Abstract
Toll-like receptor-3 (TLR3) senses double-stranded RNA, initiating signaling that activates NF-κB and interferon regulatory factor 3 (IRF-3), thereby inducing the synthesis of proinflammatory cytokines, type I interferons, and numerous interferon-stimulated genes (ISGs). This pathway has not been extensively investigated in human hepatocytes, and its role in sensing and protecting against hepatitis virus infections is uncertain. We show here that primary human hepatocytes express TLR3 and robustly upregulate ISGs upon poly(I·C) stimulation. We also show that TLR3 senses hepatitis C virus (HCV) infection when expressed in permissive hepatoma cells, acting independently of retinoic acid-inducible gene I and inducing IRF-3 activation and the synthesis of ISGs that restrict virus replication. In turn, HCV infection reduces the abundance of TRIF, an essential TLR3 adaptor, and impairs poly(I·C)-induced signaling. The induction and disruption of TLR3 signaling by HCV may be important factors in determining the outcome of infection and the ability of HCV to establish persistent infections.
Hepatitis C virus (HCV) infects many millions of people worldwide. Classified within the family Flaviviridae, HCV is a small, enveloped RNA virus that possesses a positive-sense, single-stranded RNA genome (25). The virus has a tightly restricted host range, confined to humans and chimpanzees, and is highly hepatotropic, with replication occurring predominantly, if not exclusively, in hepatocytes. Remarkably, HCV establishes lifelong, persistent infections in the majority of infected individuals. Because persistent infections are associated with progressive liver fibrosis and hepatocellular carcinoma, the virus poses a major threat to human health. Despite recent advances in the development of specific antiviral therapies, current interferon (IFN)-based treatment regimens eliminate the virus in only 50% of patients (37). Importantly, the mechanism(s) underlying the unique capacity of HCV to establish persistent infections and resist IFN-based therapy remains poorly understood.
Early events in the recognition of HCV by the infected host may be decisive in determining the outcome of infection. Cells respond to virus infections by rapidly inducing IFNs upon recognition of viral pathogen-associated molecular patterns. At least two general classes of pattern recognition receptors (PRRs), membrane-bound Toll-like receptors (TLRs) and cytosolic retinoic acid-inducible gene I (RIG-I)-like helicases, sense viral RNAs (22). Two specific PRRs, TLR3 and RIG-I, sense double-stranded RNA (dsRNA), an essential intermediate in the HCV replication cycle, and thus, they may be important in the pathogenesis of hepatitis C. TLR3 is expressed on endosomal membranes (and the plasma membranes of some cells) and thus senses dsRNA that is present in endosomal and/or extracellular compartments (34), while RIG-I senses cytosolic dsRNA. When engaged by their ligands, these PRRs trigger signaling pathways converging on a common set of IκB kinase-related kinases that activate the transcription factors IRF-3 (interferon regulatory factor 3) and/or IRF-7 and NF-κB (22). This leads subsequently to the synthesis of type I IFNs (IFN-α/β), numerous IFN-stimulated genes (ISGs), and proinflammatory cytokines. While an essential role in host protection against virus infection has been established for the RIG-I pathway, the role played by TLR3 in antiviral immunity is less certain and may differ among various families of viruses and in different infection settings (50).
Efforts to understand how HCV interacts with these innate immune sensors have been hampered by the narrow host range of HCV and the corresponding lack of small-animal models. Nonetheless, several lines of evidence suggest that HCV is capable of evading innate host responses mediated through both the RIG-I and TLR3 signaling pathways. In vitro studies indicate that the HCV serine protease, NS3/4A, is capable of cleaving cellular adaptor proteins that are essential to signal transduction through these pathways: mitochondrial antiviral signaling protein (MAVS, also known as IPS-1, VISA, or Cardif) and Toll/interleukin-1 receptor (TIR) domain-containing adaptor inducing IFN-β (TRIF, also known as TICAM-1) (27, 28, 35). The evidence that NS3/4A is capable of disrupting RIG-I signaling is especially compelling. RIG-I senses genomic HCV RNA transfected into mouse liver by hydrodynamic injection (40) and initiates signaling that activates IRF-3 and induces a cellular response that limits HCV infection in cultured cells (47). MAVS cleavage has also been observed in hepatoma cells infected with HCV, as well as in liver biopsy specimens from patients with chronic hepatitis C (33). In aggregate, these data suggest that RIG-I plays an important role in the recognition and control of HCV infection.
The role played by TLR3 in HCV infection is much less certain. TRIF shares amino acid homology (Ser-Thr-Pro-Cys-Ser) with the HCV polyprotein at the NS4B/5A site of cleavage by NS3/4A and is cleaved by NS3/4A in cell-free reactions (12, 27). Ectopically expressed TRIF is degraded in HEK293 and osteosarcoma cells expressing the protease (27). Consistent with this, poly(I·C)-induced activation of IRF-3 is inhibited in HeLa cells containing HCV RNA replicons, and endogenous TRIF abundance is reduced in these cells (27). However, others have been unable to demonstrate cleavage of ectopically expressed TRIF by NS3/4A (9). In addition, few studies have investigated whether TLR3 is expressed in an active form in human hepatocytes, the cell type HCV targets for infection. Although some hepatocyte-derived cell lines demonstrate active TLR3 signaling (26), Huh7 hepatoma cells, which are almost unique in their ability to support HCV infection in vitro, are deficient in TLR3 signaling due to a lack of TLR3 expression (26). The absence of an HCV-permissive cell line containing a functional TLR3/TRIF-dependent pathway has made it difficult to determine whether HCV infection is sensed by TLR3, and whether TLR3-mediated responses, like RIG-I-induced responses, may act to restrict HCV replication.
Here we demonstrate that TLR3 is indeed expressed in normal human hepatocytes in situ and that primary cultures of human hepatocytes possess a robust signaling pathway that induces the expression of ISGs when stimulated by poly(I·C), a widely used surrogate for dsRNA. Furthermore, we show that TLR3, expressed by a transduced gene in HCV-permissive Huh7 cells, acts to sense HCV infection, thereby triggering the establishment of a cellular antiviral state that limits HCV replication. Importantly, we show that TLR3 acts independently of RIG-I in inducing this antiviral state and that TLR3 signaling, like RIG-I signaling, is disrupted as HCV infection becomes established in these cells.
MATERIALS AND METHODS
Cells, plasmids, and viruses.
Huh7, Huh7.5, PH5CH8, and Vero cells were cultured as described previously (4, 26). Primary human hepatocyte cultures (Clonetics Ready Heps fresh human hepatocytes, which tested negative for human immunodeficiency virus type 1, HCV, and HBV) were purchased from Lonza and cultured in hepatocyte basal medium supplemented with HCM SingleQuots growth factors (Lonza). Huh7 and Huh7.5 cells expressing wild-type (WT) and mutant TLR3 were generated by retroviral gene transduction. Detailed methods are given in the supplemental material. HCV genotype 2a strain JFH1 and an intergenotypic chimeric HCV, HJ3-5, containing genotype 1a (H77) structural proteins, were produced by transfection of Huh7 or Huh7.5 cells with in vitro-transcribed JFH1 or HJ3-5 RNAs, as described previously (51, 52). Vesicular stomatitis virus (VSV) (strain Indiana) stocks were propagated and titrated in Vero cells (10). Infectious virus titers were determined by standard techniques, as described in the supplemental material.
dsRNA treatment, SeV infection, and IFN-β promoter assay.
Where indicated, cells were incubated with 50 μg/ml of poly(I·C) (Sigma) in culture medium for 7 to 18 h or were infected with 50 hemagglutinin units (HAU)/ml of Sendai virus (SeV) (strain Cantell; Charles River Laboratories) for 17 h prior to lysis, as previously described (26). IFN-β promoter activities were determined by cotransfecting cells with IFN-β-Luc and pRL-TK (an internal control for the normalization of transfection efficiency), followed by a dual luciferase assay, as described previously (8).
Type I IFN assay.
IFN activity was measured in cell culture supernatants by a microtiter bioassay based on VSV plaque reduction in Vero cells, as described previously (23).
Immunoblot analyses.
Cell lysates were prepared and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblot analysis as described previously (8, 27). The following monoclonal antibodies (MAbs) or polyclonal antibodies (pAbs) were used for immunoblotting: anti-FLAG MAb M2 and an anti-actin MAb (Sigma); a rabbit anti-ISG15 pAb (a gift from A. Haas); a rabbit anti-ISG56 pAb (generated by immunizing rabbits with a keyhole limpet hemocyanin-coupled peptide spanning amino acids 2 to 19 of human ISG56) and an anti-TLR3 MAb (Imgenex); rabbit anti-TLR3 (48), anti-MxA, and anti-SeV pAbs (gifts from I. Julkunen); rabbit anti-TLR3 pAb 900-1 (generated by immunizing rabbits with a keyhole limpet hemocyanin-coupled peptide spanning amino acids 55 to 70 of human TLR3), a rabbit anti-TRIF pAb (27), and an anti-HCV NS3 MAb (Biodesign); a rabbit anti-HCV NS5B pAb (Virogen); and peroxidase-conjugated secondary goat anti-rabbit and goat anti-mouse immunoglobulin G pAbs (Southern Biotech). Protein bands were visualized by enhanced chemiluminescence (Millipore), followed by exposure to X-ray films.
2PE and laser scanning confocal fluorescence microscopy.
Two-photon (2PE) microscopy was carried out with a custom-built nonlinear optical microscope as described previously (29). Detailed procedures for the preparation of samples and for 2PE image collection are given in the supplemental material. 2PE pseudocolor assignments and multicolor overlays were created with MetaMorph (version 7.1; Molecular Devices, Sunnyvale, CA). Contrast and brightness were enhanced in a uniform manner across 2PE images (including any related control images) using Photoshop CS2 (Adobe Systems, San Jose, CA).
RT-PCR.
Total cellular RNA was extracted by Trizol (Invitrogen) and treated with DNase I to remove contaminating DNA. Real-time reverse transcription-PCR (RT-PCR) analysis of human TLR3, ISG56, and MxA transcripts used commercially available primers and TaqMan probes according to the manufacturer's instructions (Applied Biosystems). The relative abundance of each target was determined by normalization to endogenous 18S RNA.
RESULTS
The TLR3 pathway is functional in human hepatocytes.
The ability of human hepatocytes to sense extracellular dsRNA and signal the induction of ISG expression via TLR3 has not been extensively investigated. Huh7 hepatoma cells, which support HCV infection and are customarily used for in vitro studies of HCV infection, are deficient in TLR3 expression (26). In contrast, PH5CH8 cells, which are derived from large T antigen-transformed, non-neoplastic human hepatocytes (18), support a robust response to poly(I·C) that is dependent on TLR3 expression (26). We confirmed these earlier findings, demonstrating strong induction of ISG15 and MxA expression and a dose-related increase in IFN-β promoter activity following the stimulation of PH5CH8 cells with extracellular poly(I·C) (see Fig. S1A and S1B in the supplemental material). We also observed that the lack of poly(I·C)-induced ISG expression in Huh7 cells could be reversed by transient expression of TLR3 (see Fig. S1C in the supplemental material). These results indicate that human hepatocytes have the capacity to induce an IFN response via TLR3, but they leave unanswered the question of whether the TLR3 pathway is active in hepatocytes in vivo.
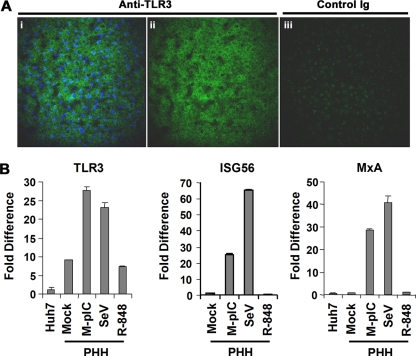
To address this issue, we examined frozen sections of liver tissue obtained from patients with chronic HCV infections who were undergoing resection for liver cancer. 2PE microscopy revealed extensive labeling of hepatocytes in normal (noncirrhotic) liver tissue when sections were incubated with an affinity-purified antibody to TLR3 (Fig. 1A, left and center). No such immunolabeling was observed in adjacent tissue sections incubated with a control immunoglobulin (Fig. 1A, right). A lower level of TLR3 expression was evident in hepatocellular carcinoma tissue (data not shown). We also demonstrated the presence of TLR3 mRNA in cultures of primary human hepatocytes, finding the basal abundance of TLR3 mRNA to be ∼9-fold higher than in Huh7 cells (Fig. 1B, left). Importantly, the addition of poly(I·C) to the culture medium resulted in a threefold increase in the TLR3 mRNA level in these cells (Fig. 1B, left). Further confirming the presence of a functional TLR3 pathway, poly(I·C) strongly activated the expression of two ISGs, ISG56 and MxA (25- and 29-fold, respectively) (Fig. 1B, center and right). Infection with SeV also stimulated the expression of TLR3, ISG56, and MxA (Fig. 1B), consistent with the presence of an active RIG pathway in hepatocytes, as expected. However, under similar conditions, R-848 did not stimulate the expression of TLR3, ISG56, or MxA, suggesting that human hepatocytes do not mount an IFN response to TLR7/TLR8 stimulation. Taken together, these data indicate that TLR3 is expressed by human hepatocytes in situ and that such cells are able to sense extracellular dsRNA and to initiate a robust ISG response.
FIG. 1.
TLR3 is expressed and functional in primary human hepatocytes. (A) TLR3 expression in human liver detected by 2PE microscopy and Qdot fluorescence. Panels i (TLR3 signal merged with a nuclear counterstain) and ii (TLR3 signal only) show a frozen section of human liver labeled with affinity-purified rabbit anti-TLR3, while panel iii shows the adjacent 12-μm-thick section labeled with a control immunoglobulin (Ig). TLR3 expression is evident in the cytoplasm of hepatocytes, with increased perinuclear abundance. (B) Poly(I·C) stimulates ISG expression in primary human hepatocyte (PHH) cultures. Cells in 12-well plates were either mock treated, stimulated with 50 μg/ml of poly(I·C) added to the culture medium (M-pIC), infected with SeV (50 HAU), or treated with 10 μM R-848 for 18 h prior to cell lysis. Extracted RNA samples were subjected to real-time RT-PCR analysis of TLR3, ISG56, and MxA expression. The changes (n-fold) in TLR3 mRNA levels were calculated by using untreated Huh7 cells as the baseline, while the changes in ISG56 and MxA transcript levels were based on transcript abundance in untreated PHHs. Note that although PHHs expressed a higher basal level of TLR3 message than Huh7 cells, basal expression of MxA was similar in PHHs and Huh7 cells.
Reconstitution of TLR3 signaling restricts HCV replication in Huh7 cells.
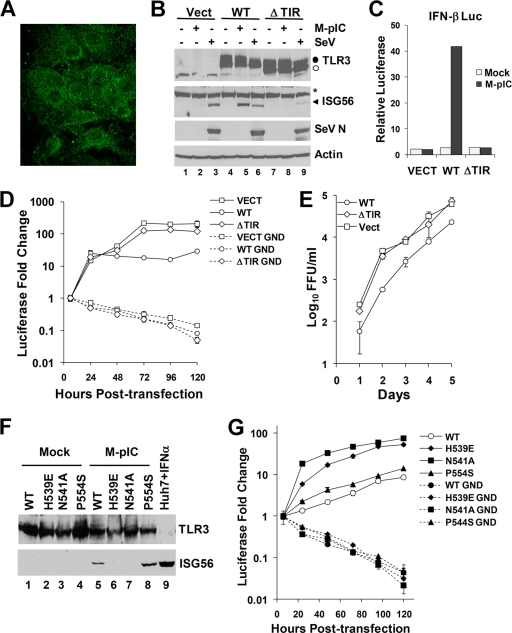
Huh7 cells are almost unique in their ability to support the replication of cell culture-derived HCV (6, 31, 51, 53, 56). Although these cells are defective in TLR3 signaling (26), we observed that this can be reconstituted by transient expression of TLR3 (see Fig. S1C in the supplemental material). Thus, to determine whether TLR3 can sense HCV infection and induce a response that limits viral replication, we used retrovirus-mediated gene transfer to establish Huh7 and Huh7.5 cell lines that stably express TLR3. Huh7.5 cells represent a Huh7 subclone that is defective in RIG-I signaling (47). As controls, we similarly transduced cells with an empty vector (Huh7-Vect cells) or with a TLR3 mutant in which the TIR domain was deleted (Huh7-ΔTIR cells) and which is thus unable to signal (1). 2PE microscopy demonstrated that reconstituted TLR3 was localized to the cytoplasm, with a perinuclear distribution, in both Huh7.5-TLR3 (Fig. 2A) and Huh7-TLR3 (data not shown) cells, a pattern similar to that observed for endogenous TLR3 in hepatocytes in situ (Fig. 1A). To confirm that the expressed TLR3 was functional, we demonstrated that poly(I·C) stimulation strongly induced the expression of ISG56 in Huh7-TLR3 cells (Fig. 2B, lane 5). In contrast, this response was absent in Huh7-Vect and Huh7-ΔTIR cells. Poly(I·C) also induced IFN-β promoter activity in Huh7-TLR3 cells but had no such effect on either control cell line (Fig. 2C). Importantly, this signaling was ablated by pretreatment of Huh7-TLR3 cells with bafilomycin A1 (data not shown), demonstrating that TLR3 signaling requires endosomal maturation in these cells, as is the case for endogenous TLR3 expressed in other cell types (34). In aggregate, these data indicate that retroviral transduction of the TLR3 gene is capable of reconstituting a normally functioning extracellular dsRNA-sensing pathway in Huh7 cells. Similar results were obtained with the RIG-I-deficient Huh7.5 cells (see below). As expected, each of the Huh7-derived cell lines responded to SeV infection, a potent stimulus of the RIG-I pathway (21, 54) (Fig. 2B), while this response was absent in TLR3-transduced Huh7.5 cells (see below).
FIG. 2.
Reconstitution of TLR3 signaling inhibits HCV replication in Huh7 hepatoma cells. (A) 2PE microscopic image demonstrating cytoplasmic localization of TLR3 (green fluorescence) expressed in retrovirus-transduced Huh7.5 cells. (B) Huh7 cells stably transduced with retroviruses expressing an empty vector (Vect) (lanes 1 to 3), WT TLR3 (lanes 4 to 6), or ΔTIR TLR3 (lanes 7 to 9) were either mock treated, stimulated with poly(I·C) (M-pIC), or infected with SeV for 18 h prior to immunoblot analysis of TLR3, ISG56, SeV N protein, and actin expression. •, full-length TLR3; ○, TLR3ΔTIR; *, nonspecific band. (C) IFN-β promoter activities in Huh7 cells stably transduced with an empty vector or with WT or ΔTIR TLR3 retroviral vectors. Cells were either mock treated (open bars) or treated with poly(I·C) (filled bars). (D) Huh7-Vect, Huh7-TLR3 (WT), and Huh7-ΔTIR cells were transfected either with the J6/JFH1 chimeric HCV luciferase replicon (49) (solid lines) or with a related, polymerase-deficient mutant replicon (GND) (dotted lines). HCV RNA replication is reflected in the change in luciferase activity (normalized to that at 6 h posttransfection, when luciferase activity results from translation of input HCV RNA only). (E) Huh7-Vect, Huh7-TLR3 (WT), and Huh7-ΔTIR cells were infected with cell culture-derived HCV (strain JFH1; MOI, 0.1), and culture supernatants were assayed for infectious virus yield at the indicated times by a focus-forming unit (FFU) assay. (F) Immunoblot analysis of TLR3 and ISG56 expression in Huh7 cells stably transduced with retroviruses expressing WT or mutant (H539E, N541A, or P554S) TLR3. Cells were either mock treated (lanes 1 to 4) or stimulated with poly(I·C) (lanes 5 to 8) for 18 h. Huh7 cells treated with IFN-α (lane 9) were included as a positive control for ISG56 expression. Endogenous TLR3 protein was not detectable in these Huh7-derived cell lines under these conditions. (G) Luciferase activities representing the replication of the J6/JFH1 chimeric HCV replicon (solid lines) versus the GND mutant replicon (dotted lines) in transfected Huh7 cells stably expressing WT TLR3 or the H539E, N541A, or P554S TLR3 mutant.
Having confirmed the signaling phenotypes of the Huh7-derived cell lines, we investigated their abilities to support the replication of a genome-length HCV (J6/JFH1) replicon RNA that encodes luciferase. This replicon RNA replicates efficiently in Huh7 cells, expressing luciferase as a measure of viral RNA amplification (49). Cells were transfected in parallel with the WT replicon and a related mutant (GND) containing a replication-lethal mutation in the NS5B RNA-dependent RNA polymerase. As expected, the mutant replicon failed to amplify when transfected into any cell line, as evidenced by diminishing luciferase activities following transfection (Fig. 2D). In contrast, the WT replicon replicated efficiently in both Huh7-Vect and Huh7-ΔTIR cells, resulting in an increase in luciferase activity over 72 h, followed by a plateau at later time points. In contrast, replication of this RNA was significantly compromised in Huh7-TLR3 cells, resulting in 5- to 12-fold lower luciferase expression than that observed with Huh7-Vect and Huh7-ΔTIR cells 72 to 120 h posttransfection (Fig. 2D). These results suggest that TLR3 expression restricts HCV RNA replication in Huh7 cells. To confirm this, we determined the yield of infectious HCV produced by these cells when challenged with cell culture-derived virus (genotype 2a, strain JFH1). When cells were infected under identical conditions, the yield of infectious HCV released into the cell culture medium was 5- to 10-fold lower in Huh7-TLR3 cells than in Huh7-Vect and Huh7-ΔTIR cells, with no difference between the latter two cell types (Fig. 2E). We conclude from these experiments that reconstitution of TLR3 expression has a negative effect on HCV replication in Huh7 cells.
dsRNA binding is required for TLR3-mediated restriction of HCV replication.
Since TLR3 and HCV RNA replication complexes are both associated with intracellular membranes (20, 43), we considered the possibility that the transduced TLR3 protein might be activated and limit HCV replication nonspecifically by rearrangement of these membranes during the formation of the viral replication complex. To investigate this possibility, we established additional Huh7 cell lines expressing TLR3 mutants that are defective in the recognition of dsRNA. TLR3 signaling commences upon engagement of the leucine-rich repeats of its ectodomain by dsRNA. Mutations near the C terminus of the ectodomain, His539 to Glu (H539E) or Asn541 to Ala (N541A), completely abrogate dsRNA binding (2, 32). An additional mutation close to the C terminus, Pro554 to Ser (P554S), was identified recently in patients with herpes simplex encephalitis and has been suggested to exert a dominant-negative effect on TLR3 signaling (55). As expected, we observed that ISG56 expression was not induced by poly(I·C) stimulation of Huh7-derived cell lines expressing either the H539E or the N541A mutant (Fig. 2F, compare lanes 6 and 7 with lane 5). However, expression of the P554S mutant efficiently complemented the TLR3 signaling defect in Huh7 cells (Fig. 2F, lane 8). This contrasts with previously published results (55), which suggest that the P554S mutation abrogates TLR3 signaling in fibroblasts. While the reason(s) for this difference is not fully understood, it is noteworthy that Zhang et al. reported the P554S mutant to be expressed as a C-terminally-truncated product (55), while the P554S mutant we expressed in Huh7 cells was of normal length (Fig. 2F).
The luciferase-expressing HCV replicon replicated efficiently following transfection into Huh7 cells expressing N541A or H539E mutant TLR3, resulting in 53- to 75-fold increases in luciferase expression, respectively (Fig. 2G). In contrast, replication was severely compromised in Huh7 cells expressing wild-type or P554S mutant TLR3, with luciferase expression increasing only 9- to 14-fold, respectively (Fig. 2G). In several replicate experiments, HCV RNA replication was always 5- to 10-fold less in cells reconstituted with functional TLR3 (i.e., WT or P554S TLR3) than in those expressing the N541A or H539E mutants, which are incapable of binding dsRNA. These results demonstrate that TLR3-mediated restriction of HCV replication requires TLR3 recognition of dsRNA and is not induced by nonspecific rearrangements of intracellular membranes. Since we also observed that the ΔTIR mutant is unable to regulate HCV replication (Fig. 2D and E), TLR3 must also engage its adaptor protein, TRIF, to initiate this antiviral response.
To ascertain the specificity of TLR3-mediated suppression of HCV replication, we compared the abilities of Huh7 cells expressing WT or mutant TLR3 to support the replication of VSV, a negative-strand RNA virus classified within the Rhabdoviridae. These results demonstrated no differences in the infectious virus yields produced by cells expressing functional or nonfunctional TLR3 mutants (see Fig. S2 in the supplemental material). Thus, while reconstitution of TLR3 signaling in Huh7 cells restricts the replication of HCV (a positive-strand RNA virus), it had no appreciable effect on VSV infection.
TLR3-mediated restriction of HCV replication is independent of RIG-I.
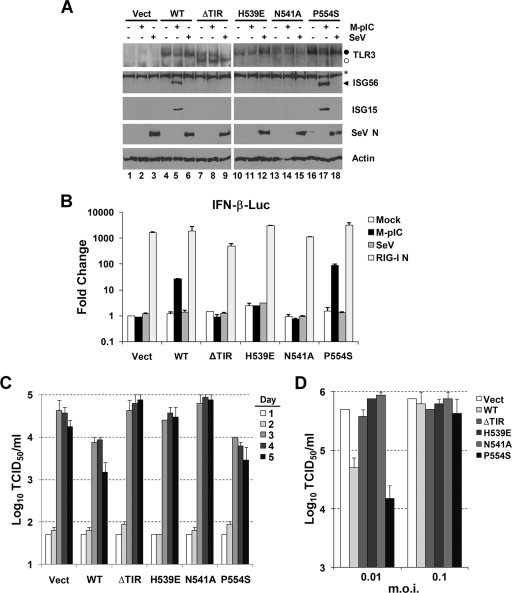
While Huh7 cells contain a functional RIG-I pathway, Huh7.5 cells, which were derived by eliminating HCV RNA from a stable Huh7 replicon cell line, express a mutant RIG-I that is defective in signaling (47). To determine whether TLR3 senses HCV and limits its replication independently of RIG-I, we established Huh7.5 cell lines expressing both WT and mutant TLR3. In findings similar to those with the Huh7-derived cell lines, Huh7.5 cells transduced to express WT TLR3 upregulated the expression of ISG56 and ISG15 when stimulated with poly(I·C) (Fig. 3A, compare lane 5 with lane 4), as did the P554S mutant (compare lane 17 with lane 16). In contrast, poly(I·C) induced neither ISG in Huh7.5 cells transduced with an empty vector (Fig. 3A, compare lane 2 with lane 1) or with ΔTIR (compare lane 8 with lane 7), H539E (compare lane 11 with lane 10), or N541A (compare lane 14 with lane 13) mutant TLR3. RIG-I signaling remained absent in all of these transduced cells, and SeV infection failed to induce ISG15 or ISG56 in any of them (Fig. 3A, lanes 3, 6, 9, 12, 15, and 18). IFN-β promoter activity was also stimulated by poly(I·C) only in Huh7.5 cells expressing WT TLR3 or the P554S mutant and was not increased in any of the cell lines following SeV infection (Fig. 3B). As expected (47), the RIG-I-null phenotype was not due to a defect downstream of RIG-I, as evidenced by the fact that ectopic expression of the constitutively active RIG-I CARD domain strongly activated the IFN-β promoter in these cells (Fig. 3B, RIG-I N). Importantly, the yield of infectious HCV released by Huh7.5 cells expressing WT TLR3 or the P554S mutant was consistently 5- to 10-fold lower than the yields from cells expressing nonfunctional (ΔTIR, H539E, or N541A) TLR3 mutants following challenge with the virus at a low multiplicity of infection (MOI) (0.001) (Fig. 3C). Similar results were obtained at an MOI of 0.01, but TLR3 expression had no apparent impact on virus yields when cells were challenged with virus at an MOI of 0.1 (Fig. 3C and D). These data confirm that the ability of TLR3 to sense HCV infection and exert a restrictive response is independent of RIG-I, which is known to mediate antiviral responses to HCV (33, 47). However, they also suggest that HCV may be capable of overwhelming the TLR3-mediated response at a high MOI.
FIG. 3.
TLR3 signaling inhibits HCV replication in cells defective in RIG-I. (A) Huh7.5 cells stably transduced with retroviruses carrying the empty vector (lanes 1 to 3), WT TLR3 (lanes 4 to 6), or the ΔTIR (lanes 7 to 9), H539E (lanes 10 to 12), N541A (lanes 13 to 15), or P554S (lanes 16 to 18) mutant TLR3 expression vector were either mock treated (lanes 1, 4, 7, 10, 13, and 16), stimulated with poly(I·C) (lanes 2, 5, 8, 11, 14, and 17), or infected with SeV (lanes 3, 6, 9, 12, 15, and 18) for 18 h prior to lysis and immunoblot analysis of TLR3, ISG56, ISG15, SeV N protein, and actin expression. (B) IFN-β promoter activities in Huh7.5 cells stably transduced with either an empty vector, WT TLR3, or a TLR3 mutant following mock treatment, stimulation with poly(I·C) (M-pIC), infection with SeV, or transfection with a vector expressing RIG-I CARD (RIG-I N). (C) Huh7.5 cells, stably transduced either with an empty vector or with a WT or mutant TLR3 expression vector, were infected with HCV (strain JFH1) at a low MOI (0.001). Culture supernatants were assayed for infectious virus yields by a 50% tissue culture infective dose (TCID50) assay on the days indicated. (D) Virus yields in cultures of TLR3-expressing Huh7.5 cells infected with HCV (strain JFH1) at higher MOIs: 0.01 (day 5 postinfection) or 0.1 (day 4 postinfection).
TLR3 senses HCV infection and triggers the expression of ISGs.
Because the ability of TLR3 to bind dsRNA appears to be indispensable for its ability to limit HCV replication (Fig. 2G and 3C and D), it is likely that TLR3 senses dsRNA produced by the virus and initiates signaling cascades in these cells that lead to an IFN response and ISG expression. To confirm this, we analyzed ISG expression in Huh7.5-TLR3 cells after infection with HCV. Immunoblotting demonstrated that both ISG15 and ISG56 expression were induced by HCV infection in cells expressing either WT or P554S mutant TLR3 (Fig. 4A, compare lanes 5 and 6 and lanes 14 and 15, respectively). In contrast, there was no induction of ISG expression in ΔTIR, H539E, or N541A cells. We also observed the translocation of IRF-3 into the nuclei of Huh7.5-TLR3 cells following infection with HCV (Fig. 4B). This translocation occurred in many, but not all, cells expressing HCV core protein, suggesting activation of this transcription factor in many infected cells. Nuclear translocation of IRF-3 was also observed in rare Huh7.5-TLR3 cells that were devoid of detectable core protein expression (Fig. 4B). We cannot exclude very early HCV infection in these cells, but these observations suggest that some uninfected cells may sense viral dsRNA released into extracellular fluids by apoptosis or other mechanisms. Importantly, nuclear translocation of IRF-3 was never observed in infected cells expressing the H539E mutant or transduced with an empty vector (Fig. 4B).
FIG. 4.
TLR3 senses HCV infection in Huh7.5 cells, which are deficient in RIG-I signaling. (A) Huh7.5 cells stably expressing WT TLR3 or various TLR3 mutants were either mock treated (lanes 3, 6, 9, 12, and 15), stimulated by the addition of 50 μg/ml poly(I·C) for 18 h (lanes 1, 4, 7, 10, and 13), or infected with cell culture-derived HCV (strain JFH1; MOI, 0.1) for 4 days (lanes 2, 5, 8, 11, and 14). Cells were lysed and subjected to immunoblot analysis for ISG56, ISG15, HCV core protein, and actin. Asterisks indicate nonspecific bands. (B) Laser scanning confocal immunofluorescence microscopy images showing HCV core protein (green) and IRF-3 (red) expression in Huh7.5 cells expressing WT TLR3, H539E mutant TLR3, or an empty vector (Vect) 2 days after infection with cell culture-derived HCV (strain JFH1). The arrow indicates a cell in which nuclear translocation of IRF-3 was evident in the absence of detectable core protein. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). “Mock” stands for mock infection.
The ISG response to HCV infection was notably weaker than that induced by poly(I·C) in cells expressing functional TLR3 (Fig. 4A, compare lanes 5 and 4 and lanes 14 and 13, respectively), suggesting either that dsRNA produced by HCV presents a less abundant or less potent trigger for TLR3 signaling or that HCV infection may interfere with TLR3 signaling (see below). We were unable to detect IFN antiviral activity in culture supernatants harvested at any time after HCV infection of Huh7.5-TLR3 cells, while overnight poly(I·C) treatment was able to induce ∼3 U/ml of IFN in these cells, near the limit of detection in a VSV plaque reduction assay. Thus, the induction of ISGs may have occurred as a primary result of IRF-3 activation following HCV infection, since activated IRF-3 directly regulates ISG56 expression and, to a lesser extent, ISG15 expression (14, 38). However, our data do not exclude the possibility that a small amount of IFN, below the limit of detection in the bioassay, also contributed to ISG induction.
HCV infection disrupts poly(I·C)-induced TLR3 signaling.
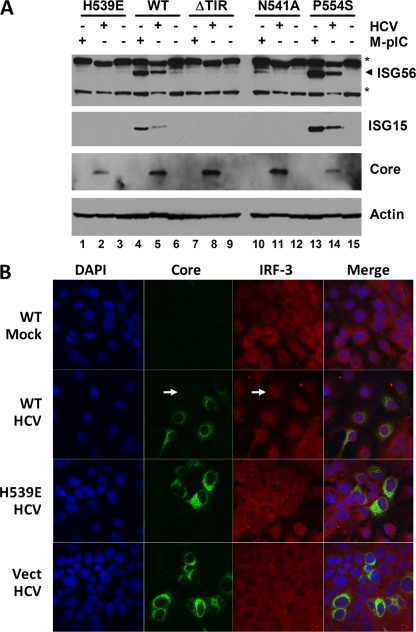
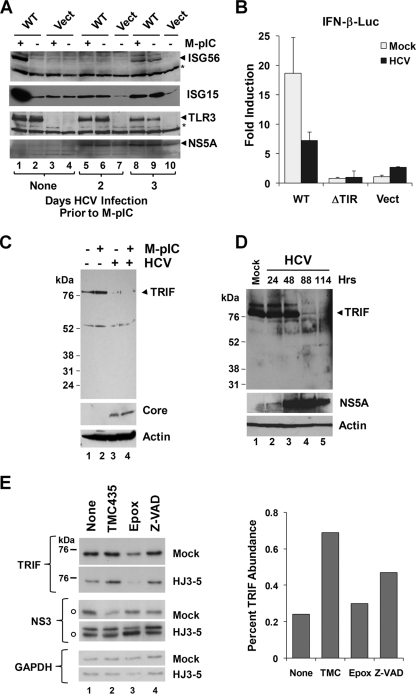
Infection of Huh7.5-TLR3 cells by HCV resulted in weak induction of ISG15 and ISG56 by days 3 to 4 (Fig. 5A, compare lanes 6 and 9 with lane 2), but it also efficiently blocked the ability of poly(I·C) to stimulate the synthesis of ISGs. This was evident as early as 2 days after infection (Fig. 5A, compare lanes 5 and 6 with lanes 1 and 2). Poly(I·C)-induced activation of the IFN-β promoter was also substantially reduced in HCV-infected Huh7.5-TLR3 cells from that in uninfected cells (Fig. 5B). Since our previous studies have shown TRIF to be cleaved by the NS3/4A protease (12, 27), we assessed TRIF abundance in these cells in an effort to gain insight into the mechanism underlying the disruption of TLR3 signaling by HCV. As shown in Fig. 5C, TRIF protein abundance was substantially reduced in HCV-infected Huh7.5-TLR3 cells (compare lanes 3 and 4 with lanes 1 and 2). Decreases were evident as early as 3 days after infection with HCV (Fig. 5D) and could not be attributed to TLR3 signaling per se, since poly(I·C) stimulation did not reduce TRIF abundance (Fig. 5C, compare lanes 2 and 1). In addition, the reduction in TRIF abundance was not due to cellular apoptosis or proteasome-mediated degradation of the adaptor protein, since it was not restored by treatment with ZVAD, a pan-caspase inhibitor, or epoxomicin, a potent inhibitor of the proteasome (Fig. 5E; see also Fig. S3 in the supplemental material). In contrast, brief treatment with a potent and specific inhibitor of HCV NS3/4A protease, TMC435350 (44), substantially restored TRIF abundance in infected cells while exerting only a minimal effect on NS3/4A abundance over a 24-h treatment period (Fig. 5E). Taken together, these data provide strong additional support for the notion that HCV has evolved the capacity to disrupt TLR3-mediated signaling as a means of evading early innate immune responses to the virus (27).
FIG. 5.
HCV infection disrupts TLR3 signaling and degrades TRIF. (A) Immunoblot analysis of ISG56, ISG15, TLR3, and HCV NS5A in Huh7.5-TLR3 (WT) or Huh7.5-Vect cells that were either mock infected (lanes 1 to 4) or infected with cell culture-derived HCV (strain JFH1) at an MOI of 0.1 (lanes 5 to 10) for the indicated times. Where indicated (M-pIC), poly(I·C) was added to the medium at 2 and 3 days post-HCV infection in lanes 5 and 8, respectively, for 18 h. Note that TLR3 expression (lanes 8 and 9) was slightly reduced at 4 days post-HCV infection. (B) Poly(I·C)-induced IFN-β promoter activities in Huh7.5-TLR3 (WT), -ΔTIR, and -Vect cells that were either mock infected or infected with cell culture-derived HCV (HJ3-5 virus; MOI, 1.0) for 4 days. Poly(I·C) stimulation was carried out for 7 h prior to cell lysis and a luciferase assay. (C) Huh7.5-TLR3 cells were either mock infected (lanes 1 and 2) or infected with HCV (strain JFH1) at an MOI of 2 (lanes 3 and 4) for 3 days; subsequently, they were either mock treated (lanes 1 and 3) or stimulated with poly(I·C) for 18 h (lanes 2 and 4) prior to immunoblot analysis of TRIF, HCV core protein, and actin. (D) Huh7.5 cells were infected with HCV (strain JFH1; MOI, 0.1) for the indicated length of time prior to lysis and immunoblot analysis of TRIF, HCV NS5A, and actin. (E) (Left) Huh7.5 cells were either mock infected or infected with HCV (HJ3-5 virus; MOI, 1) for 4 days prior to a 24-h period of treatment with TMC435350 (1 μM), epoxomicin (1 μM), or ZVAD (50 μM) or no treatment (“None”), followed by cell lysis and immunoblot analysis of TRIF, HCV NS3, and glyceraldehyde-3-phosphate dehydrogenase. Open circles indicate nonspecific bands in immunoblots. (Right) Relative abundances of TRIF in TMC435350-, epoxomicin-, and ZVAD-treated versus untreated Huh7.5 cells as determined by densitometry of the immunoblot shown in the left panel.
DISCUSSION
There is substantial controversy in the literature concerning the role played by the TLR3 pathway in the induction of antiviral immunity (50). Here we have demonstrated that TLR3 is capable of sensing HCV infection in cultured Huh7 cells and that this triggers IRF-3 activation and the expression of ISGs (Fig. 4) that limit HCV RNA replication and progeny virus yields (Fig. 2 and 3). While reconstitution of TLR3 expression was required in order to demonstrate this antiviral effect in hepatoma cells, we also present evidence that TLR3 is normally expressed by human hepatocytes in vivo and that primary hepatocytes induce an IFN response when stimulated by poly(I·C) in vitro (Fig. 1). The absence of TLR3 expression in Huh7 cells can be related to their malignant phenotype; indeed, TLR3 is expressed by PH5CH8 cells, which are derived from nonmalignant hepatocytes but do not support efficient replication of HCV (26). Strikingly, mutations that disrupt the dsRNA-binding ability of TLR3 ablated the ISG response to HCV infection (Fig. 4) and the antiviral effect against HCV (Fig. 2G and 3C) in transduced hepatoma cells, indicating that TLR3 signaling, and not simply the expression of TLR3 protein, was responsible for suppressing HCV replication. This is supported also by the absence of an antiviral response in TLR3-ΔTIR cells (Fig. 2D and E and 3C). Importantly, we demonstrated the ability of TLR3 to sense and restrict HCV replication not only in Huh7 cells but also in RIG-I-deficient Huh7.5 cells, indicating that TLR3 functions to detect and defend against HCV infection independently of RIG-I signaling. Thus, the TLR3 pathway acts in parallel to the RIG-I pathway, which is known to sense HCV RNA and to limit cellular permissiveness for HCV (33, 40, 47).
The capacities of both signaling pathways to restrict virus replication are consistent with their convergence downstream at the level of kinases responsible for the activation of IRF-3 and NF-κB (22). dsRNA engagement by TLR3 results in the activation of IRF-3-dependent type I and III IFN responses, as well as in NF-κB-dependent proinflammatory cytokine production. Both types of IFN, as well as a number of ISGs, possess antiviral activity against HCV in vitro (7, 17, 19, 39, 57), making it likely that the antiviral state induced by TLR3 signaling is dependent on the activation of IRF-3. However, the proinflammatory cytokine interleukin-1β also inhibits the replication of HCV RNA (58), and overexpression of TRIF (mimicking TLR3 signaling) inhibits HBV replication through mechanisms that are dependent on NF-κB as well as IRF-3 activation in Huh7 cells (15). Fundamental differences in the replication mechanisms of HBV and HCV warrant considerable caution in comparing these antiviral responses, but the latter findings make it clear that further studies will be required to determine the extent to which the TLR3-mediated antiviral response to HCV is dependent on the activation of IRF-3 and/or NF-κB.
Although we have shown that TLR3 senses HCV infection, the HCV pathogen-associated molecular pattern that triggers TLR3 signaling remains to be characterized. TLR3 senses dsRNA that is present within an early-late endosome compartment, and endosomal acidification has been shown to be essential for TLR3 signaling in HEK293 cells stably expressing TLR3 as well as in immature dendritic cells (34). We confirmed this requirement in Huh7.5-TLR3 cells (data not shown). Compared to poly(I·C) stimulation, HCV infection induced a delayed TLR3-dependent ISG response in these cells (Fig. 4 and 5A). This suggests that HCV may not be recognized upon viral entry and that there is a need for replication of the virus to generate an abundance of dsRNA sufficient to trigger TLR3 signaling. We observed TLR3-dependent nuclear translocation of IRF-3 in HCV-infected cells (Fig. 4B), suggesting that dsRNA is likely to be sensed within the cell in which it is produced. How viral RNA enters the endosomal compartment, where it can be engaged by TLR3 within infected cells, is uncertain. Conceivably, this could be accomplished through autophagy, as reported for TLR7 recognition of VSV infection in plasmacytoid dendritic cells (24). However, Rab5, an early endosome protein, colocalizes with the HCV RNA replication complex and is essential for HCV RNA replication (45). Thus, it is possible that TLR3 expression may be localized to intracellular membranes involved in HCV RNA replication that constitute the so-called “membranous web” in which the replication complex resides (11). Further investigation is needed to resolve these issues.
HCV has evolved a number of strategies to antagonize host innate and adaptive immune responses, thereby enabling it to establish lifelong, persistent infections (5, 13). Among these strategies, the NS3/4A protease targets the RIG-I adaptor protein, MAVS, disrupting RIG-I signaling in infected cells (28, 30, 33, 35). Although we found that HCV infection triggers ISG expression in Huh7.5-TLR3 cells, we also observed that the magnitude of this response was substantially lower than that induced by poly(I·C) (Fig. 4A and 5A). At least in part, this can be explained by the ability of HCV to disrupt, as well as to activate, TLR3 signaling. HCV infection inhibited poly(I·C) activation of the IFN-β promoter and induction of ISG15 and ISG56 synthesis in Huh7.5-TLR3 cells (Fig. 5A and B). These results may explain why we did not observe IRF-3 nuclear translocation in all infected Huh7.5-TLR3 cells (Fig. 4B) and why we observed no antiviral effect of TLR3 signaling at high MOIs (Fig. 3D). We have previously shown that TRIF is a substrate for NS3/4A proteolysis and that TLR3 signaling is disrupted in HeLa cells containing HCV replicons (12, 27). Consistent with this, we found that the abundance of TRIF was reduced in HCV-infected Huh7.5 cells (Fig. 5C and D; see also Fig. S3 in the supplemental material) but could be restored partially by short-term treatment of these cells with a potent inhibitor of NS3/4A, TMC435350 (Fig. 5E). These data suggest that NS3/4A-mediated degradation of TRIF may contribute to the suppression of TLR3 responses by HCV. However, they do not exclude a role for other HCV proteins, such as NS5A, which perturbs the phosphoinositide 3-kinase pathway (16, 46), which is important for full activation of IRF-3 downstream of TLR3 (41). Further studies will be needed to fully dissect the underlying molecular mechanisms and to determine the relative contribution of NS3/4A cleavage of TRIF, but the data presented here indicate that TLR3 signaling, like RIG-I signaling (33), is both activated and disrupted in HCV-infected hepatoma cells.
The disruption of both the RIG-I and TLR3 signaling pathways may favor HCV infection by impairing the establishment of an antiviral state within infected cells. However, disruption of TLR3 signaling may play an additional role in promoting viral persistence at the level of the host. Both the magnitude and the breadth of the adaptive T-cell response are critical determinants in the control and clearance of HCV infection (5). While MAVS is of fundamental importance in the innate intracellular response to invading viruses, it is not essential for the development of robust cytotoxic T-lymphocyte responses (3). In contrast, TLR3 is known to promote the cross-priming of T lymphocytes to viral antigens (42) and has recently been shown to be essential for type II IFN responses to coxsackievirus infection (36). Although much remains to be learned about how early innate immune responses shape the subsequent development of adaptive immunity to HCV, viral disruption of the TLR3-TRIF axis is likely to figure prominently among the HCV-host interactions that determine the outcome of infection.
Supplementary Material
Acknowledgments
This work was supported by grants R01-AI69285 (to K.L.), U19-AI40035 (to S.M.L.), and R21-AI081058 (to S.M.L.) from the National Institute of Allergy and Infectious Diseases and by grant IRG-96-152-07 from the American Cancer Society (to K.L.). K.L. was a Cain Foundation Investigator in Innate Immunity. Research in S. M. Lemon's laboratory is supported by a grant from Tibotec Pharmaceuticals.
We thank G. Vargas and T. Shilagard of the UTMB Center for Biomedical Engineering for assistance with 2PE microscopy and L. Cicalese of the UTMB Transplant Center for providing liver specimens. We are also grateful to T. Wakita for JFH1 cDNA, to M. Yi for HJ3-5 virus, to C. Rice for J6/JFH1 cDNA, Huh7.5 cells, and the anti-NS5A MAb 9E10, to N. Kato for PH5CH8 cells, to A. Haas for the anti-ISG15 antibody, to I. Julkunen for anti-TLR3 antisera, and to T.-I. Lin and Tibotec BVBA for TMC435350.
Footnotes
Published ahead of print on 22 July 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413:732-738. [DOI] [PubMed] [Google Scholar]
- 2.Bell, J. K., J. Askins, P. R. Hall, D. R. Davies, and D. M. Segal. 2006. The dsRNA binding site of human Toll-like receptor 3. Proc. Natl. Acad. Sci. USA 103:8792-8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhoj, V. G., Q. Sun, E. J. Bhoj, C. Somers, X. Chen, J. P. Torres, A. Mejias, A. M. Gomez, H. Jafri, O. Ramilo, and Z. J. Chen. 2008. MAVS and MyD88 are essential for innate immunity but not cytotoxic T lymphocyte response against respiratory syncytial virus. Proc. Natl. Acad. Sci. USA 105:14046-14051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowen, D. G., and C. M. Walker. 2005. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature 436:946-952. [DOI] [PubMed] [Google Scholar]
- 6.Cai, Z., C. Zhang, K. S. Chang, J. Jiang, B. C. Ahn, T. Wakita, T. J. Liang, and G. Luo. 2005. Robust production of infectious hepatitis C virus (HCV) from stably HCV cDNA-transfected human hepatoma cells. J. Virol. 79:13963-13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, K. S., Z. Cai, C. Zhang, G. C. Sen, B. R. Williams, and G. Luo. 2006. Replication of hepatitis C virus (HCV) RNA in mouse embryonic fibroblasts: protein kinase R (PKR)-dependent and PKR-independent mechanisms for controlling HCV RNA replication and mediating interferon activities. J. Virol. 80:7364-7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Z., R. Rijnbrand, R. K. Jangra, S. G. Devaraj, L. Qu, Y. Ma, S. M. Lemon, and K. Li. 2007. Ubiquitination and proteasomal degradation of interferon regulatory factor-3 induced by Npro from a cytopathic bovine viral diarrhea virus. Virology 366:277-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dansako, H., M. Ikeda, and N. Kato. 2007. Limited suppression of the interferon-beta production by hepatitis C virus serine protease in cultured human hepatocytes. FEBS J. 274:4161-4176. [DOI] [PubMed] [Google Scholar]
- 10.Devaraj, S. G., N. Wang, Z. Chen, M. Tseng, N. Barretto, R. Lin, C. J. Peters, C. T. Tseng, S. C. Baker, and K. Li. 2007. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 282:32208-32221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferreon, J. C., A. C. Ferreon, K. Li, and S. M. Lemon. 2005. Molecular determinants of TRIF proteolysis mediated by the hepatitis C virus NS3/4A protease. J. Biol. Chem. 280:20483-20492. [DOI] [PubMed] [Google Scholar]
- 13.Gale, M., Jr., and E. M. Foy. 2005. Evasion of intracellular host defence by hepatitis C virus. Nature 436:939-945. [DOI] [PubMed] [Google Scholar]
- 14.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76:5532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo, H., D. Jiang, D. Ma, J. Chang, A. M. Dougherty, A. Cuconati, T. M. Block, and J. T. Guo. 2009. Activation of pattern recognition receptor-mediated innate immunity inhibits the replication of hepatitis B virus in human hepatocyte-derived cells. J. Virol. 83:847-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He, Y., H. Nakao, S. L. Tan, S. J. Polyak, P. Neddermann, S. Vijaysri, B. L. Jacobs, and M. G. Katze. 2002. Subversion of cell signaling pathways by hepatitis C virus nonstructural 5A protein via interaction with Grb2 and P85 phosphatidylinositol 3-kinase. J. Virol. 76:9207-9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helbig, K. J., D. T. Lau, L. Semendric, H. A. Harley, and M. R. Beard. 2005. Analysis of ISG expression in chronic hepatitis C identifies viperin as a potential antiviral effector. Hepatology 42:702-710. [DOI] [PubMed] [Google Scholar]
- 18.Ikeda, M., K. Sugiyama, T. Mizutani, T. Tanaka, K. Tanaka, H. Sekihara, K. Shimotohno, and N. Kato. 1998. Human hepatocyte clonal cell lines that support persistent replication of hepatitis C virus. Virus Res. 56:157-167. [DOI] [PubMed] [Google Scholar]
- 19.Jiang, D., H. Guo, C. Xu, J. Chang, B. Gu, L. Wang, T. M. Block, and J. T. Guo. 2008. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J. Virol. 82:1665-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnsen, I. B., T. T. Nguyen, M. Ringdal, A. M. Tryggestad, O. Bakke, E. Lien, T. Espevik, and M. W. Anthonsen. 2006. Toll-like receptor 3 associates with c-Src tyrosine kinase on endosomes to initiate antiviral signaling. EMBO J. 25:3335-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101-105. [DOI] [PubMed] [Google Scholar]
- 22.Kawai, T., and S. Akira. 2008. Toll-like receptor and RIG-I-like receptor signaling. Ann. N. Y. Acad. Sci. 1143:1-20. [DOI] [PubMed] [Google Scholar]
- 23.Langford, M. P., D. A. Weigent, G. J. Stanton, and S. Baron. 1981. Virus plaque-reduction assay for interferon: microplaque and regular macroplaque reduction assays. Methods Enzymol. 78:339-346. [DOI] [PubMed] [Google Scholar]
- 24.Lee, H. K., J. M. Lund, B. Ramanathan, N. Mizushima, and A. Iwasaki. 2007. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 315:1398-1401. [DOI] [PubMed] [Google Scholar]
- 25.Lemon, S. M., C. Walker, M. J. Alter, and M. Yi. 2007. Hepatitis C viruses, p. 1253-1304. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 26.Li, K., Z. Chen, N. Kato, M. Gale, Jr., and S. M. Lemon. 2005. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-beta production in hepatocytes. J. Biol. Chem. 280:16739-16747. [DOI] [PubMed] [Google Scholar]
- 27.Li, K., E. Foy, J. C. Ferreon, M. Nakamura, A. C. Ferreon, M. Ikeda, S. C. Ray, M. Gale, Jr., and S. M. Lemon. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. USA 102:2992-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, X. D., L. Sun, R. B. Seth, G. Pineda, and Z. J. Chen. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. USA 102:17717-17722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang, Y., T. Shilagard, S.-Y. Xiao, N. Snyder, D. T. Lau, L. Cicalese, H. Weiss, G. Vargas, and S. M. Lemon. Visualizing hepatitis C virus infections in human liver by two-photon microscopy. Gastroenterology, in press. [DOI] [PubMed]
- 30.Lin, R., J. Lacoste, P. Nakhaei, Q. Sun, L. Yang, S. Paz, P. Wilkinson, I. Julkunen, D. Vitour, E. Meurs, and J. Hiscott. 2006. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKɛ molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J. Virol. 80:6072-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 32.Liu, L., I. Botos, Y. Wang, J. N. Leonard, J. Shiloach, D. M. Segal, and D. R. Davies. 2008. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science 320:379-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loo, Y. M., D. M. Owen, K. Li, A. K. Erickson, C. L. Johnson, P. M. Fish, D. S. Carney, T. Wang, H. Ishida, M. Yoneyama, T. Fujita, T. Saito, W. M. Lee, C. H. Hagedorn, D. T. Lau, S. A. Weinman, S. M. Lemon, and M. Gale, Jr. 2006. Viral and therapeutic control of IFN-β promoter stimulator 1 during hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 103:6001-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsumoto, M., K. Funami, M. Tanabe, H. Oshiumi, M. Shingai, Y. Seto, A. Yamamoto, and T. Seya. 2003. Subcellular localization of Toll-like receptor 3 in human dendritic cells. J. Immunol. 171:3154-3162. [DOI] [PubMed] [Google Scholar]
- 35.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 36.Negishi, H., T. Osawa, K. Ogami, X. Ouyang, S. Sakaguchi, R. Koshiba, H. Yanai, Y. Seko, H. Shitara, K. Bishop, H. Yonekawa, T. Tamura, T. Kaisho, C. Taya, T. Taniguchi, and K. Honda. 2008. A critical link between Toll-like receptor 3 and type II interferon signaling pathways in antiviral innate immunity. Proc. Natl. Acad. Sci. USA 105:20446-20451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pawlotsky, J. M., S. Chevaliez, and J. G. McHutchison. 2007. The hepatitis C virus life cycle as a target for new antiviral therapies. Gastroenterology 132:1979-1998. [DOI] [PubMed] [Google Scholar]
- 38.Peters, K. L., H. L. Smith, G. R. Stark, and G. C. Sen. 2002. IRF-3-dependent, NF-κB- and JNK-independent activation of the 561 and IFN-β genes in response to double-stranded RNA. Proc. Natl. Acad. Sci. USA 99:6322-6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robek, M. D., B. S. Boyd, and F. V. Chisari. 2005. Lambda interferon inhibits hepatitis B and C virus replication. J. Virol. 79:3851-3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito, T., D. M. Owen, F. Jiang, J. Marcotrigiano, and M. Gale, Jr. 2008. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarkar, S. N., K. L. Peters, C. P. Elco, S. Sakamoto, S. Pal, and G. C. Sen. 2004. Novel roles of TLR3 tyrosine phosphorylation and PI3 kinase in double-stranded RNA signaling. Nat. Struct. Mol. Biol. 11:1060-1067. [DOI] [PubMed] [Google Scholar]
- 42.Schulz, O., S. S. Diebold, M. Chen, T. I. Naslund, M. A. Nolte, L. Alexopoulou, Y. T. Azuma, R. A. Flavell, P. Liljestrom, and C. Reis e Sousa. 2005. Toll-like receptor 3 promotes cross-priming to virus-infected cells. Nature 433:887-892. [DOI] [PubMed] [Google Scholar]
- 43.Shi, S. T., K. J. Lee, H. Aizaki, S. B. Hwang, and M. M. Lai. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J. Virol. 77:4160-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simmen, K., O. Lenz, T. I. Lin, G. Fanning, P. Raboisson, H. de Kock, G. van't Klooster, A. Rosenquist, M. Edlund, M. Nilsson, L. Vrang, and B. Samuelsson. 2007. In vitro activity and preclinical pharmacokinetics of the HCV protease inhibitor TMC435350, poster 1390. 58th Annu. Meet. Am. Assoc. Study Liver Dis., Boston, MA, 2 to 6 November 2007.
- 45.Stone, M., S. Jia, W. D. Heo, T. Meyer, and K. V. Konan. 2007. Participation of Rab5, an early endosome protein, in hepatitis C virus RNA replication machinery. J. Virol. 81:4551-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Street, A., A. Macdonald, K. Crowder, and M. Harris. 2004. The hepatitis C virus NS5A protein activates a phosphoinositide 3-kinase-dependent survival signaling cascade. J. Biol. Chem. 279:12232-12241. [DOI] [PubMed] [Google Scholar]
- 47.Sumpter, R., Jr., Y. M. Loo, E. Foy, K. Li, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79:2689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tissari, J., J. Siren, S. Meri, I. Julkunen, and S. Matikainen. 2005. IFN-α enhances TLR3-mediated antiviral cytokine expression in human endothelial and epithelial cells by up-regulating TLR3 expression. J. Immunol. 174:4289-4294. [DOI] [PubMed] [Google Scholar]
- 49.Tscherne, D. M., C. T. Jones, M. J. Evans, B. D. Lindenbach, J. A. McKeating, and C. M. Rice. 2006. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 80:1734-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vercammen, E., J. Staal, and R. Beyaert. 2008. Sensing of viral infection and activation of innate immunity by toll-like receptor 3. Clin. Microbiol. Rev. 21:13-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yi, M., Y. Ma, J. Yates, and S. M. Lemon. 2007. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J. Virol. 81:629-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yi, M., R. A. Villanueva, D. L. Thomas, T. Wakita, and S. M. Lemon. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. USA 103:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, S. Y., E. Jouanguy, S. Ugolini, A. Smahi, G. Elain, P. Romero, D. Segal, V. Sancho-Shimizu, L. Lorenzo, A. Puel, C. Picard, A. Chapgier, S. Plancoulaine, M. Titeux, C. Cognet, H. von Bernuth, C. L. Ku, A. Casrouge, X. X. Zhang, L. Barreiro, J. Leonard, C. Hamilton, P. Lebon, B. Heron, L. Vallee, L. Quintana-Murci, A. Hovnanian, F. Rozenberg, E. Vivier, F. Geissmann, M. Tardieu, L. Abel, and J. L. Casanova. 2007. TLR3 deficiency in patients with herpes simplex encephalitis. Science 317:1522-1527. [DOI] [PubMed] [Google Scholar]
- 56.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu, H., M. Butera, D. R. Nelson, and C. Liu. 2005. Novel type I interferon IL-28A suppresses hepatitis C viral RNA replication. Virol. J. 2:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu, H., and C. Liu. 2003. Interleukin-1 inhibits hepatitis C virus subgenomic RNA replication by activation of extracellular regulated kinase pathway. J. Virol. 77:5493-5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.