Abstract
Anthrax lethal toxin causes macrophages and dendritic cells from some mouse strains to undergo caspase-1-dependent cell death. Central to this process is the NOD-like receptor Nlrp1b (Nalp1b), which detects intoxication and then self-associates to form a complex, termed an inflammasome, that is capable of activating the procaspase-1 zymogen. The nature of the signal detected directly by Nlrp1b is not known, and the mechanisms of inflammasome assembly are poorly understood. Here, we demonstrate that transfection of human fibroblasts with plasmids encoding murine Nlrp1b and procaspase-1 was sufficient to confer susceptibility to lethal toxin-mediated death on the cells. As has been observed in murine macrophages, the enzymatic activities of lethal toxin and the proteasome were both required for activation of the Nlrp1b inflammasome and this activation led to prointerleukin-1β processing. Release of interleukin-1β from cells was not dependent on cell lysis, as its secretion was not affected by an osmoprotectant that prevented the appearance of lactate dehydrogenase in the culture medium. We generated constitutively active mutants of Nlrp1b by making amino-terminal deletions to the protein and observed that the ability to activate procaspase-1 was dependent on the CARD domain, which bound procaspase-1, and a region adjacent to the CARD domain that promoted self-association. Our results demonstrate that lethal toxin can activate Nlrp1b in a nonmyeloid cell line and are consistent with work that suggests that activation induces proximity of procaspase-1.
During an anthrax infection, Bacillus anthracis secretes the proteins protective antigen (PA) and lethal factor (LF), which together form the essential virulence factor lethal toxin (LeTx) (6). PA is responsible for entry of the toxin into cells; LF is a zinc metalloprotease that cleaves mitogen-activated protein kinase kinases, thereby inhibiting the activation of downstream signaling proteins (8). LeTx kills macrophages and dendritic cells by a caspase-1-dependent cell death program known as pyroptosis (2, 9, 16, 21), although the involvement of mitogen-activated protein kinase kinase cleavage in initiating this program has not been established and it is possible that it is the cleavage of other LF substrates that triggers pyroptosis.
Pyroptotic cell death occurs when the cytosolic sensor Nlrp1b detects LeTx activity and forms a complex, known as the inflammasome, that facilitates the processing of procaspase-1 (5, 17). The Nlrp1b gene is polymorphic, and only macrophages from strains of mice that express functional alleles of Nlrp1b (allele 1 or 5) are susceptible to LeTx, while those that express allele 2, 3, or 4 are resistant to pyroptosis (5).
Nlrp1b is a member of the Nod-like receptor (NLR) family of proteins, whose members share similar domain organizations and function as sensors of pathogens or cellular damage (3, 13). Nlrp1b contains an amino-terminal NACHT domain and a central leucine-rich repeat (LRR) domain, followed by a FIIND domain and a carboxy-terminal CARD domain (5, 19). The LRR domain is thought to detect a cytosolic signal derived from LeTx activity. The nature of the signal is unknown, but the signal appears to be dependent on proteasome function, as proteasome inhibitors block the activation of the Nlrp1b inflammasome (22, 24). Detection of the signal relieves an autoinhibitory conformation of Nlrp1b to allow its oligomerization through the NACHT domain, which facilitates autoproteolysis of procaspase-1 bound to the Nlrp1b CARD domain. The role of the FIIND domain in inflammasome assembly and procaspase-1 processing is unclear (23). Caspase-1 cleaves numerous substrates, including prointerleukin-1β (pro-IL-1β), which can result in mitochondrial dysfunction and cell death (2). Processing of pro-IL-1β, however, does not appear to play a role in cell death (24).
In this report, we use a heterologous expression system to study the Nlrp1b inflammasome. Human fibroblasts transfected with murine Nlrp1b and procaspase-1 became susceptible to LeTx-mediated pyroptosis, as demonstrated by lactate dehydrogenase (LDH) release. Nlrp1b inflammasome function was also detected by processing and secretion of IL-1β. Secretion of IL-1β did not require cell lysis, because an osmoprotectant that blocked lysis did not inhibit IL-1β secretion. As has been observed in murine macrophages, the enzymatic activity of LF was required for inflammasome activation and the proteasome inhibitor MG-132 blocked its activation. We next made a series of Nlrp1b deletion mutants and observed that activation of procaspase-1 required the CARD domain but not the LRR or NACHT domain. We narrowed this activity to a fragment of Nlrp1b containing the CARD domain and 56 amino acids amino terminal to the CARD domain. This amino-terminal segment promoted the oligomerization of the CARD-containing fragment, which presumably served to bring molecules of procaspase-1 into close proximity for autoproteolysis.
MATERIALS AND METHODS
Cell culture and reagents.
HT1080 cells (ATCC) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Polyethylene glycol 200 (PEG 200), PEG 2000, and PEG 6000 (Sigma-Aldrich) were added to the culture medium at a final concentration of 15 mM. PA, LF, and LF-E687A were purified as described previously and applied to cells at a final concentration of 10−8 M (14). The proteasome inhibitor MG-132 (Calbiochem) was used at 10 μM.
Plasmid construction, cDNA cloning, and site-directed mutagenesis.
The Nlrp1b allele 1 gene was amplified, using the forward primer 5′-CGC GGA TCC TAT GGA AGA ATC CCC ACC CAA G-3′ and the reverse primer 5′-CGC CTC GAG TCA TGA TCC CAA AGA GAC CCC ACC TG-3′, from cDNA derived from RAW264.7 cells. The PCR product was digested with BamHI and XhoI and then ligated into pNTAP-A (Stratagene 240101-51).
The Nlrp1b allele 3 gene was cloned in two steps. A 5′ fragment was amplified, using the forward primer 5′-CGC GGA TCC TAT GGA AGA ATC CCC ACC CAA G-3′ and the reverse primer 5′-CGC CTC GAG TCA TGA TCC CAA AGA GAC CCC ACC TG-3′, from cDNA derived from TIB-47 cells. The PCR product was digested with the restriction enzymes BamHI and XhoI and then ligated into pNTAP-A. The second fragment of the Nlrp1b allele 3 gene was amplified by using the forward primer 5′-CGC CTC GAG GAA GTC ACC CTT CAC CTC TAC and the reverse primer 5′-CGC GGG CCC TCA TGA TCC CAA AGA GAC CCC ACC-3′. The PCR product was digested with XhoI and ApaI and then ligated into the plasmid containing the 5′ fragment of the Nlrp1b allele 3 gene.
The pro-IL-1β gene was amplified by using the forward primer 5′-CGC GAA TTC ATG GCA ACT GTT CCT GAA CTC-3′ and the reverse primer 5′-CGC CTC GAG GGA AGA CAC GGA TTC CAT GGT G-3′. The PCR product was digested with EcoRI and XhoI and then ligated into pcDNA3-HA (10).
pcDNA3-T7 was generated by inserting the T7 tag into pcDNA3 between the ApaI and NheI restriction sites. Procaspase-1 was amplified by using the forward primer 5′-CGC GGA TTC TAT GGC TGA CAA GAT CCT GAG G-3′ and the reverse primer 5′-CGC CTC GAG ATG TCC CGG GAA GAG GTA G-3′. The PCR product was digested with BamHI and XhoI and then ligated into pcDNA3-T7. QuikChange site-directed mutagenesis (Stratagene) was performed according to the manufacturer's instructions to introduce the C284A mutation into pcDNA3-pro-caspase-1-T7, using the oligonucleotide 5′-GAT CAT TAT TCA GGC AGC GCG TGG AGA GAA ACA AGG-3′ and its complement.
Nlrp1b truncation plasmids were constructed by amplifying fragments from pNTAP-Nlrp1b allele 1. The reverse primer 5′-CGC CTC GAG TCA TGA TCC CAA AGA GAC CCC ACC TG-3′ was used with the following forward primers to amplify the designated fragments: for Nlrp1b436-1233, 5′-CGC GGA TCC TGA GGA TAG TGA GGA AAG ACA C-3′; for Nlrp1b720-1233, 5′-CGC GGA TCC TGA CCT GTC CTC TCT CAG TGC C-3′; for Nlrp1b1086-1233, 5′-CGC GGA TCC TTT CCA ACT CTT CTC TGA GAT CTA C-3′; and for Nlrp1b1142-1233, 5′-CGC GGA TCC TCT GCA CTT CAT GGA CCA GCA TC-3′. Nlrp1b1-1141 was amplified by using the forward primer 5′-CGC GGA TCC TAT GGA AGA ATC CCC ACC CAA G-3′ and the reverse primer 5′-CGC CTC GAG TCA CAA GGA AGG GGC ATC TTT GAG-3′. The PCR products were digested with the restriction enzymes BamHI and XhoI, and the resulting products were ligated into pNTAP-A.
To construct the Nlrp1bΔ626-719 vector, a SalI site was introduced into pNTAP-Nlrp1b allele 1 by using the oligonucleotide 5′-CTT AAA TTC ACC GTC GAC CTG GAG GGG TTG-3′ and its complement. A second SalI site was introduced by using the oligonucleotide 5′-AGC ATC CTG TGG GTC GAC CTG TCC TCT CTC-3′ and its complement. This plasmid was digested with SalI and resolved by agarose gel electrophoresis. Gel extraction was performed, and the plasmid lacking the SalI fragment was ligated together.
Nlrp1b fragments for glutathione S-transferase (GST) fusion protein constructs were amplified with the forward primer 5′-CGC GGA TCC TTC CAA CTC TTC TCT GAG ATC TAC-3′ and the reverse primer 5′-CGC CTC GAG TCA TGA TCC CAA AGA GAC CCC ACC TG-3′ for Nlrp1b1086-1233 and the forward primer 5′-CGC GGA TCC CTG CAC TTC ATG GAC CAG CAT C-3′ and the reverse primer 5′-CGC CTC GAG TCA TGA TCC CAA AGA GAC CCC ACC TG-3′ for Nlrp1b1142-1233. The PCR products were digested with restriction enzymes BamHI and XhoI, and the resulting products were ligated into pGEX4T-1.
A six-histidine tag was inserted into pcDNA3-HA between the HindIII and BamHI restriction sites to generate pcDNA3-His-HA. Nlrp1b1086-1233 was amplified by using the forward primer 5′-CGC GGA TCC TTC CAA CTC TTC TCT GAG ATC TAC-3′ and the reverse primer 5′-CGC CTC GAG TGA TCC CAA AGA GAC CCC AC-3′. Nlrp1b1142-1233 was amplified by using the forward primer 5′-CGC GGA TCC CTG CAC TTC ATG GAC CAG CAT C-3′ and the reverse primer 5′-CGC CTC GAG TGA TCC CAA AGA GAC CCC AC-3′. The PCR products were digested with the restriction enzymes BamHI and XhoI, and the resulting products were ligated into pcDNA3-His-HA. pcDNA3-His-Nlrp1b1086-1233-HA and pcDNA3-His-Nlrp1b1142-1233-HA were digested with the restriction enzymes HindIII and XhoI. The digestion products were resolved by electrophoresis. Gel extraction was performed to isolate the inserts, which were then ligated into pcDNA3-T7 to generate pcDNA3-His-Nlrp1b1086-1233-T7 and pcDNA3-His-Nlrp1b1142-1233-T7.
IL-1β and LDH release assays.
One million HT1080 cells were seeded on a 10-cm dish the day before transfection. On the day of transfection, 1 μg each of pNTAP-Nlrp1b, pcDNA3-pro-caspase-1-T7, and pcDNA3-pro-IL-1β-HA was transfected using 9 μl of 1 mg/ml polyethylenimine, pH 7.2. Approximately 24 h after transfection, cells were treated with LF (10−8 M) and PA (10−8 M) for 3 h. The cell supernatant was mixed with 1 μl of antihemagglutinin (anti-HA) antibody (Sigma-Aldrich H9658) overnight, followed by the addition of 100 μl of protein A Sepharose (GE Healthcare) and a 2-h incubation. Proteins were eluted from the protein A Sepharose beads with sodium dodecyl sulfate (SDS) loading dye and subjected to immunoblotting using a polyclonal HA antibody (Santa Cruz sc805).
Cell pellets were harvested and then lysed with 300 μl of EBC buffer (0.5% NP-40, 20 mM Tris, pH 8, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride) for 60 min. Equivalent amounts of cell lysate protein (∼30 μg) were subjected to SDS-polyacrylamide gel electrophoresis and immunoblotted with anti-HA (Santa Cruz sc805) and anti-β-actin (Sigma-Aldrich A5441) antibodies.
Release of cytoplasmic LDH into the cell supernatant was measured by a CytoTox 96 nonradioactive cytotoxicity assay (Promega G-1780) in accordance with the manufacturer's instructions. The percentage of LDH released was calculated as 100 × (experimental LDH − spontaneous LDH)/(maximum LDH − spontaneous LDH).
Detection of TAP-tagged proteins.
One 10-cm dish of HT1080 cells was transfected with 4 μg of a plasmid encoding designated tandem affinity purification (TAP)-tagged Nlrp1b fragments. Cell pellets from each plate were lysed with 300 μl EBC buffer at 4°C for 1 h. Cell lysates from three plates were incubated with 25 μl streptavidin agarose resin (Thermo Scientific 20349) for ∼2 h. Beads were washed three times with 1 ml EBC buffer. Proteins were eluted with SDS and analyzed by immunoblotting with anti-calmodulin binding peptide antibody (Upstate 07-482).
GST fusion protein purification and in vitro binding assay.
GST, GST-Nlrp1b1086-1233, and GST-Nlrp1b1142-1233 proteins were bound to glutathione Sepharose at a concentration of ∼0.4 mg/ml in accordance with the manufacturer's instructions (GE Healthcare). Lysates were prepared from one plate of HT1080 cells transfected with the designated plasmids by sonication or incubation with 300 μl EBC buffer at 4°C for 1 h. For the procaspase-1-T7 binding assay, 40 μl of beads was incubated at 4°C for ∼2 h with 400 μl of cell lysate containing procaspase-1-C284A-T7. For the oligomerization binding assay, 40 μl of beads was incubated with 600 μl cell lysate containing either His-Nlrp1b1086-1233-HA or His-Nlrp1b1142-1233-HA. Beads were washed three times with 1 ml of cold EBC buffer. Proteins were eluted with SDS and analyzed by immunoblotting with anti-caspase-1 p10 (M20) (Santa Cruz sc514) or anti-HA (Santa Cruz sc805) antibodies.
Coimmunoprecipitation assay.
Two plates of HT1080 cells were transfected with pcDNA3-His-Nlrp1b1086-1233-T7 and pcDNA3-His-Nlrp1b1086-1233-HA or with pcDNA3-His-Nlrp1b1142-1233-HA and pcDNA3-His-Nlrp1b1142-1233-T7. Cells were lysed in 600 μl EBC buffer by sonication, and the lysates were clarified by centrifugation. Lysates were incubated with 1 μl of anti-HA antibody (Sigma-Aldrich H9658) or 1 μl of control anti-green fluorescent protein antibody (Covance MMS-118R) for 2 h, followed by the addition of 50 μl of protein A Sepharose (GE Healthcare) and a 2-h incubation. Complexes were resolved by SDS-polyacrylamide gel electrophoresis and immunoblotted using an anti-T7 antibody (Novagen 69522).
RESULTS
Activation of the Nlrp1b inflammasome in HT1080 cells.
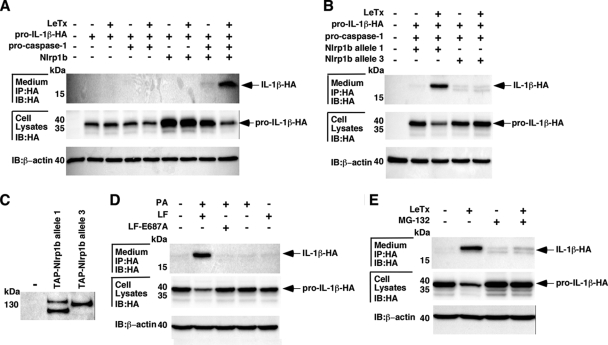
We sought to determine whether LeTx could activate the assembly of the murine Nlrp1b inflammasome in a human nonmyeloid cell line. Different combinations of plasmids encoding Nlrp1b (allele 1), procaspase-1, and pro-IL-1β were transfected into HT1080 fibroblasts. Approximately 24 h after transfection, cells were treated with LeTx (10−8 M LF and 10−8 M PA) for 3 h, and cell lysates and supernatants were then probed for HA-tagged IL-1β by immunoblotting. Increased levels of pro-IL-1β were observed in the cytosolic lysates of unintoxicated cells that had been transfected with Nlrp1b, compared to those in the lysates of cells that had not been transfected with Nlrp1b (Fig. 1A), although the significance of this observation is not clear. The 17-kDa mature form of IL-1β was detected in the supernatant of cells transfected with plasmids encoding Nlrp1b and procaspase-1 only if the cells had been treated with LeTx (Fig. 1A). The level of the 35-kDa pro-IL-1β in these cells was lower than that in the corresponding unintoxicated cells, which is consistent with pro-IL-1β being processed and secreted. Neither processing of pro-IL-1β nor secretion of IL-1β was observed when cells were transfected with plasmids encoding Nlrp1b or procaspase-1 alone. These results suggest that LeTx induces the assembly of a functional Nlrp1b inflammasome in transfected fibroblasts.
FIG. 1.
Reconstitution of the Nlrp1b inflammasome in HT1080 cells. (A) Combinations of pNTAP-Nlrp1b (1 μg), pcDNA3-pro-caspase-1-T7 (1 μg), and pcDNA3-pro-IL-1β-ΗΑ (1 μg) were transfected into HT1080 cells. Approximately 24 h after transfection, cells were treated with LeTx (10−8 M LF and 10−8 M PA) for 3 h and cell lysates were probed for HA-tagged pro-IL-1β and β-actin by immunoblotting (IB); supernatants were immunoprecipitated (IP) with anti-HA antibodies and then probed for HA-tagged IL-1β by immunoblotting. (B) Plasmids pcDNA3-pro-caspase-1-T7 (1 μg) and pcDNA3-pro-IL-1β-ΗΑ (1 μg) were cotransfected with either pNTAP-Nlrp1b allele 1 (1 μg) or allele 3 (1 μg) into HT1080 cells. Cells were treated with LeTx, and HA-tagged IL-1β was detected as described above. (C) Cells were transfected with either pNTAP-Nlrp1b allele 1 or allele 3. Approximately 24 h after transfection, cells were lysed, and TAP-tagged proteins were precipitated using streptavidin resin and subjected to Western blotting using an antibody against calmodulin binding peptide to detect the TAP tag. (D) Cells were transfected with plasmids pNTAP-Nlrp1b (1 μg), pcDNA3-pro-caspase-1-T7 (1 μg), and pcDNA3-pro-IL-1β-ΗΑ (1 μg). Transfected cells were left untreated or were treated with the indicated combinations of PA (10−8 M), LF (10−8 M), and LF-E687A (10−8 M). After 3 h, HA-tagged IL-1β was detected as described above. (E) HT1080 cells were transfected with plasmids pNTAP-Nlrp1b (1 μg), pcDNA3-pro-caspase-1-T7 (1 μg), and pcDNA3-pro-IL-1β-ΗΑ (1 μg). Transfected cells were cotreated with LeTx (10−8 M LF and 10−8 M PA) in the absence or presence of MG-132 (10 μM). After 3 h, HA-tagged IL-1β was detected as described above. Blots are representative of three independent experiments.
LeTx induces pyroptosis in macrophages expressing Nlrp1b allele 1 or 5 but not in macrophages expressing allele 2, 3, or 4 (5). To determine whether a “resistant” allele would be activated by LeTx in HT1080 cells, plasmids containing procaspase-1 and pro-IL-1β were cotransfected with a plasmid containing either allele 1 or allele 3 of Nlrp1b. The transfected cells were treated with LeTx for 3 h, and cell lysates and supernatants were probed for HA-tagged IL-1β. Consistent with allele-specific activation of the inflammasome in macrophages, LeTx caused processing and secretion of IL-1β in cells that expressed Nlrp1b allele 1 but not in those that expressed allele 3 (Fig. 1B). The expression levels of Nlrp1b allele 1 and allele 3 were similar in HT1080 cells; the presence of a higher mobility band observed for Nlrp1b allele 1 suggested the presence of a protease-sensitive site in this protein (Fig. 1C). This processing may result from expression of the protein in human cells, as the higher-mobility form was not detected in murine macrophages (data not shown).
To confirm that the enzymatic activity of LF is required for activation of the Nlrp1b inflammasome, LF-E687A, which lacks catalytic activity because of an active site mutation, was used in the assay. As above, pro-IL-1β was processed and IL-1β was secreted into the medium in the presence of wild-type LF and PA. When the cells were treated with PA and LF-E687A, PA alone, or LF alone, the level of pro-IL-1β remained constant in cells and mature IL-1β was not observed in the medium (Fig. 1D). This result indicates that the enzymatic activity of LF is required for activation of the Nlrp1b inflammasome.
Proteasome activity is involved in LeTx-mediated macrophage death (22), so to test whether the proteasome is required for activation of the Nlrp1b inflammasome in fibroblasts, transfected cells were cotreated with LeTx and the proteasome inhibitor MG-132. The mature form of IL-1β was detected in the medium of cells treated with LeTx but not in the medium of cells treated with LeTx and MG-132 (Fig. 1E). The level of pro-IL-1β in the cell lysates did not diminish in LeTx-treated cells exposed to MG-132, suggesting that proteasome activity is required for inflammasome activation.
Characterization of LeTx-induced cell death and IL-1β release.
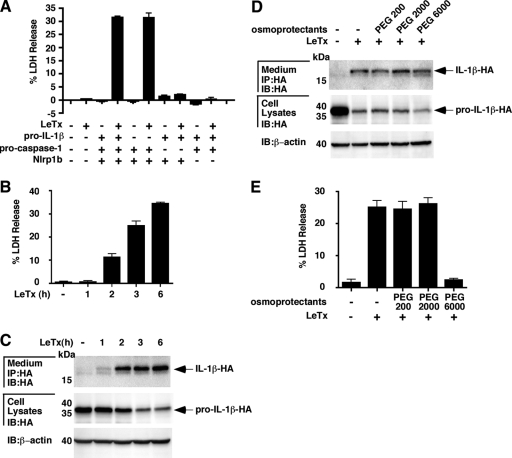
We next sought to determine whether the activation of the Nlrp1b inflammasome by LeTx caused death among HT1080 cells. Cells were transfected with different combinations of plasmids encoding Nlrp1b, procaspase-1, and pro-IL-1β. Approximately 24 h after transfection, the cells were treated with LeTx for 6 h and then the supernatants were collected and assayed for LDH activity. LDH activity was observed in the supernatants of only those cells transfected with Nlrp1b and procaspase-1 and then treated with LeTx (Fig. 2A). Cotransfection of pro-IL-1β did not increase LDH activity in the supernatant. These data suggest that activation of the Nlrp1b inflammasome leads to both IL-1β secretion and cell death but that IL-1β does not stimulate cell death.
FIG. 2.
Characterization of LeTx-induced cell death and IL-1β release. (A) Different combinations of plasmids pNTAP-Nlrp1b (1 μg), pcDNA3-pro-caspase-1-T7 (1 μg), and pcDNA3-pro-IL-1β-ΗΑ (1 μg) were transfected into HT1080 cells. Approximately 24 h after transfection, cells were treated with LeTx (10−8 M LF and 10−8 M PA) for 6 h, and supernatants were assayed for LDH activity. (B and C) Cells were transfected with plasmids pNTAP-Nlrp1b (1 μg), pcDNA3-pro-caspase-1-T7 (1 μg), and pcDNA3-pro-IL-1β-ΗΑ (1 μg). Transfected cells were left untreated or treated with LeTx (10−8 M LF and 10−8 M PA) for 1, 2, 3, or 6 h. Supernatants were assayed for LDH activity (B); supernatants and cell lysates were probed for HA-tagged IL-1β by immunoblotting (IB) (C). IP, immunoprecipitation. (D and E) Plasmids carrying pNTAP-Nlrp1b (1 μg), pcDNA3-pro-caspase-1-T7 (1 μg), and pcDNA3-pro-IL-1β-ΗΑ (1 μg) were transfected into HT1080 cells. Approximately 24 h after transfection, cells were cotreated with LeTx (10−8 M LF and 10−8 M PA) and the indicated osmoprotectants (15 mM) for 3 h, and supernatants and cell lysates were probed for HA-tagged IL-1β by immunoblotting (D); the supernatants were assayed for LDH activity (E). Results shown are representative of three independent experiments.
Previously published work has indicated that IL-1β release from LeTx-treated macrophages occurs through cell lysis rather than through a secretory system (24). To address whether IL-1β release coincides with cell death in fibroblasts, we performed time course experiments. Transfected cells were treated with LeTx for up to 6 h, and the cell supernatants were assayed for LDH activity (Fig. 2B) and IL-1β protein (Fig. 2C). The release of LDH and IL-1β occurred with similar kinetics. We next addressed whether osmoprotection of cells would prevent release of IL-1β by incubating cells with PEG. Addition of PEG 200, PEG 2000, or PEG 6000 to cells did not affect the release of IL-1β (Fig. 2D). PEG 6000 did, however, prevent the release of LDH (Fig. 2E). These data indicate that caspase-1 activity leads to the formation of membrane pores with diameters between 2.8 nm (PEG 2000) and 5 nm (PEG 6000) (7) and that IL-1β release does not require cell lysis.
The CARD domain, but not the NACHT or LRR domain, is required for inflammasome activity.
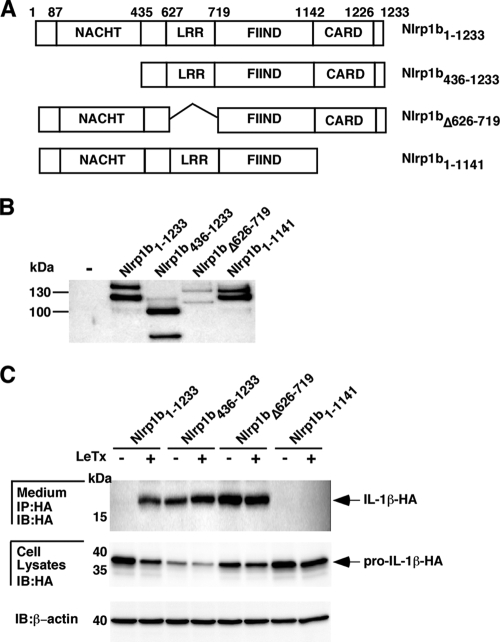
To determine the functional importance of the Nlrp1b domains, three deletion mutants were made: Nlrp1b436-1233 (deletion of the amino-terminal region and the NACHT domain), Nlrp1bΔ626-719 (LRR domain deletion), and Nlrp1b1-1141 (CARD domain deletion) (Fig. 3A and B). Each construct was cotransfected with plasmids encoding procaspase-1 and pro-IL-1β. Transfected cells were treated with LeTx for 3 h, and pro-IL-1β processing and secretion were monitored. In contrast to that of full-length Nlrp1b, expression of Nlrp1b436-1233 or Nlrp1bΔ626-719 resulted in the secretion of IL-1β in the absence of LeTx (Fig. 3C). Nlrp1b1-1141 was not able to activate caspase-1, as no active form of IL-1β was detected in the supernatant in the presence or absence of LeTx. These data suggest that the CARD domain is necessary for Nlrp1b to activate caspase-1; the NACHT and LRR domains are not necessary for caspase-1 activation and may function in autoinhibition.
FIG. 3.
Deletion analysis of Nlrp1b. (A) Domain structures of various Nlrp1b deletion constructs. (B) HT1080 cells were transfected with various TAP-tagged Nlrp1b deletion constructs. Approximately 24 h after transfection, cells were lysed and TAP-tagged proteins were precipitated using streptavidin resin and immunoblotted using antibody directed against the calmodulin binding peptide segment of the TAP tag. (C) Nlrp1b deletion constructs were cotransfected with plasmids pcDNA3-pro-caspase-1-T7 (1 μg) and pcDNA3-pro-IL-1β-ΗΑ (1 μg) into HT1080 cells. After 24 h, the cells were left untreated or were treated with LeTx (10−8 M LF and 10−8 M PA) for 3 h. Cell lysates were probed for HA-tagged pro-IL-1β and β-actin by immunoblotting (IB); supernatants were immunoprecipitated (IP) with anti-HA antibodies and then probed for HA-tagged IL-1β by immunoblotting. Blots are representative of three independent experiments.
Amino-terminal truncation mutants of Nlrp1b are constitutively active.
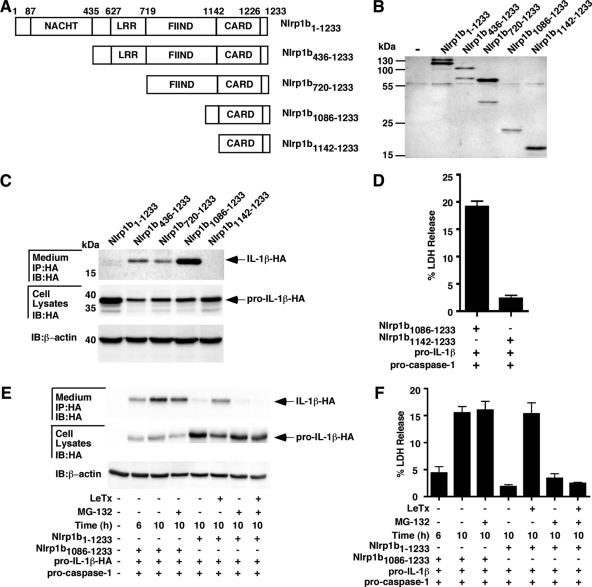
We next made a series of Nlrp1b deletion mutants to define the region that can constitutively activate caspase-1 (Fig. 4A and B). Expression of either Nlrp1b436-1233, Nlrp1b720-1233, or Nlrp1b1086-1233 caused secretion of IL-1β (Fig. 4C). In contrast, we did not detect IL-1β in the supernatant of cells transfected with the Nlrp1b1142-1233 plasmid. This correlated with the observation that expression of Nlrp1b1086-1233, but not Nlrp1b1142-1233, led to LDH release (Fig. 4D). Thus, a fragment of Nlrp1b containing the CARD domain and 56 amino acids amino terminal to the CARD domain activates caspase-1, whereas the CARD domain alone does not exhibit any detectable activity.
FIG. 4.
Amino-terminal truncation mutants of Nlrp1b are constitutively active. (A) Domain structures of Nlrp1b deletion constructs. (B) HT1080 cells were transfected with various Nlrp1b deletion constructs. Approximately 24 h after transfection, cells were lysed, and TAP-tagged proteins were precipitated using streptavidin resin and immunoblotted using antibody directed against the calmodulin binding peptide segment of the TAP tag. (C) Cells were transfected with plasmids containing different Nlrp1b fragments, procaspase-1, and pro-IL-1β. Approximately 24 h after transfection, cell lysates were probed for HA-tagged pro-IL-1β and β-actin by immunoblotting (IB); supernatants were immunoprecipitated (IP) with anti-HA antibodies and then probed for HA-tagged IL-1β by immunoblotting. (D) Cells were transfected with plasmids containing Nlrp1b fragments, procaspase-1, and pro-IL-1β. Approximately 24 h after transfection, supernatants were assayed for LDH release. (E and F) Cells were transfected with plasmids encoding the indicated proteins, and after 6 h, cells were treated with MG-132 and/or LeTx for an additional 4 h. Supernatants were probed for IL-1β-HA and LDH at either 6 h or 10 h posttransfection, as indicated. Results shown are representative of three independent experiments.
The role of the proteasome in mediating events downstream of inflammasome activation was assessed by treating cells expressing Nlrp1b1086-1233 with MG-132. IL-1β was detected in the medium of Nlrp1b1086-1233-expressing cells 6 h after transfection (Fig. 4E); the level of IL-1β was higher by 10 h posttransfection whether or not MG-132 was added to the cells at 6 h posttransfection. In contrast, IL-1β was detected in the supernatants of cells expressing full-length Nlrp1b at 10 h if toxin was added at 6 h, but no increase in IL-1β level was detected if MG-132 was added with the toxin. This lack of involvement of proteasome activity downstream of inflammasome activation was confirmed by monitoring the release of LDH; the increase in LDH activity detected in the supernatant of Nlrp1b1086-1233-expressing cells between 6 h and 10 h posttransfection was not blocked by MG-132 (Fig. 4F).
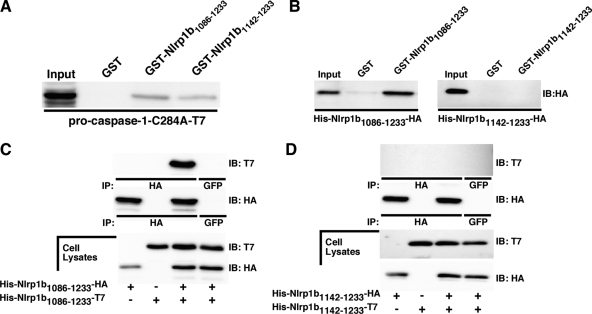
Nlrp1b1086-1233 and Nlrp1b1142-1233 interact with procaspase-1.
To determine why Nlrp1b1086-1233, but not Nlrp1b1142-1233, induced secretion of IL-1β, we performed an in vitro binding assay to determine whether the two Nlrp1b constructs could interact with procaspase-1. GST fusions of the Nlrp1b fragments were used to precipitate the catalytically inactive procaspase-1-C284A-T7 mutant from HT1080 cell lysates. Both GST-Nlrp1b1086-1233 and GST-Nlrp1b1142-1233 bound procaspase-1-C284A-T7, while the GST control did not (Fig. 5A). Although Nlrp1b1086-1233 precipitated slightly more procaspase-1-C284A-T7 than Nlrp1b1142-1233 did, this result suggests that the inability of the CARD domain construct to promote secretion of IL-1β is not due to an inability to bind procaspase-1.
FIG. 5.
Nlrp1b1086-1233 and Nlrp1b 1142-1233 interact with procaspase-1, but only Nlrp1b1086-1233 self-associates. (A) GST, GST-Nlrp1b1086-1233, and GST-Nlrp1b1142-1233 were immobilized on glutathione-Sepharose beads and incubated with mammalian cell lysates containing procaspase-1-C284A-T7. Precipitated proteins and 5% of the input lysates were subjected to Western blot analysis using anti-caspase-1 p10 antibody. (B) GST, GST-Nlrp1b1086-1233, and GST-Nlrp1b1142-1233 were immobilized on glutathione-Sepharose beads and incubated with mammalian cell lysates containing either His-Nlrp1b1086-1233-HA or His-Nlrp1b1142-1233-HA. Precipitated proteins and 5% of the input lysates were subjected to Western blot analysis using anti-HA antibodies. IB, immunoblotting. (C) HT1080 cells were transfected with His-Nlrp1b1086-1233-HA, His-Nlrp1b1086-1233-T7, or both constructs. Cells were lysed 24 h after transfection, and proteins were immunoprecipitated (IP) using anti-HA antibody, followed by immunoblotting with anti-T7 antibody. GFP, green fluorescent protein. (D) HT1080 cells were transfected with His-Nlrp1b1142-1233-HA, His-Nlrp1b1142-1233-T7, or both constructs. Cells were lysed 24 h after transfection, and proteins were immunoprecipitated using anti-HA antibody, followed by immunoblotting with anti-T7 antibody. Blots are representative of three independent experiments.
Self-association of Nlrp1b1086-1233.
The proximity model of caspase activation posits that mediator proteins activate molecules of procaspases by bringing them together to facilitate a trans proteolytic reaction (4). Thus, we speculated that Nlrp1b1086-1233, but not Nlrp1b1142-1233, would self-associate. To test this notion, GST fusion proteins were used in binding assays with HT1080 cell extracts containing either His-Nlrp1b1086-1233- HA or His-Nlrp1b1142-1233-HA. His-Nlrp1b1086-1233-HA was precipitated by GST-Nlrp1b1086-1233, but no detectable amount of His-Nlrp1b1142-1233-HA was pulled down by GST-Nlrp1b1142-1233 (Fig. 5B).
Coimmunoprecipitation experiments were then performed to confirm that Nlrp1b1086-1233 self-associates and that Nlrp1b1142-1233 does not. His-Nlrp1b1086-1233-HA was expressed with His-Nlrp1b1086-1233-T7 in HT1080 cells. The T7-tagged protein was immunoprecipitated along with HA-tagged Nlrp1b1086-1233 (Fig. 5C). In contrast, coimmunoprecipitation was not observed between the Nlrp1b1142-1233 constructs (Fig. 5D). Taken together, these experiments indicate that Nlrp1b1086-1233 associates with itself but that Nlrp1b1142-1233 does not.
DISCUSSION
Numerous studies indicate that NLRs detect microbial products or endogenous danger signals, but the identification of the molecules that directly activate NLRs has proven to be difficult (13, 15). The activation of Nlrp1b by LeTx is dependent on the proteolytic activity of the toxin, but neither the LeTx substrate nor the Nlrp1b ligand involved in activation is known. Murine dendritic cells and macrophages are the only cell types that have been shown to undergo pyroptosis upon treatment with LeTx, which could be because expression of Nlrp1b is restricted to these cell types but could also be because factors involved in activation are missing in other cell types. We have shown here that transient expression of Nlrp1b and procaspase-1 is sufficient to sensitize human fibroblasts to LeTx-induced pyroptosis. Thus, the activation pathway appears to be conserved in human cells and is not dependent on myeloid cell-specific proteins.
That this heterologous system reflects how Nlrp1b is activated in murine macrophages is supported by the observation that proteasome activity is required for inflammasome activation in both cases (Fig. 1E and 4E and F) (22). The role of the proteasome in this process is not known, but the involvement of the proteasome suggests that Nlrp1b is not a direct target of LF and rather that the proteasome might degrade a negative regulator of Nlrp1b. A second observation that indicates the fidelity of the heterologous system is the demonstration of Nlrp1b allele specificity for function. Nlrp1b allele 1, which supports pyroptosis in macrophages, supported pro-IL-1β processing in fibroblasts, whereas allele 3, found in LeTx-resistant macrophages, did not. There are a number of amino acid differences between alleles 1 and 3, making it difficult to speculate on why allele 3 is not able to detect LeTx activity and/or to assemble into a functional inflammasome. We note that the CARD domains of the two alleles are identical, indicating that the defect in allele 3 is not due to an inability to bind procaspase-1.
Activation of caspase-1 not only caused IL-1β secretion but also induced death among the HT1080 cells. Studies using murine macrophages have suggested that IL-1β release results from cell lysis, precluding the release of this cytokine as a cause of death (24). Our results indicated that IL-1β secretion does not require cell lysis, because the osmoprotectant PEG 6000 prevented LDH release but not IL-1β secretion. LDH release was dependent on the transfection of Nlrp1b and procaspase-1 but not on that of pro-IL-1β, which is consistent with the notion that IL-1β does not mediate cell death. A difference between what has been observed in macrophages and in transfected fibroblasts involves the kinetics of cell death; macrophages are killed within 60 to 90 min of toxin treatment (22, 24), whereas the transfected fibroblasts died over the course of several hours. This difference in the kinetics of cell death could be a result of differences in the expression levels of inflammasome components or downstream mediators of cell death, such as Bnip3 (11).
We demonstrated that the CARD domain of Nlrp1b was essential for inflammasome activity. This finding was not surprising, because unlike human NLRP1, Nlrp1b lacks a pyrin domain at its amino terminus to recruit procaspase-1 through the adaptor ASC (apoptosis-associated speck-like protein containing a CARD). ASC is not likely required for Nlrp1b inflammasome function, as the LeTx-sensitive RAW264.7 cell line does not express ASC at detectable levels (18) and we found that cotransfection of ASC into HT1080 cells did not stimulate inflammasome activity (data not shown).
Deletion of the LRR domain yielded a constitutively active Nlrp1b mutant. This finding is consistent with an autoinhibition model in which an interaction between the LRR domain and the NACHT domain holds Nlrp1b in an inactive conformation that is relieved when the LRR domain detects an activating signal. Deletion of the NACHT domain also yielded a constitutively active mutant indicating that oligomerization of the NACHT domain is not required for processing of pro-IL-1β, at least in the context of a truncated protein. These data agree with a study of human NLRP1 that showed that a truncation mutant that lacked the NACHT domain was able to cause cell death (12).
We made a series of truncation mutants to delimit the region of Nlrp1b required for constitutive activity. Nlrp1b1086-1233, which contains the CARD domain and the adjacent 56 amino acids, activated procaspase-1, whereas the CARD domain alone did not. Both of these constructs bound procaspase-1, demonstrating that a CARD-procaspase-1 interaction is not sufficient to activate procaspase-1. The 56 amino acids adjacent to the CARD domain promoted self-association of the truncation mutant in GST pulldown and coimmunoprecipitation experiments. These results suggest that a segment of the FIIND domain induces the close proximity of procaspase-1 molecules that promotes trans proteolysis. A similar region exists adjacent to the CARD domains of human NLRP1 and CARD8 (TUCAN/CARDINAL) (1, 20) and might also enhance the self-association of these proteins.
Acknowledgments
This research was supported by NIH grant RO1 AI067683. J.M. holds the Canada Research Chair in Bacterial Pathogenesis.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 3 August 2009.
REFERENCES
- 1.Agostini, L., F. Martinon, K. Burns, M. F. McDermott, P. N. Hawkins, and J. Tschopp. 2004. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity 20:319-325. [DOI] [PubMed] [Google Scholar]
- 2.Alileche, A., E. R. Serfass, S. M. Muehlbauer, S. A. Porcelli, and J. Brojatsch. 2005. Anthrax lethal toxin-mediated killing of human and murine dendritic cells impairs the adaptive immune response. PLoS Pathog. 1:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benko, S., D. J. Philpott, and S. E. Girardin. 2008. The microbial and danger signals that activate Nod-like receptors. Cytokine 43:368-373. [DOI] [PubMed] [Google Scholar]
- 4.Boatright, K. M., M. Renatus, F. L. Scott, S. Sperandio, H. Shin, I. M. Pedersen, J. E. Ricci, W. A. Edris, D. P. Sutherlin, D. R. Green, and G. S. Salvesen. 2003. A unified model for apical caspase activation. Mol. Cell 11:529-541. [DOI] [PubMed] [Google Scholar]
- 5.Boyden, E. D., and W. F. Dietrich. 2006. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat. Genet. 38:240-244. [DOI] [PubMed] [Google Scholar]
- 6.Collier, R. J., and J. A. Young. 2003. Anthrax toxin. Annu. Rev. Cell Dev. Biol. 19:45-70. [DOI] [PubMed] [Google Scholar]
- 7.Dacheux, D., J. Goure, J. Chabert, Y. Usson, and I. Attree. 2001. Pore-forming activity of type III system-secreted proteins leads to oncosis of Pseudomonas aeruginosa-infected macrophages. Mol. Microbiol. 40:76-85. [DOI] [PubMed] [Google Scholar]
- 8.Duesbery, N. S., C. P. Webb, S. H. Leppla, V. M. Gordon, K. R. Klimpel, T. D. Copeland, N. G. Ahn, M. K. Oskarsson, K. Fukasawa, K. D. Paull, and G. F. Vande Woude. 1998. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science 280:734-737. [DOI] [PubMed] [Google Scholar]
- 9.Fink, S. L., T. Bergsbaken, and B. T. Cookson. 2008. Anthrax lethal toxin and Salmonella elicit the common cell death pathway of caspase-1-dependent pyroptosis via distinct mechanisms. Proc. Natl. Acad. Sci. USA 105:4312-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Go, M. Y., E. M. Chow, and J. Mogridge. 2009. The cytoplasmic domain of anthrax toxin receptor 1 affects binding of the protective antigen. Infect. Immun. 77:52-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ha, S. D., D. Ng, J. Lamothe, M. A. Valvano, J. Han, and S. O. Kim. 2007. Mitochondrial proteins Bnip3 and Bnip3L are involved in anthrax lethal toxin-induced macrophage cell death. J. Biol. Chem. 282:26275-26283. [DOI] [PubMed] [Google Scholar]
- 12.Hlaing, T., R. F. Guo, K. A. Dilley, J. M. Loussia, T. A. Morrish, M. M. Shi, C. Vincenz, and P. A. Ward. 2001. Molecular cloning and characterization of DEFCAP-L and -S, two isoforms of a novel member of the mammalian Ced-4 family of apoptosis proteins. J. Biol. Chem. 276:9230-9238. [DOI] [PubMed] [Google Scholar]
- 13.Kanneganti, T. D., M. Lamkanfi, and G. Nunez. 2007. Intracellular NOD-like receptors in host defense and disease. Immunity 27:549-559. [DOI] [PubMed] [Google Scholar]
- 14.Kassam, A., S. D. Der, and J. Mogridge. 2005. Differentiation of human monocytic cell lines confers susceptibility to Bacillus anthracis lethal toxin. Cell. Microbiol. 7:281-292. [DOI] [PubMed] [Google Scholar]
- 15.Mariathasan, S., and D. M. Monack. 2007. Inflammasome adaptors and sensors: intracellular regulators of infection and inflammation. Nat. Rev. Immunol. 7:31-40. [DOI] [PubMed] [Google Scholar]
- 16.Muehlbauer, S. M., T. H. Evering, G. Bonuccelli, R. C. Squires, A. W. Ashton, S. A. Porcelli, M. P. Lisanti, and J. Brojatsch. 2007. Anthrax lethal toxin kills macrophages in a strain-specific manner by apoptosis or caspase-1-mediated necrosis. Cell Cycle 6:758-766. [DOI] [PubMed] [Google Scholar]
- 17.Nour, A. M., Y. G. Yeung, L. Santambrogio, E. D. Boyden, E. R. Stanley, and J. Brojatsch. 2009. Anthrax lethal toxin triggers the formation of a membrane-associated inflammasome complex in murine macrophages. Infect. Immun. 77:1262-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pelegrin, P., C. Barroso-Gutierrez, and A. Surprenant. 2008. P2X7 receptor differentially couples to distinct release pathways for IL-1beta in mouse macrophage. J. Immunol. 180:7147-7157. [DOI] [PubMed] [Google Scholar]
- 19.Proell, M., S. J. Riedl, J. H. Fritz, A. M. Rojas, and R. Schwarzenbacher. 2008. The Nod-like receptor (NLR) family: a tale of similarities and differences. PLoS ONE 3:e2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Razmara, M., S. M. Srinivasula, L. Wang, J. L. Poyet, B. J. Geddes, P. S. DiStefano, J. Bertin, and E. S. Alnemri. 2002. CARD-8 protein, a new CARD family member that regulates caspase-1 activation and apoptosis. J. Biol. Chem. 277:13952-13958. [DOI] [PubMed] [Google Scholar]
- 21.Reig, N., A. Jiang, R. Couture, F. S. Sutterwala, Y. Ogura, R. A. Flavell, I. Mellman, and F. G. van der Goot. 2008. Maturation modulates caspase-1-independent responses of dendritic cells to anthrax lethal toxin. Cell. Microbiol. 10:1190-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Squires, R. C., S. M. Muehlbauer, and J. Brojatsch. 2007. Proteasomes control caspase-1 activation in anthrax lethal toxin-mediated cell killing. J. Biol. Chem. 282:34260-34267. [DOI] [PubMed] [Google Scholar]
- 23.Tschopp, J., F. Martinon, and K. Burns. 2003. NALPs: a novel protein family involved in inflammation. Nat. Rev. Mol. Cell Biol. 4:95-104. [DOI] [PubMed] [Google Scholar]
- 24.Wickliffe, K. E., S. H. Leppla, and M. Moayeri. 2008. Anthrax lethal toxin-induced inflammasome formation and caspase-1 activation are late events dependent on ion fluxes and the proteasome. Cell. Microbiol. 10:332-343. [DOI] [PMC free article] [PubMed] [Google Scholar]