Abstract
Chronic infection with the gram-negative organism Pseudomonas aeruginosa is a leading cause of morbidity and mortality in human patients, despite high doses of antibiotics used to treat the various diseases this organism causes. These infections are chronic because P. aeruginosa readily forms biofilms, which are inherently resistant to antibiotics as well as the host's immune system. Our laboratory has been investigating specific mutations in P. aeruginosa that regulate biofilm bacterial susceptibility to the host. To continue our investigation of the role of genetics in bacterial biofilm host resistance, we examined P. aeruginosa biofilms that lack the flgK gene. This mutant lacks flagella, which results in defects in early biofilm development (up to 36 h). For these experiments, the flgK-disrupted strain and the parental strain (PA14) were used in a modified version of the 96-well plate microtiter assay. Biofilms were challenged with freshly isolated human leukocytes for 4 to 6 h and viable bacteria enumerated by CFU. Subsequent to the challenge, both mononuclear cells (monocytes and lymphocytes) and neutrophils, along with tumor necrosis factor alpha (TNF-α), were required for optimal killing of the flgK biofilm bacteria. We identified a cytokine cross talk network between mononuclear cells and neutrophils that was essential to the production of lactoferrin and bacterial killing. Our data suggest that TNF-α is secreted from mononuclear cells, causing neutrophil activation, resulting in the secretion of bactericidal concentrations of lactoferrin. These results extend previous studies of the importance of lactoferrin in the innate immune defense against bacterial biofilms.
Through the ability to form biofilms, bacteria change their susceptibility to antibiotics and the host's immune system, contributing to chronic infections in humans. It's estimated that biofilm infections cost $6.5 billion annually to treat (2). Moreover, most biofilm communities utilize genetically regulated processes to limit the effectiveness of a broad range of antimicrobial agents, likely resulting in even greater health care costs (54). Thus, the development of therapy that protects the host, or augments the clearance of biofilm infections, is vital to improved patient health and to reducing the cost burden of these infections. However, the immune response to biofilm infections is poorly characterized, and these mechanisms must be elucidated to direct the development of biofilm-specific therapies.
Although much of the current understanding of host-pathogen interactions came from the study of planktonic bacteria, it is now clear that bacteria in the environment, including the human environment, live more often as communities of microorganisms (biofilms) than as single-cell suspensions (12, 44, 47). Bacteria within a biofilm differ from their planktonic counterparts in a number of ways, including an inherent resistance to antimicrobial agents and to clearance by the human immune system (23, 35, 36, 40, 44, 48, 54). We recently confirmed a direct link between Pseudomonas aeruginosa biofilm genetics and resistance to components of the human innate immune system (35). During this time, other papers have been published supporting our idea that P. aeruginosa and biofilms in general manipulate the immune system to their advantage (41, 55, 56). Most recently, O'Toole and colleagues demonstrated that P. aeruginosa biofilms growing on airway epithelial cells exhibited decreased virulence in the presence of the antibiotic tobramycin (1).
The most classic example of Pseudomonas aeruginosa biofilm infections in humans are those associated with the lungs of cystic fibrosis (CF) patients, which correlate with increased morbidity and mortality (11, 13-15, 21, 22, 32, 33, 39, 53). Over three decades ago, Costerton and others demonstrated that lung material from CF patients contained P. aeruginosa likely in the form of biofilms (25, 33). More recently, Singh et al. showed that biofilms of P. aeruginosa existed in the CF lung (53). Collectively, these papers underscore the importance of biofilm formation in vivo and document the inability of the host's immune system to clear the infections.
Besides infections in the lungs of CF patients, P. aeruginosa also wreaks havoc in patients with corneal infections, burn wounds, implanted medical devices, and human immunodeficiency virus (46). Increased morbidity and mortality in these infections are linked to the ability of P. aeruginosa to form a biofilm (44, 45, 55). Schaber and colleagues (50) demonstrated the formation of biofilms on a mouse burn model after just 8 h postinoculation. In a 3-year study in Poland, P. aeruginosa was the most abundant organism isolated from burn wounds (5). Additionally, P. aeruginosa biofilm presence in chronic wounds is a likely culprit for the longevity of these infections by keeping the wound in a chronic state of inflammation (7, 31). These studies highlight the ubiquitous nature of this opportunistic pathogen and suggest a much larger role of P. aeruginosa biofilms in many chronic infections in humans.
Alginate protects biofilm bacteria against macrophage phagocytosis (35). Besides the physical process of phagocytosis and the killing of bacteria by macrophages and neutrophils, a number of other antibacterial effector molecules are important in host defense. Singh and colleagues have shown that lactoferrin (LTF) prevents biofilm development (52). Over a decade ago, superoxide production by neutrophils was shown to be decreased in biofilm versus planktonic P. aeruginosa (26, 27). Other oxygen-dependent (nitric oxide [NO]) and oxygen-independent (lysozyme and myeloperoxidase [MPO]) responses are also reduced in magnitude or function in response to P. aeruginosa biofilms (9, 29, 30). These effector molecules are important in host defense, yet their effect on biofilm virulence or development, especially how they relate mechanistically, remains understudied. Nonetheless, it is now clear that genetic regulation within the biofilm bacteria plays a large role in resistance to the human host.
In Salmonella enterica serovar Typhimurium and Escherichia coli, mutations in the flgK gene result in a nonmotile phenotype, as flgK is a vital gene involved in flagellar synthesis (24). This functional defect is also observed in P. aeruginosa. The lack of flagella in P. aeruginosa decreases the magnitude of early biofilm formation but does not prevent it (45). This defect is overcome with time as mutants lacking the flgK gene phenotypically approach wild-type biofilm levels after ∼36 h of growth. To determine if this genetic mutation affected biofilm susceptibility to components of the innate immune system, we grew biofilms of PA14 and isogenic flgK, challenged them with freshly isolated human leukocytes, and measured biofilm bacterial survival as previously described (35). In the absence of functional flagella, P. aeruginosa biofilm bacteria were killed by human leukocytes. To further dissect the interactions of the host cells with the biofilms, we developed a novel transwell biofilm killing assay that allowed for the separation of specific leukocyte populations (mononuclear cells from neutrophils) and the analysis of antimicrobial products such as superoxide, MPO, and LTF. Our data demonstrate that cytokine cross talk, as well as the production of components of the innate immune system, is important in generating mechanistic responses that in this case were able to kill biofilm bacteria that lacked intact flagella.
MATERIALS AND METHODS
Bacterial strains.
P. aeruginosa wild-type strain PA14 and a mutant of this strain lacking the flgK gene, PA14 flgK::Tn5B30(Tc) (hereby referred to as flgK throughout this paper), were used in this study. The PA14 strain was grown on Luria-Bertani (LB) agar and in LB broth for the experiments, and the flgK strain was grown on LB agar containing 10 μg/ml of tetracycline for positive selection of the bacteria carrying this mutation. The number of viable biofilm bacteria was enumerated by CFU on LB agar for both strains. To further demonstrate that the biofilm-specific killing effect was due to the absence of functional flagella, three other strains, carrying mutations in flgD, fliC, and fliF, were also tested. These mutant strains were obtained from the nonredundant P. aeruginosa PA14 mutant library developed by Liberati and colleagues (38).
Reagents.
Recombinant human gamma interferon (IFN-γ; 100 μg) was purchased from Southern Biotech (Birmingham, AL), and recombinant human tumor necrosis factor alpha (TNF-α; 50 μg) was purchased from Millipore (Billerica, MA). The cytokines were resuspended in 1× phosphate-buffered saline and distilled H2O, respectively. Human LTF (5 mg) was purchased from Sigma-Aldrich (St. Louis, MO). Hanks balanced salt solution (HBSS), 1 M HEPES buffer, and bacterial growth medium were purchased from Fisher Scientific (Pittsburgh, PA).
Growth of Pseudomonas aeruginosa.
P. aeruginosa wild-type PA14 and the flgK mutant were cryogenically stored, and aliquots were used for all experiments. PA14 and flgK were streaked on LB agar alone and LB agar with 10 μg/ml tetracycline, respectively, for 24 h at 37°C. Single colonies of each were used to inoculate 10 ml of sterile LB broth, which was incubated at 37°C under constant agitation overnight. After sufficient growth, the culture was diluted 1:100 in fresh LB broth and incubated for 3 h, until the culture reached the midpoint of exponential growth.
Static biofilm growth in microtiter plates.
Biofilms were grown in 96-well polyvinyl chloride (PVC) plates (Falcon) or in 24-well polystyrene plates (Falcon) as previously described (35). The 3-h cultures described above were further diluted 1:50 in fresh LB medium, and 100 μl of the inoculated medium was added to a sterile 96-well PVC plate or 400 μl was added to a 24-well polystyrene plate, depending on the assay. The plates were covered and incubated at 37°C with no agitation for 24 h. These biofilms were used for leukocyte killing assays as described below.
Isolation of human peripheral blood leukocytes.
Human leukocyte isolation was performed as previously described (35). Healthy human donors were used as sources for peripheral blood leukocytes after reading and signing donor consent forms (protocol approved by the NAU Human Subjects IRB). Human blood was collected into acid citrate Vacutainer tubes (Becton, Dickinson and Company, Franklin, NJ) and centrifuged for 12 min at 500 × g, and the peripheral blood leukocytes and autologous plasma were collected. The remaining erythrocytes in the buffy coat were lysed by hypotonic treatment, followed by rapid dilution in HBSS. The purified leukocytes were counted and resuspended in an appropriate volume of HBSS and 50% autologous plasma to yield a final leukocyte concentration of 5 × 107 cells/ml. The critical neutrophil concentration was previously determined (37); however, we designed our studies so that the concentration of human peripheral leukocytes was fivefold greater because we used a mixed concentration of leukocytes and because biofilms have been shown to be inherently resistant to leukocyte killing factors (35).
In order to determine human leukocyte cytokine cross talk, subpopulations of leukocytes were fractionated by use of a density gradient as previously described (35). Briefly, 10 to 15 ml of freshly collected human blood was diluted 1:2 with HBSS, underlaid first with 10 ml of 1.077 g/ml Histopaque (Sigma-Aldrich), and then 10 ml of 1.119 g/ml Histopaque (Sigma-Aldrich) in a sterile 50-ml conical. The tubes were centrifuged at 700 × g for 30 min, and the resulting 1.077 and 1.119 buffy coats containing mononuclear cells and neutrophils, respectively, were collected into separate, sterile 50-ml conicals. If there were remaining erythrocytes, they were lysed by hypotonic treatment for 10 s and rapidly diluted with HBSS to a final volume of 50 ml. The purified subpopulations of leukocytes were counted and resuspended in an appropriate volume of HBSS and pooled human serum to yield 1 × 107 cells/ml. The cells were used in a transwell biofilm killing assay, as described below. For all biofilm killing assays, 10 mM of HEPES buffer was added to maintain a constant pH throughout the assay.
Biofilm killing assay.
Biofilm challenge experiments were performed as previously described (35). To determine whether the flgK strain was more susceptible to human leukocyte killing than PA14, the biofilms were incubated with an appropriate concentration of a mixed population or purified subpopulations of leukocytes and IFN-γ at a final concentration of 50 units/ml for 4 h at 37°C. Prior to challenge by leukocytes, microtiter plates containing biofilms were inverted and the nonadherent cells were washed away. Following this, 100 μl of the mixed leukocyte population or the respective subpopulations was added in triplicate to PA14 and flgK biofilms. Control treatments included challenge with LB medium and HBSS-50% autologous plasma. Both of the control treatments were also added at a volume of 100 μl in triplicate wells. After the 4-h challenge, the PVC plates containing biofilms were inverted, gently sonicated in a Microhorn sonicator (Misonix, Farmingdale, NY), and serially diluted 10-fold in sterile deionized water. Serial dilutions were plated on LB agar in triplicate 20-μl spots and allowed to grow for 12 to 18 h at 37°C. CFU were counted and reported as CFU/ml. To determine whether TNF-α played a role in the activation of neutrophils, it was added at a final concentration of 125 pg/ml into the wells and biofilm killing was quantified by CFU/ml as described above. An analysis by trypan blue exclusion showed that leukocytes associated with biofilm bacteria did not undergo extensive cell death by this protocol (data not shown).
Transwell biofilm killing assay.
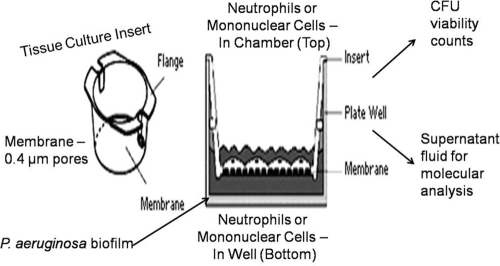
To establish the killing mechanism(s), we developed a novel transwell assay where the specific leukocyte subpopulations could be separated from one another and the supernatant fluid assayed for the presence of bioactive molecules. Biofilms were grown in 24-well polystyrene plates (Falcon) for 24 h at 37°C. Following incubation, the plates were inverted and nonadherent cells washed away. Isolated subpopulations of leukocytes (mononuclear cells and neutrophils) were obtained via the double Histopaque procedure described above. Tissue culture inserts with a pore size of 0.4 μm were placed into each well containing biofilm bacteria of the 24-well plate. The purified leukocyte subpopulations were kept separate through the use of the tissue culture inserts. Each treatment consisted of alternating subpopulations of leukocytes, at a volume of 200 μl each for 400 μl total volume, either on the top of the membrane or directly onto the biofilm bacteria (e.g., mononuclear cells on top of the membrane, neutrophils directly on the biofilm). For clarity, we have included a cartoon schematic of this experimental design in Fig. 1. Recombinant IFN-γ (50 U/ml) was added to each well to induce macrophage activation. To determine if TNF-α played a role in neutrophil activation, recombinant TNF-α was also added to some treatments at a final concentration of 125 pg/ml. Control treatments included LB medium and HBSS-50% pooled human serum. Biofilm bacteria were quantified as described above, and the number of CFU/ml was reported. In order to verify that cytokine cross talk played a role in bacterial biofilm killing, the leukocytes were treated with 1 μg/ml brefeldin A (Sigma-Aldrich), which inhibits secretory pathways in eukaryotes, including cytokine release. Briefly, the leukocytes were added to the biofilms in the presence of brefeldin A, the biofilm was challenged for 6 h, viable bacteria were serially diluted, and the CFU were counted and reported.
FIG. 1.
Schematic cartoon of the transwell biofilm killing assay. Note that cells of the innate immune system can be added either to the top chamber of the insert with the artificial membrane (no biofilm physical contact) or to the bottom chamber (physical biofilm contact) and that, because of the pore size of the inserts (0.4 μm), transmigration is extensively limited. Supernatant fluid can then be analyzed for components of innate immunity that are known for their bactericidal activity.
NO, LTF, and MPO quantification assays.
To determine which reactive moiety(ies) played a role in biofilm bacterial killing, assays were employed for various known neutrophil antibacterial compounds. NO production was quantified using a Griess reagent kit (Promega), following the manufacturer's protocol. Briefly, supernatant fluid was obtained from the biofilm treatment after 6 h of incubation and centrifuged at maximum speed for 10 min to remove leukocytes and bacterial cells. Samples were incubated with Griess reagent for 20 min and read on an Opsys MR microplate spectrophotometer (Dynex Technologies) at 550 nm. Unknown samples were compared to a standard curve. LTF and MPO production were quantified using enzyme-linked immunosorbent assay (ELISA) kits obtained from Calbiochem. The assays were run per the manufacturer's protocol. Briefly, supernatant fluid was obtained from the tissue culture insert and the 24-well plate and centrifuged at maximum speed for 10 min. The supernatant was diluted if necessary and added to the wells of the 96-well plate containing monoclonal antibodies. After incubation, the wells were washed five times with wash buffer, and a secondary antibody-horseradish peroxidase conjugate was added and then incubated for 15 min at 37°C. The wells were washed again, and substrate was added. Color change was measured on a microplate spectrophotometer (Dynex Technologies) at 405 nm (MPO) and 420 nm (LTF). Unknown samples were compared to a standard curve, and the experimental concentration of LTF or MPO was calculated.
LTF killing assay.
To determine if LTF alone was responsible for the killing of flgK biofilm bacteria, recombinant human LTF was added to the biofilm in the absence of leukocytes. The optimal concentration of LTF for biofilm killing was determined by adding 100 μl LTF at various concentrations. Control treatments included LB medium and sterile deionized H2O containing no LTF. After 6 h of incubation, viable bacteria were enumerated by CFU. After determining the optimal concentration for bacterial biofilm killing, the effects of recombinant LTF were compared to those of natural LTF. Briefly, mixed population leukocytes were isolated, and 100 μl was placed into triplicate wells containing biofilm bacteria. Also, 100 μl recombinant LTF at 1 μg/ml was added in triplicate to the biofilm. Control treatments included LB medium and 50% HBSS with 50% autologous plasma. The biofilms were challenged for 6 h at 37°C, and viable bacteria were serially diluted and the CFU counted as described above.
Statistical analysis.
An analysis was performed using JMP IN statistical software. The results are presented as means ± standard errors of the means and were analyzed using Student's t test. A P value of <0.05 was considered statistically significant.
RESULTS
Effect of human leukocytes on PA14 and flgK mutant P. aeruginosa biofilm bacteria.
P. aeruginosa biofilm bacteria that lack flagella are killed when challenged by whole-blood leukocytes, and optimal killing occurs in the presence of exogenous IFN-γ (Fig. 2). After 4 h of challenge, greater than a log reduction in viable biofilm bacteria was observed by CFU enumeration. In contrast, even in the presence of IFN-γ, biofilm bacteria from the parental strain (PA14) were not killed. The killing of flgK biofilm bacteria was significant compared to both LB treatment (positive control for growth) and HBSS plus 50% autologous plasma (leukocyte medium control showing nutrient limitation of the medium and killing activity of any bactericidal components in the human plasma). Challenging the biofilms of either the parental strain or mutant for up to 24 h did not increase the number of bacteria killed under any experimental setting (4; data not shown). Additional studies for which biofilms of PA14 and flgK were grown for 8 h, and challenged by freshly isolated human leukocytes as described above, demonstrated that the flgK biofilm bacteria were more susceptible to leukocyte killing, even at this early time point in biofilm development (data not shown). Finally, planktonic PA14 and flgK biofilm bacteria were challenged by freshly isolated human leukocytes and no differences in the amount of killed bacteria were observed (data not shown). Collectively, these studies suggest that the susceptibility of flgK bacteria to human leukocytes is a biofilm-specific paradigm. However, by the approaches we have taken here, we cannot rule out subtle impacts that the flagellar mutation may have on biofilm physiology, which may have an effect on the immune response to this flagellar negative population.
FIG. 2.
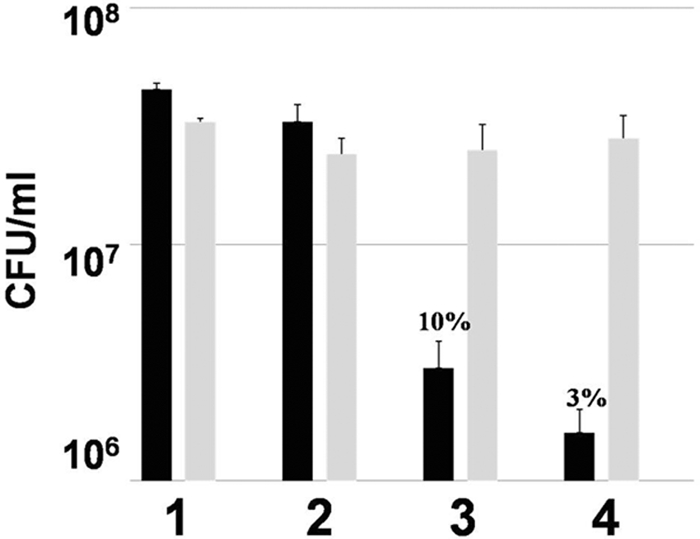
P. aeruginosa biofilm bacteria lacking flagella are killed by human leukocytes, whereas the parental strain PA14 biofilm bacteria are not. Graphic representation of CFU/ml of flgK (black bars) and PA14 (gray bars) biofilm bacteria after treatment with medium control (LB) (1), HBSS plus 50% autologous human plasma (2), human leukocytes in HBSS containing 50% autologous plasma (3), and leukocytes in HBSS containing 50% autologous plasma plus IFN-γ (50 U/ml) (4). Percent survival was calculated by normalizing the LB medium control to equal 100% survival and is listed above the appropriate treatment. *, P < 0.001 for the leukocytes plus IFN-γ treatment compared to all other treatments. Data are representative of the results from three separate experiments with variable blood donors.
Killing of flgK biofilm bacteria requires both mononuclear cells, neutrophils, and cytokines.
To determine if there was a leukocyte phenotype that was responsible for the killing of the flgK biofilm bacteria, mononuclear cells (primarily monocytes and lymphocytes) and neutrophils were separated by density gradient centrifugation. PA14 and flgK biofilm bacteria were then challenged for 4 h and viable bacteria enumerated by CFU. Subsequent to the challenge, we observed moderate killing in the presence of mononuclear cells (Fig. 3, bar 3) that was not statistically significant. Conversely, challenge by neutrophils alone resulted in an approximately 1-log reduction of viable biofilm bacteria (Fig. 3, bar 4). However, optimal killing was observed when mononuclear cells, neutrophils, and exogenous TNF-α were utilized to challenge the flgK biofilm bacteria (Fig. 3, bar 8), although neutrophils and TNF-α also induced a strong biofilm killing effect. A previous report by a group in Denmark demonstrated that P. aeruginosa, with an intact quorum-sensing system, rapidly killed human neutrophils by rhamnolipid secretion (6, 28). To determine if this occurred in our assay system, the biofilm and neutrophils were gently sonicated, the resuspended nonattached population of bacteria and cells pipetted onto a hemocytometer, and neutrophil viability tested by trypan blue exclusion. In our assay system, neutrophil viability remained >90% after 4 to 6 h of biofilm challenge (data not shown).
FIG. 3.
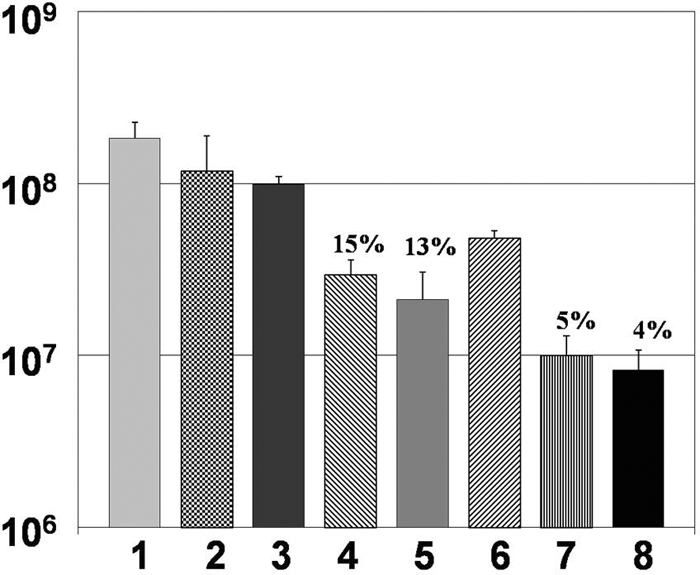
Neutrophils, in the presence of exogenous TNF-α, are primarily responsible for the killing of flgK biofilm bacteria. Graphic representation of CFU/ml of flgK biofilm bacteria after challenge by specific leukocyte subpopulations. 1, LB treatment; 2, HBSS plus 50% autologous plasma; 3, mononuclear cells; 4, neutrophils; 5, mononuclear cells plus neutrophils; 6, mononuclear cells plus TNF-α (125 pg/ml); 7, neutrophils plus TNF-α; 8, mononuclear cells, neutrophils, and TNF-α. Percent survival was calculated by normalizing the LB medium control to equal 100% survival and is listed above the appropriate treatment. *, P < 0.01 for bars 4 to 8 compared to all other treatments. Data are representative of the results from three separate experiments with variable blood donors.
Optimal killing of biofilm bacteria was dependent upon the presence of functional flagella.
To confirm the specific role of flagella in the protection of biofilms from leukocyte killing, we tested the following three additional strains carrying mutations that disrupt flagellar synthesis: the flgD, fliC, and fliF strains (38, 45). The biofilms of these three additional strains, plus the parental strain PA14, were grown and challenged by leukocytes in the presence of recombinant TNF-α and the viable bacteria determined as described above. All three of these mutants were susceptible to human leukocyte killing (Fig. 4). At least a log reduction in viable bacteria was observed for all three mutants, and the fliF mutant demonstrated hypersusceptibility to killing under these conditions (Fig. 4, bar 3). These data confirm that the absence of flagella mediates biofilm bacterial susceptibility to components of the human innate immune system. Because all of these genes are within a large operon, complementation studies would have been difficult and were not attempted. However, because the flgD gene is in the same operon as flgK, and the fliC and fliF mutations are in a distinct flagellar operon, the aggregate analysis of these mutant strains has effectively allowed us to eliminate potential issues confounding the interpretation of our findings, including secondary mutations and polarity. We are currently attempting to decipher why the fliF mutant exhibited hypersusceptibility to components of the innate immune system.
FIG. 4.
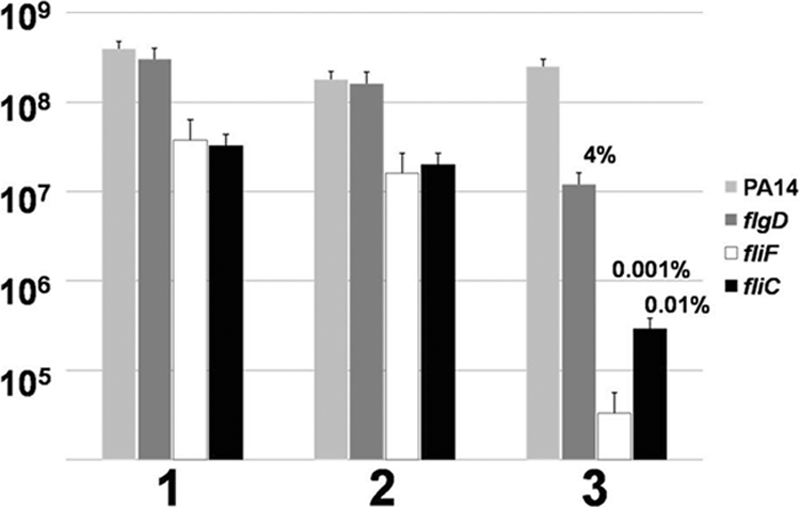
Three additional mutations in the flagellar operon lead to biofilm bacterial susceptibility. Graphic representation of CFU/ml of biofilm bacterial viability of the parental strain PA14 and mutations in flgD, fliC, and fliF after challenge by human leukocytes. 1, LB treatment; 2, HBSS plus 50% autologous plasma; 3, total leukocytes plus TNF-α. Percent survival was calculated by normalizing the LB medium control to equal 100% survival and is listed above the appropriate treatment. *, P < 0.001 for the leukocytes plus TNF-α treatment compared to all other treatments. Data are representative of the results from two separate experiments with variable blood donors.
Cytokine secretion by leukocytes is requisite for the killing of flgK biofilm bacteria.
Since the data suggested that both IFN-γ and TNF-α were important cytokines that lead to the killing of flgK biofilm bacteria, we tested whether the ability to secrete cytokines was requisite for the killing of biofilm bacteria. For these studies, the flgK biofilm bacteria were challenged with whole-blood leukocytes (mononuclear cells and neutrophils) in the presence and absence of brefeldin A, which blocks the secretion of cytokines from human leukocytes (10). As Fig. 5 shows, in the presence of brefeldin A, no biofilm bacterial killing was observed.
FIG. 5.
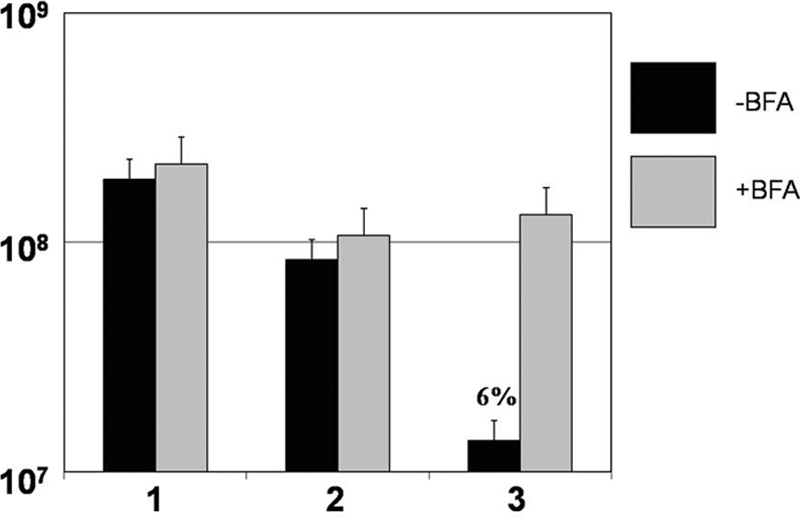
Brefeldin A, which prevents the secretion of cytokines, blocked the killing of flgK biofilm bacteria. Graphic representation of CFU/ml of flgK biofilm bacteria in the presence (gray) or absence (black) of brefeldin A. 1, LB treatment; 2, HBSS plus 50% autologous plasma; 3, leukocytes plus IFN-γ (50 U/ml). Percent survival was calculated by normalizing the LB medium control to equal 100% survival and is listed above the appropriate treatment. *, P < 0.001 for the leukocytes plus IFN-γ treatment compared to all other treatments. Data are representative of the results from three separate experiments with variable blood donors.
Cytokine cross talk between mononuclear cells and neutrophils is necessary for the killing of flgK biofilm bacteria.
Cytokine cascades occur in response to many types of stimuli, and cross talk often occurs between cells so that the effector functions of those cells are enhanced. In order to decipher if cytokine cross talk between mononuclear cells and neutrophils was required for optimal biofilm bacterial killing, we developed a novel assay that employed tissue culture transwell inserts. This assay was described in detail in Materials and Methods (Fig. 1). Briefly, by utilizing culture inserts with a pore size of 0.4 μm, we were able to physically isolate the specific leukocyte populations so that only proteins (such as cytokines) and other small molecules could diffuse through the membrane and act upon whatever cell population was present on the other side of the membrane. By utilizing this approach, we demonstrated that the maximum killing of flgK biofilm bacteria occurred when mononuclear cells were in the tissue culture insert (top), without access to the bacterial biofilm itself, and neutrophils were in direct contact with the biofilm (Fig. 6, bar 6). All combinations of leukocyte subsets and cytokines were assayed and only the most relevant combinations are shown in Fig. 6. Additionally, ELISAs were utilized to determine that TNF-α was produced by the mononuclear cell fraction and diffused across the membrane into the bottom chamber (data not shown). The difference in killing activity between the combination in lanes 5 and 6 may be attributed to subtle differences in the activity of recombinant human TNF-α versus TNF-α naturally produced from peripheral blood mononuclear cells, although this was not directly tested in our study.
FIG. 6.
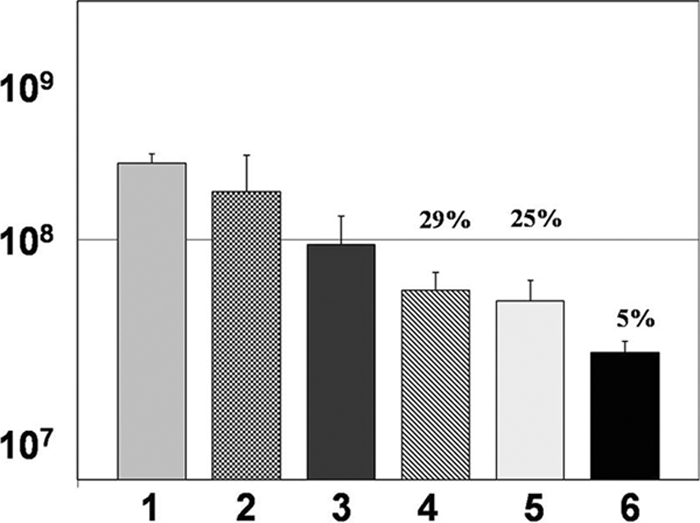
Cytokine cross talk between mononuclear cells and neutrophils leads to the killing of flgK biofilm bacteria by neutrophils. Graphic representation of CFU/ml of flgK biofilm bacteria after challenge by specific leukocyte subpopulations. 1, LB treatment; 2, HBSS plus 50% autologous plasma; 3, mononuclear cells and neutrophils plus TNF-α in the top chamber; 4, mononuclear cells and neutrophils plus IFN-γ and TNF-α in the bottom chamber; 5, mononuclear cells plus IFN-γ in the top chamber and neutrophils and TNF-α in the bottom chamber; 6, mononuclear cells plus IFN-γ in the top chamber and neutrophils on the bottom. Percent survival was calculated by normalizing the LB medium control to equal 100% survival and is listed above the appropriate treatment. *, P < 0.001 for bar 6 compared to all other treatments. Data are representative of the results from three separate experiments with variable blood donors. Refer to Materials and Methods for the details of the transwell assay.
Neutrophil LTF is induced by TNF-α and leads to the killing of flgK biofilm bacteria.
One of the other important aspects of the novel biofilm killing assay described above is that it allows for the evaluation of effector molecules from leukocytes, such as MPO, NO, and LTF. Utilizing this approach, we assayed for the presence of these innate effector compounds by standard ELISA. Interestingly, we did not observe any significant difference in the production of MPO and NO compared to wells where leukocytes were incubated in the absence of bacteria. However, we did see dramatic release of the iron-binding protein lactoferrin (Fig. 7). The largest amount of LTF was observed by ELISA in the experimental design where the optimal killing was observed (Fig. 6, bar 6), when mononuclear cells were in the tissue culture insert, in the presence of IFN-γ, and neutrophils were in direct contact with the biofilm (Fig. 7, bar 6). Endogenous LTF levels from neutrophils incubated alone, in the absence of biofilm bacteria, were ∼80 ng/ml (data not shown). The main differences between lanes 5 and 6 in this figure could be attributable to subtle activity differences between recombinant and native TNF-α. These results complemented the findings of Singh and colleagues where they demonstrated that LTF inhibited P. aeruginosa biofilm development but did not result in bacterial killing (52).
FIG. 7.
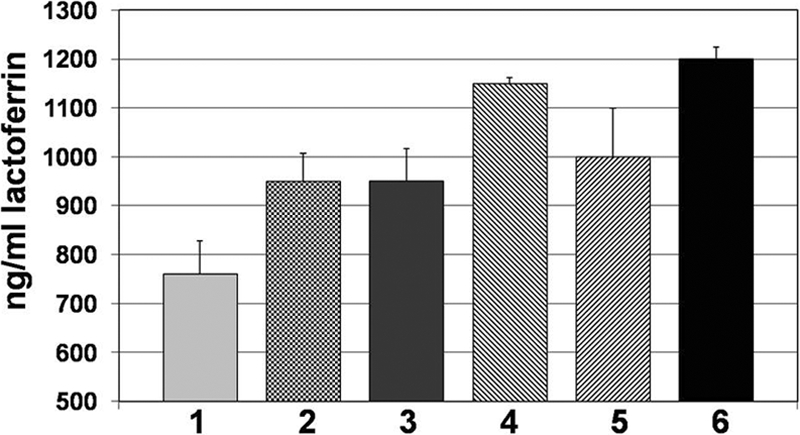
LTF, a bacteriostatic and bactericidal component of the innate immune system, is produced in response to P. aeruginosa biofilms and is associated with biofilm bacterial killing. Graphic representation of LTF (ng/ml) produced during challenge of flgK by specific leukocyte subpopulations. 1, mononuclear cells and neutrophils plus IFN-γ and TNF-α in the top chamber; 2, neutrophils plus IFN-γ in the top chamber; 3, neutrophils plus TNF-α in the top chamber; 4, mononuclear cells plus IFN-γ in the top chamber and neutrophils plus TNF-α in the bottom chamber; 5, mononuclear cells and neutrophils plus IFN-γ and TNF-α in the bottom chamber; 6, mononuclear cells plus IFN-γ in the top chamber and neutrophils in the bottom chamber. Data are representative of the results from three separate experiments with variable blood donors. Refer to Materials and Methods for the details of the transwell assay.
Purified LTF kills flgK biofilm bacteria in a dose-dependent manner.
In order to determine if the bactericidal effect of LTF was dose-dependent, we challenged flgK biofilm bacteria with increasing concentrations of recombinant human LTF. Although minimal killing was observed at 100 and 500 ng/ml of LTF, significant killing was observed when 1,000 ng/ml was used to challenge the biofilm bacteria (Fig. 8, bar 6). These data demonstrated that the concentration of LTF was important in its ability to act as a bactericidal compound. As a control for these studies, mononuclear cells, neutrophils and IFN-γ were used to challenge the flgK biofilm bacteria, which as reported above and here, also resulted in biofilm bacterial killing (Fig. 8, bar 7). The amount of LTF produced by neutrophils in these assays averaged ∼900 ng/ml in these studies (Fig. 7 and data not shown). Collectively, these data demonstrated that LTF was responsible for killing flgK biofilm bacteria and that the killing concentration of LTF was well within the physiological range of the amount produced by human neutrophils (see Discussion below).
FIG. 8.
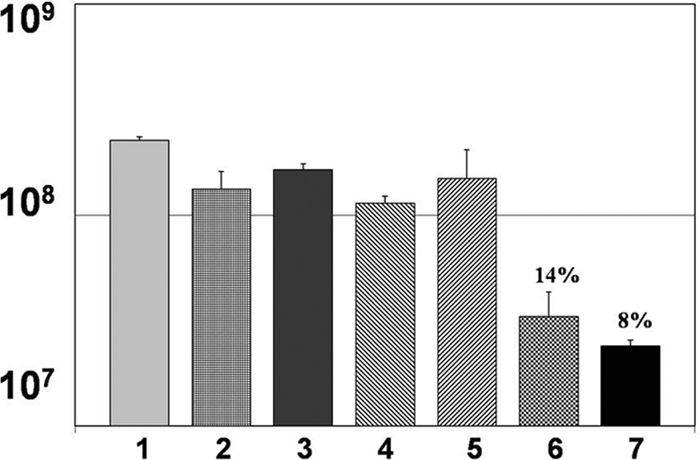
Addition of purified, exogenous LTF alone results in the killing of flgK biofilm bacteria. Graphic representation of CFU/ml of flgK biofilm bacteria after challenge by specific concentrations of purified, recombinant human LTF. 1, LB treatment; 2, water (solvent for purified LTF); 3, 50 ng/ml LTF; 4, 100 ng/ml LTF; 5, 500 ng/ml LTF; 6, 1,000 ng/ml LTF; 7, mononuclear cells and neutrophils plus IFN-γ (measured at ∼900 ng/ml LTF). Percent survival was calculated by normalizing the LB medium control to equal 100% survival and is listed above the appropriate treatment. Data are representative of the results from three separate experiments with variable blood donors.
LTF is responsible for killing flgK, but not PA14, biofilm bacteria.
Finally, we wanted to determine the extent of LTF-mediated killing of the flgK biofilm bacteria versus the parental strain, PA14. For these studies, biofilms were grown in 96-well plates and challenged with mononuclear cells, neutrophils, and IFN-γ for 4 h, and viable bacteria were enumerated by CFU. As expected, the flgK biofilm bacteria were killed by recombinant human LTF (1,000 ng/ml [Fig. 9, bar 3]) and by LTF produced by the neutrophils (Fig. 9, bar 4). However, the parental biofilm formed by strain PA14 was resistant to both the purified LTF and challenge by human leukocytes (Fig. 9). Although a reduction in viable parental biofilm bacteria was observed, this effect was not specific to LTF activity as there was no difference between LTF challenge and exposure to HBSS-50% autologous plasma. Collectively, these data represent an in-depth analysis of the innate immune response to biofilm bacteria and identify another genetic basis for biofilm resistance to human-host components.
FIG. 9.
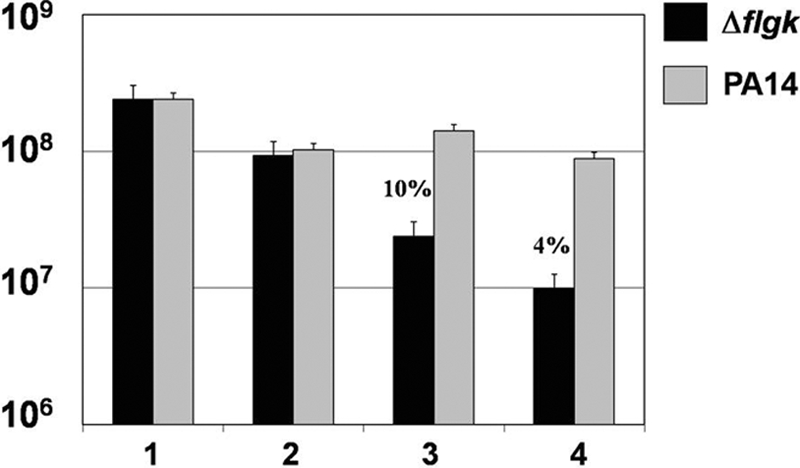
LTF is bactericidal against flgK biofilm bacteria but not that of the parental strain PA14. Graphic representation of CFU/ml of PA14 (gray) and flgK (black) biofilm bacteria after challenge by purified LTF and human leukocytes. 1, LB treatment; 2, HBSS plus 50% autologous plasma; 3, 1,000 ng/ml purified LTF; 4, mononuclear cells and neutrophils plus IFN-γ. Percent survival was calculated by normalizing the LB medium control to equal 100% survival and is listed above the appropriate treatment. Data are representative of the results from three separate experiments with variable blood donors.
DISCUSSION
Previously, we have characterized genetically mediated resistance of P. aeruginosa biofilm bacteria to human leukocytes (35). Here we have extended those studies to demonstrate that the flgK gene in P. aeruginosa is important for protection against innate immune cells and their effector products (LTF). In the presence of IFN-γ, whole-blood leukocytes were able to kill P. aeruginosa biofilm bacteria that lacked this gene. The killing of flgK biofilm bacteria required the presence of mononuclear cells, the secretion of TNF-α from this leukocyte population, and the resultant production of bactericidal concentrations of LTF from neutrophils. These data complement the observations of Singh and colleagues (52) on LTF preventing P. aeruginosa biofilm development, while showing that physiological concentrations of LTF can be bactericidal, and document the importance of cytokine cross talk between immune cells to produce antibiofilm compounds.
As with our previous study, the presence of specific cytokines was important as brefeldin A, which blocks the secretion of cytokines, absolved the killing of the flgK biofilm bacteria. Cytokine cross talk between the cells of the immune system has been known for some time (e.g., the Th1 versus Th2 paradigm) and is often vital to the generation of appropriate and protective immune responses. For this study, the release of the cytokine TNF-α by mononuclear cells was essential to neutrophil-mediated killing through the secretion of bactericidal concentrations of LTF.
During biofilm development, the presence of flagella in P. aeruginosa is tightly regulated (48, 49). As planktonic cells encounter a surface, exhibiting swarming and twitching motility, these bacteria contain functional flagella. Once firm attachment has taken place, in as little as 6 h, flagella are dramatically downregulated from the surface of the attached bacteria. However, as the biofilm life cycle continues, and mature biofilm clusters proceed to individual dispersed cells, flagella are again upregulated (48, 49). Importantly, these detached cells exhibit similar antibiotic resistance as those in the parental biofilm, and this developmental paradigm does not depend upon growth inside the CF lung (19). The targeting of the flagellum-negative population of P. aeruginosa biofilm with exogenous LTF, or by the immunomodulation of cytokines, may improve the host's ability to kill these organisms. Although the parental biofilm was not killed by physiological concentrations of purified, recombinant LTF, it may be that the induction of this defense in vivo, in the presence of fully activated immune cells, would lead to improved biofilm clearance. Biofilm populations of P. aeruginosa are heterogeneous and some do not express flagella (34, 57). By combining a targeted, adjuvant-based approach to enhance the appropriate immune response to these cells, it may be possible to reduce the bacterial load in P. aeruginosa biofilm infections of indwelling medical devices, burn wounds, chronic wounds, and potentially the lungs of CF patients.
LTF is a nonheme iron-binding glycoprotein that has both bacteriostatic and bactericidal activity (17). In a previous study, Singh and colleagues demonstrated that LTF, at relatively low concentrations, successfully prevented P. aeruginosa biofilm formation in vitro but was not bactericidal to the microorganisms (52). However, our studies on preformed P. aeruginosa biofilms demonstrated that concentrations 20 times less than the optimal subinhibitory concentration listed in their paper killed biofilm bacteria lacking intact flagella. Importantly, we characterized the amount of LTF produced by purified human neutrophils in our assays and confirmed that these physiological concentrations were responsible for biofilm bacterial killing by the addition of recombinant human LTF. At physiologically relevant killing levels (∼900 ng/ml), purified LTF resulted in an approximately 1-log reduction in viable bacteria. Presumably, as published reports have shown, LTF was active against the biofilm bacteria by binding to lipopolysaccharide causing disruption of the bacterial membrane (3, 17, 43). We are currently characterizing the effect of LTF on P. aeruginosa biofilm bacteria.
Inflammation, induced by either bacteria or other insults, causes the release of cytokines. One early inflammatory cytokine is IFN-γ, and its main cellular sources are T cells, NK cells, and monocytes/macrophages (8). T cells are the most notable secretors of IFN-γ when they encounter antigen-presenting cells and major histocompatibility complex class II. However, macrophages and NK cells secrete IFN-γ in the absence of costimulation from other immune cells. If present, neutrophils can respond to IFN-γ in a number of ways, including by the secretion of LTF from specific granules (16, 18). For the studies described here, we believe that macrophages are activated directly, potentially by the interaction of TLR4 with lipopolysaccharide or by activation with exogenous IFN-γ, and secrete TNF-α, which in turn induces neutrophils to secrete LTF, subsequently killing biofilm bacteria. Although not directly related to bacterial infection, a previous report demonstrated that lung macrophages were able to respond to external stimuli, resulting in cytokine secretion, which directly leads to secretion of the azurophilic component elastase (42).
Bacterial biofilms of Pseudomonas aeruginosa are the common cause of chronic infections in CF patients as well as other patients who exhibit some form of immune system suppression (e.g., burn wounds). Biofilm communities of P. aeruginosa have been observed in chronic wounds and burn wounds, where they are a leading cause of bacterial sepsis and mortality (5, 7, 50). Additionally, P. aeruginosa will form biofilms on implanted medical devices including prostheses, catheters, and central venous lines, among others (51). A 2005 case study by Germiller and colleagues showed prominent P. aeruginosa infection on cochlear implants affecting otherwise healthy individuals and individuals with previous cases of chronic otitis media (20). These infections are notoriously hard to treat because these communities are inherently resistant to antibiotics and to the host's immune system. By deciphering the molecular approaches that biofilms of P. aeruginosa utilize to evade components of the human immune system, we have potentially identified either biofilm-specific vaccine candidates or potential therapeutic targets for biofilm adjuvant therapy. For example, to coincide with the biofilm-specific genetic regulation of flagella in P. aeruginosa, it may be possible to elicit bactericidal concentrations of human LTF from neutrophils recruited to the infection site either by the activation of resident macrophages or through TNF-α immunotherapy. The in vivo production of LTF may then reduce the amount of biofilm colonization of the lung or other tissue resulting in a decreased disease burden for the patient. This approach has been suggested to improve chemotherapy for cancer in humans (4). Irrespective of the approach, our studies clearly indicate the importance of the human immune response to biofilm infections and suggest that in addition to the massive effort to discover novel antibiofilm antibiotics, understanding the role of the immune system in biofilm infections is vital to reducing the burden of these infections.
Acknowledgments
We thank Randall Basaraba and Mark Quinn for their helpful comments about the manuscript.
This work was supported by NAU TRIF funds, funds from Science Foundation Arizona, and funds from the NIH MARC U*Star program.
Editor: A. Camilli
Footnotes
Published ahead of print on 3 August 2009.
REFERENCES
- 1.Anderson, G. G., S. Moreau-Marquis, B. A. Stanton, and G. A. O'Toole. 2008. In vitro analysis of tobramycin-treated Pseudomonas aeruginosa biofilms on cystic fibrosis-derived airway epithelial cells. Infect. Immun. 76:1423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archibald, L. K., and R. P. Gaynes. 1997. Hospital-acquired infections in the United States. The importance of interhospital comparisons. Infect. Dis. Clin. N. Am. 11:245-255. [DOI] [PubMed] [Google Scholar]
- 3.Bellamy, W., M. Takase, K. Yamauchi, H. Wakabayashi, K. Kawase, and M. Tomita. 1992. Identification of the bactericidal domain of lactoferrin. Biochim. Biophys. Acta 1121:130-136. [DOI] [PubMed] [Google Scholar]
- 4.Bellone, M., A. Mondino, and A. Corti. 2008. Vascular targeting, chemotherapy and active immunotherapy: teaming up to attack cancer. Trends Immunol. 29:235-241. [DOI] [PubMed] [Google Scholar]
- 5.Bielecki, P., J. Glik, M. Kawecki, and V. A. Martins dos Santos. 2008. Towards understanding Pseudomonas aeruginosa burn wound infections by profiling gene expression. Biotechnol. Lett. 30:777-790. [DOI] [PubMed] [Google Scholar]
- 6.Bjarnsholt, T., P. O. Jensen, M. Burmolle, M. Hentzer, J. A. Haagensen, H. P. Hougen, H. Calum, K. G. Madsen, C. Moser, S. Molin, N. Hoiby, and M. Givskov. 2005. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 151:373-383. [DOI] [PubMed] [Google Scholar]
- 7.Bjarnsholt, T., K. Kirketerp-Moller, P. O. Jensen, K. G. Madsen, R. Phipps, K. Krogfelt, N. Hoiby, and M. Givskov. 2008. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen. 16:2-10. [DOI] [PubMed] [Google Scholar]
- 8.Boehm, U., T. Klamp, M. Groot, and J. C. Howard. 1997. Cellular responses to interferon-gamma. Annu. Rev. Immunol. 15:749-795. [DOI] [PubMed] [Google Scholar]
- 9.Chmiel, J. F., and P. B. Davis. 2003. State of the art: why do the lungs of patients with cystic fibrosis become infected and why can't they clear the infection? Respir. Res. 4:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, D. P., B. J. Luebering, and D. M. Shaut. 1998. T-lymphocyte functionality assessed by analysis of cytokine receptor expression, intracellular cytokine expression, and femtomolar detection of cytokine secretion by quantitative flow cytometry. Cytometry 33:249-255. [DOI] [PubMed] [Google Scholar]
- 11.Conway, S. P., K. G. Brownlee, M. Denton, and D. G. Peckham. 2003. Antibiotic treatment of multidrug-resistant organisms in cystic fibrosis. Am. J. Respir. Med. 2:321-332. [DOI] [PubMed] [Google Scholar]
- 12.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 13.Costerton, J. W. 2001. Cystic fibrosis pathogenesis and the role of biofilms in persistent infections. Trends Microbiol. 9:50-52. [DOI] [PubMed] [Google Scholar]
- 14.Costerton, J. W. 2002. Anaerobic biofilm infections in cystic fibrosis. Mol. Cell 10:699-700. [DOI] [PubMed] [Google Scholar]
- 15.Dasgupta, M. K., P. Zuberbuhler, A. Abbi, F. L. Harley, N. E. Brown, K. Lam, J. B. Dossetor, and J. W. Costerton. 1987. Combined evaluation of circulating immune complexes and antibodies to Pseudomonas aeruginosa as an immunologic profile in relation to pulmonary function in cystic fibrosis. J. Clin. Immunol. 7:51-58. [DOI] [PubMed] [Google Scholar]
- 16.Ellis, T. N., and B. L. Beaman. 2004. Interferon-gamma activation of polymorphonuclear neutrophil function. Immunology 112:2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farnaud, S., and R. W. Evans. 2003. Lactoferrin—a multifunctional protein with antimicrobial properties. Mol. Immunol. 40:395-405. [DOI] [PubMed] [Google Scholar]
- 18.Faurschou, M., and N. Borregaard. 2003. Neutrophil granules and secretory vesicles in inflammation. Microbes Infect. 5:1317-1327. [DOI] [PubMed] [Google Scholar]
- 19.Fux, C., S. Wilson, and P. Stoodley. 2004. Detachment characteristics and oxacillin resistance of Staphylococcus aureus biofilm emboli in an in vitro catheter infection model. J. Bacteriol. 186:4486-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Germiller, J. A., H. K. El-Kashlan, and U. K. Shah. 2005. Chronic Pseudomonas infection of cochlear implants. Otol. Neurotol. 26:196-201. [DOI] [PubMed] [Google Scholar]
- 21.Govan, J. R. W., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Govan, J. R., and J. A. Fyfe. 1978. Mucoid Pseudomonas aeruginosa and cystic fibrosis: resistance of the mucoid form to carbenicillin, flucloxacillin and tobramycin and the isolation of mucoid variants in vitro. J. Antimicrob. Chemother. 4:233-240. [DOI] [PubMed] [Google Scholar]
- 23.Hoiby, N., H. Krogh Johansen, C. Moser, Z. Song, O. Ciofu, and A. Kharazmi. 2001. Pseudomonas aeruginosa and the in vitro and in vivo biofilm mode of growth. Microbes Infect. 3:23-35. [DOI] [PubMed] [Google Scholar]
- 24.Homma, M., D. J. DeRosier, and R. M. Macnab. 1990. Flagellar hook and hook-associated proteins of Salmonella typhimurium and their relationship to other axial components of the flagellum. J. Mol. Biol. 213:819-832. [DOI] [PubMed] [Google Scholar]
- 25.Irvin, R. T., J. W. Govan, J. A. Fyfe, and J. W. Costerton. 1981. Heterogeneity of antibiotic resistance in mucoid isolates of Pseudomonas aeruginosa obtained from cystic fibrosis patients: role of outer membrane proteins. Antimicrob. Agents Chemother. 19:1056-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen, E. T., A. Kharazmi, K. Lam, J. W. Costerton, and N. Hoiby. 1990. Human polymorphonuclear leukocyte response to Pseudomonas aeruginosa grown in biofilms. Infect. Immun. 58:2383-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen, E. T., A. Kharazmi, N. Hoiby, and J. W. Costerton. 1992. Some bacterial parameters influencing the neutrophil oxidative burst response to Pseudomonas aeruginosa biofilms. APMIS 100:727-733. [PubMed] [Google Scholar]
- 28.Jensen, P. O., T. Bjarnsholt, R. Phipps, T. B. Rasmussen, H. Calum, L. Christoffersen, C. Moser, P. Williams, T. Pressler, M. Givskov, and N. Hoiby. 2007. Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology 153:1329-1338. [DOI] [PubMed] [Google Scholar]
- 29.Jesaitis, A. J., M. J. Franklin, D. Berglund, M. Sasaki, C. I. Lord, J. B. Bleazard, J. E. Duffy, H. Beyenal, and Z. Lewandowski. 2003. Compromised host defense on Pseudomonas aeruginosa biofilms: characterization of neutrophil and biofilm interactions. J. Immunol. 171:4329-4339. [DOI] [PubMed] [Google Scholar]
- 30.Kelley, T. J., and M. L. Drumm. 1998. Inducible nitric oxide synthase expression is reduced in cystic fibrosis murine and human airway epithelial cells. J. Clin. Investig. 102:1200-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirketerp-Moller, K., P. O. Jensen, M. Fazli, K. G. Madsen, J. Pederson, C. Moser, T. Tolker-Nielsen, N. Hoiby, M. Givskov, and T. Bjarnsholt. 2008. Distribution, organization, and ecology of bacteria in chronic wounds. J. Clin. Microbiol. 46:2717-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koch, C., and N. Hoiby. 2000. Diagnosis and treatment of cystic fibrosis. Respiration 67:239-247. [DOI] [PubMed] [Google Scholar]
- 33.Lam, J., R. Chan, K. Lam, and J. W. Costerton. 1980. Production of mucoid microcolonies by Pseudomonas aeruginosa with infected lungs in cystic fibrosis. Infect. Immun. 28:546-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, B., J. A. Haagensen, O. Ciofu, J. B. Andersen, N. Hoiby, and S. Molin. 2005. Heterogeneity of biofilms formed by nonmucoid Pseudomonas aeruginosa isolates from patients with cystic fibrosis. J. Clin. Microbiol. 43:5247-5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leid, J. G., C. J. Willson, M. E. Shirtliff, D. J. Hassett, M. R. Parsek, and A. K. Jeffers. 2005. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J. Immunol. 175:7512-7518. [DOI] [PubMed] [Google Scholar]
- 36.Leid, J. G., M. E. Shirtliff, J. W. Costerton, and P. Stoodley. 2002. Human leukocytes adhere, penetrate and respond to Staphylococcus aureus biofilms. Infect. Immun. 70:6339-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, Y., A. Karlin, J. D. Loike, and S. C. Silverstein. 2002. A critical concentration of neutrophils is required for effective bacterial killing in suspension. Proc. Natl. Acad. Sci. USA 99:8289-8294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liberati, N. T., J. M. Urbach, S. Miyata, D. G. Lee, E. Drenkard, G. Wu, J. Villanueva, T. Wei, and F. M. Ausubel. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA 103:2833-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mah, T. F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 41.Murga, R., J. M. Miller, and R. M. Donlan. 2001. Biofilm formation by gram-negative bacteria on central venous catheter connectors: effect of conditioning films in a laboratory model. J. Clin. Microbiol. 39:2294-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nemmar, A., B. Nemery, P. H. Hoet, N. Van Rooijen, and M. F. Hoylaerts. 2005. Silica particles enhance peripheral thrombosis: key role of lung macrophage-neutrophil cross-talk. Am. J. Respir. Crit. Care Med. 171:872-879. [DOI] [PubMed] [Google Scholar]
- 43.Orsi, N. 2004. The antimicrobial activity of lactoferrin: current status and perspectives. BioMetals 17:189-196. [DOI] [PubMed] [Google Scholar]
- 44.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 45.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 46.Rumbaugh, K. P., J. A. Griswold, and A. N. Hamood. 2000. The role of quorum sensing in the in vivo virulence of Pseudomonas aeruginosa. Microbes Infect. 2:1721-1731. [DOI] [PubMed] [Google Scholar]
- 47.Sadikot, R. T., T. S. Blackwell, J. W. Christman, and A. S. Prince. 2005. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 171:1209-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sauer, K., and A. K. Camper. 2001. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 183:6579-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaber, J. A., W. J. Trifflo, S. J. Suh, J. W. Oliver, M. C. Hastert, J. A. Griswold, M. Auer, A. N. Hamood, and K. P. Rumbaugh. 2007. Pseudomonas aeruginosa forms biofilms in acute infection independent of cell-to-cell signaling. Infect. Immun. 75:3715-3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schierholz, J. M., and J. Beuth. 2001. Implant infections: a haven for opportunistic bacteria. J. Hosp. Infect. 49:87-93. [DOI] [PubMed] [Google Scholar]
- 52.Singh, P. K., M. R. Parsek, E. P. Greenberg, and M. J. Welsh. 2002. A component of innate immunity prevents bacterial biofilm development. Nature 417:552-555. [DOI] [PubMed] [Google Scholar]
- 53.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 54.Stewart, P. S., and J. W. Costerton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 55.Walker, T. S., K. L. Tomlin, G. S. Worthen, K. R. Poch, J. G. Lieber, M. T. Saaverdra, M. B. Fessler, K. C. Malcolm, M. L. Vasil, and J. A. Nick. 2005. Enhanced Pseudomonas aeruginosa biofilm development mediated by human neutrophils. Infect. Immun. 73:3693-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, L., O. Estrada, A. Zaborina, M. Bains, L. Shen, J. E. Kohler, N. Patel, M. W. Musch, E. B. Chang, Y.-X. Fu, M. A. Jacobs, M. I. Nishimura, R. E. W. Hancock, J. R. Turner, and J. C. Alverdy. 2005. Recognition of host immune activation by Pseudomonas aeruginosa. Science 309:774-777. [DOI] [PubMed] [Google Scholar]
- 57.Yang, L., J. A. Haagensen, L. Jelsbak, H. K. Johansen, C. Sternberg, N. Hoiby, and S. Molin. 2008. In situ growth rates and biofilm development of Pseudomonas aeruginosa populations in chronic lung infections. J. Bacteriol. 190:2767-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]