Abstract
During a productive infection, species C adenovirus reprograms the host cell to promote viral translation at the expense of cellular translation. The E1B 55-kilodalton (E1B-55K) and E4 open reading frame 6 (E4orf6) proteins are important in this control of gene expression. As part of a ubiquitin-protein ligase, these viral proteins stimulate viral mRNA export, inhibit cellular mRNA export, promote viral gene expression, and direct the degradation of certain host proteins. We report here that the E1B-55K and E4orf6 proteins limited phosphorylation of eIF2α and the activation of the eIF2α kinase PKR. Phospho-eIF2α levels were observed to rise and fall at least twice during infection. The E1B-55K and E4orf6 proteins prevented a third increase at late times of infection. PKR appeared to phosphorylate eIF2α only in the absence of E1B-55K/E4orf6 function. PKR activation and eIF2α phosphorylation was unrelated to the cytoplasmic levels of the adenovirus inhibitor of PKR, VA-I RNA. Nonetheless, expression of a PKR inhibitor, the reovirus double-stranded RNA-binding protein sigma 3, prevented PKR activation and eIF2α phosphorylation. The sigma 3 protein largely corrected the defect in viral late protein synthesis associated with the E1B-55K and E4orf6 mutant viruses without affecting cytoplasmic levels of the late viral mRNA. The ubiquitin-protein ligase activity associated with the E1B-55K/E4orf6 complex was necessary to prevent activation of PKR and phosphorylation of eIF2α. These findings reveal a new contribution of the E1B-55K/E4orf6 complex to viral late protein synthesis and the existence of multiple layers of regulation imposed on eIF2α phosphorylation and PKR activation in adenovirus-infected cells.
Adenovirus is a ubiquitous virus with a double-stranded DNA (dsRNA) genome that infects cells of epithelial and lymphocytic origin. A productive infection in epithelial cells involves the temporal regulation of viral gene expression differentiated by the onset of viral DNA replication. During the late phase, which follows viral DNA replication, adenovirus promotes viral protein synthesis while inhibiting cellular protein synthesis. The inhibition of cellular protein synthesis cannot be attributed to diminished transcription, stability, or integrity of cellular mRNAs; this inhibition reflects changes in the use of cellular mRNA (3). The adenovirus E1B 55-kilodalton (E1B-55K) and E4 open reading frame 6 (E4orf6) proteins, which are synthesized during the early phase of infection, govern the use of cellular and viral mRNA during the late phase of infection. Individually, these proteins serve multiple functions throughout the infectious cycle. The E1B-55K protein directly interferes with the transcriptional activity of p53 (10). The E4orf6 protein promotes efficient viral DNA synthesis (12, 13, 28, 35), stabilizes viral late mRNAs in the nucleus (12, 13, 65, 66), and contributes to splice site selection (45, 46). Both the E1B-55K and E4orf6 proteins are oncoproteins that can block p53-dependent apoptosis (10). Furthermore, the E1B-55K and E4orf6 proteins stimulate the export of viral late mRNA, inhibit cellular mRNA export, promote efficient viral late gene expression, and direct degradation of host proteins that suppress viral replication (reviewed in reference 10). The overlapping activities of the E1B-55K and E4orf6 proteins are most likely due to their incorporation into a novel, multicomponent ubiquitin-protein ligase.
The novel E3 ubiquitin-protein ligase formed during adenovirus infection contains the E1B-55K and E4orf6 proteins, along with the cellular proteins cullin 5 (Cul5), Ring-box 1 (Rbx1), and elongins B and C (29, 58). Because the E4orf6 protein binds the elongin C moiety and the E1B-55K protein is involved in substrate recognition (11), the absence of either viral protein precludes formation of the viral E3 ubiquitin ligase. Cellular proteins targeted by the adenovirus ubiquitin-protein ligase include p53 (29, 58), members of the MRN DNA-damage recognition complex (75), DNA ligase IV (6), and integrin alpha 3 (21). Within the nucleus, the E1B-55K/E4orf6 complex is located at the periphery of viral DNA replication centers (49, 73), where it directs the preferential export of viral late mRNAs from the nucleus to the cytoplasm (5, 12, 53), while simultaneously inhibiting export of cellular mRNAs (9). Although the mechanism underlying the regulation of mRNA export is not fully understood, the ubiquitin ligase activity of the E1B-55K/E4orf6 complex is implicated in this process (17, 82).
The selective control of RNA transport by the E1B-55K/E4orf6 complex is one mechanism underlying the preferential expression of viral late genes (5, 9). Another mechanism contributing to the preferential synthesis of viral late proteins is the selective translation of viral mRNA bearing the tripartite leader. The tripartite leader is a 200-nucleotide, 5′ noncoding sequence added to most viral late mRNAs by differential splicing (1, 14). mRNAs bearing this structured sequence (86) are exported from the nucleus with greater efficiency than cellular mRNA (36). Tripartite leader-bearing mRNAs also are translated more efficiently than typical cellular mRNA in the adenovirus-infected cell (42) by a form of cap-dependent translation initiation known as ribosome shunting (22, 85). Ribosome shunting requires the cap-binding eukaryotic initiation factor 4E protein (eIF4E) (78). However, translation by ribosome shunting continues when eIF4E is underphosphorylated (33, 85, 87). By causing the dephosphorylation of eIF4E (20), the adenovirus L4-100K protein inhibits host protein synthesis (33, 87). The L4-100K protein also promotes viral late protein synthesis (30) through its ability to bind eIF4G and the tripartite leader, which in turn recruits the 40S ribosomal subunit to initiate translation by ribosome shunting (83). Although the E1B-55K/E4orf6 complex may indirectly promote viral late gene expression by promoting expression of the L4-100K gene, we show here that the E1B-55K/E4orf6 complex may play a more direct role in the regulation of translation through its action on additional cellular factors that govern translation.
Another translation factor regulated by adenovirus is eIF2. When the alpha subunit of eIF2 (eIF2α) is phosphorylated, the multisubunit eIF2 protein complex remains tightly associated with its guanine exchange factor eIF2B. This tight association prevents eIF2B from recycling hydrolyzed GDP for GTP on eIF2 (61, 63), thus diminishing translation initiation. Since the amount of eIF2B is typically less than the amount of eIF2 (47), low levels of phospho-eIF2α can inactivate eIF2B leading to the global inhibition of protein synthesis. Four serine/threonine kinases are known to phosphorylate eIF2α at serine 51. These are the heme-regulated inhibitor kinase (HRI), PKR-like endoplasmic reticulum kinase (PERK), general control nonderepressible-2 kinase (GCN2), and the dsRNA-dependent protein kinase (PKR). The eIF2α kinase that is frequently active during virus infection is PKR (25, 56).
Adenovirus expresses at least one gene that prevents PKR-mediated eIF2α phosphorylation. The virus-associated RNA molecule I, or VAI RNA, is a noncoding, 160-nucleotide RNA molecule transcribed by RNA polymerase III that is highly expressed during the late phase of infection (55, 74, 81). VAI RNA directly binds PKR and prevents its activation (44). By preventing PKR activation and subsequent eIF2α phosphorylation (41, 48, 59, 68, 69, 72), VAI RNA is believed to prevent the global inhibition of translation during an adenovirus infection (44, 69, 76, 77). It remains unclear whether or how VAI RNA contributes to ribosome shunting, which is the translation initiation mechanism used to synthesize viral late proteins (85). Another unresolved observation is that phospho-eIF2α remains high in adenovirus-infected cells at certain late times of infection (34). These issues raise the possibility that additional adenovirus gene products may cooperate to modulate the activation of PKR and consequences of eIF2α phosphorylation on viral translation.
In the present study, we show that the E1B-55K and E4orf6 proteins were required to maintain a low level of eIF2α phosphorylation and PKR activation during the late phase of infection and that these activities were correlated with the efficiency of viral late protein synthesis. Regulation of eIF2α phosphorylation and PKR activation during the late phase of infection required the Cul5-mediated E3 ubiquitin-protein ligase activity of the E1B-55K/E4orf6 complex and was unrelated to the cytoplasmic levels of VAI and VAII RNA. These results reveal a previously unknown function of the adenovirus E1B-55K/E4orf6 ubiquitin-protein ligase complex that is important during late-phase infection.
MATERIALS AND METHODS
Cell culture.
Cell culture media, cell culture supplements, and sera were obtained from Invitrogen/Life Technologies (Gaithersburg, MD) or HyClone (Logan, UT) through the Tissue Culture and Virus Vector Core Laboratory of the Comprehensive Cancer Center of Wake Forest University. Cervical carcinoma-derived HeLa cells (ATCC CCL 2; American Type Culture Collection, Manassas, VA) were maintained as monolayer cultures in Dulbecco modified Eagle minimal essential medium (DMEM) supplemented with 10% newborn calf serum. HeLa cells expressing the reovirus type 3 Dearing sigma 3 protein (σ3-HeLa) and their parental cell line, CTRL-HeLa, were kindly provided by G. Parks (Wake Forest University School of Medicine) and previously described (24). The CTRL-HeLa cells were maintained in DMEM supplemented with 10% fetal bovine serum, and σ3-HeLa cells were maintained under selection with DMEM with 10% fetal bovine serum containing 500 μg of G418/ml. Cells were screened for expression of σ3 protein with the σ3 antiserum 4F2, kindly provided by T. Dermody (Vanderbilt University School of Medicine). All cells were incubated at 37°C in a humidified atmosphere containing 5% CO2 and maintained as subconfluent monolayers achieved by passaging two to three times weekly at an appropriate dilution (1:10 to 1:20).
Viruses.
The wild-type virus used throughout the present study is the adenovirus type 5 strain dl309, a phenotypically wild-type virus that lacks the portion of the E3 region that has been shown to be dispensable for growth in tissue culture (39). The E1B-55K mutant virus dl1520 (8) contains an 827-bp deletion within the E1B-55K coding region, in addition to a premature termination codon that prevents expression of smaller splice variants of the E1B-55K protein. The E1B-55K mutant dl338 (53) contains a smaller (524-bp) deletion within the E1B-55K gene and can therefore express the protein variants not expressed by dl1520. The E1B-55K mutant virus dl110 (5) contains a 472-bp deletion within the E1B coding region that causes a frameshift and insertion of a stop codon, leading to expression of a truncated 14-kDa protein. The E4orf6 mutant virus dl355* (35) has a restored E3 region and contains a 14-bp deletion within the E4orf6 gene. The viruses ψ5, ψ5-FL (FL, full-length), and ψ5-NTD (NTD, N-terminal domain) have been previously described (82) and were kindly provided by A. Berk (University of California, Los Angeles). All viruses were grown in HEK293 cells and concentrated virus stocks prepared by sequential centrifugation through CsCl gradients as described previously (70).
Virus infectivity was determined for each of the cell lines (HeLa, CTRL-HeLa, and σ3-HeLa) used in the present study prior to infection. To perform adenovirus infections, cells were seeded ∼16 h prior to infection at a density of 5 to 10 × 104 cells/cm2. At the time of infection, cells were counted, and the appropriate amount of virus required to infect each cell with 10 PFU was diluted in adenovirus infection medium (phosphate-buffered saline [PBS] supplemented with 0.2 mM CaCl2, 2 mM MgCl2, and 2% calf serum). Virus was applied to cells in a volume one-half the normal culture volume, and the culture vessels were gently rocked for 1 h at 37°C. Virus suspension was then removed and replaced with complete culture medium, and the cells were returned to normal culture conditions.
For experiments with the ψ5, ψ5-FL, and ψ5-NTD viral constructs, cells were infected as described previously (82). Briefly, HeLa cells were infected with each Cul5 construct at a multiplicity of infection (MOI) of 50 PFU/cell for 1 h and then virus replaced with complete culture medium. At 16 h postinfection, the same cells were superinfected with dl309 or dl1520 virus at an MOI of 40 PFU/cell for 1 h. After replacement with complete culture medium, the infection was allowed to proceed for ∼30 h, at which time whole-cell lysates were collected for immunoblot analysis.
Viral late protein synthesis.
At each time point indicated, viral late protein synthesis was analyzed by first starving cells for 30 min in methionine- and cysteine-free DMEM for 30 min. Cells were then pulse-labeled for 1 h with ∼0.1 mCi of [35S]methionine (Tran35S-label; MP Biomedicals, Costa Mesa, CA)/ml in methionine- and cysteine-free DMEM supplemented with 2% serum. Whole-cell lysates were collected in the presence of hot (95°C) sodium dodecyl sulfate (SDS) protein sample buffer containing 20% SDS, 0.5 M Tris (pH 6.8), glycerol, 0.01% bromophenol blue, and 2.5% β-mercaptoethanol. Approximately 5 × 103 to 10 × 103 cells were loaded per lane of a 10% polyacrylamide gel and resolved by SDS-polyacrylamide gel electrophoresis (PAGE). PAGE gels were stained with Coomassie blue (50% methanol, 10% acetic acid, 0.25% R-250) for 1 h and then destained overnight in fixative containing 20% glacial acetic acid-7% methanol. Gels were then dried and either exposed to X-ray film and developed or analyzed by phosphorescence imaging using a Molecular Dynamics PhosphorImager. Quantification of protein was performed using ImageQuant analysis software (Molecular Dynamics, Sunnyvale, CA).
Immunoblot analysis and antibodies.
At each time point, whole-cell lysates were collected in the presence of protease and phosphatase inhibitors and resuspended in hot SDS protein sample buffer. Briefly, cells were washed with ice-cold PBS and then incubated minimally with 0.1% trypsin-1 mM EDTA in PBS on ice to disrupt the adherent monolayer. Complete cell culture medium and PBS supplemented with protease and phosphatase inhibitors (2 mM EDTA, 1 mM NaF, 1 mM sodium pyrophosphate, 1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, and 2 μM leupeptin) were added, and the cells were collected and pelleted. Cell pellets were washed twice with PBS plus inhibitors, and the final pellets resuspended in one-tenth volume PBS plus inhibitors at a concentration of 100 μl per 106 cells. Whole-cell lysates were heated at 95°C for 5 min and sonicated, and the debris was deposited by centrifugation. For poly(I:C) treatment, HeLa cells were exposed to 200 ng of poly(I:C) (#tlrl-pic; InvivoGen, San Diego, CA)/ml diluted in complete culture medium for 6, 12, and 24 h, and whole-cell lysates were collected as outlined above.
Protein from an equivalent number of cells was resolved by SDS-PAGE and then transferred to 0.2 μM nitrocellulose (Protran BA83; Whatman, Dassel, Germany). Membranes were blocked with 5% nonfat dry milk in TBS-BGT (all TBS-BGT solutions were made using a final concentration of 0.1% Tween 20 detergent). Separated and immobilized proteins were evaluated with the following antibodies according to the manufacturers' instructions: rabbit polyclonal total eIF4E (catalog no. 9742 [Cell Signaling Technology, Danvers, MA]), rabbit polyclonal phospho-eIF4E (Cell Signaling Technology catalog no. 9741), rabbit polyclonal eIF4GI (either Cell Signaling Technology catalog no. 2498 or Bethyl Laboratories, Inc., [Montgomery, TX] catalog no. A300-502A), rabbit polyclonal phospho-eIF2α(Ser51) (either Cell Signaling Technology catalog no. 9721 or Santa Cruz Biotechnology [Santa Cruz, CA] catalog no. SC-101670), rabbit polyclonal total eIF2α (Cell Signaling Technology catalog no. 9722), rabbit polyclonal eIF2Bɛ (Santa Cruz Biotechnology catalog no. SC-28854), rabbit monoclonal phospho-PKR (Thr446 or Thr451) (Epitomics [Burlingame, CA] catalog no. 1120-1 or 2283-1, respectively), rabbit polyclonal total PKR (Cell Signaling Technology catalog no. 3072), mouse monoclonal β-actin (Sigma [St. Louis, MO], clone AC-15 mouse ascites fluid), and rabbit polyclonal Mre11 (Calbiochem [San Diego, CA] catalog no. PC388). The mouse monoclonal antibody specific for the reovirus σ3 protein (antibody 4F2) was kindly provided by T. Dermody (Vanderbilt University). Rabbit antiserum specific for the Ad5 virion was raised against purified, disrupted Ad5 virions as described previously (82) and was kindly provided by A. Berk (University of California, Los Angeles). Immune complexes were visualized with horseradish peroxidase-conjugated secondary antibodies from Jackson Immunoresearch Laboratories (West Grove, PA) and subsequent incubation with SuperSignal chemiluminescent substrate from Pierce (Rockford, IL).
Cell fractionation and RNA isolation.
All solutions used for RNA purification were prepared using diethyl pyrocarbonate-treated RNase-free water. At 32 or 36 h postinfection, approximately 6 × 105 mock-infected or infected HeLa, CTRL-HeLa, or σ3-HeLa cells were scraped into ice-cold PBS and then pelleted at 4°C at 400 × g. Pellets were resuspended in PBS, transferred to a microtube, and pelleted at 4°C at 800 × g. Pellets were resuspended in isotonic buffer (10 mM NaCl, 10 mM Tris-Cl [pH 7.4], 3 mM MgCl2) supplemented with a 1/20 volume vanadyl ribonucleoside complex and incubated on ice for 5 min. While gently mixing, an equal volume of isotonic buffer supplemented with detergent (10% sodium deoxycholate, 20% Tween 40) was added, and the cells incubated on ice for an additional 5 min. After centrifugation at 4°C at 1,000 × g, the cytoplasmic fraction was collected. Cytoplasmic RNA was further purified by using TRIzol LS reagent (Life Technologies) and resuspended in 10 μM sodium acetate. RNA preparations were not subjected to any form of DNase treatment after purification. Concentrations of RNA were determined by using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific).
Quantitative reverse transcriptase real-time PCR.
Approximately 25 ng of cytoplasmic RNA was reverse transcribed using qScript cDNA SuperMix (Quanta Biosciences, Inc., Gaithersburg, MD) according to the manufacturer's instructions using an Eppendorf MasterCycler EP (Westbury, NY). Subsequently, one-tenth of the reaction volume was used for quantitative PCR using the PerfeCTa SYBR green FastMix, ROX (Quanta Biosciences, Inc., Gaithersburg, MD). Control reactions were performed that contained RNA in the absence of reverse transcriptase enzyme present in the cDNA SuperMix. Briefly, triplicate 20-μl reactions were assembled and contained cDNA or viral DNA, PerfeCTa SYBR green FastMix, forward and reverse primers (250 nM final concentrations each), and nuclease-free water. Reaction mixes were incubated using a revised protocol as follows: 50°C for 2 min, 95°C for 1 min, and 40 cycles of 95°C for 15 s, 60°C for 1 min. Reactions were performed and analyzed by using the ABI Prism 7000 sequence detection system (Applied Biosystems, Forest City, CA). The following primers were used in quantitative PCRs: VAI RNA-fwd, 5′-ACT CTT CCG TGG TCT GGT GGA TAA-3′; VAI RNA-rev, 5′-TTG TCT GAC GTC GCA CAC CT-3′; VAII RNA-fwd, 5′-TTT CCA AGG GTT GAG TCG CGG-3′; VAII RNA-rev, 5′-TGT TTC CGG AGG AAT TTG CAA GCG-3′; hexon-fwd, 5′-ACC CAT TTA ACC ACC ACC GCA ATG-3′; and hexon-rev, 5′-TAA TGC TGG CTC CGT CAA CCC TTA-3′. To present the VAI and VAII RNA results, the cycle threshold calculated for each reaction was compared to a standard curve generated with adenovirus genome concentrations ranging between 1 and 107 genomes per reaction. To present the hexon mRNA results in CTRL-HeLa and σ3-HeLa cells, the cycle threshold calculated for each reaction was compared to a standard curve generated by reverse transcription of a serial dilution of cytoplasmic extracts from wild-type virus-infected cells prepared in uninfected cell lysates.
RESULTS
The E1B-55K and E4orf6 proteins are required for efficient viral late protein synthesis.
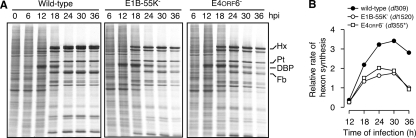
The rate of protein synthesis was measured during infection with the wild-type adenovirus and E1B-55K and E4orf6 mutant viruses. At various times after infection, HeLa cells were pulse-labeled with radioactive amino acids, and proteins were subsequently analyzed by SDS-PAGE and autoradiography (Fig. 1A). Viral protein synthesis in both mutant and wild-type virus-infected cells peaked between 24 and 30 h postinfection. As expected, the rate of viral late protein synthesis directed by the E1B-55K and E4orf6 mutant viruses was less than the rate directed by the wild-type virus. The overall rate of hexon protein synthesis was two- to threefold higher in wild-type virus-infected cells than in mutant virus-infected cells (Fig. 1B). These results are in accord with previously published reports showing that that the E1B-55K and E4orf6 proteins are required for efficient viral late protein synthesis.
FIG. 1.
The E1B-55K and E4orf6 proteins are required for efficient viral late protein synthesis. (A) HeLa cells were mock infected or infected at an MOI of 10 PFU per cell with wild-type dl309, E1B-55K mutant virus dl1520, or the E4orf6 mutant virus dl355*. At the times indicated, cells were pulse-labeled for 1 h with radioactive amino acids. Proteins from 104 cells (per lane) were resolved by SDS-PAGE and visualized by exposure to X-ray film. Labels for hexon (Hx), penton base (Pt), fiber (Fb), and E2A DNA-binding protein (DBP) are shown at the right of the image. The results are representative of at least three experiments. (B) The rate of hexon synthesis is shown as a function of time postinfection. The radioactivity present in the band corresponding to hexon at 120 kDa was quantified from an image obtained by phosphorescence imaging. The background of cellular protein synthesis was subtracted from each lane's hexon measurement. The rate of hexon synthesis measured at 36 h postinfection in cells infected with dl1520 was set to a value of 1.
A low level of eIF2α phosphorylation is maintained by the E1B-55K and E4orf6 proteins during the late phase of infection.
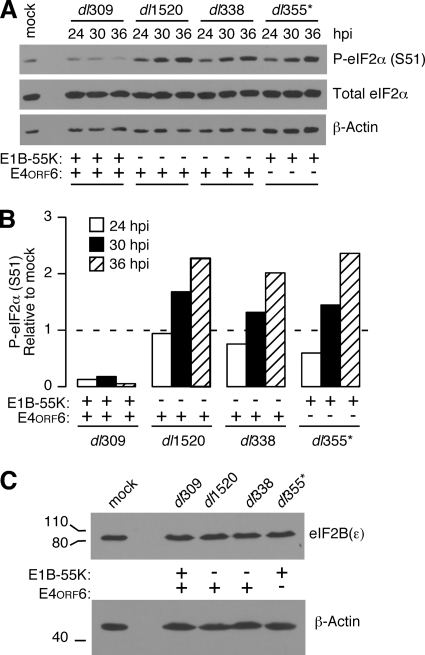
The level and phosphorylation of eIF4E, eIF4G, and eIF2α were compared among HeLa cells infected with the wild-type virus, two different E1B-55K mutant viruses, or an E4orf6 mutant virus. At various times during the late phase of infection, the total level and phosphorylation of translation factors were evaluated by immunoblotting. There were no significant differences in the total level or phosphorylation of eIF4E and eIF4G during wild-type and mutant virus infections (data not shown). However, striking differences were observed in the phosphorylation of eIF2. There was a substantial decrease in the relative phosphorylation of serine 51 on the alpha subunit of eIF2 in wild-type virus-infected cells compared to cells infected with E1B-55K and E4orf6 mutant viruses (Fig. 2A). The level of eIF2α phosphorylation in wild-type virus-infected cells was lower than that observed in mock-infected cells, but continually increased over time in cells infected with both E1B-55K and E4orf6 mutant viruses. By 36 h postinfection, there was a 7- to 10-fold increase in the phosphorylation of eIF2α in mutant virus-infected cells compared to wild-type virus-infected cells (Fig. 2B). Similar results were obtained in HeLa cells infected with a third E1B-55K mutant virus, dl110 (data not shown). These results suggest that the E1B-55K and E4orf6 proteins maintain a low level of eIF2α phosphorylation during the late phase of adenovirus infection.
FIG. 2.
A low level of eIF2α phosphorylation is maintained by the E1B-55K and E4orf6 proteins during the late phase of infection. (A) HeLa cells were infected at an MOI of 10 PFU per cell with wild-type dl309, E1B-55K mutant viruses dl1520 and dl338, or the E4orf6 mutant virus dl355*. At 24, 30, and 36 h postinfection, whole-cell lysates were collected and analyzed by immunoblotting for levels of phospho-eIF2α (S51; serine 51), total eIF2α, and β-actin. (B) The relative phosphorylation of eIF2α was calculated from the immunoblots in panel A by normalizing the level of phospho-eIF2α to the total level of eIF2α. The quantity of relative phospho-eIF2α in mock-infected cells was set to a value of 1. (C) Whole-cell lysates from mock-infected or HeLa cells infected for 36 h with dl309, dl1520, dl338, and dl355* were evaluated by immunoblotting with an antibody to the epsilon subunit of the translation factor eIF2B. An immunoblot for β-actin is shown as a loading control.
Phosphorylation of eIF2α diminishes the rate of protein synthesis by usurping the guanine exchange factor eIF2B, a factor whose cellular concentration is limiting. The level of eIF2α phosphorylation measured during E1B-55K and E4orf6 mutant virus infections would normally be associated with a near complete inhibition of protein synthesis (57). However, as shown in Fig. 1, a considerable level of protein synthesis continues during these infections. A potential mechanism that could allow continued protein synthesis in the presence of such high levels of eIF2α phosphorylation would be increased expression of the factor eIF2B. To determine whether eIF2B levels increase during adenovirus infection, the level of the largest subunit of eIF2B, ɛ, which contains the catalytic subunit of eIF2B (26), was measured by immunoblotting. However, the level of eIF2Bɛ remained constant regardless of the presence or absence of the E1B-55K and E4orf6 proteins (Fig. 2C). Therefore, it seems unlikely that a compensatory increase in eIF2B levels allowed for sustained levels of protein synthesis in cells containing high levels of phospho-eIF2α.
Phosphorylation of eIF2α rises and falls during adenovirus infection and is limited at late times of infection by the E1B-55K and E4orf6 proteins.
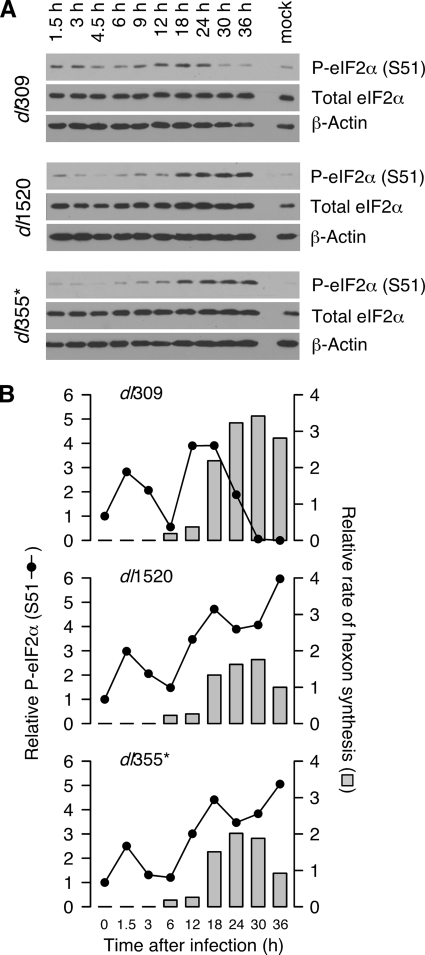
To understand when the E1B-55K and E4orf6 proteins affect eIF2α phosphorylation, eIF2α phosphorylation was measured throughout the course of either the wild-type or mutant virus infections. HeLa cells were infected with the viruses indicated in Fig. 3, and the total amount of eIF2α and level of phosphorylation on Ser51 were analyzed by immunoblotting (Fig. 3A). During the early phase of infection, phosphorylation of eIF2α was similar in both wild-type and mutant virus infections. The level of eIF2α phosphorylation increased to approximately threefold above the level found in mock-infected cells as early as 90 min postinfection. It then rapidly decreased to a level near that measured in uninfected cells by 6 h postinfection. At this time, eIF2α phosphorylation increased again and continued to increase until 12 to 18 h postinfection. However, the pattern of eIF2α phosphorylation in wild-type and mutant virus-infected cells began to differ at the onset of the late phase of infection. In cells infected with the wild-type virus, eIF2α phosphorylation began to drop at 18 h postinfection and continued dropping to levels that were nearly undetectable by 30 to 36 h. In contrast, levels of phospho-eIF2α increased steadily in cells infected with the E1B-55K or E4orf6 mutant virus, reaching a level three- to fivefold greater than that in mock-infected cells and more than 10-fold greater than that in wild-type virus-infected cells (Fig. 3B). These results indicate that the E1B-55K and E4orf6 proteins reduce phosphorylation of eIF2α during the late phase of infection, often below that measured in mock-infected cells. Although phosphorylation of eIF2α showed a multiphasic nature in multiple experiments, the molecular basis for this pattern remains unclear.
FIG. 3.
Phosphorylation of eIF2α fluctuates during adenovirus infection and is kept low at late times of infection by the E1B-55K and E4orf6 proteins. (A) Whole-cell lysates were collected at various times postinfection from HeLa cells infected at an MOI of 10 PFU/cell with dl309, dl1520, or dl355*. Protein from an equal number of cells (6 × 104) was analyzed by immunoblotting for phospho-eIF2α (S51; serine 51), total eIF2α, or β-actin as a loading control. (B) Total and phospho-eIF2α were measured from the immunoblots in panel A by densitometry, and the relative phosphorylation of eIF2α (P-eIF2α; dotted black line) was calculated by normalizing the level of phosphorylated eIF2α to the level of total eIF2α. The value of relative phospho-eIF2α measured in mock-infected cells was set to a value of 1. The rates of hexon synthesis from Fig. 1B (gray bars) are compared here with the relative phospho-eIF2α level. The value of hexon synthesis at 36 h postinfection in cells infected with dl1520 was set to a value of 1.
To determine how eIF2α phosphorylation correlated with viral late protein synthesis, the rate of hexon synthesis (reported previously in Fig. 1B) and the levels of phospho-eIF2α (Fig. 3B) were compared. First, the onset of hexon protein synthesis coincided with the precipitous drop in phospho-eIF2α observed at 6 to 9 h postinfection. Second, the elevated rates of viral late protein synthesis observed in wild-type virus-infected cells coincided with a reduction in phospho-eIF2α levels between 24 and 36 h postinfection. This analysis suggests that an inverse correlation exists between eIF2α phosphorylation and the rate of viral late protein synthesis. We suggest that the ability of the E1B-55K and E4orf6 proteins to reduce eIF2α phosphorylation during the late phase of infection may be necessary for efficient viral late protein synthesis.
The E1B-55K and E4orf6 proteins prevent PKR activation at very late times of infection.
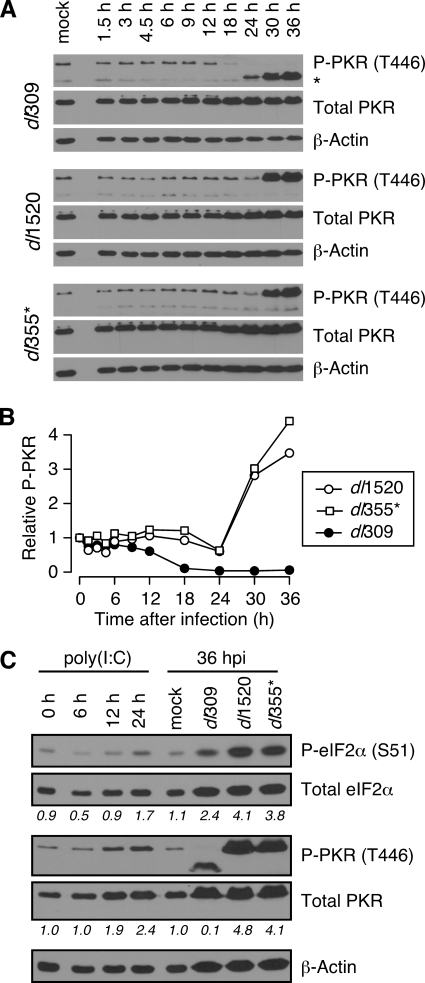
PKR is frequently the active eIF2α kinase during many virus infections (25). The results presented in Fig. 2 and 3 establish that the E1B-55K and E4orf6 proteins prevent phosphorylation of eIF2α at late times of infection. Therefore, we wanted to determine whether the E1B-55K and E4orf6 proteins also prevent activation of PKR. Phosphorylation of two residues that are critical for PKR kinase activity, threonine 446 (Thr446) and threonine 451 (Thr451) (60), was measured by immunoblotting. Similar results were obtained with antibodies for each phosphorylated residue, and only representative results for phospho-Thr446 are shown (Fig. 4A). Surprisingly, unlike the fluctuating levels of phospho-eIF2α, PKR activation remained at or near basal levels throughout the early phase of both wild-type and E1B-55K and E4orf6 mutant virus infections. It therefore seems unlikely that PKR is responsible for the increase in phospho-eIF2α observed during the early phase of infection, unless the activation of PKR is below the threshold of detection by immunoblotting.
FIG. 4.
The E1B-55K and E4orf6 proteins prevent PKR activation at very late times of infection. (A) Whole-cell lysates were collected at various times postinfection from HeLa cells infected at an MOI of 10 PFU per cell with dl309, dl1520, or dl355*. Protein from an equal number of cells (6 × 104) was analyzed by immunoblotting for phospho-PKR (T446; threonine 446), total PKR, or β-actin as a loading control. An unidentified lower-molecular-weight species is observed at very late times during wild-type, but not mutant, virus infections and can be seen in the phospho-PKR panel (*). Similar results (not shown) were seen with an antibody specific to phosphorylated threonine 451. (B) The level of phosphorylated and total PKR was measured by densitometry and the relative phosphorylation of PKR (P-PKR; phospho-PKR) was calculated by normalizing the level of phosphorylated PKR (Thr446) to the level of total PKR. The graph shows the level of relative phospho-PKR as a function of time postinfection (h). The level of phospho-PKR measured in mock-infected cells was set to a value of 1. The results presented in panels A and B are representative of three experiments using antibodies to phospho-PKR (Thr466). (C) Mock-infected HeLa cells were treated with 200 ng of poly(I:C)/ml, and whole-cell lysates were collected at 6, 12, and 24 h posttreatment. Protein from an equal number of cells (6 × 104) was analyzed by immunoblotting for phospho-PKR (Thr446), total PKR, phospho-eIF2α, total eIF2α, or β-actin as a loading control. The numbers shown beneath each lane represent the relative phosphorylation of eIF2α and PKR as quantified by densitometry. The values measured in mock-treated cells was set to a value of 1.
Like eIF2α phosphorylation, the pattern of PKR activation differed in wild-type and mutant virus-infected cells during the late phase of infection. In cells infected with the wild-type virus, PKR phosphorylation began to decrease at 12 h postinfection to levels even lower than those observed in mock-infected cells (Fig. 4B). This decrease may be a result of increased VAI RNA expression that occurs at the onset of the late phase of infection (74). Although we were unable to detect PKR phosphorylation during the late phase of wild-type adenovirus infection, we consistently observed the appearance of an unidentified immunoreactive species of approximately 58 to 60 kDa between 24 and 36 h postinfection (see the top panel of Fig. 4A) using antibodies to phospho-Thr446, but not total PKR. Although PKR can be cleaved during caspase-dependent apoptosis (62), the apoptotic fragments are smaller than those observed here, and we detect no caspase activation during these adenovirus infections (unpublished data). This suggests that the immunoreactive species is unlikely to be a proteolytic fragment of PKR, and the origin of this species remains unknown.
Conversely, in cells infected with the E1B-55K or E4orf6 mutant virus, PKR activation remained low until rising drastically between 24 and 30 h postinfection with increased phosphorylation observed at 36 h postinfection. The elevation in active PKR mirrored the increase in phospho-eIF2α measured in mutant virus-infected cells at 30 and 36 h postinfection (Fig. 3). Taken together, these results are consistent with the idea that PKR may be responsible for the high level and continued increase of eIF2α phosphorylation observed at very late times after infection with E1B-55K and E4orf6 mutant viruses. These data also indicate that the E1B-55K and E4orf6 proteins are required to prevent PKR activation and subsequent eIF2α phosphorylation at very late times of infection. In addition, these data suggest that an additional activity or kinase other than PKR must contribute to the phosphorylation of eIF2α at early times since PKR did not appear to be activated until very late times of infection.
To confirm the identify of the proteins detected by the antibodies used here and to compare the activation observed during adenovirus infection to another stimulus, infected cell lysates were analyzed in parallel with lysates obtained from cells exposed to polyinosine-poly(C) or poly(I:C), a known inducer of PKR activation and eIF2α phosphorylation. Exposure to poly(I:C) caused an increase in PKR activation over time, reaching approximately twofold that observed in mock-treated HeLa cells by 24 h posttreatment (Fig. 4C). Strikingly, the level of PKR activation achieved with poly(I:C) treatment never reached the level observed in cells infected with the E1B-55K and E4orf6 mutant viruses (approximately five- and fourfold greater, respectively, compared to mock-infected cells). Although the effect was not as pronounced as on PKR, phosphorylation of eIF2α also increased over time and was almost twofold greater than that in mock-treated cells by 24 h posttreatment. Again, the level of eIF2α phosphorylation induced by poly(I:C) treatment was lower than that in cells infected with E1B-55K and E4orf6 mutant viruses. More importantly, the proteins identified as activated PKR and phosphorylated eIF2α during poly(I:C) treatment were identical to those observed in adenovirus-infected cells. Therefore, we are confident of the identify of the proteins identified as phosphorylated eIF2α and phosphorylated PKR from cells infected with E1B-55K and E4orf6 mutant viruses.
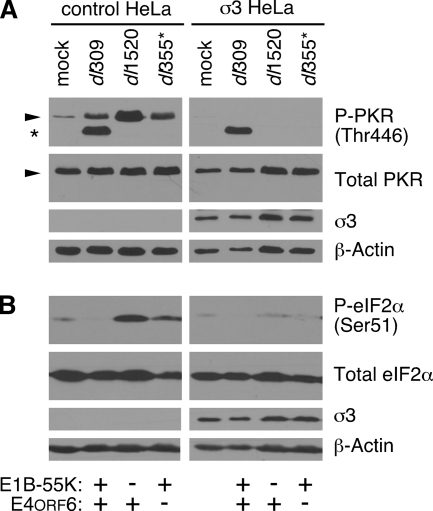
The reovirus σ3 protein prevents PKR activation and eIF2α phosphorylation during E1B-55K and E4orf6 mutant virus infections at very late times.
The E1B-55K and E4orf6 proteins prevent eIF2α phosphorylation during the late phase of infection (Fig. 3) and prevent PKR activation at very late times of infection (Fig. 4); these functions were correlated with efficient viral late protein synthesis (Fig. 3B). To determine whether the phosphorylation of PKR and eIF2α were responsible for the differential rates of viral late protein synthesis, we analyzed HeLa cells stably expressing the reovirus σ3 protein (σ3-HeLa). The reovirus σ3 protein sequesters dsRNA to prevent PKR activation and kinase activity (67, 84). We hypothesized that blocking the activation of PKR observed at very late times of infection would prevent the increase in eIF2α phosphorylation in cells infected with E1B-55K and E4orf6 mutant viruses. If the phosphorylation of these proteins suppressed viral late protein synthesis in the mutant virus-infected cells, we would anticipate that preventing PKR activation and eIF2α phosphorylation should restore levels of protein synthesis to wild-type levels during the very late phase of infection. The σ3-HeLa cells or their parental cell line (CTRL-HeLa) were mock infected or infected with wild-type virus, E1B-55K mutant virus, or E4orf6 mutant virus. After 36 h, the levels of eIF2α phosphorylation and PKR phosphorylation were evaluated (Fig. 5). Expression of the reovirus σ3 protein prevented phosphorylation of PKR during wild-type and E1B-55K and E4orf6 mutant virus infections (Fig. 5A). Curiously, the unidentified immunoreactive species first identified by the phospho-Thr446 PKR antibody in Fig. 4B was detected in wild-type-infected CTRL-HeLa cells and remained detectable when PKR phosphorylation was inhibited in σ3-HeLa cells. Phosphorylated eIF2α was nearly undetectable in σ3-HeLa cells that were infected with the E1B-55K and E4orf6 mutant viruses (Fig. 5B). These results show that stable expression of the reovirus σ3 protein is sufficient to block PKR activation at very late times postinfection and prevent eIF2α phosphorylation in cells infected with E1B-55K and E4orf6 mutant viruses. These findings also support the hypothesis that PKR mediates phosphorylation of eIF2α at very late times in cells infected with mutant viruses lacking the E1B-55K and E4orf6 proteins.
FIG. 5.
The reovirus σ3 protein prevents PKR activation and eIF2α phosphorylation during E1B-55K and E4orf6 mutant virus infections at very late times. (A) CTRL-HeLa cells and σ3-HeLa cells were mock infected or infected at an MOI of 10 PFU/cell with dl309, dl1520, or dl355*, and whole-cell lysates were collected at 36 h postinfection. Protein from an equal number of cells (6 × 104) was analyzed by immunoblot analysis for the levels of phospho-PKR (P-PKR; phospho-PKR, Thr446; threonine 446), total PKR, and β-actin. Stable expression of the reovirus sigma 3 protein (σ3) was verified using the 4F2 antibody. (B) The whole-cell lysates used in panel A were analyzed similarly by immunoblotting, but for the levels of phospho-eIF2α (S51; serine 51), total eIF2α, and β-actin. Stable expression of the reovirus sigma 3 protein (σ3) was verified using the 4F2 antibody.
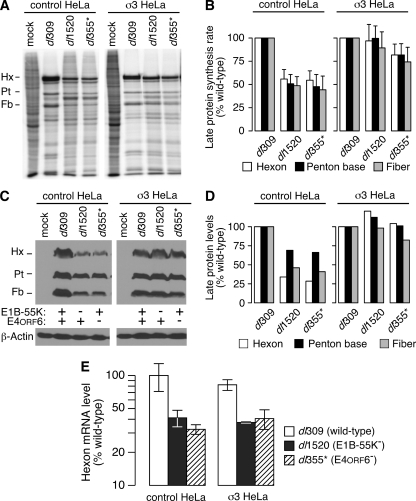
The reovirus σ3 protein corrects the defect in viral late protein synthesis during E1B-55K and E4orf6 mutant virus infections without restoring viral late mRNA export.
We next evaluated the effects of σ3 protein expression on the rate of synthesis and total accumulation of the viral late proteins hexon, penton, and fiber during E1B-55K and E4orf6 mutant virus infections. The rate of viral late protein synthesis was measured by pulse-labeling with radioactive amino acids at 36 h postinfection and subsequent quantitative phosphorescence imaging (Fig. 6A and B). The total accumulation of viral late proteins was measured by immunoblotting with an antibody specific to the adenovirus 5 virion (82) (Fig. 6C) and quantified by densitometry (Fig. 6D). Consistent with published reports (28, 53, 80), the rate of synthesis and total accumulation of viral late proteins were significantly reduced in CTRL-HeLa cells infected with either the E1B-55K mutant or the E4orf6 mutant viruses in comparison to wild-type virus-infected CTRL-HeLa cells. Strikingly, the defect in both the rate of viral late protein synthesis (Fig. 6A and B) and accumulation of viral late proteins (Fig. 6C and D) was corrected in σ3-HeLa cells infected with the E1B-55K and E4orf6 mutants. These results suggest that inhibiting PKR activation and phosphorylation of eIF2α is sufficient to restore viral late protein synthesis to wild-type levels in the absence of the E1B-55K and E4orf6 proteins at very late times.
FIG. 6.
The reovirus σ3 protein corrects the defect in viral late protein synthesis during E1B-55K and E4orf6 mutant virus infections without restoring viral late mRNA transport. (A) CTRL-HeLa and σ3-HeLa cells were either mock infected or infected at an MOI of 10 PFU per cell with dl309, dl1520, or dl355*. At 36 h postinfection, cells were pulse-labeled for 1 h with radioactive amino acids. Protein from 104 cells was loaded per lane and separated by SDS-PAGE and then visualized by phosphorescence imaging. The image shown is representative of at least three experiments. The positions of the viral late proteins hexon (Hx), penton base (Pt), and fiber (Fb) are indicated to the left of the image. (B) The radioactivity present in the bands corresponding to hexon, penton, and fiber was quantified by phosphorimaging from three independent experiments and represents the rate of synthesis of each protein. Rates of synthesis during infection with wild-type virus were set to 100% for each cell line. Error bars indicate the standard error of the mean. A two-tailed t test corrected for multiple comparisons indicates that the rates of synthesis during wild-type and mutant virus infections were significantly different in CTRL-HeLa cells (all P values were <0.03) but were indistinguishable in σ3-HeLa cells (all P values were >0.6). (C) Whole-cell lysates were collected at 36 h postinfection from CTRL-HeLa and σ3-HeLa cells that were mock infected or infected at an MOI of 10 PFU/cell with dl309, dl1520, or dl355*. Lysates were analyzed by immunoblotting for steady-state accumulation of the viral late proteins hexon, penton, and fiber by using antisera specific to the Ad5 virion. The level of β-actin protein is shown as a loading control. The immunoblot shown is representative of three experiments. (D) The total levels of hexon, penton, and fiber from the immunoblot shown in panel C were quantified by densitometry. The level of each protein in CTRL-HeLa or σ3-HeLa cells during the wild-type infection was set to 100%. (E) CTRL-HeLa and σ3-HeLa cells were infected at an MOI of 10 PFU/cell with dl309, dl1520, or dl355*, and cytoplasmic RNA was isolated at 36 h postinfection. Approximately 25 ng of purified RNA was reverse-transcribed to cDNA or incubated in the absence of reverse transcriptase and subsequently analyzed by quantitative PCR to measure the level of hexon mRNA. Cycle thresholds were compared to a standard curve generated using a titration of cytoplasmic RNA from wild-type virus-infected CTRL-HeLa cells that also underwent reverse transcription and quantitative PCR analysis. The value measured in CTRL-HeLa cells infected with wild-type virus was set to 100%. The data shown are representative of multiple experiments, and error bars represent the standard deviation between experiments. A two-tailed t test adjusted for multiple comparisons indicates there is significant variation between wild-type and mutant virus-infected cells (P < 0.005) and no difference among viruses between cell lines (P > 0.16). The results obtained in the absence of reverse transcriptase are not shown since the level of detection was negligible and generally more than 100-fold lower than the signal generated in its presence.
The E1B-55K and E4orf6 proteins promote the preferential export of viral late mRNA (5, 12, 53), and this is believed to be one mechanism by which the E1B-55K and E4orf6 proteins promote efficient viral late gene expression (5, 9). To determine whether the reovirus σ3 protein corrected the defect in viral late protein synthesis by restoring viral late RNA export to wild-type levels, we determined the levels of hexon mRNA by quantitative reverse transcriptase followed by real-time PCR. Cytoplasmic levels of hexon mRNA were measured at 36 h postinfection in CTRL-HeLa and σ3-HeLa cells that had been mock infected or infected with wild-type, E1B-55K mutant, or E4orf6 mutant viruses. As expected (5, 12, 53), CTRL-HeLa cells infected with viruses lacking the E1B-55K or E4orf6 proteins contained approximately twofold less hexon mRNA compared to wild-type virus-infected cells (Fig. 6E). We observed nearly identical and statistically indistinguishable results in σ3-HeLa cells infected with wild-type or E1B-55K and E4orf6 mutant viruses (Fig. 6E). Therefore, the ability of the reovirus σ3 protein to restore viral late protein synthesis in the absence of the E1B-55K and E4orf6 proteins cannot be explained by a restoration in viral late mRNA export. Taken together, these results suggest that the E1B-55K and E4orf6 proteins prevent PKR activation and eIF2α phosphorylation during the late phase of infection to promote efficient viral late translation.
The steady-state level of cytoplasmic VA RNA is similar in wild-type and mutant virus-infected cells during the late phase of infection.
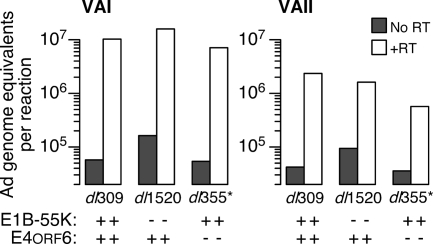
The results reported in Fig. 6 show that preventing eIF2α phosphorylation and PKR activation can significantly compensate for the absence of the E1B-55K and E4orf6 proteins at very late times of infection. Adenovirus expresses at least one gene product, VAI RNA that has been shown to prevent PKR activation and block the phosphorylation of eIF2α (2, 41, 68). Although studies by Babiss and Ginsberg (4) found that VAI RNA levels were equivalent in wild-type virus-infected and E1B-55K mutant virus-infected cells between 2 to 14 h postinfection, it remained possible that the E1B-55K and E4orf6 proteins maintained high levels of VAI RNA at very late times of infection.
Reverse transcription, followed by real time PCR, was used to quantify the level of VAI RNA and VAII RNA in the cytoplasm of infected cells at 32 and 36 h postinfection. Representative results of an analysis performed with RNA collected at 32 h postinfection are shown in Fig. 7, and equivalent results were observed at 36 h postinfection. No signal was detected in mock-infected cells. Because the PCR primers would amplify both genomic DNA and cDNA, the level of target measured in samples that lack reverse transcriptase (“No RT”) show that no more than 0.025 to 0.1% of total viral DNA present in the infected HeLa cell was recovered in the cytoplasmic fraction (Fig. 7). At 32 h postinfection, VAI RNA levels were nearly equivalent in wild-type virus-infected and mutant virus-infected HeLa cells (Fig. 7). As expected, the levels of VAII RNA were ∼10-fold less than the levels of VAI RNA in all infected cells (74). We also found no significant difference in VAII RNA levels in wild-type virus-infected cells compared to mutant virus-infected cells (Fig. 7). These results corroborate and extend those previously reported (4) and suggest that the E1B-55K and E4orf6 proteins do not influence the cytoplasmic accumulation of VAI RNA and VAII RNA during an adenovirus infection in HeLa cells.
FIG. 7.
The steady-state level of cytoplasmic VA RNA is similar in wild-type and mutant virus-infected cells during the late phase of infection. HeLa cells were mock infected or infected at an MOI of 10 PFU per cell with dl309 or dl1520. At 32 h postinfection, cytoplasmic RNA was isolated, and 25 ng of purified RNA was reverse transcribed to cDNA or incubated in the absence of reverse transcriptase (No RT; no reverse transcriptase). The cDNA was subsequently analyzed by quantitative PCR to measure the level of VAI and VAII RNA. Cycle thresholds were compared to a standard curve of viral genomic DNA equivalents, and data are presented as adenovirus genome equivalents per reaction. The data shown are representative of results from six experiments, performed at three late times postinfection using multiple primer sets.
A Cul5-mediated activity of the E1B-55K/E4orf6 complex is required to prevent eIF2α phosphorylation and PKR activation during the late phase of infection.
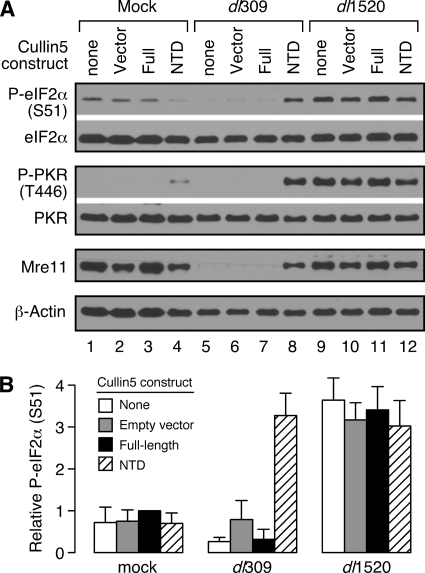
Results presented thus far indicate that both E1B-55K and E4orf6 mutant viruses share a similar phenotype with respect to viral late protein synthesis (Fig. 1), eIF2α phosphorylation (Fig. 2 and 3), PKR activation (Fig. 4), and the ability of the reovirus σ3 protein to correct the defect in viral late protein synthesis (Fig. 6). Because of these similarities, it seems likely that eIF2α phosphorylation and PKR activation are regulated by the E3 ubiquitin-protein ligase complex that includes these two viral proteins. A critical component of this E3 ubiquitin ligase is Cul5 (58). Woo and Berk created and characterized a dominant-negative Cul5 protein (82) by deleting the carboxy-terminal domain responsible for interaction with Rbx1, a protein that recruits ubiquitin-conjugating machinery (40). The remaining amino-terminal portion of Cul5 retains its ability to bind elongins B and C and the E1B-55K and E4orf6 proteins, but the complex cannot ubiquitinate targets due to the absence of Rbx1. This tool is especially valuable because it allows specific inhibition of Cul5-mediated degradation by the E1B-55K/E4orf6 complex. Expression of the dominant-negative amino-terminal domain of Cul5 prior to wild-type adenovirus infection was shown to reduce viral late protein synthesis directed by the wild-type virus to levels similar to that measured during infection with an E1B-55K mutant virus (82). This observation, taken together with results described here indicating that eIF2α phosphorylation is inversely proportional to viral late protein synthesis, prompted us to investigate whether the reduction of eIF2α phosphorylation and inhibition of PKR activation also requires the specific ubiquitin-ligase activity of the E1B-55K/E4orf6 complex.
To address this query, HeLa cells were infected with Ad5 vectors lacking an insert (ψ5-vector), expressing the full-length Cul5 (ψ5-full), or expressing the dominant-negative Cul5 amino-terminal domain (ψ5-NTD) that were described previously (82). After 16 h, the cells were infected again (superinfected) with wild-type virus or E1B-55K mutant virus for 30 h, and phosphorylation of eIF2α and PKR was evaluated by immunoblotting (Fig. 8A). Expression of the ψ5-vector and ψ5-full Cul5 constructs had virtually no effect on the expected level of phospho-eIF2α or phospho-PKR (compare lane 1 to lanes 2 and 3; lane 5 to lanes 6 and 7; and lane 9 to lanes 10 and 11 in Fig. 8A; see also Fig. 8B). For both empty-vector and full-length Cul5 controls, the levels of phospho-eIF2α and phospho-PKR were low in mock-infected and wild-type virus-infected cells but severalfold higher in cells infected with the E1B-55K mutant virus. Furthermore, expression of the ψ5-NTD dominant-negative Cul5 construct had no significant effect on phospho-eIF2α and phospho-PKR levels in mock-infected and E1B-55K mutant virus-infected cells (see lanes 4 and 12 of Fig. 8A and B).
FIG. 8.
A Cul5-mediated activity of the E1B-55K/E4orf6 complex is required to prevent eIF2α phosphorylation and PKR activation during the late phase of infection. (A) HeLa cells were mock preinfected or preinfected at an MOI of 50 PFU per cell with either ψ5 (vector), ψ5-FL (full-length Cul5), or ψ5-NTD (N-terminal domain Cul5) cullin 5 viral construct for 16 h. Cells were then either mock superinfected or superinfected at an MOI of 40 PFU/cell with dl309 or dl1520 viruses. At 30 h postinfection, whole-cell lysates were collected and analyzed by immunoblotting for phospho-eIF2α, total eIF2α, phospho-PKR (Thr446), total PKR, Mre11, or β-actin. The results shown in the figure are representative of multiple experiments. (B) The level of relative phospho-eIF2α (P-eIF2α; S51, serine 51) was calculated as described in Fig. 3. A pairwise, two-tailed t test with corrections for multiple comparisons indicates that dl309-NTD is significantly different from dl309-none (P = 0.0029), dl309-Vector (P = 0.0064), and dl309-Full (P = 0.0029). All treatments in dl1520-infected cells are statistically indistinguishable (P > 0.5), as are the values comparing dl309-NTD and all treatments in dl1520-infected cells (P > 0.99). The data represent combined results from three independent experiments. Error bars show the standard error of the mean.
In contrast, expression of the ψ5-NTD Cul5 construct had a profound effect on the status of eIF2α and PKR in wild-type virus-infected cells. Strikingly, cells that were preinfected with the ψ5-NTD construct and then superinfected with the wild-type virus strongly resembled the mutant virus, at least with respect to eIF2α phosphorylation and PKR activation (compare lane 8 to lanes 9 to 12 in Fig. 8A). When the adenoviral E3 ubiquitin-protein ligase complex was inactivated in wild-type adenovirus-infected cells, the levels of phospho-eIF2α and phospho-PKR were severalfold higher than the levels in cells with an intact adenoviral E3 ubiquitin-protein ligase complex. Furthermore, preinfection with the dominant-negative Cul5 construct lead to levels of phospho-eIF2α in wild-type virus-infected cells that were statistically indistinguishable from that measured in E1B-55K mutant virus-infected cells (Fig. 8B). Verification of the complete and specific inhibition of the E1B-55K/E4orf6 complex in wild-type virus-infected cells was shown by the failure to degrade the cellular Mre11 protein, a known target of the adenovirus E3 ubiquitin-protein ligase complex (75) (Fig. 8A). These data clearly indicate that when a major component of the ubiquitin-ligase complex is inactivated or absent, as in wild-type virus-infected cells coinfected with the ψ5-NTD vector or in cells infected with E1B-55K and E4orf6 mutant viruses, PKR is active and eIF2α is highly phosphorylated at late times after infection. We did not observe similar results when adenovirus-infected cells were treated with the broad-spectrum proteasome inhibitor MG132 (data not shown), suggesting that regulation of eIF2α phosphorylation and PKR activation by the E1B-55K and E4orf6 proteins is specific to the Cul5-containing E1B-55K/E4orf6 complex. Taken together, these results suggest that a Cul5-mediated activity, most likely formation of a functional adenoviral E3 ubiquitin-protein ligase complex, is required for low levels of eIF2α phosphorylation and PKR activation during the late phase of adenovirus infection.
DISCUSSION
In this report, we describe a new layer of regulation in the activation of PKR and phosphorylation of eIF2α during a productive adenovirus infection. At late times of infection, the E1B-55K and E4orf6 proteins maintain low levels of phospho-eIF2α (Fig. 2 and 3) and prevent PKR activation (Fig. 4). This activity was correlated with the ability of the E1B-55K and E4orf6 proteins to promote viral late protein synthesis (Fig. 1 and 3B). It seems likely that the E1B-55K and E4orf6 proteins suppress a dsRNA-mediated signal because the reovirus σ3 protein, which binds and sequesters dsRNA (38), prevented PKR activation, blocked eIF2α phosphorylation, and restored viral late protein synthesis in cells infected with the E1B-55K or E4orf6 mutant adenovirus (Fig. 5 and 6). Expression of the σ3 protein did not correct the defect in viral late mRNA accumulation (Fig. 6), leading us to conclude that the E1B-55K and E4orf6 proteins sustain viral late translation by preventing the phosphorylation of eIF2α. This is precisely the role attributed to the adenovirus inhibitor of PKR, VAI RNA (43). Nonetheless, because cytoplasmic levels of VAI RNA and VAII RNA did not differ among cells infected with the wild-type or mutant viruses (Fig. 7), it seems unlikely that the E1B-55K and E4orf6 proteins influence the synthesis or transport of the VA RNAs. Although it remains unclear whether or how the E1B-55K and E4orf6 proteins can support the function of VAI RNA, additional experiments reported here demonstrate that the ubiquitin-protein ligase formed by the E1B-55K and E4orf6 proteins with Cul5 is essential for the regulation of eIF2α phosphorylation and PKR activation (Fig. 8).
The phosphorylation of eIF2α fluctuated over the course of the infectious cycle. Levels of phospho-eIF2α increased shortly after infection and then again at the onset of viral DNA synthesis. A final, precipitous decline in eIF2α phosphorylation coincided with the onset of robust viral late protein synthesis (Fig. 3). The low levels of eIF2α phosphorylation at late times of infection required both the E1B-55K and E4orf6 proteins and their ability to form the Cul5-based E3 ubiquitin-protein ligase (Fig. 8). In the absence of either viral protein, phosphorylation of eIF2α increased to the greatest level at very late times of infection. Only the final increase in phospho-eIF2α in cells infected with the E1B-55K or E4orf6 mutant viruses appeared to be mediated by PKR, which itself was phosphorylated, and presumably activated, only at very late times in the mutant virus-infected cells. The absence of phosphorylated PKR at all other times of infection leads us to postulate that PKR does not contribute to eIF2α phosphorylation during the early phase. The kinase or kinases responsible for eIF2α phosphorylation at early times remain unknown. However, published reports support the likelihood of a kinase other than PKR acting on eIF2α between 6 and 18 h postinfection. Huang and Schneider noted that approximately one-quarter of all eIF2α protein was phosphorylated in adenovirus-infected cells at 18 h postinfection (34). Interestingly, this level of phospho-eIF2α was not reduced by the PKR inhibitor 2-aminopurine, implying that PKR did not contribute to the eIF2α phosphorylation at that time of infection. Other investigators have also observed phospho-eIF2α in wild-type virus-infected cells at both 18 and 24 h postinfection but fail to detect activated PKR in these cells (48, 69). These published findings are in agreement with the findings reported here and implicate an eIF2α kinase other than PKR acting at relatively early times in adenovirus-infected cells.
Three other eIF2α kinases have been described. These include HRI, GCN2, and PERK (56, 64). The HRI kinase phosphorylates eIF2α in response to heat shock and oxidative stress but is primarily found in erythrocytes, where it is regulated by heme. The GCN2 kinase is activated in response to amino acid starvation. The nature of these two kinases and their regulation make it unlikely that they contribute to the eIF2α phosphorylation during adenovirus infection. PERK, which phosphorylates eIF2α in response to endoplasmic reticulum stress, hypoxia, and as part of the misfolded protein response, seems a reasonable candidate for the possible early eIF2α kinase. Many viruses elicit the unfolded protein response (31). Although PERK activated by human cytomegalovirus infection did not contribute to a significant increase eIF2α phosphorylation (37), it would be informative to elucidate the signals responsible for eIF2α kinase activation. In addition, because cell extracts of Ad2-infected HeLa cells contained an activity that dephosphorylated phospho-eIF2α (72), differential regulation of eIF2α phosphatases may play a role in the multiphasic pattern of eIF2α phosphorylation observed throughout infection.
An unusual characteristic of the adenovirus-infected cell that has been noted by other investigators (43) is that protein synthesis proceeds with levels of phospho-eIF2α that should block translation (32). The transformed cell lines used in these studies may tolerate higher levels of phospho-eIF2α (16) because they contain higher levels of the limiting factor eIF2Bɛ than untransformed cells (7). Although we did not observe a compensatory increase in the level of eIF2Bɛ during adenovirus infection of HeLa cells (Fig. 2), a greater basal ratio of eIF2Bɛ to eIF2α may permit translation until phosphorylation of eIF2α rises to levels found only at very late times of infection. It is also possible that phosphorylated eIF2α resides in a functional segregated cellular compartment (23, 69), although preliminary immunofluorescence microscopy revealed no physical basis for this separation in virus-infected cells (data not shown). Continued translation in the presence of phosphorylated eIF2α may reflect reprogramming of the translational machinery by adenovirus. Adenovirus late translation proceeds by a noncanonical form of translation initiation known as ribosome shunting (22, 85) that involves modification of the cap-binding complex eIF4F by the L4-100K protein (19, 83). A recent report showed that eIF2α phosphorylation decreased the rate of ribosome shunting on the cellular cIAP2 mRNA (71). Although we did not investigate the consequences of eIF2α phosphorylation on ribosome shunting in adenovirus-infected cells, our results lead us to predict an adverse effect.
The results reported here lead us to hypothesize that the E1B-55K and E4orf6 proteins block eIF2α phosphorylation by preventing PKR activation. Consistent with this notion, the dramatic increase in phospho-eIF2α seen in the mutant virus-infected cells at 30 and 36 h postinfection (Fig. 3) coincided with an abrupt increase in PKR activation (Fig. 4). The signal for PKR activation appears to arise during the late phase of infection, since activated PKR was absent in cells infected with a VAI RNA mutant virus whose progression to the late phase was blocked by treatment with hydroxyurea or cycloheximide (48). We further postulate that the signal responsible for the activation of PKR and phosphorylation of eIF2α in adenovirus-infected cells is the well-known activator of PKR, dsRNA (25). Support for this conjecture derives from the use of HeLa cells expressing the reovirus σ3 protein. In contrast to control HeLa cells, when σ3 cells were infected with the E1B-55K or E4orf6 mutant viruses, activated PKR and phosphorylated eIF2α were nearly undetectable at late times of infection (Fig. 5). The only activity associated with the σ3 protein, in addition to serving as a viral capsid protein, is its ability bind and sequester dsRNA to prevent PKR activation (38, 84). The source of the postulated dsRNA that activates PKR in the absence of E1B-55K/E4orf6 function remains an open question. dsRNA of viral origin may arise from symmetrical transcription of the viral DNA genome during the late phase of infection (52). dsRNA of cellular origin may arise from fragments of RNA known as promoter-associated transcripts generated by spurious transcription (18, 54). Because the E1B-55K/E4orf6 complex controls nuclear mRNA export during an infection, some transcripts normally retained in the nucleus may reach the cytoplasm in the absence of E1B-55K/E4orf6 function. Accordingly, we observed an increase in intron-containing viral late mRNA in the cytoplasm of cells infected with the E1B-55K mutant virus dl338 compared to wild-type virus-infected cells (unpublished observations and see also reference 27). Although dsRNA may have the potential to activate PKR at late times of infection, additional factors may also contribute. For example, the cellular PKR-associated activator protein PACT directly binds PKR in order to promote its activation and stimulates eIF2α phosphorylation in a dsRNA-independent manner (50). Since a Cul5-mediated activity is needed to suppress PKR activation (Fig. 8), perhaps PACT is a target of the adenovirus E3 ubiquitin-protein-ligase. Regulation of PACT by viral proteins would not be unexpected in light of a report that the Us11 protein of herpes simplex virus type 1 blocks PKR activation induced by PACT (51).
The discovery that the E1B-55K/E4orf6 complex prevents PKR activation highlights a novel connection between the E1B-55K/E4orf6 complex and VAI RNA. The Cul5-associated ubiquitin ligase activity of the E1B55K/E4orf6 complex stimulates viral mRNA export (82) and is required to block PKR activation and eIF2α phosphorylation. However, the complex did not promote the cytoplasmic accumulation of either VAI or VAII RNA (Fig. 7). This leads us to question how the E1B-55K/E4orf6 complex might be necessary for VAI RNA function during an adenovirus infection even though VAI RNA can stimulate translation in vitro (43) or after transfection (76). In data not shown, Kitajewski and associates noted that a 50-fold mass excess of VAI RNA over dsRNA was required to prevent activation of PKR (41). Perhaps without E1B-55K/E4orf6 function, dysregulated mRNA export leads to duplex RNA in the cytoplasm in excess of the 50-fold requirement. Because we observe an abrupt increase in PKR activation between 24 and 30 h postinfection, this could be the time at which this balance changes. Alternatively, since VAI RNA can interact with secondary structures present in some cellular mRNA (59), perhaps cellular mRNA of this nature accumulate in the absence of E1B-55K/E4orf6 and alter the activity of VAI RNA. It is also possible that a viral late product, whose expression is enhanced by the E1B-55K/E4orf6 complex, suppresses the eIF2α phosphorylation and PKR activation. Clearly, further study is needed to understand the apparent connection between VAI RNA function and the E1B-55K and E4orf6 proteins.
Another possible link between the control of VAI RNA function and the E1B-55K/E4orf6 complex involves overexpression of the adenovirus E2A DNA-binding protein (DBP). We and others have noted high levels of DBP in cells infected with E1B-55K and E4orf6 mutant viruses (27). Wild-type virus-infected cells treated with 2-aminopurine also synthesize unusually large quantities of DBP (34). In both of these circumstances, VAI RNA is present but evidently nonfunctional. Although it is a DNA-binding protein, DBP also binds RNA during the late phase of infection (15). Perhaps the abundance of DBP during E1B-55K and E4orf6 mutant virus infections and during treatment with 2-aminopurine inactivates VAI RNA.
In summary, we have demonstrated a new role for the E1B-55K and E4orf6 proteins in the regulation of PKR activation and eIF2α phosphorylation during the late phase of adenovirus infection. Knowing the molecular mechanism of eIF2α regulation by the E1B-55K/E4orf6 complex may uncover new targets of the complex and contribute to our understanding of events that govern viral late protein synthesis. Furthermore, this information may contribute to a better understanding of the oncolytic nature of E1B-55K mutant adenoviruses. For example, it would be of interest to determine whether the abnormally high levels of activated PKR elicited by infection with the E1B-55K mutant virus precipitates pathways of cell death that may be unique to tumor-derived cells (see for example, reference 79). Finally, the work presented here serves to further highlight the complex and multifaceted nature of the E1B-55K and E4orf6 proteins and their interaction with the host cell.
Acknowledgments
We thank the following individuals for kindly providing valuable reagents: K. Clark and G. Parks (Wake Forest University School of Medicine) for the σ3-HeLa cell line and control, T. Dermody (Vanderbilt University) for the σ3 antisera 4F2, and J. Woo and A. Berk (University of California, Los Angeles) for the Cul5 constructs and the Ad5 antisera.
Cell culture reagents were provided by the Cell and Virus Vector Core Laboratory of the Comprehensive Cancer Center of Wake Forest University, which is supported in part by National Cancer Institute grant CA12197. This research was supported by grant R01 CA077342 from the National Cancer Institute.
The content of this report is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Footnotes
Published ahead of print on 15 July 2009.
REFERENCES
- 1.Akusjarvi, G., and U. Pettersson. 1979. Sequence analysis of adenovirus DNA: complete nucleotide sequence of the spliced 5′ noncoding region of adenovirus 2 hexon messenger RNA. Cell 16:841-850. [DOI] [PubMed] [Google Scholar]
- 2.Akusjarvi, G., C. Svensson, and O. Nygard. 1987. A mechanism by which adenovirus virus-associated RNAI controls translation in a transient expression assay. Mol. Cell. Biol. 7:549-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babich, A., L. T. Feldman, J. R. Nevins, J. E. Darnell, Jr., and C. Weinberger. 1983. Effect of adenovirus on metabolism of specific host mRNAs: transport control and specific translational discrimination. Mol. Cell. Biol. 3:1212-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babiss, L. E., and H. S. Ginsberg. 1984. Adenovirus type 5 early region 1b gene product is required for efficient shutoff of host protein synthesis. J. Virol. 50:202-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babiss, L. E., H. S. Ginsberg, and J. E. Darnell, Jr. 1985. Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol. Cell. Biol. 5:2552-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker, A., K. J. Rohleder, L. A. Hanakahi, and G. Ketner. 2007. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 81:7034-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balachandran, S., and G. N. Barber. 2004. Defective translational control facilitates vesicular stomatitis virus oncolysis. Cancer Cell 5:51-65. [DOI] [PubMed] [Google Scholar]
- 8.Barker, D. D., and A. J. Berk. 1987. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology 156:107-121. [DOI] [PubMed] [Google Scholar]
- 9.Beltz, G. A., and S. J. Flint. 1979. Inhibition of HeLa cell protein synthesis during adenovirus infection. Restriction of cellular messenger RNA sequences to the nucleus. J. Mol. Biol. 131:353-373. [DOI] [PubMed] [Google Scholar]
- 10.Blackford, A. N., and R. J. Grand. 2009. Adenovirus E1B-55K: a protein with multiple roles in viral infection and cell transformation. J. Virol. 83:4000-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanchette, P., C. Y. Cheng, Q. Yan, G. Ketner, D. A. Ornelles, T. Dobner, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2004. Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol. Cell. Biol. 24:9619-9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bridge, E., and G. Ketner. 1990. Interaction of adenoviral E4 and E1b products in late gene expression. Virology 174:345-353. [DOI] [PubMed] [Google Scholar]
- 13.Bridge, E., and G. Ketner. 1989. Redundant control of adenovirus late gene expression by early region 4. J. Virol. 63:631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chow, L. T., and T. R. Broker. 1978. The spliced structures of adenovirus 2 fiber message and the other late mRNAs. Cell 15:497-510. [DOI] [PubMed] [Google Scholar]
- 15.Cleghon, V. G., and D. F. Klessig. 1986. Association of the adenovirus DNA-binding protein with RNA both in vitro and in vivo. Proc. Natl. Acad. Sci. USA 83:8947-8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clemens, M. J. 2004. Targets and mechanisms for the regulation of translation in malignant transformation. Oncogene 23:3180-3188. [DOI] [PubMed] [Google Scholar]
- 17.Corbin-Lickfett, K. A., and E. Bridge. 2003. Adenovirus E4-34kDa requires active proteasomes to promote late gene expression. Virology 315:234-244. [DOI] [PubMed] [Google Scholar]
- 18.Core, L. J., J. J. Waterfall, and J. T. Lis. 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322:1845-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cuesta, R., Q. Xi, and R. J. Schneider. 2000. Adenovirus-specific translation by displacement of kinase Mnk1 from cap-initiation complex eIF4F. EMBO J. 19:3465-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuesta, R., Q. Xi, and R. J. Schneider. 2004. Structural basis for competitive inhibition of eIF4G-Mnk1 interaction by the adenovirus 100-kilodalton protein. J. Virol. 78:7707-7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dallaire, F., P. Blanchette, P. Groitl, T. Dobner, and P. E. Branton. 2009. Identification of integrin α3 as a new substrate of the adenovirus E4orf6/E1B55K E3 ubiquitin ligase complex. J. Virol. 83:5329-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dolph, P. J., J. T. Huang, and R. J. Schneider. 1990. Translation by the adenovirus tripartite leader: elements which determine independence from cap-binding protein complex. J. Virol. 64:2669-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flint, S. J., and R. A. Gonzalez. 2003. Regulation of mRNA production by the adenoviral E1B 55-kDa and E4 Orf6 proteins. Curr. Top. Microbiol. Immunol. 272:287-330. [DOI] [PubMed] [Google Scholar]
- 24.Gainey, M. D., P. J. Dillon, K. M. Clark, M. J. Manuse, and G. D. Parks. 2008. Paramyxovirus-induced shutoff of host and viral protein synthesis: role of the P and V proteins in limiting PKR activation. J. Virol. 82:828-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia, M. A., E. F. Meurs, and M. Esteban. 2007. The dsRNA protein kinase PKR: virus and cell control. Biochimie 89:799-811. [DOI] [PubMed] [Google Scholar]
- 26.Gomez, E., S. S. Mohammad, and G. D. Pavitt. 2002. Characterization of the minimal catalytic domain within eIF2B: the guanine-nucleotide exchange factor for translation initiation. EMBO J. 21:5292-5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodrum, F. D., and D. A. Ornelles. 1999. Roles for the E4 orf6, orf3, and E1B 55-kilodalton proteins in cell cycle-independent adenovirus replication. J. Virol. 73:7474-7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halbert, D. N., J. R. Cutt, and T. Shenk. 1985. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol. 56:250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harada, J. N., A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 76:9194-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayes, B. W., G. C. Telling, M. M. Myat, J. F. Williams, and S. J. Flint. 1990. The adenovirus L4 100-kilodalton protein is necessary for efficient translation of viral late mRNA species. J. Virol. 64:2732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He, B. 2006. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 13:393-403. [DOI] [PubMed] [Google Scholar]
- 32.Hershey, J. W. 1989. Protein phosphorylation controls translation rates. J. Biol. Chem. 264:20823-20826. [PubMed] [Google Scholar]
- 33.Huang, J. T., and R. J. Schneider. 1991. Adenovirus inhibition of cellular protein synthesis involves inactivation of cap-binding protein. Cell 65:271-280. [DOI] [PubMed] [Google Scholar]
- 34.Huang, J. T., and R. J. Schneider. 1990. Adenovirus inhibition of cellular protein synthesis is prevented by the drug 2-aminopurine. Proc. Natl. Acad. Sci. USA 87:7115-7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang, M. M., and P. Hearing. 1989. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J. Virol. 63:2605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang, W., and S. J. Flint. 1998. The tripartite leader sequence of subgroup C adenovirus major late mRNAs can increase the efficiency of mRNA export. J. Virol. 72:225-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Isler, J. A., A. H. Skalet, and J. C. Alwine. 2005. Human cytomegalovirus infection activates and regulates the unfolded protein response. J. Virol. 79:6890-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacobs, B. L., and J. O. Langland. 1998. Reovirus sigma 3 protein: dsRNA binding and inhibition of RNA-activated protein kinase. Curr. Top. Microbiol. Immunol. 233:185-196. [DOI] [PubMed] [Google Scholar]
- 39.Jones, N., and T. Shenk. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17:683-689. [DOI] [PubMed] [Google Scholar]
- 40.Kamura, T., M. N. Conrad, Q. Yan, R. C. Conaway, and J. W. Conaway. 1999. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 13:2928-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kitajewski, J., R. J. Schneider, B. Safer, S. M. Munemitsu, C. E. Samuel, B. Thimmappaya, and T. Shenk. 1986. Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2α kinase. Cell 45:195-200. [DOI] [PubMed] [Google Scholar]
- 42.Logan, J., and T. Shenk. 1984. Adenovirus tripartite leader sequence enhances translation of mRNAs late after infection. Proc. Natl. Acad. Sci. USA 81:3655-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathews, M. B. 1990. Control of translation in adenovirus-infected cells. Enzyme 44:250-264. [DOI] [PubMed] [Google Scholar]
- 44.Mathews, M. B., and T. Shenk. 1991. Adenovirus virus-associated RNA and translation control. J. Virol. 65:5657-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nordqvist, K., K. Ohman, and G. Akusjarvi. 1994. Human adenovirus encodes two proteins which have opposite effects on accumulation of alternatively spliced mRNAs. Mol. Cell. Biol. 14:437-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohman, K., K. Nordqvist, and G. Akusjarvi. 1993. Two adenovirus proteins with redundant activities in virus growth facilitates tripartite leader mRNA accumulation. Virology 194:50-58. [DOI] [PubMed] [Google Scholar]
- 47.Oldfield, S., B. L. Jones, D. Tanton, and C. G. Proud. 1994. Use of monoclonal antibodies to study the structure and function of eukaryotic protein synthesis initiation factor eIF-2B. Eur. J. Biochem. 221:399-410. [DOI] [PubMed] [Google Scholar]
- 48.O'Malley, R. P., T. M. Mariano, J. Siekierka, and M. B. Mathews. 1986. A mechanism for the control of protein synthesis by adenovirus VA RNAI. Cell 44:391-400. [DOI] [PubMed] [Google Scholar]
- 49.Ornelles, D. A., and T. Shenk. 1991. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J. Virol. 65:424-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patel, R. C., and G. C. Sen. 1998. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 17:4379-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peters, G. A., D. Khoo, I. Mohr, and G. C. Sen. 2002. Inhibition of PACT-mediated activation of PKR by the herpes simplex virus type 1 Us11 protein. J. Virol. 76:11054-11064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pettersson, U., and L. Philipson. 1974. Synthesis of complementary RNA sequences during productive adenovirus infection. Proc. Natl. Acad. Sci. USA 71:4887-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pilder, S., M. Moore, J. Logan, and T. Shenk. 1986. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol. Cell. Biol. 6:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Preker, P., J. Nielsen, S. Kammler, S. Lykke-Andersen, M. S. Christensen, C. K. Mapendano, M. H. Schierup, and T. H. Jensen. 2008. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322:1851-1854. [DOI] [PubMed] [Google Scholar]
- 55.Price, R., and S. Penman. 1972. A distinct RNA polymerase activity, synthesizing 5-5 s, 5 s and 4 s RNA in nuclei from adenovirus 2-infected HeLa cells. J. Mol. Biol. 70:435-450. [DOI] [PubMed] [Google Scholar]
- 56.Proud, C. G. 2005. eIF2 and the control of cell physiology. Semin. Cell Dev. Biol. 16:3-12. [DOI] [PubMed] [Google Scholar]
- 57.Proud, C. G. 1992. Protein phosphorylation in translational control. Curr. Top. Cell. Regul. 32:243-369. [DOI] [PubMed] [Google Scholar]
- 58.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reichel, P. A., W. C. Merrick, J. Siekierka, and M. B. Mathews. 1985. Regulation of a protein synthesis initiation factor by adenovirus virus-associated RNA. Nature 313:196-200. [DOI] [PubMed] [Google Scholar]
- 60.Romano, P. R., M. T. Garcia-Barrio, X. Zhang, Q. Wang, D. R. Taylor, F. Zhang, C. Herring, M. B. Mathews, J. Qin, and A. G. Hinnebusch. 1998. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2α kinases PKR and GCN2. Mol. Cell. Biol. 18:2282-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rowlands, A. G., R. Panniers, and E. C. Henshaw. 1988. The catalytic mechanism of guanine nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor 2. J. Biol. Chem. 263:5526-5533. [PubMed] [Google Scholar]
- 62.Saelens, X., M. Kalai, and P. Vandenabeele. 2001. Translation inhibition in apoptosis: caspase-dependent PKR activation and eIF2-alpha phosphorylation. J. Biol. Chem. 276:41620-41628. [DOI] [PubMed] [Google Scholar]
- 63.Safer, B. 1983. 2B or not 2B: regulation of the catalytic utilization of eIF-2. Cell 33:7-8. [DOI] [PubMed] [Google Scholar]
- 64.Samuel, C. E. 1993. The eIF-2 alpha protein kinases, regulators of translation in eukaryotes from yeasts to humans. J. Biol. Chem. 268:7603-7606. [PubMed] [Google Scholar]
- 65.Sandler, A. B., and G. Ketner. 1989. Adenovirus early region 4 is essential for normal stability of late nuclear RNAs. J. Virol. 63:624-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sandler, A. B., and G. Ketner. 1991. The metabolism of host RNAs in cells infected by an adenovirus E4 mutant. Virology 181:319-326. [DOI] [PubMed] [Google Scholar]
- 67.Schmechel, S., M. Chute, P. Skinner, R. Anderson, and L. Schiff. 1997. Preferential translation of reovirus mRNA by a sigma3-dependent mechanism. Virology 232:62-73. [DOI] [PubMed] [Google Scholar]
- 68.Schneider, R. J., B. Safer, S. M. Munemitsu, C. E. Samuel, and T. Shenk. 1985. Adenovirus VAI RNA prevents phosphorylation of the eukaryotic initiation factor 2 alpha subunit subsequent to infection. Proc. Natl. Acad. Sci. USA 82:4321-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schneider, R. J., C. Weinberger, and T. Shenk. 1984. Adenovirus VAI RNA facilitates the initiation of translation in virus-infected cells. Cell 37:291-298. [DOI] [PubMed] [Google Scholar]
- 70.Shepard, R. N., and D. A. Ornelles. 2004. Diverse roles for E4orf3 at late times of infection revealed in an E1B 55-kilodalton protein mutant background. J. Virol. 78:9924-9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sherrill, K. W., and R. E. Lloyd. 2008. Translation of cIAP2 mRNA is mediated exclusively by a stress-modulated ribosome shunt. Mol. Cell. Biol. 28:2011-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Siekierka, J., T. M. Mariano, P. A. Reichel, and M. B. Mathews. 1985. Translational control by adenovirus: lack of virus-associated RNAI during adenovirus infection results in phosphorylation of initiation factor eIF-2 and inhibition of protein synthesis. Proc. Natl. Acad. Sci. USA 82:1959-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smiley, J. K., M. A. Young, and S. J. Flint. 1990. Intranuclear location of the adenovirus type 5 E1B 55-kilodalton protein. J. Virol. 64:4558-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Soderlund, H., U. Pettersson, B. Vennstrom, L. Philipson, and M. B. Mathews. 1976. A new species of virus-coded low molecular weight RNA from cells infected with adenovirus type 2. Cell 7:585-593. [DOI] [PubMed] [Google Scholar]
- 75.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348-352. [DOI] [PubMed] [Google Scholar]
- 76.Svensson, C., and G. Akusjarvi. 1984. Adenovirus VA RNAI: a positive regulator of mRNA translation. Mol. Cell. Biol. 4:736-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thimmappaya, B., C. Weinberger, R. J. Schneider, and T. Shenk. 1982. Adenovirus VAI RNA is required for efficient translation of viral mRNAs at late times after infection. Cell 31:543-551. [DOI] [PubMed] [Google Scholar]
- 78.Thomas, A. A., G. C. Scheper, M. Kleijn, M. De Boer, and H. O. Voorma. 1992. Dependence of the adenovirus tripartite leader on the p220 subunit of eukaryotic initiation factor 4F during in vitro translation: effect of p220 cleavage by foot-and-mouth-disease-virus l-protease on in vitro translation. Eur. J. Biochem. 207:471-477. [DOI] [PubMed] [Google Scholar]
- 79.Toth, A. M., P. Devaux, R. Cattaneo, and C. E. Samuel. 2009. Protein kinase PKR mediates the apoptosis induction and growth restriction phenotypes of C protein-deficient measles virus. J. Virol. 83:961-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weinberg, D. H., and G. Ketner. 1986. Adenoviral early region 4 is required for efficient viral DNA replication and for late gene expression. J. Virol. 57:833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weinmann, R., H. J. Raskas, and R. G. Roeder. 1974. Role of DNA-dependent RNA polymerases II and III in transcription of the adenovirus genome late in productive infection. Proc. Natl. Acad. Sci. USA 71:3426-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Woo, J. L., and A. J. Berk. 2007. Adenovirus ubiquitin-protein ligase stimulates viral late mRNA nuclear export. J. Virol. 81:575-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xi, Q., R. Cuesta, and R. J. Schneider. 2004. Tethering of eIF4G to adenoviral mRNAs by viral 100K protein drives ribosome shunting. Genes Dev. 18:1997-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yue, Z., and A. J. Shatkin. 1997. Double-stranded RNA-dependent protein kinase (PKR) is regulated by reovirus structural proteins. Virology 234:364-371. [DOI] [PubMed] [Google Scholar]
- 85.Yueh, A., and R. J. Schneider. 1996. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 10:1557-1567. [DOI] [PubMed] [Google Scholar]
- 86.Zhang, Y., P. J. Dolph, and R. J. Schneider. 1989. Secondary structure analysis of adenovirus tripartite leader. J. Biol. Chem. 264:10679-10684. [PubMed] [Google Scholar]
- 87.Zhang, Y., D. Feigenblum, and R. J. Schneider. 1994. A late adenovirus factor induces eIF-4E dephosphorylation and inhibition of cell protein synthesis. J. Virol. 68:7040-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]