Abstract
Mitochondrial RNA metabolism in Trypanosoma brucei is a complex process involving both extensive RNA editing and control of RNA stability. MRP1/2 and RBP16 are two factors that have been implicated in regulating the editing and stability of specific mRNAs. These two factors exhibit similar nonspecific RNA binding and RNA-annealing activities, suggesting that some of their actions may have been previously masked by functional redundancy. Here, we examine the functional interaction of MRP1/2 and RBP16 by separate and simultaneous RNA interference and by overexpressing RBP16 in an MRP1/2-depleted background. Simultaneous depletion of these factors resulted in synthetic lethality in procyclic trypanosomes. Analysis of mitochondrial RNAs in procyclic cells revealed distinct functions for MRP1/2 and RBP16 toward edited apocytochrome b mRNA, redundant functions in stabilization of edited ATPase subunit 6 and cytochrome oxidase subunit 3 mRNAs, and concentration-dependent positive and negative functions for RBP16 toward edited RPS12 mRNAs. While simultaneous MRP1/2-RBP16 depletion had no effect on the growth of bloodstream form cells, massive adverse effects on the levels of almost all mitochondrial RNAs were observed. These studies greatly expand our knowledge regarding the functions of MRP1/2 and RBP16 and suggest that both RNA-specific and life cycle stage-specific factors impact MRP1/2 and RBP16 functions.
Mitochondrial gene regulation in kinetoplastid protozoa is a complex process entailing multiple posttranscriptional steps (24, 45, 47). For example, in Trypanosoma brucei, 12 of 18 mitochondrially encoded RNAs require editing for the creation of a translatable mRNA. Kinetoplastid editing is a unique process in which uridines (U) are posttranscriptionally added to and deleted from transcripts in a highly specific manner. During editing, the pre-mRNA associates with small trans-acting RNAs known as guide RNAs (gRNAs), which are also mitochondrially encoded and contain the information for the addition and/or deletion of a specifically set number of U residues. Editing is catalyzed by several related multiprotein complexes, termed editosomes or L complexes, which contain all of the required enzymatic activities and additional proteins important for structure and macromolecular interactions (7, 30, 31, 44). While it is associated with the editosomes, pre-mRNA is cleaved by one of several gRNA-directed endonucleases, and U residues are inserted or deleted through the actions of a terminal uridylyl transferase or U-specific exoribonuclease, respectively, using the gRNA-encoded information. Following this step, the mRNA is religated by one of two related RNA ligases. Each gRNA specifies editing at approximately 10 editing sites; thus, complete editing of an mRNA typically requires the sequential actions of dozens of gRNAs. In addition to the approximately 20 proteins contained in the core enzymatic and structural editosome, several transiently associated accessory factors are important in RNA editing (1, 11, 15, 22, 25, 35). These factors (and the subcomplexes they may be contained in) are speculated to coordinate the recruitment of gRNAs to the editosome, direct the correct binding of the gRNA to the mRNA, and regulate the overall progression of editing as it transverses 5′ along the pre-mRNA.
RNA stability is also important in the control of mitochondrial gene expression. For instance, because transcription of the mitochondrial maxicircle genome is polycistronic (20, 37) and the abundance of some never-edited RNAs is dramatically regulated during the T. brucei life cycle (42), control at the level of RNA stability is necessitated in these cases. In addition, RNA stability and editing appear to be linked, although the proteins involved and their functional interactions are less well understood than is the editing process itself. Both in vitro and in vivo experiments have demonstrated that RNAs with only a minimal amount of editing at their 3′ ends become stabilized by the addition of a poly(A) tail (10, 17). Polyadenylation is catalyzed by kPAP1, and possibly kPAP2, poly(A) polymerases (10, 18). The mechanism by which the poly(A) tail mediates stabilization of partially edited RNAs is not known, but several factors potentially involved in the stabilization of edited RNAs have recently been described, including TbRGG1 and MERS1 (15, 50).
Two of the most extensively studied mitochondrial gene regulatory factors are the RBP16 protein and the MRP1/2 complex. RBP16 is an RNA binding protein and a member of the multifunctional Y-box protein family (16). It is comprised of two distinct domains, an N-terminal cold shock domain and a C-terminal RG-rich domain. Several arginines in the RG-rich domain undergo methylation, which affects RBP16 function (13, 14). RNA interference (RNAi) studies in procyclic-form (PF) T. brucei demonstrated that RBP16 is essential for growth and for editing of the apocytochrome b (CYb) mRNA (35). The effect on CYb mRNA editing is highly specific, since other edited mRNAs are not significantly affected by RBP16 downregulation (35). RNAi-mediated depletion of RBP16 also resulted in decreased levels of two never-edited mRNAs, NADH dehydrogenase subunit 4 (ND4) and cytochrome oxidase subunit 1 (COI), indicating that RBP16 also plays a role in the stabilization of these two RNAs (35). In accordance with its multifunctional nature, RBP16 exhibits relatively nonspecific RNA binding properties and has been shown to associate with mRNA and gRNA, both in vitro and in vivo (14, 16, 26, 33). Notably, RBP16 directly stimulates in vitro RNA-editing reactions at or prior to the step of mRNA cleavage (25). We have also shown that RBP16 exhibits both gRNA/pre-mRNA-annealing activity and RNA-unwinding activity (2). Thus, RBP16 affects both the editing and stability of specific mitochondrial mRNAs, presumably through its ability to bind RNA and modulate RNA-RNA interactions.
A second mitochondrial gene regulatory protein is the heterotetrameric MRP1/2 complex (19). MRP1/2 is comprised of two molecules each of the related MRP1 and MRP2 proteins (formerly gBP21 and gBP25) (4, 41). Like RBP16, MRP1/2 exhibits relatively nonspecific RNA binding activity and promotes the annealing of complementary RNAs, including gRNA to its cognate pre-mRNA (27, 28, 41, 54). Interestingly, the in vivo phenotype of PF T. brucei depleted of MRP1/2 is very similar to that of RBP16 knockdowns, with the prominent features being the loss of CYb editing and the destabilization of both ND4 and COI never-edited mRNAs (49). While MRP1/2 is essential for the growth of PF T. brucei, MRP1 is not essential for the growth of the bloodstream form (BF) (22), although several never-edited RNAs are destabilized in this stage. In the related parasite Leishmania tarentolae, MRP1/2 is directly associated with three proteins originally termed AP1 to AP3, and this complex is associated with several editing components (4). In T. brucei, MRP1/2 has not been found physically associated with the editosome (54). It does, however, interact with several proteins involved in RNA metabolism, such as GRBC1 (AP1), GRBC2 (AP2), and MERS1 (AP3), in an RNA-dependent manner (50).
Due to the similar phenotypes exhibited by RBP16- and MRP1/2-depleted cells, as well as the similar in vitro RNA binding and -annealing activities of the two factors, we reasoned that RBP16 and MRP1/2 might perform some redundant functions in RNA editing and/or stability. Such functional redundancy could explain the somewhat limited effects of RBP16 or MRP1/2 depletion on RNA metabolism in vivo. To address the functional relationships between these two factors, we simultaneously depleted RBP16 and MRP1/2 in PF and BF T. brucei and analyzed the effects on mitochondrial RNA metabolism. Additionally, we created PF cell lines in which RBP16 was overexpressed in an MRP1/2-depleted background. These experiments revealed a surprisingly complex interplay between these two mitochondrial gene regulatory factors. In PF, simultaneous MRP1/2-RBP16 downregulation led to a more significant growth defect than that observed upon knockdown of a single factor. With regard to edited CYb RNA levels, we observed an additive effect, indicative of distinct MRP1/2 and RBP16 functions. On the other hand, simultaneous MRP1/2 and RBP16 depletion revealed redundant functions in the maintenance of edited ATPase subunit 6 (A6) and COIII RNA levels. The effect on edited A6 and COIII RNAs is apparently exerted at the level of fully edited RNA stability, thus expanding the roles for these factors in the control of mitochondrial RNA stabilization. Our studies also unexpectedly uncovered a negative role for RBP16 in the stabilization of specific RNAs, and the results are consistent with concentration-dependent effects of the protein. Finally, simultaneous MRP1/2-RBP16 depletion had no effect on BF cell growth. Nevertheless, we did observe dramatic effects on a number of mitochondrial RNAs in BF cells depleted of both factors. As the effects on a given RNA differed greatly between BF and PF life cycle stages, our results imply the involvement of stage-specific factors in MRP1/2- and RBP16-dependent processes.
MATERIALS AND METHODS
Cell cultures and RNAi.
PF T. brucei strain 29-13 (provided by George A. M. Cross, Rockefeller University), derived from the Lister 427 strain and containing integrated genes for the T7 RNA polymerase and the tetracycline repressor (52), was grown in SDM-79 medium supplemented with 10% fetal bovine serum as described previously (35). BF “single-marker” T. brucei cells, also derived from Lister 427 (also provided by George A. M. Cross), were cultured in HMI-9 medium supplemented with 10% fetal bovine serum and 10% Serum plus (35).
The creation of double- and triple-gene RNAi constructs was performed as follows. The genes encoding RBP16, MRP1, and MRP2 were PCR amplified from oligo(dT)-primed cDNA derived from PF T. brucei (strain 927 Eatro 1.1) RNA using the following primers: MRP1-5 (5′-CACCTCGAGATGATTCGACTCGCATG), MRP1-3 (5′-CCCATCGATGGTATCGCGATGTGTCA), MRP2-5 (5′-GGATCGATATGCTCCGACGTATCATCAGCCAG), MRP2-3 (5′-GTAATCGATGATTCGGTGTGAAGGTG), RBP16-5 (5′- GGTCTAGAGTCTGGACGTGGTTTTGG-3′), and RBP16-3 (5′-GGTCTAGAAAAGTCATCGCTGAAGCTCTGG-3′). The restriction sites for each primer are underlined. The resultant PCR products were each cloned into pCR2.1 (TA-Topo kit; Invitrogen). The MRP1 gene was excised from pCR-MRP1 by XhoI and ClaI restriction digestion and ligated into the XhoI-ClaI sites of the RNAi vector p2T7-177 (51), resulting in p2T7-177-MRP1. Next, RBP16 was excised from the RNAi vector pZJM-RBP16 (35) using XbaI and ligated into the XbaI site of p2T7-177, resulting in p2T7-177-MRP1-RBP16. MRP2 was then excised from the pCR2-MRP2 cloning vector using ClaI and ligated into the ClaI site of p2T7-177-MRP1-RBP16, resulting in the triple-RNAi vector p2T7-177-MRP1-MRP2-RBP16. Finally, to create a double-insert RNAi vector targeting only the MRP1/2 complex and not RBP16, the RBP16 insert was removed from p2T7-177-MRP1-MRP2-RBP16 using XbaI and religated, resulting in p2T7-177-MRP1-MRP2. p2T7-177-RBP16 was also generated by ligating the XbaI digest of pZJM-RBP16 into the XbaI digest of p2T7-177. PF and BF RNAi cell lines were generated by transfecting NotI-linearized p2T7-177-MRP1-MRP2 and p2T7-177-MRP1-MRP2-RBP16 into 29-13 PF cells or single-marker BF cells, and cells harboring these constructs were selected with 2.5 μg/ml phleomycin. PF cells harboring the pZJM-RBP16 RNAi construct have been introduced previously (35). To establish BF RNAi cells against RBP16, p2T7-177-RBP16 was transfected into BF single-marker cells and selected with phleomycin. Phleomycin-resistant clones were obtained by serial dilution and grown over the indicated periods in the absence or presence of 2.5 μg/ml tetracycline.
To generate cells for the expression of RBP16 in an MRP1/2 RNAi background, the full-length RBP16 open reading frame with a C-terminal tag was excised from p2MYC-RBP16-WT using HindIII and BamHI (13) and ligated into the HindIII-BamHI-digested pLEW100-TbAuk1-3HA-PURO vector (48), a generous contribution from C. C. Wang (University of California, San Francisco), to generate pLEW-RBP16-myc-PURO. PF RNAi cells harboring the p2T7-177-MRP1-MRP2 construct were transfected with NotI-linearized pLEW-RBP16-myc-PURO and selected with the addition of 1 μg/ml puromycin (Invivogen). Clones were generated by limiting dilution and verified by Western blotting against the C-terminal myc tag of RBP16-myc. RNAi-induced knockdown of all proteins was verified by Western blotting.
Antibodies.
Previously described polyclonal antibodies were used against RBP16, TBRGG1, and MRP2 (2, 16). Monoclonal antibodies against MRP1 were a kind gift from Kenneth Stuart (Seattle Biomedical Research Institute), and anti-HSP70 antibodies were a gift from Jay Bangs (University of Wisconsin).
qRT-PCR.
Total RNA was extracted from uninduced and induced RNAi cells using TRIzol reagent (Invitrogen) at day 2 following induction for PF cells and at day 4 for BF cells. Ten micrograms of RNA was treated with a DNA-free DNase kit (Ambion) to remove any residual DNA. The RNA was reverse transcribed using random hexamer primers and the Taq-Man reverse transcription (RT) kit (Applied Biosciences). The resultant cDNA was used in RT-PCRs using primers for various mitochondrial mRNAs as described previously (8). Twenty-five-microliter quantitative RT-PCR (qRT-PCR) reaction mixtures were established as described by Carnes et al. (8), and the cDNA was amplified using a MyiQ single-color real-time PCR detection system (Bio-Rad). The results were analyzed using iQ5 software (Bio-Rad) and compared to levels of steady-state β-tubulin RNA using the standard-curve method. Similar results were also obtained with a subset of the mRNAs using 18S rRNA as an alternate standard. The levels of each mRNA are represented as the mean and standard error of at least six separate determinations.
Gene-specific PCRs.
RT-PCR was performed essentially as described previously (40). Total RNA from uninduced and induced MRP1/2-RBP16 RNAi cells was DNase treated with the DNA-free kit (Ambion). RT reactions (20-μl mixtures) were performed by annealing 150 pmol of oligo(dT) primer to 1 μg of total RNA and incubating it at 70°C for 5 min, followed by slow cooling (30 min) to 20°C. The oligo(dT) primer was extended with Superscript II Reverse Transcriptase (Life Technologies) for 1 h at 42°C according to the manufacturer's specifications. Samples were heat inactivated for 15 min at 70°C and RNase H treated for 20 min at 37°C, followed by heat inactivation for 20 min at 60°C. Three microliters of the RT reaction mixture was used in a PCR with primers A6 5′NE (GCGAATTCAAATAAGTATTTTGATATTATTAAAG) and A63′ (ATTAACTTATTTGATCTTATTCTATAA), which anneal to the 3′ and 5′ never-edited regions of A6 mRNA, and COIII 5′NE (TTATTGAGGATTGTTTAAAATTGAATA) and COIII 3′NE (AACTTCCTACAAACTACCAATAC), which anneal to the 3′ and 5′ never-edited regions of COIII mRNA. Note that COIII 3′ NE extended 5 nucleotides (two editing sites) into the edited region. The reactions were performed at an annealing temperature of 45°C for 35 cycles. The PCR products were analyzed on a 2.5% Nu-Sieve agarose gel.
RESULTS
MRP1/2 and RBP16 display synthetic lethality.
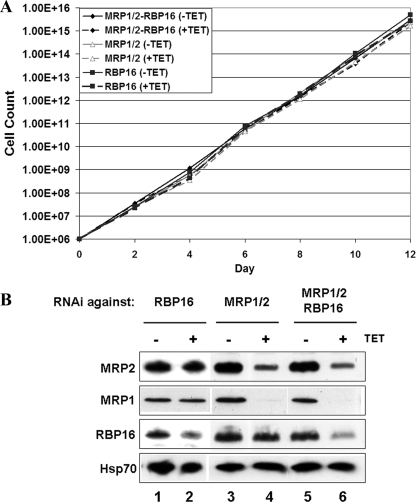
PF T. brucei cell lines in which either the MRP1/2 heterotetramer or RBP16 was depleted by RNAi have been reported previously (35, 49, 53). Individual downregulation of MRP1/2 or RBP16 resulted in very similar phenotypic effects on mitochondrial RNAs, primarily consisting of significantly decreased CYb RNA editing and decreased stability of the never-edited ND4 and COI mRNAs. In addition to their similar in vivo phenotypes, MRP1/2 and RBP16 both display the ability to bind gRNA and mRNA and to facilitate gRNA-mRNA annealing in vitro (2, 16, 19, 27, 28, 33, 41). These data led us to hypothesize that the apparently limited effects of MRP1/2 and RBP16 knockdown on mitochondrial RNA metabolism might be, at least in part, due to their abilities to perform some redundant functions in RNA editing and/or stability. To address this issue, several T. brucei cell lines were established. First, we created a cell line with an integrated triple-RNAi construct simultaneously targeting the RBP16, MRP1, and MRP2 proteins. Western blot analysis confirmed that the levels of all three proteins were substantially decreased upon tetracycline induction of RNAi (Fig. 1B). We previously established and characterized an RBP16 knockdown line (35), and for this study, as a control, we created PF MRP1/2 RNAi cells similar to those described previously (49, 53) (Fig. 1B). To further address the potentially redundant functions of MRP1/2 and RBP16, a construct that expressed RBP16-myc (13) was introduced into the MRP1/2 RNAi cell line, yielding a cell line that overexpressed RBP16 by about twofold within an MRP1/2-downregulated background (Fig. 1B). We then determined the effects of varying the amounts of these factors on cell growth, as well as on the levels of edited and never-edited mRNAs.
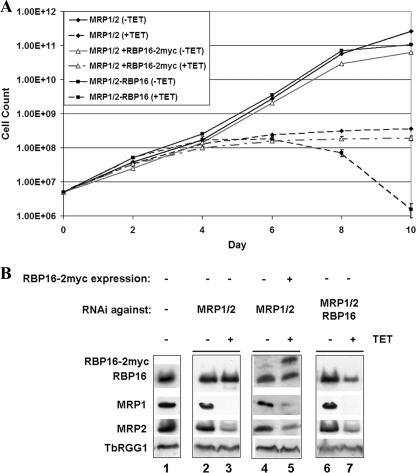
FIG. 1.
Growth effects of separately or simultaneously modulating MPR1/2 and RBP16 levels in PF cells. (A) PF T. brucei cell lines harboring RNAi constructs against both MRP1 and MRP2 (MRP1/2) or against MRP1, MRP2, and RBP16 (MRP1/2-RBP16) were grown in the absence (−TET) or presence (+TET) of 2.5 μg/ml tetracycline, and growth was monitored over 10 days. Cell lines harboring the MRP1 and MRP2 RNAi construct and simultaneously expressing exogenous RBP16-2myc (MRP1/2 + RBP16-2myc) were similarly analyzed. The averages and standard deviations of triplicate determinations are shown. (B) Protein levels (equivalent to 1 × 106 cells) in the cell lines analyzed in panel A on day 2 following tetracycline induction were determined by Western blotting using polyclonal antibodies against RBP16 and MRP2 and monoclonal antibodies against MRP1. TbRGG1 levels were analyzed as a loading control. The leftmost lane (−) contained parental 29-13 cells.
Previous findings demonstrated that downregulation of either MRP1/2 or RBP16 results in dramatically slowed growth, although neither of these knockdowns is lethal and some level of recovery was evident (35, 49, 53). Analysis of the MRP1/2 line created for this study recapitulated these results (Fig. 1A). In contrast, when the MRP1/2 complex and RBP16 were simultaneously depleted, a much more dramatic growth phenotype was observed. In the triple knockdown, cell growth slowed by day 4 following tetracycline induction of RNAi, cell death was apparent by day 8, and by day 10, few live cells were observed (Fig. 1A). This effect was evident in two different triple-knockdown RNAi clonal cell lines that were investigated (data not shown). Thus, MRP1/2 and RBP16 display synthetic lethality, suggesting that these factors act in parallel, redundant processes or that they can act in the same essential process so that the combination of the two knockdowns is lethal. We next asked whether overexpression of one of the factors could compensate for the loss of the other factor by analyzing growth in MRP1/2 RNAi cells that simultaneously overexpress RBP16 with a C-terminal two-myc tag. We previously showed that the small two-myc tag on exogenously expressed RBP16 did not alter its ability to localize properly to mitochondria or to associate with various macromolecular complexes (13, 14). In these cells, growth was virtually identical to that of the parental MRP1/2 cell line following RNAi induction, indicating that a twofold overexpression of RBP16 could not rescue growth inhibition induced by the loss of MRP1/2 (Fig. 1A). Thus, MRP1/2 may have some functions that cannot be compensated for by RBP16. Together, these genetic data suggest both distinct and overlapping functions for MRP1/2 and RBP16.
MRP1/2 and RBP16 have distinct functions in CYb mRNA editing.
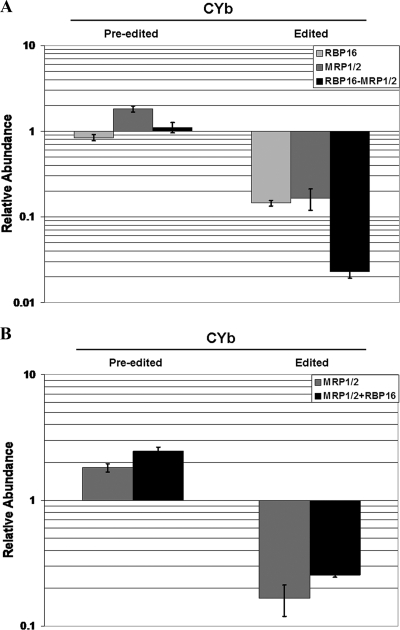
We examined the effects of separate and simultaneous MRP1/2 and RBP16 depletion on mitochondrial RNAs using qRT-PCR (8). For these studies, total RNA was extracted from each of the cell lines shown in Fig. 1 and from our previously established RBP16 knockdown line (35) on day 2 following RNAi induction. We began by examining the minimally edited CYb mRNA, which is the major RNA affected upon the depletion of RBP16 or MRP1/2 separately, as previously shown by poisoned-primer extension analyses (35, 49, 53). Figure 2A shows that the level of preedited CYb mRNA was not appreciably affected by either separate or simultaneous MRP1/2 and RBP16 depletion. As expected, PF cells with knockdown of MRP1/2 or RBP16 alone exhibited substantial loss of the fully edited form of CYb mRNA (approximately 85% decrease) (Fig. 2A). When PF cells were analyzed following simultaneous depletion of RBP16 and MRP1/2, the level of fully edited CYb mRNA decreased even more, to approximately 2% of control levels (a 98% decrease) (Fig. 2A). Therefore, the loss of both factors had an additive detrimental effect on CYb mRNA editing. If this additive effect reflects the combined actions of MRP1/2 and RBP16 in one essential process, we would expect that overexpression of one factor could at least partially compensate for depletion of the other factor. To address this, we asked whether overexpression of RBP16 could compensate for the effect of MRP1/2 depletion on edited CYb mRNA levels. As shown in Fig. 2B, no appreciable rescue of CYb fully edited mRNA levels was observed in MRP1/2 knockdown cells when RBP16 was overexpressed. From these results, we conclude that MRP1/2 and RBP16 are both required for full editing of CYb RNA and that they act through distinct and separate processes. Therefore, the separate functions of MRP1/2 and RBP16 are additive during CYb RNA editing.
FIG. 2.
MPR1/2 and RBP16 have distinct functions, and their simultaneous depletion has an additive effect on the levels of edited CYb mRNA in PF cells. (A) RNAs from the following cell lines (harvested at day 2 following tetracycline induction) were analyzed by qRT-PCR: RBP16 depleted (retaining MRP1/2), MRP1/2 depleted (retaining RBP16), and MRP1/2 and RBP16 depleted (retaining neither factor). The factors that were depleted by RNAi are indicated in the upper right corner of the graph. A value of 1 indicates no change in RNA levels in induced cells relative to uninduced cells. The error bars indicate standard deviations. (B) qRT-PCR was performed as for panel A with RNAs from the following cell lines harvested at day 2 following tetracycline induction: MRP1/2 depleted (retaining RBP16; same data as in panel A) and MRP1/2 depleted with overexpression of RBP16-2myc (containing total RBP16 at two times the wild-type levels [Fig. 1B]) (MRP1/2+RBP16).
MRP1/2 and RBP16 have redundant functions in the maintenance of edited A6 and COIII mRNAs.
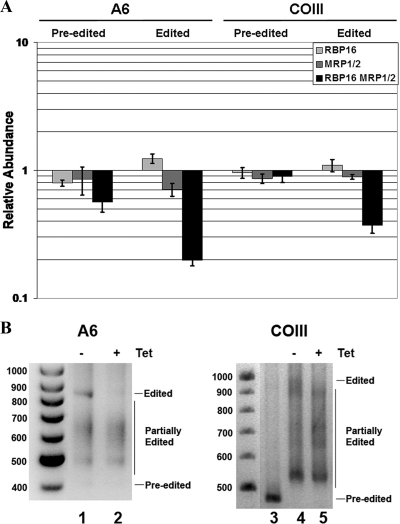
Since RNAi against both RBP16 and MRP1/2 resulted in synthetic lethality, this suggested that edited mRNAs other than CYb might be affected by simultaneous loss of both accessory factors. Effects of MRP1/2 and RBP16 on other mRNAs could be masked in the separate knockdown lines if the two factors perform redundant functions in the metabolism of mRNAs other than CYb. To investigate potential redundancy of MRP1/2 and RBP16, we analyzed two panedited mRNAs, A6 and COIII, in cells depleted either separately or simultaneously of MRP1/2 and RBP16. The separate depletion of MRP1/2 or RBP16 caused little or no reduction in the levels of preedited or edited A6 mRNA, consistent with previous reports (35, 49) (Fig. 3A, left). Similarly, neither preedited nor edited COIII mRNA was affected by the separate depletion of either factor, as reported for MRP1/2 knockdowns (49) (Fig. 3A, right). However, upon simultaneous downregulation of MRP1/2 and RBP16, both edited A6 mRNA and edited COIII mRNA exhibited very substantial decreases (90% and 75% decreases, respectively) (Fig. 3A). The preedited form of A6 mRNA also exhibited a decrease of about 40% in the triple-RNAi line (Fig. 3A), suggesting the possibility that simultaneous MRP1/2-RBP16 depletion may affect A6 RNA stability, although we cannot rule out additional effects on the editing process.
FIG. 3.
MRP1/2 and RBP16 have redundant functions, and their simultaneous depletion has a synthetic effect on the levels of edited A6 and COIII mRNAs in PF cells. (A) The same RNA populations were analyzed as in Fig. 2A. qRT-PCR was performed for preedited and edited A6, as well as preedited COIII and edited COIII RNAs. The error bars indicate standard deviations. (B) RT-PCR analysis of the entire A6 mRNA population (left, lanes 1 and 2) and the entire COIII mRNA population (right, lanes 4 and 5) in uninduced (−) or tetracycline (Tet)-induced (+) cells harboring the triple RBP16-MRP1/2 RNAi construct. RNA was analyzed by RT-PCR using primers against the 5′ and 3′ never-edited regions of the A6 and COIII mRNAs, and the products were resolved by agarose gel electrophoresis. In addition, unedited COIII DNA was amplified from genomic DNA as a size marker for completely unedited COIII mRNA (lane 3). Preedited, partially edited, and fully edited mRNA populations are indicated on the right, and size markers (in bp) are shown on the left.
To further investigate whether the dramatic decrease in edited A6 and COIII mRNAs observed upon MRP1/2-RBP16 depletion results from a defect in RNA editing or stabilization, we used a gene-specific RT-PCR approach to amplify the entire populations of both A6 and COIII mRNAs, regardless of editing status. This technique amplifies the various forms of a given mRNA using primers for its static never-edited 5′ and 3′ ends. Products of the amplification include preedited, fully edited, and all the variously sized partially edited forms of the mRNA, the last of which are typically abundant. If RNA editing is compromised, one generally observes a decrease in fully edited RNA and an increase in preedited and/or partially edited RNAs (9, 40). We first performed the gene-specific PCR on A6 mRNA from uninduced and induced MRP1/2-RBP16 RNAi cells and resolved the amplified products by agarose gel electrophoresis to visualize the extent of A6 editing (Fig. 3B, lanes 1 and 2). In the uninduced cells (lane 1), both the 403-bp preedited and the 822-bp fully edited forms of A6 were evident (Fig. 3B). In addition, several variously sized products were evident between the preedited and fully edited forms, indicative of RNAs undergoing editing. Following simultaneous RNAi against RBP16 and MRP1/2, preedited A6 mRNA was decreased slightly (Fig. 3B, lane 2), consistent with the qRT-PCR results. More importantly, we observed a complete loss of fully edited A6 mRNA (lane 2), while the partially edited RNA population remained unchanged upon MRP1/2-RBP16 depletion (Fig. 3B), strongly suggesting a specific effect on the stability of fully edited A6 mRNA. A similar, but more complex, result was observed using gene-specific RT-PCR on COIII mRNA (Fig. 3B, lanes 4 and 5). In uninduced cells, we observed amplification products at approximately the size expected for fully edited RNA (970 bp), as well as a range of partially edited products. We did not observe a band corresponding to preedited RNA (expected at 463 bp, as confirmed by PCR of genomic DNA [Fig. 3B, lane 3]). Upon MRP1/2-RBP16 depletion, we observed the disappearance of products corresponding approximately to the size of fully edited COIII RNA (compare lanes 4 and 5), similar to what was observed with A6 RNA (lane 2). Unlike A6 RNA, we also saw a modest increase in some, but not all, of the variously sized partially edited COIII RNA populations, suggesting stalling of the editing process or an increase in stabilization. Moreover, as the abundance of the partially edited RNA at approximately 880 to 890 bp was not decreased in the knockdown of MRP1/2-RBP16, this suggests that the disappearance of only the fully edited COIII mRNA results from destabilization, rather than inhibition, of editing. Together, these results establish that either MRP1/2 or RBP16 is sufficient to maintain essentially normal levels of edited A6 and COIII mRNAs, even in the absence of the other factor. The roles of MRP1/2 and RBP16 are multifaceted regarding COIII RNA, and our results suggest there may be effects on both stability and editing, two processes that are not mutually exclusive. However, the very striking and specific decrease of the fully edited A6 upon simultaneous depletion of MRP1/2 and RBP16 strongly supports a model in which the two factors perform redundant functions in the stabilization of this panedited mRNA.
MRP1/2 and RBP16 are both required for maintenance of ND4, COI, and MURF2 mRNAs, and they act in the same process.
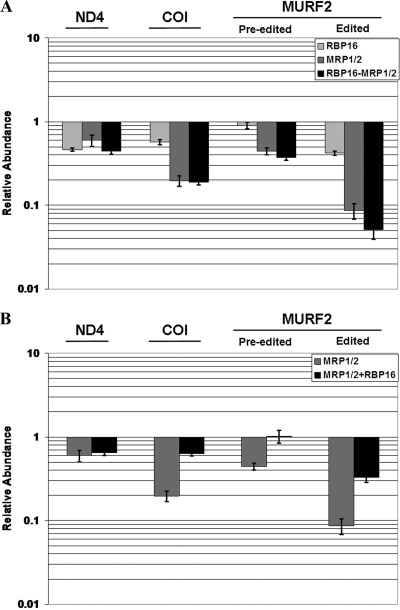
Previously, two never-edited mRNAs, ND4 and COI, were found by poisoned-primer extension to be destabilized upon loss of MRP1/2 or RBP16 (35, 49, 53). Using qRT-PCR, we confirmed that both ND4 and COI mRNAs decreased following downregulation of either MRP1/2 or RBP16, although MPR1/2 had a more dramatic effect on ND4 RNA levels than did RBP16 (Fig. 4A). We next wanted to determine whether MRP1/2 and RBP16 perform separate functions, thereby producing additive effects (Fig. 2). To this end, we measured ND4 and COI mRNA levels in the MRP1/2-RBP16 triple knockdown. In contrast to what was observed for edited CYb mRNA (Fig. 2), simultaneous depletion of MRP1/2 and RBP16 did not lead to any further appreciable destabilization of ND4 beyond what was observed with the MRP1/2 knockdown (Fig. 4A). Similarly, COI mRNA levels were not decreased in the triple knockdown compared to the MRP1/2 or RBP16 knockdowns. From these data, we conclude that MRP1/2 and RBP16 do not have additive effects in stabilizing ND4 and COI mRNAs. This suggests that their functions with respect to maintenance of these two RNAs are not separate and that MRP1/2 and RBP16 act in a single process. A similar effect was also observed when we measured both preedited and edited forms of the minimally edited MURF2 mRNA (Fig. 4A). The effects on MURF2 mRNAs were more similar to those of COI, in which depletion of MRP1/2 led to a more dramatic decrease than did depletion of RBP16 (Fig. 4A). To further define the relative roles of MRP1/2 and RBP16 in the maintenance of ND4, COI, and MURF2 mRNAs, we assessed the levels of these RNAs in MRP1/2 RNAi cells that simultaneously overexpressed RBP16 (Fig. 4B). Notably, overexpression of RBP16 was able to significantly restore the levels of COI and both preedited and edited MURF2 mRNAs in cells depleted of MRP1/2. However, RBP16 overexpression did not restore ND4 RNA levels in MRP1/2 knockdowns, consistent with previous results suggesting that the stabilization of ND4 and COI RNAs by RBP16 takes place through somewhat different mechanisms (13). In summary, simultaneous MRP1/2-RBP16 depletion has no more deleterious effect on ND4, COI, or MURF2 mRNA levels than the depletion of the most important factor (MRP1/2) alone, and overexpressed RBP16 partially compensates for MRP1/2 depletion in some cases. Together, these data suggest that MRP1/2 and RBP16 act in the same process with regard to the maintenance of this subset of mRNAs.
FIG. 4.
Both MRP1/2 and RBP16 are required for maintenance of COI, ND4, and edited MURF2 RNA levels in PF cells, and they act in the same process. (A) The same RNA populations were analyzed as in Fig. 2A. qRT-PCR was performed for COI, ND4, preedited MURF2, and edited MURF2 RNAs. (B) The same RNA populations were analyzed as in Fig. 2B. The error bars indicate standard deviations.
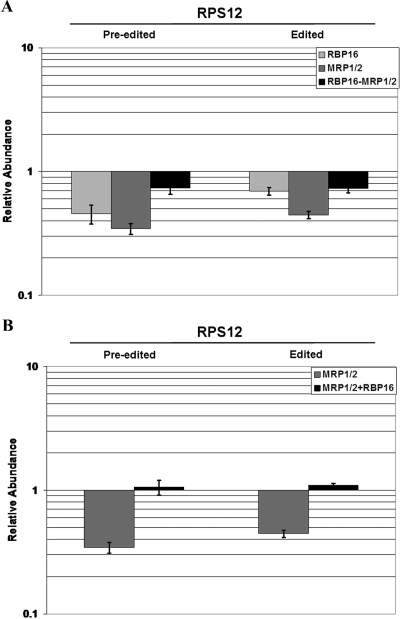
MRP1/2 and RBP16 have both related and antagonistic functions in the maintenance of edited RPS12 mRNA.
Analysis of another panedited mRNA, RPS12, revealed a more complicated interplay between MPR1/2 and RBP16. Previous findings from poisoned-primer extension experiments indicated that edited RPS12 mRNA decreases by about 50% following loss of the MRP1/2 heterotetramer while the corresponding preedited mRNA is relatively unaffected (49). The effect of RBP16 depletion on RPS12 RNA levels was not previously addressed. Our qRT-PCR analyses show that, following loss of MRP1/2 or RBP16, the levels of preedited RPS12 mRNAs are substantially decreased (65% and 55% decreases, respectively) (Fig. 5A). Edited RPS12 mRNA levels were also affected, with MRP1/2 depletion exerting a more marked effect than RBP16 depletion (55% and 30% decreases, respectively). These data suggest that both MRP1/2 and RBP16 exert positive effects on the maintenance of these mRNAs. Unexpectedly, when we examined RPS12 mRNA levels in the triple-MRP1/2-RBP16-knockdown line, both preedited and edited mRNA levels were significantly restored compared to their levels in MRP1/2 knockdown cells (Fig. 5A, compare the black bars to the dark-gray bars). Since MRP1/2 RNAi cells express wild-type amounts of RBP16 and little MRP1/2, the restoration of mRNA levels upon depletion of RBP16 in the MRP1/2 knockdown background, points to a negative effect of RBP16 on the maintenance of preedited and edited RPS12 RNA in the context of depleted MRP1/2. To gain further insight into the complex interplay of MRP1/2 and RBP16, we examined preedited and edited RPS12 mRNA levels in MRP1/2-depleted cells overexpressing RBP16. To our surprise, overexpression of RBP16 restored both forms of RPS12 mRNA to wild-type levels in the MRP1/2 knockdown (Fig. 5B). Together, these data indicate that RBP16 can exert both positive and negative effects on RPS12 mRNA levels and that these effects are concentration dependent. In part, the interplay between MRP1/2 and RBP16 toward RPS12 mRNA is similar to the scenario with COI and MURF2 RNAs in that depletion of either factor causes a loss of mRNA and RBP16 can compensate for MRP1/2 depletion, suggesting that the two factors act in the same process. However, in regard to RPS12 mRNA, an additional antagonistic effect of RBP16 was observed in the MRP1/2 RNAi background. This antagonistic effect apparently can be overcome when RBP16 is present in excess of wild-type levels. That is, in the MRP1/2 knockdown background, wild-type amounts of RBP16 preferentially act via the negative element. When overexpressed, the excess RBP16 can then feed into the positive pathway and compensate for the loss of MRP1/2.
FIG. 5.
MRP1/2 and RBP16 act in the same pathway to stabilize preedited and edited RPS12 mRNA in PF cells, but they can also have antagonistic effects in mRNA maintenance. (A) The same RNA populations were analyzed as in Fig. 2A. (B) The same RNA populations were analyzed as in Fig. 2B. The error bars indicate standard deviations.
gRNA levels are unaffected by simultaneous MRP1/2-RBP16 depletion.
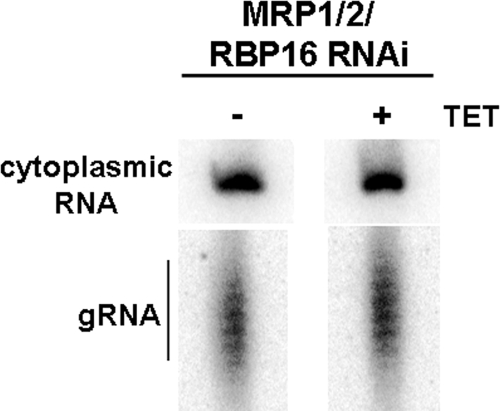
Both MRP1/2 and RBP16 can bind to various types of RNAs, including gRNAs (16, 19, 28, 33). Previous studies indicated that neither MRP1/2 nor RBP16 knockdown affects the abundance of gRNAs (35, 50). Nevertheless, the possibility remained that the decreased levels of edited mRNAs in cells in which MRP1/2 and RBP16 have been simultaneously depleted result from altered gRNA levels. To assess gRNA levels in uninduced and induced MRP1/2-RBP16 triple-knockdown cells, total RNA was labeled using guanylyltransferase and [α-32P]GTP, which labels the gRNA pool, as well as cytoplasmic RNA that can be used as a loading control (16). Figure 6 shows that total gRNA levels were unchanged in cells simultaneously depleted of MRP1/2 and RBP16, indicating that the bulk of the total gRNA population was unchanged following loss of both factors.
FIG. 6.
Simultaneous depletion of MRP1/2 and RBP16 has no effect on total gRNA levels in PF cells. Twenty micrograms of total RNA from cells depleted of both MRP1/2 and RBP16, either uninduced (−) or induced with tetracycline (TET) (+) for 2 days, was labeled using guanylyltransferase and [α-32P]GTP and resolved on a denaturing gel. A cytoplasmic RNA that undergoes labeling was used as a loading control.
Simultaneous depletion of MRP1/2 and RBP16 does not affect the growth of BF T. brucei.
To this point, we had demonstrated multiple effects of single and combined MRP1/2 and RBP16 depletion on growth and mitochondrial mRNA metabolism in PF trypanosomes. We next wanted to determine the effects of MRP1/2 and RBP16 downregulation in BF T. brucei. Neither MRP2 nor RBP16 has been investigated in this life cycle stage of the parasite. Previous studies in which both MRP1 alleles were deleted in BF T. brucei showed that MRP1 itself is not required for BF growth but that MRP1 null cells cannot differentiate into PF T. brucei, highlighting the importance of MRP1 in the insect stage (22). Nevertheless, RNA editing is essential in BF T. brucei (40), and another RNA editing accessory factor, TbRGG2, is required for growth of both life cycle stages of the parasite (11). To address the roles of MRP1/2 and RBP16 in BF T. brucei, the RNAi vectors directed against RBP16, MRP1/2, and MRP1/2-RBP16 were transfected into the single marker strain of BF T. brucei. Following establishment of clonal transfectants, we confirmed by Western blotting that the target genes were downregulated after induction of RNAi (Fig. 7B). We then examined growth in MRP1/2, RBP16, and MRP1/2-RBP16 RNAi lines. After 12 days, all three BF RNAi lines grew at rates identical to those of cells lacking RNAi induction (Fig. 7A). Therefore, unlike PF cells, BF T. brucei cells do not require MRP1/2 or RBP16 for growth under in vitro culture conditions.
FIG. 7.
Growth effects of separately or simultaneously depleting MPR1/2 and RBP16 in BF cells. (A) BF T. brucei cell lines harboring RNAi constructs against RBP16; both MRP1 and MRP2 (MRP1/2); or MRP1, MPR2, and RBP16 (MRP1/2-RBP16) were grown in the absence (−TET) or presence (+TET) of tetracycline, and growth was monitored over 12 days. (B) MRP1, MRP2, and RBP16 protein levels in the cell lines analyzed in panel A were determined by Western blotting. HSP70 protein levels were monitored as a loading control.
Separate and simultaneous depletion of MRP1/2 and RBP16 in BF T. brucei leads to multiple defects in RNA metabolism that differ from those in PF T. brucei.
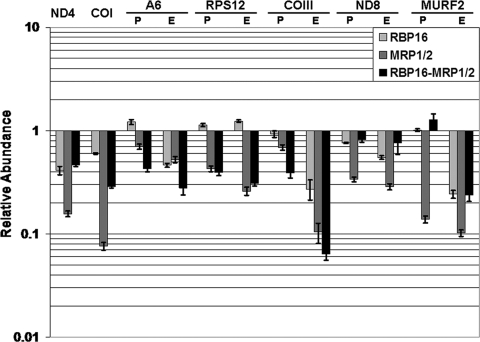
Despite the absence of a growth defect in cells downregulated for MRP1/2 and/or RBP16, the possibility remained that depletion of these factors impacts the metabolism of specific mRNAs. In support of this concept, MRP1 null BF cells exhibited decreased levels of several mRNAs, including the never-edited COI, preedited COII, and edited A6 mRNAs (22). To investigate further the roles of MRP1/2 and RBP16 in mitochondrial mRNA metabolism, we extracted RNA from uninduced and induced MRP1/2, RBP16, and MRP1/2-RBP16 cells on day 4 following tetracycline addition and quantified numerous never-edited and edited mRNAs by qRT-PCR. Surprisingly, many different mRNAs were significantly and adversely affected by the loss of MRP1/2 and/or RBP16 in BF cells (Fig. 8). In the case of the never-edited mRNAs ND4 and COI, both mRNAs decreased upon loss of RBP16 or MRP1/2, with the most severe decrease being COI mRNA levels in MRP1/2-depleted cells (∼92% decrease) (Fig. 8). Additionally, all edited mRNAs tested exhibited decreased abundance. This panel of mRNAs included panedited mRNAs (ND8, A6, RPS12, and COIII) and a minimally edited mRNA (MURF2), as well as constitutively edited mRNAs (A6 and COIII) and mRNAs whose editing is upregulated in BF cells (ND8 and RPS12). The most adversely affected mRNAs were edited MURF2 and edited COIII, which decreased 90% upon MRP1/2 downregulation (Fig. 8).
FIG. 8.
qRT-PCR analysis of mRNA levels in BF cells depleted of RBP16, MRP1/2, or both factors. RNA was harvested from the same cell lines analyzed in Fig. 7 after 4 days of growth either in the absence or presence of tetracycline. RNA levels in induced cells relative to those in uninduced cells are shown. P, preedited; E, edited. The error bars indicate standard deviations.
One striking feature of BF cells was the prevalence of both positive and negative effects of RBP16 toward a given RNA, similar to what was observed for RPS12 mRNA in PF cells (Fig. 5A). That is, ND4, COI, preedited and edited ND8, and edited MURF2 all exhibited a pattern in which depletion of either factor alone decreased RNA levels. In all cases, MRP1/2 depletion led to a more severe decrease in RNA levels than did RBP16 depletion. However, downregulation of RBP16 in the MRP1/2 depleted background significantly restored RNA levels, consistent with a negative effect of RBP16 in the absence of MRP1/2 (Fig. 8). The negative effect of RBP16 is even more apparent when one considers preedited MURF2 mRNA. Depletion of RBP16 alone had no effect on preedited MURF2 mRNA levels, while MRP1/2 depletion alone led to an 85% decrease. Remarkably, the decrease caused by MRP1/2 depletion was completely reversed by simultaneous depletion of RBP16 (Fig. 8). From these data, we conclude that MRP1/2 can protect some RNAs from RBP16-mediated decay.
A second interesting finding from these studies is that, for a given RNA, the interplay between MRP1/2 and RBP16 in BF cells differs from what is observed for the same RNA in PF cells. For example, the aforementioned negative effect of RBP16, apparent toward RPS12 in PF cells (Fig. 5A), was not manifested against this RNA in BF cells. Instead, in BF cells, RBP16 knockdown had no effect on the levels of preedited or edited RPS12 RNA in either the MRP1/2-replete or MRP1/2-depleted background (Fig. 8). Regarding ND4 and COI mRNAs, MRP1/2 and RBP16 apparently function in the same positive pathway in PF cells, while in BF cells, negative effects of RBP16 in the MRP1/2-depleted background were apparent (compare Fig. 8 and 4A). Edited COIII RNA also exhibited very different patterns in the two life cycle stages. While depletion of MRP1/2 or RBP16 alone had no effect on edited COIII RNA levels in PF cells (Fig. 3A), the downregulation of either of these factors in BF cells led to a dramatic decrease in edited COIII RNA (90% and 75%, respectively) (Fig. 8). Collectively, these data strongly imply the presence of life cycle stage-specific factors that intersect the MRP1/2 and RBP16 pathways.
DISCUSSION
Separate knockdowns or knockouts of MRP1/2 and RBP16 have previously revealed limited functions for these factors, in both cases involving primarily a dramatic effect on CYb mRNA editing and a more modest effect on the stability of some never-edited RNAs (22, 35, 49). RNAi against either factor also results in a similar growth defect in which PF cultures plateau several days following induction of RNAi but death of the culture is not observed (35, 49). In this work, we examined the effects of simultaneous MRP1/2-RBP16 depletion on mitochondrial RNA metabolism in both PF and BF T. brucei. By establishing simultaneous RNAi against both factors in PF cells, we found that cells depleted of both RBP16 and MRP1/2 were eventually cleared from the media, indicative of synthetic lethality. We also interrogated the relationship between these two factors by asking whether overexpression of RBP16 could rescue growth or RNA metabolic defects in MRP1/2-depleted cells. Together, these studies revealed a complex mixture of overlapping and distinct roles for MRP1/2 and RBP16, which differ depending on the RNA being examined and the life cycle stage.
Both RBP16 and MRP1/2 have previously been shown to be essential for editing of CYb mRNA (35, 49). Here, we have shown that these factors have separate and distinct functions and that the effects of their depletion are additive with regard to edited CYb mRNA (Fig. 2). Possible mechanisms by which MRP1/2 and RBP16 act during CYb RNA editing are suggested by structural and functional studies. Schumacher et al. (41) demonstrated that the MRP1/2 heterotetramer binds gRNA in a manner that exposes its anchor region, which is otherwise partially engaged in an intramolecular duplex. This suggests that MRP1/2 is critical for gRNA function during editing. RNA structure studies from the Koslowsky laboratory have shown that the cognate mRNA portion of the CYb/gCYb anchor duplex is engaged in an extremely long and thermodynamically stable stem-loop structure (23), which would presumably need to be unwound prior to the initiation of editing. Because RBP16 possesses RNA chaperone and RNA-unwinding activities (2), it could conceivably act to maintain the mRNA portion of the CYb anchor in an accessible conformation. Thus, a model consistent with the distinct and additive functions of MRP1/2 and RBP16 is one in which RBP16 maintains the mRNA anchor sequence in an accessible conformation, while MRP1/2 exposes the cognate gRNA anchor sequence. Additional biochemical studies will be required to validate this model. In any case, the current data indicate that MRP1/2 and RP16 do not interact with each other directly (39; C. Goulah and L. K. Read, unpublished data), again consistent with their separate functions during CYb mRNA editing.
In contrast to what was observed with CYb mRNA, we uncovered redundant roles for RBP16 and MRP1/2, as exemplified by their effects on the maintenance of edited A6 and COIII mRNAs. The levels of edited A6 and COIII mRNAs were largely unchanged in PF cells depleted of either MRP1/2 or RBP16. Only upon simultaneous downregulation of both MRP1/2 and RBP16 did the levels of fully edited A6 and COIII mRNAs display a marked decrease. RT-PCR analysis of A6 mRNAs in MRP1/2-RBP16-depleted cells demonstrated that the only edited species lost upon MRP1/2-RBP16 downregulation is apparently the fully edited A6 mRNA. The substantial population of partially edited RNAs is unaffected by MPR1/2-RBP16 depletion. These results strongly suggest that MRP1/2 and RBP16 act to stabilize fully edited A6 mRNA and do not impact the editing process itself. A similar process also occurs for COIII mRNA, where only the loss of MRP1/2 and RBP16 together results in a decrease of the fully edited RNA. However, the modest increase in RNAs corresponding to variously sized partially edited forms also suggests that loss of MRP1/2 and RBP16 may impact the editing of COIII, in addition to affecting the stability of the fully edited form. Notably, this is the first demonstration of an RNA decay pathway specific for fully edited mRNAs. A mitochondrial RNA decay pathway has been described in which edited RNAs are protected from degradation as a result of their poly(A) tails (10, 17). However, both in vitro and in vivo studies showed that polyadenylation-mediated stabilization of edited RNAs requires only minimal editing at the 3′ end of an RNA in order to afford protection from decay (10, 17). Thus, the efficient RNA decay pathway targeting fully edited A6 and COIII mRNAs that we observed here following the loss of MRP1/2 and RBP16 differs significantly from the previously described pathway in terms of substrates. Restriction of the pathway identified here to fully edited RNA suggests either that it involves specific recognition and stabilization of translatable RNAs by MRP1/2 or RBP16 upon completion of editing or that it possibly occurs in conjunction with translation. Overall, these data uncover redundant functional roles for MPR1/2 and RBP16 in stabilizing mRNAs following the editing process, as well as potential, yet specific, impacts on the editing process itself.
Past studies of MRP1/2 and RBP16 suggested that both factors exert positive effects on mitochondrial mRNA metabolism, so that depletion of either factor leads to reduced levels of some never-edited mRNAs (22, 35, 49). Thus, we were surprised to discover that RBP16 can also negatively affect the accumulation of specific RNAs. During our analysis of PF cells, we observed that both preedited and edited RPS12 mRNAs were stabilized in cells depleted of both MRP1/2 and RBP16 compared to cells expressing only RBP16 (i.e., MRP1/2-depleted cells). Similar effects were also observed for several RNAs in BF RNAi cells. These data are consistent with a model in which RBP16 can destabilize RNAs in an MRP1/2-depleted background, possibly by exposing negative cis elements or recruiting decay factors, but that this effect is blocked by MRP1/2. Counterintuitively, if we increase the RBP16 concentration about twofold in MRP1/2-depleted cells, the opposite effect is observed regarding RPS12 mRNA levels in PF cells. That is, RPS12 mRNA is completely stabilized when RBP16 is expressed above wild-type levels in the MRP1/2-depleted background. The simplest interpretation of these data is that, in an MRP1/2-depleted background, RBP16 can have both positive and negative effects on RNA stability depending on its local concentration. These results are reminiscent of what was observed for another cold shock domain-containing protein, p50, which in mammals has been shown to affect translation, as well as RNA annealing and RNA unwinding, in either a positive or negative manner depending on its local concentration (36, 46). In wild-type T. brucei, it is conceivable that local MRP1/2 and/or RBP16 concentrations differ depending on mitochondrial subcompartments or physiological conditions. The ratios of these two factors may then play an important role in the stabilization of specific RNA populations.
In BF trypanosomes, the simultaneous depletion of MRP1/2 and RBP16 had no discernible effect on the growth rate. Nevertheless, it is notable that we observed very substantial decreases in the levels of many mRNAs in these RNAi cells. It was striking that BF cells could survive in the face of such reduced RNA levels. This is particularly true as regards edited A6 mRNA, which was present at just 25 to 30% of wild-type levels. Edited A6 mRNA is thought to be essential in BF cells, since the ATPase complex is required for the maintenance of membrane potential in bloodstream T. brucei (6, 38). Work with akinetoplastic T. brucei, i.e., cells lacking kinetoplast DNA and which therefore do not undergo editing, has shown that these cells compensate for loss of the necessary A6 subunit and maintain membrane potential by a mutation in the nuclear-encoded ATPase gamma chain (21, 38). Therefore, the decrease in edited A6 mRNA in our BF cells could be compensated for by a mutation in the gamma chain of the ATPase. When we analyzed our BF RBP16-MRP1/2 RNAi cells for the gamma chain mutation reported in the petite T. brucei evansi line (21), the corresponding mutation was not evident (data not shown). However, we have not eliminated the possibility of other nuclear mutations in ATPase components that could compensate for A6 loss in these cells. Overall, our data suggest that wild-type levels of many mitochondrial RNAs are dispensable in the BF life cycle stage.
Another unexpected finding was that simultaneous MRP1/2-RBP16 depletion exerted very different effects on many mRNAs in BF compared to PF cells. Both RBP16 and MRP1/2 appear to be present at approximately equivalent protein levels during the two life cycle stages (35, 49). Together, these observations imply life cycle stage-specific factors that interact with MRP1/2 and RBP16, either stably or transiently, and modulate their functions toward specific mRNAs. It is also possible that differences in posttranslational modifications contribute to the functions of these factors. RBP16 is present in PF cells in multiple arginine-methylated forms, and both RBP16 and MRP2 are in vitro substrates for trypanosome protein arginine methyltransferases (12, 13, 32, 34). Arginine methylation has been shown to alter the functions and/or localization of RNA binding proteins (3, 5, 29, 43) and could modify the interactions of MRP1/2 and/or RBP16 with other proteins or RNAs or impact their suborganellar localization differentially during the life cycle.
In summary, the studies presented here revealed an unexpectedly complex interplay between the RNA binding factors MRP1/2 and RBP16 in their impacts on mitochondrial RNA metabolism. We uncovered a much larger role for these factors than had been previously appreciated. The observations that MRP1/2 and RBP16 have such different functions toward specific RNA populations suggests that there are multiple cis elements on their target RNAs and that additional mitochondrial proteins impact and modulate MRP1/2 and RBP16 functions.
Acknowledgments
We thank the members of the Read laboratory for many helpful discussions. We also thank George Cross for providing cell lines, C. C. Wang for the pLEW100-TbAuk1-3HA-PURO vector, and Jay Bangs and Ken Stuart for antibodies.
These studies were supported by NIH AI061580 and AI077520 grants to L.K.R. M.L.A. was supported in part by American Heart Association Postdoctoral Fellowship no. 0725813T.
Footnotes
Published ahead of print on 20 July 2009.
REFERENCES
- 1.Allen, T. E., S. Heidmann, R. Reed, P. J. Myler, H. U. Goringer, and K. D. Stuart. 1998. Association of guide RNA binding protein gBP21 with active RNA editing complexes in Trypanosoma brucei. Mol. Cell. Biol. 18:6014-6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ammerman, M. L., J. C. Fisk, and L. K. Read. 2008. gRNA/pre-mRNA annealing and RNA chaperone activities of RBP16. RNA 14:1069-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoki, K., Y. Ishii, K. Matsumoto, and M. Tsujimoto. 2002. Methylation of Xenopus CIRP2 regulates its arginine- and glycine-rich region-mediated nucleocytoplasmic distribution. Nucleic Acids Res. 30:5182-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aphasizhev, R., I. Aphasizheva, R. E. Nelson, and L. Simpson. 2003. A 100-kD complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA 9:62-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boisvert, F.-M., J. Cote, M.-C. Boulanger, P. Cleroux, F. Bachand, C. Autexier, and S. Richard. 2002. Symmetrical dimethylarginine methylation is required for the localization of SMN in Cajal bodies and pre-mRNA splicing. J. Cell Biol. 159:957-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, S. V., P. Hosking, J. Li, and N. Williams. 2006. ATP synthase is responsible for maintaining mitochondrial membrane potential in bloodstream form Trypanosoma brucei. Eukaryot. Cell 5:45-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carnes, J., J. Trotter, A. Peltan, M. Fleck, and K. Stuart. 2008. RNA editing in Trypanosoma brucei requires three different editosomes. Mol. Cell. Biol. 28:122-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carnes, J., J. R. Trotter, N. L. Ernst, A. Steinberg, and K. Stuart. 2005. An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 102:16614-16619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drozdz, M., S. S. Palazzo, R. Salavati, J. O'Rear, C. Clayton, and K. Stuart. 2002. TbMP81 is required for RNA editing in Trypanosoma brucei. EMBO J. 21:1791-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Etheridge, R. D., I. Aphasizheva, P. D. Gershon, and R. Aphasizhev. 2008. 3′ adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 27:1596-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisk, J. C., M. L. Ammerman, V. Presnyak, and L. K. Read. 2008. TbRGG2, an essential RNA editing accessory factor in two Trypanosoma brucei life cycle stages. J. Biol. Chem. 283:23016-23025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisk, J. C., J. Sayegh, C. Zurita-Lopez, S. Menon, V. Presnyak, S. G. Clarke, and L. K. Read. 2009. A type III protein arginine methyltransferase from the protozoan parasite Trypanosoma brucei. J. Biol. Chem. 284:11590-11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goulah, C. C., M. Pelletier, and L. K. Read. 2006. Arginine methylation regulates mitochondrial gene expression in Trypanosoma brucei through multiple effector proteins. RNA 12:1545-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goulah, C. C., and L. K. Read. 2007. Differential effects of arginine methylation on RBP16 mRNA binding, guide RNA (gRNA) binding, and gRNA-containing ribonucleoprotein complex (gRNP) formation. J. Biol. Chem. 282:7181-7190. [DOI] [PubMed] [Google Scholar]
- 15.Hashimi, H., A. Zikova, A. K. Panigrahi, K. D. Stuart, and J. Lukes. 2008. TbRGG1, an essential protein involved in kinetoplastid RNA metabolism that is associated with a novel multiprotein complex. RNA 14:970-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayman, M. L., and L. K. Read. 1999. Trypanosoma brucei RBP16 is a mitochondrial Y-box family protein with guide RNA binding activity. J. Biol. Chem. 274:12067-12074. [DOI] [PubMed] [Google Scholar]
- 17.Kao, C.-Y., and L. K. Read. 2005. Opposing effects of polyadenylation on the stability of edited and unedited mitochondrial RNAs in Trypanosoma brucei. Mol. Cell. Biol. 25:1634-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kao, C.-Y., and L. K. Read. 2007. Targeted depletion of a mitochondrial nucleotidyltransferase suggests the presence of multiple enzymes that polymerize mRNA 3′ tails in Trypanosoma brucei mitochondria. Mol. Biochem. Parasitol. 154:158-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koller, J., U. F. Muller, B. Schmid, A. Missel, V. Kruft, K. Stuart, and H. U. Goringer. 1997. Trypanosoma brucei gBP21. An arginine-rich mitochondrial protein that binds to guide RNA with high affinity. J. Biol. Chem. 272:3749-3757. [DOI] [PubMed] [Google Scholar]
- 20.Koslowsky, D. J., and G. Yahampath. 1997. Mitochondrial mRNA 3′ cleavage/polyadenylation and RNA editing in Trypanosoma brucei are independent events. Mol. Biochem. Parasitol. 90:81-94. [DOI] [PubMed] [Google Scholar]
- 21.Lai, D.-H., H. Hashimi, Z.-R. Lun, F. J. Ayala, and J. Lukes. 2008. Adaptations of Trypanosoma brucei to gradual loss of kinetoplast DNA: Trypanosoma equiperdum and Trypanosoma evansi are petite mutants of T. brucei. Proc. Natl. Acad. Sci. USA 105:1999-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert, L., U. F. Muller, A. E. Souza, and H. U. Goringer. 1999. The involvement of gRNA-binding protein gBP21 in RNA editing—an in vitro and in vivo analysis. Nucleic Acids Res. 27:1429-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leung, S. S., and D. J. Koslowsky. 1999. Mapping contacts between gRNA and mRNA in trypanosome RNA editing. Nucleic Acids Res. 27:778-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukes, J., H. Hashimi, and A. Zikova. 2005. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr. Genet. 48:277-299. [DOI] [PubMed] [Google Scholar]
- 25.Miller, M. M., K. Halbig, J. Cruz-Reyes, and L. K. Read. 2006. RBP16 stimulates trypanosome RNA editing in vitro at an early step in the editing reaction. RNA 12:1292-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, M. M., and L. K. Read. 2003. Trypanosoma brucei: functions of RBP16 cold shock and RGG domains in macromolecular interactions. Exp. Parasitol. 105:140-148. [DOI] [PubMed] [Google Scholar]
- 27.Muller, U. F., and H. U. Goringer. 2002. Mechanism of the gBP21-mediated RNA/RNA annealing reaction: matchmaking and charge reduction. Nucleic Acids Res. 30:447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller, U. F., L. Lambert, and H. U. Goringer. 2001. Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. EMBO J. 20:1394-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pahlich, S., R. P. Zakaryan, and H. Gehring. 2006. Protein arginine methylation: cellular functions and methods of analysis. Biochim. Biophys. Acta 1764:1890-1903. [DOI] [PubMed] [Google Scholar]
- 30.Panigrahi, A. K., N. L. Ernst, G. J. Domingo, M. Fleck, R. Salavati, and K. D. Stuart. 2006. Compositionally and functionally distinct editosomes in Trypanosoma brucei. RNA 12:1038-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panigrahi, A. K., A. Schnaufer, and K. D. Stuart. 2007. Isolation and compositional analysis of trypanosomatid editosomes. Methods Enzymol. 424:3-24. [DOI] [PubMed] [Google Scholar]
- 32.Pasternack, D. A., J. Sayegh, S. Clarke, and L. K. Read. 2007. Evolutionarily divergent type II protein arginine methyltransferase in Trypanosoma brucei. Eukaryot. Cell 6:1665-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelletier, M., M. M. Miller, and L. K. Read. 2000. RNA-binding properties of the mitochondrial Y-box protein RBP16. Nucleic Acids Res. 28:1266-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pelletier, M., D. A. Pasternack, and L. K. Read. 2005. In vitro and in vivo analysis of the major type I protein arginine methyltransferase from Trypanosoma brucei. Mol. Biochem. Parasitol. 144:206-217. [DOI] [PubMed] [Google Scholar]
- 35.Pelletier, M., and L. K. Read. 2003. RBP16 is a multifunctional gene regulatory protein involved in editing and stabilization of specific mitochondrial mRNAs in Trypanosoma brucei. RNA 9:457-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pisarev, A. V., M. A. Skabkin, A. A. Thomas, W. C. Merrick, L. P. Ovchinnikov, and I. N. Shatsky. 2002. Positive and negative effects of the major mammalian messenger ribonucleoprotein p50 on binding of 40S ribosomal subunits to the initiation codon of beta-globin mRNA. J. Biol. Chem. 277:15445-15451. [DOI] [PubMed] [Google Scholar]
- 37.Read, L. K., P. J. Myler, and K. Stuart. 1992. Extensive editing of both processed and preprocessed maxicircle CR6 transcripts in Trypanosoma brucei. J. Biol. Chem. 267:1123-1128. [PubMed] [Google Scholar]
- 38.Schnaufer, A., G. D. Clark-Walker, A. G. Steinberg, and K. Stuart. 2005. The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J. 24:4029-4040. (Erratum, 25:1175-1176, 2006.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schnaufer, A., N. L. Ernst, S. S. Palazzo, J. O'Rear, R. Salavati, and K. Stuart. 2003. Separate insertion and deletion subcomplexes of the Trypanosoma brucei RNA editing complex. Mol. Cell 12:307-319. [DOI] [PubMed] [Google Scholar]
- 40.Schnaufer, A., A. K. Panigrahi, B. Panicucci, R. P. Igo, Jr., E. Wirtz, R. Salavati, and K. Stuart. 2001. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science 291:2159-2162. (Erratum, 293:1997, 2001.) [DOI] [PubMed] [Google Scholar]
- 41.Schumacher, M. A., E. Karamooz, A. Zikova, L. Trantirek, and J. Lukes. 2006. Crystal structures of T. brucei MRP1/MRP2 guide-RNA binding complex reveal RNA matchmaking mechanism. Cell 126:701-711. [DOI] [PubMed] [Google Scholar]
- 42.Seiwert, S. D. 1995. The ins and outs of editing RNA in kinetoplastids. Parasitol. Today 11:362-368. [DOI] [PubMed] [Google Scholar]
- 43.Shen, E. C., M. F. Henry, V. H. Weiss, S. R. Valentini, P. A. Silver, and M. S. Lee. 1998. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 12:679-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simpson, L., R. Aphasizhev, G. Gao, and X. Kang. 2004. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. RNA 10:159-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simpson, L., S. Sbicego, and R. Aphasizhev. 2003. Uridine insertion/deletion RNA editing in trypanosome mitochondria: a complex business. RNA 9:265-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skabkin, M. A., V. Evdokimova, A. A. Thomas, and L. P. Ovchinnikov. 2001. The major messenger ribonucleoprotein particle protein p50 (YB-1) promotes nucleic acid strand annealing. J. Biol. Chem. 276:44841-44847. [DOI] [PubMed] [Google Scholar]
- 47.Stuart, K. D., A. Schnaufer, N. L. Ernst, and A. K. Panigrahi. 2005. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 30:97-105. [DOI] [PubMed] [Google Scholar]
- 48.Tu, X., P. Kumar, Z. Li, and C. C. Wang. 2006. An aurora kinase homologue is involved in regulating both mitosis and cytokinesis in Trypanosoma brucei. J. Biol. Chem. 281:9677-9687. [DOI] [PubMed] [Google Scholar]
- 49.Vondruskova, E., J. van den Burg, A. Zikova, N. L. Ernst, K. Stuart, R. Benne, and J. Lukes. 2005. RNA interference analyses suggest a transcript-specific regulatory role for mitochondrial RNA-binding proteins MRP1 and MRP2 in RNA editing and other RNA processing in Trypanosoma brucei. J. Biol. Chem. 280:2429-2438. [DOI] [PubMed] [Google Scholar]
- 50.Weng, J., I. Aphasizheva, R. D. Etheridge, L. Huang, X. Wang, A. M. Falick, and R. Aphasizhev. 2008. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol. Cell 32:198-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wickstead, B., K. Ersfeld, and K. Gull. 2002. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol. Biochem. Parasitol. 125:211-216. [DOI] [PubMed] [Google Scholar]
- 52.Wirtz, E., S. Leal, C. Ochatt, and G. A. Cross. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89-101. [DOI] [PubMed] [Google Scholar]
- 53.Zikova, A., E. Horakova, M. Jirk, P. Dunajcikova, and J. Lukes. 2006. The effect of down-regulation of mitochondrial RNA-binding proteins MRP1 and MRP2 on respiratory complexes in procyclic Trypanosoma brucei. Mol. Biochem. Parasitol. 149:65-73. [DOI] [PubMed] [Google Scholar]
- 54.Zikova, A., J. Kopecna, M. A. Schumacher, K. Stuart, L. Trantirek, and J. Lukes. 2008. Structure and function of the native and recombinant mitochondrial MRP1/MRP2 complex from Trypanosoma brucei. Int. J. Parasitol. 38:901-912. [DOI] [PMC free article] [PubMed] [Google Scholar]