Abstract
The interferon-induced protein kinase RNA activated (PKR) is activated after virus infection. This activation is transient during the human immunodeficiency virus type 1 (HIV-1) infection of lymphocytes, and the protein is not activated at the peak of infection. We observed that interferon-induced adenosine deaminase acting on RNA 1-p150 (ADAR1-p150) and ADAR1-p110 expression increases while the virus replicates actively. Furthermore, both forms of ADAR1 show enhanced interactions with PKR at the peak of HIV infection, suggesting a role for this protein in the regulation of PKR activation. We observed that ADAR1-p150, as previously shown for the TAR RNA binding protein (TRBP), reverses the PKR inhibition of HIV expression and production in HEK 293T cells. This activity requires the Z-DNA binding motif and the three double-stranded RNA binding domains but not the catalytic domain. In astrocytic cells, ADAR1-p150 increased HIV expression and production to an extent similar to that of TRBP. Small interfering RNAs against ADAR1-p150 moderately decreased HIV production. These results indicate that two interferon-induced proteins, ADAR1 and PKR, have antagonistic functions on HIV production. They suggest that ADAR1 and TRBP belong to a multiprotein complex that inhibits PKR during the HIV infection of lymphocytes.
The treatment of human cells by interferon (IFN) induces the expression of hundreds of IFN-stimulated genes (ISGs), some of which have antiviral activity. These genes include the 2′-5′-oligoadenylate synthetase, adenosine deaminase acting on RNA 1 (ADAR1), Mx GTPases, major histocompatibility complex classes I and II, protein kinase RNA activated (PKR), and many others (47). Among the ISGs, PKR is a key serine/threonine kinase that has antiviral and antigrowth activities (14, 32). PKR is activated by dimerization after binding to low levels of double-stranded RNA (dsRNA) through its two dsRNA binding domains (dsRBDs) (46). Once active, PKR phosphorylates a few substrates, among which the best characterized is the alpha subunit of the translation eukaryotic initiation factor 2 (eIF2α), which negatively alters the efficiency and rate of translational initiation.
PKR activation is a critical component of antiviral and cell growth pathways (19), and its importance is illustrated by numerous cellular and antiviral mechanisms aiming to counteract its response. Viral mechanisms include the expression of competitive inhibitory RNAs or viral proteins that act either by the direct inhibition of PKR, by the sequestration of dsRNA, as competitive substrates, or as translational rescuers by dephosphorylating eIF2α (19, 20). Cells also control PKR activation to limit the translational repression induced by the protein and to control cell growth. For example, the ribosomal L18, TAR RNA binding protein (TRBP), and p58IPK sequester dsRNA or prevent PKR phosphorylation (20). Inhibition by protein-protein interactions also occurs with TRBP, tRNA-dihydrouridine synthase A, and ADAR1, which bind PKR through their dsRBDs (16, 34, 35). In contrast, dsRNA, heparin, and cellular proteins MDA7, PKR activator (PACT), and E2F-1 activate PKR (26, 37-40, 49). Viruses also have adapted to the cell in which they replicate by using cellular factors to regulate PKR activation. For example, influenza virus activates p58IPK (31), herpes virus US11 inhibits PACT (44), human immunodeficiency virus (HIV) TAR RNA recruits TRBP in the proximity of PKR (13, 16, 36), and vesicular stomatitis virus (VSV) uses ADAR1 to inhibit PKR (35).
ADARs are RNA-editing enzymes that modify nuclear and viral RNAs by deamination, which convert adenosines to inosines (6). Full-length ADAR1 enzymes possess two N-terminal Z-DNA binding domains (Z-DBD), three central dsRBDs, and a C-terminal deaminase domain. Three immunologically related isoforms of ADAR1 are found in human cells: the IFN-inducible cytoplasmic 150-kDa protein and constitutively expressed 110- and 80-kDa proteins, which lack the first Z-DBD or both Z-DBDs plus the first dsRBD, respectively (50). The 150-kDa form of ADAR1 was recently shown to bind to and inhibit PKR and to increase susceptibility to VSV infection (35). Whether ADAR1 plays a role as a PKR inhibitor in other viral infections has not been explored.
HIV expression is controlled at the transcriptional, posttranscriptional, and translational levels (3, 21, 29). HIV-infected cells treated with IFN show a decreased production of HIV proteins and a reduced HIV production mainly ascribed to PKR activation (8). The HIV-1 Tat protein was shown to inhibit PKR activity by acting as a competitive substrate (30). Astrocytic cells represent an example of naturally HIV-resistant cells with high PKR activation. In these cells, TRBP is expressed in very small amounts and cannot counteract PKR activation induced by the virus (4, 5, 36). Therefore, PKR activation can become a barrier to HIV replication, but the status of PKR phosphorylation has not been studied during the viral infection of lymphocytes.
In this paper, we show that PKR is only transiently activated during the HIV infection of lymphocytic cells. The analysis of cellular factors that interact with PKR during HIV infection shows that ADAR1 plays an important role in the inhibition of the kinase function during active replication.
MATERIALS AND METHODS
Plasmid constructions and siRNA synthesis.
pCMV-ADAR1 plasmid, containing ADAR1 cDNA (residues 1 to 4058), GenBank accession no. NM_001111.3, was obtained from K. Nishikura (12). This plasmid was used as a template to generate a cloning intermediate plasmid, ADAR1-p150 (residues 1 to 3678), with an XhoI cleavage site added to the 3′ site to facilitate cloning. The ADAR1-p150 fragment was cleaved with HindIII and XhoI and subcloned into the pcDNA3.1_V5 vector (Invitrogen). This construct was used to generate the variants ADAR-p110 (residues 888 to 3678), ADAR p80 (1869 to 3678), ADAR p70 (residues 1 to 1869), and ADAR Dcat (residues 1 to 2475). All constructs were verified by sequencing. pGL2-LTR-Luc, pcDNA3-TRBP2, and pcDNA1-PKR were previously described (16, 18, 33). The small interfering RNAs (siRNAs) used in this study were NS (13), siA (35), and si4 (Qiagen SI00292320). All were synthesized by Qiagen.
Cells and transfections.
Astrocytoma cell line U251MG (5) and HEK 293T (ATCC CRL-11268) cells were maintained at 37°C in 5% CO2 in Dulbecco's modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (HyClone), 2 mM l-glutamine, and 1% penicillin-streptomycin (Invitrogen). HEK 293T cells express adenovirus sequences and simian virus 40 large T antigen (41). Jurkat T cells (ATCC TIB-152) and Jurkat-CCR5 cells (2), obtained from K. Peden, were maintained in RPMI 1640 (Invitrogen) supplemented similarly and with 0.4 mg/ml G418 (Multicell) for Jurkat-CCR5. For the transfection of HEK 293T cells and astrocytes, cells were plated at 50% confluence 24 h prior to transfection using TransIT-LT1 transfection reagent by following the manufacturer's protocol (Mirus). The transfection of HEK 293T cells with siRNAs was performed in 6-well plates using Lipofectamine 2000 (Invitrogen) as previously described (13) 24 h prior to transfection with pNL4-3 using TransIT-LT1 (Mirus). Cells were lysed at 48 h posttransfection for immunoblotting or luciferase analysis.
Transfection of HIV clones and RT assay.
For the transfection of HIV provirus, HEK 293T cells were transfected as described above with pNL4-3 or pMAL proviral DNA. Cell supernatants were collected at 48 h posttransfection and assayed by a standard reverse transcriptase (RT) assay (7), except that the reaction mixture was spotted onto a DEAE filtermat (PerkinElmer). After five washes in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and twice in 95% ethanol, the filter mat was air dried and read using a Microbeta scintillation counter (PerkinElmer). These supernatants were used for the infection of Jurkat or Jurkat-CCR5 cells.
HIV-1 viral infection.
For each infection, 107 Jurkat or Jurkat-CCR5 cells were infected with HIV cell supernatant corresponding to 2.5 × 106 cpm, as measured by a standard RT assay in a final volume of 5 ml RPMI medium (Invitrogen), supplemented as described above, and incubated for 2 h at 37°C with mixing every 30 min. Ten milliliters of RPMI medium was then added to the cell-virus mixture, transferred to a T75 flask, and incubated overnight at 37°C. Another 15 ml of medium was added, and the cell culture was maintained at 37°C for 25 days. The cells were fed every other day (or every 3 days when their growth was not sufficient) by replacing 12 ml of supernatant with fresh medium and maintaining the cell density between 2.5 × 106 and 1 × 107 cells/ml. Supernatant and cell samples were collected at different times and assayed for RT activity, immunoblotting, and immunoprecipitation (IP).
Immunoblotting.
HEK 293T, U251MG, or Jurkat T cells extracts were prepared, separated, and transferred for immunoblotting as previously described (27). Membranes were blocked for 1 h in 5% nonfat dry milk and Tris-buffered saline-0.1% Tween 20 (TBST) or 5% bovine serum albumin (BSA) and 0.1% TBST for anti-PKR-pT451 antibody (Biosource). Membranes were incubated overnight at 4°C with the primary antibody. After five washes in TBST, membranes were incubated with horseradish peroxidase-conjugated secondary goat anti-rabbit or goat anti-mouse antibody (GE Healthcare). Anti-phosphorylated PKR (P-PKR) was used first in 3% BSA-TBST, and the membranes were washed overnight in TBST and reused to detect other proteins. The bands were visualized using ECL (GE Healthcare). Primary antibodies used for immunoblotting in 5% milk-TBST were monoclonal anti-PKR 71-10 (28) obtained from A. Hovanessian at a 1/500 dilution, anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (Santa Cruz) at a 1/2,500 dilution, anti-HIVp24 183-H12-5C (9), anti-V5 (Invitrogen), anti-actin (Chemicon) at a 1/5,000 dilution, polyclonal anti-P-PKR, anti-human ADAR1 (a kind gift from B. Bass) at a 1/1,000 dilution, and anti-TRBPjbx as previously described (15). Actin or GAPDH was probed on each separate blot. Where indicated, the bands were quantified by densitometry analysis as described previously (17).
IP.
HIV-infected and mock-infected Jurkat T cells were washed twice with ice-cold phosphate-buffered saline (PBS) and lysed in the cold lysis buffer with protease inhibitors. For each IP, 50 μl of protein G agarose fast-flow compact beads (Sigma) were washed with ice-cold PBS and left rotating at 4°C for 4 h with 8 μg anti-PKR 70-10 or 5 μg anti-human ADAR1 antibody. Cell extracts (2.5 mg) were added to the beads for overnight incubation at 4°C. The beads were washed five times with 1 ml of ice-cold PBS and resuspended in sodium dodecyl sulfate (SDS) loading dye. Bound proteins were eluted by boiling the beads for 5 min and were fractionated by SDS-10% polyacrylamide gel electrophoresis (SDS-10% PAGE). The immunoprecipitates were analyzed by Western blot analysis using appropriate antibodies.
RESULTS
PKR activation is inhibited during active HIV replication.
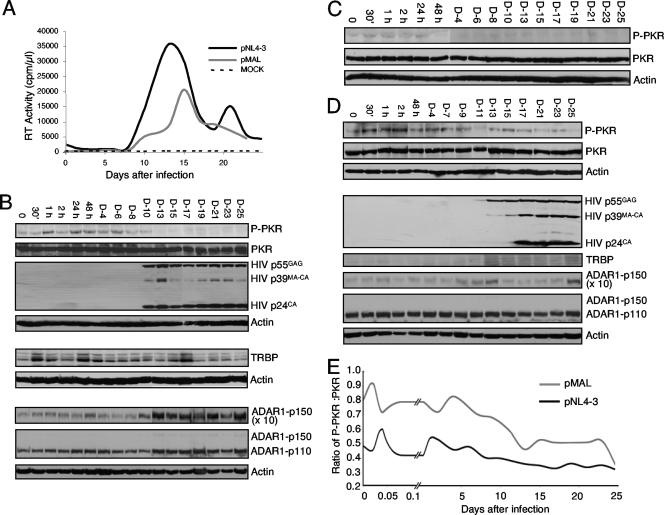
PKR becomes activated after the transfection of HIV molecular clones in nonproductively infected astrocytes but not in productively infected HeLa cells (36). To determine if PKR becomes activated in lymphocytes, we analyzed its phosphorylation during viral infection. Jurkat cells were infected by HIV pNL4-3, a strain that uses CXCR4 as a coreceptor (1, 43), and viral kinetics were monitored by RT assay on the culture medium for 25 days (Fig. 1A). Cell extracts were analyzed for PKR expression and phosphorylation (Fig. 1B). We observed that PKR was transiently phosphorylated up to day 6. This phosphorylation decreased at days 8 to 10 and no longer was observed after day 12. This PKR activation correlated with the appearance of RT activity that became visible at day 10 and showed a peak at days 11 to 15, corresponding to active viral expression and production, as shown by immunoblotting against HIV p24 antibody revealing p55GAG expression (Fig. 1B). This correlation suggests that PKR activation is inhibited during active viral replication.
FIG. 1.
PKR is transiently activated after HIV infection and is inhibited during active HIV replication. (A) HIV-1 pNL4-3 and pMAL infection kinetics. Jurkat cells were mock infected (dotted line) or were infected with HIV pNL4-3 (black line). Jurkat-CCR5 cells were infected with HIV-1 pMAL (gray line). Aliquots of cell supernatants were collected at different times and assayed for RT activity. (B) Protein expression of pNL4-3-infected Jurkat cells. For the upper four blots, 250 μg of whole-cell extracts from pNL4-3-infected Jurkat cells were subjected to SDS-10% PAGE and blotted with anti-P-PKR, anti-PKR, anti-HIV-p24, and anti-actin antibodies as indicated. For the middle two blots, 250-μg aliquots of the same extracts were subjected to a similar SDS-PAGE and blotted with anti-TRBPjbx and anti-actin antibodies as indicated. For the lower three blots, 250-μg aliquots of the same extracts were subjected to SDS-7.5% PAGE and blotted with anti-ADAR1 and anti-actin antibodies as indicated. The exposure was 10 times longer for ADAR1-p150 than for ADAR1-p110 and ADAR1-p150 where indicated. D-4, day 4. (C) Protein expression of mock-infected Jurkat cells. Aliquots (250 μg) of whole-cell extracts from mock-infected Jurkat cells were subjected to SDS-10% PAGE and blotted as described for panel B with the indicated antibodies. (D) Protein expression of pMAL-infected Jurkat-CCR5 cells. Aliquots (250 μg) of whole-cell extracts from pMAL-infected Jurkat-CCR5 cells were subjected to SDS-10% PAGE and SDS-7.5% PAGE and blotted as described for panel B with the indicated antibodies. (E) Ratio of P-PKR/PKR during HIV infection. The band intensity was digitized using Adobe Photoshop software from the bands shown in panel B for pNL4-3 and in panel D for pMAL. The P-PKR/PKR ratio was calculated by dividing the P-PKR intensity by the total PKR intensity of each band.
Because TRBP has been demonstrated to be a strong inhibitor of PKR in the context of HIV expression (8, 13, 16, 36), we verified if an increase of its expression could explain the lack of PKR activation, but no correlation between TRBP levels and PKR activation was observed (Fig. 1B). ADAR1 is also a PKR inhibitor in HEK 293T cells and during VSV infection (35), and therefore its expression was verified on the same extracts. Surprisingly, a strong increase of both ADAR1-p110 and ADAR1-p150 forms correlated with the appearance of HIV p55GAG expression and the decrease of P-PKR (Fig. 1B). Cell extract analysis from a mock infection of Jurkat cells performed under the same conditions showed no PKR activation, indicating that besides HIV infection, no other parameter influenced PKR activation (Fig. 1C).
pMAL is an HIV strain that uses CCR5 as a coreceptor and can replicate in lymphocytes that express the appropriate coreceptor (42, 43). We infected Jurkat-CCR5 cells (2) with pMAL and monitored the viral kinetics (Fig. 1A). RT assays show an overall lower activity compared to that of pNL4-3 in this setting. Viral production peaked at day 15 and was half the level of production of pNL4-3. pMAL-infected Jurkat cell extract analysis showed that PKR was phosphorylated up to day 9 and then remained weakly activated throughout the infection (Fig. 1D). In this infection, TRBP and ADAR1-p150 were moderately increased after day 13, suggesting a contribution of both cellular proteins. The calculated P-PKR/PKR ratio measured during the infection by pNL4-3 and pMAL reflected an overall decrease of PKR activation during active HIV replication (Fig. 1E).
HIV infection of lymphocytes increases ADAR1-PKR interactions.
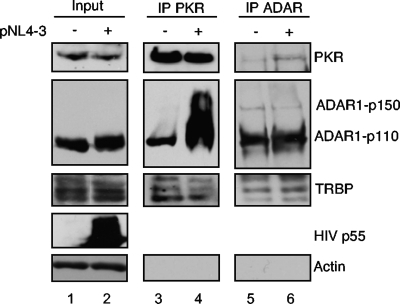
Because we observed an increase in ADAR1-p150 and ADAR1-p110 expression during the HIV infection of Jurkat cells, we evaluated whether the interaction of these proteins with PKR reflected these modifications. Cell lysates from mock-infected and HIV pNL4-3-infected Jurkat T cells at the peak of infection were immunoprecipitated with an anti-PKR antibody, and the associated proteins were analyzed using antibodies against PKR, ADAR, and TRBP (Fig. 2). Whereas TRBP-PKR interactions were not changed by viral infection, we observed a dramatic increase in the binding of both the cytoplasmic full-length and the nuclear spliced form of ADAR1 to PKR in the presence of replicating HIV. This result suggested a role of the protein in enhancing HIV replication by controlling PKR activation. Furthermore, the reverse IP with the ADAR antibody showed that ADAR1-p150, ADAR1-p110, PKR, and TRBP were immunoprecipitated and ADAR-PKR interactions were increased in the presence of HIV (Fig. 2, upper right).
FIG. 2.
ADAR1-PKR interaction increases during HIV-1 infection. Jurkat cells were mock infected or were infected with HIV pNL4-3. Cell lysates collected at day 15 (peak of infection) were immunoprecipitated with anti-PKR or anti-ADAR1. Aliquots (250 μg) of proteins from each lysate (input; lanes 1 and 2) and PKR (lanes 3 and 4)- or ADAR (lanes 5 and 6)-immunoprecipitated complexes were run on SDS-10% PAGE and blotted using anti-PKR, anti-ADAR1, anti-TRBPjbx, anti-HIV-p24, and anti-actin.
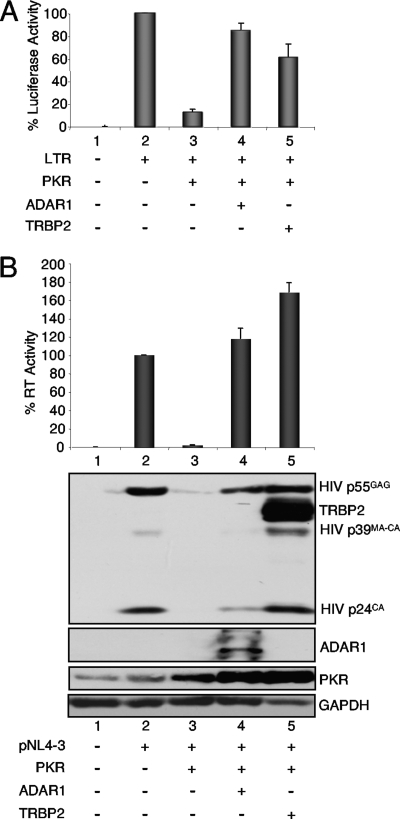
ADAR1-p150 reverses the PKR inhibition of HIV-1 LTR expression and viral production.
Because ADAR1 has been shown to inhibit PKR activation (35), and because it may have a similar role during HIV replication, we verified if the protein was able to reverse the PKR inhibition of HIV long terminal repeat (LTR) expression. A similar role previously has been attributed to TRBP (16), and we therefore compared ADAR1 and TRBP in the same assay (Fig. 3A). In this context, both ADAR1 and TRBP reversed the PKR-mediated inhibition of the HIV-1 LTR, suggesting a similar activity. ADAR1 also was compared to TRBP in the context of HIV-1 production (Fig. 3B). As previously described, the transfection of a PKR-expressing vector inhibited HIV expression in HEK 293T cells, and TRBP reversed this effect (36). In this assay, ADAR1 had the same activity as TRBP, strongly suggesting that ADAR1 is also a cellular inhibitor of PKR during HIV replication.
FIG. 3.
ADAR1-p150 and TRBP2 reverse PKR inhibition of HIV expression and virus production. (A) ADAR1-p150 and TRBP2 reverse PKR inhibition of HIV LTR expression. HEK 293T cells were transfected with 0.05 μg of pGL2-LTR-Luc (lanes 2 to 5), 0.5 μg of pcDNA1-PKR (lanes 3 to 5), and 1 μg pCMV-ADAR1-V5 p150 (lane 4) or 1 μg pcDNA3-TRBP2 (lane 5). Empty plasmids pcDNA1 and pcDNA3.1_V5 or pcDNA3 were added to reach the same amount of transfected DNA. The percent luciferase activity is the ratio between the luciferase level in the presence of PKR and that of either ADAR1 or TRBP2 versus that of LTR-luciferase alone. Shown are the averages from four independent transfections ± standard error of the mean. (B) ADAR1 and TRBP reverse PKR inhibited HIV-1 expression. HEK 293T cells were transfected with 2 μg pNL4-3 (lanes 2 to 5), 0.5 μg pcDNA1-PKR (lanes 3 to 5), and 1.5 μg of pCMV-ADAR1 (lane 4) or pcDNA3-TRBP2 (lane 5). Shown are the averages from four independent transfections ± standard error of the mean. (Top) RT assay from cell supernatants normalized to 100% in the absence of PKR or dsRBPs. (Bottom) Aliquots (250 μg) of each cell extract were analyzed by immunoblotting against HIV p24, ADAR1, TRBP, PKR, or GAPDH as indicated. TRBP was blotted before HIV p24 and appears on the same blot. GAPDH was used instead of actin, which runs close to TRBP.
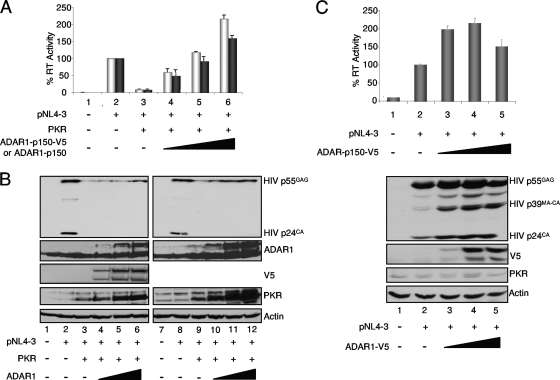
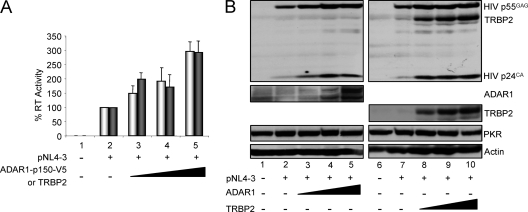
ADAR1-p150 increases HIV production in the presence and in the absence of overexpressed PKR.
To adequately monitor the expression and the activity of ADAR1-p150 and to compare it to mutant forms, we constructed a plasmid expressing a tagged protein with V5 in its C terminus. We first determined if this tagged protein had the same activity as the untagged form for PKR inhibition in the context of active HIV production. HEK 293T cells were transfected with PKR and pCMV-ADAR1-p150 or pcDNA3-ADAR1-p150-V5 (Fig. 4). Cell culture supernatants and lysates were collected and assayed for RT activity and protein expression, respectively. In the context of a 10-fold inhibition of HIV expression by PKR, the addition of either form of ADAR-p150 reversed this inhibition completely and added an additional twofold increase above the original level. This result indicates that the long form of ADAR1 is a powerful PKR inhibitor, whether it is tagged or not (Fig. 4A). The expression of HIV p55GAG protein confirmed the restoration of viral protein production with ADAR1 (Fig. 4B). Unexpectedly, although we transfected the same amount of PKR-expressing vector, PKR expression increased with ADAR1 plasmid transfection. To determine if part of the increased HIV production could be ascribed to a PKR-independent activity, the same experiment was performed with ADAR1-V5 in the absence of exogenous PKR. In this case, a maximum of a twofold increase in HIV production was observed with increasing amounts of ADAR1-p150-V5, with a slight decrease at the highest concentration (Fig. 4C). We noted that increased ADAR1 concentrations had no effect on endogenous PKR expression.
FIG. 4.
ADAR1-p150 increases HIV production in the presence and in the absence of overexpressed PKR. (A) ADAR1 and ADAR1-V5 activity on PKR-inhibited virus production. HEK 293T cells were transfected alone (lane 1) or with 2 μg pNL4-3 (lanes 2 to 6), 0.5 μg pcDNA1-PKR (lanes 3 to 6), 0.5 μg (lane 4), 1.0 μg (lane 5), or 1.5 μg (lane 6) of ADAR1-p150-V5 (light gray) or ADAR1-p150 (dark gray). Forty-eight hours posttransfection, supernatants were collected for RT assays and cell lysates were generated. Shown are the averages from four independent transfections ± standard error of the mean. (B) ADAR1 and ADAR1-V5 activity on PKR-inhibited HIV protein expression. Aliquots (250 μg) of cell extracts produced in panel A alone and with ADAR1 p150-V5 (lanes 1 to 6) or ADAR1 p150 (lanes 7 to 12) were subjected to SDS-10% PAGE and blotted with anti-HIV-p24, anti-ADAR1, anti-V5, anti-PKR, and anti-actin antibodies as indicated. (C) ADAR1-V5 activity on HIV expression and virus production. HEK 293T cells were transfected alone (lane 1) or with 2 μg pNL4-3 (lanes 2 to 5), 0.5 μg (lane 3), 1.0 μg (lane 4), or 1.5 μg (lane 5) of ADAR1-p150-V5. Forty-eight hours posttransfection, supernatants were collected for RT assay (top) and cell lysates were generated. Shown are the averages from three independent transfections ± standard error of the mean. Aliquots (250 μg) of cell extracts were subjected to SDS-10% PAGE and blotted with anti-HIV-p24, anti-ADAR1, anti-V5, and anti-actin antibodies as indicated.
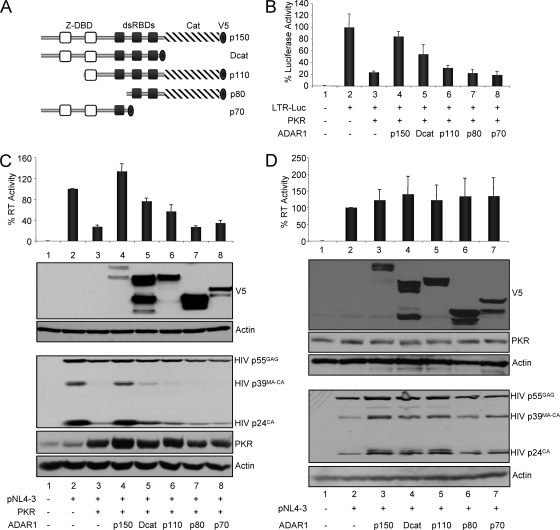
ADAR1 inhibition of PKR requires the three dsRBDs and is not due to deaminase function.
To test which part of ADAR1 is responsible for the inhibition of PKR, full-length ADAR1 (p150), the truncated variants p110 and p80 (50), a mutant deleted in the catalytic deaminase function (Dcat), and a mutant with only the Z-DBDs and dsRBD1 (p70) were tagged with the V5 epitope at their C termini and were transiently transfected into HEK 293T cells (Fig. 5A). To determine the ability of the ADAR1 variants and mutants to reverse PKR activity, first they were assayed in an HIV-1-LTR luciferase assay as described for Fig. 3A (Fig. 5B). Under conditions in which PKR inhibited the LTR activity fivefold, ADAR1-p150 restored this activity almost completely, Dcat increased it threefold relative to that of PKR only, and p110 increased it by less than twofold relative to that of PKR only; however, ADAR p80 and p70 mutants failed to restore luciferase expression. The wild type and mutants were then tested on the rescue of HIV production inhibited by PKR (Fig. 5C). In this case, HIV production was fully restored by ADAR1-p150, whereas a threefold and twofold increase relative to the level with PKR was observed with Dcat and p110, respectively. Similarly to these results, p80 and p70 had no or very mild activity in this context. These results were confirmed by immunoblotting against HIV p24, which showed a complete rescue of viral protein expression with ADAR1-p150 and a partial one with Dcat and p110. The expression of the different ADAR1 variants and mutants show that they all were expressed, but they exhibit some variations in their levels of expression. Although the full-length p150 form was weakly expressed, it had the strongest activity, suggesting that its real activity is much more potent than that seen in the luciferase and RT assays. Overall, these results suggest that the deaminase function is not required for PKR inhibition, but that the integrity of the three dsRBDs is necessary.
FIG. 5.
ADAR1 inhibition of PKR requires the dsRBDs but not the deaminase function. (A) Schematic of naturally existing variants and mutant forms of ADAR1 tagged with V5. ADAR1-p150 is the full-length protein (amino acids 1 to 3678). Mutants and variants are ADAR1 Dcat (amino acids 1 to 2475), p110 (888 to 3678), p80 (1869 to 3678), and p70 (1 to 1869). The DNA binding domain (Z-DBD), dsRBDs, the catalytic domain (Cat), and the V5 tag are indicated. (B) Activity of ADAR1 and ADAR1 mutants on PKR inhibited HIV-1 LTR expression. HEK 293T cells were transfected with 0.10 μg of pGL2-LTR-Luc (lanes 2 to 8), 0.10 μg of pcDNA1-PKR (lanes 3 to 8), and with 1 μg ADAR1-p150 (lane 4), Dcat (lane 5), p110 (lane 6), p80 (lane 7), and p70 (lane 8). Empty plasmids pcDNA1 and pcDNA3.1_V5 were added to reach the same amount of transfected DNA. The percent luciferase activity is the ratio between the luciferase level in the presence of PKR and different ADAR1 mutants versus that of LTR-Luc alone. Shown are the averages from three independent transfections ± standard error of the mean. (C) Activity of ADAR1 and ADAR1 mutants on PKR inhibited HIV-1 production. HEK 293T cells were transfected with 2.0 μg of pNL4-3 (lanes 2 to 8), 0.50 μg of pcDNA1-PKR (lanes 3 to 8), and with 1.5 μg ADAR1 p150 (lane 4), Dcat (lane 5), p110 (lane 6), p80 (lane 7), and p70 (lane 8). Empty plasmids pcDNA1 and pcDNA3.1_V5 were added to reach the same amount of transfected DNA. (Top) Percent RT activity is the ratio between the RT level in the presence of PKR and different ADAR1 variants versus that of pNL4-3 alone. Shown are the averages from five independent transfections ± standard error of the mean. (Bottom) Immunoblot of cell extracts of a representative experiment from the same transfected cells using antibodies against V5, HIV p24, PKR, and actin. (D) Activity of ADAR1 and ADAR1 mutants on HIV-1 production. HEK 293T cells were transfected with 2.0 μg of pNL4-3 (lanes 2 to 7) and with 1.5 μg ADAR1-p150 (lane 3), Dcat (lane 4), p110 (lane 5), p80 (lane 6), and p70 (lane 7). Empty plasmids pcDNA1 and pcDNA3.1_V5 were added to reach the same amount of transfected DNA. (Top) Percent RT activity is the ratio between the RT level in the presence of different ADAR1 variants versus that of pNL4-3 alone. Shown are the averages of three independent transfections ± standard error of the mean. (Bottom) Immunoblot of cell extracts of a representative experiment from the same transfected cells using antibodies against V5, PKR, HIV p24, and actin.
It was suggested recently that the overexpression of ADAR1 or ADAR2, another member of the ADAR family, increases HIV production in the absence of PKR transfection, and this activity was ascribed to the deaminase function (45). To determine if this was also the case in our assay, we verified the activity of our constructs in the absence of transfected PKR (Fig. 5D). We observed a twofold increase in HIV p55GAG expression and only a mild increase (30 to 40%) in RT activity with wild-type ADAR1 and ADAR1 mutants. This increase also occurred with the mutant deleted in the catalytic domain, suggesting that the deaminase function does not affect HIV production in our assay. As shown in Fig. 3 and 4, the transfection of the various ADAR1-expressing plasmids increased the expression of transfected PKR (Fig. 5C) but not that of endogenous PKR (Fig. 5D).
ADAR1 increases HIV-1 production in astrocytes.
Astrocytes provide a model in which HIV replication is very low, due in part to a high level of PKR activation that prevents viral translation (22, 36). This high level of PKR activation is due to the weak activity of the TRBP promoter. This induces the production of only very small amounts of TRBP, which are unable to counteract PKR activation (4, 5). To determine if ADAR1 also contributes to PKR inhibition in this cellular context, we analyzed the activity of ADAR1-p150-V5 and compared it to that of TRBP on HIV expression and production in the U251MG astrocytic cells (Fig. 6). In these cells, ADAR1 and TRBP induced increases in HIV production of up to threefold and a similar increase in p55GAG expression. This result is compatible with ADAR1 activity as an inhibitor of endogenous PKR activation.
FIG. 6.
ADAR1 and TRBP increase HIV-1 virus production in astrocytes. (A) ADAR1 and TRBP increase pNL4-3 virus production in astrocytes. U251MG cells were transfected alone (lane 1) or with 2 μg pNL4-3 (lanes 2 to 5) and 0.5 μg (lane 3), 1.0 μg (lane 4), or 1.5 μg (lane 5) of pcDNA3-ADAR1-p150-V5 (light gray) or pcDNA3-TRBP2 (dark gray). Empty corresponding plasmids were added to reach the same amount of transfected DNA. The percent RT activity is the ratio between the activity in the presence of pNL4-3 and ADAR1 or TRBP versus that of pNL4-3 alone in the cell supernatant. Shown are the averages from four independent transfections ± standard error of the mean. (B) ADAR1 and TRBP increase HIV protein expression in astrocytes. Aliquots (250 μg) of cell extracts produced for panel A without and with ADAR1-p150-V5 (lanes 1 to 5) or TRBP2 (lanes 6 to 10) were subjected to SDS-10% PAGE and blotted with anti-HIV-p24, anti-ADAR1, anti-TRBP, anti-PKR, and anti-actin antibodies as indicated.
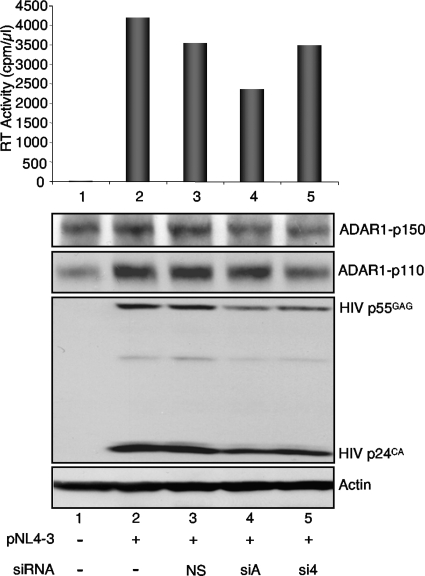
Inhibition of ADAR1-p150 expression decreases HIV expression.
A previous study showed that an siRNA against ADAR1-p150 only (siA) decreased the long form of the protein and decreased VSV production correlated with increased PKR activation (35). Another study showed that an siRNA against both ADAR1-p150 and ADAR1-p110 decreased HIV expression but was not correlated with PKR activation (45). To further determine the role of ADAR1 in HIV replication, we used siA and an siRNA targeting both forms (si4; Qiagen), and we analyzed their effect on viral expression and production (Fig. 7). The activity of siA mildly decreased ADAR1-p150, whereas si4 decreased both forms. Despite the mild activity of siA, virus expression was significantly decreased in cells, and virus production was decreased by 30%. In contrast, si4 reduced HIV gag expression moderately and HIV virus production very weakly.
FIG. 7.
Decrease of ADAR1-p150 expression affects HIV production. HEK 293T cells were not transfected (lane 1) or were transfected alone (lane 2) or with 14 nM of siNS (lane 3), siA (lane 4), or si4 (lane 5). They were transfected 24 h later with 2.0 μg of pNL4-3 (lanes 2 to 5). (Top) RT activity of the cell supernatant is represented on the graph. (Bottom) Immunoblot of 150 μg of cell extracts from the same transfected cells using antibodies against ADAR1, HIV p24, and actin.
DISCUSSION
Although IFN is able to inhibit HIV production in cell culture (8), the IFN produced in plasmocytoid dendritic cells during HIV infection does not eliminate the virus in patients and, in the long term, contributes to pathogenesis (23-25, 48). This in vivo inefficacy could be due to an inadequate innate immune response during the first days of infection, but the activity of the ISGs on virus replication in lymphocytes during this time frame has been poorly investigated. Because PKR is one of the main ISGs that can inhibit HIV production in cell culture, we wanted to determine if PKR is activated or not during the HIV infection of lymphocytes. We found that PKR becomes phosphorylated soon after HIV infection. This is followed by an inactivation of PKR, which correlates with HIV replication (Fig. 1). These results suggest that the innate immune response mediated by PKR is fully functional but is only transiently active. Because in astrocytes PKR activation is an important barrier to HIV expression and replication (36), we thought that HIV might specifically replicate in cells where PKR activation is repressed.
We studied the expression of ISGs and the role of various PKR inhibitors, and we observed that the expression of ADAR1-p150 and ADAR1-p110 forms is enhanced during HIV infection, which correlates with increased HIV replication and increased PKR binding (Fig. 1 and 2). Because ADAR1 is an ISG, one could expect that this protein contributes to a cell response against virus replication. While these results were being collected, ADAR1 was isolated in a two-hybrid screen using PKR as the bait (M. Bonnet and E. Meurs, data not shown), and ADAR1 was shown to be a PKR inhibitor in the context of VSV infection (35). Both studies showed that the first dsRBD in ADAR1 is the domain that binds PKR. Results of the two-hybrid screen showed that the five isolated clones all express amino acids 503 to 556 within dsRBD1. Similarly to TRBP, ADAR1 was able to reverse PKR inhibition in cells expressing only the HIV LTR or with active HIV replication, indicating that its increased binding to PKR at the peak of HIV infection contributes to the enhancement of HIV replication in lymphocytes (Fig. 3 and 4A). The analysis of ADAR variants' and mutants' activity further demonstrates that the integrity of the three dsRBDs and at least one Z-DNA binding domain, but not the catalytic domain, are necessary for PKR inhibition (Fig. 5).
Because PKR is not expressed from an overexpressed plasmid in HIV-replicating cells, we evaluated the activity of ADAR1 in the absence of exogenous PKR. In this context, ADAR1-p150 increased HIV expression and production, but this effect was only about twofold (Fig. 4C and 5D). A similar activity was recently observed in an independent study and showed that the deaminase function was responsible for this increased HIV production (45). However, we observed similar results with our Dcat mutant and ADAR1-p150 in the absence of PKR, which does not support the same conclusion (Fig. 5D). In addition, the activity of the Dcat in the presence of exogenous PKR (Fig. 5B and C) suggests a main activity by inactivating endogenous PKR as well. The discrepancy between this previous study and our results is currently unexplained but could be due to a difference in experimental settings. Similarly to the previous study, siRNAs that decreased both ADAR1-p110 and ADAR1-p150 decreased HIV expression, but this effect was stronger with a specific inhibition of the p150 form (siA) (Fig. 7).
The similar activities of ADAR1 and TRBP in increasing viral production in astrocytes further suggests that ADAR1 replaces TRBP in regard to PKR inhibition and the enhancement of HIV production in astrocytes (Fig. 6). The similar effects of siRNAs against ADAR1 (Fig. 7) and siRNAs against TRBP (13) confirm this hypothesis. Taken together, our results suggest that the main activity of the IFN-induced ADAR1-p150 isoform is to counteract the PKR inhibition of HIV expression. They indicate that two IFN-induced proteins can have opposite effects, which ultimately contributes to the enhancement of HIV replication in lymphocytes.
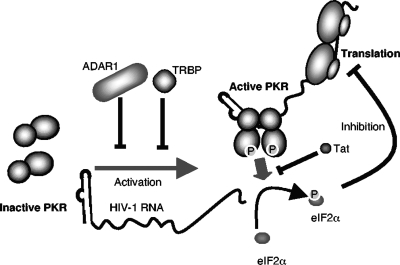
Previous results have shown that the virus itself can counteract PKR activation with its Tat protein, which acts as a competitive substrate (10, 11, 30). Taken together, previous and present results suggest that PKR inhibition during HIV replication is mediated both by the viral Tat protein and by host factors ADAR1 and TRBP (Fig. 8). We also have shown recently that in addition to direct PKR inhibition, TRBP binds to and inhibits the PKR activator PACT (15, 27). This function could further contribute to PKR inhibition during HIV replication. It is also possible that in the experiments that use HEK 293T cells, the presence of the virus-associated RNA I and simian virus 40 large T antigen also contributes to PKR inhibition (Fig. 3 to 5 and 7), but this presence did not prevent the PKR and ADAR1 activities observed here. Overall, HIV uses at least three different mechanisms to counteract PKR activation: (i) it produces the Tat protein during the early steps of its replication, (ii) it has evolved to replicate specifically in cells that express high levels of TRBP, and (iii) it increases the synthesis of the ADAR1-p150 isoform either directly or indirectly through IFN induction. Further studies of PKR binding factors during HIV infection may reveal additional proteins that contribute to PKR inactivation and enhanced virus replication.
FIG. 8.
Schematic representation of the regulation of HIV translation by PKR and the contribution of host and viral factors. The viral TAR RNA contributes to PKR activation by inducing its phosphorylation, which in turn phosphorylates eIF2α and consequently inhibits HIV translation. During HIV replication, the cellular proteins TRBP and ADAR1 prevent or inhibit PKR phosphorylation, whereas the viral protein Tat prevents eIF2α phosphorylation. All three proteins contribute to increased HIV translation.
Acknowledgments
We thank B. L. Bass' laboratory for the gift of ADAR1 antibody, K. Nishikura for the gift of the ADAR1-p150 plasmid, and K. Peden and E. Berger for the gift of Jurkat-CCR5 cells. We are grateful to Sylvanne Daniels and Robert Scarborough for helpful discussions and comments on the manuscript.
This work was supported in part by grants MOP77747 and HOP93434 from the Canadian Institutes for Health Research and by grant 019508 from the Canadian Foundation for AIDS Research (to A.G.). A.G. was a recipient of a Hugh and Helen McPherson Memorial Salary Award.
Footnotes
Published ahead of print on 15 July 2009.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 3.Bannwarth, S., and A. Gatignol. 2005. HIV-1 TAR RNA: the target of molecular interactions between the virus and its host. Curr. HIV Res. 3:61-71. [DOI] [PubMed] [Google Scholar]
- 4.Bannwarth, S., S. Lainé, A. Daher, N. Grandvaux, G. Clerzius, A. C. Leblanc, J. Hiscott, and A. Gatignol. 2006. Cell-specific regulation of TRBP1 promoter by NF-Y transcription factor in lymphocytes and astrocytes. J. Mol. Biol. 355:898-910. [DOI] [PubMed] [Google Scholar]
- 5.Bannwarth, S., L. Talakoub, F. Letourneur, M. Duarte, D. F. Purcell, J. Hiscott, and A. Gatignol. 2001. Organization of the human tarbp2 gene reveals two promoters that are repressed in an astrocytic cell line. J. Biol. Chem. 276:48803-48813. [DOI] [PubMed] [Google Scholar]
- 6.Bass, B. L. 2002. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71:817-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Battisti, P. L., A. Daher, S. Bannwarth, J. Voortman, K. W. Peden, J. Hiscott, A. J. Mouland, R. Benarous, and A. Gatignol. 2003. Additive activity between the trans-activation response RNA-binding protein, TRBP2, and cyclin T1 on HIV type 1 expression and viral production in murine cells. AIDS Res. Hum. Retrovir. 19:767-778. [DOI] [PubMed] [Google Scholar]
- 8.Benkirane, M., C. Neuveut, R. F. Chun, S. M. Smith, C. E. Samuel, A. Gatignol, and K. T. Jeang. 1997. Oncogenic potential of TAR RNA binding protein TRBP and its regulatory interaction with RNA-dependent protein kinase PKR. EMBO J. 16:611-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bounou, S., J. E. Leclerc, and M. J. Tremblay. 2002. Presence of host ICAM-1 in laboratory and clinical strains of human immunodeficiency virus type 1 increases virus infectivity and CD4+ T-cell depletion in human lymphoid tissue, a major site of replication in vivo. J. Virol. 76:1004-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brand, S. R., R. Kobayashi, and M. B. Mathews. 1997. The Tat protein of human immunodeficiency virus type 1 is a substrate and inhibitor of the interferon-induced, virally activated protein kinase, PKR. J. Biol. Chem. 272:8388-8395. [DOI] [PubMed] [Google Scholar]
- 11.Cai, R., B. Carpick, R. F. Chun, K. T. Jeang, and B. R. Williams. 2000. HIV-I TAT inhibits PKR activity by both RNA-dependent and RNA-independent mechanisms. Arch. Biochem. Biophys. 373:361-367. [DOI] [PubMed] [Google Scholar]
- 12.Chen, C. X., D. S. Cho, Q. Wang, F. Lai, K. C. Carter, and K. Nishikura. 2000. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA 6:755-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen, H. S., A. Daher, K. J. Soye, L. B. Frankel, M. R. Alexander, S. Lainé, S. Bannwarth, C. L. Ong, S. W. Chung, S. M. Campbell, D. F. Purcell, and A. Gatignol. 2007. Small interfering RNAs against the TAR RNA binding protein, TRBP, a Dicer cofactor, inhibit human immunodeficiency virus type 1 long terminal repeat expression and viral production. J. Virol. 81:5121-5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clemens, M. J., J. W. Hershey, A. C. Hovanessian, B. C. Jacobs, M. G. Katze, R. J. Kaufman, P. Lengyel, C. E. Samuel, G. C. Sen, and B. R. Williams. 1993. PKR: proposed nomenclature for the RNA-dependent protein kinase induced by interferon. J. Interferon Res. 13:241. [DOI] [PubMed] [Google Scholar]
- 15.Daher, A., G. Laraki, M. Singh, C. E. Melendez-Peña, S. Bannwarth, A. H. Peters, E. F. Meurs, R. E. Braun, R. C. Patel, and A. Gatignol. 2009. TRBP control of PACT-induced phosphorylation of PKR is reversed by stress. Mol. Cell. Biol. 29:254-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daher, A., M. Longuet, D. Dorin, F. Bois, E. Segeral, S. Bannwarth, P. L. Battisti, D. F. Purcell, R. Benarous, C. Vaquero, E. F. Meurs, and A. Gatignol. 2001. Two dimerization domains in the trans-activation response RNA-binding protein (TRBP) individually reverse the protein kinase R inhibition of HIV-1 long terminal repeat expression. J. Biol. Chem. 276:33899-33905. [DOI] [PubMed] [Google Scholar]
- 17.Daniels, S. M., C. E. Melendez-Pena, R. J. Scarborough, A. Daher, H. S. Christensen, M. El Far, D. F. Purcell, S. Laine, and A. Gatignol. 2009. Characterization of the TRBP domain required for dicer interaction and function in RNA interference. BMC Mol. Biol. 10:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duarte, M., K. Graham, A. Daher, P. L. Battisti, S. Bannwarth, E. Segeral, K. T. Jeang, and A. Gatignol. 2000. Characterization of TRBP1 and TRBP2. Stable stem-loop structure at the 5′ end of TRBP2 mRNA resembles HIV-1 TAR and is not found in its processed pseudogene. J. Biomed. Sci. 7:494-506. [DOI] [PubMed] [Google Scholar]
- 19.García, M. A., J. Gil, I. Ventoso, S. Guerra, E. Domingo, C. Rivas, and M. Esteban. 2006. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 70:1032-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García, M. A., E. F. Meurs, and M. Esteban. 2007. The dsRNA protein kinase PKR: virus and cell control. Biochimie 89:799-811. [DOI] [PubMed] [Google Scholar]
- 21.Gatignol, A. 2007. Transcription of HIV: Tat and cellular chromatin. Adv. Pharmacol. 55:137-159. [DOI] [PubMed] [Google Scholar]
- 22.Gorry, P. R., J. L. Howard, M. J. Churchill, J. L. Anderson, A. Cunningham, D. Adrian, D. A. McPhee, and D. F. Purcell. 1999. Diminished production of human immunodeficiency virus type 1 in astrocytes results from inefficient translation of gag, env, and nef mRNAs despite efficient expression of Tat and Rev. J. Virol. 73:352-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardy, A. W., D. R. Graham, G. M. Shearer, and J. P. Herbeuval. 2007. HIV turns plasmacytoid dendritic cells (pDC) into TRAIL-expressing killer pDC and down-regulates HIV coreceptors by Toll-like receptor 7-induced IFN-alpha. Proc. Natl. Acad. Sci. USA 104:17453-17458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herbeuval, J. P., and G. M. Shearer. 2007. HIV-1 immunopathogenesis: how good interferon turns bad. Clin. Immunol. 123:121-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hosmalin, A., and P. Lebon. 2006. Type I interferon production in HIV-infected patients. J. Leukoc. Biol. 80:984-993. [DOI] [PubMed] [Google Scholar]
- 26.Ito, T., M. Yang, and W. S. May. 1999. RAX, a cellular activator for double-stranded RNA-dependent protein kinase during stress signaling. J. Biol. Chem. 274:15427-15432. [DOI] [PubMed] [Google Scholar]
- 27.Laraki, G., G. Clerzius, A. Daher, C. Melendez-Pena, S. Daniels, and A. Gatignol. 2008. Interactions between the double-stranded RNA-binding proteins TRBP and PACT define the Medipal domain that mediates protein-protein interactions. RNA Biol. 5:92-103. [DOI] [PubMed] [Google Scholar]
- 28.Laurent, A. G., B. Krust, J. Galabru, J. Svab, and A. G. Hovanessian. 1985. Monoclonal antibodies to an interferon-induced Mr 68,000 protein and their use for the detection of double-stranded RNA-dependent protein kinase in human cells. Proc. Natl. Acad. Sci. USA 82:4341-4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLaren, M., K. Marsh, and A. Cochrane. 2008. Modulating HIV-1 RNA processing and utilization. Front. Biosci. 13:5693-5707. [DOI] [PubMed] [Google Scholar]
- 30.McMillan, N. A., R. F. Chun, D. P. Siderovski, J. Galabru, W. M. Toone, C. E. Samuel, T. W. Mak, A. G. Hovanessian, K. T. Jeang, and B. R. Williams. 1995. HIV-1 Tat directly interacts with the interferon-induced, double-stranded RNA-dependent kinase, PKR. Virology 213:413-424. [DOI] [PubMed] [Google Scholar]
- 31.Melville, M. W., S. L. Tan, M. Wambach, J. Song, R. I. Morimoto, and M. G. Katze. 1999. The cellular inhibitor of the PKR protein kinase, P58(IPK), is an influenza virus-activated co-chaperone that modulates heat shock protein 70 activity. J. Biol. Chem. 274:3797-3803. [DOI] [PubMed] [Google Scholar]
- 32.Meurs, E., K. Chong, J. Galabru, N. S. Thomas, I. M. Kerr, B. R. Williams, and A. G. Hovanessian. 1990. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell 62:379-390. [DOI] [PubMed] [Google Scholar]
- 33.Meurs, E. F., N. McMillan, B. R. Williams, A. G. Hovanessian, and P. J. Southern. 1995. Human PKR transfected into murine cells stimulates expression of genes under control of the HIV1 or HTLV-I LTR. Virology 214:653-659 (Erratum, 247:125, 1998.) [DOI] [PubMed] [Google Scholar]
- 34.Mittelstadt, M., A. Frump, T. Khuu, V. Fowlkes, I. Handy, C. V. Patel, and R. C. Patel. 2008. Interaction of human tRNA-dihydrouridine synthase-2 with interferon-induced protein kinase PKR. Nucleic Acids Res. 36:998-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nie, Y., G. L. Hammond, and J. H. Yang. 2007. Double-stranded RNA deaminase ADAR1 increases host susceptibility to virus infection. J. Virol. 81:917-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ong, C. L., J. C. Thorpe, P. R. Gorry, S. Bannwarth, A. Jaworowski, J. L. Howard, S. Chung, S. Campbell, H. S. Christensen, G. Clerzius, A. J. Mouland, A. Gatignol, and D. F. Purcell. 2005. Low TRBP levels support an innate human immunodeficiency virus type 1 resistance in astrocytes by enhancing the PKR antiviral response. J. Virol. 79:12763-12772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pataer, A., S. A. Vorburger, G. N. Barber, S. Chada, A. M. Mhashilkar, H. Zou-Yang, A. L. Stewart, S. Balachandran, J. A. Roth, K. K. Hunt, and S. G. Swisher. 2002. Adenoviral transfer of the melanoma differentiation-associated gene 7 (mda7) induces apoptosis of lung cancer cells via up-regulation of the double-stranded RNA-dependent protein kinase (PKR). Cancer Res. 62:2239-2243. [PubMed] [Google Scholar]
- 38.Pataer, A., S. A. Vorburger, S. Chada, S. Balachandran, G. N. Barber, J. A. Roth, K. K. Hunt, and S. G. Swisher. 2005. Melanoma differentiation-associated gene-7 protein physically associates with the double-stranded RNA-activated protein kinase PKR. Mol. Ther. 11:717-723. [DOI] [PubMed] [Google Scholar]
- 39.Patel, C. V., I. Handy, T. Goldsmith, and R. C. Patel. 2000. PACT, a stress-modulated cellular activator of interferon-induced double-stranded RNA-activated protein kinase, PKR. J. Biol. Chem. 275:37993-37998. [DOI] [PubMed] [Google Scholar]
- 40.Patel, R. C., and G. C. Sen. 1998. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 17:4379-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peden, K., M. Emerman, and L. Montagnier. 1991. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology 185:661-672. [DOI] [PubMed] [Google Scholar]
- 43.Peden, K. W., and J. M. Farber. 2000. Coreceptors for human immunodeficiency virus and simian immunodeficiency virus. Adv. Pharmacol. 48:409-478. [DOI] [PubMed] [Google Scholar]
- 44.Peters, G. A., D. Khoo, I. Mohr, and G. C. Sen. 2002. Inhibition of PACT-mediated activation of PKR by the herpes simplex virus type 1 Us11 protein. J. Virol. 76:11054-11064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phuphuakrat, A., R. Kraiwong, C. Boonarkart, D. Lauhakirti, T. H. Lee, and P. Auewarakul. 2008. Double-stranded RNA adenosine deaminases enhance expression of human immunodeficiency virus type 1 proteins. J. Virol. 82:10864-10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sadler, A. J., and B. R. Williams. 2007. Structure and function of the protein kinase R. Curr. Top. Microbiol. Immunol. 316:253-292. [DOI] [PubMed] [Google Scholar]
- 47.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt, B., B. M. Ashlock, H. Foster, S. H. Fujimura, and J. A. Levy. 2005. HIV-infected cells are major inducers of plasmacytoid dendritic cell interferon production, maturation, and migration. Virology 343:256-266. [DOI] [PubMed] [Google Scholar]
- 49.Vorburger, S. A., A. Pataer, K. Yoshida, G. N. Barber, W. Xia, P. Chiao, L. M. Ellis, M. C. Hung, S. G. Swisher, and K. K. Hunt. 2002. Role for the double-stranded RNA activated protein kinase PKR in E2F-1-induced apoptosis. Oncogene 21:6278-6288. [DOI] [PubMed] [Google Scholar]
- 50.Yang, J. H., Y. Nie, Q. Zhao, Y. Su, M. Pypaert, H. Su, and R. Rabinovici. 2003. Intracellular localization of differentially regulated RNA-specific adenosine deaminase isoforms in inflammation. J. Biol. Chem. 278:45833-45842. [DOI] [PubMed] [Google Scholar]