Abstract
Avian reovirus sigmaA is a double-stranded RNA (dsRNA)-binding protein that has been shown to stabilize viral core particles and to protect the virus against the antiviral action of interferon. To continue with the characterization of this viral protein, we have investigated its intracellular distribution in avian cells. Most sigmaA accumulates into cytoplasmic viral factories of infected cells, and yet a significant fraction was detected in the nucleolus. The protein also localizes in the nucleolus of transfected cells, suggesting that nucleolar targeting is not facilitated by the viral infection or by viral factors. Assays performed in both intact cells and digitonin-permeabilized cells demonstrate that sigmaA is able to enter the nucleus via a nucleoporin-dependent nondiffusional mechanism that does not require added cytosolic factors or energy input. These results indicate that sigmaA by itself is able to penetrate into the nucleus using a process that is mechanistically different from the classical nuclear localization signal/importin pathway. On the other hand, two sigmaA arginines that are necessary for dsRNA binding are also required for nucleolar localization, suggesting that dsRNA-binding and nucleolar targeting are intimately linked properties of the viral protein.
Avian reoviruses are members of the Orthoreovirus genus, one of the 12 genera of the Reoviridae family. These agents, which are ubiquitous in commercial poultry, induce several disease conditions that lead to important economic losses in the poultry industry. Avian reoviruses are nonenveloped viruses that replicate in the cytoplasm of infected cells and that induce fusion of the host cells. They contain a genome of 10 linear double stranded-RNA (dsRNA) segments encased within two concentric protein shells. Avian reoviruses express at least 10 different structural proteins (lambdaA, -B, and -C; muA, -B, -BC, and -BN; and sigmaA, -B, and -C) and four nonstructural proteins (muNS, sigmaNS, p10, and p17) (for a recent review on avian reovirus, see reference 6 and references therein).
Avian reovirus replication starts with the extracellular attachment of viral particles to the host cell, which is mediated by specific interactions between the outer-capsid protein sigmaC with still-unknown cell surface receptors (43). The virus penetrates by receptor-mediated endocytosis, and the acidification of virus-containing endosomes promotes virus uncoating (14, 37). Uncoated viral cores are then able to cross the endosomal membrane and reach the cytoplasm, where a core-associated RNA polymerase catalyzes the synthesis of all 10 viral mRNAs, which display a dual function: to program viral protein synthesis at the ribosomes and to serve as templates for the production of dsRNA minus strands. Minus-strand synthesis and virus morphogenesis occurs within globular cytoplasmic inclusions, termed viral factories, which are initially formed by the nonstructural protein muNS (60, 61). Core assembly occurs within the first 30 min after the synthesis of its protein components and cores are subsequently coated by outer-capsid polypeptides over the next 30 min to generate mature reovirions (reviewed in reference 7).
Protein sigmaA, which is encoded by the S2 genome segment, is a major component of the inner capsid shell and acts as a clamp on the outside of this shell to stabilize the subcore particles formed by protein lambdaA (74). On the other hand, sigmaA binds dsRNA very tightly, and this activity appears to play a key role in the resistance of avian reovirus to the antiviral action of interferon (42, 71). Experimental evidence suggests that sigmaA provides interferon resistance by preventing the activation of the interferon-inducible and dsRNA-dependent protein kinase PKR (22). The crystal structure of a bacterially expressed recombinant sigmaA has been recently solved. The protein self-assembles as two short double helical hexamers, and mutational analysis suggests that sigmaA cooperatively binds to the outside of the dsRNA helix (24).
In the present study we have investigated the subcellular localization of sigmaA in avian cells. Our results unexpectedly revealed that sigmaA targets the nucleolus of infected and transfected cells. Experiments performed with digitonin-permeabilized cells further showed that sigmaA translocates into the nucleus by a nondiffusional and nonclassical import pathway, which does not require the addition of exogenous cytosolic factors or energy input. We also found that those sigmaA point mutants previously shown to be unable to bind dsRNA are also unable to target the nucleolus, suggesting that dsRNA binding and nucleolar targeting are linked activities of the sigmaA protein.
MATERIALS AND METHODS
Cells, viruses, antibodies, and reagents.
Primary cultures of chicken embryo fibroblasts (CEF) were prepared from 9- to 10-day-old chicken embryos and grown in medium 199 supplemented with 10% tryptose phosphate broth and 5% calf serum. Strain S1133 of avian reovirus was grown on semiconfluent monolayers of primary CEF as previously described (23).
The generation of both polyclonal antiserum and a monoclonal antibody against sigmaA has been previously described (22, 42). The production of polyclonal antisera against the nonstructural viral proteins p17 and muNS was described previously (11, 61). The monoclonal antibody against chicken nucleolin was a generous gift from Elena Nigg. Anti-vimentin (mouse monoclonal, clone VIM-13.2), and goat anti-rabbit and anti-mouse peroxidase-conjugated antibodies were purchased from Sigma-Aldrich (Madrid, Spain). The mouse monoclonal antibody FK2 against conjugated ubiquitin was from Biomol International L.P. (Exeter, United Kingdom). Anti-fibrillarin (mouse monoclonal, clone AFBN01) and anti-dynein (mouse monoclonal sc-13524) were purchased from Cytoskeleton, Inc. (Denver, CO), and from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), respectively. Alexa Fluor 594-goat anti-mouse (red; catalog no. A11005) Alexa Fluor 594-goat anti-rabbit (red; catalog no. A11012), Alexa Fluor 488-goat anti-mouse (green; catalog no. A11001), and Alexa-Fluor 488-goat anti-rabbit (green; catalog no. A11008) fluorescent-conjugated secondary antibodies, as well as anti-COX4 monoclonal antibody (catalog no. A21348) were purchased from Invitrogen (Barcelona, Spain). Digitonin, wheat germ agglutinin (WGA), and Mowiol were purchased from Calbiochem (Darmstadt, Germany). All other reagents used in the present study were from Sigma-Aldrich.
Plasmids.
The generation of plasmids pMal-sigmaA (for bacterial expression of maltose-binding protein (MBP) fused to the amino terminus of sigmaA), pcDNA3.1-sigmaA (for eukaryotic expression of full-length sigmaA) and pCIneo-muNS (for eukaryotic expression of full-length muNS) has been described (22, 61). The plasmid for bacterial expression of GST-NLS-EGFP (72) was a generous gift from Yoshihiro Yoneda and William Hall. To generate the recombinant plasmid that expresses in eukaryotic cells enhanced green fluorescent protein (EGFP) fused to the N terminus of sigmaA (EGFP-sigmaA), the sigmaA encoding sequence of the pcDNA3.1-sigmaA plasmid was cut with restriction enzymes BglII and HindIII and inserted into the BglII and HindIII sites of the pEGFP-C1 vector (BD Biosciences, Madrid, Spain). The pcDNA3.1-sigmaA plasmid and the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) were used according to the manufacturer's specifications to generate recombinant plasmids that express R155A, R273A, and R134,135A mutant versions of sigmaA. The following mutagenic oligonucleotide primers were used. For the production of sigmaA(R155A), the sense primer was 5′-GCTGCTTTCTGCCATGGCAGCTGGTCCTGTTCTC-3′ and the antisense primer was 5′-GAGAACAGGACCAGCTGCCATGGCAGAAAGCAGC-3′; for the production of sigmaA(R273A), the sense primer was 5′-GCTTACGTGTGTGCTGCGTCTCCCGACTGGAAC-3′ and the antisense primer was 5′-GTTCCAGTCGGGAGACGCAGCACACACGTAAGC-3′; and for the production of sigmaA(R134,135A), the sense primer was 5′-CCCTAGATGGGCAAACGCAGCTCGTGAGCTGCAATC-3′ and the antisense primer was 5′-GATTGCAGCTCACGAGCTGCGTTTGCCCATCTAGGG-3′. The correctness of the constructs was assessed by plasmid sequencing and by Western blot analysis of the expressed proteins (data not shown).
Infections, transfections, and fluorescence microscopy.
The infection of CEF by avian reovirus has been described (23). Transfections of cell monolayers were done with the Lipofectamine (Invitrogen), according to the manufacturer's instructions. Transfected cells were incubated at 37°C for 24 h, unless otherwise stated. For indirect immunofluorescence microscopy, cell monolayers were grown on coverslips and subsequently infected or transfected. At the indicated times, monolayers were washed twice with phosphate-buffered saline (PBS) and fixed either for 10 min at room temperature in 4% paraformaldehyde in PBS, or for 15 min at −20°C in 100% methanol. Paraformaldehyde-fixed cells were washed twice with PBS, incubated for 3 min in permeabilizing buffer (0.5% Triton X-100 in PBS), and then blocked in PBS containing 2% bovine serum albumin for 30 min at room temperature. Methanol-fixed cells were washed twice with PBS and then blocked for 30 min at room temperature in PBS containing 2% bovine serum albumin. All fixed cells were subsequently incubated for 1 h at room temperature with primary antibodies diluted in blocking buffer. After three washes with PBS, the cells were incubated for 30 min with secondary antibodies and DAPI (4′,6′-diamidino-2-phenylindole). Cell-containing coverslips were then washed six times with PBS and mounted on glass slides. Images were obtained with an Olympus DP-71 digital camera mounted on an Olympus BX51 fluorescence microscope. Images were also obtained by sequential scanning with a Leica TCS SP2 confocal microscopy using a 100× 1.3 oil immersion objective. Images were processed with Adobe Photoshop (Adobe Systems, California).
Subcellular fractionation and immunoblot analysis.
Avian reovirus-infected CEF (300 × 106 cells; 10 PFU/cell) were washed with prewarmed PBS, trypsinized at 10 h postinfection, and then collected by low-speed centrifugation. The pelleted cells were washed twice with cold PBS, resuspended in 5 ml of 10 mM HEPES-KOH (pH 7.9), 10 mM KCl, 1.5 mM MgCl2, and 0.5 mM dithiothreitol (DTT), and incubated for 5 min on ice. The suspension was subjected to Dounce homogenization (10 strokes with a tight pestle) followed by low-speed centrifugation. The resulting supernatant was considered the cytosolic fraction. The nuclear pellet was resuspended in 3 ml of 0.25 mM sucrose and 10 mM MgCl2, layered over 3 ml of a solution containing 0.35 M sucrose and 0.5 mM MgCl2, and centrifuged at 4°C for 5 min at 1,430 × g. The resulting pellet was resuspended in 3 ml of 0.25 mM sucrose and 10 mM MgCl2, sonicated (6 × 10 s bursts), layered over a 3-ml solution containing 0.88 M sucrose and 0.5 mM MgCl2, and centrifuged at 4°C for 10 min at 2,800 × g. The supernatant was considered the nucleoplasmic fraction. The nucleolar pellet was resuspended in 0.5 ml of 0.35 M sucrose and 0.5 mM MgCl2, and centrifuged at 4°C for 2 min at 2,000 × g. The pellet, containing purified nucleoli, was resuspended in 0.5 ml of 0.35 M sucrose and 0.5 mM MgCl2. The fraction-containing solutions were stored at −80°C.
For Western blot analysis, cell extracts were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and proteins in unfixed gels were transferred to polyvinylidene difluoride (Immobilon-P; Millipore, Madrid, Spain) for 1 h at 100 mA in a semidry blotting apparatus (Bio-Rad, Richmond, CA). Protein bands were detected with specific antibodies using the HRP detection system (Millipore).
Bacterial expression and purification of recombinant proteins.
Expression of MBP-sigmaA in the XL1-Blue bacteria, purification of the fused protein, its cleavage by protease factor Xa, and purification of sigmaA were performed as described previously (27). The expression and purification of GST-NLS-EGFP has been described (47, 72).
Digitonin permeabilization and import assays.
To obtain cytosolic extracts as a source of soluble import factors, CEF monolayers (∼4 × 107 cells) were washed with ice-cold PBS and scraped off the plate into 5 ml of cold PBS. The cell suspension was centrifuged at 600 × g for 10 min at 4°C, and pelleted cells were washed with cytosolic buffer (50 mM HEPES-KOH [pH 7.4], 50 mM KCl, 2 mM MgCl2, 5 mM EGTA, 1 mM DTT, and 1 μg of aprotinin, leupeptin, and pepstatin A/ml) and resuspended in 0.1 ml of cytosolic buffer. The cell suspension was flash-frozen in liquid nitrogen, and the sample was then placed at 37°C until thawed. The resulting cell lysate was centrifuged at 16,000 × g for 15 min at 4°C in a refrigerated microfuge, and the supernatants were flash-frozen in liquid nitrogen and stored at −80°C.
For permeabilization, CEF cells plated on glass slides and grown up to 70% confluence were washed twice with ice-cold transport buffer (25 mM HEPES-KOH [pH 7.3], 125 mM potassium acetate, 2 mM magnesium acetate, 5 mM sodium acetate, 1 mM EGTA, and 1 μg of aprotinin, leupeptin, and pepstatin A/ml) and permeabilized with digitonin (25 μg/ml) for 3 min on ice. Cells were then washed twice with ice-cold transport buffer containing 10 μg of bovine serum albumin/ml. The standard reaction mixtures (50 μl) contained purified import substrates (0.2 mg/ml) dissolved in complete transport solution. This solution contained transport buffer supplemented with an ATP regeneration system (0.5 mM ATP and GTP, 12.5 mM glucose, 10 mM phosphocreatine, and 0.3 U of creatine phosphokinase/ml) as a source of energy and CEF cytosolic extracts (30 μl) as a source of soluble import factors. The import reactions were performed for 30 min at 30°C unless otherwise indicated. Cells were then washed twice with transport buffer and fixed for fluorescence analysis.
When indicated, digitonin-permeabilized CEF were incubated as follows before performing import assays. (i) For hypertonic buffer treatment, samples were incubated for 5 min at room temperature with transport buffer supplemented with 1 M KCl. (ii) For hypotonic buffer treatment, samples were incubated for 2 min on ice with hypotonic buffer (10 mM HEPES-KOH [pH 7.3], 10 mM KCl, 1.5 mM MgCl2, 1 mM DTT, and 0.05% Triton X-100). (iii) For WGA treatment, samples were incubated for 15 min at room temperature with transport buffer containing 0.5 μg of WGA/ml. (iv) For apyrase treatment, samples were incubated for 15 min at room temperature with transport buffer containing 25 U of apyrase/ml and 1 mM CaCl2. (v) For NEM treatment, samples were incubated for 10 min at room temperature with transport buffer containing 5 mM NEM and then for 5 min with transport buffer containing 10 mM DTT to inactivate the alkylating activity of the NEM. (vi) For GMP-PNP treatment, samples were incubated for 20 min at room temperature with 1 mM GMP-PNP. (vii) Finally, for controls for apyrase and NEM treatments, samples were incubated for 15 min with transport buffer containing either 1 mM CaCl2 or 10 mM DTT. We observed that these treatments had no adverse effects on nuclear import of the protein substrates used in the present study (not shown).
RESULTS
SigmaA is present in the cytoplasm and nucleolus of avian reovirus-infected CEF.
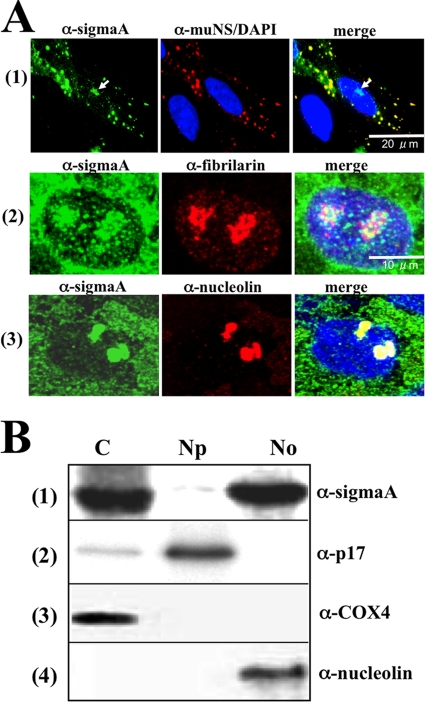
To determine the intracellular localization of protein sigmaA in infected cells, confocal analysis of avian reovirus S1133-infected CEF was performed using an anti-sigmaA monoclonal antibody. Polyclonal antibodies against the nonstructural muNS protein were also used to visualize the avian reovirus factories. Most sigmaA staining was detected in the cytoplasm at 8 h postinfection, colocalizing with muNS within the viral factories (Fig. 1A, row 1). This was an anticipated result, since sigmaA is a structural protein and therefore must be present within viral factories for incorporating into progeny viral particles. Unexpectedly, sigmaA staining was also detected in the nucleus, concentrated within distinct nuclear foci that resembled nucleoli (Fig. 1A, marked with an arrow on the left and right panels of row 1). To confirm that these foci were nucleoli, dual staining for sigmaA and the nucleolar proteins fibrillarin or nucleolin was next performed. Visualization of the infected cells with a confocal microscope revealed that nuclear sigmaA colocalized with both fibrillarin (Fig. 1A, row 2) and nucleolin (Fig. 1A, row 3), confirming that sigmaA targets the nucleolus of avian reovirus-infected cells. Similar results were obtained when CEF monolayers were infected with the avian reovirus isolates 1733 and 2408 or when infecting the stable chicken cell line DF-1 with these viruses (results not shown).
FIG. 1.
Intracellular protein distribution in infected cells. (A) Semiconfluent monolayers of CEF were infected with 20 PFU of avian reovirus S1133/cell for 8 h. The cells were fixed with methanol and subsequently incubated with DAPI, with a mouse monoclonal antibody to sigmaA (α-sigmaA) and a rabbit polyclonal antibody to muNS (top panel), or with rabbit polyclonal antiserum to sigmaA and monoclonal antibodies to fibrillarin (middle panel) and nucleolin (bottom panel). The cells were then immunostained with Alexa Fluor 594-goat anti-rabbit serum and Alexa Fluor 448-goat anti-mouse serum. The sigmaA protein is stained green; muNS, fibrillarin, and nucleolin are stained red; and nuclei are stained blue. Stained cells were visualized by confocal microscopy. The white arrows in row 1 point to nucleolar sigmaA. (B) Western blot analysis of the cytoplasmic (C), nucleoplasmic (Np), and nucleolar (No) fractions obtained from infected cells as indicated in Materials and Methods. The membrane filters were probed with specific antibodies against sigmaA (row 1), p17 (row 2), COX4 (row 3), and nucleolin (row 4).
To rule out the possibility that nucleolar sigmaA staining was an artifact of the immunofluorescence fixation conditions, as has been reported for other nucleic acid-binding proteins (40), its subcellular distribution was also determined by an alternative approach. Avian reovirus-infected cells were biochemically fractionated as indicated in Materials and Methods, and the resulting cytoplasmic, nuclear, and nucleolar extracts were subjected to Western blot analysis (Fig. 1B). The reliability of the fractionating method was assessed by probing the blots with antibodies against the cytoplasmic protein COX-4, the nucleolar protein nucleolin, and the nucleoplasmic avian reovirus protein p17 (11). COX-4 was exclusively detected in the cytoplasmic fraction, nucleolin in the nucleolar fraction, and p17 was highly enriched in the nucleoplasmic fraction (Fig. 1B). Once the purity of the nucleolar fraction was confirmed, immunoblot analysis for sigmaA revealed that the protein is present in the cytoplasmic and nucleolar fractions of avian reovirus-infected cells (Fig. 1B, top panel), which is in agreement with the immunofluorescence analysis.
Time course analysis of avian reovirus-infected cells revealed that sigmaA was already present in both the cytoplasm and nucleolus at 6 h postinfection, when the protein was first detected by immunofluorescence microscopy, and its intracellular distribution did not change during the course of the infection (results not shown).
Protein sigmaA also localizes to the nucleoli of transfected avian cells.
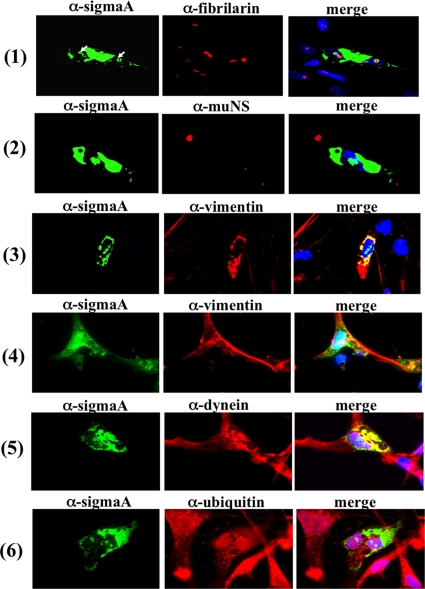
To determine whether other avian reovirus proteins and/or the changes induced in the host by the viral infection are involved in the nucleolar localization of sigmaA, the subcellular distribution of this protein was next examined in transfected avian cells. For this, the sigmaA-encoding plasmid pcDNA3.1-sigmaA was introduced by lipofection into a CEF cell monolayer, and the cells were then fixed and stained with antibodies to sigmaA and fibrillarin. Visualization of the cells with a confocal microscope revealed that, as in infected cells, sigmaA localized to the cytoplasm and nucleoli of transfected cells (Fig. 2A, row 1). Immunofluorescence analysis of cells coexpressing sigmaA and muNS revealed that the two proteins do not colocalize in the cytoplasm of the dually transfected cells (Fig. 2, row 2), supporting our previous observation that sigmaA does not associate with muNS (60).
FIG. 2.
Intracellular protein distribution in transfected cells. Semiconfluent monolayers of CEF cells were transfected with the pcDNA3.1-sigmaA plasmid by the use of Lipofectamine, and 24 h later the cells were fixed, stained with DAPI (blue), and immunostained with antibodies to sigmaA (green) and against each of the endogenous proteins indicated on top of the panels (red). Stained cells were visualized by fluorescence microscopy. The cells shown in row 4 were incubated from 6 to 24 h after transfection with a 50 μM concentration of the vimentin-cage-formation inhibitor KN93. The white arrows in row 1 point to nucleolar sigmaA.
The sigmaA signal detected in the cytoplasm of the transfected cells was not uniformly distributed but accumulated into perinuclear cytoplasmic inclusions that resembled aggresomes. Aggresomes are phase-dense cytoplasmic inclusions into which aggregated and/or misfolded proteins are sequestered when the capacity of the intracellular protein degradation machinery is exceeded. The proteins that form aggresomes are often polyubiquitinated, as a proteasomal degradation signal for the target protein. Formation of aggresomes is usually accompanied by redistribution of the intermediate filament protein vimentin to form cages that surround the aggresome core. Furthermore, aggresomes usually contain dynein, which is used as a motor for retrograde transport along microtubules (reviewed in reference 67). To assess whether the cytoplasmic sigmaA inclusions are aggresomes, dual staining with antibodies against sigmaA and either vimentin or dynein was performed. The results revealed that vimentin forms elongated fibers in cells that do not express sigmaA but is redistributed to form cages that surround sigmaA inclusions in cells that express the viral protein (Fig. 2, row 3). However, vimentin rearrangement in the cells expressing sigmaA was not observed upon incubation with the calcium calmodulin kinase II inhibitor KN93 (Fig. 2, row 4), which inhibits vimentin cage formation (57). Cytoplasmic sigmaA inclusions were also found to contain dynein (Fig. 2, row 5). These results suggest that the sigmaA-containing inclusions observed in the cytoplasm of transfected cells are aggresomes. However, immunofluorescence analysis of the transfected cells with antibodies to ubiquitin revealed that sigmaA-derived aggresomes and nucleolar sigmaA are not polyubiquitinated (Fig. 2, row 6), suggesting that the sigmaA protein expressed in transfected cells is not ubiquitinated.
SigmaA penetrates into the nucleus by selective translocation through the NPC.
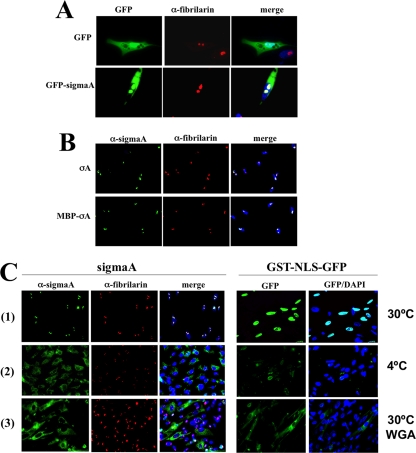
Proteins are imported into the nucleus through the nuclear pore complexes (NPCs), which are large channels that permit the passage of materials in two ways, passive diffusion and selective translocation. Proteins of less than 40 to 60 kDa can trespass the nuclear pores and penetrate into the nucleus by passive diffusion. In contrast, selective translocation is a highly selective process that requires specific interactions between the translocating species and NPC components, allowing the fast transport of even large proteins (for recent reviews, see references 56 and 58). Because sigmaA has a deduced molecular mass of 46 kDa and because this protein appears to be monomeric in solution (24), it is possible that this protein diffuse passively through the nuclear pores. To test this possibility, we first examined the intracellular localization of a protein construct generated by fusing EGFP to the sigmaA amino terminus (EGFP-sigmaA), since the molecular mass of this construct (∼73 kDa) should preclude it from entering the nucleus efficiently by passive diffusion. The results shown in Fig. 3A revealed that, while EGFP was uniformly distributed throughout the cell, EGFP-sigmaA localized to the cytoplasm and nucleolus. This result suggests that sigmaA penetrates into the nucleus by selective translocation through the NPC. To confirm this suggestion, we next used a digitonin-permeabilized cell-free assay that accurately recapitulates nuclear protein import in living cells (1). For a standard import assay, adherent CEF cells were permeabilized with digitonin, and purified import substrates were added to the permeabilized cells dissolved in a complete transport solution, which contained transport buffer supplemented with an energy source and CEF cytosolic extracts (as a source of soluble import factors), as described in Materials and Methods. We initially compared the import of purified sigmaA with that of a construct consisting of MBP fused to the amino terminus of sigmaA. As with sigmaA, MBP-sigmaA also possesses dsRNA binding activity (data not shown), suggesting that the MBP tag has no adverse effect on sigmaA function. Purified GST-NLS-EGFP, which contains the nuclear localization signal (NLS) of the simian virus 40 T antigen carboxy terminal to glutathione S-transferase (GST) and amino terminal to EGFP, was also used in this assay as a positive import substrate, because it is transported into the nucleus by an active nondiffusional mechanism and also because it can be detected by fluorescence without using antibodies (47, 62). The reliability of the in vitro import assay was confirmed by the observation that both GST-NLS-EGFP and sigmaA migrated to the nucleus of permeabilized cells and became concentrated into the expected compartment, the former in the nucleoplasm and the latter in the nucleolus (Fig. 3C, row 1). Our finding that MBP-sigmaA, which is large enough (∼90 kDa) to exceed the passive diffusion limit of NPCs, was able to reach the nucleus and concentrate into the nucleolus of permeabilized cells (Fig. 3B, row 2) confirms that sigmaA does not enter the nucleus by passive diffusion. Our results further demonstrate that sigmaA is able to transport and concentrate a protein (EGFP or MBP) attached to its amino terminus into the nucleolus.
FIG. 3.
SigmaA does not enter the nucleus by passive diffusion. (A) Semiconfluent CEF monolayers were transfected with the plasmids pEGFP (top panel) or pEGFP-sigmaA (bottom panel) for 24 h. The cells were then fixed and stained with DAPI (blue) and with antibodies to fibrillarin (red). Stained cells were visualized by fluorescence microscopy. (B) Digitonin-permeabilized CEF cells were incubated for 30 min at 30°C with purified sigmaA (top panel) or MBP-sigmaA (bottom panel) dissolved in complete transport solution. The cells were then washed, fixed, stained with DAPI (blue), and immunostained with antibodies to sigmaA (green) and fibrillarin (red). The stained cells were visualized by fluorescence microscopy. (C) CEF monolayers were permeabilized with digitonin at room temperature (rows 1 and 3) or at 4°C (row 2). The cells shown in row 3 were incubated with 500 μg of WGA/ml for 15 min at room temperature. Import assays were subsequently performed with purified sigmaA (left panels) or GST-NLS-GFP (right panels) at 30°C (rows 1 and 3) or at 4°C (row 2). The cells were stained with DAPI (blue) and immunostained for sigmaA (green) and fibrillarin (red).
Two further assays were performed to rule out the possibility that sigmaA can gain access to the nucleus by passive diffusion. Since nuclear import by passive diffusion is not inhibited at low temperatures (56), we investigated the capacity of purified sigmaA to reach the nucleus of chilled digitonin-permeabilized cells. Both sigmaA and GST-NLS-EGFP accumulated in the nucleus when the assay was performed at 30°C (Fig. 3C, row 1) but remained confined to the cytoplasm of most cells when the assay was carried out at 4°C (Fig. 3C, compare rows 1 and 2). Furthermore, nuclear import of the two proteins was restored when the import assays were shifted back to 30°C (data not shown). In a second assay, we assessed whether active nucleoporins are required for sigmaA nuclear import. Many nucleoporins of the NPC contain O-linked N-acetylglucosamines, and they become inactivated by incubation with WGA. Thus, WGA treatment blocks nuclear import through the NPC by all receptor-mediated pathways without restricting passive diffusion (73). The incubation of digitonin-permeabilized CEF cells with WGA prevented sigmaA and GST-NLS-EGFP from reaching the nucleus; each of the two proteins accumulated in the cytoplasm of most cells (Fig. 3C, row 3), demonstrating that functional nucleoporins are required for the nuclear import of these proteins. Our finding that nuclear import of sigmaA and GST-NLS-EGFP is blocked by both chilling and WGA also demonstrates that the nuclear envelope of the avian cells was not disrupted during digitonin permeabilization. Taken together, our data indicate that sigmaA reaches the nucleus by a nucleoporin-dependent selective transport pathway and not by passive diffusion.
Nucleolar targeting and dsRNA binding are intimately related sigmaA activities.
Most proteins that enter the nucleus by a nondiffusional pathway contain specific sequences, designated NLSs, which are recognized in the cytoplasm by soluble nuclear transport receptors termed importins. Although there are many different types of NLSs, most nuclear proteins contain the so-called canonical or classical NLS that comes in two varieties: monopartite NLS that contains one continuous array of basic amino acid residues and bipartite NLS that possesses two basic regions separated by an unconserved linker region of 10 to 12 amino acid residues (reviewed in reference 38). In order to map specific sigmaA regions displaying NLS activity, we first tried to analyze the karyophilic properties of truncated sigmaA versions expressed in transfected cells. Unfortunately, the deletion of just a few residues from either of the two sigmaA ends generated insoluble proteins that, probably because of their insolubility, no longer bound dsRNA when expressed in an in vitro translation system (not shown).
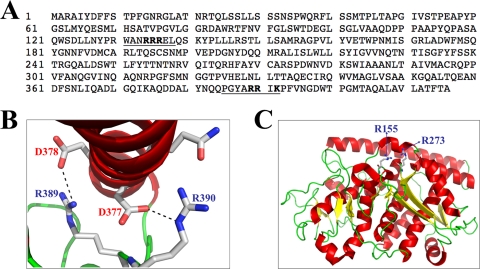
A close examination of the amino acid sequence of sigmaA revealed the presence of two basic regions: 131WANRRREL138 and 385PGYARRIK392 (underlined in Fig. 4A), which share similarities with functional monopartite cNLSs of different nuclear proteins and which are highly conserved among the sigmaA proteins from different avian reovirus isolates, from muscovy duck reovirus and from the fusogenic mammalian Nelson Bay reovirus (data not shown). However, these regions are not conserved in the sigma 2 proteins from nonfusogenic mammalian reoviruses (24). Examination of the recently identified sigmaA crystal structure revealed that the putative NLS region between sigmaA positions 385 and 392 is not likely a functional NLS, because surface exposure of its basic residues is occluded by an alpha helix (24) (Fig. 4B). Furthermore, its basic arginine residues at positions 389 and 390 form salt bridges with aspartate residues at positions 378 and 379, respectively (Fig. 4B), suggesting both that they are not able to interact with importins and that they play a role in maintaining the three-dimensional protein structure. This latter suggestion is supported by our finding that changing arginines 389 and 390 to alanines, either together or individually, generated sigmaA mutants that were expressed in bacteria in insoluble form (results not shown), whereas nonmutated sigmaA was expressed as a soluble protein. Because of this, we did not test the NLS activity of this sigmaA region. On the other hand, a positively charged patch has been identified on the surface of the sigmaA structure. This patch only contains two basic amino acids, Arg155 and Arg273, and the mutation of each of these residues has been shown to abolish dsRNA binding (24) (Fig. 4C).
FIG. 4.
Putative NLS sequences. (A) Deduced primary amino acid sequence of the avian reovirus S1133 sigmaA protein. Putative monopartite NLSs are underlined and basic residues within these sequences are depicted in boldface type. (B) Crystal structure of the sigmaA region containing arginine residues R389 and R390. Note that surface exposure of these residues is prevented by an alpha-helix and that these residues form salt bridges with the aspartic acid residues at positions 377 and 378. (C) Crystal structure of a sigmaA monomer. The location of the arginine residues at positions 155 and 273 is indicated.
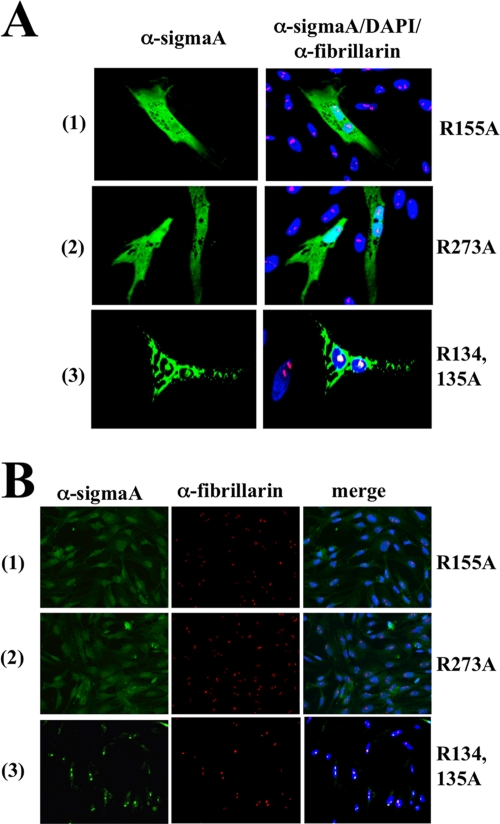
To assess the importance of the region 131WANRRREL138, as well as of the arginine residues R155 and R273, in sigmaA nucleolar targeting activity, we generated recombinant plasmids encoding sigmaA versions that contained alanine substitutions for the arginine residues R134 (R134A), R135 (R135A), R155 (R155A), and R273 (R273A). Visualization of CEF cells transfected with these plasmids revealed that while the sigmaA versions R134A and R135A were still able to target the nucleolus (data not shown), the versions R155A and R273A were uniformly distributed throughout the nucleus and cytoplasm but excluded from the nucleolus (Fig. 5A, rows 1 and 2). These results demonstrate that R155 and R273, but not R134 and R135, are specifically required for sigmaA to target the nucleolus and also suggest that the region 131WANRRREL138 is not a functional NLS. To confirm the latter suggestion, we generated a recombinant plasmid that expresses alanine substitutions for the two arginine residues R134 and R135 (R134,135A). As with the single mutants R134A and R135A, the double mutant R134,135A was also found to target the nucleolus (Fig. 5A, row 3), thus implying that the region 131WANRRREL138 does not mediate sigmaA nuclear import. The nucleolar targeting capacity of these sigmaA mutants was further tested by performing import assays in digitonin-permeabilized cells. The results revealed that, while the double mutant R134,135A retained the capacity of nonmutated sigmaA to reach the nucleus and concentrate into the nucleolus (Fig. 5B, row 3), the single mutants R155A and R273A were detected in the nucleus and cytoplasm but did not target the nucleolus (Fig. 5B, rows 1 and 2). The observation that the sigmaA versions R134A, R135A, and R134,135A bind dsRNA and localize to the nucleolus, whereas the versions R155A and R273A do not bind dsRNA and do not target the nucleolus, suggests that dsRNA binding and nucleolar targeting are two intimately related activities of the sigmaA protein. Our data further suggest that the arginine residues R155 and R273 form part of a nucleolar localization and/or retention sequence. Alternatively, these residues could be components of a nonclassical NLS that, by being recognized by cytoplasmic nuclear transporters, allows sigmaA to migrate into the nucleus in a facilitated fashion. However, this hypothesis is rather unlikely, since the mutation of each of these residues generates sigmaA versions that are still able to enter the nucleus.
FIG. 5.
Intracellular distribution and nuclear import of sigmaA mutants. (A) Semiconfluent monolayers of CEF cells were transfected for 24 h with plasmids that express the following sigmaA mutants: the arginine residue at position 155 was replaced by alanine (R155A) (row 1); the arginine residue at position 273 was replaced by alanine (R273A) (row 2); and the arginine residues at positions 134 and 135 were replaced by alanines (R134,135A) (row 3). (B) Import assays in digitonin-permeabilized cells were performed using as import substrates the sigmaA mutants depicted at the left of the figure. The cells were then fixed, stained with DAPI (blue), and immunostained for sigmaA (green) and fibrillarin (red).
SigmaA does not use the classical NLS/importin nuclear transport pathway.
Most nuclear proteins use the conventional or classical nuclear transport pathway to reach the nucleus. The NLSs of these proteins are recognized by importins and the importin-cargo complex docks at the NPC and translocates through the central transport channel by a mechanism called facilitated translocation. Once in the nucleoplasm, the GTP-bound state of the Ran GTPase promotes the dissociation of the importin-protein complex, releasing the free nuclear protein in the nucleoplasm, and the cargo-free importin is recycled back to the cytoplasm to start another round of nuclear import (reviewed in references 38, 56, and 58). In contrast, the nuclear import of a number of proteins has been reported to be accomplished by carrier- and Ran-independent mechanisms (Table 1).
TABLE 1.
Proteins that can enter the nucleus without the aid of cytosolic factors
| Proteina | Protein nuclear import dependenceb
|
Nucleoporin(s)c | Source or reference | ||||
|---|---|---|---|---|---|---|---|
| CF | Ran | Ene | Chi | WGA | |||
| Group 1 | |||||||
| SigmaA | I | I | I | D | D | ? | This study |
| HIV-1 Vpr | I | I | I | D | D | POM 121/Nsp1p | 31, 63 |
| HIV-1 Integrase | I | I | I | D | D | Nup153 | 13, 68 |
| HIV-1 Tat | I | I | D | ? | ? | ? | 15 |
| HTLV-1 Tax | I | I | I | D | D | Nup62 | 62 |
| Group 2 | |||||||
| Importin-α | I | I | I | D | D | Nup1p, Nup2p, Nup135 | 47, 48 |
| Importin-β1 | I | I | I | I | D | Nup62 | 26, 33, 34 |
| Transportin-I | I | I | I | D | D | ? | 49 |
| Exportin-t | I | I | I | D | D | Nup153, Nup214, Nup358 | 36 |
| Crm1 | I | I | I | I | D | ? | 75 |
| RCC1 | I | I | I | D | I | ? | 50 |
| Group 3 | |||||||
| β-Catenin | I | I | I | D | I | ? | 18, 35, 72 |
| ERK 2 | I | I | I | D | D | Nup153, Nup214 | 44, 66 |
| NHP6A | I | I | I | I | D | ? | 70 |
| PKC α | I | I | I | D | I | ? | 55, 64 |
| SMAD2-4 | I | I | ? | D | D | Nup214 | 69 |
| L5 | I | I | ? | D | D | ? | 52 |
| STAT1,3,5 | I | ? | I | D | D | Nup153, Nup214 | 41, 76 |
| hnRNP K | I | I | D | D | D | Nup153, Nup214 | 46 |
| U1A and U2B″ | I | I | D | D | D | ? | 28 |
| IκBα | I | I | D | D | D | ? | 53 |
| PU.1 | I | D | D | D | D | Nup62, Nup135 | 77 |
| Hsp104 | I | ? | ? | ? | ? | Nup57, Nup116 | 59 |
The proteins have been separated into three groups. Viral proteins are depicted in group 1, proteins implicated in nuclear transport are in group 2, and other cellular proteins are in group 3.
Protein nuclear import dependence on cytosolic factors (CF), RanGTPase (Ran), energy (Ene), chilling (Chi), and WGA preincubation (WGA) is indicated as follows: I, independent; D, dependent; and ?, not assayed or shown.
Nucleoporins are listed that have been identified to directly interact with the indicated proteins.
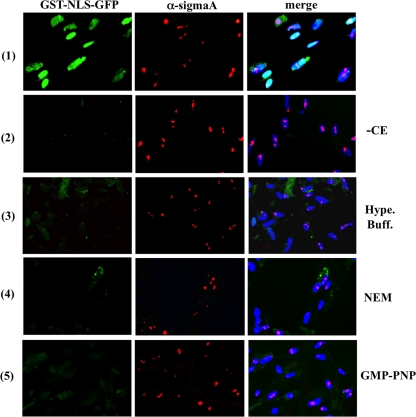
To assess whether sigmaA is imported into the nucleus by the classical pathway or by nonfacilitated transport, we examined the effect that omitting the cytosolic extract from the standard transport solution has on the capacity of both sigmaA and GST-NLS-EGFP to reach the nucleus, when the two proteins are added together to digitonin-permeabilized cells. The results showed that, in the absence of added extract, GST-NLS-EGFP was no longer able to enter the nucleus of permeabilized CEF, whereas sigmaA fully retained its capacity to reach the nucleus and accumulate into the nucleolus (Fig. 6, compare rows 1 and 2). These findings demonstrate that nuclear import of sigmaA does not require the soluble factors that are lost during the digitonin permeabilization step. However, the possibility still exists that sigmaA has a low importin requirement and that inefficient extraction of cytosolic proteins during digitonin permeabilization might leave behind enough intracellular importins as to promote sigmaA nuclear entry. To try to remove or inactivate putative residual endogenous importins, digitonin-permeabilized cells were subjected to three different treatments before performing cytosol-free nuclear import assays. In the first treatment, digitonin-permeabilized cells were incubated with hypotonic and hypertonic buffers, incubations that have been previously shown to efficiently remove both soluble and nuclear pore-attached factors from permeabilized cells (16, 32). In the second approach, digitonin-permeabilized cells were treated with N-ethylmaleimide (NEM), a compound that has been shown to block the classical nuclear import pathway, because it alkylates specific importin-β cysteine residues, and as a result, the covalently modified importin is unable to transport proteins to the nucleus because it is no longer able to bind importin-α or nucleoporins (10). In the third and final approach, digitonin-permeabilized cells were incubated with GMP-PNP, a nonhydrolizable GTP analog that blocks classical nuclear import of NLS-containing substrates by binding to and inhibiting the activity of the Ran-GTPase (18, 45). The results shown in Fig. 6 revealed that incubation of digitonin-permeabilized CEF with either of the two buffers (hypertonic-buffer-treated cells are shown in row 3), with NEM (row 4), or with GMP-PNP (row 5) did not significantly modify the capacity of sigmaA or MBP-sigmaA (data not shown) to penetrate into the nucleus and accumulate in the nucleolus. Nuclear import of sigmaA, but not of GST-NLS-GFP, was also observed when the proteins were added to permeabilized cells in complete transport solution supplemented with NEM or GMP-PNP (data not shown). Notably, we did not observe any difference in sigmaA nuclear import efficiency in the reactions containing cytoplasmic extracts relative to those lacking the extracts (Fig. 6, compare the middle panel of row 1 with the middle panels of rows 2 to 5). Taken together, these results indicate that sigmaA itself contains the necessary and sufficient information to cross the NPC without the aid of cytosolic factors, which in turn suggests that this protein does not use the classical NLS/importin pathway for nuclear translocation. These findings further suggest that the arginine residues at sigmaA positions 155 and 273 form part of a nucleolar localization/retention signal and not of carrier-dependent NLS.
FIG. 6.
Cytosolic factor requirements. The import substrates GST-NLS-EGFP and sigmaA were dissolved together in complete transport solution (row 1) or in transport solution lacking the cytosolic extract (rows 2 to 5). The resulting solutions were added to digitonin-permeabilized cells that had been mock-incubated (rows 1 and 2) or incubated at room temperature with hypertonic buffer (row 3), NEM (row 4), and GMP-PNP (row 5) as indicated in Materials and Methods. After 30 min at 30°C, the cells were fixed, stained with DAPI (blue) and immunostained for sigmaA (red). The cells were visualized by fluorescence microscopy, and GST-NLS-GFP is stained in green.
Nuclear import occurs in the absence of an exogenously added energy supply.
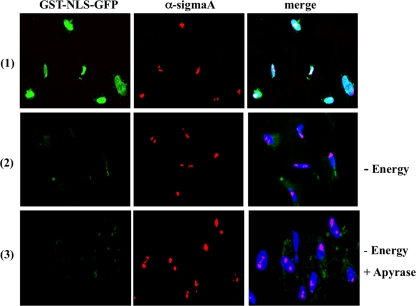
The results shown thus far demonstrate that sigmaA does not require importins for nuclear translocation. Many proteins that also target the nucleus in an importin-unassisted manner do not either require an input of energy for nuclear migration (Table 1). To determine the energy requirements for sigmaA import, we examined the capacity of this protein to reach the nucleus of permeabilized cells when the assay is performed in transport buffer devoid of an energy supply. GST-NLS-EGFP was used as a suitable negative control protein, since it has been shown to reach the nucleus of digitonin-permeabilized cells in an importin- and energy-dependent fashion (47, 62). Removal of the energy source dramatically reduced the nuclear targeting ability of GST-NLS-EGFP but did not significantly affect the capacity of sigmaA to concentrate into the nucleolus (Fig. 7, compare rows 1 and 2), when the two proteins were added together to permeabilized cells. To rule out the possibility that sigmaA utilizes the residual ATP left behind after digitonin permeabilization, an import assay was next performed on apyrase-treated cells since this enzyme has been shown to readily diminish free ATP and GTP levels from permeabilized cells (31). The effectiveness of the ATP-depleting enzyme was assessed by showing that GST-NLS-EGFP was unable to enter the nucleus of digitonin-permeabilized cells when added in a standard transport solution supplemented with apyrase (not shown). However, sigmaA and MBP-sigmaA were still able to migrate and concentrate into the nucleolus of apyrase-treated cells when the import assay was performed in either transport buffer alone or transport buffer supplemented with cytosolic extracts (Fig. 7, row 3 shows the sigmaA assay with added extracts), suggesting that sigmaA is imported into the nucleus by a mechanism that does not require ATP or its hydrolysis.
FIG. 7.
Energy requirements. The import substrates GST-NLS-EGFP and sigmaA were dissolved together in complete transport solution (row 1) or in transport solution lacking the energy-regenerating system (rows 2 and 3). The resulting solutions were added to digitonin-permeabilized cells that had been mock incubated (rows 1 and 2) or incubated for 15 min at room temperature with a solution containing 25 U of apyrase/ml and 1 mM CaCl2 (row 3). After 30 min at 30°C, the cells were fixed, stained with DAPI (blue) and immunostained for sigmaA (red). The cells were visualized by fluorescence microscopy, and GST-NLS-GFP is stained green.
DISCUSSION
In the first part of this study we have investigated the subcellular distribution of avian reovirus protein sigmaA in avian cells. Because avian reoviruses are cytoplasmic replicating viruses and also because sigmaA is a structural component of viral cores, we anticipated that sigmaA should concentrate in cytoplasmic viral factories of infected cells, which are the sites where avian reovirus morphogenesis takes place (60). Our results confirmed that prediction, since most sigmaA was found colocalizing with muNS within the viral factories. Unexpectedly, a sigmaA fraction was detected in the nucleolus of infected cells, which was confirmed by different experimental approaches: (i) confocal microscopy of infected and transfected cells revealed that sigmaA colocalizes with the nucleolar proteins fibrillarin and nucleolin; (ii) immunoblot analysis of subcellular fractions from infected cells showed that sigmaA is present in the cytoplasmic and nucleolar, but not nucleoplasmic, fractions; (iii) a recombinant sigmaA protein expressed in bacteria migrates into the nucleus of digitonin-permeabilized cells and accumulates in the nucleolus; and (iv) two sigmaA point mutants that do not bind dsRNA are unable to target the nucleolus. Furthermore, our finding that this protein is present in the nucleolus of both infected and uninfected cells indicates that sigmaA nucleolar targeting is not promoted by viral factors or by the changes induced by the viral infection within the host cell.
In clear contrast with the nucleolar situation, it seems that the cytoplasmic localization of sigmaA is indeed influenced by the viral infection since this protein accumulates in the viral factories of infected cells but in the perinuclear inclusions of transfected cells. Fluorescence analysis of cells costained with sigmaA and either vimentin or dynein suggests that cytoplasmic sigmaA forms nonubiquitinated aggresomes in transfected cells, supporting the previously published suggestion that aggresomes can be formed by soluble, nonubiquitinated proteins (20). A plausible explanation is that sigmaA, by being a capsomer-forming protein, tends to aggregate when expressed alone in transfected cells, but its association with viral factors and its recruitment into viral factories prevents sigmaA aggregation in infected cells. A likely factor candidate would be the still-unidentified viral protein that recruits sigmaA to the viral factories.
The nucleolus is a nonenveloped dynamic subnuclear structure that is maintained by the accumulation of rRNA and nucleolar proteins such as nucleolin, fibrillarin, and nucleoplasmin. The main nucleolar function is to produce ribosomal subunits, although recent studies have also implicated the nucleolus in several different processes such as nucleocytoplasmic mRNA transport, cell cycle control, DNA repair, apoptosis, aging, etc. (reviewed in reference 8). That a cytoplasmic replicating virus expresses a nucleolar targeting protein was a rather unexpected finding, but a subsequent bibliographic search revealed that this situation is not without precedent, since several other RNA cytoplasmic replicating viruses, such as coronavirus and poliovirus, express proteins that have been similarly detected within this compartment (reviewed in references 29 and 30). Furthermore, the expression of a nucleolar protein by another member of the Orthoreovirus genus has also been reported. Thus, the nonstructural sigma1S protein, which is encoded by the second open reading frame of the mammalian reovirus S1 gene, was detected in the nucleolus of Cos cells that had been transfected with the S1 gene (4). However, we were unable to find further properties and/or activities that could be shared by sigma1S and sigmaA. On the other hand, sigmaA is not the only avian reovirus protein with nuclear targeting activity, since we have previously demonstrated that the nonstructural protein p17, encoded by the second cistron of the avian reovirus genome segment S1, accumulates in the nucleoplasm of infected and transfected cells but is excluded from the nucleolus (11).
In the second part of the present study we investigated the mechanism by which sigmaA is imported into the nucleus and accumulates in the nucleolus. Several lines of evidence revealed that sigmaA does not reach the nucleus by passive diffusion, and the inhibitory effect of WGA implies that it is transported through the NPC channel by a mechanism that requires nucleoporin activity. Recent studies suggest that the inner tunnel of the NPC is a three-dimensional sievelike barrier formed by hydrophobic intermolecular contacts between individual repeat units of the nucleoporin phenylalanine-glycine-rich repeat (FG repeats) domains. Molecules with translocating activity should possess multiple binding sites for FG repeats and should be able to transiently dissociate adjacent inter-repeat contacts as they cross the permeability barrier, although inter-repeat contacts must be reformed just after cargo passage to regenerate gating barrier activity (19).
We were unable to find a classical functional NLS in the sigmaA sequence, and our results revealed that sigmaA does not require cytosolic factors for entering the nucleus of digitonin-permeabilized cells. This and the fact that sigmaA is also able to reach the nucleus of GMP-PNP-treated permeabilized cells suggests that this protein is imported into the nucleus in an importin- and Ran-independent manner, as has been reported for a number of other viral and cellular proteins (Table 1). Thus, it appears that sigmaA and the proteins shown in Table 1 are by themselves able to interact with nucleoporins at the cytoplasmic face of the NPC and to dissociate adjacent hydrophobic contacts formed by FG-rich nucleoporin repeats for translocating through the inner pore channel and reach the nucleoplasm. Indeed, many of the proteins that are able to cross the NPC in a nonfacilitated manner have been shown to interact directly with specific nucleoporins (Table 1), suggesting that nucleoporin association is a common feature of carrier-independent nuclear import. The interaction of these proteins with NPC components might alter or damage the structure of the NPC, which in turn could change the normal nucleocytoplasmic transport and/or nucleocytoplasmic protein distribution of the host cell. Accordingly, several viruses and specific viral proteins have been reported to target NPC components as a strategy for promoting viral replication (3, 5, 12, 25, 39, 51, 63).
Our finding that sigmaA, but not GST-NLS-EGFP, is able to gain access to the nucleus of digitonin-permeabilized cells in the absence of an exogenously added energy source and after apyrase or GMP-PNP treatments suggests that sigmaA is imported by an energy-independent mechanism that does not require nucleoside triphosphate hydrolysis. This in turn reinforces the notion that sigmaA uses an importin- and Ran-independent mechanism, since all import pathways known to date that depend on transport receptors are known to require metabolic energy (56). The sigmaA energy-independent import pathway, which is also displayed by many of the proteins shown in Table 1, may be similar to the so-called facilitated diffusion mechanism that has been proposed to require specific low-affinity interactions with nucleoporins and to be blocked by chilling (65). However, mechanistic differences in the import pathways used by these carrier- and energy-independent proteins do appear to exist, since the nuclear import of sigmaA and most of the proteins shown in Table 1 is inhibited by both WGA and chilling, whereas the nuclear import of CRM1, importin β, and NHP6A is not inhibited by chilling and that of RCC1, β-catenin and protein kinase C is not prevented by WGA treatment (Table 1). Furthermore, importin β-family molecules competitively inhibit the nuclear import of some of these proteins but not of others (47). It is worth noting that many of the proteins shown in Table 1 indeed contain functional NLSs and, in addition to be able to enter the nucleus by a nonfacilitated NPC-mediated pathway, they can also migrate into the nucleus by a classical receptor-mediated mechanism. They are believed to use the former mechanism to continue to access the nucleus when the importin-mediated pathway is disrupted. It would be interesting to determine whether sigmaA interacts with specific importins for entering the nucleus by a receptor-mediated mechanism. It will also be of interest to determine the effect of dominant-negative importin β and Ran mutants on sigmaA nuclear import. However, we did not investigate the effects of these molecules in the present study because the avian genes of these proteins have not been cloned and also because the effects that their mammalian counterparts could exert on sigmaA nuclear import in avian cells may not have physiological relevance. We are, however, planning to investigate the effects of the mammalian proteins on sigmaA localization and nuclear import in mammalian cells, a study that is currently being carried out in our laboratory.
Recent evidence suggests that the NPC passage per se is a fully reversible and energy-independent process, but that energy is required for active and vectorial release of the cargo in the destination compartment (17, 49). Although the self-translocating activity of sigmaA might allow this protein to cross the NPC and reach the nucleoplasm in a nonfacilitated and energy-independent fashion, the reversibility of the NPC translocation process would preclude sigmaA from accumulating in the nucleus. Since rRNA, the main nucleolar RNA component, possesses a relatively high content of duplex structures, and since the binding of sigmaA to dsRNA is independent of RNA sequence, our finding that dsRNA binding and nucleolar targeting are intimately related sigmaA activities suggests that the association of sigmaA with duplex structures of rRNA is the driving force that removes sigmaA from the nucleoplasm and allows its accumulation in the nucleolus, as has been reported for other nucleolar RNA-binding proteins (12, 21, 23, 54, 59). Experiments are in progress in our laboratory to confirm this hypothesis.
The question remains as to whether sigmaA could function as an exogenous nucleolar transport receptor that associates with cellular cytoplasmic factors and escorts them to the nucleolus in a piggyback fashion. Three observations appear to support this assumption: (i) sigmaA is able to guide to the nucleolus a protein covalently attached to its amino terminus, EGFP and MBP (Fig. 3A and B); (ii) many of the proteins that are also able to enter the nucleus by carrier- and energy-independent mechanisms are receptors involved in nuclear import and export (Table 1); and (iii) it has been recently reported that two proteins that enter the nucleus by a similar carrier- and energy-independent mechanism, β-catenin and the HTLV-1 Tax, have a carrier function, in that the former is able to transport LEF/TCF (2), and the latter the NF-κB subunit p65 (62), to the nucleus. If sigmaA associates with cellular factors in the cytoplasm, the self-translocating activity of the viral protein could permit the sigmaA-factor complex to cross the NPC and reach the nucleoplasm. Once there, the complex would move toward the nucleolus because of the affinity of sigmaA for nucleolar components; the interaction of sigmaA with nucleolar components could cause dissociation of the complex, resulting in the retention of sigmaA in the nucleolus and the releasing of the sigmaA-escorted factor in the nucleus. Nuclear sequestration of cytoplasmic factors via sigmaA association might be a strategy used by avian reoviruses to alter the normal nuclear-cytoplasmic distribution of cellular proteins, which might deregulate cellular functions and enhance viral replication.
Additional studies will be required to obtain a detailed understanding of the mechanisms used by sigmaA to enter the nucleus and accumulate in the nucleolus, as well as to decipher the role that its nucleolar targeting plays on cellular metabolism and viral replication. The identification of cellular sigmaA-binding factors in the cytoplasm, the NPC and the nucleolus are also key questions to be elucidated.
Acknowledgments
We thank Laboratorios Intervet (Salamanca, Spain) for providing the pathogen-free embryonated eggs and Elena Nigg, William Hall, and Yoshihiro Yoneda for their generosity in providing plasmids and antibodies used in this study. We are also grateful to Rebeca Menaya and Leticia Barcia for their excellent technical assistance and to Pablo Guardado for help with the illustrations in Fig. 4.
This study was supported by grants from the Spanish Ministerio de Ciencia y Tecnología (BFU2004-05641/BMC and BFU2007-61330/BMC) and the Xunta de Galicia (08CSA009203PR). L.V.-I. and I.L.-S. were recipients of predoctoral fellowships from the FPI and FPU programs of the Spanish Ministerio de Ciencia y Tecnología.
Footnotes
Published ahead of print on 29 July 2009.
REFERENCES
- 1.Adam, S. A., R. S. Marr, and L. Gerace. 1990. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111:807-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asally, M., and Y. Yoneda. 2005. Beta-catenin can act as a nuclear import receptor for its partner transcription factor, lymphocyte enhancer factor-1 (lef-1). Exp. Cell Res. 308:357-363. [DOI] [PubMed] [Google Scholar]
- 3.Atasheva, S., N. Garmashova, I. Frolov, and E. Frolova. 2008. Venezuelan equine encephalitis virus capsid protein inhibits nuclear import in mammalian but not in mosquito cells. J. Virol. 82:4028-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belli, B. A., and C. E. Samuel. 1991. Biosynthesis of reovirus-specified polypeptides: expression of reovirus S1-encoded sigma 1NS protein in transfected and infected cells as measured with serotype specific polyclonal antibody. Virology 185:698-709. [DOI] [PubMed] [Google Scholar]
- 5.Belov, G. A., P. V. Lidsky, O. V. Mikitas, D. Egger, K. A. Lukyanov, K. Bienz, and V. I. Agol. 2004. Bidirectional increase in permeability of nuclear envelope upon poliovirus infection and accompanying alterations of nuclear pores. J. Virol. 78:10166-10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benavente, J., and J. Martinez-Costas. 2007. Avian reovirus: structure and biology. Virus Res. 123:105-119. [DOI] [PubMed] [Google Scholar]
- 7.Benavente, J., and J. Martinez-Costas. 2006. Early steps in avian reovirus morphogenesis. Curr. Top. Microbiol. Immunol. 309:67-85. [DOI] [PubMed] [Google Scholar]
- 8.Boisvert, F. M., S. van Koningsbruggen, J. Navascués, and A. I. Lamond. 2007. The multifunctional nucleolus. Nat. Rev. Mol. Cell. Biol. 8:574-585. [DOI] [PubMed] [Google Scholar]
- 9.Carmo-Fonseca, M., L. Mendes-Soares, and I. Campos. 2002. To be or not to be in the nucleolus. Nat. Cell Biol. 2:E107-E112. [DOI] [PubMed] [Google Scholar]
- 10.Chi, N. C., and S. A. Adam. 1997. Functional domains in nuclear import factor p97 for binding the nuclear localization sequence receptor and the nuclear pore. Mol. Biol. Cell 8:945-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costas, C., J. Martínez-Costas, G. Bodelon, and J. Benavente. 2005. The second open reading frame of the avian reovirus S1 gene encodes a transcription-dependent and CRM1-independent nucleocytoplasmic shuttling protein. J. Virol. 79:2141-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delhaye, S., V. van Pesch, and T. Michiels. 2004. The leader protein of Theiler's virus interferes with nucleocytoplasmic trafficking of cellular proteins. J. Virol. 78:4357-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Depienne, C., A. Mousnier, H. Leh, E. Le Rouizic, D. Dormont, S. Benichou, and C. Dargemont. 2001. Characterization of the nuclear import pathway for HIV-1 integrase. J. Biol. Chem. 276:18102-18107. [DOI] [PubMed] [Google Scholar]
- 14.Duncan, R. 1996. The low pH-dependent entry of avian reovirus is accompanied by two specific cleavages of the major outer capsid protein mu2C. Virology 219:179-189. [DOI] [PubMed] [Google Scholar]
- 15.Efthymiadis, A., L. J. Briggs, and D. A. Jans. 1998. The HIV-1 Tat nuclear localization sequence confers novel nuclear import properties. J. Biol. Chem. 273:1623-1628. [DOI] [PubMed] [Google Scholar]
- 16.Elbi, C., D. A. Walter, G. Romero, W. P. Sullivan, D. O. Toft, and G. L. Hager. 2004. Molecular chaperones function as steroid receptor nuclear mobility factors. Proc. Natl. Acad. Sci. USA 101:2876-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Englmeier, L., J. C. Olivo, and I. W. Mattaj. 1999. Receptor-mediated substrate translocation through the nuclear pore complex without nucleotide triphosphate hydrolysis. Curr. Biol. 9:30-41. [DOI] [PubMed] [Google Scholar]
- 18.Fagotto, F., U. Glück, and B. M. Gumbiner. 1998. Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of β-catenin. Curr. Biol. 8:181-190. [DOI] [PubMed] [Google Scholar]
- 19.Frey, S., and D. Görlich. 2007. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 130:512-523. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Mata, R., Z. Bebök, E. J. Sorscher, and E. S. Sztul. 1999. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 146:1239-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghorbel, S., U. Sinha-Datta, M. Dundr, M. Brown, and G. Franchini. 2006. Human T-cell leukaemia virus type I p30 nuclear/nucleolar retention is mediated through interactions with RNA and a constituent of the 60 S ribosomal subunit. J. Biol. Chem. 281:37150-37158. [DOI] [PubMed] [Google Scholar]
- 22.González-López, C., J. Martínez-Costas, M. Esteban, and J. Benavente. 2003. Evidence that avian reovirus σA protein is an inhibitor of the double-stranded RNA-dependent protein kinase. J. Gen. Virol. 84:1629-1639. [DOI] [PubMed] [Google Scholar]
- 23.Grande, A., and J. Benavente. 2000. Optimal conditions for the growth, purification, and storage of avian reovirus S1133. J. Virol. Methods 85:43-54. [DOI] [PubMed] [Google Scholar]
- 24.Guardado-Calvo, P., L. Vazquez-Iglesias, J. Martinez-Costas, A. L. Llamas-Sainz, G. Schoehn, G. C. Fox, L. Hermo-Parrado, J. Benavente, and M. J. van Raaij. 2008. Crystal structure of the avian reovirus inner capsid protein σA. J. Virol. 82:11208-11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gustin, K. E. 2003. Inhibition of nucleo-cytoplasmic trafficking by RNA viruses: targeting the nuclear pore complex. Virus Res. 95:35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harel, A., and D. J. Forbes. 2004. Importin beta: conducting a much larger cellular symphony. Mol. Cell 16:319-330. [DOI] [PubMed] [Google Scholar]
- 27.Hermo-Parrado, X. L., P. Guardado-Calvo, A. L. Llamas-Saiz, C. Costas, J. Martinez-Costas, J. Benavente, and M. J. van Raaij. 2007. Crystallization of the avian reovirus double-stranded RNA-binding and core protein sigmaA. Acta Crystallogr. F. 63:426-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hetzer, M., and I. W. Mattaj. 2000. An ATP-dependent, Ran-independent mechanism for nuclear import of the U1A and U2B″ spliceosome proteins. J. Cell Biol. 148:293-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiscox, J. A. 2007. RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 5:119-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hiscox, J. A. 2003. The interaction of animal cytoplasmic RNA viruses with the nucleus to facilitate replication. Virus Res. 95:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenkins, Y., M. McEntee, K. Weis, and W. C. Greene. 1998. Characterization of HIV-1 vpr nuclear import: analysis of signals and pathways. J. Cell Biol. 143:875-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kodiha, M., P. Banski, D. Ho-Wo-Cheong, and U. Stochaj. 2008. Dissection of the molecular mechanisms that control the nuclear accumulation of transport factors importin-α and CAS in stressed cells. Cell Mol. Life Sci. 65:1756-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kose, S., N. Imamoto, T. Tachibana, T. Shimamoto, and Y. Moneda. 1997. Ran-unassisted nuclear migration of a 97 kD component of nuclear pore-targeting complex. J. Cell Biol. 139:841-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kose, S., N. Imamoto, and Y. Yoneda. 1999. Distinct energy requirements for nuclear import and export of importin β in living cells. FEBS Lett. 463:327-330. [DOI] [PubMed] [Google Scholar]
- 35.Krieghoff, E., J. Behrens, and B. Mayr. 2006. Nucleo-cytoplasmic distribution of beta-catenin is regulated by retention. J. Cell Sci. 119:1453-1463. [DOI] [PubMed] [Google Scholar]
- 36.Kuersten, S., G. J. Arts, T. C. Walther, L. Englmeier, and I. W. Mattaj. 2002. Steady-state nuclear localization of exportin-t involves RanGTP binding and two distinct nuclear pore complex interaction domains. Mol. Cell. Biol. 22:5708-5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Labrada, L., G. Bodelón, J. Viñuela, and J. Benavente. 2002. Avian reoviruses cause apoptosis in cultured cells: viral uncoating, but not viral gene expression, is required for apoptosis induction. J. Virol. 76:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lange, A., R. A. Mills, C. J. Lange, M. Stewart, S. E. Devine, and A. H. Corbett. 2007. Classical nuclear localization signals: definition, function, and interaction with importin α. J. Biol. Chem. 282:5101-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lidsky, P. V., S. Hato, M. V. Bardina, A. G. Aminev, A. C. Palmenberg, E. V. Sheval, V. Y. Polyakov, F. J. van Kuppeveld, and V. I. Agol. Nucleocytoplasmic traffic disorder induced by cardioviruses. J. Virol. 80:2705-2717. [DOI] [PMC free article] [PubMed]
- 40.Lundberg, M., and M. Johansson. 2002. Positively charged DNA-binding proteins cause apparent cell membrane translocation. Biochem. Biophys. Res. Commun. 291:367-371. [DOI] [PubMed] [Google Scholar]
- 41.Marg, A., Y. Shan, T. Meyer, T. Meissner, M. Brandenburg, and U. Vinkemeier. 2004. Nucleocytoplasmic shuttling by nucleoporins Nup153 and Nup214 and CRM1-dependent nuclear export control the subcellular distribution of latent Stat1. J. Cell Biol. 165:823-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez-Costas, J., C. Gonzalez-Lopez, V. N. Vakharia, and J. Benavente. 2000. Possible involvement of the double-stranded RNA-binding core protein sigmaA in the resistance of avian reovirus to interferon. J. Virol. 74:1124-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez-Costas, J., A. Grande, R. Varela, C. Garcia-Martinez, and J. Benavente. 1997. Protein architecture of avian reovirus S1133 and identification of the cell attachment protein. J. Virol. 71:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsubayashi, Y., M. Fukuda, and E. Nishida. 2001. Evidence for existence of a nuclear pore complex-mediated, cytosol-independent pathway of nuclear translocation of ERK MAP kinase in permeabilized cells. J. Biol. Chem. 276:41755-41760. [DOI] [PubMed] [Google Scholar]
- 45.Melchior, F., B. Paschal, J. Evans, and L. Gerace. 1993. Inhibition of the nuclear protein import by nonhydrolizable analogues of GTP and identification of the small GTPaseRan/TC4 as an essential transport factor. J. Cell Biol. 112:1649-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Michael, W. M., P. S. Eder, and G. Dreyfuss. 1997. The K nuclear shuttling domain. A novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 16:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miyamoto, Y., M. Hieda, M. T. Harreman, M. Fukumoto, T. Saiwaki, A. E. Hotel, A. N. Corbett, and Y. Yoneda. 2002. Importin α can migrate into the nucleus in an importin β- and Ran-independent manner. EMBO J. 21:5833-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moroianu, J., G. Blobel, and A. Radu. 1997. RanGTP-mediated nuclear export of karyopherin α involves its interaction with the nucleoporin Nup153. Proc. Natl. Acad. Sci. USA 94:9699-9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakielny, S., and G. Dreyfuss. 1998. Import and export of the nuclear protein import receptor transportin by a mechanism of GTP hydrolysis. Curr. Biol. 8:89-95. [DOI] [PubMed] [Google Scholar]
- 50.Nemergut, M. E., and I. G. Macara. 2000. Nuclear import of the Ran exchange factor, RCC1, is mediated by at least two distinct mechanisms. J. Cell Biol. 149:835-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park, N., P. Katikaneni, T. Skern, and K. E. Gustin. 2008. Differential targeting of nuclear pore complex proteins in poliovirus-infected cells. J. Virol. 82:1647-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rudt, F., and T. Pieler. 2001. Cytosolic import factor- and Ran-independent nuclear transport of ribosomal protein L5. Eur. J. Cell Biol. 80:661-668. [DOI] [PubMed] [Google Scholar]
- 53.Sachdev, S., S. Bagchi, D. D. Zhang, A. C. Mings, and M. Hannink. 2000. Nuclear import of IκBα is accomplished by a Ran-independent transport pathway. Mol. Cell. Biol. 20:1571-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sansam, C., K. S. Wells, and R. B. Emeson. 2003. Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc. Natl. Acad. Sci. USA 100:14018-14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmalz, D., F. Hucho, and K. Buchne. 1998. Nuclear import of protein kinase C occurs by a mechanism distinct from the mechanism used by proteins with a classical nuclear localization signal. J. Cell Sci. 111:1823-1830. [DOI] [PubMed] [Google Scholar]
- 56.Sorokin, A. V., E. R. Kim, and L. P. Ovchinnikov. 2007. Nucleocytoplasmic transport of proteins. Biochemistry 72:1439-1457. [DOI] [PubMed] [Google Scholar]
- 57.Stefanovic, S., M. Windsor, K.-I. Nagata, M. Inagaki, and T. Wileman. Vimentin rearrangement during African swine fever virus infection involves retrograde transport along microtubules and phosphorylation of vimentin by calcium calmodulin kinase II. J. Virol. 79:11766-11775. [DOI] [PMC free article] [PubMed]
- 58.Stewart, M. 2007. Molecular mechanisms of the nuclear protein import cycle. Mol. Cell. Biol. 8:195-208. [DOI] [PubMed] [Google Scholar]
- 59.Tkach, J. M., and J. R. Glover. 2008. Nucleocytoplasmic trafficking of the molecular chaperone Hsp104 in unstressed and heat-shocked cells. Traffic 9:39-56. [DOI] [PubMed] [Google Scholar]
- 60.Tourís-Otero, F., M. Cortez-San Martín, J. Martínez-Costas, and J. Benavente. 2004. Avian reovirus morphogenesis occurs within viral factories and begins with the selective recruitment of σNS and λA to μNS inclusions. J. Mol. Biol. 341:361-374. [DOI] [PubMed] [Google Scholar]
- 61.Tourís-Otero, F., J. Martínez-Costas, V. N. Vakharia, and J. Benavente. 2004. Avian reovirus nonstructural protein μNS forms viroplasm-like inclusions and recruits σNS to these structures. Virology 319:94-106. [DOI] [PubMed] [Google Scholar]
- 62.Tsuji, T., N. Sheehy, V. W. Gautier, H. Haykawa, H. Sawa, and W. W. Hall. 2007. The nuclear import of the human T lymphotropic virus type I (HTLV-1) Tax protein is carrier- and energy-independent. J. Biol. Chem. 282:13857-13883. [DOI] [PubMed] [Google Scholar]
- 63.Vodicka, M. A., D. M. Koepp, P. A. Silver, and M. Emerman. 1998. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 12:175-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wagner, S., C. Harteneck, F. Hucho, and K. Buchner. 2000. Analysis of the subcellular distribution of protein kinase Cα using PKC-EGFP fusion proteins. Exp. Cell Res. 258:204-214. [DOI] [PubMed] [Google Scholar]
- 65.Wente, S. R. 2000. Gatekeepers of the nucleus. Science 288:1374-1377. [DOI] [PubMed] [Google Scholar]
- 66.Whitehurst, A. W., J. L. Wilsbacher, Y. You, K. Luby-Phelps, M. S. Moore, and M. H. Cobb. 2002. ERK2 enters the nucleus by a carrier-independent mechanism. Proc. Natl. Acad. Sci. USA 99:7496-7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wileman, T. 2007. Aggresomes and pericentriolar sites of virus assembly: cellular defense or viral design? Annu. Rev. Microbiol. 61:149-167. [DOI] [PubMed] [Google Scholar]
- 68.Woodward, C. L., S. Prakobwanakit, S. Mosessian, and S. A. Chow. 15 April 2009. Integrase interacts with nucleoporin NUP153 in mediating the nuclear import of human immunodeficiency virus type 1. J. Virol. [Epub ahead of print.] [DOI] [PMC free article] [PubMed]
- 69.Xu, L., C. Alarcon, S. Col, and J. Massague. 2003. Distinct domain utilization by Smad3 and Smad4 for nucleoporin interaction and nuclear import. J. Biol. Chem. 278:42569-42577. [DOI] [PubMed] [Google Scholar]
- 70.Yen, Y. M., P. M. Roberts, and R. C. Johnson. 2001. Nuclear localization of the Saccharomyces cerevisiae HMG protein NHP6A occurs by a Ran-independent nonclassical pathway. Traffic 2:449-464. [DOI] [PubMed] [Google Scholar]
- 71.Yin, H. S., J. H. Shien, and L. H. Lee. 2000. Synthesis in Escherichia coli of avian reovirus core protein σA and its dsRNA-binding activity. Virology 266:33-41. [DOI] [PubMed] [Google Scholar]
- 72.Yokoya, F., M. Imamoto, T. Tachibana, and Y. Yoneda. 1999. β-Catenin can be transported into the nucleus in a Ran-unassisted manner. Mol. Biol. Cell 10:1119-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yoneda, Y., N. Imamoto-Sonobe, M. Yamaizumi, and T. Uchida. 1987. Reversible inhibition of protein import into the nucleus by wheat germ agglutinin injected into cultured cells. Exp. Cell Res. 173:586-595. [DOI] [PubMed] [Google Scholar]
- 74.Zhang, X., J. Tang, S. B. Walker, D. O'Hara, M. L. Nibert, R. Duncan, and T. S. Baker. 2005. Structure of avian orthoreovirus virion by electron cryomicroscopy and image reconstruction. Virology 343:25-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, X., M. Yamada, N. Mabuchi, and H. Shida. 2003. Cellular requirements for CRM1 import and export. J. Biochem. 134:759-764. [DOI] [PubMed] [Google Scholar]
- 76.Zheng, R., Y. Aoki, M. Yoshida, K. Arai, and S. Watanabe. 2002. Stat5b shuttles between cytoplasm and nucleus in a cytokine-dependent and -independent manner. J. Immunol. 168:4567-4575. [DOI] [PubMed] [Google Scholar]
- 77.Zhong, H., A. Takeda, R. Nazari, H. Shio, G. Blobel, and N. R. Yaseen. 2005. Carrier-independent nuclear import of the transcription factor PU.1 via RanGTP-stimulated binding to Nup153. J. Biol. Chem. 280:10675-10682. [DOI] [PubMed] [Google Scholar]