Scholar]