Abstract
The family Arenaviridae includes a number of highly pathogenic viruses that are responsible for acute hemorrhagic fevers in humans. Genetic diversity among arenavirus species in their respective rodent hosts supports the continued emergence of new pathogens. In the absence of available vaccines or therapeutic agents, the hemorrhagic fever arenaviruses remain a serious public health and biodefense concern. Arenaviruses are enveloped virions that assemble and bud from the plasma membrane. In this study, we have characterized the microdomain organization of the virus envelope glycoprotein (GPC) on the cell surface by using immunogold electron microscopy. We find that Junín virus (JUNV) GPC clusters into discrete microdomains of 120 to 160 nm in diameter and that this property of GPC is independent of its myristoylation and of coexpression with the virus matrix protein Z. In cells infected with the Candid#1 strain of JUNV, and in purified Candid#1 virions, these GPC microdomains are soluble in cold Triton X-100 detergent and are thus distinct from conventional lipid rafts, which are utilized by numerous other viruses for assembly. Virion morphogenesis ultimately requires colocalization of viral components, yet our dual-label immunogold staining studies failed to reveal a spatial association of Z with GPC microdomains. This observation may reflect either rapid Z-dependent budding of virus-like particles upon coassociation or a requirement for additional viral components in the assembly process. Together, these results provide new insight into the molecular basis for arenavirus morphogenesis.
The Arenaviridae are a diverse group of rodent-borne viruses, some of which are responsible for severe acute hemorrhagic fevers in humans. Lassa fever virus (LASV) infects as many as 300,000 persons annually in western Africa (45), and at least four virus species, including Junín virus (JUNV), are recognized as causing fatal disease in the Americas. The genetic diversity of arenaviruses in their respective rodent hosts provides a vast reservoir for the continual emergence of new viruses (1, 18). At present, there are no licensed vaccines to prevent arenavirus infection and no effective therapies. Therefore, the hemorrhagic fever arenaviruses remain an urgent public health concern.
Arenaviruses are enveloped viruses, with a bisegmented RNA genome that encodes the ambisense expression of four proteins. The nucleoprotein (N) and the multidomain RNA-dependent RNA polymerase (L) make up the ribonucleoprotein core of the virion and together are sufficient for the replication and transcription of the RNA genome (30, 35, 40). The small matrix protein (Z) is myristoylated and associates with the inner leaflet of the plasma membrane to drive the formation and budding of virion particles (16, 51, 65). The virus envelope glycoprotein (GPC) is trafficked to the surfaces of infected cells for incorporation into budding virions and mediates the entry of the virus into its host cell. The pathogenic New World arenaviruses, such as JUNV, bind the host cell transferrin receptor 1 (TfR1) through the G1 receptor-binding subunit of GPC (1, 54), whereupon the virus is endocytosed. The Old World arenaviruses, such as LASV and lymphocytic choriomeningitis virus (LCMV), as well as several nonpathogenic New World viruses, use the cellular α-dystroglycan protein for uptake into the endosome (15, 64). Fusion of the viral and cellular membranes is activated by low pH in the maturing endosome and is promoted by the transmembrane fusion subunit of GPC (G2) (19, 71). This fusion process allows entry into the cytosol, where viral replication and transcription ensue.
The key role of GPC in virus entry underscores its potential as a target for antiviral intervention (9, 33, 34, 72). GPC is synthesized as a precursor polypeptide that undergoes two proteolytic cleavage events to form the mature envelope glycoprotein complex (13). The nascent polypeptide is directed to the endoplasmic reticulum via its signal peptide, which is then cleaved from the G1-G2 precursor by the cellular signal peptidase (22, 74). Unlike conventional signal peptides, however, the GPC signal peptide is stable and forms an essential element of the mature complex (21, 77). The 58-amino-acid stable signal peptide (SSP) is myristoylated and spans the membrane twice, with both the N and C termini in the cytosol (3). Curiously, the SSP can be expressed in trans and will associate with the G1-G2 precursor to reconstitute the functional complex (21, 77). The SSP associates with the cytoplasmic domain of G2, likely through an intersubunit zinc finger structure (73). Among its several roles, the SSP is required for the transit of the complex through the Golgi apparatus (2, 21). SSP binding masks dibasic endoplasmic reticulum retrieval motifs in the cytoplasmic domain of G2 to permit transport to the cell surface (2). In addition, the interaction between the short ectodomain loop of the SSP and the ectodomain of G2 is known to modulate pH-induced activation of GPC-mediated membrane fusion (58, 75-77).
A second proteolytic cleavage of GPC occurs in the Golgi apparatus to generate the mature G1 and G2 subunits (14). This cleavage is mediated by the cellular site-1-protease/subtilisin kexin isozyme 1 protease (S1P/SKI-1) and confers on the complex the ability to promote membrane fusion (7, 32, 37, 55). The requirement for proteolytic activation is a hallmark of the class I viral fusion proteins, which include influenza virus hemagglutinin (HA), retrovirus Env, and GPC (24, 26, 71). Cleavage of the G1-G2 precursor is not essential for transport to the plasma membrane, yet only the fully mature complex is found on virion particles (32, 37, 77). The basis for this selective incorporation into the virion is unknown. A similar segregation of mature and immature glycoprotein has been reported for human immunodeficiency virus type 1 (HIV-1) Env (20).
The processes whereby viral components are trafficked, sorted, and assembled into virion particles are multiple and complex (reference 60 and references therein). One organizing principle involves the virus's use of microdomains on the cell surface as localized platforms for assembly. Membrane microdomains are defined as regions of a membrane containing a membrane component (e.g., protein or lipid) at a density higher than the average density in the membrane as a whole. The best characterized of these microdomains are the so-called lipid rafts, cholesterol-rich membrane domains that serve as scaffolds for a number of cellular functions, including protein sorting, supramolecular assembly, and signal transduction (10, 62, 63). While the term “lipid raft” likely subsumes a variety of membrane domains, common features include their small size (10 to 200 nm) and dynamic nature, as well as their liquid-ordered structure that is stabilized by cholesterol and sphingolipids (53). The latter feature contributes to the relative resistance of lipid rafts to solubilization in cold nonionic detergents, such as Triton X-100 (TX-100) (11, 46), and thus provides an operational definition of these difficult-to-study lipid raft structures.
In many instances, enveloped viruses utilize these detergent-resistant membrane microdomains (DRMs) for assembly and budding (references 47, 50, and 60 and references therein). Influenza virus HA, for example, is intrinsically targeted to DRMs and from there recruits the matrix protein and other viral components for morphogenesis (17, 59, 78). Lipid rafts are also implicated in the life cycles of HIV-1 (49), paramyxoviruses (4, 43), and filoviruses (6). In contrast, the G glycoprotein of vesicular stomatitis virus (VSV) is excluded from DRMs (29, 70, 78) but nonetheless assembles into distinct microdomains on the cell surface (12, 67).
The membrane requirements for arenavirus assembly have not been examined in detail, but several considerations suggest that lipid rafts might be involved. The fact that the arenavirus Z and GPC proteins are both myristoylated raises the possibility that they may cotraffic to specific membrane domains for assembly. In many instances, protein acylation is critical for association with lipid rafts (39, 46), and this may be the case for these arenavirus proteins. The matrix protein Z, for example, fails to associate with the plasma membrane in the absence of myristoylation (16, 66). Furthermore, the nonmyristoylated G2A mutant of GPC is trafficked to and accumulates on the cell surface, but there it suffers a major deficiency in membrane fusion activity (58, 77). This fusion defect is similar to that reported for a mutant of influenza virus HA that fails to associate with lipid rafts (68). The reduced fusion activity of the mutant HA is attributed to its relatively sparse density in lipid rafts, on which virus particles are assembled (68).
We have investigated whether arenaviruses utilize lipid rafts for assembly. By using cold-detergent extraction and electron microscopy, we find that the mature JUNV GPC complex does not fractionate with DRMs yet assembles on the cell surface into nonraft microdomains that measure 120 to 160 nm. GPC clustering is independent of N-terminal myristoylation in the SSP. Interestingly, Z does not colocalize with GPC microdomains in cotransfected cells, suggesting that additional signals may operate in virus-infected cells to recruit viral components to GPC microdomains for assembly and budding.
MATERIALS AND METHODS
Molecular and viral reagents.
The GPC cDNA from the pathogenic JUNV strain MC2 (27) was introduced into Vero cells by Lipofectamine 2000 (Invitrogen)-mediated transfection of a pcDNA3.1-based (Invitrogen) plasmid (77). Expression was driven by the bacteriophage T7 promoter in the vector, and T7 RNA polymerase was provided by concomitant infection with the recombinant vaccinia virus vTF7-3 (25, 77). The attenuated Candid#1 strain of JUNV (5, 42) was provided by Robert Tesh (WHO Reference Center for Arboviruses) and was propagated in Vero cells. The Z protein cDNA from the Candid#1 virus (28) was kindly provided by Victor Romanowski (National University of La Plata, La Plata, Argentina) and was subsequently engineered to encode a C-terminal S peptide (Spep) tag (31, 77). The GPC and Z proteins of MC2 and Candid#1 are essentially identical, each containing two conservative amino acid substitutions among 485 and 94 residues, respectively.
The G1-directed monoclonal antibody (MAb) BE08 (57) was obtained from the NIH Biodefense and Emerging Infections Research Resources Repository. The G2-directed MAb F106G3 was generated against an Escherichia coli-expressed JUNV G2 ectodomain protein and was provided by Jody D. Berry, Ute Ströher, and Heinz Feldmann (National Microbiology Laboratory, Winnipeg, Canada). Biotinylated S protein was purchased from Novagen (Madison, WI). Streptavidin and secondary antibodies (Fab fragments) conjugated to Alexa Fluor (AF), and the Vibrant lipid raft labeling kit containing AF555-conjugated cholera toxin subunit B (CTB) and anti-CTB rabbit serum, were purchased from Molecular Probes (Eugene, OR). A goat anti-mouse immunoglobulin antibody conjugated with 12-nm-diameter colloidal gold particles was purchased from Jackson ImmunoResearch (West Grove, PA), and streptavidin conjugated with 6-nm-diameter colloidal gold particles was purchased from Electron Microscopy Sciences (Hatfield, PA).
Confocal microscopy.
Vero cells were infected with vTF7-3 and transfected with plasmids encoding JUNV GPC or Z on chambered coverglasses (Lab-Tek II) as previously described (3). The cultures were maintained in a medium containing 10 μM cytosine arabinoside to prevent vaccinia virus replication and were used for immunolabeling 6 h postinfection to minimize cytopathic effect. Cells were fixed for 1 h in cold 2% formaldehyde in phosphate-buffered saline (PBS) prior to incubation with AF555-conjugated CTB in PBS containing 5% fetal bovine serum (FBS). In many studies, live cells were stained with AF555-conjugated CTB and subsequently incubated with a rabbit antiserum against CTB in order to patch lipid rafts. Following fixation, reactive formaldehyde was quenched in the cold using 50 mM Tris HCl (pH 7.4). Further incubations with the anti-G1 specific MAb BE08 and the AF488-conjugated secondary goat anti-mouse immunoglobulin G Fab fragment were carried out in cold PBS containing 5% FBS, and ultimately in cold PBS. Slow Fade Gold (Molecular Probes) was used to limit photobleaching. In control studies, the distribution of GPC on the cell surface was similar when formaldehyde-fixed cells were permeabilized prior to incubation with MAb BE08 (see below). Thus, it is likely that the native membrane distribution of GPC is stabilized by formaldehyde fixation.
To examine cultures infected with the Candid#1 strain of JUNV, infected Vero cells were plated onto chambered coverslips 4 days postinfection and were subsequently incubated with AF555-conjugated CTB and anti-CTB antiserum, fixed in cold 2% formaldehyde, and labeled with anti-G1 MAb BE08 and the AF488-conjugated secondary Fab. Labeling was visualized using an inverted Nikon TE-300 microscope and a Bio-Rad Radiance 2000 confocal laser scanning microscopy system. Images were merged using Lasersharp software (Bio-Rad).
Membrane fractionation.
Vero cells infected with Candid#1 virus were solubilized in cold PBS lysis buffer containing 1 mM EDTA, 1 mM EGTA, and 1% TX-100 (44). Cells were incubated on ice for 1 h with occasional mixing to ensure complete segregation of soluble and insoluble membranes. Ten percent of this crude lysate was reserved and treated with Benzonase nuclease (Novagen) for subsequent analysis. The remainder was subjected to high-speed centrifugation (21,000 × g for 10 min at 4°C) in order to pellet DRMs. These were subsequently resuspended in cold PBS lysis buffer, and 10% was reserved and treated with Benzonase for 30 min on ice. The 10% aliquots of the crude lysate and DRMs, as well as 10% of the detergent-soluble cleared supernatant, were subjected to peptide-N-glycosidase F (PNGase F; New England Biolabs) digestion to improve antigenicity in the subsequent Western blot analysis. Deglycosylated polypeptides were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions (NuPAGE 4 to 12% Bis-Tris gel with morpholineethanesulfonic acid running buffer; Invitrogen) and were transferred to a nitrocellulose membrane (Invitrogen). The membrane was treated in Tris HCl-buffered saline containing 0.1% Tween 20 and 5% dry milk and was probed using the G2-directed MAb F106G3. Signal was detected with a horseradish peroxidase-conjugated anti-mouse immunoglobulin antibody (Cappel) and SuperSignal West Pico chemiluminescence (Pierce). Using these methods (44), the cellular proteins caveolin and TfR segregate appropriately into detergent-insoluble and detergent-soluble fractions, respectively.
Candid#1 virions were purified by ultracentrifugation through a 20% sucrose cushion and were solubilized in cold PBS lysis buffer (containing 1 mM EDTA, 1 mM EGTA, and 1% TX-100). Half of the crude lysate was treated with Benzonase, while the other half was subjected to high-speed centrifugation in order to isolate DRMs, which were then resuspended and treated with Benzonase. The crude lysate, DRMs, and high-speed supernatant were treated with PNGase F, and the polypeptides were resolved by SDS-PAGE and subjected to Western blot analysis using the G2-directed MAb F106G3.
Density gradient analysis of DRMs.
Vero cells infected with Candid#1 virus were solubilized in cold PBS lysis buffer, and the crude lysate was cleared by high-speed centrifugation. Aliquots of both the crude lysate and the detergent-soluble supernatant were reserved as before for subsequent analysis. The detergent-insoluble membranes were resuspended in cold buffer B (38 mM each of the potassium salts of aspartic acid, glutamic acid, and gluconic acid, 20 mM morpholinepropanesulfonic acid [MOPS; pH 7.1], 10 mM potassium bicarbonate, 0.5 mM magnesium carbonate, 1 mM EDTA, 1 mM EGTA) also containing protease, phosphatase inhibitors, and glutathione (44) and were adjusted to 1% TX-100. This suspension was treated with Benzonase for 30 min on ice, adjusted to 49% OptiPrep (Sigma) using a 60% OptiPrep stock solution, and subjected to flotation equilibrium density gradient centrifugation (44). Five hundred microliters was placed in a polyallomer ultracentrifuge tube and overlaid with a gradient of 48 to 15% OptiPrep in cold buffer B (without TX-100). Samples were centrifuged to equilibrium using an SW55 Ti rotor (100,000 × g for 18 h at 4°C), and fractions of ∼200 μl each were manually collected from the bottom of the tube. The refractive indices of the fractions were determined and used to calculate the respective densities (44). Proteins were concentrated by overnight precipitation in 10% cold trichloroacetic acid and were collected by high-speed centrifugation. These pellets were washed in cold acetone, air dried, solubilized in 50 μl of 7 M urea sample buffer (7 M urea, 125 mM Tris HCl [pH 6.95], 2% SDS, 0.1 mM dithiothreitol, 1 mM EDTA, 0.1% bromophenol blue), and heated to 55°C for 15 min. Samples were resolved by SDS-PAGE (10% Bis-Tris gel) and transferred to nitrocellulose membranes as previously described (44). GPC was detected using the G2-directed MAb F106G3. In parallel gradients, an antibody directed to caveolin 1 (Abcam Technologies, Cambridge, MA) was used to identify fractions containing caveolae and lipid rafts. An antibody specific for TfR was purchased from Chemicon (Temecula, CA).
Electron microscopy analysis.
Vero cells expressing recombinant wild-type or G2A GPC were fixed with 2% cold formaldehyde and treated on ice with 5 μg/ml of digitonin in cytoplasmic buffer (20 mM HEPES [pH 7.3], 110 mM potassium acetate, 5 mM sodium chloride, 2 mM magnesium chloride, and 5 mM EGTA) for 20 min to selectively permeabilize the plasma membrane (3). Cells were then incubated in cytoplasmic buffer containing 5% dialyzed FBS, followed by the G1-specific MAb BE08 and a secondary antibody conjugated to 12-nm-diameter gold particles. Cells were then scraped, fixed in 2.5% glutaraldehyde, and processed for electron microscopy as previously described (12). Thin sections were lightly stained in order to better visualize gold particles. Negative controls included mock-transfected cells, in which no immunoreactivity could be detected. For double labeling of Z and GPC, GPC was again visualized with anti-G1 antibody and 12-nm-diameter gold particles, while Z was detected using biotinylated S protein (Novagen), which binds an Spep affinity tag (31) appended to the C terminus of Z, and streptavidin conjugated to 6-nm-diameter gold particles. Thin sections were scanned at low magnification to identify transfected cells that had positive immunoreactivity, and arbitrary regions of plasma membrane were imaged at high magnification (×43,000). Micrographs from 25 individual cells were collected, digitized, and analyzed for the distribution of gold particles as described previously (12).
RESULTS
Confocal microscopy reveals GPC clustering on the cell surface.
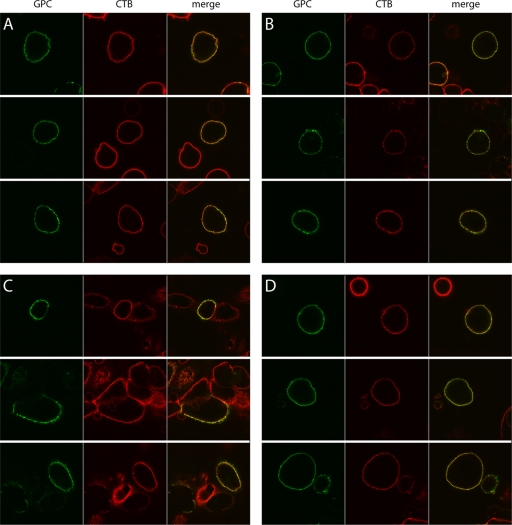
Our observation that myristoylation of the SSP is important for GPC-mediated membrane fusion led us to hypothesize that lipid rafts might play a role in arenavirus assembly (76). We thus sought to determine the distribution of GPC on the plasma membrane and its relationship to lipid raft microdomains by confocal microscopy. Vero cells expressing wild-type GPC were fixed in cold 2% formaldehyde and incubated with a G1-specific MAb (BE08 [57]) and an AF488-conjugated secondary antibody. Lipid rafts were in turn identified using AF555-conjugated CTB, which binds to the raft-associated GM1 ganglioside. At the level of resolution of confocal microscopy, GPC staining appeared as a green “string of beads” along the cell surface (Fig. 1A, left), consistent with GPC clustering into membrane microdomains. CTB staining (red) was largely continuous throughout the plasma membrane (Fig. 1A, center) due to the widespread distribution of submicroscopic lipid rafts, and thus the GPC “beads” appeared predominantly yellow in the merged images (Fig. 1A, right). Without being able to distinguish individual lipid rafts, we could not use the overlay of GPC and CTB signals to infer the localization of GPC within lipid rafts.
FIG. 1.
Confocal microscopic analysis of GPC distribution on the cell surface. Intact cells either expressing wild-type GPC (A and B) or the nonmyristoylated G2A mutant (D) or infected with Candid#1 virus (C) were stained with CTB (red) to visualize lipid rafts and with the G1-specific MAb BE08 (green). In panels B through D, bound CTB was cross-linked using a rabbit antiserum to induce patching of lipid rafts prior to fixation and incubation with MAb BE08. The cells are displayed as green (left), red (center), and merged (right) images. Three representative micrographs are shown in each panel in order to illustrate the variability in the appearance of the GPC “string of beads” and the limitations in resolving isolated GPC and CTB-labeled lipid raft regions.
In order to resolve discrete lipid rafts, we used rabbit anti-CTB serum to cross-link individual rafts in order to form larger patches (29). The cells were then fixed and stained for GPC as described above. If GPC is raft associated, it should patch with lipid rafts and appear yellow in the merged images. GPC that is not associated with these lipid rafts would appear green in the remaining membrane. While CTB cross-linking allowed us to form some discrete lipid raft patches (Fig. 1B, center), much of the cell surface remained densely labeled. In these unresolved regions of the membrane, the GPC clusters remained yellow and uninformative (Fig. 1B, right). Importantly, however, the visible patching of lipid rafts in other regions of the membrane now allowed us to detect multiple instances in which GPC clusters remained green. By aggregating rafts into distinct patches, we exposed GPC clusters that did not colocalize with CTB staining, suggesting that at least some GPC may not be associated with lipid raft microdomains.
GPC clusters on the surfaces of Candid#1-infected cells.
To determine the relevance of GPC clustering in viral infection, we examined expression in cells infected with the attenuated Candid#1 strain of JUNV (5, 42). In these experiments, live cells were first incubated in the cold with AF555-conjugated CTB and rabbit anti-CTB antiserum in order to patch lipid rafts; then they were fixed and labeled with a G1-specific antibody. Infected cells comprised ∼10% of the culture and displayed large amounts of GPC on their surfaces (Fig. 1C, left). In many cases, this prevented the detection of discrete clusters. Nonetheless, regions containing lesser amounts of GPC showed individual clusters spread across the cell surface, comparable to the “string-of-beads” appearance in cells expressing recombinant GPC. In these regions, overlap with CTB patches for the most part remained, although clear islands of green fluorescence were also visible (Fig. 1C, right). Taken together, confocal microscopy studies of the cell surface distribution of GPC in virus-infected and recombinant GPC-expressing cells indicated that GPC intrinsically forms discrete clusters on the cell surface. This tendency to cluster did not require other viral proteins or virus infection.
GPC is not associated with detergent-resistant lipid rafts in virus-infected cells.
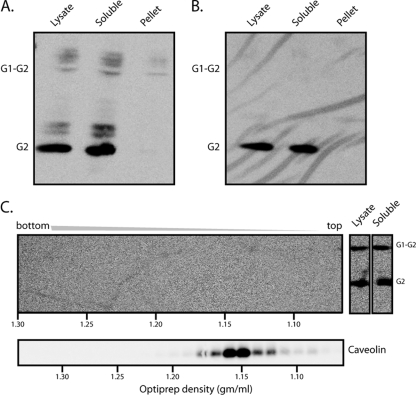
To further explore the relationship between GPC clusters and rafts, we turned to the biochemical definition of lipid rafts as DRMs (53). Specifically, we examined the partitioning of GPC from Candid#1-infected cells into detergent-soluble and detergent-insoluble membranes. Cells were disrupted in cold buffer containing 1% TX-100, and lysates were subjected to high-speed centrifugation in order to generate soluble and pelleted membrane fractions. Equivalent aliquots (10%) of each fraction were incubated with PNGase F to generate deglycosylated polypeptides (77). This treatment was found to enhance detection by the G2-specific MAb F106G3 in Western blot analysis, which was used to determine the distribution of GPC. As shown in Fig. 2A, the vast majority of the GPC in Candid#1-infected cells, and all of the mature complex, was found in the detergent-soluble lysate. Only a trace of the uncleaved G1-G2 precursor was evident in the pelleted, detergent-insoluble membrane fraction.
FIG. 2.
GPC segregates with the TX-100-soluble membrane in Candid#1-infected cells and virions. (A) Cells infected with Candid#1 were lysed in cold buffer containing 1% TX-100, and insoluble membranes were separated by high-speed centrifugation. Equal aliquots of the total lysate as well as of the high-speed supernatant (soluble) and the insoluble membrane pellet were incubated with PNGase F and subjected to Western blot analysis using the G2-specific MAb F106G3. Appropriate segregation of detergent-soluble (TfR) and detergent-insoluble (caveolin) cell markers was confirmed in other studies. (B) Candid#1 virions were purified from the cell culture medium by ultracentrifugation through a buffer containing 20% sucrose and were lysed in cold buffer containing 1% TX-100. Equal aliquots of the virion lysate and the detergent-soluble and -insoluble membrane fractions were deglycosylated and analyzed as described for panel A. The G1-G2 precursor and the mature G2 subunit are indicated. Incomplete deglycosylation products from cell lysates are also visible. (C) (Top) The detergent-insoluble fraction from cell lysates prepared as described for panel A was resuspended in TX-100-containing lysis buffer and subjected to OptiPrep density gradient ultracentrifugation in order to float detergent-resistant membranes. Fractions from the resulting gradient (left), as well as aliquots of the total-cell lysate and the high-speed supernatant containing detergent-soluble membranes (right), were analyzed as described for panel A. The positions of G2 and the G1-G2 precursor are indicated. (Bottom) Caveolin-containing lipid rafts were detected in parallel gradients by Western blot analysis. Buoyant densities calculated from the refractive indices of fractions (44) are plotted along the horizontal axes to standardize individual gradients.
This finding was corroborated by studies in which the detergent-insoluble membranes from Candid#1 virus-infected cells were resuspended in a buffer containing 1% TX-100, and equilibrium density gradient ultracentrifugation was performed to float intact membranes. In this manner, we sought to purify and concentrate detergent-insoluble membranes in order to further confirm the absence of GPC in lipid rafts, and to provide a positive control for the isolation of lipid rafts by analyzing gradient fractions for the lipid raft marker caveolin. Following ultracentrifugation, the gradients were fractionated, and each fraction was treated with PNGase F and subjected to Western blot analysis. In parallel gradients, caveolin was detected in fractions whose density corresponded to that of lipid rafts (Fig. 2C, bottom). However, no GPC was found in these or any other detergent-resistant membrane-containing fractions (Fig. 2C, top). These findings extend those from our microscopy studies and indicate that GPC does not associate appreciably with lipid rafts.
Cell membranes are dynamic, and raft structures may be transient, so we wanted to determine whether the Candid#1 virion itself contained a detergent-resistant membrane. Virions from the cell culture supernatant were first purified by ultracentrifugation through a 20% sucrose pad and then disrupted in cold buffer containing 1% TX-100. Detergent-insoluble membranes were isolated by high-speed centrifugation, and soluble and pelleted fractions were treated with PNGase F and subjected to Western blot analysis using the G2-specific MAb F106G3 (Fig. 2B). No GPC was detected in the detergent-insoluble membranes, and all the virion GPC was in the soluble-membrane fraction. In contrast to the infected cells (examples in Fig. 2A and C), the uncleaved G1-G2 precursor was excluded from virus particles (32, 37, 77).
GPC clustering does not require myristoylation.
The studies described above suggest that detergent-resistant lipid rafts are not involved in GPC localization on the cell surface. Nonetheless, confocal microscopy clearly shows a “string-of-beads” distribution, consistent with some form of clustering. We were therefore interested in determining whether this pattern might be dependent on GPC myristoylation. Cells expressing the nonmyristoylated G2A mutant were examined by confocal microscopy and showed a beaded distribution of GPC similar to that of the wild type (Fig. 1D, left). At this level of resolution, myristoylation was not required for directing GPC to distinct clusters on the cell surface. As with the wild type, cross-linking with CTB allowed the resolution of occasional G2A GPC clusters that were not associated with lipid rafts (Fig. 1D, right).
Electron microscopy demonstrates the presence of GPC in membrane microdomains.
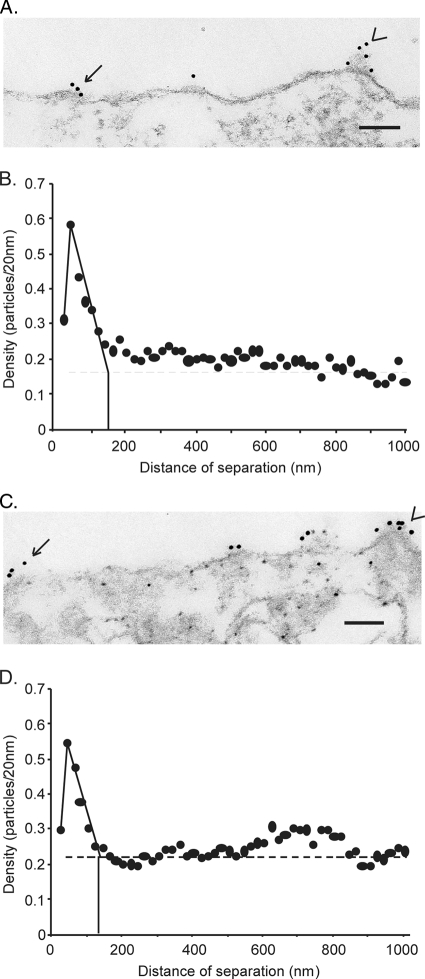
Because the size of typical membrane microdomains is at or below the limit of resolution of confocal microscopy (<200 nm), we further analyzed the distribution of GPC on the cell surface by using immunogold electron microscopy. Vero cells expressing recombinant GPC were fixed, permeabilized with 5 μg/ml of digitonin (as part of subsequent studies, described below), and incubated with the G1-specific MAb BE08 and a secondary antibody conjugated to 12-nm-diameter gold particles. Digitonin selectively solubilizes the plasma membrane, leaving intracellular membrane structures intact. The anti-G1 antibody thus binds only on the cell surface, since intracellular G1 is luminal and is not accessible (3). Cells were then scraped, fixed in 2.5% glutaraldehyde, and processed for electron microscopy as previously described (12). Plasma membrane-containing regions of 25 individual transfected cells were arbitrarily chosen for analysis. A representative micrograph of GPC labeling is shown in Fig. 3A. Labeling was often observed to occur in clusters of three to eight gold particles, supporting the notion that GPC is organized into discrete membrane microdomains. In many micrographs, the GPC clusters showed outward curvature of the membrane and a thickening of the underlying structure (Fig. 3A), features reminiscent of nascent budding domains. These images were surprising in that the virus matrix protein Z, which is sufficient for the budding of virus-like particles (51, 65), was not expressed in these cells. These structures suggest that GPC may itself contribute to membrane curvature in support of virion budding.
FIG. 3.
Analysis of wild-type and G2A GPC distribution in plasma membrane microdomains. Vero cells were transfected with plasmids encoding wild-type (A and B) or G2A (C and D) GPC and were subsequently fixed, permeabilized, and labeled with mouse anti-G1 MAb BE08 followed by a goat anti-mouse immunoglobulin G Fab fragment conjugated with 12-nm-diameter colloidal gold particles. (A and C) Representative electron micrographs. Arrows indicate clusters of 12-nm-diameter gold particles. Arrowheads indicate examples of outward curvature of the membrane containing GPC. Bars, 100 nm. (B and D) Pairwise distances between all particles in 25 micrographs were calculated from the collected measurements of distances between particles along the trace of the plasma membrane. The density was determined as the number of gold particles in each 20-nm increment, normalized to the total number of particles analyzed. The normalized density of particles per 20 nm was plotted on the y axis against the distance between particles on the x axis in a histogram. The average size of the membrane microdomains was estimated as the x intercept (vertical solid line) where the extrapolated slope intersects the average density of GPC in the plasma membrane (horizontal dashed line).
The extent of GPC clustering on the plasma membrane was quantified from digitized images by analyzing the distribution of interparticle distances (12). Briefly, distances between all pairs of gold particles were measured along the trace of the plasma membrane, collected into 20-nm increments, and plotted as a histogram with the distance of separation on the x axis and the number of particles in each 20-nm increment of distance on the y axis. The y axis values were further normalized for the number of particles analyzed in order to yield a normalized particle density. This histogram was used to determine whether interparticle distances were randomly distributed or grouped around short, raft-like dimensions (12).
Normalized particle densities for GPC are plotted in Fig. 3B and were maximal for short distances of separation (21 to 40 nm), indicating that GPC is organized in clusters. The normalized density was reduced at lower distances of separation (1 to 20 nm) because one 12-nm-diameter gold particle, which has a “halo” of bound protein, can interfere with the binding of another gold particle within this distance. This interference phenomenon implies that each gold cluster includes multiple oligomeric envelope spikes, which are known to be separated by 10 nm on the virion surface (48). As interparticle distances increased beyond 40 nm, the normalized density declined to that predicted for a random distribution of particles (Fig. 3B). By extrapolating the steep decline in particle density beyond 40 nm to the calculated random density (solid lines), we can approximate the average dimension of the GPC microdomains as 120 to 160 nm. This size is similar to the dimensions described for microdomains of the VSV G glycoprotein (100 to 150 nm) (12) and smaller than those of the influenza virus HA (325 to 500 nm) (38).
We had previously speculated that the significant fusion deficiency in the G2A mutant of GPC might result from its inability to concentrate in specific membrane microdomains and thereby to cooperate with adjacent envelope spikes for efficient membrane fusion (77). To investigate this hypothesis, we examined the distribution of the nonmyristoylated mutant on the cell surface. As shown in Fig. 3C and D, G2A GPC retained the ability to cluster on the plasma membrane. Indeed, the maximal particle density, a measure of the tendency to cluster, and the extrapolated diameter of the G2A GPC microdomains (140 to 160 nm) were remarkably similar to those of the wild type. Membrane structures consistent with nascent budding domains were also observed. Thus, myristoylation of the SSP is not required for GPC assembly into membrane microdomains; clustering appears to be an intrinsic property of the GPC protein. Furthermore, the reduced efficiency of cell-cell fusion by the G2A mutant does not appear to be related to differences in the distribution of GPC on the plasma membrane (77).
Coexpression of Z does not alter GPC clustering.
Expression of the matrix protein Z is sufficient for the assembly and budding of virus-like particles (51, 65), and coexpression with GPC and other viral components in reverse-genetic systems gives rise to virions capable of GPC-mediated entry into target cells (36; S. S. Agnihothram and J. H. Nunberg, unpublished data). Recent evidence suggests that the Z and GPC proteins interact directly in virion assembly (16, 48). We therefore wanted to investigate whether the distribution of GPC on the plasma membrane might be affected by coexpression of Z. In these studies, GPC was again visualized with an anti-G1 antibody labeled with 12-nm-diameter gold particles, and because no specific antibodies directed to Z are available, coexpression of the matrix protein was detected using an Spep affinity tag (31) appended to the C terminus of Z. This construct had been shown to support the budding of virus-like particles when coexpressed with GPC (J. York and S. S. Agnihothram, unpublished data), and analogously tagged Z proteins have been used successfully in reverse-genetic studies with LCMV (52). Cotransfected cells were fixed with formaldehyde, and the plasma membranes were selectively permeabilized using digitonin (3) in order to allow the biotinylated S protein (Novagen) to access the cytosolic Spep tag. The Z protein was detected by electron microscopy using streptavidin conjugated to 6-nm-diameter gold particles.
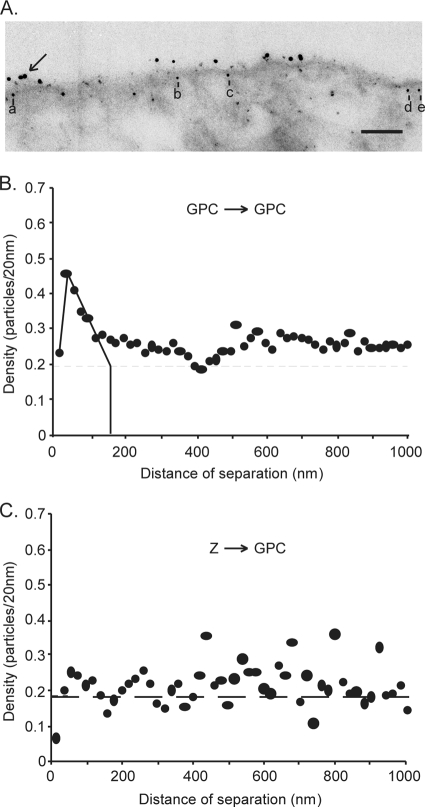
Figure 4A shows a representative electron micrograph of dual-labeled cells. GPC, labeled with 12-nm-diameter gold particles, was distributed in small clusters similar to those in the absence of Z (Fig. 3A). The Z protein, labeled with 6-nm-diameter gold particles, was distributed throughout the cytoplasm of transfected cells but appeared to be concentrated near the plasma membrane. This perimembrane localization of Z was also noted upon labeling of the same cells for confocal microscopy (Agnihothram and Nunberg, unpublished) and upon expression of the LCMV Z protein (16), but not in the case of LASV (23). Six-nanometer-diameter gold particles within 30 nm of the trace of the plasma membrane were judged to be associated with the membrane itself (12, 67). For example, the 6-nm-diameter gold particles labeled a to e in Fig. 4A are considered to associate with the plasma membrane. Electron micrographs from 25 individual cells expressing both GPC and Z were quantified in order to determine the distribution of gold particles on the plasma membrane. As shown in Fig. 4B, the overall clustering of GPC was largely unchanged by the coexpression of Z. The average dimensions of the GPC microdomains (130 to 160 nm) were identical to those in the absence of Z.
FIG. 4.
Analysis of the organization of GPC in the presence of Z. Vero cells were cotransfected with plasmids encoding GPC and Spep-tagged Z. Cells were fixed, permeabilized, and double-labeled first with the mouse anti-G1 MAb BE08 and biotinylated S protein and then with a secondary antibody conjugated with 12-nm-diameter colloidal gold particles and streptavidin conjugated with 6-nm-diameter colloidal gold particles. (A) Representative electron micrograph of cells double-labeled for GPC and Z. The arrow indicates a cluster of 12-nm-diameter gold particles, and the letters a to e indicate 6-nm-diameter gold particles within 30 nm of the trace of the plasma membrane, which were judged to be associated with the membrane itself. Bar, 100 nm. (B) Pairwise distances between all 12-nm-diameter gold particles (GPC), analyzed as described for Fig. 3. (C) Measurements were collected between each 6-nm-diameter gold particle (Z) within 30 nm of the trace of the plasma membrane and every 12-nm-diameter gold particle (GPC). The density was calculated as the number of 12-nm-diameter gold particles in 20-nm distance increments from each 6-nm-diameter gold particle and was normalized to the number of gold particles analyzed. The normalized density of 12-nm-diameter particles/20 nm was plotted on the y axis against the distance from the 6-nm-diameter particles on the x axis in a histogram. The average density of GPC labeling in the plasma membrane is indicated by the horizontal dashed line.
Z does not colocalize with GPC microdomains.
The intrinsic property of GPC to cluster into discrete microdomains raised the possibility that Z may be recruited to GPC microdomains for virion assembly. To test this hypothesis, we determined the distribution of distances from every 6-nm gold particle (Z) to every 12-nm particle (GPC) in the 25 micrographs. Colocalization would be indicated by a tendency of Z and GPC gold particles to be in close proximity, as indicated by a high normalized density of 12-nm-diameter particles within short distances of separation from 6-nm-diameter particles. This analysis, however, revealed only a random distribution of GPC relative to Z (Fig. 4C). Despite the requirement that Z and GPC ultimately coassemble to generate infectious virions, we found no evidence for colocalization in the present studies.
DISCUSSION
Virus assembly necessarily includes colocalization of virion structural components at the site of virus budding. For many enveloped viruses, specialized membrane microdomains called lipid rafts provide a physical platform for this process (references 47, 50, and 60 and references therein). These cholesterol-rich and liquid-ordered membrane structures range in size from 10 to 200 nm on the plasma membrane and are often characterized by their resistance to solubilization in cold nonionic detergents. We have used this biochemical property of lipid rafts to demonstrate that the arenavirus envelope glycoprotein GPC does not associate with DRMs in cells infected with the Candid#1 strain of JUNV. All the GPC protein in the infected cells, as well as in purified Candid#1 virions, is solubilized by extraction in cold 1% TX-100 detergent. Nonetheless, by using quantitative immunogold electron microscopy, we have shown that GPC does indeed assemble into discrete microdomains of 120 to 160 nm on the plasma membrane.
Several lines of evidence had suggested that lipid rafts play a role in arenavirus assembly. First, depletion of cellular cholesterol by methyl-β-cyclodextrin has been reported to inhibit arenavirus replication (61, 69), and because the arenavirus receptors TfR1 and α-dystroglycan (15, 54, 64) do not themselves associate with lipid rafts, we surmised that the requirement for cholesterol might arise in virus egress. Second, myristoylation is essential for both GPC and the Z matrix protein (52, 77), and the addition of a saturated fatty acid moiety in raft proteins is often required for association (39, 46). However, the myristoylated wild-type JUNV GPC is clearly not associated with lipid rafts. Indeed, we show that myristoylation is dispensable for GPC clustering into nonraft membrane microdomains. The cell surface microdomains formed by the nonmyristoylated G2A mutant of GPC are indistinguishable from those of the wild type. Therefore, the ability of the arenavirus GPC to localize to discrete, and presumably specialized, membrane microdomains is independent of myristate addition to the SSP.
We report that GPC does not utilize cholesterol-rich lipid rafts as the basis for its assembly into membrane microdomains. This finding distinguishes the arenaviruses from the many enveloped viruses that do require DRMs for virion morphogenesis, e.g., influenza virus, HIV-1, paramyxoviruses, and filoviruses (references 47, 50, and 60 and references therein). In this regard, GPC is similar to the well-studied G glycoprotein of VSV, which is likewise excluded from DRMs (29, 70, 78) and utilizes detergent-soluble membrane microdomains as a platform for assembly (12, 67). The significance of this distinction between membrane sites for virion assembly is currently unclear. Indeed, the difference between detergent-resistant and detergent-soluble membrane microdomains may reflect quantitative rather than qualitative differences in lipid composition. Although VSV serves as the prototype for viruses that utilize nonraft microdomains for assembly, its envelope is nonetheless enriched in sphingomyelin and cholesterol (41).
We and others have previously shown that the membrane fusion activity of the nonmyristoylated G2A GPC is reduced by 70 to 80% (58, 77). This defect was thought to reflect an inability of the G2A mutant to cluster into specific membrane microdomains (77). Because the successful completion of membrane fusion likely involves cooperative interactions among multiple envelope spikes, a suboptimal concentration at the site of membrane fusion might be expected to compromise fusion efficiency, as it does for influenza virus HA (68). However, this does not appear to be the explanation for the fusion deficiency in the G2A mutant of GPC. Since the interaction between the SSP and G2 is critical for membrane fusion activity (72, 75, 76), we propose that structural changes due to the absence of myristic acid, or the alanine substitution itself, may be transduced across the membrane to perturb membrane fusion. The G2A mutation might equally reduce the efficiency of fusion by affecting monomer interactions in what are likely oligomeric envelope glycoprotein spikes (48).
The intrinsic tendency of GPC to cluster into 120- to 160-nm microdomains on the plasma membrane is largely unaffected by coexpression of the Z matrix protein. Although the maximal density of GPC on the membrane, a measure of the tendency of GPC to cluster, was somewhat reduced upon expression of Z (peak density, 0.45 units versus 0.6 units without Z), the overall dimensions of the microdomains were unchanged. Consistent with the lack of effect of Z expression, we could not detect any significant spatial correlation between plasma membrane-associated Z protein and either individual or clustered GPC molecules. If Z and GPC are interacting in these experiments, as they must in viral morphogenesis, we could not detect that association in our analysis.
Because our G1-directed antibody does not distinguish between the precursor and mature forms of the protein, it is possible that a biologically relevant interaction between Z and a subset of mature GPC molecules might be missed. Immature envelope glycoproteins are thought to be segregated at the cell surface to ensure that only the functional protein is incorporated into virion particles (20, 32, 37, 77), and our analysis would be blind to this distinction. It is also possible that the interaction between Z and GPC leads to rapid budding of virus-like particles and that this transient interaction is not captured in our images. Alternatively, the association between Z and GPC during virus assembly may require additional viral information. This model has been invoked in the assembly of VSV, in which the G glycoprotein and the M matrix protein initially cluster onto separate nonraft microdomains on the cell surface (67). Coalescence of the respective microdomains at the site of VSV budding is proposed to be initiated by the viral nucleoprotein (67). In our studies, the labeling of Z was too sparse to allow us to determine whether or not the arenavirus matrix protein is itself clustered into microdomains.
It is intriguing that GPC clusters were often detected on membrane structures reminiscent of nascent budding domains (Fig. 3) despite the absence of the matrix protein, which is recognized to mediate the budding of virus-like particles (51, 65). These structures suggest that GPC may contribute to the establishment of membrane curvature at the assembly platform, either directly or through the recruitment of host cell proteins.
Our present description of arenavirus GPC assembly onto discrete membrane microdomains provides further support for the general notion that virion assembly is often anchored around specialized patches on the plasma membrane. It is not known whether these specialized regions of the membrane exist prior to virus infection or whether they are structured by the virus proteins. In many viruses, the envelope glycoprotein appears central in establishing a platform for the recruitment of virion components, and the virus matrix protein is recognized to be critical for the ultimate budding and release of the virus particle. In some viruses, the envelope glycoprotein may itself be able to promote virion budding (17). In others, membrane microdomains used for viral budding are established by the matrix protein (8, 56). Whereas the logistics of morphogenesis appear to differ among viruses, the requirement for a specialized patch of membrane to nucleate this process is common to many enveloped viruses and, if susceptible to specific pharmacologic intervention, may provide a generalized target for antiviral therapy.
ADDENDUM IN PROOF
Casabona et al. (J. C. Casabona, J. M. Levingston Macleod, M. E. Loureiro, G. A. Gomez, and N. Lopez, J. Virol. 83:7029-7039, 2009) recently reported that coexpression of the arenavirus nucleoprotein with Z substantially enhances the incorporation of GPC into virus-like particles. This would be consistent with our observation that GPC and Z are not substantially associated at the plasma membrane in the absence of nucleoprotein.
Acknowledgments
We are grateful to Jody D. Berry (National Microbiology Laboratory, Winnipeg, Manitoba, Canada) for providing the G2-specific MAb F106G3. The G1-specific MAb BE08 was obtained from the CDC through the NIH Biodefense and Emerging Infections Research Resources Program. We thank Meg Trahey (University of Montana) for editorial suggestions, and S.S.A. also thanks Sheyla Keefe for technical assistance.
This research was supported by Public Health Service grants AI074818 and AI065357 (Rocky Mountain Regional Center of Excellence for Biodefense and Emerging Infectious Diseases) to J.H.N. and Public Health Service grant AI015892 to D.S.L. The electron microscopy analysis was performed in the Cellular Imaging Core Laboratory, supported in part by a Core grant for the Comprehensive Cancer Center of Wake Forest University (CA12197) from the National Cancer Institute.
Footnotes
Published ahead of print on 22 July 2009.
REFERENCES
- 1.Abraham, J., J. A. Kwong, C. G. Albariño, J. G. Lu, S. R. Radoshitzky, J. Salazar-Bravo, M. Farzan, C. F. Spiropoulou, and H. Choe. 2009. Host-species transferrin receptor 1 orthologs are cellular receptors for nonpathogenic New World clade B arenaviruses. PLoS Pathog. 5:e1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agnihothram, S. S., J. York, and J. H. Nunberg. 2006. Role of the stable signal peptide and cytoplasmic domain of G2 in regulating intracellular transport of the Junín virus envelope glycoprotein complex. J. Virol. 80:5189-5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agnihothram, S. S., J. York, M. Trahey, and J. H. Nunberg. 2007. Bitopic membrane topology of the stable signal peptide in the tripartite Junín virus GP-C envelope glycoprotein complex. J. Virol. 81:4331-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ali, A., and D. P. Nayak. 2000. Assembly of Sendai virus: M protein interacts with F and HN proteins and with the cytoplasmic tail and transmembrane domain of F protein. Virology 276:289-303. [DOI] [PubMed] [Google Scholar]
- 5.Barrera Oro, J. G., and K. T. McKee, Jr. 1991. Toward a vaccine against Argentine hemorrhagic fever. Bull. Pan Am. Health Organ. 25:118-126. [PubMed] [Google Scholar]
- 6.Bavari, S., C. M. Bosio, E. Wiegand, G. Ruthel, A. B. Will, T. W. Geisbert, M. Hevey, C. Schmaljohn, A. Schmaljohn, and M. J. Aman. 2002. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J. Exp. Med. 195:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beyer, W. R., D. Popplau, W. Garten, D. von Laer, and O. Lenz. 2003. Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. J. Virol. 77:2866-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharya, J., A. Repik, and P. R. Clapham. 2006. Gag regulates association of human immunodeficiency virus type 1 envelope with detergent-resistant membranes. J. Virol. 80:5292-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolken, T. C., S. Laquerre, Y. Zhang, T. R. Bailey, D. C. Pevear, S. S. Kickner, L. E. Sperzel, K. F. Jones, T. K. Warren, S. A. Lund, D. L. Kirkwood-Watts, D. S. King, A. C. Shurtleff, M. C. Guttieri, Y. Deng, M. Bleam, and D. E. Hruby. 2006. Identification and characterization of potent small molecule inhibitor of hemorrhagic fever New World arenaviruses. Antivir. Res. 69:86-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, D. A., and E. London. 1998. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 14:111-136. [DOI] [PubMed] [Google Scholar]
- 11.Brown, D. A., and J. K. Rose. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68:533-544. [DOI] [PubMed] [Google Scholar]
- 12.Brown, E. L., and D. S. Lyles. 2003. A novel method for analysis of membrane microdomains: vesicular stomatitis virus glycoprotein microdomains change in size during infection, and those outside of budding sites resemble sites of virus budding. Virology 310:343-358. [DOI] [PubMed] [Google Scholar]
- 13.Buchmeier, M. J. 2002. Arenaviruses: protein structure and function. Curr. Top. Microbiol. Immunol. 262:159-173. [DOI] [PubMed] [Google Scholar]
- 14.Buchmeier, M. J., J. H. Elder, and M. B. Oldstone. 1978. Protein structure of lymphocytic choriomeningitis virus: identification of the virus structural and cell associated polypeptides. Virology 89:133-145. [DOI] [PubMed] [Google Scholar]
- 15.Cao, W., M. D. Henry, P. Borrow, H. Yamada, J. H. Elder, E. V. Ravkov, S. T. Nichol, R. W. Compans, K. P. Campbell, and M. B. A. Oldstone. 1998. Identification of α-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science 282:2079-2081. [DOI] [PubMed] [Google Scholar]
- 16.Capul, A. A., M. Perez, E. Burke, S. Kunz, M. J. Buchmeier, and J. C. de la Torre. 2007. Arenavirus Z-GP association requires Z myristoylation but not functional RING or L domains. J. Virol. 81:9451-9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, B. J., G. P. Leser, E. Morita, and R. A. Lamb. 2007. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J. Virol. 81:7111-7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delgado, S., B. R. Erickson, R. Agudo, P. J. Blair, E. Vallejo, C. G. Albariño, J. Vargas, J. A. Comer, P. E. Rollin, T. G. Ksiazek, J. G. Olson, and S. T. Nichol. 2008. Chapare virus, a newly discovered arenavirus isolated from a fatal hemorrhagic fever case in Bolivia. PLoS Pathog. 4:e1000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Simone, C., M. A. Zandonatti, and M. J. Buchmeier. 1994. Acidic pH triggers LCMV membrane fusion activity and conformational change in the glycoprotein spike. Virology 198:455-465. [DOI] [PubMed] [Google Scholar]
- 20.Dubay, J. W., S. R. Dubay, H.-J. Shin, and E. Hunter. 1995. Analysis of the cleavage site of the human immunodeficiency virus type 1 glycoprotein: requirement of precursor cleavage for glycoprotein incorporation. J. Virol. 69:4675-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eichler, R., O. Lenz, T. Strecker, M. Eickmann, H. D. Klenk, and W. Garten. 2003. Identification of Lassa virus glycoprotein signal peptide as a trans-acting maturation factor. EMBO Rep. 4:1084-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eichler, R., O. Lenz, T. Strecker, and W. Garten. 2003. Signal peptide of Lassa virus glycoprotein GP-C exhibits an unusual length. FEBS Lett. 538:203-206. [DOI] [PubMed] [Google Scholar]
- 23.Eichler, R., T. Strecker, L. Kolesnikova, J. ter Meulen, W. Weissenhorn, S. Becker, H. D. Klenk, W. Garten, and O. Lenz. 2004. Characterization of the Lassa virus matrix protein Z: electron microscopic study of virus-like particles and interaction with the nucleoprotein (NP). Virus Res. 100:249-255. [DOI] [PubMed] [Google Scholar]
- 24.Eschli, B., K. Quirin, A. Wepf, J. Weber, R. Zinkernagel, and H. Hengartner. 2006. Identification of an N-terminal trimeric coiled-coil core within arenavirus glycoprotein 2 permits assignment to class I viral fusion proteins. J. Virol. 80:5897-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallaher, W. R., C. DiSimone, and M. J. Buchmeier. 2001. The viral transmembrane superfamily: possible divergence of Arenavirus and Filovirus glycoproteins from a common RNA virus ancestor. BMC Microbiol. 1:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghiringhelli, P. D., R. V. Rivera-Pomar, M. E. Lozano, O. Grau, and V. Romanowski. 1991. Molecular organization of Junín virus S RNA: complete nucleotide sequence, relationship with other members of the Arenaviridae and unusual secondary structures. J. Gen. Virol. 72:2129-2141. [DOI] [PubMed] [Google Scholar]
- 28.Goñi, S. E., J. A. Iserte, A. M. Ambrosio, V. Romanowski, P. D. Ghiringhelli, and M. E. Lozano. 2006. Genomic features of attenuated Junín virus vaccine strain candidate. Virus Genes 32:37-41. [DOI] [PubMed] [Google Scholar]
- 29.Harder, T., P. Scheiffele, P. Verkade, and K. Simons. 1998. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J. Cell Biol. 141:929-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hass, M., U. Golnitz, S. Muller, B. Becker-Ziaja, and S. Gunther. 2004. Replicon system for Lassa virus. J. Virol. 78:13793-13803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, J.-S., and R. T. Raines. 1993. Ribonuclease S-peptide as a carrier in fusion proteins. Protein Sci. 2:348-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kunz, S., K. H. Edelmann, J.-C. de la Torre, R. Gorney, and M. B. A. Oldstone. 2003. Mechanisms for lymphocytic choriomeningitis virus glycoprotein cleavage, transport, and incorporation into virions. Virology 314:168-178. [DOI] [PubMed] [Google Scholar]
- 33.Larson, R. A., D. Dai, V. T. Hosack, Y. Tan, T. C. Bolken, D. E. Hruby, and S. M. Amberg. 2008. Identification of a broad-spectrum arenavirus entry inhibitor. J. Virol. 82:10768-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, A. M., J. M. Rojek, C. F. Spiropoulou, A. T. Gundersen, W. Jin, A. Shaginian, J. York, J. H. Nunberg, D. L. Boger, M. B. A. Oldstone, and S. Kunz. 2008. Unique small molecule entry inhibitors of hemorrhagic fever arenaviruses. J. Biol. Chem. 283:18734-18742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, K. J., I. S. Novella, M. N. Teng, M. B. Oldstone, and J. C. de La Torre. 2000. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J. Virol. 74:3470-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, K. J., M. Perez, D. D. Pinschewer, and J. C. de la Torre. 2002. Identification of the lymphocytic choriomeningitis virus (LCMV) proteins required to rescue LCMV RNA analogs into LCMV-like particles. J. Virol. 76:6393-6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lenz, O., J. ter Meulen, H.-D. Klenk, N. G. Seidah, and W. Garten. 2001. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc. Natl. Acad. Sci. USA 98:12701-12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leser, G. P., and R. A. Lamb. 2005. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology 342:215-227. [DOI] [PubMed] [Google Scholar]
- 39.Lindwasser, O. W., and M. D. Resh. 2001. Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. J. Virol. 75:7913-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.López, N., R. Jacamo, and M. T. Franze-Fernandez. 2001. Transcription and RNA replication of Tacaribe virus genome and antigenome analogs require N and L proteins: Z protein is an inhibitor of these processes. J. Virol. 75:12241-12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luan, P., L. Yang, and M. Glaser. 1995. Formation of membrane domains created during the budding of vesicular stomatitis virus. A model for selective lipid and protein sorting in biological membranes. Biochemistry 34:9874-9883. [DOI] [PubMed] [Google Scholar]
- 42.Maiztegui, J. I., K. T. McKee, Jr., J. G. Barrera Oro, L. H. Harrison, P. H. Gibbs, M. R. Feuillade, D. A. Enria, A. M. Briggiler, S. C. Levis, A. M. Ambrosio, N. A. Halsey, C. J. Peters, et al. 1998. Protective efficacy of a live attenuated vaccine against Argentine hemorrhagic fever. J. Infect. Dis. 177:277-283. [DOI] [PubMed] [Google Scholar]
- 43.Manié, S. N., S. Debreyne, S. Vincent, and D. Gerlier. 2000. Measles virus structural components are enriched into lipid raft microdomains: a potential cellular location for virus assembly. J. Virol. 74:305-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCaffrey, G., J. Welker, J. Scott, L. van der Salm, and M. L. Grimes. 5 March 2009. High-resolution fractionation of signaling endosomes containing different receptors. Traffic. [Epub ahead of print.] doi: 10.1111/j.1600-0854.2009.00909.x. [DOI] [PMC free article] [PubMed]
- 45.McCormick, J. B., P. A. Webb, J. W. Krebs, K. M. Johnson, and E. S. Smith. 1987. A prospective study of the epidemiology and ecology of Lassa fever. J. Infect. Dis. 155:437-444. [DOI] [PubMed] [Google Scholar]
- 46.Melkonian, K. A., A. G. Ostermeyer, J. Z. Chen, M. G. Roth, and D. A. Brown. 1999. Role of lipid modifications in targeting proteins to detergent-resistant membrane rafts. Many raft proteins are acylated, while few are prenylated. J. Biol. Chem. 274:3910-3917. [DOI] [PubMed] [Google Scholar]
- 47.Nayak, D. P., E. K. Hui, and S. Barman. 2004. Assembly and budding of influenza virus. Virus Res. 106:147-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neuman, B. W., B. D. Adair, J. W. Burns, R. A. Milligan, M. J. Buchmeier, and M. Yeager. 2005. Complementarity in the supramolecular design of arenaviruses and retroviruses revealed by electron cryomicroscopy and image analysis. J. Virol. 79:3822-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen, D. H., and J. E. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ono, A., and E. O. Freed. 2005. Role of lipid rafts in virus replication. Adv. Virus Res. 64:311-358. [DOI] [PubMed] [Google Scholar]
- 51.Perez, M., R. C. Craven, and J. C. de la Torre. 2003. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc. Natl. Acad. Sci. USA 100:12978-12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perez, M., D. L. Greenwald, and J. C. de la Torre. 2004. Myristoylation of the RING finger Z protein is essential for arenavirus budding. J. Virol. 78:11443-11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pike, L. J. 2006. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res. 47:1597-1598. [DOI] [PubMed] [Google Scholar]
- 54.Radoshitzky, S. R., J. Abraham, C. F. Spiropoulou, J. H. Kuhn, D. Nguyen, W. Li, J. Nagel, P. J. Schmidt, J. H. Nunberg, N. C. Andrews, M. Farzan, and H. Choe. 2007. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 446:92-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rojek, J. M., A. M. Lee, N. Nguyen, C. F. Spiropoulou, and S. Kunz. 2008. Site 1 protease is required for proteolytic processing of the glycoproteins of the South American hemorrhagic fever viruses Junín, Machupo, and Guanarito. J. Virol. 82:6045-6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saad, J. S., J. Miller, J. Tai, A. Kim, R. H. Ghanam, and M. F. Summers. 2006. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. USA 103:11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanchez, A., D. Y. Pifat, R. H. Kenyon, C. J. Peters, J. B. McCormick, and M. P. Kiley. 1989. Junín virus monoclonal antibodies: characterization and cross-reactivity with other arenaviruses. J. Gen. Virol. 70:1125-1132. [DOI] [PubMed] [Google Scholar]
- 58.Saunders, A. A., J. P. Ting, J. Meisner, B. W. Neuman, M. Perez, J. C. de la Torre, and M. J. Buchmeier. 2007. Mapping the landscape of the lymphocytic choriomeningitis virus stable signal peptide reveals novel functional domains. J. Virol. 81:5649-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scheiffele, P., A. Rietveld, T. Wilk, and K. Simons. 1999. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 274:2038-2044. [DOI] [PubMed] [Google Scholar]
- 60.Schmitt, A. P., and R. A. Lamb. 2004. Escaping from the cell: assembly and budding of negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283:145-196. [DOI] [PubMed] [Google Scholar]
- 61.Shah, W. A., H. Peng, and S. Carbonetto. 2006. Role of non-raft cholesterol in lymphocytic choriomeningitis virus infection via α-dystroglycan. J. Gen. Virol. 87:673-678. [DOI] [PubMed] [Google Scholar]
- 62.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature 387:569-572. [DOI] [PubMed] [Google Scholar]
- 63.Simons, K., and D. Toomre. 2000. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1:31-39. [DOI] [PubMed] [Google Scholar]
- 64.Spiropoulou, C. F., S. Kunz, P. E. Rollin, K. P. Campbell, and M. B. A. Oldstone. 2002. New World arenavirus clade C, but not clade A and B viruses, utilizes α-dystroglycan as its major receptor. J. Virol. 76:5140-5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strecker, T., R. Eichler, J. ter Meulen, W. Weissenhorn, H. D. Klenk, W. Garten, and O. Lenz. 2003. Lassa virus Z protein is a matrix protein and sufficient for the release of virus-like particles. J. Virol. 77:10700-10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strecker, T., A. Maisa, S. Daffis, R. Eichler, O. Lenz, and W. Garten. 2006. The role of myristoylation in the membrane association of the Lassa virus matrix protein Z. Virol. J. 3:e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Swinteck, B. D., and D. S. Lyles. 2008. Plasma membrane microdomains containing vesicular stomatitis virus M protein are separate from microdomains containing G protein and nucleocapsids. J. Virol. 82:5536-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takeda, M., G. P. Leser, C. J. Russell, and R. A. Lamb. 2003. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl. Acad. Sci. USA 100:14610-14617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vela, E. M., L. Zhang, T. M. Colpitts, R. A. Davey, and J. F. Aronson. 2007. Arenavirus entry occurs through a cholesterol-dependent, non-caveolar, clathrin-mediated endocytic mechanism. Virology 369:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vincent, S., D. Gerlier, and S. N. Manié. 2000. Measles virus assembly within membrane rafts. J. Virol. 74:9911-9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.York, J., S. S. Agnihothram, V. Romanowski, and J. H. Nunberg. 2005. Genetic analysis of heptad-repeat regions in the G2 fusion subunit of the Junín arenavirus envelope glycoprotein. Virology 343:267-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.York, J., D. Dai, S. A. Amberg, and J. H. Nunberg. 2008. pH-induced activation of arenavirus membrane fusion is antagonized by small-molecule inhibitors. J. Virol. 82:10932-10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.York, J., and J. H. Nunberg. 2007. A novel zinc-binding domain is essential for formation of the functional Junín virus envelope glycoprotein complex. J. Virol. 81:13385-13391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.York, J., and J. H. Nunberg. 2007. Distinct requirements for signal peptidase processing and function of the stable signal peptide (SSP) subunit in the Junín virus envelope glycoprotein. Virology 359:72-81. [DOI] [PubMed] [Google Scholar]
- 75.York, J., and J. H. Nunberg. 2009. Intersubunit interactions modulate pH-induced activation of membrane fusion by the Junín virus envelope glycoprotein GPC. J. Virol. 83:4121-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.York, J., and J. H. Nunberg. 2006. Role of the stable signal peptide of the Junín arenavirus envelope glycoprotein in pH-dependent membrane fusion. J. Virol. 80:7775-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.York, J., V. Romanowski, M. Lu, and J. H. Nunberg. 2004. The signal peptide of the Junín arenavirus envelope glycoprotein is myristoylated and forms an essential subunit of the mature G1-G2 complex. J. Virol. 78:10783-10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang, J., A. Pekosz, and R. A. Lamb. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 74:4634-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]