Abstract
We investigated the spatial and temporal response of the murine gut microbiome to infection with Citrobacter rodentium, an attaching-and-effacing bacterium that provokes innate and adaptive immune responses, resulting in transient bacterial colitis. Previous studies have suggested that C. rodentium-induced inflammation is associated with an increased abundance of Enterobacteriaceae. We report here a deeper analysis of this model using DNA bar coding and 454 pyrosequencing to characterize 101,894 partial 16S rRNA gene sequences from 85 microbial samples from tissue-adhered and luminal bacteria of the cecum, proximal colon, and distal colon, which allowed us to identify previously unappreciated spatial and kinetic changes in multiple bacterial lineages. The deep sequencing data revealed that C. rodentium was most abundantly associated with the cecal mucosa at day 9 postinfection and then diminished in abundance, providing the first reported use of deep sequencing to track a pathogen in vivo through the course of infection. Notable changes were associated with both the mucosally adhered and luminal microbiota at both day 9 and day 14 postinfection. Alterations in abundance were seen for Proteobacteria, Deferribacteres, Clostridia, and others; however, changes in Enterobacteriaceae could be accounted for by the presence of C. rodentium itself, which is a member of this family. The Lactobacillus group decreased in abundance during infection, which may be important for pathogenesis because members of this lineage modulate the composition of the gut microbiota and are used as probiotics. Thus, deep sequencing provides previously inaccessible information on how Citrobacter infection and clearance reshapes the gut microbial community in space and time.
The human intestine is home to some 100 trillion microorganisms. The density of bacterial cells in the colon has been estimated at 1011 to 1012 per ml, which make the colon one of the most densely populated microbial habitats known on Earth (22, 42). The pooled genome size of the intestinal bacteria is estimated to exceed the size of the human nuclear genome by 2 orders of magnitude (22, 43). Mutualistic bacteria allow digestion of otherwise undigestible foods, protect against epithelial injury, regulate fat metabolism, boost intestinal angiogenesis, and promote proper development of the immune system (see reference 43 and references therein). The great majority of these gut bacterial species have not been cultured outside the mammalian host, so most species are little studied. Recent reports have surveyed the intestinal microbiota using DNA sequencing of uncultured communities (9, 21, 29, 34), microarray-based methods (31, 32), and targeted full-genome sequencing (12). Many of these studies used 16S rRNA sequences to query the different bacterial taxa, since 16S sequences are similar enough to allow amplification of most bacterial species with single primer pairs but diverse enough to allow assignment of taxa to relatively fine taxonomic levels (30). These and other studies have revealed that major bacterial phyla in the mammalian gut include Bacteroidetes, Firmicutes, Proteobacteria, Actinobacteria, and Fusobacteria.
Inappropriate innate and adaptive immune responses to the commensal gut bacteria are implicated in the pathogenesis of inflammatory bowel disease (IBD; e.g., see references 6, 11, and 37). The development of IBD in rodent models can be suppressed by raising animals in a germfree environment, where commensal bacteria are absent, but can be induced in IBD-prone animals after colonization. Both animal models of IBD and human IBD patients are characterized by an increase in mucosal adherence and translocation of bacteria, T-cell responses to bacterial antigens, and amelioration with antibiotic treatment.
A few studies have begun to characterize the extensive variation in the human-associated microbiota at different anatomic sites (2, 9, 13, 29). For example, populations of 16S rRNA gene sequences have been isolated from the mouth (33), stomach (2), six major subdivisions of the colon (cecum, ascending colon, transverse colon, descending colon, sigmoid colon, and rectum), and stool (9, 13). A recent comparison among human individuals indicate extreme variation in gastrointestinal (GI) bacterial communities, to the point where it is unclear whether a “core” human microbiome even exists (43). In comparison, laboratory mice show much more similarity between individual animals, due at least in part to the homogeneity of diet and genotype possible with these models.
Relatively few studies have investigated the spatial and temporal effects of immune activation on the gut microbiota, which is critical to understanding the factors leading to disregulation during IBD. In one study (27), inflammation was induced in mice by infection with Citrobacter rodentium as a model for enteropathogenic Escherichia coli infection in humans or by chemical means. Studies using fluorescence in situ hybridization and Sanger sequencing suggested that Enterobacteriaceae were increased in abundance in the colonic microbiota with C. rodentium infection. An increase in Enterobacteriaceae was also suggested to be associated with chemical induction of inflammation by treatment with dextran sodium sulfate or deletion of the regulatory cytokine IL-10 but not with infection with Campylobacter jejuni, which does not cause inflammation. In humans, a variety of changes in gut microbial communities have been proposed to accompany GI inflammation, but the generality of observations to date is uncertain, particularly because diet has profound effects on gut microbial composition and has not been controlled in human (21, 39) or macaque (21, 29) studies.
Given that alterations in gut microbial communities are widely expected to be associated with disease states but that the nature of these transitions are mostly unknown, we investigated how infection with C. rodentium affects the spatial and temporal dynamics of the full gut microbial community. We used DNA bar coding (3, 4, 14, 15) and pyrosequencing (28) to characterize a total of 85 microbial communities, 48 from infected mice and 37 from uninfected mice, by sequencing 102,398 gene segments from the 16S rRNA gene. Luminal contents and tissue-associated bacteria obtained from the cecum, proximal colon, and distal colon were compared. Samples were compared at day 9 after infection, the time of maximal counts of Citrobacter adhered to cecal tissue, and at day 14, as the infection begins to resolve, allowing a comprehensive analysis of community-wide effects of infection. In the present study, we observed an increase in Enterobacteriaceae upon infection, as was previously reported (27), but in contrast to the previous report, the increase was accounted for by colonization with C. rodentium itself, which could be quantified in the deep-sequencing data. A statistically rigorous analysis revealed alterations in the Proteobacteria, Deferribacteres, Clostridia, and Lactobacillus lineages accompanying infection, and their distributions differed in time postinfection (p.i.) and in space within the gut. Thus, this comprehensive study of the progression of a bacterial infection revealed unanticipated and extensive changes in bacterial community structure in space and time.
MATERIALS AND METHODS
Animals, bacterial infections, cytokine responses, histological analysis, and sample acquisition.
Twenty C57BL/6 mice in three litters (both males and females) were obtained from Charles River Laboratory. Animals were housed as littermates from birth and throughout the study. Four-week-old animals were orally gavaged on day 0 with 5 × 108 C. rodentium CFU, strain DBS100 (formerly C. freundii biotype 4280 [provided by Bruce Vallance, University of British Columbia, Vancouver, British Columbia, Canada]). Mice were fed acidified water and autoclaved LabDiet 5010 mouse chow (LabDiet). Areas of the large intestine areas were defined by anatomical criteria, i.e., the cecum, and the proximal (1-cm portion immediately following the cecum) or distal (the last 1-cm portion of the large intestine immediately adjacent to the anus) colon. The expression of Ifnγ, Il-22, Il-17a, and Il-17f was analyzed by intestinal tissue RNA isolation using an RNeasy kit (Qiagen, Inc., Valencia, CA), followed by cDNA generation and real-time PCR using SYBR green technology (Applied Biosystems, Foster City, CA) and transcript-specific primers (Qiagen). For real-time PCR analysis of gene expression, RNA was isolated from a section of the mid-colon. The primers used are specific to the cytokine transcripts for gamma interferon (IFN-γ), interleukin-22 (IL-22), IL-17a, and IL-17f and were either purchased from Qiagen (IFN-γ [QT01038821] and IL-22 [QT00128324]) or synthesized (IL-17a, forward [5′-TCCAGAAGGCCCTCAGACTA-3′] and reverse [5′-TTCATTGCGGTGGAGAGTC-3′]; IL-17f, forward [5′-CCTGGATAACACTGTGAGA-3′] and reverse [5′-AATTCACGTGGGACAGAAATG-3′]).
To determine the number of C. rodentium CFU per gram of stool, four C57BL/6 mice obtained from Charles River Laboratory were orally gavaged on day 0 with 5 × 108 nalidixic acid-resistant C. rodentium CFU, and pellets were collected at days 0, 6, 11, 20, and 27. Pellets were homogenized and plated at appropriate dilutions on LB-Nal plates. CFU were enumerated after overnight incubation at 37C.
Recovery of 16S DNA was analyzed by quantitative TaqMan PCR using universal 16S primers to determine the numbers of 16S DNA copies per microgram of total DNA. Comparison of infected and uninfected animals showed no decreases in abundance upon infection, in contrast to a published report (27). The only statistically significant differences were for 2 of the 12 samples (tissue associated from the distal colon [day 9] and luminal contents from the distal colon [day 9]), where the number of 16S copies actually increased after Citrobacter infection.
For histological analysis, IKKβ “floxed” mice on a mixed B6×129 background or C57BL/6 mice obtained from Charles River Laboratory (cecum and colon samples, respectively) were orally gavaged on day 0 with 5 × 108 C. rodentium CFU and euthanized at day 13 or 14 p.i. Paraffin-embedded 4% paraformaldehyde-fixed cecal and colon tissue sections were stained with hematoxylin and eosin (H&E). To separate luminal from mucosal attached bacteria, luminal content was removed by sterile squeezing from 1-cm sections of intestine (luminal bacteria). The intestinal samples were then cut open and gently washed by shaking the tissue in sterile phosphate-buffered saline. Whole DNA was isolated from these washed tissue samples (mucosal attached bacteria). For histological analyses, samples were collected from either commercially purchased C57BL/6 mice or B6 mice in which an IKKβ “floxed” allele was inserted. IKKβ “floxed” mice were generated using the 129 embryonic stem cell line and backcrossed at least six times to a C57BL/6 background. In all of our studies with these mice, they were immunologically and pathologically indistinguishable from C57BL/6 mice. Stool samples were obtained at the time of voiding (fresh from the mice). Pellets were frozen and −80°C for up to several weeks. DNA was extracted from tissue frozen at −80°C.
All experiments were performed following the guidelines of the University of Pennsylvania Institutional Animal Care and Use Committee.
DNA manipulations.
Total DNA from stool samples was extracted from frozen samples by using a QIAamp DNA stool minikit (Qiagen) according to the manufacturer's protocol for pathogen detection. Total DNA extracted from tissue samples was obtained by using the DNeasy blood and tissue kit (Qiagen) according to the protocol for DNA extraction of gram-positive bacteria.
The 16S rRNA gene fragment used in the present study was obtained as previously described (29). Briefly, each sample was amplified by using a bar-coded primer pair (3, 14, 15) based on the universal bacterial primers BSF8 and BSR357. These primers were chosen on the basis of extensive optimization studies that compared segments of the 16S gene for (i) the ability to recover known clustering among bacterial communities and (ii) support accurate phylogenetic placement. In addition to the eight-nucleotide bar code, the primers used to obtain the 16S rRNA gene fragment were also modified to contain the 454 Life Sciences primers A and B.
A 50-μl PCR was carried out using in the AmpliTaq System (Applied Biosystems). The amplification system used AmpliTaq Buffer II and contained 1.5 mM MgCl2, 0.05% Triton X-100, 100 μM concentrations of each deoxyribonucleoside triphosphates, 0.4 μM concentrations of each primer, 0.1 μg of bovine serum albumin/μl, and 2.5 U of AmpliTaq DNA polymerase. A total of 100 ng of template DNA was used for stool samples and 500 ng for tissue samples (to account for mouse DNA extracted together with the bacterial DNA). Amplification was carried out using the following cycling parameters: 5 min denaturing at 95°C, followed by 20 cycles (for stool samples) or 25 cycles (for tissue samples) of 30 s at 95°C (denaturing), 30 s at 56°C (annealing), and 90 s at 72°C (elongation), with a final extension at 72°C for 7 min. Four independent PCRs were performed for each sample, along with a no-template negative control.
Each PCR product was gel purified from a 0.8% agarose gel. DNA was isolated by using a QIAquick gel extraction kit (Qiagen). A 100-ng portion of each of the 100 gel-purified DNAs was added to a master pool of DNA, which was evaluated by pyrosequencing. Of the 120 samples available, 98 were successfully amplified and sequenced; 85 of these yielded 100 sequences or more for analysis. All 20 distal colon tissue samples failed to produce PCR products that were visible in the agarose gel.
Bioinformatic analysis.
Sequences were assessed for quality based upon the following criteria: (i) they were required to show a perfect match to the bar code and 16S rRNA gene primer, (iii) they were at least 50 nucleotides in length after removal of the primer sequences, (iii) they had no more than two undetermined bases in the sequence read; and (iv) at least a 75% match was found to a previously determined 16S rRNA gene sequence after alignment with NAST (7). After the application of these criteria, 101,894 sequences were available for analysis, yielding approximately 1,200 sequences per sample. Luminal content samples consistently yielded approximately twice the amount of sequences compared to the tissue samples.
For all analyses unless otherwise specified, the sequences were inserted into the 16S rRNA gene tree (17) using parsimony insertion implemented in ARB (26). The taxonomic assignment for each sequence was exported from the ARB tree and converted into the accepted nomenclatural taxonomy. In some cases, assignments in ARB were compared to the Ribosome Database Project (RDP), with generally good though not perfect agreement. The tree was also used to perform the Unifrac analysis to compared global community structure (23-25). To test for separation along a given principal coordinate, the Mann-Whitney test was used to compare mean principal coordinate values.
To allow unambiguous identification of the C. rodentium sequences in our pyrosequencing data, we amplified the 16S rRNA gene from the Citrobacter strain examined here by using the primers BSF8 and BSR1541 and determined the sequence using the Sanger method. All sequences assigned to the Enterobacteriaceae family, as well as the Citrobacter sequence and other Enterobacteriaceae sequences available on RDP (trimmed to the length of the longest pyrosequence in the Enterobacteriaceae family), were further analyzed using the PHYLIP package (10). Sequences were aligned, and a distance matrix was constructed using DNADIST. The distance matrix was used to construct a neighbor-joining based tree using NEIGHBOR.
Comparison of mean proportions (see Table 2) was carried out by using the Mann-Whitney test.
TABLE 2.
Statistical analysis of bacterial lineages changing in abundance during infection, treating each mouse as a data point
| Days p.i. | Location | Site | Level | Lineage | P | Change upon infection |
|---|---|---|---|---|---|---|
| 9 | Tissue | Cecum | Class | Clostridia | 0.0253 | Down |
| Proximal colon | Phylum | Deferribacteres | 0.0339 | Up | ||
| Class | Deferribacteres | 0.0339 | Up | |||
| Family | Deferribacteraceae | 0.0339 | Up | |||
| Clostridiaceae | 0.0339 | Up | ||||
| Luminal content | Cecum | Phylum | Deferribacteres | 0.0275 | Up | |
| Class | Deferribacteres | 0.0275 | Up | |||
| Family | Deferribacteraceae | 0.0275 | Up | |||
| Clostridiaceae | 0.0143 | Up | ||||
| Distal colon | Phylum | Proteobacteria | 0.0143 | Up | ||
| Class | Gammaproteobacteria | 0.0275 | Up | |||
| Bacilli | 0.0500 | Down | ||||
| Clostridia | 0.0275 | Up | ||||
| Family | Other Proteobacteria | 0.0275 | Up | |||
| Clostridiaceae | 0.0275 | Up | ||||
| Lachnospiraceae | 0.0500 | Up | ||||
| Lactobacillaceae | 0.0500 | Down | ||||
| Other Firmicutes | 0.0143 | Up | ||||
| 14 | Luminal content | Cecum | Phylum | Deferribacteres | 0.0283 | Up |
| Other Bacteria | 0.0163 | Up | ||||
| Class | Deferribacteres | 0.0283 | Up | |||
| Other Bacteria | 0.0163 | Up | ||||
| Family | Deferribacteraceae | 0.0283 | Up | |||
| Other Bacteria | 0.0163 | Up | ||||
| Proximal colon | Phylum | Deferribacteres | 0.0500 | Up | ||
| Class | Deferribacteres | 0.0500 | Up | |||
| Family | Deferribacteraceae | 0.0500 | Up | |||
| Other Firmicutes | 0.0500 | Up | ||||
| Distal colon | Phylum | Other Bacteria | 0.0500 | Up | ||
| Family | Other Bacteria | 0.0500 | Up |
Nucleotide sequence accession numbers.
All 16S rRNA gene sequences produced in this study have been deposited in GenBank under accession numbers GQ527174 to GQ576477
RESULTS
C. rodentium infection elicits robust expression of proinflammatory cytokines and transient bacterial colitis.
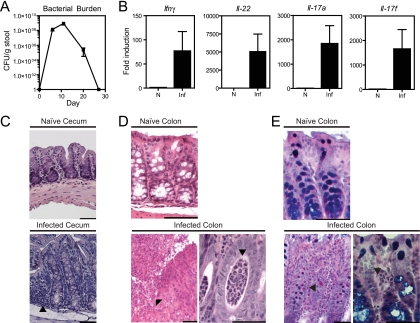
To examine the kinetics of bacterial burden over the course of infection, C57BL/6 mice were infected with nalidixic acid-resistant C. rodentium, stool samples were collected at various times, and CFU were enumerated on LB-Nal plates (Fig. 1A). The bacterial burden peaked by day 11, fell significantly by day 20, and was undetectable by day 27, indicating clearance kinetics similar to those published previously (1). To characterize the inflammatory changes associated with C. rodentium infection, mice were infected with Citrobacter rodentium DBS100 (formerly C. freundii biotype 4280 [provided by Bruce Vallance]) per os or mock infected as controls. Mice were sacrificed on day 13 or 14 p.i. for immunological, pathological, and microbiological analyses.
FIG. 1.
C. rodentium infection in mice. (A) Kinetics of C. rodentium colonization of adult C57BL/6 mice. The C. rodentium burden per gram of stool was measured by plating for nalidixic acid-resistant colonies after infection with C. rodentium containing a nalidixic acid resistance marker. No nalidixic acid-resistant colonies were detected in the absence of C. rodentium infection. (B) Cytokine expression on day 8 as assessed by real-time PCR in intestinal samples from naive (N) and C. rodentium-infected (Inf) mice. The results represent the fold induction over naive WT mice ± the standard error of the mean. (C) Photomicrographs of H&E-stained sections of cecum obtained from naive and C. rodentium-infected mice at day 13. An image of an infected cecum demonstrates a diminution crypt elongation, mononuclear infiltrates (arrow), and an activated epithelium accompanying infection compared to the naive image. (D) Photomicrographs of H&E-stained sections of colon obtained from naive and C. rodentium-infected mice at day 14. A low-magnification image of the infected colon (left) shows focal ulceration of the mucosa (arrow), crypt elongation, and an inflammatory infiltrate composed predominantly of neutrophils. In the high-magnification image of the infected colon (right), crypt abscesses are focally present (arrow) and the epithelium remains reactive. Bar, 50 μm (all images). (E) Periodic acid-Schiff staining shows altered architecture in an infected colon.
Consistent with previous studies, quantitative reverse transcription-PCR analysis revealed that mRNA levels of IL-17A, IL-17F, IL-22, and IFN-γ in the gut peaked on day 8 p.i. and subsequently declined by day 11 with resolution of infection (Fig. 1B and data not shown) (44). Histological analysis of cecal tissue obtained from infected mice showed characteristic changes in the brush border by day 13 p.i., including elongation of the crypts, a mononuclear cell infiltrate, and a reactive epithelial layer compared to mock-infected controls (Fig. 1C). Intestinal mucosa in the colon also demonstrated heightened activation of epithelial cells, as well as focal ulcerations, a prominent neutrophilia, and crypt abscesses (Fig. 1D). Periodic acid-Schiff staining showed characteristic goblet cell loss accompanying infection (Fig. 1E). Collectively, these findings indicate that C. rodentium infection elicits a robust proinflammatory cytokine response that peaks by day 8 and is associated with the development of bacterial colitis by day 14.
Deep-sequencing analysis and distribution of C. rodentium sequence reads.
To determine whether C. rodentium infection and the anti-C. rodentium immune response influenced the structure or stability of gut microbial communities, we used pyrosequencing of 16S DNA segments. Nine mice were analyzed at 9 days p.i. (five infected and four controls), and eleven mice were analyzed at 14 days p.i. (five infected and six controls). Luminal contents were isolated from the cecum, proximal colon, and distal colon of each mouse. Tissue samples were also harvested from each of these sites for analysis of mucosal attached communities. DNA was extracted from each sample, PCR amplified using 16S primers, and subjected to pyrosequencing. The 16S sequence reads were then aligned by using the NAST and GreenGenes servers (7) and inserted into a well-characterized phylogenetic tree (17) using ARB (26), allowing phylogenetic placement of each sequence read.
Sequence reads for C. rodentium could be distinguished unambiguously in the pyrosequence data, allowing the dissemination of the infection to be monitored in time and space (Table 1). C. rodentium sequences were detected only in the infected samples and not in the naive controls (P < 0.0001). By far the largest accumulation of C. rodentium sequences was detected in mucosally adhered samples from the cecum at day 9 p.i., where C. rodentium 16S sequences reached 2.79% of all sequences. Smaller numbers of sequences were found in communities adhered to the proximal colon and in the luminal contents of the cecum, proximal colon, and distal colon. By day 14 p.i., C. rodentium constituted a smaller proportion of sequences at all sites, indicating control of the infection, although infection at the cecum still constituted 0.91% of sequences.
TABLE 1.
Spatial and temporal recovery of Enterobacteriaceae sequences
| Infection state | Days p.i. | Sample
|
% Content (count/total)
|
||
|---|---|---|---|---|---|
| Type | Location | Citrobacter | Other Enterobacteriaceae | ||
| Infected | 9 | Luminal content | Cecum | 0.23 (24/10,430) | 0 (0/10,430) |
| Proximal colon | 0.05 (4/7,960) | 0.01 (1/7,960) | |||
| Distal colon | 0.69 (59/8,548) | 0 (0/8,548) | |||
| Tissue | Cecum | 2.79 (103/3,687) | 0 (0/3,687) | ||
| Proximal colon | 0.04 (1/2,311) | 0.04 (1/2,311) | |||
| 14 | Luminal content | Cecum | 0 (0/6,488) | 0 (0/6,488) | |
| Proximal colon | 0 (0/5,110) | 0.05 (3/5,110) | |||
| Distal colon | 0.02 (1/4,608) | 0.04 (2/4,608) | |||
| Tissue | Cecum | 0.91 (29/3,170) | 0.06 (2/3,170) | ||
| Proximal colon | 0 (0/4,389) | 0.06 (3/4,389) | |||
| Naive | 9 | Luminal content | Cecum | 0 (0/4,993) | 0.02 (1/4,993) |
| Proximal colon | 0 (0/3,388) | 0.02 (1/3,388) | |||
| Distal colon | 0 (0/3,197) | 0.06 (2/3,197) | |||
| Tissue | Cecum | 0 (0/2,048) | 0 (0/2,048) | ||
| Proximal colon | 0 (0/1,037) | 0.09 (1/1,037) | |||
| 14 | Luminal content | Cecum | 0 (0/11,899) | 0.16 (20/11,899) | |
| Proximal colon | 0 (0/6,418) | 0.01 (1/6,418) | |||
| Distal colon | 0 (0/6,856) | 0.83 (57/6,856) | |||
| Tissue | Cecum | 0 (0/896) | 0 (0/896) | ||
| Proximal colon | 0 (0/4,936) | 0.02 (1/4,936) | |||
Table 1 also summarizes counts of Enterobacteriaceae sequences that do not correspond to C. rodentium (which is itself a member of the Enterobacteriaceae family). No obvious differences were seen between infected and uninfected samples. In fact, the greatest number of sequences detected was in the distal colon of an uninfected control mouse. Thus, we did not observe a generalized bloom of Enterobacteriaceae as previously reported (27); rather, the alterations in Enterobacteriaceae appear to have been due to colonization by C. rodentium itself.
UniFrac analysis discloses changes in global community structure accompanying C. rodentium infection.
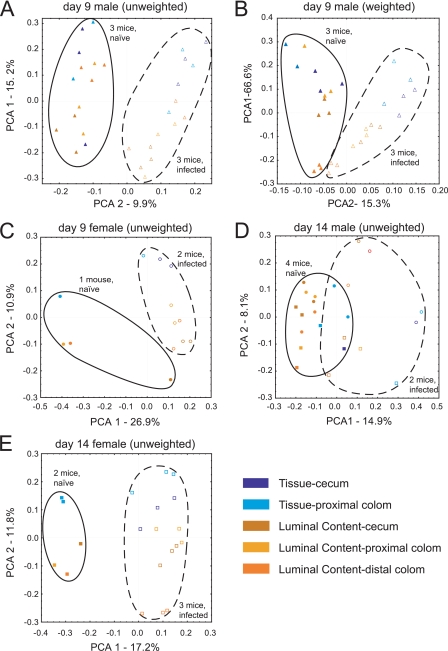
As a first step in analyzing the global effects of C. rodentium infection we compared microbial communities using UniFrac (23-25), which quantifies the similarities among microbial communities based on phylogenetic distances (Fig. 2). To compare two communities, sequences from both communities are placed on a common phylogenetic tree generated using ARB (26). The fraction of the branch length on the tree unique to each community is then measured. This provides an objective measure of community similarity based on the amount of shared evolutionary history. To compare multiple communities, distances between all pairs of communities are computed to generate a distance matrix, and principal coordinate analysis is used to plot communities in a scatter plot along orthogonal axes of maximal variance. Such scatter plots can be generated, taking into account the abundance, as well as the presence of each taxa (weighted UniFrac), or using only the presence or absence information (unweighted UniFrac). The two methods thus address different questions: weighted analysis allows differences in proportional representation of community members to be assessed, whereas unweighted analysis discloses changes in community composition. In Fig. 2 we present examples of both weighted and unweighted analysis. The specific bacterial taxa that change significantly in abundance during infection are discussed below (see Table 2).
FIG. 2.
Global effects of C. rodentium infection, separated by time after infection and the sex of the mice compared. Points from naive (solid symbols) and infected (open symbols) animals are enclosed in ovals. Each litter is represented by a different symbol (circles, litter A; squares, litter B; triangles, litter C). Blue and yellow indicate tissue and luminal content samples, respectively. Axes show the principal coordinate number and the percent of variation explained. (A) Data for males at day 9 p.i., using unweighted UniFrac. The comparison between luminal contents and tissue samples achieved P < 0.0007 on PCA1; the comparison between naive versus infected achieved significance of P < 0.0001 on PCA2. (B) Same as in panel A, but using weighted UniFrac. The comparison between luminal contents and tissue samples achieved P < 0.0001 on PCA1; the comparison between naive and infected mice achieved a significance of P < 0.0001 on PCA2. (C) Data for day 9 females, unweighted UniFrac. The comparison between luminal contents and tissue samples achieved a significance of P < 0.0028 on PCA2; the comparison between naive and infected achieved a significance of P < 0.0112 on PCA1. (D) Data for day 14 males, unweighted UniFrac. The comparison between luminal contents and tissue samples achieved a significance of P < 0.0149 on PCA1; the comparison between naive and infected achieved a significance of P < 0.0001 on PCA1. (E) Data for day 14 females, unweighted UniFrac. The comparison between luminal contents and tissue samples achieved a significance of P < 0.0001 on PCA2; the comparison between naive and infected achieved a significance of P < 0.0001 on PCA1.
A variety of factors are known to influence the murine gut microbiota, including gender and caging history. The comparison in Fig. 2A and B eliminates these variables by focusing on males from a single litter in which three uninfected mice could be compared to three infected mice at day 9 p.i.
In the unweighted analysis (presence or absence information only), the first principal coordinate (15% of sample variation) separated the samples by tissue-associated versus luminal communities (Fig. 2A). The second principal coordinate (10% of sample variation) separated samples by C. rodentium infection state. A nonparametric test of the difference in means along the second principal coordinate achieved high-level significance (P < 0.0001), indicating that the C. rodentium infection caused a community-wide change in the microbial taxa present.
For weighted UniFrac (Fig. 2B), which takes into account abundance information, the first principal coordinate explained a relatively high proportion of the variation in the sample (67%). Again, this first principal coordinate separated the communities by sample type of origin, with tissue-associated communities separating from luminal contents. The second principal coordinate, which accounted for 15% of the variation, separated communities by infection status (P < 0.0001), indicating that the proportional representation of the different types of bacteria also changed. The tissue samples were more widely separated along principal coordinate 2 than the samples from luminal contents, a finding consistent with the idea that C. rodentium infection and associated inflammation most strongly affect tissue-associated bacteria. Collectively, Fig. 2A and B indicate that both the amounts and the types of gut bacteria detected differed due to the infection status and sample type.
Infected and uninfected animals from two additional litters were also available for comparison. Figure 2C shows females infected for 9 days and controls compared using unweighted UniFrac. Here, too, clustering by infection status and sample type is evident. Analysis after 14 days of infection (Fig. 2D and E) showed that distinctive communities persisted in infected versus uninfected animals up to this time point, even though the infection was beginning to resolve. The effects of infection were evident in both males (Fig. 2D) and females (Fig. 2E).
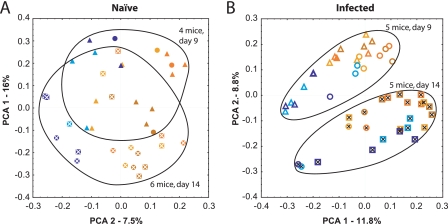
The temporal progression could also be assessed using unweighted UniFrac, comparing the uninfected (Fig. 3A) and infected (Fig. 3B) animals. For C. rodentium-infected animals from all three litters (Fig. 3B), communities from 9 days p.i. clustered separately from those from 14 days p.i., and the separation was highly significant (P < 10−7 on PCA2). Separation over time was much less prominent for the uninfected controls (Fig. 3A), although it did achieve significance (P = 0.0002 on PCA1). The mice at the different time points were necessarily from different litters, and littermates are known to harbor relatively similar microbiomes (20), so the slight difference seen in the naive controls is not surprising. Together, these findings indicate that the temporal progression of the C. rodentium infection was accompanied by global changes in the composition of the gut microbiota.
FIG. 3.
Temporal progression of C. rodentium infection, showing involvement of multiple tissues, analyzed using unweighted UniFrac. Each litter is represented by a different symbol (circles, litter A; squares, litter B; triangles, litter C). Blues and yellows indicate tissue and luminal content samples, respectively. Axes show the principal coordinate number, and the percentage of variation is explained. Points from naive (solid symbols) (A) and infected (open symbols) (B) animals are enclosed in ovals, and day 14 samples are criss-crossed. P values for the comparison of day 9 versus day 14 were as follows: (A) PCA1, P < 0.0002, and PCA2, P < 0.201; and (B) PCA1, P < 0.0001, and PCA2, P < 0.0001. For comparisons of luminal contents and tissue samples, the findings in both panels A and B achieved a significance of P < 0.001 on PCA2.
Bacterial communities attached to the gut mucosa show changes in multiple lineages accompanying infection.
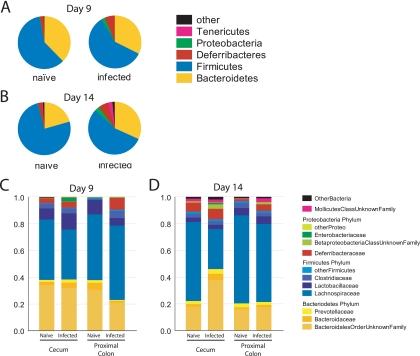
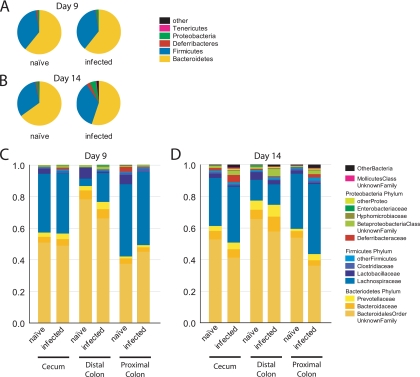
We first present an analysis of bacterial taxa in communities adhered to the gut mucosa, since this is the where C. rodentium infection was most developed and where the greatest differences were detected in the UniFrac analysis (Fig. 4). Tissue-associated samples were pooled and analyzed at the phylum level for day 9 (Fig. 4A) or day 14 (Fig. 4B) samples. Samples were pooled over litters and genders of the mice. For all samples, the Bacteroidetes and Firmicutes comprised at least 85% of the sample. The next most abundant phyla were the Proteobacteria and the little-studied Deferribacteres. Infection resulted in an outgrowth of minor groups, including the Deferribacteres, some Proteobacteria, and Tenericutes (the Tenericutes are a newly formed phylum composed of what was previously the Mollicutes class of the Firmicutes). The extent of the expansion of the lower abundance phyla was notable at day 9 but more pronounced by day 14.
FIG. 4.
Tissue-associated bacteria for naive and C. rodentium-infected mice, showing classification at the phylum level. (A) Day 9 p.i.; (B) day 14 p.i. The group marked “Other” includes Actinobacteria, Cyanobacteria, Nitrospira, Spirochaetes, candidate phylum TM7, and unclassified Bacteria. (C and D) Tissue-associated bacteria, obtained from the cecum and proximal colon, from infected and naive mice at day 9 (C) and day 14 p.i. (D) with C. rodentium. Bacterial sequences were classified at the family level. Deferribacteraceae and Mollicutes are the sole families from the phyla Deferribacteres and Tenericutes, respectively. The minor groups were compressed as follows. “OtherBacteria” is comprised of the phyla Actinobacteria, Nitrospira, TM7, and unclassified Bacteria. “OtherProteo” contains sequences from Hyphomicrobiaceae, Comamonadaceae, Oxalobacteraceae, Helicobacteraceae, and unclassified Alphaproteobacteria. “OtherFirmicutes” contains sequences from Eubacteriaceae, Staphylococcaceae, Aerococcaceae, Erysipelotrichaceae, Peptococcaceae, Acidaminococcaceae, the unclassified Clostridiales order, and unclassified Firmicutes.
The deep-sequencing data allowed quantitative analysis of lineages at a finer taxonomic resolution (Fig. 4C and D). In each plot, the tissue-associated bacteria are separated into the cecum and proximal colon (sequences from the tissue-adhered samples from the distal colon were too few in number for analysis). Analysis of the cecal samples allows the minor classes expanded after infection to be identified primarily as Bacilli (Firmicutes) and Gammaproteobacteria, which correspond mainly to C. rodentium itself. In the proximal colon, a bloom of Deferribacteres was quite prominent. By day 14, a variety of groups had increased in abundance in the cecum, including a particularly prominent bloom of Betaproteobacteria. In the proximal colon, blooms of Deferribacteres, Betaproteobacteria, and Mollicutes were prominent. The Enterobacteriaceae increased in number, as reported previously (27), but in our study these corresponded the bloom of C. rodentium itself, rather than another member of the Enterobacteriaceae. The Lactobacillaceae, a group suggested to ameliorate intestinal inflammation (41), were seen to be diminished in abundance adhered to the proximal colon in the day 9 sample. Collectively, these data specify the changes in the tissue-associated bacteria accompanying infection in space and time.
Exposure to C. rodentium results in dynamic changes in luminally dwelling microbiota.
As discussed above, C. rodentium is an attaching-and-effacing bacterium that forms pedestals on the luminal surface of intestinal epithelial cells. We describe above changes in the surface-attached microbial communities. We also sought to determine whether more global changes in microbial communities could also be identified in the luminal contents of the gut.
We found that the bacteria in luminal contents at all levels of the GI tract in uninfected and infected mice differed from the tissue-associated bacteria in the ratio of Firmicutes to Bacteroidetes: the ratio was <1 for all tissue-associated samples but >1 for luminal contents (compare Fig. 4 and 5). The representation of minor groups also varied, with the Deferribacteres group more abundant in the tissue-associated samples.
FIG. 5.
Luminal-content bacteria from naive and C. rodentium-infected mice. (A and B) Classification at the phylum level. (A) Day 9 p.i.; (B) day 14 p.i. The “Other” group includes sequences classified as Actinobacteria, Cyanobacteria, Nitrospira, Spirochaetes, candidate phylum TM7, and unclassified Bacteria. (C and D) Family level analysis of luminal content bacteria from the cecum, distal colon, and proximal colon in infected and naive mice at day 9 (C) and (D) day 14 p.i. with C. rodentium. Bacterial sequences were classified at the Family level. Deferribacteraceae and Mollicutes are the sole families from phyla Deferribacteres and Tenericutes respectively. For simplicity of presentation, the minor groups were compressed as follows: OtherBacteria is comprised of the phyla Actinobacteria, Nitrospira, TM7, and unclassified Bacteria. OtherProteo contains sequences from unclassified Alphaproteobacteria, the unclassified Desulfovibrionales order, Moraxellaceae, Comamonadaceae, Oxalobacteraceae, unclassified Proteobacteria, Burkholderiaceae, and an unclassified Bdellovibrionales order. Other Firmicutes contains sequences from an unclassified Clostridiales order, unclassified Firmicutes, Peptostreptococcaceae, Streptococcaceae, Leuconostocaceae, Eubacteriaceae, Erysipelotrichaceae, Aerococcaceae, Syntrophomonadaceae, Paenibacillaceae, Staphylococcaceae, and Acidaminococcaceae.
The microbial phyla of the lumen showed little difference associated with infection at day 9 (Fig. 5A), but by day 14 there was a notable expansion of minor phyla (Fig. 5B), including Proteobacteria and Deferribacteres. A finer analysis of taxa affected in the lumen of the cecum, distal colon, and proximal colon at the family level for day 9 (Fig. 5C) and day 14 (Fig. 5D) showed that many groups were altered in abundance by infection. This took place at both times and was not apparent in the phylum-level analysis because the composition actually changed within the Firmicutes phylum. Of these, the Lactobacillaceae were particularly strongly affected, showing considerably reduced abundance accompanying infection in all of the samples of luminal contents at both time points. Among other groups that were affected by the infection were the Deferribacteraceae, Betaproteobacteria, and Alphaproteobacteria, all of which increase both at day 9 and day 14 p.i., and Mollicutes, which increase substantially at day 14 p.i. Thus, these studies demonstrate previously unappreciated changes in both luminal and tissue-adhered bacterial communities accompanying C. rodentium infection.
Statistical analysis of affected lineages.
Because of the large number of samples analyzed by pyrosequencing, it was possible to carry out a rigorous statistical analysis of the lineages that were altered by C. rodentium infection, treating each mouse as a single measurement of proportions. This is in contrast to less rigorous studies in which many community samples are pooled and statistical analysis carried out on the pool (27), since in such a comparison animals could differ radically from each other, but this would be lost upon pooling.
Table 2 presents a nonparametric comparison in which the collection of proportions for each bacterial lineage, measured within each mouse, is compared between infected and uninfected mice. This shows that the Deferribacteres phylum and its single class and family consistently increased in abundance upon infection in both tissue-associated and luminal content samples. The Proteobacteria phylum increased in abundance in luminal contents in day 9 p.i. Several lineages of Firmicutes changed in abundance in both tissue-associated and luminal content samples. The Lactobacillaceae family decreased in abundance in the luminal contents of the distal colon at day 9 p.i. As a control, the uninfected control samples were compared over the two time points (days 9 and 14), and no significant changes in proportion were detected. Thus, thanks to the number of samples available for analysis, it was possible to document changes in microbiome composition that were consistently associated with infection over all animals tested.
DISCUSSION
We used deep sequencing of 16S rRNA gene segments to quantify the response of the gut microbiome to C. rodentium infection and the associated inflammatory response. This strategy allowed us for the first time to track an intestinal pathogen in space and time while monitoring the consequences for the full gut microbial community as well. We found distinctive communities in the three regions of the gut analyzed, and the communities differed between luminal and tissue-associated bacteria. In contrast to a previous report (27), we did not observe a general increase in Enterobacteriaceae lineages apart from C. rodentium itself, but we did detect alterations in abundance for Proteobacteria, Deferribacteres, Clostridia, and Lactobacillaceae. To our knowledge, this was the first use of 454/Roche deep sequencing to track the community-wide effects of an enteric pathogen in space and time. These findings specify bacterial groups that change their proportion in the community in response to C. rodentium infection and which may affect pathogenesis and clearance.
The 454 pyrosequencing approach used here differed from many previously published studies. Sogin and coworkers emphasized the use of 454 sequencing to study the “rare biosphere” by sequencing deeply into a relatively small number of samples (16, 38). Others have used 454 pyrosequencing for resequencing of full genomes (28). Our approach relies on DNA bar coding to characterize large numbers of samples, but using comparatively modest numbers of 16S rRNA gene sequence reads, in the range of 1,200 per sample. This then allows statistical analysis over a large number of samples where each is treated as a separate measurement of proportions (Table 2). Similar approaches were used in other studies (8, 29, 40). Collecting data in this fashion allows the use of 454 pyrosequencing as a tool for hypothesis testing, in this case asking whether significant global differences could be detected consistently in C. rodentium-infected versus uninfected mice. A limitation is that rare bacteria will not be sampled efficiently, but effects on the most abundant groups are well resolved.
Early during infection, C. rodentium colonizes the cecum, but at the earliest time point of our analysis (day 9 p.i.) significant alterations in the composition of microbial communities accompanied infection in the proximal and distal colon as well. Given the capacity of bacteria to express antimicrobial products, changes in microbial diversity could be the result of direct competition between C. rodentium and resident bacterial communities (5). Alternatively, the robust innate and adaptive immune responses elicited by C. rodentium infection could have nonspecific effects on resident bacterial communities. Consistent with this possibility, we and others have observed elevated expression of IL-17, IL-22, and IFN-γ after infection (Fig. 1) (44). These proinflammatory cytokines can induce the expression of antimicrobial peptides in intestinal epithelial cells, including REGIII proteins (44), that could have potent nonspecific effects on C. rodentium and resident microbial communities. The observation that C. rodentium infection and the associated host inflammatory response correlate with alterations in attached and luminal microbial communities throughout the GI tract supports a model wherein C. rodentium-specific lymphocytes are primed in the draining lymph nodes and recirculate to the gut microenvironment, where they are widely distributed throughout the cecum and colon. As a result, the widespread C. rodentium-induced inflammatory response could explain the global changes in the microbiota we have observed after infection.
In a previous study of C. rodentium-infected animals from Lupp et al. (27), which relied on fluorescence in situ hybridization and small numbers of near-full-length 16S sequences (a total of 688 over multiple time points, treatments, and controls), significant evidence was obtained for a bloom of Enterobacteriaceae, and significant increases were seen here for this group in cecum-associated communities. However, in our data, this bloom could be attributed to the added C. rodentium itself, which is a member of the Enterobacteriaceae. Other members of the Enterobacteriaceae family could be quantified in our data and did not increase in abundance with infection. Thus, our data do not support the model of Lupp et al. that inflammation provokes a nonspecific outgrowth of Enterobacteriaceae. We note, however, that the gut microbiota present in mice prior to infection differed between the two studies, which may have affected the response to infection (discussed below).
We found that the Lactobacillaceae group was diminished in abundance during C. rodentium infection, a finding of interest because some Lactobacillus species have been studied as probiotics for use in the treatment of GI disease (reviewed in reference 41). Lactobacillus is a facultative anaerobic or microaerophilic gram-positive bacteria that converts sugars to lactic acid. The resulting acidification has been suggested to suppress the growth of some harmful bacterial lineages. We found that C. rodentium infection resulted in a reduction in the proportion of Lactobacillus in microbial communities. Whether or not the specific Lactobacillaceae lineages identified here by 16S sequences here are in fact beneficial would require further study and is unknown. However, these findings suggest a possible mechanism of pathogenesis, in which the bloom of C. rodentium resulted in reduction of the commensal Lactobacillus, thereby allowing the proliferation of more harmful bacterial lineages.
An unexpected aspect of the data was the bloom of Deferribacteres accompanying C. rodentium infection. These bacteriaes are members of the eight bacteria comprising the Altererd Schaedler Flora, which is used to reconstitute germfree rodents with defined communities (35), but in general the group is little studied. Deferribacteres have been associated with periodontal diseases in humans (18, 19, 36). The linkage of this bacterial group to colonization of the gut mucosa during inflammation raises the possibility that it contributes to pathogenesis in inflammatory conditions. Effects on Deferribacteres were not seen in a previous study (27), despite the fact that these bacteria were prominent in our data in both infected and uninfected animals. The reason for this difference is unknown. One conjecture is that mice in the two colonies studied were seeded with different gut microbial communities initially due to the happenstance of their husbandry, and that the divergent starting flora led to divergent blooms in response to C. rodentium infection and associated inflammation. It will be of considerable interest to investigate further how the initial microbial community structure influences the subsequent community-wide responses to infection.
Acknowledgments
We thank Jeff Weiser and members of the Bushman and Artis laboratories for helpful discussions.
This study was supported in part by the Penn Genome Frontiers Institute and a grant with the Pennsylvania Department of Health. Research in the Artis lab is supported by the National Institutes of Health (NIH), AI61570, AI074878 (D.A.), T32 AI060516 Training in Bacteriology (D.A.H.), the Burroughs Wellcome Fund (Investigator in Pathogenesis of Infectious Disease Award [D.A.]), the Crohns and Colitis Foundation of America's William and Shelby Model Family Foundation Research Award (D.A.), pilot grants from the University of Pennsylvania (University Research Foundation Award, Veterinary Center for Infectious Diseases Pilot Grant [D.A.]), and the Pilot Feasibility Program of the National Institute of Diabetes and Digestive Kidney Diseases DK50306. We also acknowledge NIH instrument grant S10RR024525.
The Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 27 July 2009.
REFERENCES
- 1.Bergstrom, K. S., J. A. Guttman, M. Rumi, C. Ma, S. Bouzari, M. A. Khan, D. L. Gibson, A. W. Vogl, and B. A. Vallance. 2008. Modulation of intestinal goblet cell function during infection by an attaching and effacing bacterial pathogen. Infect. Immun. 76:796-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bik, E. M., P. B. Eckburg, S. R. Gill, K. E. Nelson, E. A. Purdom, F. Francois, G. Perez-Perez, M. J. Blaser, and D. A. Relman. 2006. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. USA 103:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binladen, J., M. T. Gilbert, J. P. Bollback, F. Panitz, C. Bendixen, R. Nielsen, and E. Willerslev. 2007. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS ONE 2:e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Church, G., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawid, S., A. M. Roche, and J. N. Weiser. 2007. The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect. Immun. 75:443-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Hertogh, G., J. Aerssens, R. de Hoogt, P. Peeters, P. Verhasselt, P. Van Eyken, N. Ectors, S. Vermeire, P. Rutgeerts, B. Coulie, and K. Geboes. 2006. Validation of 16S rDNA sequencing in microdissected bowel biopsies from Crohn's disease patients to assess bacterial flora diversity. J. Pathol. 209:532-539. [DOI] [PubMed] [Google Scholar]
- 7.DeSantis, T. Z., Jr., P. Hugenholtz, K. Keller, E. L. Brodie, N. Larsen, Y. M. Piceno, R. Phan, and G. L. Andersen. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:W394-W399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dethlefsen, L., S. Huse, M. L. Sogin, and D. A. Relman. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6:e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package. Cladistics 5:164-166. [Google Scholar]
- 11.Frank, D. N., A. L. St Amand, R. A. Feldman, E. C. Boedeker, N. Harpaz, and N. R. Pace. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 104:13780-13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraser, C. M., J. A. Eisen, and S. L. Salzberg. 2000. Microbial genome sequencing. Nature 406:799-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gill, S. R., M. Pop, R. T. Deboy, P. B. Eckburg, P. J. Turnbaugh, B. S. Samuel, J. I. Gordon, D. A. Relman, C. M. Fraser-Liggett, and K. E. Nelson. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamady, M., J. J. Walker, J. K. Harris, N. J. Gold, and R. Knight. 2008. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods 5:235-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann, C., N. Minkah, J. Leipzig, G. Wang, M. Q. Arens, P. Tebas, and F. D. Bushman. 2007. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 35:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber, J. A., D. B. Welch, H. G. Morrison, S. M. Huse, P. R. Neal, D. A. Butterfield, and M. L. Sogin. 2007. Microbial population structures in the deep marine biosphere. Science 318:97-100. [DOI] [PubMed] [Google Scholar]
- 17.Hugenholtz, P. 2002. Exploring prokaryotic diversity in the genomic era. Genome Biol. 3:REVIEWS0003. [DOI] [PMC free article] [PubMed]
- 18.Hutter, G., U. Schlagenhauf, G. Valenza, M. Horn, S. Burgemeister, H. Claus, and U. Vogel. 2003. Molecular analysis of bacteria in periodontitis: evaluation of clone libraries, novel phylotypes and putative pathogens. Microbiology 149:67-75. [DOI] [PubMed] [Google Scholar]
- 19.Kumar, P. S., A. L. Griffen, J. A. Barton, B. J. Paster, M. L. Moeschberger, and E. J. Leys. 2003. New bacterial species associated with chronic periodontitis. J. Dent. Res. 82:338-344. [DOI] [PubMed] [Google Scholar]
- 20.Ley, R. E., F. Backhed, P. Turnbaugh, C. A. Lozupone, R. D. Knight, and J. I. Gordon. 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 102:11070-11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ley, R. E., M. Hamady, C. Lozupone, P. J. Turnbaugh, R. R. Ramey, J. S. Bircher, M. L. Schlegel, T. A. Tucker, M. D. Schrenzel, R. Knight, and J. I. Gordon. 2008. Evolution of mammals and their gut microbes. Science 320:1647-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ley, R. E., D. A. Peterson, and J. I. Gordon. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837-844. [DOI] [PubMed] [Google Scholar]
- 23.Lozupone, C., M. Hamady, and R. Knight. 2006. UniFrac-an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinform. 7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lozupone, C., and R. Knight. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lozupone, C. A., M. Hamady, S. T. Kelley, and R. Knight. 2007. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73:1576-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lupp, C., M. L. Robertson, M. E. Wickham, I. Sekirov, O. L. Champion, E. C. Gaynor, and B. B. Finlay. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2:204. [DOI] [PubMed] [Google Scholar]
- 28.Margulies, M., M. Egholm, W. E. Altman, S. Attiya, J. S. Bader, L. A. Bemben, J. Berka, M. S. Braverman, Y. J. Chen, Z. Chen, S. B. Dewell, L. Du, J. M. Fierro, X. V. Gomes, B. C. Godwin, W. He, S. Helgesen, C. H. Ho, G. P. Irzyk, S. C. Jando, M. L. Alenquer, T. P. Jarvie, K. B. Jirage, J. B. Kim, J. R. Knight, J. R. Lanza, J. H. Leamon, S. M. Lefkowitz, M. Lei, J. Li, K. L. Lohman, H. Lu, V. B. Makhijani, K. E. McDade, M. P. McKenna, E. W. Myers, E. Nickerson, J. R. Nobile, R. Plant, B. P. Puc, M. T. Ronan, G. T. Roth, G. J. Sarkis, J. F. Simons, J. W. Simpson, M. Srinivasan, K. R. Tartaro, A. Tomasz, K. A. Vogt, G. A. Volkmer, S. H. Wang, Y. Wang, M. P. Weiner, P. Yu, R. F. Begley, and J. M. Rothberg. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKenna, P., C. Hoffmann, N. Minkah, P. P. Aye, A. Lackner, Z. Liu, C. A. Lozupone, M. Hamady, R. Knight, and F. D. Bushman. 2008. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 4:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pace, N. R., G. J. Olsen, and C. R. Woese. 1986. Ribosomal RNA phylogeny and the primary lines of evolutionary descent. Cell 45:325-326. [DOI] [PubMed] [Google Scholar]
- 31.Palmer, C., E. M. Bik, D. B. Digiulio, D. A. Relman, and P. O. Brown. 2007. Development of the human infant intestinal microbiota. PLoS Biol. 5:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmer, C., E. M. Bik, M. B. Eisen, P. B. Eckburg, T. R. Sana, P. K. Wolber, D. A. Relman, and P. O. Brown. 2006. Rapid quantitative profiling of complex microbial populations. Nucleic Acids Res. 10:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, L. V. A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rawls, J. F., M. A. Mahowald, R. E. Ley, and J. I. Gordon. 2006. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127:423-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robertson, B. R., J. L. O'Rourke, B. A. Neilan, P. Vandamme, S. L. On, J. G. Fox, and A. Lee. 2005. Mucispirillum schaedleri gen. nov., sp. nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. Int. J. Syst. Evol. Microbiol. 55:1199-1204. [DOI] [PubMed] [Google Scholar]
- 36.Saito, D., T. Leonardo Rde, J. L. Rodrigues, S. M. Tsai, J. F. Hofling, and R. B. Goncalves. 2006. Identification of bacteria in endodontic infections by sequence analysis of 16S rDNA clone libraries. J. Med. Microbiol. 55:101-107. [DOI] [PubMed] [Google Scholar]
- 37.Scanlan, P. D., F. Shanahan, C. O'Mahony, and J. R. Marchesi. 2006. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn's disease. J. Clin. Microbiol. 44:3980-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sogin, M. L., H. G. Morrison, J. A. Huber, D. M. Welch, S. M. Huse, P. R. Neal, J. M. Arrieta, and G. J. Herndl. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. USA 103:12115-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turnbaugh, P. J., F. Backhed, L. Fulton, and J. I. Gordon. 2008. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3:213-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turnbaugh, P. J., M. Hamady, T. Yatsunenko, B. L. Cantarel, A. Duncan, R. E. Ley, M. L. Sogin, W. J. Jones, B. A. Roe, J. P. Affourtit, M. Egholm, B. Henrissat, A. C. Heath, R. Knight, and J. I. Gordon. 2009. A core gut microbiome in obese and lean twins. Nature 457:480-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ventura, M., S. O'Flaherty, M. J. Claesson, F. Turroni, T. R. Klaenhammer, D. van Sinderen, and P. W. O'Toole. 2009. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat. Rev. Microbiol. 7:61-71. [DOI] [PubMed] [Google Scholar]
- 42.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaneveld, J., P. J. Turnbaugh, C. Lozupone, R. E. Ley, M. Hamady, J. I. Gordon, and R. Knight. 2008. Host-bacterial coevolution and the search for new drug targets. Curr. Opin. Chem. Biol. 12:109-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng, Y., P. A. Valdez, D. M. Danilenko, Y. Hu, S. M. Sa, Q. Gong, A. R. Abbas, Z. Modrusan, N. Ghilardi, F. J. de Sauvage, and W. Ouyang. 2008. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14:282-289. [DOI] [PubMed] [Google Scholar]