Abstract
E3 ubiquitin ligases, which target specific molecules for proteolytic destruction, have emerged as key regulators of immune functions. Several E3 ubiquitin ligases, including c-Cbl, Cbl-b, GRAIL, Itch, and Nedd4, have been shown to negatively regulate T-cell activation. Here, we report that the HECT-type E3 ligase AIP2 positively regulates T-cell activation. Ectopic expression of AIP2 in mouse primary T cells enhances their proliferation and interleukin-2 production by suppressing the apoptosis of T cells. AIP2 interacts with and promotes ubiquitin-mediated degradation of EGR2, a zinc finger transcription factor that has been found to regulate Fas ligand (FasL) expression during activation-induced T-cell death. Suppression of AIP2 expression by small RNA interference upregulates EGR2, inhibits EGR2 ubiquitination and FasL expression, and enhances the apoptosis of T cells. Therefore, AIP2 regulates activation-induced T-cell death by suppressing EGR2-mediated FasL expression via the ubiquitin pathway.
Protein ubiquitination, a process that covalently conjugates ubiquitin to the lysine sites of a target protein, has emerged as a crucial regulatory mechanism in immune functions in mammals. E3 ubiquitin ligases determine the specificities of ubiquitination by recruiting substrate proteins. There are two main classes of E3 ligases, as follows: proteins with a catalytic domain that is homologous to the E6-AP carboxyl terminus (HECT) (19) and proteins with a really interesting new gene (RING) finger domain (20). Several E3 ubiquitin ligases have been shown to have important roles in T-cell activation and anergy induction (24, 32). The RING-type E3 ubiquitin ligases Cbl, Cbl-b, and GRAIL and the HECT-type E3 ligases Itch and NEDD4 have been identified as important regulators of T-cell development, activation, differentiation, and tolerance (1, 2, 5, 12, 18, 33).
AIP2, also known as WW domain-containing protein 2 (WWP2), is a member of the atrophin interaction protein (AIP) family (41). AIP family members are characterized by the presence of a catalytic HECT domain that facilitates ubiquitin ligation to substrate proteins. In addition, members of this family of E3 ligases contain multiple WW domains that mediate binding to PPXY motifs (“X” refers to any amino acid), and an NH2-terminal C2 domain (calcium/lipid binding) that may be important for the protein subcellular localization (27). AIP2 is widely expressed in most of the tissues and organs of both humans and mice (29), (3). The physiological functions of AIP2 are largely unknown. Several in vitro studies have suggested that AIP2 is involved in downregulating epithelial sodium channels by directly targeting all three subunits of the sodium channel for ubiquitination (36). AIP2 is also one of the host factors that regulate the budding process of retroviruses (28). Recently, Xu et al. reported that AIP2 is an E3 ubiquitin ligase of the transcription factor Oct-4 (43). Thus, AIP2 may be involved in embryonic development functioning to degrade Oct-4 via the ubiquitin pathway.
EGR2, a member of the early growth response (EGR) protein family, has been isolated as a transcription factor (4, 21, 23, 41, 42). A hallmark of the EGR family of transcription factors is a DNA-binding domain consisting of three Cys2His2 zinc finger motifs (21, 23). This domain is known to bind the sequence GCGGGGCG. There are four members of the egr family genes: EGR1 (Krox24, NGFI-A), EGR2 (Krox20), EGR3 (PILOT), and EGR4 (NGFI-C) (34). EGR2 has been most widely studied in the context of the nervous system, and targeted mutation in mice results in early lethality concurrent to defects in hindbrain patterning, peripheral nerve myelination, and bone formation (15). It has been shown that EGR2 transactivation is dependent on members of the NFAT family in T lymphocytes (35). Recent studies demonstrated that EGR2 is one of the key negative regulators of T-cell activation and activation-induced apoptosis and is involved in T-cell anergy induction (17, 37).
Here, we report that AIP2 positively regulates the activation of T cells by suppressing activation-induced apoptosis. AIP2 promotes EGR2 degradation and inhibits EGR2-driven Fas ligand (FasL) expression in T cells. These results reveal a previously uncharacterized mechanism underlying protein ubiquitination in T lymphocytes.
MATERIALS AND METHODS
Cell line, antibodies, and reagents.
Human embryonic kidney 293 (HEK293) T cells were maintained in Dulbecco's modification of Eagle's medium (Invitrogen Life Technologies, San Diego, CA) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 200 μg/ml streptomycin, and 0.25 μg/ml amphotericin B. Polyclonal antibodies against AIP2 or the epitope tag hemagglutinin (HA) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-CD3 and anti-CD28 were obtained from eBioscience (San Diego, CA). The anti-actin and anti-Flag antibodies were obtained from Sigma (St. Louis, MO). The anti-EGR2 antibody was obtained from Covance (Princeton, NJ). The anti-ubiquitin antibody was purchased from Boston Biochem, Inc. (Boston, MA).
Plasmids.
An HA-tagged ubiquitin expression plasmid was used, as reported previously (13). The full-length human AIP2 expression plasmid was a kind gift from Paul Bieniasz (The Rockefeller University) (28). To generate truncated mutants, fragments of the AIP2 cDNA were amplified by PCR using linker primers that introduced an EcoRI site at the 5′ end and an XbaI site at the 3′ end. PCR products were digested with EcoRI and XbaI and ligated into the pEF/myc/hisB expression vector (Invitrogen). The EGR2 expression plasmid is a generous gift from Jeffrey Milbrandt (Washington University) (6). Point mutations were generated by PCR. AIP2 small interfering RNA (siRNA) expression plasmids were generated by subcloning AIP2 fragments (see Fig. 6A) into pLentiLox 3.7 plasmids. All the newly generated plasmids in this study were verified by DNA sequencing.
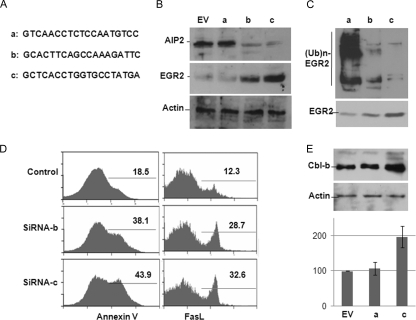
FIG. 6.
AIP2 knockdown upregulates FasL expression in T cells. (A) The selected sequences for AIP2 siRNA. (B) Mouse CD4+ T cells were infected with lentiviruses that carry empty control vector or each of these three API2 siRNAs. The expression of AIP2 (top) and EGR2 (middle) were analyzed by Western blotting. Actin expression was analyzed as a control (bottom). (C) EGR2 ubiquitination in AIP2 knockdown or control T cells was determined by immunoprecipitation of EGR2 and Western blotting with anti-ubiquitin (Ub) antibody. (D) Mouse CD4+ T cells were infected with control or AIP2 siRNA viruses. Infected cells were restimulated with anti-CD3 plus anti-CD28, and the percentages of apoptotic cells (left panels) and FasL expression cells (right panels) were analyzed by flow cytometry. (E) Whole-cell lysates of AIP2 knockdown and control cells were subjected to Western blot analysis of Cbl-b (top) and actin (middle) protein expression. The mRNA levels of Cbl-b in these cells were analyzed by real-time reverse transcriptase PCR in parallel (bottom). Error bars indicate data from three independent experiments (means ± standard deviations).
Isolating mouse primary T cells and retroviral infection.
T cells were isolated from the lymph nodes and spleens of 4- to 6-week-old C57BL/6 mice. All mice were maintained under specific pathogen-free conditions at the University of Missouri (Columbia, MO), and all animals were used in accordance with the strict guidelines of the Institutional Animal Care Committee. CD4+ T cells were purified by negative selection with the CD4+ T-cell isolation kit (Miltenyi Biotec, Auburn, CA). These cells were maintained in RPMI media (Invitrogen Life Technologies, San Diego, CA) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 200 μg/ml streptomycin, and 0.25 μg/ml amphotericin and stimulated with anti-CD3 plus anti-CD28 (5 μg/ml each) for 24 h before infection. Retrovirus was prepared by transfection of pMIG-AIP2 into the packaging cells PlatE, as described previously (31). Supernatants were used for the infection of preactivated CD4+ T cells. During infection, Polybrene (10 μg/ml), anti-CD3, anti-CD28, and interleukin-2 (IL-2) (20 U/ml) were added. Two days after infection, green fluorescent protein-positive (GFP+) cells were sorted and used for analysis.
T-cell proliferation assay and IL-2 production.
Mouse primary T cells (1 × 105 cells in 300 μl/well) were stimulated with anti-CD3 or anti-CD3 plus anti-CD28 for 48 h. Supernatants (100 μl) from each well were collected for analysis of IL-2 by enzyme-linked immunosorbent assay. Cells were pulsed with 1 μCi/ml of [3H]thymidine and incubated for an additional 16 h. [3H]thymidine incorporation was measured with a scintillation counter.
Yeast two-hybrid screening of AIP2-interacting proteins.
The yeast pGilda plasmid encoding a cDNA fragment of the AIP2 WW domains was fused to the LexA DNA-binding domain (pBD-AIP2/WW). A human Jurkat T-cell cDNA library in the pJG4-5 vector, which expresses the Escherichia coli B42 transactivating domain (pAD), was purchased from OriGene Technologies, Inc. (Rockville, MD). Library screening by the yeast two-hybrid method was performed according to the manufacturer's instructions. Briefly, the yeast EGY48 strain of Saccharomyces cerevisiae (MAT trp1 ura3 his leu2::plexApo6-leu2) was first transformed with the lacZ reporter plasmid p8op-lacZ to create EGY48[p8op-lacZ]. The latter yeast was transformed with the cDNA library. Colonies that formed on leucine-deficient synthetic dextrose plates containing 2% galactose and 1% raffinose were removed and grown in fresh synthetic dextrose plates for 2 to 4 days, then transferred to a piece of Whatman filter paper, and tested for activation of the lacZ reporter gene using the β-galactosidase colony-lift filter assay. From an initial screening of ∼2 × 106 transformants, 58 colonies that transactivated the LEU2 reporter gene were identified. Of these, 37 colonies were also positive for β-galactosidase when color was scored after 1 h. Isolated plasmid DNAs from these LEU+/lacZ+ clones were again cotransformed with the AIP2/WW bait (or with empty pGilda as a negative control) to eliminate false positives and confirm two-hybrid interactions.
Transfection, immunoprecipitation, and Western blotting.
Transient transfections were performed by using Lipofectamine (Invitrogen), according to the manufacturer's instructions, with 6-cm dishes and 2 to 5 μg of total DNA per transfection. Transfected cells were pelleted and resuspended in Nonidet P-40 lysis buffer (1% Nonidet P-40, 20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 5 mM sodium piperidinedithiocarbamate, 2 mM Na3VO4, and 10 μg/ml each of aprotinin and leupeptin). Cells were lysed for 10 min at 4°C, and insoluble materials were removed by centrifugation at 15,000 × g (4°C, 10 min). For immunoprecipitation, lysates (∼1 × 107 cells) were mixed with antibodies (1 μg) for 2 h, followed by the addition of 30 μl of protein G-Sepharose beads (Santa Cruz Biotechnology) for an additional 2 h at 4°C. Immunoprecipitates were washed four times with Nonidet P-40 lysis buffer and boiled in 20 μl of 2× Laemmli's buffer. Samples were subjected to 8% or 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis analysis and electrotransferred onto polyvinylidene difluoride membranes (Millipore). Membranes were probed with the indicated primary antibodies (usually 1 μg/ml), followed by horseradish peroxidase-conjugated secondary antibodies. Membranes were then washed and visualized with an enhanced chemiluminescence detection system (ECL kit; Amersham Pharmacia Biotech). When necessary, membranes were stripped by incubation in stripping buffer (62.5 mM Tris-HCl [pH 5.7], 100 mM 2-mercaptoethanol, and 2% SDS) for 1 h at 70°C with constant agitation, washed, and then reprobed with other antibodies as indicated.
In vitro protein-protein interaction assay.
The cDNA fragment corresponding to four WW domains of AIP2 (amino acids 281 to 500) was subcloned into pGEX-6P glutathione S-transferase (GST) fusion protein expression vector and transformed into E. coli BL21 (Invitrogen). The GST-AIP2 WW protein in the lysates of bacteria was purified using glutathione agarose beads (Sigma). Similarly, EGR2 cDNA was subcloned into a His-tagged protein expression plasmid, pET-28a(+), and the His-EGR2 fusion protein was purified using His beads. Purified proteins were mixed in binding buffer (250 mM NaCl, 10 mM Tris [pH 7.5], 1 mM dithiothreitol) at 5 μg/ml each and incubated for 1 h on ice, followed by addition of GST beads for an additional 30 min of incubation. Beads were then washed three times with binding buffer and subjected to SDS-polyacrylamide gel electrophoresis and Western blotting analysis.
RESULTS
AIP2 enhances the activation of mouse primary T cells.
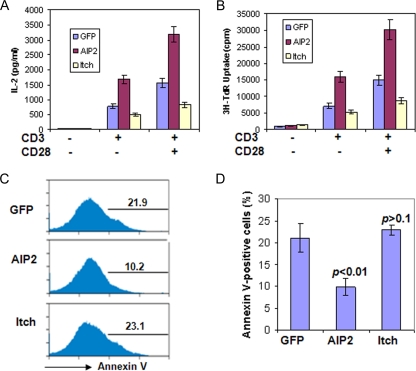
We have previously demonstrated that Itch, a HECT-type E3 ubiquitin ligase, is a negative regulator of T-cell activation and Th2 differentiation (10). To determine the involvement of the HECT-type E3 ligases in T-cell functions, we investigated the effects of seven HECT-type E3 ligases on the activation of mouse primary T cells. We found that ectopic expression of AIP2 significantly enhanced the proliferation and IL-2 production of T cells. Expression of the control Itch in mouse primary T cells inhibited T-cell activation (Fig. 1A and B). Therefore, unlike Itch, which inhibits T-cell activation, our results suggest that AIP2 is a positive regulator of T cells.
FIG. 1.
AIP2 expression enhances T-cell activation. (A) Mouse primary T cells were infected with retrovirus carrying AIP2, Itch, or GFP only as a control. Infected cells were cultivated with anti-CD3 or anti-CD3 plus anti-CD28 for 2 days. Partial supernatants were collected from each well for the analysis of IL-2 production by enzyme-linked immunosorbent assay. (B) Cells were then chased with 0.5 μCi [3H]thymidine (3H-TdR) for an additional 12 h, and the [3H]thymidine incorporation was analyzed. Error bars represent data from three independent experiments (means ± standard deviations). (C) Infected cells were stained with phycoerythrin-conjugated annexin V. GFP-positive cells were gated for the analysis of annexin V, and representative data are shown. (D) The average percentages of annexin V-positive cells from three independent experiments are shown. The Student t test was used for the statistic analysis.
To further investigate how AIP2 promotes T-cell activation, we analyzed the effects of AIP2 expression on the apoptosis of CD4+ T cells. As shown in Fig. 1C, AIP2 expression significantly inhibited the apoptosis of mouse CD4+ T cells, while expression of the control Itch had little effect on T-cell apoptosis (Fig. 1C and D). In addition, AIP2 expression increased the percentage of IL-2+ cells but not the level of IL-2 in each cell (data not shown). Therefore, AIP2 appears to facilitate T-cell activation by suppressing activation-induced cell death.
A previous report indicated that AIP2 is highly expressed in mouse spleens, as demonstrated by Northern blotting (43). Here, we further analyzed the expression patterns of AIP2 in mouse immune tissues, including bone marrow, thymus, spleen, and lymph node, by Western blotting. The AIP2 protein was detected at very low levels in mouse bone marrow, while it is highly expressed in the thymus, spleen, and lymph nodes (data not shown). AIP2 is also highly expressed in naïve CD4+ T cells. Stimulation of those T cells with anti-CD3 or anti-CD28 did not significantly induce AIP2 expression (data not shown). These results indicate that AIP2 is well expressed in mouse immune tissues and T cells, suggesting that AIP2 may be involved in the regulation of the immune functions in mice.
AIP2 interacts with and promotes ubiquitination of EGR2 in vitro.
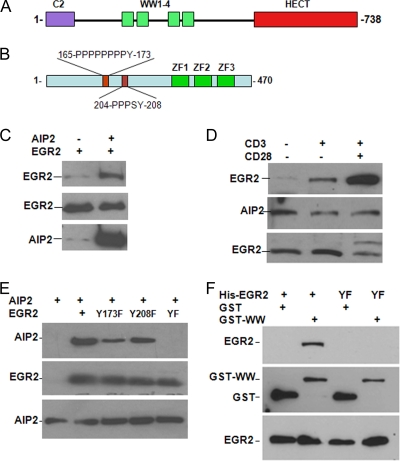
To determine the molecular mechanisms of AIP2 in regulating T-cell activation, we used a yeast two-hybrid screening approach to identify the potential target proteins of this E3 ligase. For this purpose, we subcloned the DNA fragment corresponding to these four WW domains of AIP2 into the pBD vector and used it as bait (Fig. 2A). A cDNA library generated from Jurkat T-cell mRNA was used. About 2 × 106 clones were screened, and 37 positive clones were obtained. Of those 37 clones, 3 were cDNA fragments of the N terminus of EGR2. Analysis of these three fragments revealed two PPXY motifs (Fig. 2B) that were previously demonstrated to bind specifically to the WW domains (26), suggesting that EGR2 interacts with AIP2.
FIG. 2.
AIP2 interacts with EGR2. (A and B) Domain structures of AIP2 and EGR2. (C) An EGR2 expression plasmid was transfected with or without the AIP2 plasmid into HEK293 cells. AIP2 in the lysates of transfected cells was immunoprecipitated with anti-AIP2 antibody. Binding EGR2 to AIP2 was determined with anti-EGR2 antibody (top). The same membrane was reprobed with anti-AIP2 antibody (middle). Protein levels of EGR2 in whole-cell lysates were analyzed as a control (bottom). (D) Purified primary T cells were stimulated with anti-CD3 or anti-CD3 plus anti-CD28 for 2 h. Interactions occurring in those cells were analyzed, as shown in panel A. (E) Myc-tagged AIP2 expression plasmids were cotransfected with Flag-EGR2 or each of its YF mutants into HEK293 cells. The interaction of AIP2 with EGR2 or its YF mutants was determined by coimmunoprecipitation of EGR2 with anti-Flag antibody and Western blotting with anti-Myc antibody (top). Protein expression of EGR2 (middle) and AIP2 (bottom) was analyzed by Western blotting using anti-Myc and anti-Flag, respectively. (F) GST or GST-WW proteins (5 μg) were incubated with His-EGR2 or His-EGR2/YF proteins. GST or GST-WW proteins were pulled down by adding GST-Sepharose beads. Binding of EGR2 was detected with anti-EGR2 antibody (top). The same membrane was reprobed with anti-GST antibodies (middle). The levels of EGR2 in the whole reaction mixtures were determined by Western blotting using anti-EGR2 antibody (bottom).
We then confirmed the interaction of AIP2 with EGR2. An EGR2 expression plasmid was cotransfected with or without an AIP2 expression plasmid into HEK293 cells. The AIP2 protein in the lysates of transfected cells was immunoprecipitated with anti-AIP2 antibody, and the binding of EGR2 in the immunoprecipitates was detected with anti-EGR2 antibody. As shown in Fig. 2C, a strong EGR2 band was detected in the anti-AIP2 immunoprecipitate when AIP2 was cotransfected, indicating that AIP2 interacts with EGR2. A weak EGR2 band was detected when AIP2 was not cotransfected because endogenous AIP2 protein expression is very low. These results suggest that AIP2 interacts with EGR2. In support of this, we also found that AIP2 colocalized with EGR2 when these two proteins were coexpressed in HEK293 cells (data not shown).
Next, we analyzed whether AIP2 interacts with EGR2 in mouse primary T cells. A weak EGR2 band was detected in the anti-AIP2 immunoprecipitate from the lysates of naïve T cells. Stimulation of T cells with anti-CD3 enhanced the interaction of EGR2 with AIP2, which was further enhanced by anti-CD28 stimulation (Fig. 2D). These results indicate that AIP2 interacts with EGR2, and their interaction is regulated by T-cell receptor (TCR)/CD28 signals in mouse primary T cells. Unfortunately, we were unable to detect the colocalization of AIP2 with EGR2 because the commercial anti-AIP2 antibody did not work for immunostaining the AIP protein in T cells.
AIP2 contains four WW domains that were found specifically to recognize PPXY motifs. Given that the EGR2 protein contains two PPXY motifs (Fig. 2B), we hypothesized that the WW domains of AIP2 and the PPXY motifs of EGR2 mediate their interaction. Mutation of a single PPXY motif of EGR2 only partially reduced its interaction with AIP2. However, when both PPXY motifs were mutated, their interaction was completely abolished (Fig. 2E). We then tested whether the WW domains of AIP2 mediates its interaction with EGR2. The GST-WW fusion protein but not the GST control interacted with His-EGR2 (see Fig. 4B). Mutation of the tyrosines in those two PPXY motifs of EGR2 completely abolished its interaction with GST-WW fusion proteins (Fig. 2F). These results indicate that the WW domain of AIP2 and the PPXY motifs of EGR2 mediate their interaction.
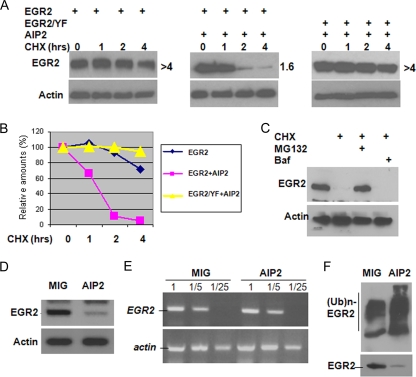
FIG. 4.
AIP2 expression promotes EGR2 degradation. (A) Flag-tagged EGR2 or its YF mutant expression plasmids were cotransfected with or without Myc-AIP2 expression plasmids into HEK293 cells. Transfected cells were treated with cycloheximide (CHX; 50 μg/ml) for different amounts of time, as indicated. The protein stability of EGR2 was examined by Western blotting using anti-Flag antibody (top panels). Protein levels of actin were determined as loading controls (bottom panels). (B) The densities of each EGR2 band were quantified using NIH Image 1.63 software. The half-lives of EGR2 and its YF mutant were calculated as shown in panel A. (C) AIP2 and EGR2 expression plasmids were cotransfected into HEK293T cells. Cells were treated with or without cycloheximide in the absence or presence of MG132 or with bafilomycin A (Baf) for 4 h. EGR2 protein levels were analyzed by Western blotting (top). The same membrane was reprobed with anti-actin antibody as a control (bottom). (D) Mouse primary T cells were infected with retrovirus carrying control vector (MIG) or with MIG-AIP2. Protein levels of EGR2 in infected T cells were analyzed by Western blotting. (E) Total RNA in these infected T cells shown in panel C was isolated, and the levels of EGR2 mRNA were determined by reverse transcriptase PCR. (F) T cells infected with virus carrying the MIG empty vector or MIG-AIP2 were cultured with anti-CD3 plus anti-CD28 for 2 days. EGR2 ubiquitination was detected by immunoprecipitation with anti-EGR2 antibody and Western blotting with anti-Ub antibody (top). Protein expression of EGR2 was analyzed with anti-EGR2 antibody (bottom).
AIP2 catalyzes ubiquitination of EGR2.
As an E3 ubiquitin ligase, AIP2 potentially induces ubiquitin conjugation onto its interaction partners. We thus tested whether AIP2 can mediate EGR2 ubiquitination. We transfected Flag-tagged EGR2 and HA-tagged ubiquitin without or with Myc-tagged AIP2 into HEK293 cells. The EGR2 protein was immunoprecipitated with anti-Flag antibody, and the conjugation of ubiquitin to EGR2 was detected with anti-HA antibody. Coexpression of AIP2 promoted EGR2 ubiquitination, and EGR2 ubiquitination was not detectable without AIP2 cotransfection (Fig. 3A). Together with our findings that AIP2 interacts with EGR2 (Fig. 2), these results suggest that AIP2 is an E3 ubiquitin ligase of EGR2.
FIG. 3.
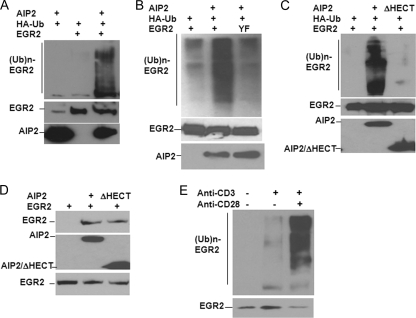
AIP2 is an E3 ubiquitin ligase of EGR2. (A) Myc-AIP2, Flag-EGR2, and HA-ubiquitin (Ub) expression plasmids were transfected into HEK293 cells. The EGR2 protein was immunoprecipitated with anti-Flag antibody. Ubiquitin conjugation onto EGR2 was detected by Western blotting using anti-HA antibody (top). The same membrane was reprobed with anti-EGR2 antibody (middle). Protein expression of AIP2 in whole-cell lysates was detected with anti-Myc antibody (bottom). (B) Flag-tagged EGR2 or its YF mutant was cotransfected with HA-ubiquitin and AIP2. Ubiquitination of EGR2 or its mutant was determined as described in the legend to panel A. (C) Flag-EGR2 and HA-ubiquitin expression plasmids were cotransfected into HEK293 cells with Myc-tagged AIP2 or with the AIP2/ΔHECT mutant. Ubiquitination of EGR2 was determined as described in the legend to panel A. (D) The interaction of Myc-AIP2 or Myc-AIP2/ΔHECT with Flag-EGR2 was analyzed by immunoprecipitation of Myc-tagged AIP2 or AIP2/ΔHECT with anti-Myc antibody and Western blotting with anti-EGR2 antibody (top). The expression levels of AIP2, AIP2/ΔHECT (middle), and EGR2 (bottom) were determined as controls. (E) Mouse primary T cells were stimulated with anti-CD3 or anti-CD3 plus anti-CD28 for 24 h. EGR2 ubiquitination was detected by immunoprecipitation of EGR2 from the lysates of stimulated T cells and Western blotting with anti-ubiquitin antibody (top). The expression of EGR2 was analyzed as a control (bottom).
The interaction of an E3 ubiquitin ligase with its substrate proteins, either direct or indirect, is required for ubiquitin conjugation (13). We next tested whether the mutation of these two PPXY motifs of EGR2, which abolished its interaction with AIP2, affected AIP2-mediated ubiquitination. As shown in Fig. 3B, AIP2 overexpression significantly enhanced EGR2 ubiquitination, but its overexpression failed to promote ubiquitin conjugation onto the tyrosine-to-phenylalanine mutant of EGR2 (EGR2/YF), indicating the interaction between AIP2 and EGR2 is required for AIP2-mediated EGR2 ubiquitination.
The HECT domain has been demonstrated to harbor E3 ligase activity, which recruits ubiquitin-carrying E2s and transfers ubiquitin to the lysine sites of a substrate protein (19). We thus tested whether the HECT domain of AIP2 is required for EGR2 ubiquitination. Deletion of the HECT domain of AIP2 completely abolished its ability to promote EGR2 ubiquitination (Fig. 3C), while the interaction of AIP2 with EGR2 was not affected by deletion of the HECT domain (Fig. 3D). Thus, AIP2-mediated EGR2 ubiquitination requires the HECT E3 ligase domain.
Since the activation signal mediated by TCR/CD28 stimuli facilitates AIP2/EGR2 interaction in T cells (Fig. 2D), we then asked whether EGR2 ubiquitination is increased in activated T cells. Indeed, anti-CD3 plus anti-CD28 stimulation significantly enhanced EGR2 ubiquitination, while TCR stimulation alone has modest effects (Fig. 3E). Therefore, AIP2-mediated EGR2 ubiquitination is regulated by T-cell activation signals.
AIP2 promotes EGR2 degradation.
Ubiquitin conjugation to protein substrates can induce their degradation via the proteasome or lysosome; therefore, we tested whether AIP2 induces EGR2 degradation. When overexpressed in HEK293 cells, EGR2 was very stable, with a half-life of more than 4 h. One possible reason for this is that endogenous AIP2 protein levels in HEK293 cells are very low (Fig. 2A). When AIP2 was coexpressed, the EGR2 protein was quickly degraded, with a half-life of 1.6 h, indicating that AIP2 promotes EGR2 degradation. Overexpression of AIP2 did not promote the degradation of the EGR2/YF mutant (Fig. 4A and B). The proteasome is involved mainly in AIP2-mediated EGR2 degradation because the proteasomal-specific inhibitor MG132, but not the lysosomal inhibitor bafilomycin A, rescued EGR2 from degradation (Fig. 4C). Together with our findings that the EGR2/YF mutation abolished AIP2-mediated ubiquitination, these results indicate that AIP2 promotes EGR2 degradation via the ubiquitin pathway.
To test whether AIP2 enhances T-cell activation by promoting EGR2 degradation, we determined the protein expression levels of EGR2 in mouse primary T cells when AIP2 is overexpressed. Figure 4D shows that protein levels of EGR2 were dramatically reduced in T cells in which AIP2 was overexpressed, suggesting that AIP2 downregulates EGR2 expression. Protein expression can be regulated at both mRNA and posttranslational levels. To further investigate the molecular mechanisms underlying AIP2 suppression of EGR2 expression, we examined whether expression of AIP2 affects EGR2 mRNA levels. As shown in Fig. 4E, mRNA levels were not changed in those T cells in which AIP2 was overexpressed compared to its levels in T cells infected with control virus, suggesting that AIP2 inhibits EGR2 expression at the posttranslational level in T cells. To support this, we further demonstrated that AIP2 expression enhanced EGR2 ubiquitination in T cells (Fig. 4F).
AIP2 inhibits FasL expression in T cells.
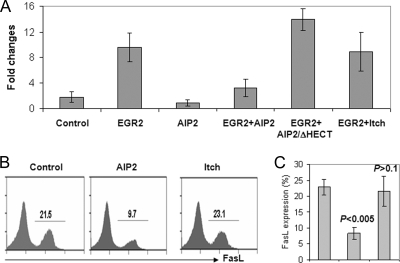
Studies have demonstrated that EGR2 regulates activation-induced T-cell death by promoting FasL expression (7-9, 30, 35, 44, 45). Together with our findings here that AIP2 expression inhibits T-cell apoptosis and promotes EGR2 degradation, we hypothesized that AIP2 suppressed FasL expression by degrading its transcription factor EGR2. Indeed, expression of AIP2, but not its E3 ligase domain deletion mutant or Itch, significantly inhibited EGR2-driven FasL transcription, as monitored by luciferase assay (Fig. 5A). Furthermore, ectopic expression of AIP2, which enhances T-cell activation and reduces activation-induced T-cell death (Fig. 1A and C), significantly inhibited FasL expression in mouse primary T cells (Fig. 5B and C). Therefore, AIP2 enhances T-cell activation and attenuates a suppressor of activation-induced T-cell death by suppressing EGR2-driven FasL expression.
FIG. 5.
AIP2 inhibits FasL expression in T cells. (A) FasL luciferase, AIP2, and EGR2 expression plasmids were cotransfected into HEK293 cells, with combinations as indicated. Luciferase activities were determined. Error bars represent data from three independent experiments. (B) Mouse primary T cells were infected with retroviruses that carry the pMIG control plasmid, pMIG-AIP2, or pMIG-Itch. Infected cells were restimulated with anti-CD3 plus antiCD28 for 24 h. FasL expression in the gated GFP+ cells was analyzed by flow cytometry. (C) The percentage of FasL+ cells, as analyzed in panel B, is shown. Error bars indicate data from four independent experiments (means ± standard deviations).
Knockdown of AIP2 expression promotes activation-induced T-cell death.
To further delineate the functional roles of AIP2 in T-cell function, we used a small RNA interference approach to suppress AIP2 expression in T cells. Out of three selected 19-nucleotide cDNA fragments corresponding to the open reading frame of AIP2 (Fig. 6A), two were found to significantly inhibit AIP2 expression in mouse primary T cells, with 85% and 93% suppressive efficiencies, respectively, and, as a consequence, increased EGR2 protein levels in T cells. As a control, actin protein levels were not affected by AIP2 siRNA (Fig. 6B). These increased EGR2 protein levels by AIP2 suppression are due to the reduced EGR2 ubiquitination (Fig. 6C). Together with our findings that AIP2 overexpression induces EGR2 ubiquitination and promotes EGR2 degradation (Fig. 3 and 4), we conclude that AIP2 E3 ubiquitin ligase is responsible for EGR2 protein degradation in T cells.
As a transcription factor for FasL expression in T cells, increased EGR2 protein expression by AIP2 knockdown presumably facilitates activation-induced T-cell death. Indeed, AIP2 knockdown increased the percentage of apoptotic cells from 18% to 43% of control cells (P < 0.01). This elevated death of T cells seems likely to be caused by increased FasL expression, since more than 38% of CD4+ T cells express FasL when AIP2 expression was knocked down, while only 12% of FasL-positive cells were detected in control T cells (Fig. 6D). The expression levels of FasL, but not cell death, are also slightly increased in rested T cells with AIP2 knockdown (data not shown). Therefore, AIP2 knockdown enhances activation-induced cell death by upregulating FasL expression.
EGR2 has been found as a negative regulator for T-cell activation by upregulating the E3 ubiquitin ligase Cbl-b expression upon TCR stimulation (37). Therefore, in addition to promoting FasL-mediated activation-induced apoptosis, AIP2 may inhibit the activation and production of IL-2 by suppressing EGR2-mediated Cbl-b transcription. In fact, Cbl-b protein expression was indeed increased by AIP2 knockdown in T cells, suggesting that AIP2 inhibits Cbl-b protein expression in T cells. As an E3 ubiquitin ligase, AIP2 may suppress Cbl-b protein expression at the posttranscriptional level. However, results from real-time PCR analysis indicated a similar increase of Cbl-b mRNA in AIP2 knockdown T cells. Together with the finding of increased EGR2 expression in AIP2 knockdown T cells, these results suggest that AIP2 suppresses EGR2-mediated Cbl-b transcription.
DISCUSSION
In this report, we have found that the E3 ubiquitin ligase AIP2 facilitates T-cell activation and inhibits activation-induced T-cell apoptosis. AIP2 suppresses T-cell apoptosis by promoting the degradation of EGR2, a transcription factor that is responsible for FasL expression. These findings uncovered a previously uncharacterized mechanism underlying AIP2 in regulation of T-cell function in mammals.
Activation-induced cell death is one crucial mechanism that maintains homeostasis in the peripheral lymphoid tissues after clonal expansion. Fas and FasL-mediated apoptosis play an important role in deleting clonally expanded T cells that are no longer useful (16). Mice deficient in Fas or FasL show defects in peripheral T-cell deletion and eventually develop autoimmune disorders (40). Several transcription factors, including NFAT, AP-1, NF-κB, and the EGR family proteins, have been found to regulate the expression of FasL during the activation-induced death of T cells (22, 25, 39). Our studies here demonstrate that AIP2 regulates FasL expression by catalyzing EGR2 for ubiquitination and degradation, which defines a new mechanism in regulating activation-induced T-cell death. EGR2 functions as a negative regulator for T-cell activation by transcription of both FasL and the E3 ubiquitin ligase Cbl-b (37). Indeed, AIP2 appears to promote T-cell activation by downregulating the transcription of EGR2 target genes, both FasL and Cbl-b, and maybe others.
EGR2 was initially identified as a protein with three zinc fingers, whose expression is activated during G0/G1 transition in cultured cells (4). Recent studies have demonstrated that EGR2 regulates activation-induced T-cell death by promoting FasL expression (7-9, 30, 35, 44, 45). In addition, EGR2 regulates the development and activation of T lymphocytes in mice (32). The expression of EGR2 is highly upregulated during the CD4-CD8 stage and is involved in regulation of pre-TCR expression during T-cell development. Ectopic expression of EGR2 inhibits T-cell activation in both Jurkat and mouse primary T cells, and the knockdown of its expression enhances antigen-specific T-cell activation (17). Upregulation of EGR2 is one of the important mechanisms in the induction of T-cell tolerance, as the reversion of T-cell tolerance by IL-2 is accompanied by a quick degradation of EGR2 (17, 37). Our findings demonstrate that AIP2 is involved in T-cell activation and death by promoting EGR2 ubiquitination and degradation. It will be interesting to further analyze whether AIP2-targeted EGR2 degradation is involved in T-cell tolerance.
The HECT-type E3 ubiquitin ligases, with few exceptions, recruit substrates using their WW domains. The WW domain refers to one of the distinguishing features of the domain: the presence of two highly conserved tryptophan (W) residues spaced by 20 to 22 amino acids (38). Previous studies have demonstrated that the WW domain specifically recognizes the proline-rich, tyrosine-containing motif PPXY (26). For example, the WW domains of Itch, another member of the NEDD4 family E3 ubiquitin ligase, specifically bind to the PPXY motif of either c-Jun or JunB (10, 11, 13, 14). There are two PPXY motifs in EGR2, and the mutation of each partially abolished its interaction with AIP2. However, the mutation of each of the EGR2 PPXY motifs reduced its interaction with AIP2 differently, suggesting that WW domains may favor one PPXY motif rather than another. The tyrosine of the PPXY motif has been found to be phosphorylated by tyrosine kinases, and this phosphorylation regulates its reorganization by WW domains (13). Interestingly, our data here clearly show that the AIP2/EGR2 interaction is regulated by TCR/CD28 signaling. Our laboratory is currently investigating how AIP2-catalyzed EGR2 ubiquitination is regulated in T cells.
E3 ubiquitin ligases have diverse functions in the activation of T lymphocytes. Two HECT-type E3 ubiquitin ligases, Itch and Nedd4, have been shown to play crucial roles in immune regulation (10, 24). Unlike Itch and Nedd4, which suppress T-cell activation, AIP2 enhances IL-2 production as well as T-cell proliferation by suppressing activation-induced T-cell apoptosis. Further investigation of AIP2 in regulating T-cell function in vivo, such as using AIP2 mutant mice, will provide a better understanding of the importance of AIP2-mediated ubiquitination in normal immune responses.
Acknowledgments
We thank Paul Bieniasz (The Rockefeller University) for the AIP2 expression plasmids and Jeffrey Milbrandt (Washington University) for the EGR2 expression plasmid.
We have no financial conflict of interest.
Footnotes
Published ahead of print on 3 August 2009.
REFERENCES
- 1.Anandasabapathy, N., G. S. Ford, D. Bloom, C. Holness, V. Paragas, C. Seroogy, H. Skrenta, M. Hollenhorst, C. G. Fathman, and L. Soares. 2003. GRAIL: an E3 ubiquitin ligase that inhibits cytokine gene transcription is expressed in anergic CD4+ T cells. Immunity 18:535-547. [DOI] [PubMed] [Google Scholar]
- 2.Bachmaier, K., C. Krawczyk, I. Kozieradzki, Y. Y. Kong, T. Sasaki, A. Oliveira-dos-Santos, S. Mariathasan, D. Bouchard, A. Wakeham, A. Itie, J. Le, P. S. Ohashi, I. Sarosi, H. Nishina, S. Lipkowitz, and J. M. Penninger. 2000. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature 403:211-216. [DOI] [PubMed] [Google Scholar]
- 3.Carattino, M. D., R. S. Edinger, H. J. Grieser, R. Wise, D. Neumann, U. Schlattner, J. P. Johnson, T. R. Kleyman, and K. R. Hallows. 2005. Epithelial sodium channel inhibition by AMP-activated protein kinase in oocytes and polarized renal epithelial cells. J. Biol. Chem. 280:17608-17616. [DOI] [PubMed] [Google Scholar]
- 4.Chavrier, P., M. Zerial, P. Lemaire, J. Almendral, R. Bravo, and P. Charnay. 1988. A gene encoding a protein with zinc fingers is activated during G0/G1 transition in cultured cells. EMBO J. 7:29-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiang, Y. J., H. K. Kole, K. Brown, M. Naramura, S. Fukuhara, R. J. Hu, I. K. Jang, J. S. Gutkind, E. Shevach, and H. Gu. 2000. Cbl-b regulates the CD28 dependence of T-cell activation. Nature 403:216-220. [DOI] [PubMed] [Google Scholar]
- 6.Crosby, S. D., R. A. Veile, H. Donis-Keller, J. M. Baraban, R. V. Bhat, K. S. Simburger, and J. Milbrandt. 1992. Neural-specific expression, genomic structure, and chromosomal localization of the gene encoding the zinc-finger transcription factor NGFI-C. Proc. Natl. Acad. Sci. USA 89:4739-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delgado, M., and D. Ganea. 2001. Vasoactive intestinal peptide and pituitary adenylate cyclase-activating polypeptide inhibit expression of Fas ligand in activated T lymphocytes by regulating c-Myc, NF-kappa B, NF-AT, and early growth factors 2/3. J. Immunol. 166:1028-1040. [DOI] [PubMed] [Google Scholar]
- 8.Droin, N. M., M. J. Pinkoski, E. Dejardin, and D. R. Green. 2003. Egr family members regulate nonlymphoid expression of Fas ligand, TRAIL, and tumor necrosis factor during immune responses. Mol. Cell. Biol. 23:7638-7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dzialo-Hatton, R., J. Milbrandt, R. D. Hockett, Jr., and C. T. Weaver. 2001. Differential expression of Fas ligand in Th1 and Th2 cells is regulated by early growth response gene and NF-AT family members. J. Immunol. 166:4534-4542. [DOI] [PubMed] [Google Scholar]
- 10.Fang, D., C. Elly, B. Gao, N. Fang, Y. Altman, C. Joazeiro, T. Hunter, N. Copeland, N. Jenkins, and Y. C. Liu. 2002. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat. Immunol. 3:281-287. [DOI] [PubMed] [Google Scholar]
- 11.Fang, D., and T. K. Kerppola. 2004. Ubiquitin-mediated fluorescence complementation reveals that Jun ubiquitinated by Itch/AIP4 is localized to lysosomes. Proc. Natl. Acad. Sci. USA 101:14782-14787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang, D., H. Y. Wang, N. Fang, Y. Altman, C. Elly, and Y. C. Liu. 2001. Cbl-b, a RING-type E3 ubiquitin ligase, targets phosphatidylinositol 3-kinase for ubiquitination in T cells. J. Biol. Chem. 276:4872-4878. [DOI] [PubMed] [Google Scholar]
- 13.Gao, B., S. M. Lee, and D. Fang. 2006. The tyrosine kinase c-Abl protects c-Jun from ubiquitination-mediated degradation in T cells. J. Biol. Chem. 281:29711-29718. [DOI] [PubMed] [Google Scholar]
- 14.Gao, M., T. Labuda, Y. Xia, E. Gallagher, D. Fang, Y. C. Liu, and M. Karin. 2004. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science 306:271-275. [DOI] [PubMed] [Google Scholar]
- 15.Gillian, A. L., and J. Svaren. 2004. The Ddx20/DP103 dead box protein represses transcriptional activation by Egr2/Krox-20. J. Biol. Chem. 279:9056-9063. [DOI] [PubMed] [Google Scholar]
- 16.Green, D. R., N. Droin, and M. Pinkoski. 2003. Activation-induced cell death in T cells. Immunol. Rev. 193:70-81. [DOI] [PubMed] [Google Scholar]
- 17.Harris, J. E., K. D. Bishop, N. E. Phillips, J. P. Mordes, D. L. Greiner, A. A. Rossini, and M. P. Czech. 2004. Early growth response gene-2, a zinc-finger transcription factor, is required for full induction of clonal anergy in CD4+ T cells. J. Immunol. 173:7331-7338. [DOI] [PubMed] [Google Scholar]
- 18.Heissmeyer, V., F. Macian, S. H. Im, R. Varma, S. Feske, K. Venuprasad, H. Gu, Y. C. Liu, M. L. Dustin, and A. Rao. 2004. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat. Immunol. 5:255-265. [DOI] [PubMed] [Google Scholar]
- 19.Huibregtse, J. M., M. Scheffner, S. Beaudenon, and P. M. Howley. 1995. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA 92:2563-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson, P. K., A. G. Eldridge, E. Freed, L. Furstenthal, J. Y. Hsu, B. K. Kaiser, and J. D. Reimann. 2000. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 10:429-439. [DOI] [PubMed] [Google Scholar]
- 21.Joseph, L. J., M. M. Le Beau, G. A. Jamieson, Jr., S. Acharya, T. B. Shows, J. D. Rowley, and V. P. Sukhatme. 1988. Molecular cloning, sequencing, and mapping of EGR2, a human early growth response gene encoding a protein with “zinc-binding finger” structure. Proc. Natl. Acad. Sci. USA 85:7164-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kavurma, M. M., and L. M. Khachigian. 2003. Signaling and transcriptional control of Fas ligand gene expression. Cell Death Differ. 10:36-44. [DOI] [PubMed] [Google Scholar]
- 23.Lemaire, P., O. Revelant, R. Bravo, and P. Charnay. 1988. Two mouse genes encoding potential transcription factors with identical DNA-binding domains are activated by growth factors in cultured cells. Proc. Natl. Acad. Sci. USA 85:4691-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, Y. C., J. Penninger, and M. Karin. 2005. Immunity by ubiquitylation: a reversible process of modification. Nat. Rev. Immunol. 5:941-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch, D. H., M. L. Watson, M. R. Alderson, P. R. Baum, R. E. Miller, T. Tough, M. Gibson, T. Davis-Smith, C. A. Smith, K. Hunter, et al. 1994. The mouse Fas-ligand gene is mutated in gld mice and is part of a TNF family gene cluster. Immunity 1:131-136. [DOI] [PubMed] [Google Scholar]
- 26.Macias, M. J., M. Hyvonen, E. Baraldi, J. Schultz, M. Sudol, M. Saraste, and H. Oschkinat. 1996. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature 382:646-649. [DOI] [PubMed] [Google Scholar]
- 27.Macias, M. J., S. Wiesner, and M. Sudol. 2002. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 513:30-37. [DOI] [PubMed] [Google Scholar]
- 28.Martin-Serrano, J., S. W. Eastman, W. Chung, and P. D. Bieniasz. 2005. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J. Cell Biol. 168:89-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonald, F. J., A. H. Western, J. D. McNeil, B. C. Thomas, D. R. Olson, and P. M. Snyder. 2002. Ubiquitin-protein ligase WWP2 binds to and downregulates the epithelial Na(+) channel. Am. J. Physiol. Renal Physiol. 283:F431-F436. [DOI] [PubMed] [Google Scholar]
- 30.Mittelstadt, P. R., and J. D. Ashwell. 1999. Role of Egr-2 in up-regulation of Fas ligand in normal T cells and aberrant double-negative lpr and gld T cells. J. Biol. Chem. 274:3222-3227. [DOI] [PubMed] [Google Scholar]
- 31.Morita, S., T. Kojima, and T. Kitamura. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7:1063-1066. [DOI] [PubMed] [Google Scholar]
- 32.Mueller, D. L. 2004. E3 ubiquitin ligases as T cell anergy factors. Nat. Immunol. 5:883-890. [DOI] [PubMed] [Google Scholar]
- 33.Naramura, M., H. K. Kole, R. J. Hu, and H. Gu. 1998. Altered thymic positive selection and intracellular signals in Cbl-deficient mice. Proc. Natl. Acad. Sci. USA 95:15547-15552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Donovan, K. J., W. G. Tourtellotte, J. Millbrandt, and J. M. Baraban. 1999. The EGR family of transcription-regulatory factors: progress at the interface of molecular and systems neuroscience. Trends Neurosci. 22:167-173. [DOI] [PubMed] [Google Scholar]
- 35.Rengarajan, J., P. R. Mittelstadt, H. W. Mages, A. J. Gerth, R. A. Kroczek, J. D. Ashwell, and L. H. Glimcher. 2000. Sequential involvement of NFAT and Egr transcription factors in FasL regulation. Immunity 12:293-300. [DOI] [PubMed] [Google Scholar]
- 36.Rougier, J. S., M. X. van Bemmelen, M. C. Bruce, T. Jespersen, B. Gavillet, F. Apotheloz, S. Cordonier, O. Staub, D. Rotin, and H. Abriel. 2005. Molecular determinants of voltage-gated sodium channel regulation by the Nedd4/Nedd4-like proteins. Am. J. Physiol. Cell Physiol. 288:C692-C701. [DOI] [PubMed] [Google Scholar]
- 37.Safford, M., S. Collins, M. A. Lutz, A. Allen, C. T. Huang, J. Kowalski, A. Blackford, M. R. Horton, C. Drake, R. H. Schwartz, and J. D. Powell. 2005. Egr-2 and Egr-3 are negative regulators of T cell activation. Nat. Immunol. 6:472-480. [DOI] [PubMed] [Google Scholar]
- 38.Sudol, M. 1996. The WW module competes with the SH3 domain? Trends Biochem. Sci. 21:161-163. [PubMed] [Google Scholar]
- 39.Takahashi, T., M. Tanaka, C. I. Brannan, N. A. Jenkins, N. G. Copeland, T. Suda, and S. Nagata. 1994. Generalized lymphoproliferative disease in mice, caused by a point mutation in the Fas ligand. Cell 76:969-976. [DOI] [PubMed] [Google Scholar]
- 40.Watanabe-Fukunaga, R., C. I. Brannan, N. G. Copeland, N. A. Jenkins, and S. Nagata. 1992. Lymphoproliferation disorder in mice explained by defects in Fas antigen that mediates apoptosis. Nature 356:314-317. [DOI] [PubMed] [Google Scholar]
- 41.Wood, J. D., J. Yuan, R. L. Margolis, V. Colomer, K. Duan, J. Kushi, Z. Kaminsky, J. J. Kleiderlein, A. H. Sharp, and C. A. Ross. 1998. Atrophin-1, the DRPLA gene product, interacts with two families of WW domain-containing proteins. Mol. Cell. Neurosci. 11:149-160. [DOI] [PubMed] [Google Scholar]
- 42.Wu, J., L. Joseph, V. P. Sukhatme, and K. K. Kidd. 1988. A HindIII polymorphism identified by the human early growth response gene 2 (EGR2) on chromosome 10. Nucleic Acids Res. 16:11855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu, H. M., B. Liao, Q. J. Zhang, B. B. Wang, H. Li, X. M. Zhong, H. Z. Sheng, Y. X. Zhao, Y. M. Zhao, and Y. Jin. 2004. Wwp2, an E3 ubiquitin ligase that targets transcription factor Oct-4 for ubiquitination. J. Biol. Chem. 279:23495-23503. [DOI] [PubMed] [Google Scholar]
- 44.Yang, Y., B. Dong, P. R. Mittelstadt, H. Xiao, and J. D. Ashwell. 2002. HIV Tat binds Egr proteins and enhances Egr-dependent transactivation of the Fas ligand promoter. J. Biol. Chem. 277:19482-19487. [DOI] [PubMed] [Google Scholar]
- 45.Yoo, Y. G., and M. O. Lee. 2004. Hepatitis B virus X protein induces expression of Fas ligand gene through enhancing transcriptional activity of early growth response factor. J. Biol. Chem. 279:36242-36249. [DOI] [PubMed] [Google Scholar]