Abstract
Forkhead box class O (FoxO) transcription factors are a family of conserved proteins that regulate the cellular responses to various stimuli, such as energy deprivation, stress, and developmental cues. FoxO proteins are important mediators of the insulin signaling pathway, adjusting growth and metabolism to nutrient availability. Insulin signaling acts together with the glucagon-stimulated cAMP signaling pathway to orchestrate the organism response to various nutritional conditions. In this study, we demonstrate that Drosophila melanogaster FoxO (dFoxO) regulates cAMP signaling by directly inducing the expression of an adenylate cyclase gene, ac76e. Interestingly, ac76e is expressed in a highly restricted pattern throughout fly development, limited to the corpus allatum (CA), gastric cecum, and malpighian tubules. dFoxO activation of AC76E in the CA increases starvation resistance and limits growth. Our results unravel a new role for dFoxO, integrating cAMP and insulin signaling to adapt organism growth to the existing nutritional conditions.
Forkhead box class O (FoxO) proteins belong to the large superfamily of forkhead box transcription factors. FoxO family members include orthologs from diverse species, such as the fruit fly, worm, and mammals, where they regulate conserved cellular and physiological processes ranging from apoptosis to stress resistance and growth (1). FoxO proteins are key players in the well-conserved insulin signaling pathway that mediates the responses of energy homeostasis depending on the availability of nutrients. In mammals, upon hypoglycemia and subsequent attenuation of circulating insulin, FoxO1 is localized in the hepatocyte nucleus, where it drives a pattern of gene expression devoted to activation of gluconeogenesis and lipid catabolism (38). In contrast, in situations of abundant nutrient availability, activated insulin signaling leads to Akt-mediated FoxO phosphorylation, which leads to FoxO cytoplasmic localization and inactivation (3, 5, 23, 32). FoxO is also involved in the regulation of proliferation in peripheral tissues by activating genes such as p27kip1 and p21Cip1 (26, 30). Consequently, FoxO is acting in a cell-autonomous and -nonautonomous manner by adjusting metabolism and growth to the prevailing nutritional conditions. The importance of FoxO in this setting is further highlighted by the finding that in the model organism Drosophila melanogaster, 28% of the nutrient-regulated genes were found to also be regulated by Drosophila FoxO (dFoxO) (12). This result suggests that the transcriptional mediators responding to nutrient shortage might be well conserved between distant phyla.
The activity of hepatic FoxO1 is complemented by cyclic AMP (cAMP) signaling, which is stimulated by glucagon. This signaling pathway is regulated by protein kinase A and the cAMP response element binding protein (CREB) (15). Together, FoxO1 and CREB ensure mobilization of energy reserves and allow maintenance of correct blood glucose levels under fasting conditions (11, 21). An analogous system has recently been demonstrated to operate in the fruit fly, where insulin-like peptides and a glucagon-like adipokinetic hormone in concert regulate the expression of alpha-glucosidase enzyme in adipose tissue (6). This finding emphasizes the conserved mechanisms underlying nutrient-induced signaling in these distantly related species. Hence, Drosophila is a very good model to use to study the systemic regulation of energy metabolism and growth.
Adenylate cyclases (AC) are a group of conserved enzymes catalyzing the conversion of ATP to cyclic AMP (cAMP) (16). In mammals, nine AC genes exist. Their corresponding proteins differ in their sensitivities to regulatory inputs such as growth factors, Ca2+/calmodulin binding, and protein kinase C phosphorylation (16). In Drosophila, there are 10 AC genes (7). The best-characterized Drosophila AC gene is rutabaga (rut), implicated in learning and memory (31) and in stress resistance and life span determination (35). The expression of rut is concentrated in brain (14), whereas the expression of ac78c, for example, is restricted to embryogenesis (8). In spite of the central importance of cAMP signaling in various key cellular processes, relatively little is known about the biology of Drosophila AC. In addition, it is unknown how the expression of these enzymes is regulated in Drosophila. Given the evident similarity in the diversity of the AC family with mammals (8), Drosophila makes an ideal model to use to discern the functional roles of differentially regulated AC proteins.
A systems biology approach was recently applied to demonstrate the key role of dFoxO in regulating gene expression upon nutrient deprivation (12). Whereas the role of some of the target genes is obvious, such as those encoding enzymes devoted to energy metabolism and growth, a number of FoxO target genes have unknown functions. In this study, we have identified a Drosophila AC gene, ac76e, as a direct transcriptional target of dFoxO in vitro and in vivo. We show that dFoxO activation of AC76E results in an increase of cAMP levels that ultimately modulates developmental growth and starvation resistance.
MATERIALS AND METHODS
Band shift analysis and chromatin immunoprecipitation (ChIP).
For analysis of the ac76e promoter, DNA fragments spanning −638 to −495 (probe 3), −531 to −389 (probe 2), and −422 to −281 (probe 1) from the transcription start site were amplified by PCR. Recombinant, purified dFoxO (100 nM or 200 nM) (27) was mixed with [32P]dCTP-labeled PCR products (80 nM) under reaction conditions described by Coleman and Pugh (9). The DNA complexes were resolved on native polyacrylamide (5.3%). In the cold competitor experiments, the concentrations of the unlabeled probe ranged between 1 and 8 μM. The primers for the probes were the following: probe 1, CGATTGTATTTATTTTGACTTGATGCCATG and GGAAAAAGTAAATACTAAAAGGTAAACACAAAAAGG; probe 2, CCTTTTTGTGTTTACCTTTTAGTATTTACTTTTTCC and CAATAAACTCAAATAAATATTACTGATCCGTC; probe 3, GACGGATCAGTAATATTTATTTGAGTTTATTGC and GATATCTATTATATGTCATAATAATTTGTTACCAGAAAGC; positive control (dInR), TAATACGACTCACTATAGG and CCCTTTAGTGAGGGTTAATT; negative control (pBs), GCAAATTCATCAATAGTTTTTGTTG and CACCGGTATCAGTGGCGGAATTG.
ChIP was performed as described by Puig et al. (27) by cross-linking S2 cells expressing dFoxO-A3 or wild-type dFoxO (dFoxO-WT) with 0.1% formaldehyde. For immunoprecipitation, anti-dFoxO-specific antibodies were used (27). Coimmunoprecipitated DNA was amplified by PCR, using the primers for probe 2 (see above) in the presence of [32P]dCTP. The PCR products were resolved by polyacrylamide (6%) and visualized by using a phosphorimager (Fujifilm). The band intensities were quantified by AIDA image analyzer software.
Constructs and Drosophila strains.
pMT dFoxO-A3 and pMT dFoxO-WT have been described (27). The ac76e promoter regions were generated by PCR, and the products were cloned into the pGL3 basic vector (Promega). The expressed-sequence-tag clone GH26260 contains a partial ac76e sequence coding for amino acids 433 to 1307. The N-terminal sequence was generated by PCR, and the full-length sequence was cloned into the pMT vector (Invitrogen). The integrity of the obtained full-length coding sequence was verified by sequencing.
Tubulin-Gal4 was obtained from Bloomington. The foxo null foxo25 strain and its littermate control, EP-147, are described in reference 20 and DI-11-Gal4 in reference 2. To produce UAS-AC76E transgenic flies, the coding region of ac76e tagged with a three-hemagglutinin (HA)-tag sequence in its C terminus was cloned into the p[UAS] vector. Transgenic flies were obtained by injecting the p[UAS]-AC76E-3xHA constructs into w1118 Drosophila embryos. Two independent transgenic lines harboring the insertion in the second and third chromosomes, respectively, were utilized in the experiments.
Cell culture, transfection, RNA interference (RNAi), and quantitative reverse transcription-PCR (qPCR).
Drosophila S2 cells were maintained, transfected, and treated with double-stranded RNA in M3 medium (Sigma) supplemented with insect medium supplement (Sigma), 2% fetal bovine serum, penicillin, and streptomycin. The cells were transfected with the Effectene transfection reagent (Qiagen) according to the manufacturer's protocol. For cAMP measurements, a competitive enzyme immunoassay was used (Assay Designs). Before lysis, the cells were treated for 30 min with forskolin (Fluka) and 3-isobutyl-1-methylxanthine (Fluka) in concentrations of 50 μM and 100 μM, respectively. For luciferase activity measurements (Promega), the cells were lysed in passive lysis buffer (Promega) and luciferase values were normalized to the total protein content of the lysates as measured by the Bradford reagent (Bio-Rad). The antibodies used were anti-V5 (1:5,000; Invitrogen), anti-HA (1:3,000; Roche), anti-α-tubulin (1:10,000; Sigma), anti-green fluorescent protein (GFP) (1:500; Santa Cruz Biotechnology), and secondary rabbit anti-mouse immunoglobulin G antibody conjugated with horseradish peroxidase (1:5,000; Upstate Biotechnology) and Alexa Fluor568-rabbit anti-mouse immunoglobulin G (1:1,000; Molecular Probes).
RNA interference and qPCR were performed as described previously (25). The primers used to synthesize AC76E double-stranded RNA were GGTCATGTCACACGCAATCGTCC and CACCCTGGCGATCCAGAGTGTG. The primers used for the AC76E qPCR were CAGGATGAATGACGCCCTTTCGG and ATGGACACAACACATGCCAGCAGC.
RNA in situ hybridization.
Antisense and sense RNA probes were generated by an in vitro transcription reaction using T7/T3-dependent RNA polymerase (Promega) in the presence of digoxigenin-UTP (Roche). The probes were hybridized to formaldehyde-fixed larval/adult tissue for 16 h at 55°C in hybridizing solution containing 50% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 50 μg/ml heparin, 0.1% Tween 20, and 100 μg/ml salmon sperm DNA. After washing of excess probe, the hybridization was visualized by using alkaline phosphatase-conjugated anti-digoxigenin antibody (1:3,000; Roche). mRNA in situ hybridizations to embryos were performed as described previously (19). The primers for the ac76e probe were T3 plus GCAACAGCGAATCTGATTGTTAAGGC and T7 plus GGGCTCCGCTGTTGGAGATGC. The primers for the dfoxo probe were T3 plus ATGATGGACGGCTACGCGCAG and T7 plus TGCACCCAGGATGGTGGCG.
Body weight and starvation resistance measurement.
For body weight and starvation resistance measurements, flies were reared the following way: flies laid eggs on apple juice plates supplemented with yeast paste for 8 h. Subsequently, the yeast was removed from the plates and the eggs developed at 25°C. The next day, hatched larvae were removed from the plate. After an additional 2 to 4 h, first-instar larvae were collected and transferred to fresh yeast paste until they reached the second-instar stage. At that time the balancer marker tubby is distinguishable, and larvae with the correct genotype were collected. To prevent crowding and to ensure equal growing conditions, 100 second-instar stage larvae were collected per vial with standard fly food. The flies developed at 25°C, and the emerging pupae were scored in 4- to 12-h intervals. The body weight of the AC76E-overexpressing flies was measured at 330 h after egg laying (AEL) from females and males kept in the same vial. The body weight of the homozygous P-element insertion lines was measured 3 days after eclosing. Body weight was measured in groups of 10 snap-frozen flies with a Mettler AE160 precision scale. For the starvation resistance measurement, flies were collected 0 to 16 h from eclosing, matured for 3 days in groups of 25 females and 25 males per vial, and then pooled for the experiment. The flies were transferred to 0.5% agarose in phosphate-buffered saline (PBS) for starvation, and dead flies were scored in 2- to 8-h intervals. The final number of female and male files in each experiment varied depending on how many flies were collected at eclosion, but this number was enough to allow us to determine statistical significance. The starvation resistance experiments were performed twice for both sexes with similar results. We used the Kaplan-Meier log rank test to estimate statistical differences of survival between treatments. To measure ac76e and 4ebp expression upon starvation, the flies were reared as described above. Three-day-old females were transferred either to standard fly food supplemented with yeast paste or to 0.5% agarose in PBS. After 24 h, total RNA was extracted from the flies and gene expression was measured by qPCR.
JHIII measurement.
The larvae were staged as described above. Approximately 200 larvae were picked at 96 h AEL, homogenized, and extracted for JHIII by using iso-octane. Before extraction, the samples were measured for their protein concentrations to ensure equal sample sizes. In addition, prior to the extraction the samples were spiked with an internal methoprene acid control. The JHIII titer was then measured by a liquid chromatography-tandem mass spectrometry method. For the complete description of the method, see materials and methods in the supplemental material.
RESULTS
ac76e is a direct target of dFoxO.
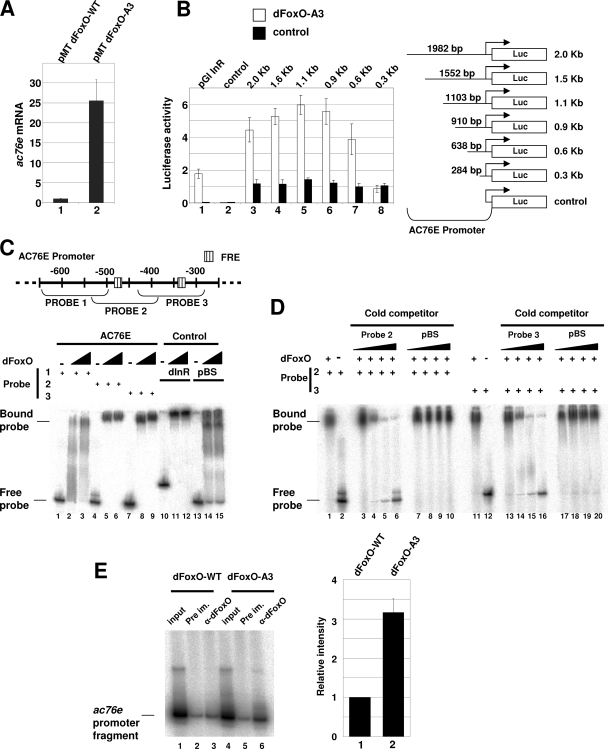
We identified ac76e in a microarray screening as one of the genes regulated by dFoxO in S2 cells (27). Overexpression of dFoxO-A3, a constitutively nuclear dFoxO protein insensitive to insulin (27), resulted in a roughly 20-fold increase in ac76e expression as measured by the microarray system (data not shown). We first confirmed the microarray result by qPCR and found that upon overexpression of dFoxO-A3 in S2 cells, ac76e was upregulated ∼25-fold compared to the negative control overexpressing dFoxO-WT, which is inhibited by insulin (27) (Fig. 1A). We next studied if dFoxO can activate gene expression from the ac76e promoter. Several promoter fragments, ranging from 2.0 kb to 0.3 kb, containing the ac76e transcription start site, were cloned into a luciferase reporter and were independently cotransfected in S2 cells together with the dFoxO-A3 expression plasmid. Luciferase activity was elevated four- to sixfold compared to that of the empty vector control (Fig. 1B, bars 3 to 7). This analysis revealed a ∼350-bp region (−281 to −638 relative to the start site) required for reporter induction (Fig. 1B, compare bars 7 and 8). This region was found to contain two putative FoxO recognition elements (FRE) (Fig. 1C). These results suggest that dFoxO activates ac76e expression by directly binding its promoter and activating gene transcription.
FIG. 1.
dFoxO directly activates transcription of ac76e. (A) qPCR assay showed a 25-fold increase in ac76e mRNA upon dFoxO-A3 overexpression (bar 2) compared to the dFoxO-WT control level (bar 1) in S2 cells. (B) Luciferase assays showed activation of different ac76e promoter fragments upon cotransfection with dFoxO-A3 compared to results for the empty vector control. Bars in lane 1, positive control with dinr promoter; bars in 2, negative control with empty pGl vector. Bars in lanes 3 to 8, dFoxO-A3 cotransfections with different ac76e promoter fragments. (C) Schematic representation of the ac76e promoter showing the putative FREs (striped boxes) in the region from −300 to −600 bp upstream of the transcription start site. The probes used in band shift experiments (below) are indicated. Recombinant dFoxO binds to probes 2 (lanes 4 to 6) and 3 (lanes 7 to 9). Lanes 10 to 12 show the positive control dinr promoter. Lanes 13 to 15 show the negative control pBS. (D) The specificity of the binding to these promoter regions was confirmed by competition experiments. Cold competitors for probes 2 (lanes 1 to 6) and 3 (lanes 11 to 16) reversed the band shift, whereas the negative control, pBS, did not (lanes 7 to 10 for probe 2 and lanes 17 to 20 for probe 3). (E) dFoxO binds specifically to ac76e promoter in vivo. ChIP of cross-linked extracts of S2 cells expressing dFoxO-WT (lanes 1 to 3) or dFoxO-A3 (lanes 4 to 6) grown in the presence of insulin. Relative intensities of lanes 3 and 6 as quantified with a phosphorimager (bars 1 and 2, respectively) are shown. The error bars represent standard deviations from three independent measurements. DNA fragments were detected by incorporating [P32]dCTP in the PCR mix.
We next asked if dFoxO can bind to this DNA fragment in vitro. We divided the 350-bp DNA fragment into three smaller fragments of ∼120 bp each and assessed whether dFoxO could bind any of them by performing band shift experiments with purified recombinant dFoxO. Interestingly, we found that two of the fragments, containing the putative FREs, were efficiently bound by dFoxO, whereas the negative control was not (Fig. 1C). The specificity of the binding was confirmed by competition assays where a cold competitor probe was added to the in vitro binding reaction mixtures (Fig. 1D). Addition of the cold competitor probe reversed the band shift of these two fragments (Fig. 1D, lanes 1 to 6 and 11 to 16), whereas the negative control did not (Fig. 1D, lanes 7 to 10 and 17 to 20). These results show that dFoxO binds efficiently the ac76e promoter in vitro.
Finally, we performed a ChIP experiment to investigate whether dFoxO binds to the ac76e promoter in vivo. S2 cells overexpressing either dFoxO-A3 or dFoxO-WT were grown in the presence of insulin (which inhibits dFoxO-WT but not dFoxO-A3), cross-linked with formaldehyde, and immunoprecipitated with anti-dFoxO. After reversal of the cross-links, DNA was recovered and PCR was performed with primers specific for the FRE-containing region (probe 2 in Fig. 1C). Figure 1E shows that dFoxO-A3 can bind specifically to the ac76e promoter in vivo (compare lanes 3 and 6; quantification is shown at the right).
The results presented above demonstrate that dFoxO is able to activate transcription from the ac76e promoter by specifically binding to the promoter in a region from ∼340 bp to ∼450 bp upstream of the transcription start site. We therefore conclude that ac76e is a direct transcriptional target of dFoxO.
AC76E is a mediator of dFoxO-induced cAMP elevation.
According to the Drosophila gene annotation database Flybase, the ac76e gene region spans approximately 40 kb in chromosome 3L, contains 20 exons, and encodes a protein of 1,307 amino acids. Sequence analysis of the AC76E protein reveals that it contains the characteristic domains of a typical transmembrane adenylate cyclase: two predicted membrane-spanning regions, each having six transmembrane helices, followed by AC catalytic domains (16) (see Fig. S1 in the supplemental material). Alignment of the AC76E protein sequence with human AC2 reveals conservation in the catalytic domains between these enzymes from distantly related species (see Fig. S1 in the supplemental material).
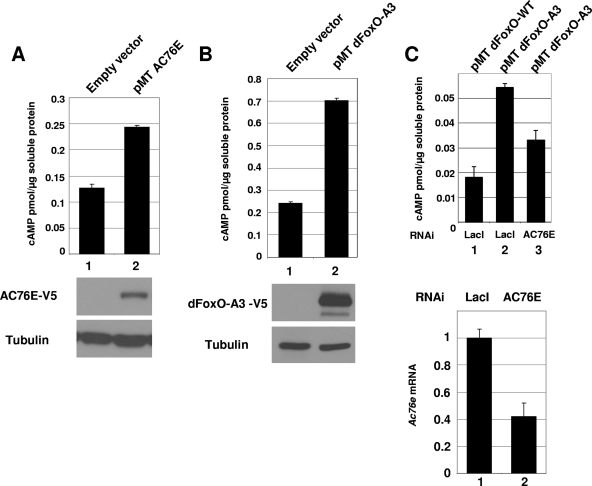
We cloned the full-length AC76E open reading frame into an expression vector, overexpressed the protein in S2 cells, and measured the cAMP concentration from the cell extract in the presence of a diterpene forskolin, a known AC activator (29). We used forskolin to get better sensitivity in our assays, but it is not required for cAMP activation by AC76E through FOXO (Fig. 2C). We found that upon AC76E overexpression, the cAMP concentration was elevated twofold compared to that of the empty vector control, indicating that AC76E possesses enzymatic activity in vivo (Fig. 2A, lanes/bars 1 and 2). Similarly, overexpression of dFoxO-A3 alone was able to increase the cellular cAMP level (Fig. 2B, lanes/bars 1 and 2). Simultaneous treatment of the cells overexpressing dFoxO-A3 with AC76E double-stranded RNA was able to attenuate the induction of cAMP driven by dFoxO-A3, indicating that at least part of this effect is mediated through AC76E (Fig. 2C, bars 2 and 3). Interestingly, the effect of the AC76E RNAi was detected only without forskolin, suggesting that under our experimental conditions the addition of forskolin stimulated other adenylate cyclases to an extent great enough to compensate for the AC76E knockdown.
FIG. 2.
AC76E is a functional adenylate cyclase and mediates dFoxO-regulated cAMP levels in S2 cells. (A) Overexpression of AC76E in S2 cells (bar/lane 2) increases the cAMP concentration compared to results for the empty vector control (bar/lane 1). Forskolin was added to both samples. (B) Overexpression of dFoxO-A3 alone was sufficient to increase the cAMP concentration in S2 cells compared to results for the empty vector control (lanes/bars 1 and 2). Forskolin was added to both samples. (C) The increase in the cAMP concentration upon dFoxO-A3 overexpression was attenuated by RNAi against AC76E (upper histogram, bars 2 and 3). No forskolin was added. The efficiency of the knockdown was measured by qPCR for AC76E mRNA (lower histogram, bars 1 and 2). The error bars in each histogram represent standard deviations from at least two independent measurements.
ac76e expression during development.
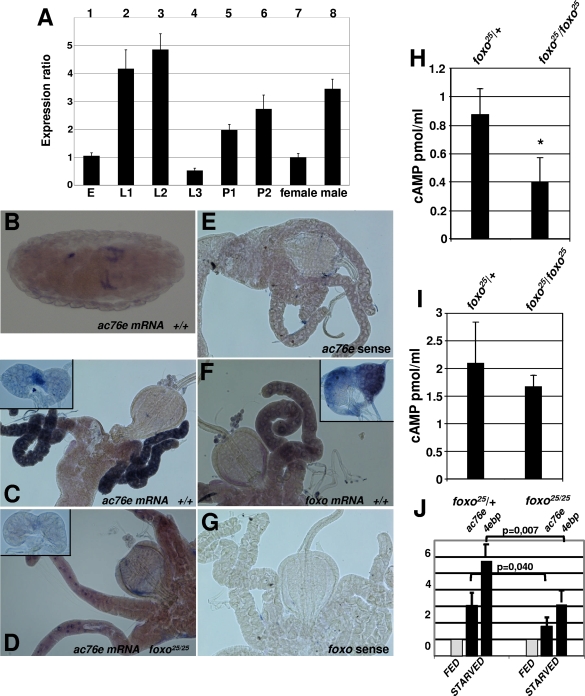
We surveyed the relative temporal expression of ac76e during fly development by qPCR analysis. The expression is highest during the larval growth period in the first- and second-instar stages, followed by attenuation of the expression at the third-instar stage and by another expression peak during pupal stages (Fig. 3A, bars 1 to 6). Interestingly, ac76e is expressed in a sex-dependent manner in adult flies, with males having a threefold-higher level of expression than females (Fig. 3A, bars 7 and 8). There are some discrepancies between our data and ac76e expression as defined in the fly atlas. However, the data in the fly atlas must be interpreted with caution, since it was obtained by microarray analysis (10). The spatial ac76e gene expression was monitored by using RNA in situ hybridizations during embryogenesis, in third-instar larvae, and in adult tissue. During embryogenesis, the ac76e mRNA in situ signal was detectable from stage 16. Four tubule-like structures and a spot in the hindgut region were stained (Fig. 3B). At third instar, distinct tissue-specific expression was detected in the gastric cecum, in the corpus allatum (CA) of the ring gland, and in the lower ureter of the malpighian tubules (Fig. 3C; see also Fig. S2 in the supplemental material). In addition, weak ubiquitous expression was observed in imaginal discs and salivary glands (data not shown). In adult flies, ac76e staining revealed a continuity of the CA expression and a male-specific expression in reproductive tissues (see Fig. S2 in the supplemental material). The latter could explain the difference between male and female adult ac76e expression observed by qPCR analysis (Fig. 3A).
FIG. 3.
Expression of ac76e during fly development. (A) The temporal expression level of ac76e was measured by qPCR analysis in different stages of fly development: E, embryo; L1, first instar; L2, second instar; L3, third instar; P1, pupa 1; P2, pupa 2; female, adult female; male, adult male. (B to G) dFoxO regulates ac76e expression in third-instar gastric cecum and CA: mRNA in situ staining of stage 16 embryo (B), third-instar gastric cecum, and ring gland (C to G). ac76e in situ with antisense (B to D) or sense (E) probe or dfoxo in situ with anti-sense (F) or sense (G) probe is shown. Fly strains used were EP-147/EP-147 (+/+) and the homozygous mutant dFoxO (dfoxo25/dfoxo25). (H to I) cAMP levels measured from third-instar ring gland (H) or gastric cecum (I) from dfoxo25/dfoxo25 and dfoxo25/+ animals. (J) Expression analysis of ac76e and 4ebp from fed and starved dfoxo25/dfoxo25 and dfoxo25/+ animals. The P values are derived from a one-tailed Student t test. The error bars represent standard deviations from at least three independent measurements. *, P < 0.05 (Student's t test).
dFoxO regulates transcription of ac76e in flies.
We wanted to explore the dependency of ac76e expression on dFoxO. The expression patterns of dfoxo and ac76e in third-instar larvae reveal a colocalization of these two genes in the gastric cecum and CA (Fig. 3, compare panels C and F). Parallel ac76e in situ stainings in the homozygous foxo25 null mutant and its littermate control strain, EP-147 (20), demonstrated a clear reduction in the ac76e in situ signal in the foxo null flies both in gastric ceca and in the CA (Fig. 3, compare panels C and D). These results show that ac76e expression is regulated by dFoxO in these tissues. Further evidence supporting the role of dFoxO as a mediator of cAMP signaling came from measurements of cAMP levels in vivo. Homozygous foxo25 tissue exhibited reduced levels of cAMP compared to control heterozygous foxo25/+ animals. This observation was especially evident for the ring gland (Fig. 3H), whereas only a minor decrease was observed in the gastric cecum (Fig. 3I). To reveal the physiological condition whereupon ac76e expression is regulated by dFoxO, we starved the homozygous foxo25 and control heterozygous foxo25/+ animals and measured the ac76e expression by qPCR. As a control, we measured the expression of the 4ebp gene, a known dFoxO target (20, 27). Upon starvation, the level of ac76e mRNA is elevated, similar to results for the 4ebp control, and this is compromised in the homozygous foxo25 flies (Fig. 3J). Taken together, these results demonstrate that ac76e is a target of dFoxO in vivo and that this regulation is induced in response to nutrient shortage.
ac76e expression in CA regulates pupa formation, size, and starvation resistance.
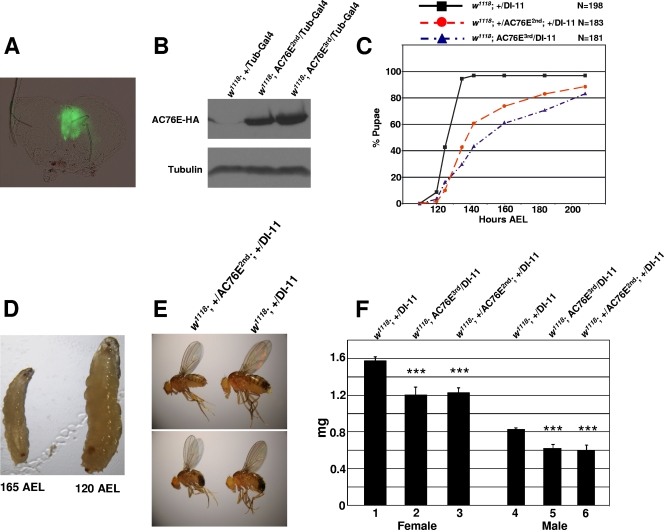
Insulin signaling has been shown to modulate Drosophila size, life span, and reproduction (4, 33). These phenotypes are at least partly regulated through the CA, which is an endocrine tissue known to regulate dipteran development (2, 18). Since the ac76e gene is expressed in this tissue under dFoxO regulation (Fig. 3C and D), we investigated the possibility that modulating its expression affects organism development. We employed a highly specific CA Gal4 line, DI-11, to drive AC76E overexpression (2) (Fig. 4A; see also Fig. S2 in the supplemental material). To avoid nonspecific side effects of P-element insertions, we overexpressed AC76E from two independent P-element insertion lines by using the UAS-Gal4 system. The AC76E proteins were tagged with HA sequence at their carboxy-terminal ends (Fig. 4B). Overexpression of AC76E in the CA delayed pupa formation (Fig. 4C). Whereas the control flies pupariated at approximately 125 h AEL, the average pupa forming time for flies overexpressing AC76E was 142 h AEL (Fig. 4C). Interestingly, a significant fraction (∼35%) of the individuals were still larvae as late as 165 h AEL, and some of them continued to wander up to 215 h AEL. These larvae were reduced in size compared to the control wandering larvae and, correspondingly, yielded smaller adults (Fig. 4D and E). The average adult weight was measured at 330 h AEL and was found to be significantly reduced in both sexes (Fig. 4F). Additional control experiments ruled out the possibility that the size phenotype was produced by the P-element insertion itself (see Fig. S3 in the supplemental material).
FIG. 4.
Overexpression of AC76E in the CA delays pupa formation and reduces adult body weight. (A) DI-11-Gal4 drives expression specifically in the CA as shown by GFP activity in a DI-11-Gal4/UAS-GFP third-instar ring gland. (B) Western blot from whole adult fly extracts showing the expression of the UAS-AC76E-HA P-element insertions driven by tubulin-Gal4. (C) Pupa formation in flies of genotypes w1118; +/DI-11-Gal4, w1118; UAS-AC76E3rd/DI-11-Gal4, and w1118; +/UAS-AC76E2nd; +/DI-11-Gal4. (D) Left: a 165-h-AEL larva of genotype w1118; +/UAS-AC76E2nd; +/DI-11-Gal4; right: a 120-h-AEL larva of genotype w1118; DI-11-Gal4/+. (E) Adult flies of genotypes w1118; DI-11-Gal4/+ (right) and w1118; +/UAS-AC76E2nd; +/DI-11-Gal4 (left). The AC76E-overexpressing flies on the left originate from larvae still wandering at 165 h AEL. (F) Average body weight measured from females (bars 1 to 3) or males (bars 4 to 6) of w1118; DI-11-Gal4/+ and w1118; DI-11-Gal4/UAS-AC76E flies at 330 AEL. The error bars represent standard deviations from at least three independent measurements. ***, P < 0.001 (Student's t test).
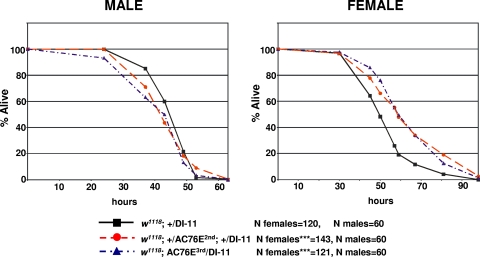
We next asked if overexpression of AC76E could affect the fly response to nutritional deprivation. Adult flies of genotypes w1118; +/DI-11-Gal4 and w1118; UAS-AC76E/DI-11-Gal4 were subjected to starvation by incubating them in 0.5% agarose in PBS. Overexpressing AC76E in the CA results in a small but significant increase in starvation resistance in females but not in males, as determined by the Kaplan-Meier log rank test (Fig. 5). This result has been confirmed in two independent experiments. Altogether, our results demonstrate that AC76E has an important function in the regulation of organism size and stress resistance, which is consistent with the role of dFoxO as a key regulator of these processes.
FIG. 5.
Overexpression of AC76E in the CA increases starvation resistance in females. Survival of 3-day-old adult males and females of genotype w1118; +/DI-11-Gal4, w1118; UAS-AC76E3rd/DI-11-Gal4, or w1118; +/UAS-AC76E2nd; +/DI-11-Gal4 in 0.5% agarose in PBS was measured. ***, P < 0.001 (Kaplan-Meier log rank test). A representative experiment is shown.
DISCUSSION
In this study, we showed that dFoxO activates transcription of the ac76e gene by binding directly to its promoter. As a result, dFoxO regulates cellular cAMP levels, which has a critical effect in reducing organism size and increasing starvation resistance through its action in the CA, a central regulator of size and development (2, 18).
It has been previously shown that dFoxO has a critical role in systemic regulation of growth. This regulation occurs through inhibition of expression of Drosophila insulin-like peptide 2 (dilp2) in the insulin-producing cells in the fly brain (13, 17, 37). Under suboptimal growing conditions, i.e., lack of nutrients or elevated stress, low insulin signaling results in dFoxO activation in insulin-producing cells, which leads to direct inhibition of DILP2 synthesis. Reduced DILP2 synthesis causes a reduction in systemic insulin signaling, resulting in dFoxO activation in peripheral tissues. This causes subsequent growth inhibition and increased stress resistance through activation of dFoxO downstream targets like 4EBP and MnSOD (20, 22, 27, 34).
Here we have identified an additional mechanism by which Drosophila regulates its size and stress resistance upon dFoxO activation: modulation of cAMP levels in the CA by direct activation of ac76e. Importantly, we have shown that activation of AC76E by dFoxO not only inhibits fly size but also delays development. We hypothesize that in this way the organism coordinates growth and development to better adapt to the prevailing environmental conditions. It is therefore tempting to speculate that during periods of nutritional deprivation or elevated stress, the developing larvae retard their growth partially through an ac76e-dependent mechanism by eliciting a systemic signal from the CA. A strong candidate for such a signal is juvenile hormone (JH). JH synthesis has been shown to be inhibited in cultured Drosophila corpus allata by increased levels of cAMP and Ca2+ (28). On the other hand, Tatar and collaborators (33) demonstrated that inr mutant flies are dwarfs and defective in JH synthesis. The dwarf flies were long lived, and the addition of synthetic JH to food was able to reverse this phenotype. Interestingly, Belgacem and Martin (2) showed that defective insulin signaling in the CA recapitulates the dwarf fly phenotype, highlighting the critical role that this endocrine tissue has in growth control. Thus, we hypothesized that under low insulin signaling, activation of AC76E in the CA by dFoxO would increase cAMP levels and result in inhibition of JH production. To our surprise, we observed that the total JHIII titer was in fact slightly increased in the AC76E-overexpressing larvae (data not shown). Even though we were not able to measure all the JH forms, this result suggests that it is not the lack of JH which is causing the observed small-fly phenotype. Hence, an additional mechanism, independent of JH, might exist to control this phenotype. Whatever the mechanism is, our results show that in addition to its known role as a cell-autonomous growth inhibitor, dFoxO has a critical role in adjusting developmental growth for the environmental conditions.
In Drosophila, several studies implicated independently FoxO and cAMP signaling in the regulation of stress resistance and life span. Tong and collaborators (35) reported that core components of the cAMP signaling pathway, adenylate cyclase Rutabaga, cAMP phosphodiesterase Dunce, and protein kinase A, mediate the neurofibromatosis-1-dependent resistance to reactive oxygen species and life span extension. Further, it was demonstrated that feeding flies with the cAMP analogs dibutyryl-cAMP and 8-bromo-cAMP increases life span (35). In addition, Wang and collaborators (36) showed that the Drosophila CREB coactivator TORC promotes resistance to reactive oxygen species. Finally, overexpression and loss of function of dfoxo result in increase in life span and reduced stress resistance, respectively (13, 17, 20, 24). Taken together, these findings suggest a connection between FoxO and cAMP signaling in maintaining cellular homeostasis upon stress. We have confirmed this link by showing that dFoxO directly activates AC76E expression and regulates cAMP levels. We also found that female flies overexpressing ac76e in the CA display elevated starvation resistance. There are two possible mechanisms to explain the increased resistance to a complete lack of nutrients: (i) during development and/or adult life, flies have accumulated more energy reserves, or (ii) during starvation, flies consume less energy per time unit for somatic maintenance than controls. Since flies overexpressing ac76e in the CA were found to be reduced in size, the latter is a more likely explanation. The mechanism behind the observed sexual dimorphism in starvation resistance is not known. Interestingly, Belgacem and Martin (2) showed that differential insulin signaling in the CA regulates sexual dimorphism in locomotor activity. Males have an overall higher expression of ac76e (Fig. 3A), which could be related to a higher expression of ac76e in male genitalia (see Fig. S2E to H in the supplemental material). However, in situ staining for ac76e in the CA does not show any differences in expression between males and females, so we attribute the sex-specific starvation resistance to differences in how cAMP signaling in the CA is relayed in both sexes (2). Further investigation will be needed to address the sexual differences in starvation resistance.
In conclusion, we have demonstrated that dFoxO is a key player in modulating cAMP signaling by regulating the expression of the ac76e gene. We also showed that this regulation, which takes place in the CA, has a critical role in determining organism size, developmental growth, and starvation resistance.
Supplementary Material
Acknowledgments
We thank Terhi Vihervaara for her help in band shifts and ChIP experiments. We thank Osamu Shimmi and Ville Hietakangas for comments on the manuscript. We are grateful to Jean-René Martin for providing us with the DI-11 fly stock.
This work was supported by the Helsinki Graduate School in Biotechnology and Molecular Biology and the Finnish Cultural Foundation (J.M.) and by grants from the Finnish Diabetes Association, Novo Nordisk, and the Juselius Foundation (to O.P.).
Footnotes
Published ahead of print on 3 August 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Arden, K. C. 2008. FOXO animal models reveal a variety of diverse roles for FOXO transcription factors. Oncogene 27:2345-2350. [DOI] [PubMed] [Google Scholar]
- 2.Belgacem, Y. H., and J. R. Martin. 2007. Hmgcr in the corpus allatum controls sexual dimorphism of locomotor activity and body size via the insulin pathway in Drosophila. PLoS ONE 2:e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biggs, W. H., III, J. Meisenhelder, T. Hunter, W. K. Cavenee, and K. C. Arden. 1999. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc. Natl. Acad. Sci. USA 96:7421-7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broughton, S. J., M. D. Piper, T. Ikeya, T. M. Bass, J. Jacobson, Y. Driege, P. Martinez, E. Hafen, D. J. Withers, S. J. Leevers, and L. Partridge. 2005. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc. Natl. Acad. Sci. USA 102:3105-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunet, A., A. Bonni, M. J. Zigmond, M. Z. Lin, P. Juo, L. S. Hu, M. J. Anderson, K. C. Arden, J. Blenis, and M. E. Greenberg. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96:857-868. [DOI] [PubMed] [Google Scholar]
- 6.Buch, S., C. Melcher, M. Bauer, J. Katzenberger, and M. J. Pankratz. 2008. Opposing effects of dietary protein and sugar regulate a transcriptional target of Drosophila insulin-like peptide signaling. Cell Metab. 7:321-332. [DOI] [PubMed] [Google Scholar]
- 7.Cann, M. J., E. Chung, and L. R. Levin. 2000. A new family of adenylyl cyclase genes in the male germline of Drosophila melanogaster. Dev. Genes Evol. 210:200-206. [DOI] [PubMed] [Google Scholar]
- 8.Cann, M. J., and L. R. Levin. 2000. Restricted expression of a truncated adenylyl cyclase in the cephalic furrow of Drosophila melanogaster. Dev. Genes Evol. 210:34-40. [DOI] [PubMed] [Google Scholar]
- 9.Coleman, R. A., and B. F. Pugh. 1997. Slow dimer dissociation of the TATA binding protein dictates the kinetics of DNA binding. Proc. Natl. Acad. Sci. USA 94:7221-7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai, M., P. Wang, A. D. Boyd, G. Kostov, B. Athey, E. G. Jones, W. E. Bunney, R. M. Myers, T. P. Speed, H. Akil, S. J. Watson, and F. Meng. 2005. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 33:e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dentin, R., Y. Liu, S. H. Koo, S. Hedrick, T. Vargas, J. Heredia, J. Yates, III, and M. Montminy. 2007. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature 449:366-369. [DOI] [PubMed] [Google Scholar]
- 12.Gershman, B., O. Puig, L. Hang, R. M. Peitzsch, M. Tatar, and R. S. Garofalo. 2007. High-resolution dynamics of the transcriptional response to nutrition in Drosophila: a key role for dFOXO. Physiol. Genomics 29:24-34. [DOI] [PubMed] [Google Scholar]
- 13.Giannakou, M. E., M. Goss, M. A. Junger, E. Hafen, S. J. Leevers, and L. Partridge. 2004. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science 305:361. [DOI] [PubMed] [Google Scholar]
- 14.Han, P. L., L. R. Levin, R. R. Reed, and R. L. Davis. 1992. Preferential expression of the Drosophila rutabaga gene in mushroom bodies, neural centers for learning in insects. Neuron 9:619-627. [DOI] [PubMed] [Google Scholar]
- 15.Herzig, S., F. Long, U. S. Jhala, S. Hedrick, R. Quinn, A. Bauer, D. Rudolph, G. Schutz, C. Yoon, P. Puigserver, B. Spiegelman, and M. Montminy. 2001. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413:179-183. [DOI] [PubMed] [Google Scholar]
- 16.Hurley, J. H. 1999. Structure, mechanism, and regulation of mammalian adenylyl cyclase. J. Biol. Chem. 274:7599-7602. [DOI] [PubMed] [Google Scholar]
- 17.Hwangbo, D. S., B. Gershman, M. P. Tu, M. Palmer, and M. Tatar. 2004. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429:562-566. [DOI] [PubMed] [Google Scholar]
- 18.Jones, D., and G. Jones. 2007. Farnesoid secretions of dipteran ring glands: what we do know and what we can know. Insect Biochem. Mol. Biol. 37:771-798. [DOI] [PubMed] [Google Scholar]
- 19.Jowett, T. 1999. Analysis of protein and gene expression. Methods Cell Biol. 59:63-85. [DOI] [PubMed] [Google Scholar]
- 20.Junger, M. A., F. Rintelen, H. Stocker, J. D. Wasserman, M. Vegh, T. Radimerski, M. E. Greenberg, and E. Hafen. 2003. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol. 2:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koo, S. H., L. Flechner, L. Qi, X. Zhang, R. A. Screaton, S. Jeffries, S. Hedrick, W. Xu, F. Boussouar, P. Brindle, H. Takemori, and M. Montminy. 2005. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 437:1109-1111. [DOI] [PubMed] [Google Scholar]
- 22.Kops, G. J., T. B. Dansen, P. E. Polderman, I. Saarloos, K. W. Wirtz, P. J. Coffer, T. T. Huang, J. L. Bos, R. H. Medema, and B. M. Burgering. 2002. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature 419:316-321. [DOI] [PubMed] [Google Scholar]
- 23.Kops, G. J., N. D. de Ruiter, A. M. Vries-Smits, D. R. Powell, J. L. Bos, and B. M. Burgering. 1999. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature 398:630-634. [DOI] [PubMed] [Google Scholar]
- 24.Kramer, J. M., J. D. Slade, and B. E. Staveley. 2008. foxo is required for resistance to amino acid starvation in Drosophila. Genome 51:668-672. [DOI] [PubMed] [Google Scholar]
- 25.Mattila, J., J. Kallijarvi, and O. Puig. 2008. RNAi screening for kinases and phosphatases identifies FoxO regulators. Proc. Natl. Acad. Sci. USA 105:14873-14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medema, R. H., G. J. Kops, J. L. Bos, and B. M. Burgering. 2000. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404:782-787. [DOI] [PubMed] [Google Scholar]
- 27.Puig, O., M. T. Marr, M. L. Ruhf, and R. Tjian. 2003. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 17:2006-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richard, D. S., S. W. Applebaum, and L. I. Gilbert. 1990. Allatostatic regulation of juvenile hormone production in vitro by the ring gland of Drosophila melanogaster. Mol. Cell Endocrinol. 68:153-161. [DOI] [PubMed] [Google Scholar]
- 29.Seamon, K. B., W. Padgett, and J. W. Daly. 1981. Forskolin: unique diterpene activator of adenylate cyclase in membranes and in intact cells. Proc. Natl. Acad. Sci. USA 78:3363-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seoane, J., H. V. Le, L. Shen, S. A. Anderson, and J. Massague. 2004. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117:211-223. [DOI] [PubMed] [Google Scholar]
- 31.Skoulakis, E. M., and S. Grammenoudi. 2006. Dunces and da Vincis: the genetics of learning and memory in Drosophila. Cell Mol. Life Sci. 63:975-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang, E. D., G. Nunez, F. G. Barr, and K. L. Guan. 1999. Negative regulation of the forkhead transcription factor FKHR by Akt. J. Biol. Chem. 274:16741-16746. [DOI] [PubMed] [Google Scholar]
- 33.Tatar, M., A. Kopelman, D. Epstein, M. P. Tu, C. M. Yin, and R. S. Garofalo. 2001. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292:107-110. [DOI] [PubMed] [Google Scholar]
- 34.Tettweiler, G., M. Miron, M. Jenkins, N. Sonenberg, and P. F. Lasko. 2005. Starvation and oxidative stress resistance in Drosophila are mediated through the eIF4E-binding protein, d4E-BP. Genes Dev. 19:1840-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tong, J. J., S. E. Schriner, D. McCleary, B. J. Day, and D. C. Wallace. 2007. Life extension through neurofibromin mitochondrial regulation and antioxidant therapy for neurofibromatosis-1 in Drosophila melanogaster. Nat. Genet. 39:476-485. [DOI] [PubMed] [Google Scholar]
- 36.Wang, B., J. Goode, J. Best, J. Meltzer, P. E. Schilman, J. Chen, D. Garza, J. B. Thomas, and M. Montminy. 2008. The insulin-regulated CREB coactivator TORC promotes stress resistance in Drosophila. Cell Metab. 7:434-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, M. C., D. Bohmann, and H. Jasper. 2005. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell 121:115-125. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, W., S. Patil, B. Chauhan, S. Guo, D. R. Powell, J. Le, A. Klotsas, R. Matika, X. Xiao, R. Franks, K. A. Heidenreich, M. P. Sajan, R. V. Farese, D. B. Stolz, P. Tso, S. H. Koo, M. Montminy, and T. G. Unterman. 2006. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J. Biol. Chem. 281:10105-10117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.