Abstract
The ability of some typical enteropathogenic Escherichia coli (EPEC) strains to adhere to, invade, and increase interleukin-8 (IL-8) production in intestinal epithelial cells in vitro has been demonstrated. However, few studies regarding these aspects have been performed with atypical EPEC (aEPEC) strains, which are emerging enteropathogens in Brazil. In this study, we evaluated a selected aEPEC strain (1711-4) of serotype O51:H40, the most prevalent aEPEC serotype in Brazil, in regard to its ability to adhere to and invade Caco-2 and T84 cells and to elicit IL-8 production in Caco-2 cells. The role of flagella in aEPEC 1711-4 adhesion, invasion, and IL-8 production was investigated by performing the same experiments with an isogenic aEPEC mutant unable to produce flagellin (FliC), the flagellum protein subunit. We demonstrated that this mutant (fliC mutant) had a marked decrease in the ability to adhere to T84 cells and invade both T84 and Caco-2 cells in gentamicin protection assays and by transmission electron microscopy. In addition, the aEPEC 1711-4 fliC mutant had a reduced ability to stimulate IL-8 production by Caco-2 cells in early (3-h) but not in late (24-h) infections. Our findings demonstrate that flagella of aEPEC 1711-4 are required for efficient adhesion, invasion, and early but not late IL-8 production in intestinal epithelial cells in vitro.
Typical enteropathogenic Escherichia coli (tEPEC) strains are a well-known cause of diarrhea in infants in developing countries (14, 27, 45) whereas atypical EPEC (aEPEC) strains are emerging enteropathogens (1, 3, 10, 14, 27, 28, 33, 36, 45). tEPEC and aEPEC produce a histopathological lesion on enterocytes that is known as the attaching and effacing (AE) lesion. This lesion is characterized by effacement of microvilli, intimate adherence between the bacterium and the cell membrane, and actin accumulation underneath adherent bacteria, forming a pedestal-like structure. Intimate adherence is mediated by intimin (an outer membrane bacterial adhesin) and its receptor Tir (translocated intimin receptor), which is translocated into the host cell by a type III secretion system (19). The proteins involved in the establishment of AE lesions are encoded on the chromosomal pathogenicity island called the locus of enterocyte effacement (LEE) (24). A functional LEE region is also found in enterohemorrhagic E. coli (EHEC) and probably plays a significant role in disease since patients with hemolytic uremic syndrome develop a strong antibody response to various LEE-encoded proteins, although the major virulence factors in EHEC are the Shiga toxins (19). In addition, only tEPEC strains possess the EPEC adherence factor plasmid, which encodes localized adhesion on cultured epithelial cells mediated by the bundle-forming pilus (Bfp) and a transcriptional activator (Per) that upregulates the expression of the bfp operon and the LEE genes (19, 27).
It has been shown that tEPEC flagella mediate adhesion and microcolony formation on cultured enterocytes, and they are also implicated in the virulence of EHEC and other enteropathogens (2, 4, 12, 21, 26, 44). However, the role of this structure in different aspects of aEPEC virulence, as adhesion and invasion, has not been evaluated. Invasion of eukaryotic cells results from a complex interplay between the host and the pathogen. Although aEPEC is not considered a classical invasive pathogen, the presence of some aEPEC strains inside HeLa or Caco-2 cells has been reported, but the role of flagella in this process has not been evaluated (15, 34, 37, 40).
Flagellin (FliC), the protein encoded by the fliC gene, is the main component of the flagella. Purified flagellin induces interleukin-8 (IL-8) production and Toll-like receptor 5 (TLR5) activation in enterocyte monolayers infected with tEPEC and enteroaggregative E. coli (12, 42). Rogers et al. (35) showed that flagellin H21, extracted from a Shiga toxin-producing E. coli (STEC) strain, stimulated IL-8 production in the enterocyte lineage HCT-8, while Zhou et al. (49) demonstrated that flagellins H6 and H34 of tEPEC, H7 of STEC, and flagellin of E. coli K-12 stimulated IL-8 production in T84 monolayers. At least two distinct mechanisms of IL-8 induction were recognized in tEPEC and Salmonella enterica serovar Typhimurium: interaction of flagellin with TLR5, which is located on the basolateral surface of the intestinal epithelium, and secretion of effector proteins through the type III secretion system (11, 41). However, there is no information regarding the ability of aEPEC flagellin to elicit an inflammatory response in vitro.
In a survey undertaken in the city of São Paulo, Brazil, aEPEC strains were isolated from fecal samples of children with diarrhea. These strains were subsequently characterized for various phenotypic and genotypic properties (25, 46). Although they belonged to a large diversity of serotypes, O51:H40 was the most frequent (10.1%) (13). Unpublished experimental evidence from our group demonstrates that a motile aEPEC strain (O51:H40), but not a nonmotile aEPEC strain, elicits an intense inflammatory reaction in a rabbit ileal loop model. The aim of this work was to study the role of flagella in the virulence of aEPEC in vitro. We evaluated the aEPEC O51:H40 strain 1711-4 and its flagellin-deficient isogenic mutant in regard their ability to adhere to and invade Caco-2 and T84 intestinal cells and to elicit IL-8 production in Caco-2 monolayers.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are detailed in Table 1. Bacterial suspensions were obtained after overnight incubation in ambient air at 37°C in 5 ml of Luria-Bertani broth (LB). Strains harboring antibiotic resistance were cultivated in LB containing 50 μg/ml of kanamycin (Km), 60 μg/ml of zeocin (Zeo), and/or 100 μg/ml of apramycin (Apra).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and characteristics | Source or reference |
|---|---|---|
| Strains | ||
| 1711-4 | aEPEC O51:H40; wild type | 13 |
| 1711-4 ΔfliC | fliC::zeo (Zeor) | This study |
| C1P1-8 | 1711-4 ΔfliC carrying pZEKmfliC (Zeor Kmr) | This study |
| C1GFP | 1711-4 ΔfliC carrying pZEKmGFP (Zeor Kmr) | This study |
| MC4160 malTΔ224::zeo (F+) | Source of zeocin cassette | Gift from J. M. Ghigo |
| Plasmids | ||
| pKOBEG-Apra | Unpublished derivative (Aprar) of pKOBEG plasmid carrying the λ phage red operon | 5 |
| pZE21-MCS2 | Kmr | 22 |
| pZEKmfliC | fliC gene from 1711-4 cloned into pZE21-MCS2 | This study |
| pZEKmGFP | Green fluorescent protein-encoding gene inserted into pZE21-MCS2 | Gift from J. M. Ghigo |
Cell lines and culture.
Human colorectal adenocarcinoma epithelial cells (Caco-2) were cultivated in Dulbecco's minimal Eagle medium (DMEM) containing a source of glutamine (Glutamax DMEM; GIBCO-BRL) supplemented with 20% heat-inactivated fetal bovine serum (GIBCO-BRL) and 1% nonessential amino acids (32). Cells were incubated at 37°C with 10% CO2. Human colorectal adenocarcinoma T84 cells were cultivated in DMEM-F12 1:1 Glutamax medium (GIBCO-BRL) supplemented with 10% fetal bovine serum at 37°C in a 5% CO2-95% air atmosphere.
Cell association and invasion assays.
Monolayers were prepared by seeding six-well plates (Falcon-Becton Dickinson Labware) with 105 cells in each well and then incubated for 7 to 10 days in order to allow cellular differentiation. Adherence and invasion assays were performed as previously described, with minimal modifications (17). Briefly, each well was infected with 40 μl of an overnight static bacterial culture. In order to synchronize the infection processes, plates were immediately centrifuged for 5 min at 730 × g. After 3 h of incubation at 37°C, supernatants were collected for IL-8 dosage, and monolayers were washed three times with phosphate-buffered saline (pH 7.4) (PBS) to remove nonadherent bacteria. To evaluate adhesiveness, a group of monolayers were lysed with 1% Triton X-100. Cell-associated bacteria were counted by plating samples on LB agar plates. The number of internalized bacteria was evaluated by performing the same procedures and an additional treatment with 100 μg/ml gentamicin (Sigma) for 2 hours. Monolayers were washed three times with PBS and lysed with 1% Triton X-100, and lysates were then plated on LB agar. Cell-association indices (AX) were calculated using the equation (AB/INOC) × 100, where AB is the number of cell-associated bacteria and INOC is the number of bacteria in the initial inoculum. Invasion indices (INVX) were calculated using the equation (ICB/AB) × 100, where ICB is the number of intracellular bacteria. All tests were performed three times in triplicate.
Construction of an aEPEC 1711-4 mutant deficient in flagella.
A ΔfliC derivative of 1711-4 was constructed by the one-step allelic exchange recombination method as described earlier (5, 6, 30). Primers containing a 40-bp region homologous to the 5′ (fliCH40.zeo-5) and 3′ (fliCH40.zeo-3) extremities of the fliC gene and specific sequences for the Zeo resistance-encoding gene were used to amplify the Zeo cassette (Table 2). Takara LA Taq DNA polymerase (Takara Bio Inc.) and a PCR thermocycler system (Applied Biosystems) were used for DNA amplification. Amplicons were purified from agarose gel using the GeneClean Turbo kit (Q-Biogene) and quantified (BioPhotometer; Eppendorf) before they were electroporated into competent aEPEC 1711-4 cells containing the pKOBEG-Apra plasmid. Recombinant bacteria were selected on Zeo-containing LB agar plates. The allelic replacement of fliC by the Zeo cassette was verified by PCR using the fliCH40.ext-5 and fliCH40.ext-3 primers (Table 2).
TABLE 2.
Primers used for construction, verification, and complementation of mutations
| Function and designation | Primer sequencea |
|---|---|
| Allelic exchange | |
| fliCH40.zeo-5 | 5′-GCCCAATAACATCAAGTTGTAATTGATAAGGAAAAGATCATGGTCATCGCTTGCATTAGAAAG |
| fliCH40.zeo-3 | 5′-AACCCCGCCGGTGGCGGGGTTTGAGCGATAAGTGTAAATTAGAATGATGCAGAGATGTAAG |
| Verification | |
| fliCH40.ext-5 | 5′-ATTCAGGTGCCGATACAAGG |
| fliCH40.ext-3 | 5′-CGGCATGATTATCCGTTTCT |
| Complementation | |
| PrfliCH40F | 5′-AAATTTCTCGAGACCTAATTCCTTTTGATTGCAAAC |
| fliCH40R | 5′-AAATTTTCTAGACAGGGGTTATTTGGGGGTTA |
| fliD amplification and sequencing | |
| fliD-ext-5L | 5′-CGTCAACCCTGTTATCGTCTG |
| fliD401 | 5′-CCGCCTTGTTGAATGGTG |
| fliD333 | 5′-CACCACCAGAGACGATACGA |
| fliD1085 | 5′-AGTTTGTCGGCATCCAGTTC |
| fliD891 | 5′-GCATCTACGGCGGTGTAT |
| fliD-ext-3L | 5′-AAACAGGCTCGCTCTAACCA |
| M13F | 5′-GTAAAACGACGGCCAG |
| M13R | 5′-CAGGAAACAGCTATGAC |
Underlined bases correspond to XhoI or XbaI restriction enzyme recognition sequences.
Complementation of the 1711-4 ΔfliC mutant.
Plasmid pZE21-MCS2 (22) was digested with XbaI and XhoI, removing the polylinker and the upstream promoter. The flagellin-encoding gene fliC with its own promoter from the 1711-4 wild-type strain was cloned between these restriction sites, resulting in plasmid pZEKmfliC (Table 1). A DNA fragment was amplified using Expand high-fidelity PCR system kit (Roche Applied Science) with primers fliCH40F and fliCH40R (Table 2) and addition of 5% dimethyl sulfoxide (Sigma Aldrich). Amplification conditions were 94°C for 4 min, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 90 s and a final extension at 72°C for 7 min. The PCR product was digested with 1 U of both XbaI and XhoI restriction enzymes (Roche Applied Science), purified with the QIAquick gel extraction kit (Qiagen), ligated to the XbaI- and XhoI-digested vector with 1 U of T4 DNA ligase (Roche Applied Science), and transformed in E. coli MC1061 prior to transformation in the 1711-4 ΔfliC strain. The recombinant plasmid was named pZEKmfliC (Table 1). Motility restoration of the complemented strain was checked as described below. Plasmid pZEKmGFP (pZE21-MCS2 containing the green fluorescent protein gene) was used to transform the 1711-4 ΔfliC mutant. The transformant harboring this plasmid was designated C1GFP and was used as a mock control (Table 1).
Motility test.
E. coli strains were cultivated overnight in LB without shaking. A 2-μl sample was seeded into a thin 0.35% agar plate and incubated overnight at 37°C. Motility was evidenced by a spread of bacterial growth around the initial inoculum as evidenced by turbidity enhancement. Nonmotile strains grew only at the inoculation point.
IL-8 secretion by intestinal epithelial cell (IEC) lines.
Cell culture supernatants were collected at 3 or 24 h postinfection. Monolayers used for the evaluation of IL-8 concentration at 24 h after infection were maintained in the presence of 10 μg/ml of gentamicin. Supernatants were centrifuged for 5 min at 10,000 × g, transferred to a new vial, and stored at −80°C. IL-8 concentrations were measured using the human IL-8 Duo Set enzyme-linked immunosorbent assay development system kit (R&D Systems).
SEM.
Confluent Caco-2 monolayers were infected as described for adhesion and invasion assays. Cells were rinsed twice in 1× PBS buffer, fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2) overnight at 4°C or 30 min at room temperature, washed three times in 0.2 M cacodylate buffer for 5 min each, postfixed for 1 h in 1% (wt/vol) osmium in 0.2 M cacodylate buffer, and finally rinsed with distilled water. Samples were dehydrated through washing with a graded series of ethanol solutions (25, 50, 75, 95, and 100%) (5 min each), followed by critical-point drying with CO2 in a CPD Baltec apparatus. The dried specimens were mounted on stubs with carbon tape and ions sputtered with 15-nm gold-palladium using a Gatan 681 high-resolution ion beam coat. Analysis of the secondary electron image was performed on a JSM 6700F JEOL microscope with a field emission gun operating at 5 kV. For flagellum identification, immunogold labeling was performed with rabbit anti-E. coli flagellum H40. Three hours after infection, monolayers were rinsed with PBS, fixed with 4% formaldehyde-0.1% glutaraldehyde for 15 min, and then washed with PBS. Monolayers were incubated in 50 mM NH4Cl and then with 0.1% bovine serum albumin in PBS. Preparations were incubated with a 1:100 dilution of anti-flagellin H40 antibody in PBS overnight at 4°C and washed with PBS before addition of 10-nm gold bead-coupled suspension. Unbounded beads were removed by rinsing with PBS before monolayers were processed for scanning electron microscopy (SEM) as described above, except that they were sputtered with a 15-nm layer of carbon and observed with a backscattered electron detector.
Transmission electron microscopy.
Confluent Caco-2 cells were infected as described for adhesion and invasion assays. Preparations were rinsed in 1× PBS buffer, fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2), and kept overnight at 4°C. After being rinsed with 0.1 M cacodylate buffer, preparations were postfixed in 1% OsO4, dehydrated through a series of graded ethanol solutions and propylene oxide, and embedded in araldite resin. Blocks were polymerized at 60°C for 48 h. Ultrathin sections were stained with 2.0% uranyl acetate aqueous solution and 2.5% lead citrate and examined under a transmission electron microscope (LEO 906E; Zeiss) at 80 kV.
Sequencing of the fliD gene.
The fliD gene was amplified with primers fliD-ext-5L, fliD401 fliD333, fliD1085, fliD89, and fliD-ext-3L (Table 2). A total of 5 μl of genomic DNA suspension (30 μg/ml) was added to 45 μl of a PCR mixture containing 50 mM KCl, 20 mM Tris-HCl (pH 8.4), 2.5 mM MgCl2, 200 μM each 2′-deoxynucleoside 5′-triphosphate, 1 μM each primer, and 1.0 U of Platinum Taq DNA polymerase (Invitrogen). The PCR mixture was heated at 95°C for 1 min and then subjected to 35 cycles of denaturation at 94°C for 30s, 55°C for 30 s, and 72°C for 60 s, with a final single step of 72°C for 5 min. PCR products were separated by 1.5% agarose gel electrophoresis, bands where cut from the gel, and DNA was extracted and ligated into pCR-XL-TOPO vector (Invitrogen). Cloning reaction products were used to transform chemically competent TOP10 E. coli (Invitrogen). Transformants were selected on LB agar containing 50 μg/ml of Km. Inserts were amplified and sequenced with primers M13F and M13R (Invitrogen). Amplicons were purified with GFX PCR DNA and a Gel Band purification kit (GE) and sequenced in an ABI PRISM 3100 sequencer with a BigDye Terminator cycle sequencing kit (Applied Biosystems). Sequences were assembled with DNA Baser program version 2.75 (Heracle Software) and then compared with those deposited in the GenBank database using BLAST (Basic Local Alignment Tool) (http://www.ncbi.nlm.nih.gov/BLAST).
Statistical analysis.
The Shapiro-Wilk test was applied to determine if variables fit in a normal distribution. The Kruskal-Wallis test and Bonferroni error protection were applied to verify the statistical significance of differences in AX and INVX among strains. Analysis of variance (ANOVA) and Tukey's post hoc test were applied to evaluate the differences between IL-8 results. Data were reported as mean ± standard deviation (SD) or median and interquartile range (IQR). A statistical significance of 5% was applied (α = 0.05). All tests were performed with Analyze-it for Microsoft Excel version 2.07 (Analyze-it Software, Ltd.).
Nucleotide sequence accession number.
The GenBank database accession number generated in this study is FJ598057 (fliD).
RESULTS
Flagella play a role in adhesion of aEPEC 1711-4 to IEC monolayers.
The ability of aEPEC 1711-4 to adhere to IECs was investigated. The median (IQR) AX, which represents the percentage of inoculated bacteria that adhered to and/or invaded eukaryotic cells until 3 hours after infection, was significantly higher in T84 (4.7% [4.1 to 5.8%]) than in Caco-2 (0.3% [0.2 to 1.1%]) monolayers (P < 0.001 [main value and Bonferroni contrast result]). To determine whether flagella played a role in such adhesion, an isogenic aEPEC 1711-4 mutant deficient in flagella (1711-4 ΔfliC) was constructed. Lack of flagellum expression in this mutant was confirmed by SEM (Fig. 1C) and by motility testing in semisolid agar (Fig. 2B). In contrast to the results obtained with the wild-type aEPEC 1711-4 strain, no statistically significant difference was observed when comparing the median (IQR) AXs of 1711-4 ΔfliC in Caco-2 (0.6% [0.4 to 0.8%]) and T84 (0.8% [0.4 to 1.3%]) monolayers (P = 1.00). An increase in the median AX was observed in the interaction of the fliC-deficient mutant with Caco-2 monolayers compared to the wild-type strain. This difference was not statistically significant (P = 1.00). In contrast, the 1711-4 ΔfliC mutant showed a significant reduction in median AX in T84 monolayers (P = 0.003) compared to the wild-type strain, suggesting a role of flagella in the adhesion of this aEPEC strain to this cell lineage.
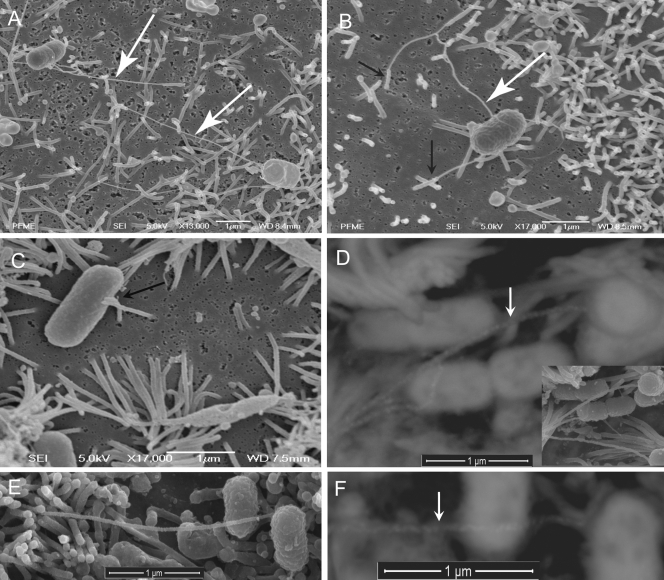
FIG. 1.
Interaction of aEPEC wild-type 1711-4, the FliC-deficient mutant, or the C1P1-8 complemented mutant with Caco-2 monolayers. (A and B) SEM of monolayers infected with wild-type aEPEC 1711-4, evidencing interaction between the flagella (white arrows) and microvillus (black arrows). Note interaction of the microvillus with the tips of flagella. The microvillus also interacts with the bacterial cell body. (C) SEM showing interaction of isogenic mutant 1711-4 ΔfliC with Caco-2 cells. Note the absence of flagella and interaction of the microvillus tip with the bacterial cell body (black arrow). (D) SEM with backscattered electron detection of an immunogold preparation with rabbit anti-E. coli H40 serum. Caco-2 cells were infected with wild-type aEPEC 1711-4. Note labeled flagella (white arrow). On the lower right is a secondary electron image of the same microscopic field. (E) SEM showing interaction of the C1P1-8 complemented isogenic mutant with Caco-2 cells. (F) SEM with backscattered electron detection of an immunogold preparation with rabbit anti-E. coli H40 serum. Caco-2 cells were infected with the complemented mutant C1P1-8. Note labeled flagella (white arrow).
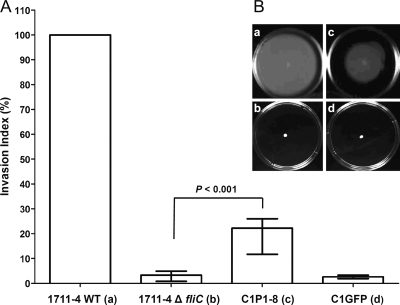
FIG. 2.
Intact flagella are required for efficient invasion of Caco-2 cells. (A) Relative median INVX (error bars represent IQRs) of aEPEC wild-type (WT) 1711-4, the fliC-deficient mutant 1711-4 ΔfliC, and isogenic deletion strains complemented with pZEKmfliC (C1P1-8) or pZEKmGFP (C1GFP). The wild-type strain INVX was assumed to be 100%. See text for exact values. (B) Motility test in nutrient agar with 0.35% agarose. Note the absence of motility in strains 1711-4 ΔfliC (b) and C1GFP (d) and restoration of motility in strain C1P1-8 (c).
As the median AXs of the 1711-4 ΔfliC mutant strain were similar in both cell lineages used, SEM of infected monolayers was performed only with the Caco-2 cells. Monolayers infected with the wild-type aEPEC 1711-4 strain evidenced direct contact between the bacterial cell body and the microvillus tip (Fig. 1A and B), forming a microvillus cluster (Fig. 1B). Of note, SEM also evidenced interaction of the wild-type aEPEC 1711-4 flagellum cap and the tip of the microvillus (Fig. 1A and B). In SEM immunogold assays with rabbit anti-E. coli H40 serum, these structures were clearly labeled, confirming that they are in fact flagella (Fig. 1D). In contrast, only direct interaction of the microvillus tip with the bacterial cell body was observed with the fliC-deficient mutant (Fig. 1C).
The flagellum-deficient aEPEC mutant 1711-4 has a decreased ability to invade IECs.
Since wild-type aEPEC 1711-4 adhered to IECs, the capacity of this strain to invade Caco-2 and T84 cell monolayers was investigated using the gentamicin protection assay. Both cell lineages were invaded by aEPEC 1711-4. With the wild-type strain, the median (IQR) INVX obtained in assays with T84 cells (10.7% [9.9 to 15.4%]) was similar to that observed with Caco-2 cells (15.1% [9.9 to 20.8%]) (P = 1.00). When the 1711-4 ΔfliC strain was tested, the lowest median (IQR) INVX (0.08% [0.06 to 0.14%]) was observed with Caco-2 cells. Compared to the median INVX of the wild-type strain, this difference was statistically significant (P < 0.001). The median (IQR) INVX of the 1711-4 ΔfliC strain (2.2% [1.9 to 4.8%]) in assays with T84 cells was significantly lower than that observed for the wild-type strain (P = 0.02). These data strongly evidenced a role of flagella in internalization of the wild-type 1711-4 strain into both Caco-2 and T84 cells.
Complementation of the 1711-4 ΔfliC mutant for flagellin production partially restores its capacity to invade Caco-2 cells.
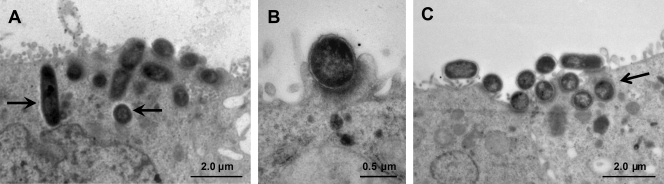
Since the most clear significant reduction in the invasion capacity of the 1711-4 ΔfliC mutant was observed in assays with Caco-2 monolayers, subsequent investigations were performed only with this cell lineage. The 1711-4 ΔfliC mutant was complemented with a plasmid carrying the complete fliC gene (coding for flagellin H40) or a mock plasmid and then compared to the wild-type strain. Motility was restored in the 1711-4 ΔfliC mutant complemented with fliC (strain C1P1-8) (Fig. 2B) but not in the same strain containing the mock plasmid (strain C1GFP). Transmission electron microscopy analysis of monolayers infected with wild-type aEPEC 1711-4 or C1P1-8 confirmed that both strains invaded Caco-2 cells (Fig. 3). Although the intracellular localization of the 1711-4 ΔfliC mutant was detected with the gentamicin protection assay, this strain could not be visualized within Caco-2 cells, probably due to the low number of intracellular bacteria (Fig. 3).
FIG. 3.
Transmission electron microscopy of Caco-2 cells infected with wild-type aEPEC 1711-4 (A), 1711-4 ΔfliC (B), or C1P1-8 (C). Both the wild type and the complemented mutant (C1P1-8) were observed in the intracellular compartment (A and C, arrows). The fliC-deficient mutant was not detected in the intracellular compartment. Note that both the fliC mutant and the complemented strain retain the ability of the parental strain to produce AE lesions as revealed by pedestal formation (B).
In gentamicin protection assays, the complemented strain C1P1-8 showed a partial but statistically significant restoration of the ability to invade Caco-2 monolayers, since the median (IQR) INVX was 5.8% (3.1 to 6.8%) (P < 0.001), while the strain containing the mock plasmid (C1GFP) showed no increase in median (IQR) INVX (0.7% [0.5 to 0.9%]) (P = 1.00) compared to strain 1711-4 ΔfliC. Assuming the median INVX obtained with the wild-type strain to be 100%, the median (IQR) INVX obtained with 1711-4 ΔfliC, C1P1-8, and C1GFP strains would be 3% (0.8 to 4.9%), 22% (11.7 to 26.0%), and 2% (1.9 to 3.3%), respectively (Fig. 2A).
The fliC-deficient mutant has a reduced ability to stimulate IL-8 production only in the early stage of infection of Caco-2 cells.
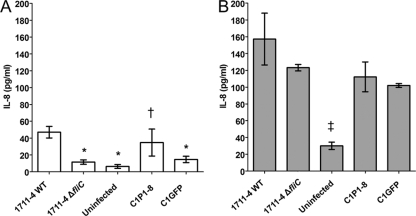
In order to evaluate the role of flagella in the stimulation of IL-8 production during infection of Caco-2 cells, the concentration of this inflammatory mediator in supernatants was determined at 3 and 24 h after infection with wild-type aEPEC 1711-4, 1711-4 ΔfliC, C1P1-8, or C1GFP. Three hours after infection, a significant decrease (P < 0.001; 95% confidence interval [CI] = 17.9 to 52.7 pg/ml) in the mean (± SD) IL-8 production was noted with the fliC-deficient mutant (12 ± 2.6 pg/ml) compared to the wild-type strain (47 ± 6.9 pg/ml) (Fig. 4). Complementation of this mutant with a plasmid encoding FliC partially restored its ability to stimulate IL-8 production (35 ± 16.1 pg/ml), and this difference was statistically significant (95% CI = 4.3 to 41.8 pg/ml). In contrast, no significant increase (95% CI = −22.2 to 15.4 pg/ml) was observed when strain C1GFP (fliC-deficient mutant complemented with a mock plasmid) was tested (15 ± 3.8 pg/ml). Twenty-four hours after infection, all strains induced average levels of IL-8 that were 2 to 5 times higher than that observed in uninfected monolayers (30 ± 4.5 pg/ml). These differences were statistically significant (P < 0.001). However, there was no statistically significant difference (95% CI = −1.3 to 69.6 pg/ml) between average IL-8 levels induced by the wild-type strain (157 ± 30.9 pg/ml) and 1711-4 ΔfliC (123 ± 4.0 pg/ml). Although strains C1P1-8 and C1GFP induced average IL-8 levels that were 29% and 35%, respectively, lower than that induced by the wild-type strain, these differences were not statistically significant. In fact, the average induced IL-8 levels of strains C1P1-8 and C1GFP were, respectively, 112 pg/ml (95% CI = 47.1 to 118.0 pg/ml) and 102 pg/ml (95% CI = 36.8 to 107.7 pg/ml). These levels were significantly higher than that observed in uninfected monolayers. These data suggest a predominantly flagellin-independent stimulation of IL-8 production at late stages after infection (24 h).
FIG. 4.
IL-8 concentration in supernatants of Caco-2 cell cultures infected with wild-type (WT) aEPEC 1711-4, its isogenic fliC mutant, and the complemented strains at 3 h (A) and 24 h (B) after infection or with no infection. Data are expressed as the mean ± SD from triplicate experiments. *, statistically significant difference compared to the WT strain 1711-4 (P < 0.001 by ANOVA). †, statistically significant difference compared to the 1711-4 ΔfliC strain (95% CI, 4.3 to 41.8 pg/ml). ‡, statistically significant difference compared to each infected monolayer (P < 0.001 by ANOVA).
The FliD proteins from aEPEC 1711-4 and the nonpathogenic strain E. coli K-12 share the same amino acid sequence.
When comparing the nucleotide sequence of the fliD gene from aEPEC strain 1711-4 to those available at GenBank, the highest similarity index, 99.6% (1401/1407), was obtained with sequences pertaining to E. coli K-12 (accession numbers CP000948.1, AP009048.1, and U00096.2). Their deduced amino acid sequences were identical.
DISCUSSION
In this study we showed that aEPEC 1711-4 (serotype O51: H40) was able to adhere in vitro to two different enterocyte lineages (Caco-2 and T84 cells). Interestingly, this strain adhered more efficiently to T84 cells than to Caco-2 cells. In contrast, Eaves-Pyles et al. (8) showed that an adherent invasive E. coli O83:H1 strain adhered better in Caco-2 cells than in T84 cells. As suggested by Girón et al. (12), this difference could be due to antigenic heterogeneity and, consequently, distinct adhesive properties of flagella and/or to the presence of other adherence factors.
Girón et al. (12) also demonstrated that H2 and H6 flagella purified from tEPEC but not H7 flagella purified from STEC (O157:H7) bound to HeLa cells. It has also been demonstrated that flagella of Salmonella enterica (7), Yersinia enterocolitica (48), E. coli strains pathogenic for birds (20), and adherent invasive E. coli strains (8) are involved in enterocyte adhesion in vitro. Likewise, we showed that flagella of aEPEC 1711-4 contribute to bacterial adhesion to IECs as evidenced by the lower AX of the aEPEC 1711-4 ΔfliC mutant in T84 monolayers. In addition, our SEM images suggest that interaction of aEPEC 1711-4 flagella occurred preferentially between the flagellum cap and the microvillus tip (Fig. 1A and B). Considering the small surface area of both the flagellum cap and microvillus tip compared to the entire Caco-2 cell surface, there is probably a strong affinity between the two structures. These findings are in accordance with those previously reported by Tasteyre et al. (44), who demonstrated that radiolabeled cultured Vero cells bound to a high degree to recombinant FliD and weakly to crude flagellar preparations or recombinant FliC from Clostridium difficile. The flagellum cap protein, FliD, is essential for flagellum assembly, and its central portion, which forms a lid, is exposed at the tip of the flagellum (23, 47). Mutants deficient in FliC, as is the case in our work, are expected to express the flagellar hook with the FliD protein at its end exposed at the bacterial wall. This may explain our SEM findings evidencing the microvillus tip in close contact with the fliC-deficient derivative cell body in a topography that could correspond to the flagellar hook. Although our SEM data could suggest that FliD rather than FliC would play a role in the interaction of aEPEC 1711-4 with IECs, the finding that the FliD protein produced by this strain has the same amino acid sequence found in E. coli K-12, a nonvirulent strain, does not support this hypothesis. We cannot exclude the possibility that adhesion of the flagellar cap to the microvilli tip could also be mediated by an adhesin secreted by a two-partner secretion pathway, as is the case for EtpA, found in enterotoxigenic E. coli (38). EtpA is plasmid mediated and to date has been described only in enterotoxigenic E. coli, although no aEPEC strains have been evaluated regarding the presence of this protein (9).
In this study we also demonstrated that the aEPEC 1711-4 ΔfliC mutant invaded IECs significantly less than the wild-type aEPEC strain. Anchoring mediated by flagella probably is also required for efficient enterocyte invasion by aEPEC 1711-4, since addition of fliC to the fliC-deficient mutant partially restored its invasion capability. This partial restoration is most probably due to low efficiency of fliC expression when located in the plasmid under the control of its own promoter and not to fliC overexpression due to plasmid copy number, since wild-type strains and their derivatives have the same growth rate (data not shown). These findings thus suggest that wild-type aEPEC 1711-4 flagella are necessary for efficient adhesion to and invasion of Caco-2 and T84 cells. Likewise, flagella were shown to be necessary for both association and invasion of human brain microvascular endothelial cells by a meningitis-associated E. coli O18:K1:H7 strain (29). Similarly, the ability of E. coli O83:H1 to adhere to and invade Caco-2BBe or T84 monolayers was shown to be dependent on these structures (8). Flagella have also been involved in the internalization of uropathogenic E. coli into collecting duct cells in vitro (30).
In this work we found that the aEPEC 1711-4 ΔfliC mutant induced lower levels of IL-8 production in Caco-2 cell culture supernatants than the wild-type strain at the earlier stage of infection (3 hours). These levels were partially restored in the 1711-4 ΔfliC mutant complemented with fliC (C1P1-8 strain) but not in the same mutant complemented with the mock plasmid (C1GFP strain). However, the fliC mutant still induced IL-8 levels at least 2 times higher than that of uninfected monolayers at 24 h postinfection. Therefore, wild-type aEPEC 1711-4 may stimulate IL-8 production in a flagellin-dependent manner at an earlier stage of infection and by a flagellin-independent pathway at a later stage of infection (24 h). However, when interpreting results from experimental procedures that evaluate the IL-8 concentration in cells cultivated in vitro, one might consider which portion of the cell monolayer was directly exposed to the bacterial suspension. Gewirtz et al. (11) demonstrated that TLR5, the main receptor for flagellin, is expressed on the basolateral rather than the apical surface of polarized T84 cells. Moreover, they showed that flagellin added at the apical cell surface achieves TLR5 receptors after translocation through the cytoplasm. Although to our knowledge there is no report in the literature on the topology of TLR5 in Caco-2 cells, Ruchaud-Sparagano et al. (39) demonstrated that the apical infection of Caco-2 cells by tEPEC strain E2348/69 induced IL-8 secretion to a lesser extent than the basolateral infection. IL-8 concentrations reported by these authors at 2, 3, 4, and 6 hours of infection with flagellated tEPEC are similar to those that we obtained with wild-type aEPEC 1711-4 at 3 hours after infection. Our findings of similar IL-8 levels in Caco-2 cells infected with the wild-type strain or fliC-deficient mutant after 24 h of infection could be due to the interaction of eukaryotic cell membrane receptor TLR4 and the bacterial lipopolysaccharide. Huang et al. (16) described that IL-8 levels in Caco-2 culture supernatants stimulated with lipopolysaccharide were shown to be irrelevant at 3 h but significant after 24 h of infection, with mean levels of 75 pg/ml. However, the data of that study may not be comparable to ours, since those authors used 14- to 15-day-old Caco-2 cultures while we used 7- to 10-day-old cultures. Caco-2 monolayers are known to achieve confluence on day 6 and stationary phase on day 9 after subculture (31). Since we obtained higher mean IL-8 levels (102 to 157 pg/ml) and our experiments were performed with 7- to 10-day-old monolayers, we cannot exclude the possibility that other factors may have stimulated IL-8 secretion. One such factor could be DNA CpG from dead bacteria inside late endosomes, recognized by TLR3 or TLR9 (18, 43). As these receptors are able to identify diverse chemical structures, another possible explanation of our findings could be that an effector protein, secreted by wild-type aEPEC 1711-4 in the intracellular compartment, would interact with TLR3 or TLR9 and stimulate IL-8 production also independently of flagella.
The data presented in this study strongly suggest a role for flagella in both adhesion to and invasion of aEPEC 1711-4 into IECs and in early but not late stimulation of IL-8 production after Caco-2 cell infection. Even though differentiated Caco-2 cells are often used as a model for human small intestinal cells, the in vivo significance of our findings remains to be established. In the complex intestinal environment, flagella could contribute not only to traverse the mucus layer and reach the epithelial surface but also in the early stages of interaction with enterocytes, acting as an anchoring device. Moreover, flagella could contribute in vivo to entrance into enterocytes, favoring aEPEC escape from the immune response.
Acknowledgments
S. C. F. Sampaio is supported by a scholarship from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and received a grant from the Colégio Doutoral Franco Brasileiro during her stay in France. C. Pichon is supported by Programme Transversal de Recherche grant PTR165 from Institut Pasteur. Work in the laboratory of T. A. T. Gomes was supported by a grant from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP; grant no. 05/59128-0) and Programa de Apoio a Núcleos de Excelência, PRONEX MCT/CNPq/FAPERJ.
We thank Jean-Marc Ghigo (Unité de Génétique des Biofilms, Departement de Microbiologie, Institut Pasteur) for providing strains and plasmids. We thank E. F. Haapalainen and R. S. Coimbra (Centro de Microscopia Eletrônica, UNIFESP) and M. Brumatti (Departamento de Minas e Petróleo, Universidade de São Paulo) for assistance with SEM immunogold preparations.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 20 July 2009.
REFERENCES
- 1.Afset, J. E., K. Bergh, and L. Bevanger. 2003. High prevalence of atypical enteropathogenic Escherichia coli (EPEC) in Norwegian children with diarrhoea. J. Med. Microbiol. 52:1015-1019. [DOI] [PubMed] [Google Scholar]
- 2.Allen-Vercoe, E., and M. J. Woodward. 1999. The role of flagella, but not fimbriae, in the adherence of Salmonella enterica serotype Enteritidis to chick gut explant. J. Med. Microbiol. 48:771-780. [DOI] [PubMed] [Google Scholar]
- 3.Araujo, J. M., G. F. Tabarelli, K. R. Aranda, S. H. Fabbricotti, U. Fagundes-Neto, C. M. Mendes, and I. C. Scaletsky. 2007. Typical enteroaggregative and atypical enteropathogenic types of Escherichia coli are the most prevalent diarrhea-associated pathotypes among Brazilian children. J. Clin. Microbiol. 45:3396-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attridge, S. R., and D. Rowley. 1983. The role of the flagellum in the adherence of Vibrio cholerae. J. Infect. Dis. 147:864-872. [DOI] [PubMed] [Google Scholar]
- 5.Chaveroche, M. K., J. M. Ghigo, and C. d'Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dibb-Fuller, M. P., E. Allen-Vercoe, C. J. Thorns, and M. J. Woodward. 1999. Fimbriae- and flagella-mediated association with and invasion of cultured epithelial cells by Salmonella enteritidis. Microbiology 145:1023-1031. [DOI] [PubMed] [Google Scholar]
- 8.Eaves-Pyles, T., C. A. Allen, J. Taormina, A. Swidsinski, C. B. Tutt, G. Eric Jezek, M. Islas-Islas, and A. G. Torres. 2008. Escherichia coli isolated from a Crohn's disease patient adheres, invades, and induces inflammatory responses in polarized intestinal epithelial cells. Int. J. Med. Microbiol. 298:397-409. [DOI] [PubMed] [Google Scholar]
- 9.Fleckenstein, J. M., K. Roy, J. F. Fischer, and M. Burkitt. 2006. Identification of a two-partner secretion locus of enterotoxigenic Escherichia coli. Infect. Immun. 74:2245-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franzolin, M. R., R. C. Alves, R. Keller, T. A. Gomes, L. Beutin, M. L. Barreto, C. Milroy, A. Strina, H. Ribeiro, and L. R. Trabulsi. 2005. Prevalence of diarrheagenic Escherichia coli in children with diarrhea in Salvador, Bahia, Brazil. Mem. Inst. Oswaldo Cruz 100:359-363. [DOI] [PubMed] [Google Scholar]
- 11.Gewirtz, A. T., T. A. Navas, S. Lyons, P. J. Godowski, and J. L. Madara. 2001. Bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J. Immunol. 167:1882-1885. [DOI] [PubMed] [Google Scholar]
- 12.Giron, J. A., A. G. Torres, E. Freer, and J. B. Kaper. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44:361-379. [DOI] [PubMed] [Google Scholar]
- 13.Gomes, T. A. T., K. Irino, D. M. Girao, V. B. Girao, B. E. Guth, T. M. Vaz, F. C. Moreira, S. H. Chinarelli, and M. A. Vieira. 2004. Emerging enteropathogenic Escherichia coli strains? Emerg. Infect. Dis. 10:1851-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomes, T. A. T., V. Rassi, K. L. MacDonald, S. R. Ramos, L. R. Trabulsi, M. A. Vieira, B. E. Guth, J. A. Candeias, C. Ivey, M. R. Toledo, et al. 1991. Enteropathogens associated with acute diarrheal disease in urban infants in Sao Paulo, Brazil. J. Infect. Dis. 164:331-337. [DOI] [PubMed] [Google Scholar]
- 15.Hernandes, R. T., R. M. Silva, S. M. Carneiro, F. A. Salvador, M. C. Fernandes, A. C. Padovan, D. Yamamoto, R. A. Mortara, W. P. Elias, M. R. da Silva Briones, and T. A. Gomes. 2008. The localized adherence pattern of an atypical enteropathogenic Escherichia coli is mediated by intimin omicron and unexpectedly promotes HeLa cell invasion. Cell. Microbiol. 10:415-425. [DOI] [PubMed] [Google Scholar]
- 16.Huang, Y., N. Li, K. Liboni, and J. Neu. 2003. Glutamine decreases lipopolysaccharide-induced IL-8 production in Caco-2 cells through a non-NF-kappaB p50 mechanism. Cytokine 22:77-83. [DOI] [PubMed] [Google Scholar]
- 17.Jouve, M., M. I. Garcia, P. Courcoux, A. Labigne, P. Gounon, and C. Le Bouguenec. 1997. Adhesion to and invasion of HeLa cells by pathogenic Escherichia coli carrying the afa-3 gene cluster are mediated by the AfaE and AfaD proteins, respectively. Infect. Immun. 65:4082-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kajita, E., T. Nishiya, and S. Miwa. 2006. The transmembrane domain directs TLR9 to intracellular compartments that contain TLR3. Biochem. Biophys. Res. Commun. 343:578-584. [DOI] [PubMed] [Google Scholar]
- 19.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 20.La Ragione, R. M., A. R. Sayers, and M. J. Woodward. 2000. The role of fimbriae and flagella in the colonization, invasion and persistence of Escherichia coli O78:K80 in the day-old-chick model. Epidemiol. Infect. 124:351-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luck, S. N., L. Badea, V. Bennett-Wood, R. Robins-Browne, and E. L. Hartland. 2006. Contribution of FliC to epithelial cell invasion by enterohemorrhagic Escherichia coli O113:H21. Infect. Immun. 74:6999-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lutz, R., and H. Bujard. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Majander, K., T. K. Korhonen, and B. Westerlund-Wikstrom. 2005. Simultaneous display of multiple foreign peptides in the FliD capping and FliC filament proteins of the Escherichia coli flagellum. Appl. Environ. Microbiol. 71:4263-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moreira, F. C., M. A. Vieira, A. J. Ferreira, D. M. Girao, T. M. Vaz, A. C. Rosa, T. Knobl, K. Irino, E. Freymuller, and T. A. Gomes. 2008. Escherichia coli strains of serotype O51:H40 comprise typical and atypical enteropathogenic E. coli strains and are potentially diarrheagenic. J. Clin. Microbiol. 46:1462-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morooka, T., A. Umeda, and K. Amako. 1985. Motility as an intestinal colonization factor for Campylobacter jejuni. J. Gen. Microbiol. 131:1973-1980. [DOI] [PubMed] [Google Scholar]
- 27.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen, R. N., L. S. Taylor, M. Tauschek, and R. M. Robins-Browne. 2006. Atypical enteropathogenic Escherichia coli infection and prolonged diarrhea in children. Emerg. Infect. Dis. 12:597-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parthasarathy, G., Y. Yao, and K. S. Kim. 2007. Flagella promote Escherichia coli K1 association with and invasion of human brain microvascular endothelial cells. Infect. Immun. 75:2937-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pichon, C., C. Hechard, L. du Merle, C. Chaudray, I. Bonne, S. Guadagnini, A. Vandewalle, and C. Le Bouguenec. 2009. Uropathogenic Escherichia coli AL511 requires flagellum to enter renal collecting duct cells. Cell. Microbiol. 11:616-628. [DOI] [PubMed] [Google Scholar]
- 31.Pinto, M., S. Robine-Leon, M. Appay, M. Kedinger, N. Triadou, E. Dussalx, B. Lacroix, P. Simon-Assmann, K. Haffen, J. Fogh, and A. Zweibaum. 1983. Enterocyte-like differentiation and polarization of the human colon carcinoma cell line Caco-2 in culture. Biol. Cell 47:323-330. [Google Scholar]
- 32.Plancon, L., L. Du Merle, S. Le Friec, P. Gounon, M. Jouve, J. Guignot, A. Servin, and C. Le Bouguenec. 2003. Recognition of the cellular beta1-chain integrin by the bacterial AfaD invasin is implicated in the internalization of afa-expressing pathogenic Escherichia coli strains. Cell. Microbiol. 5:681-693. [DOI] [PubMed] [Google Scholar]
- 33.Regua-Mangia, A. H., T. A. T. Gomes, M. A. M. Vieira, J. R. Andrade, K. Irino, and L. M. Teixeira. 2004. Frequency and characteristics of diarrhoeagenic Escherichia coli strains isolated from children with and without diarrhoea in Rio de Janeiro, Brazil. J. Infect. 48:161-167. [DOI] [PubMed] [Google Scholar]
- 34.Robins-Browne, R. M., A. M. Bordun, M. Tauschek, V. R. Bennett-Wood, J. Russell, F. Oppedisano, N. A. Lister, K. A. Bettelheim, C. K. Fairley, M. I. Sinclair, and M. E. Hellard. 2004. Escherichia coli and community-acquired gastroenteritis, Melbourne, Australia. Emerg. Infect. Dis. 10:1797-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogers, T. J., A. W. Paton, S. R. McColl, and J. C. Paton. 2003. Enhanced CXC chemokine responses of human colonic epithelial cells to locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli. Infect. Immun. 71:5623-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosa, A. C., A. T. Mariano, A. M. Pereira, A. Tibana, T. A. T. Gomes, and J. R. Andrade. 1998. Enteropathogenicity markers in Escherichia coli isolated from infants with acute diarrhoea and healthy controls in Rio de Janeiro, Brazil. J. Med. Microbiol. 47:781-790. [DOI] [PubMed] [Google Scholar]
- 37.Rosa, A. C., M. A. Vieira, A. Tibana, T. A. T. Gomes, and J. R. Andrade. 2001. Interactions of Escherichia coli strains of non-EPEC serogroups that carry eae and lack the EAF and stx gene sequences with undifferentiated and differentiated intestinal human Caco-2 cells. FEMS Microbiol. Lett. 200:117-122. [DOI] [PubMed] [Google Scholar]
- 38.Roy, K., G. M. Hilliard, D. J. Hamilton, J. Luo, M. M. Ostmann, and J. M. Fleckenstein. 2009. Enterotoxigenic Escherichia coli EtpA mediates adhesion between flagella and host cells. Nature 457:594-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruchaud-Sparagano, M. H., M. Maresca, and B. Kenny. 2007. Enteropathogenic Escherichia coli (EPEC) inactivate innate immune responses prior to compromising epithelial barrier function. Cell. Microbiol. 9:1909-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scaletsky, I. C., M. Z. Pedroso, and U. Fagundes-Neto. 1996. Attaching and effacing enteropathogenic Escherichia coli O18ab invades epithelial cells and causes persistent diarrhea. Infect. Immun. 64:4876-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma, R., S. Tesfay, F. L. Tomson, R. P. Kanteti, V. K. Viswanathan, and G. Hecht. 2006. Balance of bacterial pro- and anti-inflammatory mediators dictates net effect of enteropathogenic Escherichia coli on intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 290:G685-G694. [DOI] [PubMed] [Google Scholar]
- 42.Steiner, T. S., J. P. Nataro, C. E. Poteet-Smith, J. A. Smith, and R. L. Guerrant. 2000. Enteroaggregative Escherichia coli expresses a novel flagellin that causes IL-8 release from intestinal epithelial cells. J. Clin. Investig. 105:1769-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeda, K., T. Kaisho, and S. Akira. 2003. Toll-like receptors. Annu. Rev. Immunol. 21:335-376. [DOI] [PubMed] [Google Scholar]
- 44.Tasteyre, A., M. C. Barc, A. Collignon, H. Boureau, and T. Karjalainen. 2001. Role of FliC and FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect. Immun. 69:7937-7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trabulsi, L. R., R. Keller, and T. A. T. Gomes. 2002. Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8:508-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vieira, M. A. M., J. R. Andrade, L. R. Trabulsi, A. C. Rosa, A. M. Dias, S. R. Ramos, G. Frankel, and T. A. T. Gomes. 2001. Phenotypic and genotypic characteristics of Escherichia coli strains of non-enteropathogenic E. coli (EPEC) serogroups that carry EAE and lack the EPEC adherence factor and Shiga toxin DNA probe sequences. J. Infect. Dis. 183:762-772. [DOI] [PubMed] [Google Scholar]
- 47.Yonekura, K., S. Maki, D. G. Morgan, D. J. DeRosier, F. Vonderviszt, K. Imada, and K. Namba. 2000. The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science 290:2148-2152. [DOI] [PubMed] [Google Scholar]
- 48.Young, G. M., J. L. Badger, and V. L. Miller. 2000. Motility is required to initiate host cell invasion by Yersinia enterocolitica. Infect. Immun. 68:4323-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou, X., J. A. Giron, A. G. Torres, J. A. Crawford, E. Negrete, S. N. Vogel, and J. B. Kaper. 2003. Flagellin of enteropathogenic Escherichia coli stimulates interleukin-8 production in T84 cells. Infect. Immun. 71:2120-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]