The capacity of viruses to successfully infect the immunocompetent host to cause disease argues in favor of virus-encoded functions that specifically target components of the immune system so as to orchestrate an environment that limits the capacity of the host immune response to clear infection. In this respect, many viruses have evolved to coexist with the host immune system by developing an arsenal of strategies to avoid immune surveillance and elimination from the host. These include viruses which have acquired homologs of cellular cytokines or cytokine receptors as a strategy to limit host immune recognition. Cellular interleukin-10 (IL-10) is a pleiotropic immunomodulatory cytokine produced by a wide variety of cells, including monocytes, macrophages, T and B lymphocytes, dendritic cells (DC), keratinocytes, epithelial cells, and mast cells. The properties of IL-10 have been comprehensively reviewed elsewhere (12, 69, 70, 72, 77) and so will not be covered in detail here, but the key features of this cytokine relate mainly to its capacity to exert potent immunosuppressive functions on the expression of a range of cytokines and chemokines (2, 16, 23), as well as the repression of major histocompatibility complex (MHC) molecules and costimulatory molecules (17, 104) and the maturation and function of DC (69). The immunosuppressive properties of IL-10 are primarily restricted to cells of the myeloid lineage (69). In contrast, IL-10 has been shown to exert a stimulatory effect on B lymphocytes (15, 28, 87), mast cells (99), thymocytes (64), and CD8+ T cells (31, 88, 90), highlighting the cell-type-dependent immunomodulatory properties of this cytokine. The immunomodulatory functions manifested by IL-10 require engagement of this cytokine with its cell surface bound receptor. The IL-10 receptor (IL-10R) consists of two different subunits (IL-10R1 and IL-10R2) (52, 60). IL-10 binds first to IL-10R1 with high affinity, and the resulting intermediate IL-10/IL-10R1 complex then binds with lower affinity to IL-10R2. The resulting active signaling complex induces the JAK/Stat signal transduction pathway (69, 72). In the context of viral pathogenesis, infections with a number of different viruses have been documented to upregulate the expression of IL-10, and in some cases, this upregulation has been shown to enhance infection by suppressing the immune function, suggesting that the far-reaching effects of this cytokine have many advantages for invading pathogens (3, 4, 18, 30, 45, 81, 82, 108, 111).
IL-10-like open reading frames (ORF) have been identified by sequence homology in multiple members of the Herpesvirales and Poxviridae, all but one of which infect mammalian hosts (Table 1). The one exception, cyprinid herpesvirus 3, is a member of the Alloherpesviridae family of herpesviruses (order, Herpesvirales), which has the common carp (Cyprinus carpio) as its normal host (1). Otherwise, all of the herpesviruses identified to date as encoding IL-10-like ORF, including human cytomegalovirus (HCMV) and Epstein-Barr virus (EBV), are found only in the Beta- and Gammaherpesvirinae subfamilies of the Herpesviridae. No examples of the Alphaherpesvirinae that encode IL-10 homologs have been identified. Of the identified members of the Herpesviridae that encode IL-10 homologs, all but two, equid herpesvirus 2 and ovine herpesvirus 2 (OvHV2) (85, 96, 97), are confined to primate hosts (Table 1). All of the identified Poxviridae that encode IL-10 infect ruminants, including orf virus ([ORFV] sheep and goats), sheeppox virus ([SPPV] sheep), goatpox virus ([GPV] goats), and lumpy skin disease virus ([LSDV] cattle). The genomes of SPPV, GPV, and LSDV are 96 to 97% identical at the nucleic acid level (48, 101). Another member of the Poxviridae that has monkeys as its normal host, yaba-like disease virus, encodes an ORF (Y134R) that exhibits sequence homology to the IL-10-related cytokines IL-19, IL-20, and IL-24 (57). These viral IL-10 (vIL-10) homologs range in size from 139 to 191 amino acids (aa), bracketing the range in sizes for cellular IL-10 proteins (176 to 180 aa).
TABLE 1.
Viruses with IL-10-like or IL-10-related ORF
| Order | Family | Subfamily/genus | Virus (common abbreviation)a | ORF (GenBank accession no.) | Length (aa) | % Identity to host IL-10 (host) | Reference |
|---|---|---|---|---|---|---|---|
| Herpesvirales | Herpesviridae | Betaherpesvirinae/Cytomegalovirus | Human HV 5 (HCMV) | UL111A/cmvIL-10 (AAF36285) | 175 | 27 | 51, 57 |
| Human HV 5 (HCMV) | UL111A/LAcmvIL-10 (FJ997279) | 139 | 27 | 40 | |||
| Rhesus CMV (McHV3) | UL111A/vIL-10 (AAF59907) | 189 | 25 | 57 | |||
| African green monkey CMV (AGMCMV) | vIL-10 (AAF63435) | 185 | 27 (Sooty mangabey) | 57 | |||
| Baboon CMV (BaCMV) | vIL-10 (AAF63436) | 191 | 29 | 57 | |||
| Gammaherpesvirinae/Lymphocryptovirus | Human HV 4 (EBV) | BCRF1/vIL-10 (P03180) | 170 | 90 | 67 | ||
| Rhesus LCV (McHV4) | BCRF1 (AAK95412) | 177 | 84 | 80 | |||
| Gammaherpesvirinae/Percavirus | Equine HV 2 (EHV2) | IL-10 (AAB26148) | 179 | 84 | 81, 93 | ||
| Gammaherpesvirinae/Macavirus | Ovine HV 2 (OvHV2) | IL-10 (ABB22222) | 182 | 41 | 92 | ||
| Alloherpesviridae | Unassigned | Koi HV (CyHV3) | IL-10 (YP_001096169) | 179 | 20 | 1 | |
| Poxviridae | Chordopoxvirinae/Parapoxvirus | Orf virus (ORFV) | ORFV-IL-10 (AAC57332) | 186 | 91 | 25 | |
| Chordopoxvirinae/Capripoxvirus | Sheeppox virus (SPPV) | IL-10 (NP_659579) | 168 | 38 | 96 | ||
| Chordopoxvirinae/Capripoxvirus | Goatpox virus (GPV) | IL-10 (YP_001293197) | 170 | 45 | 96 | ||
| Chordopoxvirinae/Capripoxvirus | Lumpy skin disease virus (LSDV) | IL-10 (AAN02729) | 171 | 39 | 46 |
HV, herpesvirus; LCV, lymphocryptovirus.
The role of virus-encoded IL-10 homologs is likely to provide a tool to enable modulation of the local immune response so as to enhance the capacity to replicate, disseminate, and/or persist in an otherwise immunocompetent individual. In fact, there is emerging evidence that virus-encoded IL-10 homologs function in this capacity in a variety of settings. This review will cover those viruses which have thus far been identified as encoding homologs of IL-10. The similarities of each homolog to the IL-10 of the natural host species will be presented together with their biological functions (where known) and the role they may play in viral pathogenesis and evasion of the host immune response.
EBV
EBV-encoded BCRF1, the first vIL-10 homolog described, was identified shortly after the discovery of the cellular cytokine. IL-10 was initially known as CSIF, or cytokine synthesis inhibitory factor, an activity produced by murine TH2 cells that inhibited cytokine production by TH1 cells (22). When the mouse and human cDNAs for CSIF/IL-10 were sequenced, the protein products were found to have 70% amino acid sequence identity with an uncharacterized ORF of EBV, BCRF1 (71). BCRF1 refers to BamHI-C fragment rightward reading frame 1, and when the BCRF1 gene was cloned and expressed, the resulting 17-kDa protein inhibited synthesis of gamma interferon (IFN-γ) from a stimulated mouse TH1 clone and from human peripheral blood mononuclear cells (PBMC) (35). Cytokine production was inhibited by BCRF1 regardless of whether PBMC were stimulated with phytohemagglutinin, IL-12, or anti-CD3 antibodies, suggesting that EBV IL-10 (ebvIL-10) could act on multiple cell types and inhibit cytokine synthesis by both T cells and NK cells (35).
In parallel, numerous studies were conducted to document the biological activities of both human IL-10 (hIL-10) and ebvIL-10. Both cytokines were found to have broad immunosuppressive properties, including inhibition of inflammatory cytokine production (16, 35, 36, 103), deactivation of macrophages (73, 74), and inhibition of T-cell proliferation via reduction of MHC class II molecules on the surfaces of monocytes (17). Immunostimulatory functions were also noted; both hIL-10 and ebvIL-10 promoted proliferation and differentiation of B cells, as well as stimulating immunoglobulin production (15, 28, 87). However, it soon became apparent that the viral cytokine had not retained all properties of cellular IL-10. While human and murine IL-10 costimulated mouse thymocyte proliferation and mast cell proliferation and promoted upregulation of MHC class II surface expression on B cells, ebvIL-10 lacked these functions (28, 64, 103). Further examination also revealed that the ability to inhibit IL-2 production by a CD4 T-cell clone was greatly reduced in comparison with that of the cellular IL-10s and that ebvIL-10 had an approximately 1,000-fold-lower affinity for the cellular IL-10R than did hIL-10 (59).
Since the viral gene was most likely captured from the host and evolved over time to retain properties that were most beneficial in promoting virus persistence, these differences in biologic activity were not completely unexpected. Examination of the primary amino acid sequence and the crystal structure suggested possible reasons for these functional differences. In the primary sequence, an isoleucine at position 87 of murine and PhIL-10 was found to be essential for stimulatory activity on thymocytes, mast cells, and alloantigenic responses (19). Substitution of an alanine at this position in hIL-10 resulted in loss of stimulatory activities, while immunosuppressive functions were not affected. The wild-type ebvIL-10 protein normally contains an alanine at this position, and when a mutation substituting isoleucine for the alanine was made, the viral cytokine gained the ability to stimulate proliferation of MC/9 cells, although the proliferation was not as robust as that observed with wild-type hIL-10 (19). These results demonstrate that even a single amino acid can impact a cytokine's functional repertoire.
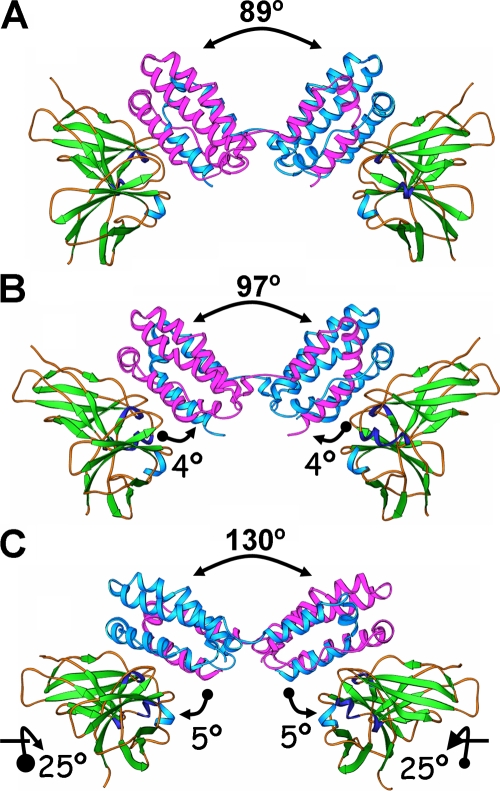
With the exception of the N-terminal 20 amino acids (the signal peptide), the human and ebvIL-10 sequences are highly conserved (35). Determination of the crystal structure demonstrated that like hIL-10, ebvIL-10 forms a homodimer consisting of two domains (113). Each monomer contains six helices, with four helices in one domain and two in the other, with the two monomers connected, forming an axis of twofold symmetry. Subtle changes in two loop structures and the resulting change in orientation of the ebvIL-10 dimer to the cellular IL-10R are believed to be responsible for the decreased binding affinity and limited functionality of the viral cytokine (110) (Fig. 1).
FIG. 1.
Structure of hIL-10 and vIL-10 homologs bound to the hIL-10R. hIL-10 (A), ebvIL-10 (B), and cmvIL-10 (C) dimers bound to soluble IL-10R1 (sIL-10R1) are shown. The interdomain angles of each IL-10 are shown at the top of each complex. The rotation (in degrees) of each IL-10 domain angle on the surface of sIL-10R1 is denoted by arrows where the movement is from the circle to the arrowhead. For the cmvIL-10/sIL-10R1 complex, the rotation of each sIL-10R1 relative to the hIL-10/sIL-10R1 complex is also shown.
Because ebvIL-10 lacks several immunostimulatory functions of the cellular cytokine, there has been great interest in investigating the therapeutic potential of the viral cytokine, particularly for autoimmune diseases or conditions associated with chronic inflammation. In animal models, ebvIL-10 was found to be effective in inhibiting collagen-induced arthritis (49, 50, 63), suppressing autoimmune diabetes (109), ameliorating symptoms of necrotizing pancreatitis (67), and improving survival of sepsis (75). In particular, ebvIL-10 has distinct advantages over hIL-10 in transplantation models, and the viral cytokine has consistently enhanced graft acceptance and survival in various models (10, 51, 69, 78, 89).
Exploitation of IL-10 is a common theme among intracellular pathogens (82), as the immunosuppressive properties of the cytokine may facilitate escape from immune detection. However, for EBV, it is not entirely clear whether the target of ebvIL-10 is mainly antigen-presenting cells to enable additional time for the virus to establish latency, or whether effects on B cells are necessary for transformation. Although BCRF1 was initially characterized as a late gene (37), other reports suggest that it is expressed within 6 to 9 h of infection (68, 100) and show an essential role for ebvIL-10 in B-cell transformation (39, 68). Regardless, the ability of this viral cytokine to promote B-cell proliferation and differentiation would most likely increase the number of latently infected cells in circulation in the host.
HCMV
vIL-10 EXPRESSION DURING PRODUCTIVE HCMV INFECTION
The first identification of a homolog of IL-10 expressed by HCMV was reported almost simultaneously by two separate groups in 2000. The Barry and Pestka groups isolated cDNA from the HCMV UL111A region in productively infected MRC-5 and HEL 299 cells, respectively (54, 61). Further characterization revealed the presence of a transcript (denoted cmvIL-10) generated from three exons and two introns that was predicted to encode a protein of 175 aa with homology to hIL-10. The cmvIL-10 protein contains a signal peptide, and studies of cells transfected with cDNA expression constructs or productively infected with HCMV confirmed that cmvIL-10 protein is secreted into culture supernatants (8, 43, 54, 94). In addition, a putative N-linked glycosylation site was identified at Asn-151-X-Thr-153 (54), and Western blot analyses supported the presence of both nonglycosylated (∼21 kDa) and larger glycosylated forms of cmvIL-10 (8, 43, 54, 58, 94).
Based upon comparison of amino acid sequence, cmvIL-10 shares only 27% identity with hIL-10, in contrast to ebvIL-10, which is >80% identical (54, 61, 71). Despite this low homology to hIL-10, cmvIL-10 can bind to the hIL-10R (47, 54). The crystal structure of cmvIL-10 bound to the extracellular fragment of the ligand binding subunit (IL-10R1) of hIL-10R has been resolved (47). Like hIL-10, cmvIL-10 forms a homodimer that binds two hIL-10R1 subunits, although analysis of this interaction has revealed some differences between cmvIL-10 and hIL-10. In contrast to that of hIL-10, the cmvIL-10 homodimer is linked via a disulfide bond formed between the two cysteine residues on corresponding protein molecules (position 59 in cleaved proteins or position 78 in proteins with the signal peptide still attached). The interdomain angle of cmvIL-10 is ∼130° (compared to ∼89° in hIL-10), resulting in reorganization of IL-10R1 when bound to the cmvIL-10 homodimer compared to the hIL-10/IL-10R1 complex (47, 110) (Fig. 1). Importantly, though, despite the low amino acid homology and the difference in the interdomain angles of cmvIL-10 and hIL-10 homodimers, cmvIL-10 binds strongly to IL-10R1, with a slightly greater affinity than even hIL-10 and a much greater affinity than ebvIL-10 (47, 59).
While cmvIL-10 efficiently binds to hIL-10R1, two common variants of this receptor, each containing single-nucleotide polymorphisms, were less susceptible to cmvIL-10-induced signaling (32). IL-10R1 variants may therefore affect the capacity of HCMV to escape from the host immune response, although studies to evaluate the effect of IL-10R1 variants on the clinical course of HCMV infection of different individuals will be required to test this hypothesis. Due to the larger interdomain angle and slightly different rotation of the cmvIL-10 dimer, the cmvIL-10/IL-10R1 complex changes its orientation, which affects the IL-10R2 binding site. As a result, the cmvIL-10/IL-10R1 complex shows greater affinity for soluble IL-10R2 and binds 3 to 5 times as much IL-10R2 as the hIL-10/IL-10R1 and ebvIL-10/IL-10R1 complexes (110).
The possibility that differences in these structural changes unique to cmvIL-10 may alter the range of biological properties encoded by cmvIL-10 has been previously raised (47), and this does appear to be the case for at least one other vIL-10 homolog, as ebvIL-10 is unable to stimulate thymocyte or mast cell proliferation or B-cell MHC class II expression, whereas hIL-10 can (28, 64, 103). To date, however, the repertoire of immunomodulatory properties of hIL-10 has been shown to also be exerted by cmvIL-10. These include both immunosuppressive and immunostimulatory functions which are manifested in a cell-type-dependent manner.
PBMC proliferation and production of proinflammatory cytokines IL-1α, IL-6, granulocyte-macrophage colony-stimulating factor, and tumor necrosis factor alpha (TNF-α) in lipopolysaccharide (LPS)-treated PMBC and monocytes is inhibited by recombinant cmvIL-10 protein or the supernatant from cells transfected with a cmvIL-10 expression construct (94). In the same study, these treatments were also reported to decrease MHC class I and MHC class II expression by monocytes but increase the nonclassical MHC class I molecule HLA-G, which can confer protection from natural killer cell-mediated lysis of MHC class I-negative cells (86). The link between the binding of cmvIL-10 to the hIL-10R and the functional properties of cmvIL-10 was provided by experiments utilizing antibodies that block the hIL-10R. Neutralizing antibody to the hIL-10R blocks the capacity of recombinant cmvIL-10 to either inhibit PBMC proliferation or downregulate MHC class II by monocytes (44, 94). This vIL-10 homolog has also been shown to upregulate the expression of Fcγ receptors and stimulate phagocytosis of immunoglobulin G-opsonized erythrocytes by human monocytes, with one interpretation of these results being that cmvIL-10 skews monocyte differentiation toward a more phagocytic phenotype and away from an antigen presentation phenotype (40).
The signaling pathway utilized by cmvIL-10 in monocytes appears to be similar to that used by hIL-10, where the classic IL-10R/JAK1/Stat3 pathway was found to be triggered by both cmvIL-10 and hIL-10 (92). Phosphorylation of Stat3 (but not Stat1) was readily detectable in monocytes treated with cmvIL-10 and hIL-10, but Stat3 phosphorylation was prevented in the presence of the JAK1 inhibitor, indicating that binding of cmvIL-10 to the IL-10R leads to JAK1-dependent Stat3 activation. Surprisingly, the JAK1/Stat3 pathway did not appear to play a significant role in the control of TNF-α suppression by cmvIL-10 in LPS-induced monocytes. No significant increase in TNF-α levels was observed when monocytes were preincubated with the JAK1 inhibitor, indicating that the JAK1/Stat3 pathway may not be the only signaling pathway utilized in inhibiting cytokine synthesis in monocytes. Indeed, further investigation demonstrated that Stat3 phosphorylation was also achieved by interaction with phosphoinositide 3-kinase (PI3K), but phosphorylation occurred at a different position in the Stat3 sequence, where S727 was phosphorylated by PI3K, whereas Y705 was phosphorylated by JAK1 (92). TNF-α expression was somewhat restored by the addition of PI3K inhibitors, indicating that PI3K-mediated Stat3 phosphorylation plays a role in the induction of proinflammatory cytokine inhibition by cmvIL-10 in monocytes. However, the reversal of TNF-α inhibition by the blockage of PI3K was only partial, thus indicating that cmvIL-10 may use other IL-10R-signaling pathways or even utilize other receptors to accomplish immunosuppression of proinflammatory cytokines.
While the study by Spencer et al. (94) reported reduced MHC class I surface expression on LPS-stimulated human monocytes exposed to recombinant cmvIL-10, Pepperl-Klindworth et al. (76) did not observe a function for cmvIL-10 in the context of virus infection and its impact on MHC class I-restricted antigen presentation by bystander (uninfected) cells. Incubation of the myeloid progenitor cell line THP-1 with recombinant cmvIL-10 protein combined with the supernatants from human fibroblasts productively infected with a cmvIL-10 deletion virus did not impair MHC class I-restricted antigen presentation by the THP-1 cells. In addition, there was no difference in MHC class I-restricted antigen presentation by an EBV-transformed B-cell line, JY-LCL, when these cells were coincubated with fibroblasts productively infected with either parental or vIL-10 deletion viruses. These experiments suggest that in the context of productive HCMV infection, cmvIL-10 does not impair MHC class I-restricted antigen presentation by uninfected bystander cells, the implication being that the milieu of cytokines secreted by productively infected cells affects the net immunomodulatory impact of secreted cmvIL-10 on MHC class I function by bystander cells. Alternatively, the number of surface MHC class I molecules on bystander cells may be reduced by cmvIL-10, but the number remaining may still be sufficient for antigen presentation. These possibilities remain to be explored, as does the determination of whether productive infection influences the capacity of secreted cmvIL-10 to modulate MHC class I-restricted function by primary monocytes and other myeloid cell types.
There is strong evidence that cmvIL-10 negatively affects DC function. It has been shown that cmvIL-10 prevents LPS-induced maturation and proinflammatory cytokine production of immature DC treated with culture supernatants from HCMV-infected cells but not culture supernatants from cells infected with a cmvIL-10 deletion virus (8). In addition, although cmvIL-10 enhanced the migration of mature DC, it also greatly suppressed their cytokine production. The prolonged effect of cmvIL-10 on activated DC was demonstrated by IL-12 production which could not be reestablished even after DC restimulation with the CD40 ligand in the absence of cmvIL-10 (8). Raftery et al. (80) evaluated various effects of cmvIL-10 on immature, LPS-induced, and mature DC, showing strong activation of a mediator of IL-10 response, Stat3, in immature DC as well as upregulation of a lectin receptor, DC-SIGN, on immature but not on mature cells. One of the reported functions of DC-SIGN is as a coreceptor for HCMV entry (34), raising the possibility that cmvIL-10 may also aid viral entry by increasing DC-SIGN expression on immature DC, although this function remains to be shown in any experimental setting. This study also reported a partial blockage of MHC class I and class II upregulation accompanied by an increase in expression of the nonclassical MHC molecule HLA-DM. Upregulation of the costimulatory molecules CD40, CD80, CD86, B7-DC, and B7-H1 was also prevented by cmvIL-10 on LPS-stimulated DC, and cmvIL-10 increased apoptosis associated with DC maturation by blocking upregulation of antiapoptotic long-form cellular FLIP. Continuing work from the same group demonstrated that the number of transcripts, as well as cell surface expression of the CD1 group of molecules (nonclassical major MHC class I molecules), was also inhibited by cmvIL-10 in DC (79). Together, these studies highlight a role for cmvIL-10 in repressing a critical arm of the immune response by shaping the phenotype of DC.
In addition, a recent study demonstrated that cmvIL-10, like IL-10, can directly suppress synthesis of type I IFNs in plasmacytoid DC, which represent, under normal circumstances, the predominant source of type I IFNs (6). Plasmacytoid DC are highly resistant to infection with HCMV, suggesting that cmvIL-10 acts in trans to potently inhibit type I IFN production. Since type I IFNs block infection of susceptible cells by HCMV in a dose-dependent manner, the results further suggest that secretion of cmvIL-10 by infected cells may facilitate the systemic spread of HCMV during the earliest stages of infection and attenuate the development of antiviral immune responses.
The impact of cmvIL-10 has also been assessed on microglial cells (resident macrophages exclusive to the brain) to shed light on the potential subversion of neuroimmune responses during HCMV encephalitis. Pretreatment of these cells with recombinant cmvIL-10 protein prior to HCMV infection resulted in a striking decrease in the protein levels of a potent antiviral molecule, CXC chemokine ligand 10 (CXCL10), in infected microglial cells. This chemokine is strongly induced upon infection with HCMV, which leads to increased chemotaxis of activated T cells toward infected cells (9). The treatment of microglial macrophages with conditioned media from astrocytes productively infected with HCMV in the presence or absence of anti-IL-10R neutralizing antibody showed a decrease of CXCL10 levels in samples where cellular IL-10Rs were not blocked by this antibody, further highlighting a role of cmvIL-10 in the control of CXCL10 expression by microglial cells (9).
In the context of congenital infection, which remains one of the most clinically significant aspects of HCMV infection, Yamamoto-Tabata et al. (108) demonstrated that recombinant cmvIL-10 increased hIL-10 expression by cytotrophoblasts and decreased matrix metalloproteinase activity in endothelial cells and cytotrophoblasts and impaired endothelial cell migration in wound closure assays and cytotrophoblast invasiveness of Matrigel. These are critical components required for proper development of the placental-uterine interface, raising the possibility that cmvIL-10 expressed during HCMV infection of pregnant women may impair cytotrophoblast remodeling of the uterine vasculature and consequently limit fetal growth. This finding provides stimulus for a more detailed analysis of the role cmvIL-10 may play in congenital HCMV infection.
In an example of the potential to harness the immunosuppressive properties of cmvIL-10 for a therapeutic purpose, the impact of cmvIL-10 on the foreign body reaction (FBR) to implanted biomaterial has been assessed. The use of biomaterials such as temporary scaffolds for structural support to assist in the delivery of cells or soluble mediators to repair damaged organs is complicated by the host FBR, an inflammatory reaction to degrade the foreign biomaterial. In this setting, cmvIL-10 has been reported to impair the progression of the FBR to hexamethylene diisocyanate-cross-linked dermal sheep collagen discs loaded with recombinant cmvIL-10 and implanted into rats, suggesting that cmvIL-10 may be a useful adjunct in biomaterials for regenerative medicine (102).
Apart from its immunomodulatory properties on myeloid cells, cmvIL-10 has been shown to induce the growth and differentiation of a human B-cell lymphoma Daudi cell line, increasing the number of cells by 60% compared to the number of untreated cells (93). cmvIL-10 also increased the levels of hIL-10 and, to a smaller extent, IL-10R produced by B cells and did not alter surface MHC class II levels. Thus, like hIL-10, cmvIL-10 can act in either an immunosuppressive or an immunostimulatory capacity, depending on the cell type to which it is exposed.
vIL-10 EXPRESSION DURING LATENT HCMV INFECTION
HCMV confronts the challenge of latently infecting myeloid cells that are themselves components of the immune system. The capacity of HCMV to establish and maintain latency in these cells in the face of a robust host immune system argues for a dynamic interplay between virus and host, where HCMV encodes immunomodulatory functions that ensure its survival during this phase of infection. In both an experimental model of latent infection utilizing primary human myeloid progenitor cells and in mononuclear cells from healthy bone marrow and mobilized peripheral blood donors, transcriptional activity from the UL111A gene region has been detected by reverse transcription-PCR (42). The transcript was designated latency associated cmvIL-10 (LAcmvIL-10). The precise initiation site of LAcmvIL-10 transcription was isolated to a 38-bp region between nucleotides 159577 and 159615 (AD169 strain), as opposed to the initiation site of cmvIL-10 transcription, which has been mapped to nucleotide position 159642. Nonetheless, the initiation of translation of both LAcmvIL-10 and cmvIL-10 is predicted to occur at the same methionine at nucleotide position 159678, and the termination site of LAcmvIL-10 transcription is shared with that of cmvIL-10 at nucleotide position 160430. The major difference between the LAcmvIL-10 transcript and the cmvIL-10 transcript lies in differential splicing; LAcmvIL-10 retains only the first of the two introns found in cmvIL-10, resulting in an in-frame stop codon at nucleotide position 160171 (AD169 strain). As a consequence, the LAcmvIL-10 protein shares the first 127 amino acids with cmvIL-10 but then diverges for its final 12 amino acids at the C terminus, resulting in a truncated protein of 139 amino acids (42). At the protein level, LAcmvIL-10 shares 27% identity and 46% similarity with hIL-10.
The assessment of biological properties of LAcmvIL-10 has thus far been carried out mainly using recombinant proteins generated from either bacterial or mammalian cell expression systems. LAcmvIL-10 reduces cell surface MHC class II expression by monocytes and more-primitive myeloid progenitor cells in a manner comparable to that of cmvIL-10. Both LAcmvIL-10 and cmvIL-10 reduce the levels of transcription of components of the MHC class II biosynthesis pathway, including the HLA-DRα chain, HLA-DRβ chain, and the invariant chain. Transcription of the master regulator of MHC class II, the class II transactivator (CIITA), which controls expression of these MHC class II components, is also reduced by LAcmvIL-10 and cmvIL-10, pointing to a mechanism of MHC class II downregulation at the level of the transcriptional activity of CIITA (44). In the same study, MHC class II was also observed to accumulate in punctate cytoplasmic vesicles in a small proportion (3 to 5%) of monocytes treated with LAcmvIL-10 or cmvIL-10, suggesting that in addition to control at the level of transcription of CIITA, LAcmvIL-10 and cmvIL-10 may exert a repressive effect on MHC class II at the posttranslational level during assembly and/or transport to the cell surface. In this respect, hIL-10 exposure to primary human monocytes has been reported to induce expression of the membrane-associated RING-CH (MARCH) ubiquitin ligase MARCH1, which in turn acts as a posttranslational inhibitor of MHC class II expression (98). It is therefore possible that any posttranslational control of MHC class II by cmvIL-10 and/or LAcmvIL-10 could be manifested via upregulation of MARCH1, and this is an area that warrants further investigation.
Although both LAcmvIL-10 and cmvIL-10 downmodulate MHC class II on myeloid lineage cells, LAcmvIL-10 does not appear to share the full repertoire of immunmodulatory functions encoded by cmvIL-10. The immunosuppressive effects of cmvIL-10 on DC maturation (8, 80) were not replicated by LAcmvIL-10 on monocyte-derived DC stimulated with LPS (44). Furthermore, unlike cmvIL-10 and hIL-10, LAcmvIL-10 was unable to stimulate the proliferation of the Daudi B cell line (93), nor was it able to enhance Fcγ receptor-mediated phagocytosis by human monocytes (40). These findings demonstrate that LAcmvIL-10 retains only a small subset of the biological properties of cmvIL-10 (and hIL-10).
Whether cmvIL-10 is expressed during latency remains to be determined, but transcripts predicted to encode LAcmvIL-10 are expressed during productive infection of human fibroblasts (43). The functional contribution of LAcmvIL-10 during productive infection in unclear, but expression of latency-associated viral transcripts during both latent and productive phases of infection has been reported for other HCMV transcripts, including the major immediate-early region latent transcripts (CLTs) and transcripts from the UL138 gene (29, 62), as well as latency-associated transcripts expressed by other herpesviruses, herpes simplex virus type 1, varicella-zoster virus, EBV, and pseudorabies virus (11, 14, 46, 56, 91, 95, 106, 107).
Clues to the mechanistic basis for the differences in functions encoded by LAcmvIL-10 and cmvIL-10 can be found by analysis of their receptor usage. cmvIL-10 functions by binding and signaling through the hIL-10R (44, 54, 94). This function is highlighted by the induction of phosphorylation of Stat3, which is a well-documented signaling outcome following binding of hIL-10 or cmvIL-10 to the hIL-10R (20, 21, 53, 83, 92), as well as by the capacity to block immunosuppressive functions of cmvIL-10 by pretreating monocytes with neutralizing antibodies to hIL-10R (44, 94). In contrast, treatment of monocytes with LAcmvIL-10 does not induce Stat3 phosphorylation, and LAcmvIL-10 retains the capacity to downregulate MHC class II in the presence of hIL-10R-neutralizing antibodies (44, 94). Thus, either LAcmvIL-10 binds and signals through a different, as yet unidentified receptor, or it still binds to the hIL-10R but does so in a manner which differs from that of cmvIL-10 and hIL-10. Jones et al. (47) resolved the crystal structure of cmvIL-10 bound to hIL-10R1 and identified several regions in cmvIL-10 important for binding to the hIL-10R. It was shown that that cmvIL-10 uses essentially the same structural epitope as that described for hIL-10, with helix A, the AB loop, and helix F contacting IL-10R1. In the case of LAcmvIL-10, the absence of amino acid residues coded by exon 3 of the UL111A transcript results in the loss of the C-terminal helices E/F, which are present in cmvIL-10. It has therefore been postulated that the truncation in LAcmvIL-10 may result in a diminished capacity for LAcmvIL-10 to dimerize or signal through the IL-10R complex, accounting for the observed differences in the repertoire of immunomodulatory functions encoded by LAcmvIL-10 and cmvIL-10 (44).
In addition to functional studies utilizing recombinant vIL-10 proteins, it has recently been reported that the HCMV UL111A gene can function to modulate immune function in the context of latent infection of primary myeloid progenitor cells (10a). Utilizing a UL111A delection virus which cannot express any vIL-10, the authors showed that in both allogeneic and autologous settings, CD4+ T-cell recognition of latently infected myeloid progenitors was regulated by this viral gene and that this was likely a consequence of vIL 10-mediated suppression of the capacity of latently infected cells to present antigenic peptides via MHC class II. This study provides the first evidence that a vIL-10 protein(s) expressed during latency from the UL111A gene can render latently infected cells refractory to CD4+ T-cell recognition, implicating this viral gene in maintenance of latency in the immunocompetent human host.
ADDITIONAL HCMV IL-10 ISOFORMS
In addition to cmvIL-10 and LAcmvIL-10 transcripts, five other, less-abundant vIL-10-like RNAs have been reported during productive HCMV infection of MRC-5 fibroblast cells (58). In this study, the start and stop sites of transcription were not defined, and cDNA was amplified from the position of the methionine used to initiate cmvIL-10 and LAcmvIL-10 translation at nucleotide position 159678, but based on alternate splicing patterns these transcripts were predicted to encode novel vIL-10 homologs. Proteins expressed from cDNA constructs from these transcripts reacted with anti-cmvIL-10 antibody, and some evidence of protein expression of several of these isoforms was detected in productively infected MRC-5 cells. None of the putative new isoforms were able to induce Stat3 phosphorylation, but the authors contend that all vIL-10 isoforms are able to form heterodimers with hIL-10 and that at least in the case of cmvIL-10, interaction with hIL-10 may enhance the capacity of hIL-10 to induce Stat3 phosphorylation. Additional experiments will be required to strengthen these conclusions, and it remains to be determined whether these smaller products were not a consequence of the degradation of the full-length cmvIL-10 protein, but this study points to there being greater complexity in expression from the UL111A gene region during productive infection.
vIL-10 HOMOLOGS ENCODED BY NONHUMAN VIRUSES
Although a number of nonhuman viruses have been identified as encoding IL-10 homologs (Table 1), the biological properties of most of them remain to be determined. This section summarizes what is currently known about the functions of those homologs that have been subjected to some degree of functional characterization, namely, ORFV, OvHV2, and rhesus CMV (RhCMV).
Recombinant ORFV vIL-10, which has been extensively studied (24-26, 33, 38, 55, 66, 105), inhibits TNF-α and IL-8 synthesis in activated ovine and murine macrophages and stimulates ovine mast cell proliferation at a level of activity indistinguishable from that of ovine IL-10. Since ORFV vIL-10 is 91% identical to ovine IL-10, comparable functional activity is not unexpected. However, a similar phenotype is observed for OvHV2 vIL-10, which retains only 41% amino acid identity to that of its ovine IL-10. The vIL-10 of OvHV2 stimulates mast cell proliferation and inhibits IL-8 production from LPS-activated macrophages (41). As in other studies, there is nothing that separates the phenotype of OvHV2 vIL-10 from that of ovine IL-10. The importance of these functions to the natural histories of ORFV and OvHV2 has been demonstrated by analysis of engineered viral variants in which the vIL-10 gene has been deleted.
The ORFV vIL-10 gene was disrupted and partially deleted by engineering a lacZ cassette into the vIL-10 coding region within the viral genome (24). Recovered virus (vIL-10ko) was used to inoculate naïve sheep by scarification in parallel with groups of sheep inoculated with either the parental unmodified virus or “rescued” virus in which the fidelity of the vIL-10 gene mutated with the lacZ cassette had been restored. In all cases, the severity of the virus-induced lesions was reduced in the vIL-10ko-inoculated sheep compared to that of both those inoculated with the parental virus and those inoculated with the “rescued” viruses. The severity of the lesions was reduced over a range of inoculation titers (104 to 106 PFU), and statistically significant reductions were observed at every time point following inoculation with 105 and 106 PFU of virus. Statistical significance was observed at a single time point following inoculation with 104 PFU, corresponding to the time of greatest lesion severity in the animals inoculated with the wild type. The loss of vIL-10 expression resulting from the lacZ insertion resulted in a 100- to 1,000-fold reduction in lesion severity. The authors of this study concluded that “the vorfIL-10 gene is important in the establishment of infection by ORFV and that innate responses (e.g., IFNs and TNF) could be important in inhibiting ORFV replication during the early stages of infection.” This statement effectively captures what may be the most salient aspect of vIL-10 functionality in the context of productive replication, namely, the manipulation of the earliest virus-host interactions to enable an immune environment promoting the successful completion of the virus's infectious life cycle.
To date, RhCMV-encoded IL-10 has been subjected only to limited published functional analysis. In a study of DNA vaccination of rhesus macaques, delivery of a construct expressing RhCMV IL-10 did not alter the protective effects of DNA vaccination with constructs expressing variants of two RhCMV-immunogenic viral proteins, gB and pp65-2 (112). The vIL-10 plasmid construct stimulated weak antibody and cellular responses after three genetic immunizations. Notable anamnestic responses were observed after challenge, demonstrating that immunization with the vIL-10 plasmid primed memory immune cells. RhCMV vIL-10 is immunogenic during natural infection; vIL-10-specific T-cell responses are detected in long-term-infected macaques, and degranulation detected by flow cytometry reveals the phenotypic heterogeneity of degranulating CMV-specific CD8+ T lymphocytes in rhesus macaques (5).
DIVERGENCE AND CONSERVATION OF vIL-10 WITH HOST IL-10
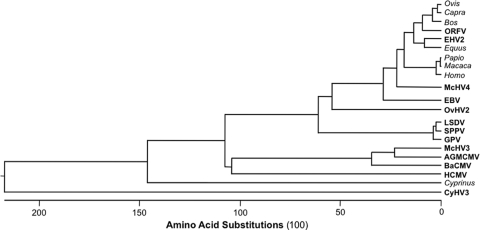
The progenitor Herpesviridae is estimated to have originated 200 to 240 million years ago (65), and it is generally thought that descendent members coevolved with their hosts during species radiation. Consistent with this, all but one (OvHV2) of the vIL-10 proteins of the Gammaherpesvirinae retain high sequence identity with the IL-10 protein of their respective hosts (84 to 91%). In marked contrast, OvHV2 and the Betaherpesvirinae vIL-10 proteins are highly divergent from the IL-10 protein of their host, retaining only 41% (OvHV2) and 25 to 29% amino acid identity, respectively (Table 1 and Fig. 2) (41, 61). The vIL-10 ORFs of HCMV, RhCMV, ORFV, and OvHV2 are not hypervariable regions among different isolates of the viruses; independent isolates exhibit 98 to 100% sequence identity (26, 27, 41, 61). This indicates that the marked sequence divergence between a vIL-10 and its host's IL-10 probably occurred early during species radiation. Another way to appreciate the extent of genetic drift is that the vIL-10 proteins of HCMV and RhCMV are as divergent from their host's IL-10 as they are from each other. The selective forces driving this genetic drift and the reasons why it appears to be limited to the Betaherpesvirinae are unknown. One potential explanation that can be excluded is that divergence represents compensatory genetic drift in the viral ligand as a result of divergence in the cellular IL-10Rs. In fact, the high-affinity IL-10Rs (IL-10R1) in humans and rhesus macaques (Macaca mulatta) (GenBank accession numbers AAA17896 and XP_001092736, respectively) are 90% identical. Moreover, if compensatory drift was the mechanistic basis for the divergence of the Betaherpesvirinae vIL-10 proteins, then it would be expected that there would be comparable drift in the vIL-10-containing Gammaherpesvirinae infecting the same host. Both EBV and macacine herpesvirus 4 are strongly conserved with their IL-10 homologs (71, 84). It is noteworthy that within the Cytomegalovirus genus of Betaherpesvirinae, the closest evolutionary relative to HCMV, i.e., chimp CMV, does not encode an IL-10 homolog (13). The vIL-10 gene is found in the same genomic location in the monkey CMVs (macacine herpesvirus 3, African green monkey CMV, and baboon CMV) as in HCMV. Therefore, the lack of vIL-10 in chimp CMV indicates a deletion of this locus at some point after the split of the great apes (Hominidae) and the Old World monkeys (Cercopithecidae). The progenitors of fish and mammals split 400 million years ago, and it is not surprising that the extent of amino acid substitutions in the IL-10 proteins of the common carp and mammals reflect this ancient evolutionary separation. The vIL-10 of cyprinid herpesvirus 3 has undergone extensive divergence relative to its host, suggesting that selective forces similar to those acting on the Betaherpesvirinae may have been at work for this fish herpesvirus. A comparable pattern of genetic conservation and drift is apparent within the Poxviridae; three closely-related viruses (SPPV, GPV, and LSDV) express vIL-10 proteins highly divergent from their host's IL-10 (38 to 45% sequence identity), while the vIL-10 of ORFV is 80% identical to ovine IL-10.
FIG. 2.
Alignment of cellular and vIL-10 proteins. Proteins were aligned by the ClustalW method, using the MegAlign software program (Lasergene, DNASTAR). Cellular and vIL-10 host names are shown in italic and bold fonts, respectively. Nucleotide sequence accession numbers for the indicated genera or viruses are as follows: Ovis (sheep; GenBank accession number NP_001009327), Capra (goat; ABI20513), Bos (bovine; NP_776513), ORFV (AAC57332), equine herpesvirus 2 ([EHV2] AAB26148), Equus (horse; NP_001075959), Papio (baboon; Q5Q0V6), Macaca (rhesus macaque [Macaca mulatta]; NP_001038192), Homo sapiens (human; P22301), macacine herpesvirus 4 [McHV4]/rhesus lymphocryptovirus (AAK95412), EBV/human herpesvirus 4 (P03180), OvHV2 (ABB22222), LSDV (AAN02729), SPPV (NP_659579), GPV (GPV Pellor; YP_001293197), McHV3/RhCMV (AAF59907), African green monkey CMV ([AGMCMV] AAF63435), baboon CMV ([BaCMV] AAF63436), HCMV/human herpesvirus 5 (AAF36285), Cyprinus (common carp [Cyprinus carpio]; BAC76885), cyprinid herpesvirus 3 ([CyHV3] YP_001096169).
A more-likely explanation for the higher rate of amino acid substitutions with particular vIL-10 proteins compared to that with host IL-10 is that functional changes associated with change in the protein sequence conferred some selective advantage over retention of the sequence of a transduced host gene. The vIL-10 genes of HCMV and the monkey CMVs consist of three and four exons, respectively, whereas the IL-10 gene is composed of five exons. Despite the loss of one and two introns in the vIL-10 genes of monkey CMV and HCMV, respectively, the remaining exon/exon boundaries are absolutely conserved with those of IL-10. Similarly, the splicing pattern of OvHV2 is identical to that of ovine IL-10 (five exons). The splicing patterns are consistent with the interpretation that the vIL-10 ORF represent the remnants of an original transduced IL-10 gene. Accordingly, accumulation of amino acid changes in the transduced IL-10 protein would convert what was initially a “self” protein expressed from the viral genome into more of a “nonself” protein. Since vIL-10 is immunogenic in an infected host, presumably selective advantages conferred by alteration in protein sequence exceeded the costs associated with increased immune recognition by the host. It is reasonable to predict that the divergence of vIL-10 proteins from host IL-10 was a positive selection for the alteration of vIL-10 functionality, and this is supported by studies of ebvIL-10 and the latency-associated homolog encoded by HCMV (LAcmvIL-10), which demonstrate differences from hIL-10 in the repertoire of biological functions and/or the mechanism of action (28, 44, 59, 64, 93, 103). However, in vitro assays have shown that the phenotype of full-length HCMV IL-10 (cmvIL-10) mimics that of hIL-10 in terms of the functional alterations to various cell types, signaling pathways, and changes in gene expression (7, 8, 94), and the same is true for the limited data available for other vIL-10 proteins.
CONCLUDING REMARKS
An important role of the potent immunomodulatory cytokine IL-10 in virus infections has become increasingly clear in recent years, and the acquisition of IL-10-like sequences by viruses represents a remarkable evolutionary step which likely equips the virus with a powerful tool to modulate host immune function. A range of both human and nonhuman viruses have been shown to encode IL-10 homologs, and the number will undoubtedly grow as new sequence information becomes available. Whether in acute, persistent, or latent infection settings, the expression of virus-encoded homologs of IL-10 would serve as an efficient means to interfere with multiple immune components to ensure successful infection of the host. To date, all of the virus-encoded IL-10 homologs tested have demonstrated immunomodulatory properties, although these homologs do not always mimic the full range of properties encompassed by their cellular counterparts, nor by each other, highlighting the complexity of this means of virus-encoded immune control. While there is still much to be understood about the full extent of the direct and indirect impacts of vIL-10 on the host immune response to virus infections, their discovery and the functional studies performed so far provide a fascinating insight into the capacity of some viruses to acquire a potent weapon with the potential to significantly influence the course of infection.
Acknowledgments
The authors thank Mark Walter of the University of Alabama at Birmingham for providing the structures of hIL-10, ebvIL-10, and cmvIL-10 bound to the hIL-10R, which are shown in Fig. 1.
Biography
 Barry Slobedman completed his Ph.D. in 1995 at the University of Adelaide in Australia, studying HSV latency. From 1996 to 1999 he trained as a Postdoctoral Fellow in the Department of Microbiology and Immunology at Stanford University, examining molecular aspects of latent HCMV infection of myeloid lineage cells. He returned to Australia to establish a research group in the Centre for Virus Research (CVR) at the Westmead Millennium Institute (WMI) and University of Sydney in 2000, where he has continued to work on HCMV latency and pathogenesis. A major aim of his research program is to identify both viral and host cell factors that contribute to the capacity of HCMV to establish, maintain, and reactivate from latency in the human host. He is currently a Senior Research Fellow, University of Sydney, and Deputy Director of CVR at WMI.
Barry Slobedman completed his Ph.D. in 1995 at the University of Adelaide in Australia, studying HSV latency. From 1996 to 1999 he trained as a Postdoctoral Fellow in the Department of Microbiology and Immunology at Stanford University, examining molecular aspects of latent HCMV infection of myeloid lineage cells. He returned to Australia to establish a research group in the Centre for Virus Research (CVR) at the Westmead Millennium Institute (WMI) and University of Sydney in 2000, where he has continued to work on HCMV latency and pathogenesis. A major aim of his research program is to identify both viral and host cell factors that contribute to the capacity of HCMV to establish, maintain, and reactivate from latency in the human host. He is currently a Senior Research Fellow, University of Sydney, and Deputy Director of CVR at WMI.
Peter A. Barry completed his Ph.D. in 1985 at the University of Utah. From 1998 to 2001 he was an Assistant Professor at the Center for Comparative Medicine (CCM), Department of Medical Pathology and Laboratory Medicine (DOPLM), University of California, Davis (UC Davis). From 1999 to 2007, he was a Staff Scientist at the California National Primate Research Center (CNPRC). From 2001 to 2007 he was an Associate Professor at the CCM, DOPLM, UC Davis. Since 2007, he has been a Professor at the CCM, Department of Medical Pathology, Infectious Diseases Unit Leader, CNPRC, and Vice Chair for Research, DOPLM, UC Davis. The main emphasis of his research addresses the mechanisms of RhCMV persistence and pathogenesis. A major aim is to identify viral determinants of latency, persistence, and pathogenesis in rhesus macaques. The goals of his lab are (i) to define mechanisms by which RhCMV establishes a persistent infection, (ii) to characterize immunological and virological determinants of pathogenesis, (iii) to identify immunological determinants of protective immune responses, and (iv) to design novel immunobased strategies that limit and/or prevent RhCMV infection and disease.
 Juliet V. Spencer completed her graduate research on HSV-1 capsid assembly in Jay C. Brown's laboratory at the University of Virginia (UVA). She then joined Thomas J. Braciale's group in UVA's Carter Immunology Center as a Parker B. Francis Fellow and studied the immune response to influenza virus infection. She continued her postdoctoral training at ChemoCentryx, Inc., where she focused on the validation of viral chemokine receptors as potential drug targets. Now an Associate Professor of Biology at the University of San Francisco, Dr. Spencer's research focuses on immune evasion strategies of HCMV with the aim of understanding how the immune system is manipulated to prevent virus clearance and allow the establishment of latency.
Juliet V. Spencer completed her graduate research on HSV-1 capsid assembly in Jay C. Brown's laboratory at the University of Virginia (UVA). She then joined Thomas J. Braciale's group in UVA's Carter Immunology Center as a Parker B. Francis Fellow and studied the immune response to influenza virus infection. She continued her postdoctoral training at ChemoCentryx, Inc., where she focused on the validation of viral chemokine receptors as potential drug targets. Now an Associate Professor of Biology at the University of San Francisco, Dr. Spencer's research focuses on immune evasion strategies of HCMV with the aim of understanding how the immune system is manipulated to prevent virus clearance and allow the establishment of latency.
 Selmir Avdic completed his Bachelor of Science degree with Honors at the University of Technology in Sydney, Australia, in 2003. He went on to undertake Ph.D. studies on the modulation of immune function by HCMV and is currently a final-year graduate student at the Centre for Virus Research, Westmead Millennium Institute and University of Sydney. His main area of research focuses on the immunomodulatory properties of vIL-10 homologs encoded by HCMV during the productive and latent phases of infection.
Selmir Avdic completed his Bachelor of Science degree with Honors at the University of Technology in Sydney, Australia, in 2003. He went on to undertake Ph.D. studies on the modulation of immune function by HCMV and is currently a final-year graduate student at the Centre for Virus Research, Westmead Millennium Institute and University of Sydney. His main area of research focuses on the immunomodulatory properties of vIL-10 homologs encoded by HCMV during the productive and latent phases of infection.
 Allison Abendroth completed her Ph.D. in 1996 on the immunobiology of HSV at the University of Adelaide, Australia. Postdoctoral training (1996-1999) at Stanford University with Ann Arvin enabled her to further her interest in herpesvirus immunobiology and pathogenesis. On returning to Australia, she established the Varicella-Zoster Virus (VZV) Research Group at the Westmead Millennium Institute and University of Sydney. She was subsequently appointed as a Senior Lecturer in Immunology at the University of Sydney in the Department of Infectious Diseases and Immunology. In addition to her work on HCMV-encoded IL-10, her research focuses on VZV immunobiology with the goal of determining how VZV affects specialized human cells in order to make advances in antiviral therapies as well improve vaccine design for the treatment or prevention of VZV disease and the crippling complication of postherpetic neuralgia following herpes zoster.
Allison Abendroth completed her Ph.D. in 1996 on the immunobiology of HSV at the University of Adelaide, Australia. Postdoctoral training (1996-1999) at Stanford University with Ann Arvin enabled her to further her interest in herpesvirus immunobiology and pathogenesis. On returning to Australia, she established the Varicella-Zoster Virus (VZV) Research Group at the Westmead Millennium Institute and University of Sydney. She was subsequently appointed as a Senior Lecturer in Immunology at the University of Sydney in the Department of Infectious Diseases and Immunology. In addition to her work on HCMV-encoded IL-10, her research focuses on VZV immunobiology with the goal of determining how VZV affects specialized human cells in order to make advances in antiviral therapies as well improve vaccine design for the treatment or prevention of VZV disease and the crippling complication of postherpetic neuralgia following herpes zoster.
Footnotes
Published ahead of print on 29 July 2009.
REFERENCES
- 1.Aoki, T., I. Hirono, K. Kurokawa, H. Fukuda, R. Nahary, A. Eldar, A. J. Davison, T. B. Waltzek, H. Bercovier, and R. P. Hedrick. 2007. Genome sequences of three koi herpesvirus isolates representing the expanding distribution of an emerging disease threatening koi and common carp worldwide. J. Virol. 81:5058-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkman, N., M. John, G. Roesems, P. J. Jose, P. J. Barnes, and K. F. Chung. 1995. Inhibition of macrophage inflammatory protein-1 alpha expression by IL-10. Differential sensitivities in human blood monocytes and alveolar macrophages. J. Immunol. 155:4412-4418. [PubMed] [Google Scholar]
- 3.Brady, M. T., A. J. MacDonald, A. G. Rowan, and K. H. Mills. 2003. Hepatitis C virus non-structural protein 4 suppresses Th1 responses by stimulating IL-10 production from monocytes. Eur. J. Immunol. 33:3448-3457. [DOI] [PubMed] [Google Scholar]
- 4.Brockman, M. A., D. S. Kwon, D. P. Tighe, D. F. Pavlik, P. C. Rosato, J. Sela, F. Porichis, S. Le Gall, M. T. Waring, K. Moss, H. Jessen, F. Pereyra, D. G. Kavanagh, B. D. Walker, and D. E. Kaufmann. 2009. IL-10 is upregulated in multiple cell types during viremic HIV infection and reversibly inhibits virus-specific T cells. Blood 114:346-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, K. S., and A. Kaur. 2007. Flow cytometric detection of degranulation reveals phenotypic heterogeneity of degranulating CMV-specific CD8+ T lymphocytes in rhesus macaques. J. Immunol. Methods 325:20-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, W. L., P. A. Barry, R. Szubin, D. Wang, and N. Baumgarth. 2009. Human cytomegalovirus suppresses type I interferon secretion by plasmacytoid dendritic cells through its interleukin 10 homolog. Virology 390:330-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, W. L., N. Baumgarth, M. K. Eberhardt, C. Y. Lee, C. A. Baron, J. P. Gregg, and P. A. Barry. 2007. Exposure of myeloid dendritic cells to exogenous or endogenous IL-10 during maturation determines their longevity. J. Immunol. 178:7794-7804. [DOI] [PubMed] [Google Scholar]
- 8.Chang, W. L., N. Baumgarth, D. Yu, and P. A. Barry. 2004. Human cytomegalovirus-encoded interleukin-10 homolog inhibits maturation of dendritic cells and alters their functionality. J. Virol. 78:8720-8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheeran, M. C., S. Hu, W. S. Sheng, P. K. Peterson, and J. R. Lokensgard. 2003. CXCL10 production from cytomegalovirus-stimulated microglia is regulated by both human and viral interleukin-10. J. Virol. 77:4502-4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, B., M. H. Kapturczak, R. Joseph, J. F. George, M. Campbell-Thompson, C. H. Wasserfall, M. A. Atkinson, C. C. Tisher, T. R. Flotte, A. Agarwal, and S. Chen. 2007. Adeno-associated viral vector-mediated interleukin-10 prolongs allograft survival in a rat kidney transplantation model. Am. J. Transplant. 7:1112-1120. [DOI] [PubMed] [Google Scholar]
- 10a.Cheung, A. K. L., D. J. Gottlieb, B. Plachter, S. Pepperl-Klindworth, S. Avdic, A. L. Cunningham, A. Abendroth, B. Slobedman. 25 August 2009, posting date. The role of the human cytomegalovirus ULIIIA gene in downregulating CD4+ T cell recognition of latently infected cells: implications for virus elimination during latency. Blood doi: 10.1182/blood-2008-12-197111. [DOI] [PubMed]
- 11.Cohrs, R. J., J. Wischer, C. Essman, and D. H. Gilden. 2002. Characterization of varicella-zoster virus gene 21 and 29 proteins in infected cells. J. Virol. 76:7228-7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couper, K. N., D. G. Blount, and E. M. Riley. 2008. IL-10: the master regulator of immunity to infection. J. Immunol. 180:5771-5777. [DOI] [PubMed] [Google Scholar]
- 13.Davison, A. J., A. Dolan, P. Akter, C. Addison, D. J. Dargan, D. J. Alcendor, D. J. McGeoch, and G. S. Hayward. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 84:17-28. [DOI] [PubMed] [Google Scholar]
- 14.Debrus, S., C. Sadzot-Delvaux, A. F. Nikkels, J. Piette, and B. Rentier. 1995. Varicella-zoster virus gene 63 encodes an immediate-early protein that is abundantly expressed during latency. J. Virol. 69:3240-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Defrance, T., B. Vanbervliet, F. Briere, I. Durand, F. Rousset, and J. Banchereau. 1992. Interleukin 10 and transforming growth factor beta cooperate to induce anti-CD40-activated naive human B cells to secrete immunoglobulin A. J. Exp. Med. 175:671-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Waal Malefyt, R., J. Abrams, B. Bennett, C. G. Figdor, and J. E. de Vries. 1991. Interleukin 10 (IL-10) inhibits cytokine synthesis by human monocytes: an autoregulatory role of IL-10 produced by monocytes. J. Exp. Med. 174:1209-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Waal Malefyt, R., J. Haanen, H. Spits, M. G. Roncarolo, A. te Velde, C. Figdor, K. Johnson, R. Kastelein, H. Yssel, and J. E. de Vries. 1991. Interleukin 10 (IL-10) and viral IL-10 strongly reduce antigen-specific human T cell proliferation by diminishing the antigen-presenting capacity of monocytes via downregulation of class II major histocompatibility complex expression. J. Exp. Med. 174:915-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Díaz-San Segundo, F., T. Rodriguez-Calvo, A. de Avila, and N. Sevilla. 21 May 2009. Immunosuppression during acute infection with foot-and-mouth disease virus in swine is mediated by IL-10. PLoS ONE 4:e5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding, Y., L. Qin, S. V. Kotenko, S. Pestka, and J. S. Bromberg. 2000. A single amino acid determines the immunostimulatory activity of interleukin 10. J. Exp. Med. 191:213-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donnelly, R. P., H. Dickensheets, and D. S. Finbloom. 1999. The interleukin-10 signal transduction pathway and regulation of gene expression in mononuclear phagocytes. J. Interferon Cytokine Res. 19:563-573. [DOI] [PubMed] [Google Scholar]
- 21.Finbloom, D. S., and K. D. Winestock. 1995. IL-10 induces the tyrosine phosphorylation of tyk2 and Jak1 and the differential assembly of STAT1 alpha and STAT3 complexes in human T cells and monocytes. J. Immunol. 155:1079-1090. [PubMed] [Google Scholar]
- 22.Fiorentino, D. F., M. W. Bond, and T. R. Mosmann. 1989. Two types of mouse T helper cell. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. J. Exp. Med. 170:2081-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiorentino, D. F., A. Zlotnik, T. R. Mosmann, M. Howard, and A. O'Garra. 1991. IL-10 inhibits cytokine production by activated macrophages. J. Immunol. 147:3815-3822. [PubMed] [Google Scholar]
- 24.Fleming, S. B., I. E. Anderson, J. Thomson, D. L. Deane, C. J. McInnes, C. A. McCaughan, A. A. Mercer, and D. M. Haig. 2007. Infection with recombinant orf viruses demonstrates that the viral interleukin-10 is a virulence factor. J. Gen. Virol. 88:1922-1927. [DOI] [PubMed] [Google Scholar]
- 25.Fleming, S. B., D. M. Haig, P. Nettleton, H. W. Reid, C. A. McCaughan, L. M. Wise, and A. Mercer. 2000. Sequence and functional analysis of a homolog of interleukin-10 encoded by the parapoxvirus orf virus. Virus Genes 21:85-95. [DOI] [PubMed] [Google Scholar]
- 26.Fleming, S. B., C. A. McCaughan, A. E. Andrews, A. D. Nash, and A. A. Mercer. 1997. A homolog of interleukin-10 is encoded by the poxvirus orf virus. J. Virol. 71:4857-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garrigue, I., M. F. Corte, N. Magnin, L. Couzi, S. Capdepont, C. Rio, P. Merville, J. Dechanet-Merville, H. Fleury, and M. E. Lafon. 2007. Variability of UL18, UL40, UL111a and US3 immunomodulatory genes among human cytomegalovirus clinical isolates from renal transplant recipients. J. Clin. Virol. 40:120-128. [DOI] [PubMed] [Google Scholar]
- 28.Go, N. F., B. E. Castle, R. Barrett, R. Kastelein, W. Dang, T. R. Mosmann, K. W. Moore, and M. Howard. 1990. Interleukin 10, a novel B cell stimulatory factor: unresponsiveness of X chromosome-linked immunodeficiency B cells. J. Exp. Med. 172:1625-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodrum, F., M. Reeves, J. Sinclair, K. High, and T. Shenk. 2007. Human cytomegalovirus sequences expressed in latently infected individuals promote a latent infection in vitro. Blood 110:937-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Granelli-Piperno, A., A. Golebiowska, C. Trumpfheller, F. P. Siegal, and R. M. Steinman. 2004. HIV-1-infected monocyte-derived dendritic cells do not undergo maturation but can elicit IL-10 production and T cell regulation. Proc. Natl. Acad. Sci. USA 101:7669-7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Groux, H., M. Bigler, J. E. de Vries, and M. G. Roncarolo. 1998. Inhibitory and stimulatory effects of IL-10 on human CD8+ T cells. J. Immunol. 160:3188-3193. [PubMed] [Google Scholar]
- 32.Gruber, S. G., M. Gloria Luciani, P. Grundtner, A. Zdanov, and C. Gasche. 2008. Differential signaling of cmvIL-10 through common variants of the IL-10 receptor 1. Eur. J. Immunol. 38:3365-3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haig, D. M., J. Thomson, C. J. McInnes, D. L. Deane, I. E. Anderson, C. A. McCaughan, W. Imlach, A. A. Mercer, C. J. Howard, and S. B. Fleming. 2002. A comparison of the anti-inflammatory and immuno-stimulatory activities of orf virus and ovine interleukin-10. Virus Res. 90:303-316. [DOI] [PubMed] [Google Scholar]
- 34.Halary, F., A. Amara, H. Lortat-Jacob, M. Messerle, T. Delaunay, C. Houles, F. Fieschi, F. Arenzana-Seisdedos, J. F. Moreau, and J. Dechanet-Merville. 2002. Human cytomegalovirus binding to DC-SIGN is required for dendritic cell infection and target cell trans-infection. Immunity 17:653-664. [DOI] [PubMed] [Google Scholar]
- 35.Hsu, D. H., R. de Waal Malefyt, D. F. Fiorentino, M. N. Dang, P. Vieira, J. de Vries, H. Spits, T. R. Mosmann, and K. W. Moore. 1990. Expression of interleukin-10 activity by Epstein-Barr virus protein BCRF1. Science 250:830-832. [DOI] [PubMed] [Google Scholar]
- 36.Hsu, D. H., K. W. Moore, and H. Spits. 1992. Differential effects of IL-4 and IL-10 on IL-2-induced IFN-gamma synthesis and lymphokine-activated killer activity. Int. Immunol. 4:563-569. [DOI] [PubMed] [Google Scholar]
- 37.Hudson, G. S., A. T. Bankier, S. C. Satchwell, and B. G. Barrell. 1985. The short unique region of the B95-8 Epstein-Barr virus genome. Virology 147:81-98. [DOI] [PubMed] [Google Scholar]
- 38.Imlach, W., C. A. McCaughan, A. A. Mercer, D. Haig, and S. B. Fleming. 2002. Orf virus-encoded interleukin-10 stimulates the proliferation of murine mast cells and inhibits cytokine synthesis in murine peritoneal macrophages. J. Gen. Virol. 83:1049-1058. [DOI] [PubMed] [Google Scholar]
- 39.Irons, R. D., and A. T. Le. 2008. Dithiocarbamates and viral IL-10 collaborate in the immortalization and evasion of immune response in EBV-infected human B lymphocytes. Chem. Biol. Interact. 172:81-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaworowski, A., W.-J. Cheng, C. L. Westhorpe, A. Abendroth, S. M. Crowe, and B. Slobedman. 2009. Enhanced monocyte Fc phagocytosis by a homologue of interleukin-10 encoded by human cytomegalovirus. Virology 391:20-24. [DOI] [PubMed] [Google Scholar]
- 41.Jayawardane, G., G. C. Russell, J. Thomson, D. Deane, H. Cox, D. Gatherer, M. Ackermann, D. M. Haig, and J. P. Stewart. 2008. A captured viral interleukin 10 gene with cellular exon structure. J. Gen. Virol. 89:2447-2455. [DOI] [PubMed] [Google Scholar]
- 42.Jenkins, C., A. Abendroth, and B. Slobedman. 2004. A novel viral transcript with homology to human interleukin-10 is expressed during latent human cytomegalovirus infection. J. Virol. 78:1440-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jenkins, C., W. Garcia, A. Abendroth, and B. Slobedman. 2008. Expression of a human cytomegalovirus latency-associated homolog of interleukin-10 during the productive phase of infection. Virology 370:285-294. [DOI] [PubMed] [Google Scholar]
- 44.Jenkins, C., W. Garcia, M. J. Godwin, J. V. Spencer, J. L. Stern, A. Abendroth, and B. Slobedman. 2008. Immunomodulatory properties of a viral homolog of human interleukin-10 expressed by human cytomegalovirus during the latent phase of infection. J. Virol. 82:3736-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ji, J., G. K. Sahu, V. L. Braciale, and M. W. Cloyd. 2005. HIV-1 induces IL-10 production in human monocytes via a CD4-independent pathway. Int. Immunol. 17:729-736. [DOI] [PubMed] [Google Scholar]
- 46.Jin, L., and G. Scherba. 1999. Expression of the pseudorabies virus latency-associated transcript gene during productive infection of cultured cells. J. Virol. 73:9781-9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones, B. C., N. J. Logsdon, K. Josephson, J. Cook, P. A. Barry, and M. R. Walter. 2002. Crystal structure of human cytomegalovirus IL-10 bound to soluble human IL-10R1. Proc. Natl. Acad. Sci. USA 99:9404-9409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kara, P. D., C. L. Afonso, D. B. Wallace, G. F. Kutish, C. Abolnik, Z. Lu, F. T. Vreede, L. C. Taljaard, A. Zsak, G. J. Viljoen, and D. L. Rock. 2003. Comparative sequence analysis of the South African vaccine strain and two virulent field isolates of lumpy skin disease virus. Arch. Virol. 148:1335-1356. [DOI] [PubMed] [Google Scholar]
- 49.Keravala, A., E. R. Lechman, J. Nash, Z. Mi, and P. D. Robbins. 2006. Human, viral or mutant human IL-10 expressed after local adenovirus-mediated gene transfer are equally effective in ameliorating disease pathology in a rabbit knee model of antigen-induced arthritis. Arthritis Res. Ther. 8:R91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim, K. N., S. Watanabe, Y. Ma, S. Thornton, E. H. Giannini, and R. Hirsch. 2000. Viral IL-10 and soluble TNF receptor act synergistically to inhibit collagen-induced arthritis following adenovirus-mediated gene transfer. J. Immunol. 164:1576-1581. [DOI] [PubMed] [Google Scholar]
- 51.Kim, Y. H., D. G. Lim, Y. M. Wee, J. H. Kim, C. O. Yun, M. Y. Choi, Y. H. Park, S. C. Kim, and D. J. Han. 2008. Viral IL-10 gene transfer prolongs rat islet allograft survival. Cell Transplant. 17:609-618. [DOI] [PubMed] [Google Scholar]
- 52.Kotenko, S. V., C. D. Krause, L. S. Izotova, B. P. Pollack, W. Wu, and S. Pestka. 1997. Identification and functional characterization of a second chain of the interleukin-10 receptor complex. EMBO J. 16:5894-5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kotenko, S. V., and S. Pestka. 2000. Jak-Stat signal transduction pathway through the eyes of cytokine class II receptor complexes. Oncogene 19:2557-2565. [DOI] [PubMed] [Google Scholar]
- 54.Kotenko, S. V., S. Saccani, L. S. Izotova, O. V. Mirochnitchenko, and S. Pestka. 2000. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proc. Natl. Acad. Sci. USA 97:1695-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lateef, Z., S. Fleming, G. Halliday, L. Faulkner, A. Mercer, and M. Baird. 2003. Orf virus-encoded interleukin-10 inhibits maturation, antigen presentation and migration of murine dendritic cells. J. Gen. Virol. 84:1101-1109. [DOI] [PubMed] [Google Scholar]
- 56.Lear, A. L., M. Rowe, M. G. Kurilla, S. Lee, S. Henderson, E. Kieff, and A. B. Rickinson. 1992. The Epstein-Barr virus (EBV) nuclear antigen 1 BamHI F promoter is activated on entry of EBV-transformed B cells into the lytic cycle. J. Virol. 66:7461-7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee, H. J., K. Essani, and G. L. Smith. 2001. The genome sequence of Yaba-like disease virus, a yatapoxvirus. Virology 281:170-192. [DOI] [PubMed] [Google Scholar]
- 58.Lin, Y. L., P. C. Chang, Y. Wang, and M. Li. 2008. Identification of novel viral interleukin-10 isoforms of human cytomegalovirus AD169. Virus Res. 131:213-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu, Y., R. de Waal Malefyt, F. Briere, C. Parham, J. M. Bridon, J. Banchereau, K. W. Moore, and J. Xu. 1997. The EBV IL-10 homologue is a selective agonist with impaired binding to the IL-10 receptor. J. Immunol. 158:604-613. [PubMed] [Google Scholar]
- 60.Liu, Y., S. H. Wei, A. S. Ho, R. de Waal Malefyt, and K. W. Moore. 1994. Expression cloning and characterization of a human IL-10 receptor. J. Immunol. 152:1821-1829. [PubMed] [Google Scholar]
- 61.Lockridge, K. M., S. S. Zhou, R. H. Kravitz, J. L. Johnson, E. T. Sawai, E. L. Blewett, and P. A. Barry. 2000. Primate cytomegaloviruses encode and express an IL-10-like protein. Virology 268:272-280. [DOI] [PubMed] [Google Scholar]
- 62.Lunetta, J. M., and J. A. Wiedeman. 2000. Latency-associated sense transcripts are expressed during in vitro human cytomegalovirus productive infection. Virology 278:467-476. [DOI] [PubMed] [Google Scholar]
- 63.Ma, Y., S. Thornton, L. E. Duwel, G. P. Boivin, E. H. Giannini, J. M. Leiden, J. A. Bluestone, and R. Hirsch. 1998. Inhibition of collagen-induced arthritis in mice by viral IL-10 gene transfer. J. Immunol. 161:1516-1524. [PubMed] [Google Scholar]
- 64.MacNeil, I. A., T. Suda, K. W. Moore, T. R. Mosmann, and A. Zlotnik. 1990. IL-10, a novel growth cofactor for mature and immature T cells. J. Immunol. 145:4167-4173. [PubMed] [Google Scholar]
- 65.McGeoch, D. J., S. Cook, A. Dolan, F. E. Jamieson, and E. A. Telford. 1995. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J. Mol. Biol. 247:443-458. [DOI] [PubMed] [Google Scholar]
- 66.Mercer, A. A., N. Ueda, S. M. Friederichs, K. Hofmann, K. M. Fraser, T. Bateman, and S. B. Fleming. 2006. Comparative analysis of genome sequences of three isolates of Orf virus reveals unexpected sequence variation. Virus Res. 116:146-158. [DOI] [PubMed] [Google Scholar]
- 67.Minter, R. M., M. A. Ferry, M. E. Murday, C. L. Tannahill, F. R. Bahjat, C. Oberholzer, A. Oberholzer, D. LaFace, B. Hutchins, S. Wen, J. Shinoda, E. M. Copeland III, and L. L. Moldawer. 2001. Adenoviral delivery of human and viral IL-10 in murine sepsis. J. Immunol. 167:1053-1059. [DOI] [PubMed] [Google Scholar]
- 68.Miyazaki, I., R. K. Cheung, and H. M. Dosch. 1993. Viral interleukin 10 is critical for the induction of B cell growth transformation by Epstein-Barr virus. J. Exp. Med. 178:439-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moore, K. W., R. de Waal Malefyt, R. L. Coffman, and A. O'Garra. 2001. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 19:683-765. [DOI] [PubMed] [Google Scholar]
- 70.Moore, K. W., A. O'Garra, R. de Waal Malefyt, P. Vieira, and T. R. Mosmann. 1993. Interleukin-10. Annu. Rev. Immunol. 11:165-190. [DOI] [PubMed] [Google Scholar]
- 71.Moore, K. W., P. Vieira, D. F. Fiorentino, M. L. Trounstine, T. A. Khan, and T. R. Mosmann. 1990. Homology of cytokine synthesis inhibitory factor (IL-10) to the Epstein-Barr virus gene BCRFI. Science 248:1230-1234. [DOI] [PubMed] [Google Scholar]
- 72.Mosser, D. M., and X. Zhang. 2008. Interleukin-10: new perspectives on an old cytokine. Immunol. Rev. 226:205-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Niiro, H., T. Otsuka, M. Abe, H. Satoh, T. Ogo, T. Nakano, Y. Furukawa, and Y. Niho. 1992. Epstein-Barr virus BCRF1 gene product (viral interleukin 10) inhibits superoxide anion production by human monocytes. Lymphokine Cytokine Res. 11:209-214. [PubMed] [Google Scholar]
- 74.Niiro, H., T. Otsuka, S. Kuga, Y. Nemoto, M. Abe, N. Hara, T. Nakano, T. Ogo, and Y. Niho. 1994. IL-10 inhibits prostaglandin E2 production by lipopolysaccharide-stimulated monocytes. Int. Immunol. 6:661-664. [DOI] [PubMed] [Google Scholar]
- 75.Oberholzer, C., A. Oberholzer, F. R. Bahjat, R. M. Minter, C. L. Tannahill, A. Abouhamze, D. LaFace, B. Hutchins, M. J. Clare-Salzler, and L. L. Moldawer. 2001. Targeted adenovirus-induced expression of IL-10 decreases thymic apoptosis and improves survival in murine sepsis. Proc. Natl. Acad. Sci. USA 98:11503-11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pepperl-Klindworth, S., K. Besold, N. Frankenberg, M. Farkas, J. Kuball, M. Theobald, and B. Plachter. 2006. Cytomegalovirus interleukin-10 expression in infected cells does not impair MHC class I restricted peptide presentation on bystanding antigen-presenting cells. Viral Immunol. 19:92-101. [DOI] [PubMed] [Google Scholar]
- 77.Pestka, S., C. D. Krause, D. Sarkar, M. R. Walter, Y. Shi, and P. B. Fisher. 2004. Interleukin-10 and related cytokines and receptors. Annu. Rev. Immunol. 22:929-979. [DOI] [PubMed] [Google Scholar]
- 78.Qin, L., K. D. Chavin, Y. Ding, H. Tahara, J. P. Favaro, J. E. Woodward, T. Suzuki, P. D. Robbins, M. T. Lotze, and J. S. Bromberg. 1996. Retrovirus-mediated transfer of viral IL-10 gene prolongs murine cardiac allograft survival. J. Immunol. 156:2316-2323. [PubMed] [Google Scholar]
- 79.Raftery, M. J., M. Hitzler, F. Winau, T. Giese, B. Plachter, S. H. Kaufmann, and G. Schonrich. 2008. Inhibition of CD1 antigen presentation by human cytomegalovirus. J. Virol. 82:4308-4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Raftery, M. J., D. Wieland, S. Gronewald, A. A. Kraus, T. Giese, and G. Schonrich. 2004. Shaping phenotype, function, and survival of dendritic cells by cytomegalovirus-encoded IL-10. J. Immunol. 173:3383-3391. [DOI] [PubMed] [Google Scholar]
- 81.Redpath, S., A. Angulo, N. R. Gascoigne, and P. Ghazal. 1999. Murine cytomegalovirus infection down-regulates MHC class II expression on macrophages by induction of IL-10. J. Immunol. 162:6701-6707. [PubMed] [Google Scholar]
- 82.Redpath, S., P. Ghazal, and N. R. Gascoigne. 2001. Hijacking and exploitation of IL-10 by intracellular pathogens. Trends Microbiol. 9:86-92. [DOI] [PubMed] [Google Scholar]
- 83.Riley, J. K., K. Takeda, S. Akira, and R. D. Schreiber. 1999. Interleukin-10 receptor signaling through the JAK-STAT pathway. Requirement for two distinct receptor-derived signals for anti-inflammatory action. J. Biol. Chem. 274:16513-16521. [DOI] [PubMed] [Google Scholar]
- 84.Rivailler, P., H. Jiang, Y. G. Cho, C. Quink, and F. Wang. 2002. Complete nucleotide sequence of the rhesus lymphocryptovirus: genetic validation for an Epstein-Barr virus animal model. J. Virol. 76:421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rode, H.-J., W. Janssen, A. Rosen-Wolff, J. J. Bugert, P. Thein, Y. Becker, and G. Darai. 1993. The genome of equine herpesvirus type 2 harbors an interleukin-10 (IL-10)-like gene. Virus Genes 7:111. [DOI] [PubMed] [Google Scholar]
- 86.Rouas-Freiss, N., R. M. Goncalves, C. Menier, J. Dausset, and E. D. Carosella. 1997. Direct evidence to support the role of HLA-G in protecting the fetus from maternal uterine natural killer cytolysis. Proc. Natl. Acad. Sci. USA 94:11520-11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rousset, F., E. Garcia, T. Defrance, C. Peronne, N. Vezzio, D. H. Hsu, R. Kastelein, K. W. Moore, and J. Banchereau. 1992. Interleukin 10 is a potent growth and differentiation factor for activated human B lymphocytes. Proc. Natl. Acad. Sci. USA 89:1890-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rowbottom, A. W., M. A. Lepper, R. J. Garland, C. V. Cox, and E. G. Corley. 1999. Interleukin-10-induced CD8 cell proliferation. Immunology 98:80-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Salgar, S. K., D. Yang, P. Ruiz, J. Miller, and A. G. Tzakis. 2004. Viral interleukin-10-engineered autologous hematopoietic stem cell therapy: a novel gene therapy approach to prevent graft rejection. Hum. Gene Ther. 15:131-144. [DOI] [PubMed] [Google Scholar]
- 90.Santin, A. D., P. L. Hermonat, A. Ravaggi, S. Bellone, S. Pecorelli, J. J. Roman, G. P. Parham, and M. J. Cannon. 2000. Interleukin-10 increases Th1 cytokine production and cytotoxic potential in human papillomavirus-specific CD8+ cytotoxic T lymphocytes. J. Virol. 74:4729-4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schaefer, B. C., J. L. Strominger, and S. H. Speck. 1995. Redefining the Epstein-Barr virus-encoded nuclear antigen EBNA-1 gene promoter and transcription initiation site in group I Burkitt lymphoma cell lines. Proc. Natl. Acad. Sci. USA 92:10565-10569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Spencer, J. V. 2007. The cytomegalovirus homolog of interleukin-10 requires phosphatidylinositol 3-kinase activity for inhibition of cytokine synthesis in monocytes. J. Virol. 81:2083-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Spencer, J. V., J. Cadaoas, P. R. Castillo, V. Saini, and B. Slobedman. 2008. Stimulation of B lymphocytes by cmvIL-10 but not LAcmvIL-10. Virology 374:164-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Spencer, J. V., K. M. Lockridge, P. A. Barry, G. Lin, M. Tsang, M. E. Penfold, and T. J. Schall. 2002. Potent immunosuppressive activities of cytomegalovirus-encoded interleukin-10. J. Virol. 76:1285-1292. (Erratum, 76:3585.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Spivack, J. G., and N. W. Fraser. 1987. Detection of herpes simplex virus type 1 transcripts during latent infection in mice. J. Virol. 61:3841-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Taus, N. S., D. R. Herndon, D. L. Traul, J. P. Stewart, M. Ackermann, H. Li, D. P. Knowles, G. S. Lewis, and K. A. Brayton. 2007. Comparison of ovine herpesvirus 2 genomes isolated from domestic sheep (Ovis aries) and a clinically affected cow (Bos bovis). J. Gen. Virol. 88:40-45. [DOI] [PubMed] [Google Scholar]
- 97.Telford, E. A., M. S. Watson, H. C. Aird, J. Perry, and A. J. Davison. 1995. The DNA sequence of equine herpesvirus 2. J. Mol. Biol. 249:520-528. [DOI] [PubMed] [Google Scholar]
- 98.Thibodeau, J., M. C. Bourgeois-Daigneault, G. Huppe, J. Tremblay, A. Aumont, M. Houde, E. Bartee, A. Brunet, M. E. Gauvreau, A. de Gassart, E. Gatti, M. Baril, M. Cloutier, S. Bontron, K. Fruh, D. Lamarre, and V. Steimle. 2008. Interleukin-10-induced MARCH1 mediates intracellular sequestration of MHC class II in monocytes. Eur. J. Immunol. 38:1225-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Thompson-Snipes, L., V. Dhar, M. W. Bond, T. R. Mosmann, K. W. Moore, and D. M. Rennick. 1991. Interleukin 10: a novel stimulatory factor for mast cells and their progenitors. J. Exp. Med. 173:507-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Touitou, R., C. Cochet, and I. Joab. 1996. Transcriptional analysis of the Epstein-Barr virus interleukin-10 homologue during the lytic cycle. J. Gen. Virol. 77:1163-1168. [DOI] [PubMed] [Google Scholar]
- 101.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, J. H. Sur, N. T. Sandybaev, U. Z. Kerembekova, V. L. Zaitsev, G. F. Kutish, and D. L. Rock. 2002. The genomes of sheeppox and goatpox viruses. J. Virol. 76:6054-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.van Putten, S. M., M. Wubben, W. E. Hennink, M. J. van Luyn, and M. C. Harmsen. 2009. The downmodulation of the foreign body reaction by cytomegalovirus encoded interleukin-10. Biomaterials 30:730-735. [DOI] [PubMed] [Google Scholar]
- 103.Vieira, P., R. de Waal-Malefyt, M. N. Dang, K. E. Johnson, R. Kastelein, D. F. Fiorentino, J. E. deVries, M. G. Roncarolo, T. R. Mosmann, and K. W. Moore. 1991. Isolation and expression of human cytokine synthesis inhibitory factor cDNA clones: homology to Epstein-Barr virus open reading frame BCRFI. Proc. Natl. Acad. Sci. USA 88:1172-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Willems, F., A. Marchant, J. P. Delville, C. Gerard, A. Delvaux, T. Velu, M. de Boer, and M. Goldman. 1994. Interleukin-10 inhibits B7 and intercellular adhesion molecule-1 expression on human monocytes. Eur. J. Immunol. 24:1007-1009. [DOI] [PubMed] [Google Scholar]
- 105.Wise, L., C. McCaughan, C. K. Tan, A. A. Mercer, and S. B. Fleming. 2007. Orf virus interleukin-10 inhibits cytokine synthesis in activated human THP-1 monocytes, but only partially impairs their proliferation. J. Gen. Virol. 88:1677-1682. [DOI] [PubMed] [Google Scholar]
- 106.Xia, D., S. Srinivas, H. Sato, L. Pesnicak, S. E. Straus, and J. I. Cohen. 2003. Varicella-zoster virus open reading frame 21, which is expressed during latency, is essential for virus replication but dispensable for establishment of latency. J. Virol. 77:1211-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xia, D., and S. E. Straus. 1999. Transcript mapping and transregulatory behavior of varicella-zoster virus gene 21, a latency-associated gene. Virology 258:304-313. [DOI] [PubMed] [Google Scholar]
- 108.Yamamoto-Tabata, T., S. McDonagh, H. T. Chang, S. Fisher, and L. Pereira. 2004. Human cytomegalovirus interleukin-10 downregulates metalloproteinase activity and impairs endothelial cell migration and placental cytotrophoblast invasiveness in vitro. J. Virol. 78:2831-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang, Z., M. Chen, R. Wu, L. B. Fialkow, J. S. Bromberg, M. McDuffie, A. Naji, and J. L. Nadler. 2002. Suppression of autoimmune diabetes by viral IL-10 gene transfer. J. Immunol. 168:6479-6485. [DOI] [PubMed] [Google Scholar]
- 110.Yoon, S. I., B. C. Jones, N. J. Logsdon, and M. R. Walter. 2005. Same structure, different function crystal structure of the Epstein-Barr virus IL-10 bound to the soluble IL-10R1 chain. Structure 13:551-564. [DOI] [PubMed] [Google Scholar]
- 111.Yu, X. L., Y. M. Cheng, B. S. Shi, F. X. Qian, F. B. Wang, X. N. Liu, H. Y. Yang, Q. N. Xu, T. K. Qi, L. J. Zha, Z. H. Yuan, and R. Ghildyal. 2008. Measles virus infection in adults induces production of IL-10 and is associated with increased CD4+ CD25+ regulatory T cells. J. Immunol. 181:7356-7366. [DOI] [PubMed] [Google Scholar]
- 112.Yue, Y., A. Kaur, M. K. Eberhardt, N. Kassis, S. S. Zhou, A. F. Tarantal, and P. A. Barry. 2007. Immunogenicity and protective efficacy of DNA vaccines expressing rhesus cytomegalovirus glycoprotein B, phosphoprotein 65-2, and viral interleukin-10 in rhesus macaques. J. Virol. 81:1095-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zdanov, A., C. Schalk-Hihi, S. Menon, K. W. Moore, and A. Wlodawer. 1997. Crystal structure of Epstein-Barr virus protein BCRF1, a homolog of cellular interleukin-10. J. Mol. Biol. 268:460-467. [DOI] [PubMed] [Google Scholar]