Abstract
Like human immunodeficiency virus type 1 (HIV-1), most simian immunodeficiency virus (SIV) strains use CCR5 to establish infection. However, while HIV-1 can acquire the ability to use CXCR4, SIVs that utilize CXCR4 have rarely been reported. To explore possible barriers against SIV coreceptor switching, we derived an R5X4 variant, termed 239-ST1, from the R5 clone SIVmac239 by serially passaging virus in CD4+ CXCR4+ CCR5− SupT1 cells. A 239-ST1 env clone, designated 239-ST1.2-32, used CXCR4 and CCR5 in cell-cell fusion and reporter virus infection assays and conferred the ability for rapid, cytopathic infection of SupT1 cells to SIVmac239. Viral replication was inhibitable by the CXCR4-specific antagonist AMD3100, and replication was abrogated in a novel CXCR4− SupT1 line. Surprisingly, parental SIVmac239 exhibited low-level replication in SupT1 cells that was not observed in CXCR4− SupT1 cells. Only two mutations in the 239-ST1.2-32 Env, K47E in the C1 domain and L328W in the V3 loop, were required for CXCR4 use in cell-cell fusion assays, although two other V3 changes, N316K and I324M, improved CXCR4 use in infection assays. An Env cytoplasmic tail truncation, acquired during propagation of 239-ST1 in SupT1 cells, was not required. Compared with SIVmac239, 239-ST1.2-32 was more sensitive to neutralization by five of seven serum and plasma samples from SIVmac239-infected rhesus macaques and was approximately 50-fold more sensitive to soluble CD4. Thus, SIVmac239 can acquire the ability to use CXCR4 with high efficiency, but the changes required for this phenotype may be distinct from those for HIV-1 CXCR4 use. This finding, along with the increased neutralization sensitivity of this CXCR4-using SIV, suggests a mechanism that could select strongly against this phenotype in vivo.
Simian immunodeficiency viruses (SIVs) share many structural and biological features with human immunodeficiency virus (HIV), including target cell entry via interactions of the viral envelope glycoprotein (Env) with CD4 and a chemokine coreceptor. For HIV, the most important coreceptors in vivo are CCR5 (2, 13, 19, 21, 22) and CXCR4 (30). HIV type 1 (HIV-1) strains that use only CCR5 (R5 viruses) predominate during the early stages of infection and are critical for transmission (84, 90), as evidenced by the finding that individuals lacking a functional CCR5 protein due to a homozygous 32-bp deletion in the CCR5 gene (ccr5-Δ32) are largely resistant to HIV-1 infection (16, 54, 82). Although R5 viruses generally persist in late-stage disease, viruses that can use CXCR4, either exclusively (X4 viruses) or in addition to CCR5 (R5X4 viruses), emerge in approximately 50% of subtype B-infected individuals (15, 43). This coreceptor switch is associated with a more rapid decline in peripheral blood CD4+ T cells and a faster progression to AIDS (15, 43, 77), although it is unclear if CXCR4-using viruses are a cause or a consequence of progressing immunodeficiency. Like HIV, the vast majority of SIVs use CCR5 to establish infection (11, 12, 45). However, although CXCR4-using SIVs have been reported (47, 52, 65, 68, 69), their occurrence is rare, especially in models of pathogenic infection, where only one CXCR4-using SIV has been identified (17, 60, 71).
This paucity of CXCR4-using SIVs is surprising for several reasons. First, SIV Envs tend to be more promiscuous than HIV-1 Envs and frequently use alternative coreceptors in addition to CCR5, including GPR1, GPR15, CXCR6, and CCR8 (20, 27, 29, 80, 81, 92) but not CXCR4. Second, HIV-2, which is more closely related to SIVmac than to HIV-1 (56, 57), commonly uses CXCR4 in vitro and in vivo (3, 28, 33, 58, 59, 67). Third, rhesus CXCR4 is ∼98% identical to human CXCR4 in amino acid sequence and can function as a coreceptor for HIV-1 in vitro (12). Finally, chimeric simian-human immunodeficiency viruses (SHIVs) that contain X4 HIV Envs on an SIV core can replicate to high levels in vivo and cause disease in rhesus macaques (39, 86). Moreover, it was recently shown that coreceptor switching can occur in rhesus macaques infected with an R5 SHIV (35). Thus, there does not appear to be any block per se against the use of rhesus CXCR4 as an entry coreceptor either in vitro or in vivo, suggesting that SIV is less capable of adapting to use CXCR4 and/or that mutations required for CXCR4 utilization may lead to a virus that is less fit and/or more susceptible to immune control in this host.
For HIV-1, the Env determinants for CXCR4 use have been well documented and often involve the acquisition of positively charged amino acids in the V3 loop (18, 32, 87), particularly at positions 11, 24, and 25 (6, 18, 31, 32, 38, 75). Although the SIVmac239 V3 loop is a critical determinant for Env-coreceptor interactions (44, 63, 72), attempts to create an X4 SIVmac239 by introducing positively charged residues into the V3 loop (63) or by inserting a V3 loop from X4 HIV-1 (44) have been unsuccessful. SIVmac155T3, the only CXCR4-using variant of SIVmac that has been identified to date, was isolated from a rhesus macaque with advanced disease and contains additional positively charged residues in V3, although the determinants for CXCR4 use have not been determined (60, 71).
Given questions concerning the possible determinants for and/or barriers to coreceptor switching in SIV, we sought to derive a CXCR4-using variant of the well-characterized pathogenic R5 SIV clone SIVmac239. Here we show that SIVmac239 could indeed acquire CXCR4 utilization when it was adapted in vitro for high-efficiency replication in the CXCR4+ CCR5− human SupT1 cell line. An env clone from this virus could use CXCR4 in cell-cell fusion and reporter virus infection assays and conferred CXCR4 tropism to a replication-competent SIV. Although V3 mutations were important for CXCR4 use, an L328W change at the V3 crown rather than the acquisition of positively charged residues was required, as was an unusual K47E mutation in the conserved C1 domain of gp120. These changes also caused the highly neutralization-resistant SIVmac239 strain to become more neutralization sensitive to sera and plasmas from SIVmac239-infected animals, and particularly to soluble CD4. These results indicate that mutations distinct from those typically seen for HIV-1 may be required for SIVmac to gain CXCR4 utilization and suggest that these changes render this virus more susceptible to humoral immune control. Collectively, our findings indicate that there are likely to be strong viral and host selection pressures against CXCR4 use that may contribute to the paucity of X4 coreceptor switching for SIVmac in vivo.
MATERIALS AND METHODS
Cells.
The human SupT1 T-lymphoblastoid cell line was maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, and 2 mM penicillin-streptomycin (RPMI-Complete). SupT1 cells that stably express human CCR5 (SupCCR5) (60) were maintained in RPMI-Complete with 300 ng/ml puromycin. CXCR4 knockout SupT1 cells (SupX4− cells) were generated using zinc finger nucleases (ZFNs; Sangamo BioSciences) (64, 70) targeting the CXCR4 gene (J. Wang et al., unpublished data). Following electroporation with a construct encoding a pair of CXCR4-specific ZFNs (pVAX-73EL-2a-70KK), CXCR4− cells were isolated using fluorescence-activated cell sorting (FACS) and the CXCR4 extracellular loop 2-directed monoclonal antibody 12G5. CXCR4− cells were then single-cell cloned by limiting dilution (clone A66), checked by PCR amplification and genomic DNA sequencing for disruption of the CXCR4 alleles, checked by flow cytometry for absent CXCR4 expression, and maintained in RPMI-Complete. SupX4− cells were engineered to stably express CCR5 (SupX4−R5+ cells) or CXCR4 (SupX4+ cells) by transduction with a CCR5- or CXCR4-containing pELNS replication-defective lentiviral vector, generated as previously described (76). Following transduction, coreceptor-positive cells were sorted by FACS with the anti-CCR5 antibody 2D7 or the anti-CXCR4 antibody 12G5. Coreceptor-positive cells were then single-cell cloned by limiting dilution, checked by flow cytometry for CCR5 or CXCR4 expression, and maintained in RPMI-Complete. The canine thymocyte cell line Cf2Th (kindly provided by Dana Gabuzda, Harvard University), the Japanese quail fibrosarcoma cell line QT6, the human embryonic kidney cell line 293T, and the human astrocytoma cell line NP-2/CD4/CCR5 were cultured in Dulbecco's modified Eagle medium supplemented with 10% FBS, 2 mM glutamine, and 2 mM penicillin-streptomycin.
Env cloning, plasmid construction, and mutagenesis.
To isolate adapted env clones from infected SupT1 cultures, genomic DNA was prepared using a QIAamp DNA Mini kit (Qiagen) according to the manufacturer's instructions, and env sequences were PCR amplified using HotStarTaq (Invitrogen) and primers that flank the SIVmac239 env gene. PCR products were then Topo TA cloned into pCR2.1 (Invitrogen) and screened for env inserts, using restriction analysis and DNA sequencing. Mutant env genes were created using a QuikChange site-directed mutagenesis kit (Stratagene) following the manufacturer's protocol. To generate recombinant molecular clones of the SIVmac239 genome containing adapted and mutant env genes, the previously described pVP-2 nef open construct, which contains the 3′ half of the viral genome, first had a BsmI restriction site in the vector region flanking the 3′ end of the genome ablated (pVP-2/BsmI−) using QuikChange mutagenesis. An internal 1.7-kb BsmI fragment containing the desired env mutations was then ligated with BsmI-digested pVP-2/BsmI−. The presence of the appropriate env gene was confirmed using restriction analysis and DNA sequencing. To generate molecular clones with a truncated cytoplasmic tail (CT), QuikChange mutagenesis was used to introduce a stop codon at position Q734 in Env in pVP-2/BsmI−. Full-length genome constructs were then generated by linearizing env-containing pVP-2/BsmI− constructs with SphI and ligating them with the 5′ genome half from the SphI-digested p239SpSp5′ construct (obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Ronald Desrosiers). The identities of the recombinant clones were confirmed using restriction analysis and DNA sequencing. For the generation of luciferase reporter viruses, SIV env genes with a premature stop codon in the CT coding region (Q734Stop) in pCR2.1 were digested with KpnI and XbaI and cloned into the similarly digested pCDNA3.1(−) expression construct. Plasmid pHSPG-R3A, containing the HIV-1R3A envelope, used in cell-cell fusion assays and luciferase reporter virus assays, has been described previously (62). Expression constructs containing CD4, CCR5, and CXCR4 and a reporter plasmid encoding luciferase under the control of a T7 promoter have been described previously (79).
Cell-cell fusion assay.
To quantify the ability of viral Envs to use a given coreceptor for cell-cell fusion, we used a previously described cell-cell fusion assay (26, 79). Briefly, effector QT6 cells were generated by first infecting cells with the recombinant vaccinia virus strain VTF1.1, expressing T7 polymerase (1), at a multiplicity of infection of 10 for 1 h at 37°C and then transfecting the cells for 5 h with the appropriate env expression vector, using the standard calcium phosphate method. Following transfection, effector cells were incubated overnight at 32°C in the presence of rifampin (rifampicin) at a concentration of 100 μg/ml. Target QT6 cells were generated by transfecting cells with the desired receptor expression vectors and a T7-luciferase reporter construct by the standard calcium phosphate method for 5 h, followed by overnight expression at 37°C. Effector cells were then added to target cells in the presence of 100 μg/ml rifampin and 100 nM cytosine arabinoside, and cell-cell fusion was assessed 7 to 8 h later by lysing cells with 0.5% Triton X-100-phosphate-buffered saline, adding luciferase substrate (Promega), and quantifying luciferase activity with a Thermo LabSystems Luminoskan Ascent luminometer. For AMD3100 inhibition experiments, serial dilutions of AMD3100 (obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH) were added to target cells at the time of addition of effector cells, and inhibition of fusion was measured as the reduction in luciferase activity compared with that of untreated controls.
Luciferase reporter viruses.
Luciferase reporter viruses were generated by cotransfecting 293T cells with a plasmid encoding the NL4-3-based luciferase virus backbone (pNL-luc-E−R−) (10, 14) and the appropriate env expression vector for 5 h by the standard calcium phosphate method. Cell supernatants were collected at 48 h posttransfection and stored at −80°C. Virus concentrations were determined by an enzyme-linked immunosorbent assay for the viral p24 antigen (PerkinElmer). Cf2Th target cells were prepared by transfecting cells with the desired receptor expression vectors for 4 h, using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The cells were then spin infected for 1 h at 1,300 rpm with 10 ng of p24 equivalents of reporter virus at 48 h posttransfection and subsequently incubated at 37°C for 72 h. The cells were then lysed with 0.5% Triton X-100-phosphate-buffered saline, and infection was quantified by adding luciferase substrate (Promega) and measuring luciferase activity with a Thermo LabSystems Luminoskan Ascent luminometer.
Replication-competent infection assays.
To generate molecularly cloned viruses, 293T cells were transfected with full-length viral genome constructs for 5 h by the standard calcium phosphate method. Cell supernatants were collected at 72 h posttransfection and stored at −80°C. The 239-ST1 swarm was generated in productively infected SupT1 cells. Infected cell supernatants were collected and stored at −80°C upon the first observation of cytopathic effects in the cultures. Virus concentrations were determined by enzyme-linked immunosorbent assay for the viral p27 antigen (Zeptometrics). SupT1, SupCCR5, SupX4−, SupX4−R5+, and SupX4+ cells were infected with equivalent amounts of p27-containing virus. Following an overnight incubation at 37°C, infected cells were washed in RPMI supplemented with 5% FBS to remove excess virus, and viral replication was monitored by measuring viral reverse transcriptase (RT) activity in the culture supernatants. For AMD3100 inhibition experiments, target cells were incubated with various concentrations of AMD3100 at 37°C for 1 h prior to the addition of virus. AMD3100 was maintained in the culture at the desired concentration throughout the course of the infection. Inhibition of infection was measured as the reduction in RT activity in the culture supernatant compared with that of an untreated control.
Neutralization assays.
We tested the sensitivity of luciferase reporter viruses bearing Envs of interest to neutralization by sera or plasmas from SIVmac239-infected rhesus macaques (kindly provided by Preston Marx, Tulane National Primate Research Center, and Guido Silvestri, University of Pennsylvania) or to soluble CD4 (sCD4). Equivalent amounts of p24-containing luciferase reporter viruses were incubated for 1 h at 37°C with various dilutions of serum, plasma, or sCD4 and then used to spin infect NP-2/CD4/CCR5 cells for 1 h at 1,300 rpm. Subsequent to spin infection, the cells were incubated at 37°C for 72 h and then lysed with 0.5% Triton X-100-phosphate-buffered saline. Infection was quantified by adding luciferase substrate (Promega) and measuring luciferase activity with a Thermo LabSystems Luminoskan Ascent luminometer. Neutralization was measured as the reduction in luciferase activity compared with that of untreated controls. To validate our results, we used sCD4 in the single-cycle TZM-bl neutralization assay as described previously (53).
Nucleotide sequence accession numbers.
The 239-ST1.2-32 env sequence from this article has been deposited in GenBank under accession number FJ838783. The parental SIVmac239 genome sequence is available under GenBank accession number M33262.
RESULTS
Adaptation of SIVmac239 to SupT1 cells.
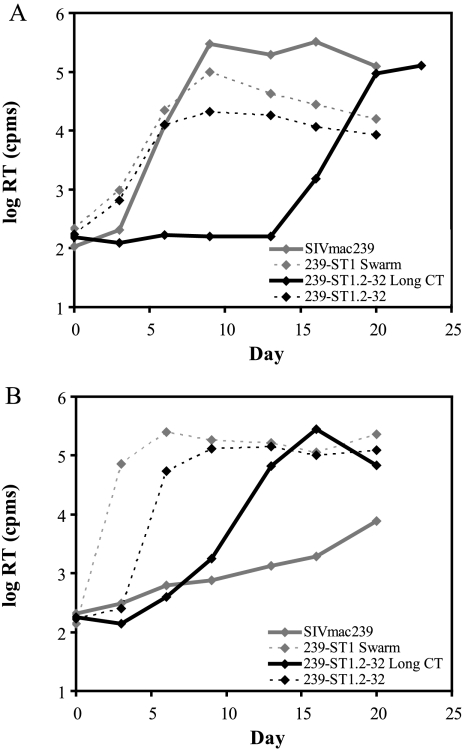
We attempted to derive a CXCR4-using variant of SIV by inoculating SIVmac239 into SupT1, a CXCR4+ CCR5− human T-lymphoblastoid cell line that is highly permissive for X4 HIV-1 and -2 replication (28, 36). In SupT1 cells that were engineered to stably express CCR5 (SupCCR5 cells) (60), SIVmac239 infection occurred rapidly, with extensive syncytium formation and cell killing and a high level of RT activity in the culture supernatant (Fig. 1A). In contrast, in parental SupT1 cells, SIVmac239 was poorly infectious, exhibiting a slow but progressive increase in RT activity over several days (Fig. 1B), without cytopathic effects and with only 5% of cells being positive for p27gag by immunofluorescence microscopy on day 34 (not shown). We maintained this SIVmac239-infected SupT1 culture for 45 days and then serially passaged virus-containing supernatant onto uninfected SupT1 cells, resulting in an increasingly efficient infection with syncytium formation and cell death. After 17 cell-free passages, virus-containing supernatant from this culture, designated 239-ST1, was harvested for evaluation. When the ability of this 239-ST1 viral swarm to infect SupT1 cells was compared with that of parental SIVmac239, a striking difference was noted in infection kinetics, with 239-ST1 exhibiting a high peak of RT activity (>2.5 × 105 cpm) at 6 days postinoculation (Fig. 1B) and all cells becoming p27gag positive by immunofluorescence microscopy (not shown).
FIG. 1.
Replication of SIVmac239 variant 239-ST1. Growth curves are shown for parental SIVmac239, the SupT1 cell-adapted 239-ST1 swarm, and recombinant SIVmac239 bearing the 239-ST1.2-32 Env clone with a full-length (Long CT) or a truncated CT in CD4+ CXCR4+ CCR5+ SupCCR5 cells (A) and CD4+ CXCR4+ CCR5− SupT1 cells (B). RT activity in culture supernatants was measured at the indicated time points. Results from a representative experiment are shown.
To determine if the efficient replication of 239-ST1 in SupT1 cells mapped to env, we PCR amplified env clones from the genomic DNA of 239-ST1-infected cells. One clone, designated 239-ST1.2-32, was inserted into an SIVmac239 backbone, generating a recombinant virus that replicated in SupT1 cells with rapid kinetics, reaching an RT peak of >1.3 × 105 cpm by day 9 (Fig. 1B). As expected, the CT of this clone contained a premature stop codon (see Fig. 4), which has been reported to occur when SIVmac is propagated in human lymphoid cells (46). Since several studies have shown that for SIVmac a CT truncation alone can increase fusogenicity (78, 93), growth kinetics (8, 93), and Env content on virions (42, 93) and that a full-length CT is restored when short-tailed SIVmac is grown in rhesus macaque cells (46), we corrected the premature stop codon in 239-ST1.2-32, designating the new virus 239-ST1.2-32 Long CT, to determine if a CT truncation was required for efficient replication in SupT1 cells. Although this virus demonstrated delayed kinetics compared with 239-ST1.2-32, it still replicated efficiently in SupT1 cells, reaching an RT peak of >2.7 × 105 cpm by day 16 (Fig. 1B). Surprisingly, growth of the 239-ST1.2-32 Long CT virus was markedly delayed in SupCCR5 cells (compare Fig. 1A and B), though this delay was not observed for 239-ST1.2-32 with a truncated CT. Because of its ability to confer rapid growth kinetics in CXCR4+ CCR5− SupT1 cells, the 239-ST1.2-32 env gene was selected for further evaluation of its coreceptor usage. For clarity, 239-ST1.2-32 refers to the Env with a truncated CT, as originally cloned from the 239-ST1-infected culture, and 239-ST1.2-32 Long CT refers to the Env with a restored full-length CT.
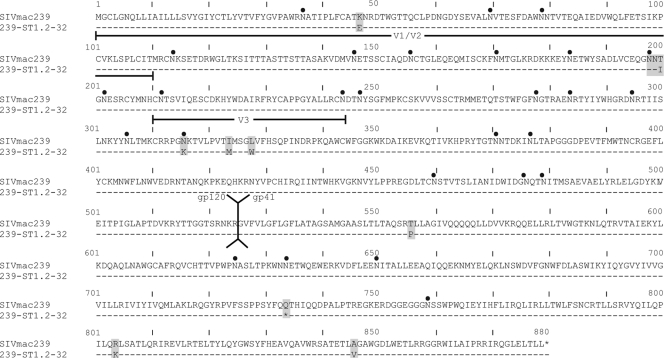
FIG. 4.
Alignment of Env sequences for SIVmac239 and 239-ST1.2-32. The indicated Env domains are based on the locations of the corresponding domains in HIV-1 Env. Locations of putative N-linked glycosylation sites are indicated by black dots. Stop codons are indicated by asterisks. The recombinant SIVmac239 virus containing the 239-ST1.2-32 Env clone used in these studies did not include the stop codon at position 734 or the two downstream mutations.
239-ST1.2-32 acquired the ability to utilize CXCR4.
We evaluated the coreceptor usage of 239-ST1.2-32 in a cell-cell fusion assay with QT6 target cells coexpressing CD4 with either CCR5 or CXCR4 to determine if 239-ST1.2-32 had gained the ability to use CXCR4. While the parental SIVmac239 Env could use only CCR5, 239-ST1.2-32 induced fusion with both CCR5- and CXCR4-expressing cells (Fig. 2A), with CCR5-mediated fusion levels at 137% of those of parental SIVmac239 and CXCR4-mediated fusion levels at 47% of those of R5X4 HIV-1R3A (61). CXCR4-dependent fusion for 239-ST1.2-32 was inhibitable by the CXCR4-specific antagonist AMD3100, with a 50% inhibitory concentration of 18 nM, which was approximately 10-fold less than that for HIV-1R3A (Fig. 2B). To assess CXCR4 use in the context of viral infection, AMD3100 sensitivity was also evaluated for a replication-competent SIV containing the 239-ST1.2-32 Long CT Env (Fig. 2C). SupT1 and SupCCR5 cells were inoculated with the 239-ST1.2-32 Long CT virus and SIVmac239, respectively, in the presence or absence of increasing concentrations of AMD3100, and RT activity was determined on day 14. 239-ST1.2-32 infection of SupT1 cells was completely inhibited by 100 and 1,000 nM AMD3100, while as expected, AMD3100 had no effect on SIVmac239 infection of SupCCR5 cells. Taken together, these data indicate that the 239-ST1.2-32 Env can utilize CXCR4 to induce cell-cell fusion and to mediate entry of a replication-competent virus.
FIG. 2.
Coreceptor utilization of 239-ST1.2-32. (A) Fusion activity in a cell-cell fusion assay of 239-ST1.2-32. Percent fusion was calculated by using luciferase activity normalized to SIVmac239 fusion with CD4+ CCR5+ QT6 cells or to HIV-1R3A fusion with CD4+ CXCR4+ QT6 cells. Background fusion levels with QT6 cells expressing only CD4 were subtracted prior to normalization. The data shown are means of three experiments plus the standard errors of the means (SEM). (B) Inhibition of 239-ST1.2-32 fusion by AMD3100. Percent fusion was calculated by using luciferase activity normalized to fusion in the absence of inhibitor. The data shown are means of three experiments ± SEM. (C) Inhibition of 239-ST1.2-32 Long CT infection by AMD3100. SupCCR5 and SupT1 cells were infected with SIVmac239 and 239-ST1.2-32 Long CT, respectively, in the presence of the indicated concentrations of AMD3100. RT activity in the culture supernatants was measured at 14 days postinfection. Results from a representative experiment are shown.
239-ST1.2-32 cannot infect SupT1 cells lacking CXCR4.
To further validate that 239-ST1.2-32 could use CXCR4 in the context of a replication-competent virus, we used engineered ZFNs targeted to CXCR4 to generate a novel CXCR4− SupT1 cell line (Wang et al., unpublished). This approach has been used successfully to stably disrupt endogenous genes in mammalian cells, including CCR5 in human lymphocytes, rendering them resistant to infection by R5 HIV-1 isolates (64, 70). SupT1 cells that were electroporated with a construct encoding a pair of CXCR4-specific ZFNs, sorted by FACS for absent CXCR4 expression, and cloned by limiting dilution had a disruption in all CXCR4 alleles, as determined by PCR amplification and sequencing of genomic DNA, and failed to express surface CXCR4 (unpublished results). One clone, designated SupX4−, was used to evaluate the recombinant SIV containing the 239-ST1.2-32 Long CT Env. As shown in Fig. 3A, 239-ST1.2-32 Long CT replicated rapidly in the parental SupT1 cells but was unable to grow in SupX4− cells. These cells were also completely resistant to R5X4 HIV-1R3A and X4 HIV-1HxB (not shown). When SupX4− cells were engineered to stably express CCR5 (SupX4−R5+ cells) or CXCR4 (SupX4+ cells), 239-ST1.2-32 replication was restored, demonstrating that these ZFN-treated cells were still capable of supporting viral replication. Thus, the ability of 239-ST1.2-32 to mediate high-efficiency infection of SupT1 cells clearly correlated with CXCR4 expression.
FIG. 3.
Use of CXCR4− SupT1 cells to assess CXCR4 use. Growth curves are shown for 239-ST1.2-32 Long CT (A) and SIVmac239 (B) in parental SupT1 cells, CD4+ CXCR4− CCR5− SupX4− cells, CD4+ CXCR4− CCR5+ SupX4−R5+ cells, and SupX4− cells that were complemented with stable CXCR4 expression (SupX4+ cells). RT activity in culture supernatants was measured at the indicated time points. Results from a representative experiment are shown for each panel.
SIVmac239 exhibits low-level usage of CXCR4 in viral infection assays.
The availability of a CXCR4− SupT1 cell line gave us the opportunity to determine if the low level of SIVmac239 replication in SupT1 cells that we initially observed (Fig. 1B), which enabled us to derive R5X4 239-ST1, was also mediated by CXCR4. SIVmac239 uses CCR5 as its primary entry coreceptor (11, 12, 45), although the use of several alternative coreceptors has also been described (20, 27, 29, 92). Because SupT1 cells do not express CCR5, parental SIVmac239 could apparently use another coreceptor on these cells, albeit inefficiently. To address this issue, we evaluated SIVmac239 infection of SupT1 and SupX4− cells. Surprisingly, although SIVmac239 again exhibited a slow and noncytopathic infection of SupT1 cells, it was unable to infect SupX4− cells, as determined by RT activity (Fig. 3B), immunofluorescence microscopy, and the inability to PCR amplify SIV gag and env genes (not shown). SupX4− cells were fully permissive for SIVmac239 infection when engineered to express CCR5 (SupX4−R5+ cells), again indicating that there was no intrinsic block to SIV replication in these ZFN-treated cells. Remarkably, SupX4− cells engineered to express CXCR4 (SupX4+ cells) also supported a noncytopathic low level of SIVmac239 replication. These results indicate that although SIVmac239 exhibits no detectable CXCR4 use in cell-cell fusion assays (25, 80) (Fig. 2A) or in infection assays using Env-pseudotyped reporter viruses (11, 12, 20, 29) (see Fig. 6A), it can utilize CXCR4 to infect SupT1 cells. Thus, our initial adaptation of SIVmac239 for rapid infection of SupT1 cells, from which 239-ST1 was derived, resulted from the improvement of a preexisting, albeit inefficient, use of CXCR4 by SIVmac239.
FIG. 6.
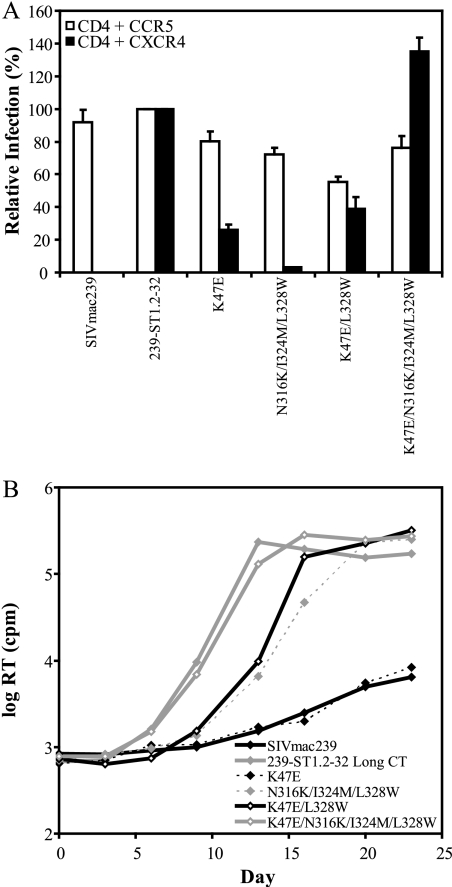
Importance of 239-ST1.2-32 mutations for coreceptor use by infectious viruses. (A) Infectivities of the indicated Envs are shown, using a single-cycle luciferase reporter virus on transfected Cf2Th target cells. Percent infection was calculated by using luciferase activity normalized to SIVmac239 fusion with CD4+ CCR5+ Cf2Th cells or to HIV-1R3A fusion with CD4+ CXCR4+ Cf2Th cells. Background infection levels with Cf2Th cells expressing only CD4 were subtracted prior to normalization. The data shown are means of three experiments plus SEM. (B) Growth curves obtained with SupT1 cells for replication-competent SIVmac239 containing the indicated Env mutations or the 239-ST1.2-32 Long CT Env. RT activity in culture supernatants was measured at the indicated time points. Results from a representative experiment are shown.
Determinants of CXCR4 use for 239-ST1.2-32 Env in cell-cell fusion assays.
Sequencing of the 239-ST1.2-32 env clone revealed nine amino acid mutations compared with parental SIVmac239 (Fig. 4). In gp120, there were the following five amino acid changes: K47E in C1; T200I in V1/V2; and N316K, I324M, and L328W in V3. Two of these mutations, T200I and N316K, resulted in the loss of predicted N-linked glycosylation sites. In gp41, there were the following four amino acid changes: T556P in the extracellular domain and Q734Stop, R804K, and A846V in the CT. Because the R840K and A846V mutations occurred downstream of the premature stop codon at position 734, they were not evaluated further.
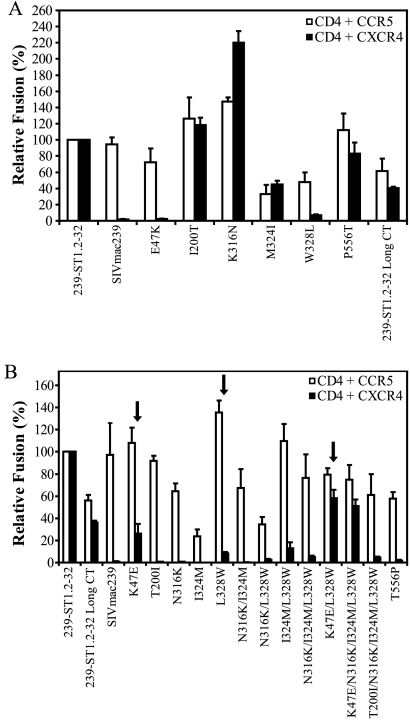
To identify Env determinants for CXCR4 use, we first used site-directed mutagenesis to correct each mutation in the 239-ST1.2-32 Env and then evaluated coreceptor usage in a cell-cell fusion assay (Fig. 5A). When the K47E and L328W mutations were each reverted (i.e., E47K and W328L, respectively), striking reductions in CXCR4-dependent fusion were produced compared with parental 239-ST1.2-32 (98% and 93%, respectively). Although reverting the I324M mutation or restoring a full-length CT (i.e., Long CT Env) reduced CXCR4-dependent fusion by approximately 50%, a similar reduction was noted for CCR5-dependent fusion. Surprisingly, correcting the N316K mutation in V3 actually enhanced CXCR4-mediated fusion, even though the N316K mutation had increased the net positive charge in V3 and removed a putative N-linked glycosylation site, both of which have been associated with CXCR4 use for HIV-1 (18, 32, 73, 87).
FIG. 5.
239-ST1.2-32 determinants of CXCR4 use in cell-cell fusion assays. (A) Fusion activities of 239-ST1.2-32 Envs with individual mutations reverted back to the corresponding residues in SIVmac239. (B) Fusion activities of SIVmac239 Envs containing 239-ST1.2-32 mutations alone and in combination. The 239-ST1.2-32 Env with a full-length CT (Long CT) was included for comparison with the SIVmac239 mutant Envs, which contain full-length tails. Percent fusion was calculated by using luciferase activity normalized to 239-ST1.2-32 fusion with CD4+ CCR5+ or CD4+ CXCR4+ QT6 cells. Background fusion levels with QT6 cells expressing only CD4 were subtracted prior to normalization. The data shown are means of three experiments plus SEM.
We next introduced 239-ST1.2-32 mutations individually and in combination into the SIVmac239 Env (Fig. 5B). For comparison, we included the 239-ST1.2-32 Long CT Env, since the loss of the stop codon at position 734 decreased overall fusion levels for both CXCR4 and CCR5 (Fig. 5A) and the SIVmac239 Env into which the 239-ST1.2-32 mutations were introduced has a full-length CT. Only two single mutations, K47E and L328W, conferred CXCR4-dependent fusion (Fig. 5B, arrows), with the K47E mutant showing 26% of the CXCR4 use of 239-ST1.2-32 and 72% of the CXCR4 use of 239-ST1.2-32 Long CT. The L328W mutation had a more modest effect, conferring CXCR4 use at 8% of the level of 239-ST1.2-32 and 22% of that of 239-ST1.2-32 with a full-length CT. As noted above, reverting either the K47E or L328W mutation in 239-ST1.2-32 substantially reduced CXCR4-mediated fusion. Interestingly, the K47E mutation is located in a conserved region of gp120 that has not previously been shown to be a determinant of coreceptor utilization. When introduced in combination, the K47E and L328W mutations had an additive effect, conferring CXCR4-dependent fusion at a level 56% of that of 239-ST1.2-32 and 161% of that of 239-ST1.2-32 Long CT. The other two V3 changes, N316K and I324M, did not confer CXCR4-mediated fusion, either individually or in combinations with the L328W and K47E mutations, nor did the T200I or T556P mutation. Collectively, these studies indicate that in the context of a cell-cell fusion assay, the K47E and L328W changes are key mutations for conferring CXCR4 use to SIVmac239 Env.
Determinants of CXCR4 use for 239-ST1.2-32 Env in virus infection.
To evaluate the determinants for CXCR4 use in the context of an infectious virus, we first generated Env-pseudotyped reporter viruses, using SIVmac239 Envs containing 239-ST1.2-32 mutations, and quantified infection in Cf2Th cells transfected with CD4 and either CCR5 or CXCR4 (Fig. 6A). To improve overall infection levels, all Envs used in these reporter virus infection assays contained a truncated CT. As in cell-cell fusion assays, SIVmac239 could use only CCR5, while 239-ST1.2-32 could use both CCR5 and CXCR4. The K47E mutation conferred a CXCR4-dependent infection that was 29% of that of 239-ST1.2-32. The three V3 loop changes (N316K, I324M, and L328W) in combination had a minimal effect (<5% of the infection level of 239-ST1.2-32), even though they included the L328W mutation, which was required for CXCR4 use in cell-cell fusion. However, when combined with the K47E mutation, these mutations increased CXCR4 use to a level that was comparable to or even greater than that of 239-ST1.2-32. When only the K47E and L328W mutations were introduced, they produced CXCR4 use at 39% of that of 239-ST1.2-32, demonstrating that while both mutations were important, the other changes in V3 (N316K and I324M) contributed to CXCR4 use, but only when the K47E mutation was present. Notably, all Envs continued to exhibit CCR5-dependent infection, with levels of >55% of those of SIVmac239, indicating that they were expressed and functional.
These mutant Envs were next introduced into the full-length SIVmac239 genome, and viral growth kinetics in SupT1 cells were evaluated (Fig. 6B). As seen previously (Fig. 1A), 239-ST1.2-32 Long CT replicated in SupT1 cells rapidly, with a peak RT activity of >2.3 × 105 cpm by day 13 postinoculation, while SIVmac239 infection occurred slowly, with <6.5 × 103 cpm by day 23. Although the K47E mutation was critical for CXCR4-mediated fusion and infection by reporter viruses, as a single mutation it was unable to increase the rate of SupT1 infection. However, when the K47E mutation was combined with the L328W mutation in V3, a clear acceleration in infection kinetics was evident, with an RT level of >1.6 × 105 cpm by day 16. A combination of the three V3 mutations (N316K, I324M, and L328W) produced a modest increase, but when these mutations were combined with the K47E mutation, infection was rapid and comparable to that with parental 239-ST1.2-32. Collectively, these findings demonstrate that the ability of 239-ST1.2-32 to use CXCR4 and to mediate SupT1 infection with rapid kinetics could be conferred fully to SIVmac239 by the K47E mutation in combination with the three V3 mutations, among which the L328W mutation was particularly important.
Increased neutralization sensitivity of 239-ST1.2-32.
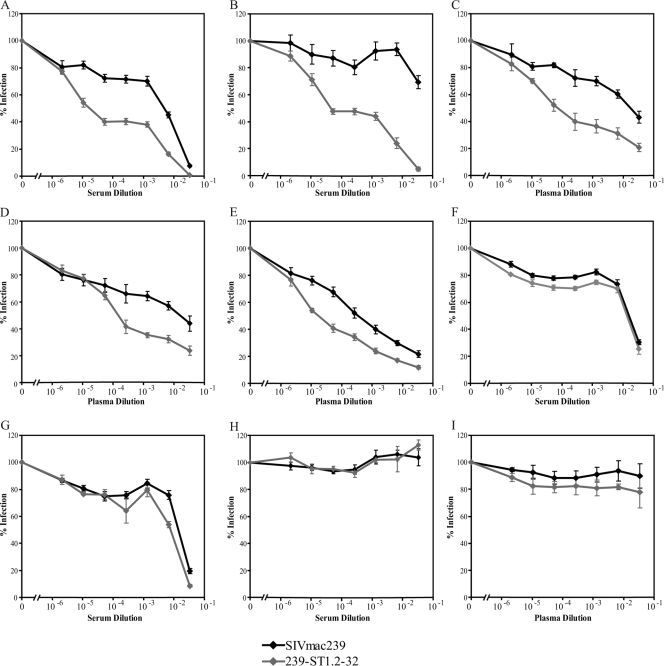
SIVmac239 is highly resistant to neutralization by sera and plasmas from SIV-infected rhesus macaques (5, 40, 59). Having derived an R5X4 variant of SIVmac239, we sought to determine if this change in tropism affected the virus's resistance to neutralization. Neutralization sensitivity was determined for 239-ST1.2-32 and SIVmac239, using Env-pseudotyped reporter viruses on NP-2 cells coexpressing CD4 and CCR5, but not CXCR4, in the presence or absence of serial dilutions of sera (Fig. 7A, B, F, and G,) or plasmas (Fig. 7C to E) from seven SIVmac239-infected rhesus macaques. For five samples (Fig. 7A to E), 239-ST1.2-32 showed enhanced neutralization sensitivity, with 50% neutralization achieved at reciprocal dilutions that were increased approximately 1 to 2.5 log relative to those with SIVmac239. In contrast, no differences were seen between 239-ST1.2-32 and SIVmac239 for sera from two rapid progressor animals that progressed to AIDS in 4 to 6 months (Fig. 7F and G). When serum or plasma from an uninfected rhesus macaque was used (Fig. 7H and I), <20% inhibition was observed at the highest concentration and there was no difference between SIVmac239 and 239-ST1.2-32, indicating that the enhanced neutralization sensitivity of 239-ST1.2-32 was specific for host immune responses to SIVmac239.
FIG. 7.
Sensitivity of viruses to sera or plasmas from SIVmac239-infected rhesus macaques. Neutralization of SIVmac239- and 239-ST1.2-32-containing single-cycle reporter viruses by the indicated dilutions of serum (A, B, F, G) or plasma (C to E) from seven SIVmac239-infected rhesus macaques or by the indicated dilutions of serum (H) or plasma (I) from uninfected rhesus macaques was determined on NP-2/CD4/CCR5 cells. Percent infection was calculated using luciferase activity normalized to that for infection in the absence of serum or plasma. The data shown are means of three experiments plus SEM.
Lastly, we also determined the neutralization sensitivities of SIVmac239 and 239-ST1.2-32 to sCD4, which has been shown to neutralize SIVmac239 infection of CD4+ cells (83). Remarkably, a striking increase in neutralization sensitivity (>50-fold) was observed for 239-ST1.2-32, with a 50% inhibitory concentration of 0.2 μg/ml, compared with 10.8 μg/ml for SIVmac239 (Fig. 8). Similar results were seen when sCD4 sensitivity was determined on Tzm-bl target cells (not shown). Thus, the acquisition of CXCR4 use by SIVmac239 was associated with an increased sensitivity to humoral immune responses and with a marked increase in sensitivity to sCD4.
FIG. 8.
Sensitivity to neutralization by sCD4. Neutralization of SIVmac239- and 239-ST1.2-32-containing single-cycle reporter viruses by sCD4 was determined on NP-2/CD4/CCR5 cells. Percent infection was calculated using luciferase activity normalized to that for infection in the absence of sCD4. The data shown are means of three experiments plus SEM.
DISCUSSION
Although HIV-1 and HIV-2 have been well documented to acquire CXCR4 use in vivo (15, 43, 85), typically in association with a rapid decline in peripheral blood CD4 cells and the onset of AIDS, SIVs rarely use CXCR4, with most examples having been described for nonpathogenic infection models (47, 52, 65, 68, 69). It is unclear if there are intrinsic barriers to CXCR4 use by SIV, possibly because of inherent differences in Env structure, and/or if there are host selection pressures that prevent X4/R5X4 viruses from emerging even in the setting of advanced immunodeficiency. In this study, we describe the in vitro derivation of 239-ST1, an R5X4 variant of the R5 clone SIVmac239 which acquired the ability to infect CXCR4+ CCR5− SupT1 cells with rapid kinetics. An env clone from this virus, 239-ST1.2-32, used CXCR4 with high efficiency in cell-cell fusion and reporter virus infection assays and conferred CXCR4 use to a replicating virus when cloned into an SIVmac239 backbone. This 239-ST1.2-32 Env-containing virus was inhibitable by AMD3100 and was unable to infect SupT1 cells lacking CXCR4, clearly showing that SIVmac239 is capable of acquiring efficient CXCR4 use in vitro. Interestingly, even though the parental SIVmac239 Env could not use CXCR4 in cell-cell fusion or reporter virus infection assays, it did exhibit low-level CXCR4-dependent replication on CCR5− SupT1 cells, as demonstrated by its inability to replicate in a CXCR4− SupT1 line (Fig. 3) (Wang et al., unpublished data). Thus, SIVmac239 exhibited a previously unrecognized ability to use CXCR4, at least on SupT1 cells, indicating that the derivation of 239-ST1 resulted from adaptations that improved upon preexisting CXCR4 use rather than from switching to CXCR4 from an alternative coreceptor.
In contrast to X4 HIV-1 strains, where CXCR4 use is closely linked to the acquisition of positively charged residues in the V3 loop (18, 32, 87), the critical determinants of CXCR4 use for 239-ST1.2-32 mapped to a negatively charged K47E mutation in the C1 domain and an L328W mutation at the tip of V3. Although two other V3 changes, N316K and I324M, improved CXCR4 use, they were insufficient to confer X4 tropism either alone or in combination. Interestingly, the N316K mutation resulted in the loss of an N-linked glycosylation site, which has been implicated as a determinant of CXCR4 use for some HIV-1 isolates (73), and mutations at I324 have been observed to modulate interactions of SIVmac with its coreceptors (72). Notably, the 239-ST1.2-32 V3 loop did not contain positively charged residues at position 11, 24, or 25, which were associated with X4 tropism for >90% of HIV-1 isolates in a previous study (6). The critical role for the K47E mutation, a change in a conserved domain outside V3, in conferring CXCR4 use suggests that different structural requirements may exist for CXCR4 use by SIVmac compared to those for HIV-1. Although Milush et al. reported that env clones from an SIVmm isolate that depleted CXCR4+ CCR5− lymphocytes in vivo and exhibited CXCR4 use in cell-cell fusion assays did acquire positive charges in V3 at positions 11, 24, and/or 25, the structural requirements for these or other changes were not determined (65). SIVmac155T3, a unique variant of SIVmac239 that acquired CXCR4 use in vivo (17, 60, 71), gained a single positively charged residue in V3, at position 19 (V329K), but remarkably, it also acquired the K47E mutation observed in 239-ST1.2-32 (T. Kodama, personal communication), suggesting an important role for this change for both in vivo- and in vitro-derived CXCR4-using SIVmac variants.
The mechanism by which the K47E mutation contributes to CXCR4 use is unclear, particularly since the C1 domain has not been implicated previously as a determinant for coreceptor specificity in HIV or SIV Envs. Earlier mutagenesis studies have implicated both the N and C termini of HIV-1 gp120 as domains that interact with gp41 (34, 48, 91), and it is possible that the K47E mutation could play a role in facilitating Env triggering after CD4 or coreceptor engagement have occurred, lowering the energy barrier for the release of gp41 and the ensuing conformational changes that are required for fusion to occur (23). Alternatively, the C1 domain has not been included in crystal structures of gp120 monomers from HIV-1 or SIVmac (9, 37, 49, 50), and it remains possible that this mutation could directly or indirectly affect CXCR4 interactions by exposing the coreceptor binding site on the gp120 core within a monomer or in the context of the Env trimer or by altering the orientation of the V3 loop to facilitate an interaction with CXCR4.
Having been derived via long-term propagation in SupT1 cells, 239-ST1 developed a premature termination codon in the CT, at position 734, which has been well described for SIVmac isolates grown in human cells (46). The reason for the occurrence of this mutation, located immediately proximal to the second exons for tat and rev, which reside in overlapping reading frames with the gp41 CT coding region, is unclear, but premature termination of the Env CT has been shown to influence fusion kinetics (78, 93), viral growth kinetics (8, 93), and Env surface expression (42, 93). However, although the stop codon in the 239-ST1.2-32 Env clone increased fusion levels and the kinetics of viral replication, this change was clearly not required for CXCR4 use in cell-cell fusion assays or for infection by replication-competent viruses.
The acquisition of CXCR4 use by the 239-ST1.2-32 Env correlated with a marked increase in its neutralization sensitivity, relative to that of SIVmac239, to sera and plasmas from SIVmac239-infected rhesus macaques and to sCD4. These findings are interesting given the extraordinary neutralization resistance of SIVmac239 (24, 40, 41, 74) and suggest that strong humoral selection pressures in rhesus macaques may play a role in suppressing the emergence of this phenotype. Although earlier studies concluded that the neutralization sensitivity of HIV-1 Envs did not correlate with coreceptor use (51, 66, 89), more recent studies have indicated that CXCR4-using HIV-1 strains may be more sensitive than R5 HIV-1 strains (4, 7). In particular, the V3 loop of X4 HIV-1 isolates may be more exposed and, as a result, more susceptible to anti-V3 antibodies (55). However, given that a neutralization-sensitive X4 SHIV arose in an R5 SHIV-infected rhesus macaque (88), it is apparent that neutralization sensitivity per se is not a barrier to the evolution of CXCR4 utilization in nonhuman primates. It is possible that the structural changes underlying CXCR4 use for SIVmac239, in addition to its neutralization sensitivity, impart fitness costs for this phenotype in vivo. Indeed, the increased sensitivity to sCD4 could reflect a more exposed CD4 binding site that could have consequences for the ability of this virus to evade host immune responses. Studies to understand the determinants and the mechanism for this effect, as well as their biological consequences, are in progress.
239-ST1.2-32 should be a useful tool to assess viral and host selection pressures on CXCR4 use in vivo. Given that its replication and CXCR4 use do not require a truncated CT, it should be possible to assess the pathogenesis and evolution of coreceptor usage of 239-ST1.2-32 Long CT in rhesus macaques. The purely X4-tropic SIVmac239 variant SIVmac155T3, derived from the lymph node of an animal with advanced immunodeficiency (17, 60), exhibited a distinct phenotype in vivo, causing the depletion of CXCR4+ CCR5− naïve CD4 cells while sparing the CCR5+ memory cells (particularly in tissues). Remarkably, this virus failed to cause any clinical disease (71). Because 239-ST1.2-32 is R5X4 tropic and would be expected to have a broader host cell range, it might prove to be more pathogenic than SIVmac155T3. In addition, SIVmac155T3 has 22 amino acid mutations in gp120 (60), making it difficult to identify the specific determinants for X4 tropism and their subsequent evolution in vivo, whereas 239-ST1.2-32 has only five changes in gp120, with now clearly defined determinants for CXCR4 use. Thus, in addition to assessing the pathogenicity of 239-ST1.2-32 Long CT in rhesus macaques, it will be interesting to determine if and how CXCR4 use is selected against in the context of a host immune response and to what extent these changes correspond to effects on the virus's neutralization sensitivity, as noted above. Studies to assess these issues are currently in progress.
Acknowledgments
We thank Meredith Hunter, Preston Marx, Jessica Engram, and Guido Silvestri for rhesus macaque sera and plasmas and Dana Gabuzda for Cf2Th cells. We also thank Max Richardson and James Riley for the pELNS lentiviral vector. Technological support for p24 and p27 assays was provided by the Viral and Molecular Core of the Penn Center for AIDS Research. We also thank Philip Gregory, Edward Rebar, Jeffrey Miller, Lei Zhang, Sarah Hinkley, and colleagues at Sangamo BioSciences for reagents and helpful discussions during the generation of CXCR4-negative SupT1 cells.
This work was supported by National Institutes of Health grants AI-49784 (to J.A.H.) and AI-30034 (to D.C.M.). G.Q.D. was supported by grant T32 AI-07632.
Footnotes
Published ahead of print on 15 July 2009.
REFERENCES
- 1.Alexander, W. A., B. Moss, and T. R. Fuerst. 1992. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J. Virol. 66:2934-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 3.Bron, R., P. J. Klasse, D. Wilkinson, P. R. Clapham, A. Pelchen-Matthews, C. Power, T. N. Wells, J. Kim, S. C. Peiper, J. A. Hoxie, and M. Marsh. 1997. Promiscuous use of CC and CXC chemokine receptors in cell-to-cell fusion mediated by a human immunodeficiency virus type 2 envelope protein. J. Virol. 71:8405-8415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bunnik, E. M., E. D. Quakkelaar, A. C. van Nuenen, B. Boeser-Nunnink, and H. Schuitemaker. 2007. Increased neutralization sensitivity of recently emerged CXCR4-using human immunodeficiency virus type 1 strains compared to coexisting CCR5-using variants from the same patient. J. Virol. 81:525-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burns, D. P., C. Collignon, and R. C. Desrosiers. 1993. Simian immunodeficiency virus mutants resistant to serum neutralization arise during persistent infection of rhesus monkeys. J. Virol. 67:4104-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardozo, T., T. Kimura, S. Philpott, B. Weiser, H. Burger, and S. Zolla-Pazner. 2007. Structural basis for coreceptor selectivity by the HIV type 1 V3 loop. AIDS Res. Hum. Retrovir. 23:415-426. [DOI] [PubMed] [Google Scholar]
- 7.Cavacini, L. A., M. Duval, J. Robinson, and M. R. Posner. 2002. Interactions of human antibodies, epitope exposure, antibody binding and neutralization of primary isolate HIV-1 virions. AIDS 16:2409-2417. [DOI] [PubMed] [Google Scholar]
- 8.Chakrabarti, L., M. Emerman, P. Tiollais, and P. Sonigo. 1989. The cytoplasmic domain of simian immunodeficiency virus transmembrane protein modulates infectivity. J. Virol. 63:4395-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, B., E. M. Vogan, H. Gong, J. J. Skehel, D. C. Wiley, and S. C. Harrison. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834-841. [DOI] [PubMed] [Google Scholar]
- 10.Chen, B. K., K. Saksela, R. Andino, and D. Baltimore. 1994. Distinct modes of human immunodeficiency virus type 1 proviral latency revealed by superinfection of nonproductively infected cell lines with recombinant luciferase-encoding viruses. J. Virol. 68:654-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, Z., A. Gettie, D. D. Ho, and P. A. Marx. 1998. Primary SIVsm isolates use the CCR5 coreceptor from sooty mangabeys naturally infected in west Africa: a comparison of coreceptor usage of primary SIVsm, HIV-2, and SIVmac. Virology 246:113-124. [DOI] [PubMed] [Google Scholar]
- 12.Chen, Z., P. Zhou, D. D. Ho, N. R. Landau, and P. A. Marx. 1997. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J. Virol. 71:2705-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 14.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 15.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dean, M., M. Carrington, C. Winkler, G. A. Huttley, M. W. Smith, R. Allikmets, J. J. Goedert, S. P. Buchbinder, E. Vittinghoff, E. Gomperts, S. Donfield, D. Vlahov, R. Kaslow, A. Saah, C. Rinaldo, R. Detels, S. J. O'Brien, et al. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science 273:1856-1862. [DOI] [PubMed] [Google Scholar]
- 17.DeGottardi, M. Q., S. K. Lew, M. Piatak, Jr., B. Jia, Y. Feng, S. J. Lee, J. M. Brenchley, D. C. Douek, T. Kodama, J. D. Lifson, and D. T. Evans. 2008. Comparison of plasma viremia and antibody responses in macaques inoculated with envelope variants of single-cycle simian immunodeficiency virus differing in infectivity and cellular tropism. J. Virol. 82:321-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Jong, J. J., A. De Ronde, W. Keulen, M. Tersmette, and J. Goudsmit. 1992. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J. Virol. 66:6777-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 20.Deng, H. K., D. Unutmaz, V. N. KewalRamani, and D. R. Littman. 1997. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature 388:296-300. [DOI] [PubMed] [Google Scholar]
- 21.Doranz, B. J., J. Rucker, Y. Yi, R. J. Smyth, M. Samson, S. C. Peiper, M. Parmentier, R. G. Collman, and R. W. Doms. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149-1158. [DOI] [PubMed] [Google Scholar]
- 22.Dragic, T., V. Litwin, G. P. Allaway, S. R. Martin, Y. Huang, K. A. Nagashima, C. Cayanan, P. J. Maddon, R. A. Koup, J. P. Moore, and W. A. Paxton. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667-673. [DOI] [PubMed] [Google Scholar]
- 23.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 24.Edinger, A. L., M. Ahuja, T. Sung, K. C. Baxter, B. Haggarty, R. W. Doms, and J. A. Hoxie. 2000. Characterization and epitope mapping of neutralizing monoclonal antibodies produced by immunization with oligomeric simian immunodeficiency virus envelope protein. J. Virol. 74:7922-7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edinger, A. L., A. Amedee, K. Miller, B. J. Doranz, M. Endres, M. Sharron, M. Samson, Z. H. Lu, J. E. Clements, M. Murphey-Corb, S. C. Peiper, M. Parmentier, C. C. Broder, and R. W. Doms. 1997. Differential utilization of CCR5 by macrophage and T cell tropic simian immunodeficiency virus strains. Proc. Natl. Acad. Sci. USA 94:4005-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edinger, A. L., and R. W. Doms. 1999. A cell-cell fusion assay to monitor HIV-1 Env interactions with chemokine receptors. Methods Mol. Med. 17:41-49. [DOI] [PubMed] [Google Scholar]
- 27.Edinger, A. L., T. L. Hoffman, M. Sharron, B. Lee, B. O'Dowd, and R. W. Doms. 1998. Use of GPR1, GPR15, and STRL33 as coreceptors by diverse human immunodeficiency virus type 1 and simian immunodeficiency virus envelope proteins. Virology 249:367-378. [DOI] [PubMed] [Google Scholar]
- 28.Endres, M. J., P. R. Clapham, M. Marsh, M. Ahuja, J. D. Turner, A. McKnight, J. F. Thomas, B. Stoebenau-Haggarty, S. Choe, P. J. Vance, T. N. Wells, C. A. Power, S. S. Sutterwala, R. W. Doms, N. R. Landau, and J. A. Hoxie. 1996. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell 87:745-756. [DOI] [PubMed] [Google Scholar]
- 29.Farzan, M., H. Choe, K. Martin, L. Marcon, W. Hofmann, G. Karlsson, Y. Sun, P. Barrett, N. Marchand, N. Sullivan, N. Gerard, C. Gerard, and J. Sodroski. 1997. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J. Exp. Med. 186:405-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 31.Fouchier, R. A., M. Brouwer, S. M. Broersen, and H. Schuitemaker. 1995. Simple determination of human immunodeficiency virus type 1 syncytium-inducing V3 genotype by PCR. J. Clin. Microbiol. 33:906-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fouchier, R. A., M. Groenink, N. A. Kootstra, M. Tersmette, H. G. Huisman, F. Miedema, and H. Schuitemaker. 1992. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J. Virol. 66:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guillon, C., M. E. van der Ende, P. H. Boers, R. A. Gruters, M. Schutten, and A. D. Osterhaus. 1998. Coreceptor usage of human immunodeficiency virus type 2 primary isolates and biological clones is broad and does not correlate with their syncytium-inducing capacities. J. Virol. 72:6260-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helseth, E., U. Olshevsky, C. Furman, and J. Sodroski. 1991. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J. Virol. 65:2119-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho, S. H., S. Tasca, L. Shek, A. Li, A. Gettie, J. Blanchard, D. Boden, and C. Cheng-Mayer. 2007. Coreceptor switch in R5-tropic simian/human immunodeficiency virus-infected macaques. J. Virol. 81:8621-8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoxie, J. A., J. D. Alpers, J. L. Rackowski, K. Huebner, B. S. Haggarty, A. J. Cedarbaum, and J. C. Reed. 1986. Alterations in T4 (CD4) protein and mRNA synthesis in cells infected with HIV. Science 234:1123-1127. [DOI] [PubMed] [Google Scholar]
- 37.Huang, C. C., M. Tang, M. Y. Zhang, S. Majeed, E. Montabana, R. L. Stanfield, D. S. Dimitrov, B. Korber, J. Sodroski, I. A. Wilson, R. Wyatt, and P. D. Kwong. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jensen, M. A., F. S. Li, A. B. van't Wout, D. C. Nickle, D. Shriner, H. X. He, S. McLaughlin, R. Shankarappa, J. B. Margolick, and J. I. Mullins. 2003. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J. Virol. 77:13376-13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joag, S. V., Z. Li, L. Foresman, E. B. Stephens, L. J. Zhao, I. Adany, D. M. Pinson, H. M. McClure, and O. Narayan. 1996. Chimeric simian/human immunodeficiency virus that causes progressive loss of CD4+ T cells and AIDS in pig-tailed macaques. J. Virol. 70:3189-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson, W. E., J. Morgan, J. Reitter, B. A. Puffer, S. Czajak, R. W. Doms, and R. C. Desrosiers. 2002. A replication-competent, neutralization-sensitive variant of simian immunodeficiency virus lacking 100 amino acids of envelope. J. Virol. 76:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson, W. E., H. Sanford, L. Schwall, D. R. Burton, P. W. Parren, J. E. Robinson, and R. C. Desrosiers. 2003. Assorted mutations in the envelope gene of simian immunodeficiency virus lead to loss of neutralization resistance against antibodies representing a broad spectrum of specificities. J. Virol. 77:9993-10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnston, P. B., J. W. Dubay, and E. Hunter. 1993. Truncations of the simian immunodeficiency virus transmembrane protein confer expanded virus host range by removing a block to virus entry into cells. J. Virol. 67:3077-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karlsson, A., K. Parsmyr, E. Sandstrom, E. M. Fenyo, and J. Albert. 1994. MT-2 cell tropism as prognostic marker for disease progression in human immunodeficiency virus type 1 infection. J. Clin. Microbiol. 32:364-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirchhoff, F., K. Mori, and R. C. Desrosiers. 1994. The “V3” domain is a determinant of simian immunodeficiency virus cell tropism. J. Virol. 68:3682-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirchhoff, F., S. Pohlmann, M. Hamacher, R. E. Means, T. Kraus, K. Uberla, and P. Di Marzio. 1997. Simian immunodeficiency virus variants with differential T-cell and macrophage tropism use CCR5 and an unidentified cofactor expressed in CEMx174 cells for efficient entry. J. Virol. 71:6509-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kodama, T., D. P. Wooley, Y. M. Naidu, H. W. Kestler III, M. D. Daniel, Y. Li, and R. C. Desrosiers. 1989. Significance of premature stop codons in env of simian immunodeficiency virus. J. Virol. 63:4709-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Konig, R. R., E. Flory, S. Steidl, J. Neumann, C. Coulibaly, E. Holznagel, S. Holzammer, S. Norley, and K. Cichutek. 2002. Engineered CD4- and CXCR4-using simian immunodeficiency virus from African green monkeys is neutralization sensitive and replicates in nonstimulated lymphocytes. J. Virol. 76:10627-10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kowalski, M., J. Potz, L. Basiripour, T. Dorfman, W. C. Goh, E. Terwilliger, A. Dayton, C. Rosen, W. Haseltine, and J. Sodroski. 1987. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science 237:1351-1355. [DOI] [PubMed] [Google Scholar]
- 49.Kwong, P. D., R. Wyatt, S. Majeed, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure 8:1329-1339. [DOI] [PubMed] [Google Scholar]
- 50.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.LaCasse, R. A., K. E. Follis, T. Moudgil, M. Trahey, J. M. Binley, V. Planelles, S. Zolla-Pazner, and J. H. Nunberg. 1998. Coreceptor utilization by human immunodeficiency virus type 1 is not a primary determinant of neutralization sensitivity. J. Virol. 72:2491-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lauren, A., D. Vodros, R. Thorstensson, and E. M. Fenyo. 2006. Comparative studies on mucosal and intravenous transmission of simian immunodeficiency virus (SIVsm): evolution of coreceptor use varies with pathogenic outcome. J. Gen. Virol. 87:581-594. [DOI] [PubMed] [Google Scholar]
- 53.Li, M., F. Gao, J. R. Mascola, L. Stamatatos, V. R. Polonis, M. Koutsoukos, G. Voss, P. Goepfert, P. Gilbert, K. M. Greene, M. Bilska, D. L. Kothe, J. F. Salazar-Gonzalez, X. Wei, J. M. Decker, B. H. Hahn, and D. C. Montefiori. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu, R., W. A. Paxton, S. Choe, D. Ceradini, S. R. Martin, R. Horuk, M. E. MacDonald, H. Stuhlmann, R. A. Koup, and N. R. Landau. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367-377. [DOI] [PubMed] [Google Scholar]
- 55.Lusso, P., P. L. Earl, F. Sironi, F. Santoro, C. Ripamonti, G. Scarlatti, R. Longhi, E. A. Berger, and S. E. Burastero. 2005. Cryptic nature of a conserved, CD4-inducible V3 loop neutralization epitope in the native envelope glycoprotein oligomer of CCR5-restricted, but not CXCR4-using, primary human immunodeficiency virus type 1 strains. J. Virol. 79:6957-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mansfield, K. G., N. W. Lerch, M. B. Gardner, and A. A. Lackner. 1995. Origins of simian immunodeficiency virus infection in macaques at the New England Regional Primate Research Center. J. Med. Primatol. 24:116-122. [DOI] [PubMed] [Google Scholar]
- 57.Marx, P. A., Y. Li, N. W. Lerche, S. Sutjipto, A. Gettie, J. A. Yee, B. H. Brotman, A. M. Prince, A. Hanson, and R. G. Webster. 1991. Isolation of a simian immunodeficiency virus related to human immunodeficiency virus type 2 from a West African pet sooty mangabey. J. Virol. 65:4480-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McKnight, A., D. Wilkinson, G. Simmons, S. Talbot, L. Picard, M. Ahuja, M. Marsh, J. A. Hoxie, and P. R. Clapham. 1997. Inhibition of human immunodeficiency virus fusion by a monoclonal antibody to a coreceptor (CXCR4) is both cell type and virus strain dependent. J. Virol. 71:1692-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Means, R. E., T. Greenough, and R. C. Desrosiers. 1997. Neutralization sensitivity of cell culture-passaged simian immunodeficiency virus. J. Virol. 71:7895-7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Means, R. E., T. Matthews, J. A. Hoxie, M. H. Malim, T. Kodama, and R. C. Desrosiers. 2001. Ability of the V3 loop of simian immunodeficiency virus to serve as a target for antibody-mediated neutralization: correlation of neutralization sensitivity, growth in macrophages, and decreased dependence on CD4. J. Virol. 75:3903-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meissner, E. G., V. M. Coffield, and L. Su. 2005. Thymic pathogenicity of an HIV-1 envelope is associated with increased CXCR4 binding efficiency and V5-gp41-dependent activity, but not V1/V2-associated CD4 binding efficiency and viral entry. Virology 336:184-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meissner, E. G., K. M. Duus, F. Gao, X. F. Yu, and L. Su. 2004. Characterization of a thymus-tropic HIV-1 isolate from a rapid progressor: role of the envelope. Virology 328:74-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meister, S., C. Otto, A. Papkalla, M. Krumbiegel, S. Pohlmann, and F. Kirchhoff. 2001. Basic amino acid residues in the V3 loop of simian immunodeficiency virus envelope alter viral coreceptor tropism and infectivity but do not allow efficient utilization of CXCR4 as entry cofactor. Virology 284:287-296. [DOI] [PubMed] [Google Scholar]
- 64.Miller, J. C., M. C. Holmes, J. Wang, D. Y. Guschin, Y. L. Lee, I. Rupniewski, C. M. Beausejour, A. J. Waite, N. S. Wang, K. A. Kim, P. D. Gregory, C. O. Pabo, and E. J. Rebar. 2007. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 25:778-785. [DOI] [PubMed] [Google Scholar]
- 65.Milush, J. M., J. D. Reeves, S. N. Gordon, D. Zhou, A. Muthukumar, D. A. Kosub, E. Chacko, L. D. Giavedoni, C. C. Ibegbu, K. S. Cole, J. L. Miamidian, M. Paiardini, A. P. Barry, S. I. Staprans, G. Silvestri, and D. L. Sodora. 2007. Virally induced CD4+ T cell depletion is not sufficient to induce AIDS in a natural host. J. Immunol. 179:3047-3056. [DOI] [PubMed] [Google Scholar]
- 66.Montefiori, D. C., R. G. Collman, T. R. Fouts, J. Y. Zhou, M. Bilska, J. A. Hoxie, J. P. Moore, and D. P. Bolognesi. 1998. Evidence that antibody-mediated neutralization of human immunodeficiency virus type 1 by sera from infected individuals is independent of coreceptor usage. J. Virol. 72:1886-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morner, A., A. Bjorndal, J. Albert, V. N. Kewalramani, D. R. Littman, R. Inoue, R. Thorstensson, E. M. Fenyo, and E. Bjorling. 1999. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 73:2343-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Owen, S. M., S. Masciotra, F. Novembre, J. Yee, W. M. Switzer, M. Ostyula, and R. B. Lal. 2000. Simian immunodeficiency viruses of diverse origin can use CXCR4 as a coreceptor for entry into human cells. J. Virol. 74:5702-5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pandrea, I., C. Kornfeld, M. J. Ploquin, C. Apetrei, A. Faye, P. Rouquet, P. Roques, F. Simon, F. Barre-Sinoussi, M. C. Muller-Trutwin, and O. M. Diop. 2005. Impact of viral factors on very early in vivo replication profiles in simian immunodeficiency virus SIVagm-infected African green monkeys. J. Virol. 79:6249-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perez, E. E., J. Wang, J. C. Miller, Y. Jouvenot, K. A. Kim, O. Liu, N. Wang, G. Lee, V. V. Bartsevich, Y. L. Lee, D. Y. Guschin, I. Rupniewski, A. J. Waite, C. Carpenito, R. G. Carroll, J. S. Orange, F. D. Urnov, E. J. Rebar, D. Ando, P. D. Gregory, J. L. Riley, M. C. Holmes, and C. H. June. 2008. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 26:808-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Picker, L. J., S. I. Hagen, R. Lum, E. F. Reed-Inderbitzin, L. M. Daly, A. W. Sylwester, J. M. Walker, D. C. Siess, M. Piatak, Jr., C. Wang, D. B. Allison, V. C. Maino, J. D. Lifson, T. Kodama, and M. K. Axthelm. 2004. Insufficient production and tissue delivery of CD4+ memory T cells in rapidly progressive simian immunodeficiency virus infection. J. Exp. Med. 200:1299-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pohlmann, S., C. Davis, S. Meister, G. J. Leslie, C. Otto, J. D. Reeves, B. A. Puffer, A. Papkalla, M. Krumbiegel, A. Marzi, S. Lorenz, J. Munch, R. W. Doms, and F. Kirchhoff. 2004. Amino acid 324 in the simian immunodeficiency virus SIVmac V3 loop can confer CD4 independence and modulate the interaction with CCR5 and alternative coreceptors. J. Virol. 78:3223-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pollakis, G., S. Kang, A. Kliphuis, M. I. Chalaby, J. Goudsmit, and W. A. Paxton. 2001. N-linked glycosylation of the HIV type-1 gp120 envelope glycoprotein as a major determinant of CCR5 and CXCR4 coreceptor utilization. J. Biol. Chem. 276:13433-13441. [DOI] [PubMed] [Google Scholar]
- 74.Reitter, J. N., and R. C. Desrosiers. 1998. Identification of replication-competent strains of simian immunodeficiency virus lacking multiple attachment sites for N-linked carbohydrates in variable regions 1 and 2 of the surface envelope protein. J. Virol. 72:5399-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Resch, W., N. Hoffman, and R. Swanstrom. 2001. Improved success of phenotype prediction of the human immunodeficiency virus type 1 from envelope variable loop 3 sequence using neural networks. Virology 288:51-62. [DOI] [PubMed] [Google Scholar]
- 76.Richardson, M. W., R. G. Carroll, M. Stremlau, N. Korokhov, L. M. Humeau, G. Silvestri, J. Sodroski, and J. L. Riley. 2008. Mode of transmission affects the sensitivity of human immunodeficiency virus type 1 to restriction by rhesus TRIM5alpha. J. Virol. 82:11117-11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Richman, D. D., and S. A. Bozzette. 1994. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J. Infect. Dis. 169:968-974. [DOI] [PubMed] [Google Scholar]
- 78.Ritter, G. D., Jr., M. J. Mulligan, S. L. Lydy, and R. W. Compans. 1993. Cell fusion activity of the simian immunodeficiency virus envelope protein is modulated by the intracytoplasmic domain. Virology 197:255-264. [DOI] [PubMed] [Google Scholar]
- 79.Rucker, J., B. J. Doranz, A. L. Edinger, D. Long, J. F. Berson, and R. W. Doms. 1997. Cell-cell fusion assay to study role of chemokine receptors in human immunodeficiency virus type 1 entry. Methods Enzymol. 288:118-133. [DOI] [PubMed] [Google Scholar]
- 80.Rucker, J., A. L. Edinger, M. Sharron, M. Samson, B. Lee, J. F. Berson, Y. Yi, B. Margulies, R. G. Collman, B. J. Doranz, M. Parmentier, and R. W. Doms. 1997. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J. Virol. 71:8999-9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Samson, M., A. L. Edinger, P. Stordeur, J. Rucker, V. Verhasselt, M. Sharron, C. Govaerts, C. Mollereau, G. Vassart, R. W. Doms, and M. Parmentier. 1998. ChemR23, a putative chemoattractant receptor, is expressed in monocyte-derived dendritic cells and macrophages and is a coreceptor for SIV and some primary HIV-1 strains. Eur. J. Immunol. 28:1689-1700. [DOI] [PubMed] [Google Scholar]
- 82.Samson, M., F. Libert, B. J. Doranz, J. Rucker, C. Liesnard, C. M. Farber, S. Saragosti, C. Lapoumeroulie, J. Cognaux, C. Forceille, G. Muyldermans, C. Verhofstede, G. Burtonboy, M. Georges, T. Imai, S. Rana, Y. Yi, R. J. Smyth, R. G. Collman, R. W. Doms, G. Vassart, and M. Parmentier. 1996. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722-725. [DOI] [PubMed] [Google Scholar]
- 83.Schenten, D., L. Marcon, G. B. Karlsson, C. Parolin, T. Kodama, N. Gerard, and J. Sodroski. 1999. Effects of soluble CD4 on simian immunodeficiency virus infection of CD4-positive and CD4-negative cells. J. Virol. 73:5373-5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. de Goede, R. P. van Steenwijk, J. M. Lange, J. K. Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 66:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shi, Y., E. Brandin, E. Vincic, M. Jansson, A. Blaxhult, K. Gyllensten, L. Moberg, C. Brostrom, E. M. Fenyo, and J. Albert. 2005. Evolution of human immunodeficiency virus type 2 coreceptor usage, autologous neutralization, envelope sequence and glycosylation. J. Gen. Virol. 86:3385-3396. [DOI] [PubMed] [Google Scholar]
- 86.Shibata, R., F. Maldarelli, C. Siemon, T. Matano, M. Parta, G. Miller, T. Fredrickson, and M. A. Martin. 1997. Infection and pathogenicity of chimeric simian-human immunodeficiency viruses in macaques: determinants of high virus loads and CD4 cell killing. J. Infect. Dis. 176:362-373. [DOI] [PubMed] [Google Scholar]
- 87.Shioda, T., S. Oka, S. Ida, K. Nokihara, H. Toriyoshi, S. Mori, Y. Takebe, S. Kimura, K. Shimada, and Y. Nagai. 1994. A naturally occurring single basic amino acid substitution in the V3 region of the human immunodeficiency virus type 1 Env protein alters the cellular host range and antigenic structure of the virus. J. Virol. 68:7689-7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tasca, S., S. H. Ho, and C. Cheng-Mayer. 2008. R5X4 viruses are evolutionary, functional, and antigenic intermediates in the pathway of a simian-human immunodeficiency virus coreceptor switch. J. Virol. 82:7089-7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Trkola, A., T. Ketas, V. N. Kewalramani, F. Endorf, J. M. Binley, H. Katinger, J. Robinson, D. R. Littman, and J. P. Moore. 1998. Neutralization sensitivity of human immunodeficiency virus type 1 primary isolates to antibodies and CD4-based reagents is independent of coreceptor usage. J. Virol. 72:1876-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van't Wout, A. B., N. A. Kootstra, G. A. Mulder-Kampinga, N. Albrecht-van Lent, H. J. Scherpbier, J. Veenstra, K. Boer, R. A. Coutinho, F. Miedema, and H. Schuitemaker. 1994. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J. Clin. Investig. 94:2060-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wyatt, R., E. Desjardin, U. Olshevsky, C. Nixon, J. Binley, V. Olshevsky, and J. Sodroski. 1997. Analysis of the interaction of the human immunodeficiency virus type 1 gp120 envelope glycoprotein with the gp41 transmembrane glycoprotein. J. Virol. 71:9722-9731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang, Y., B. Lou, R. B. Lal, A. Gettie, P. A. Marx, and J. P. Moore. 2000. Use of inhibitors to evaluate coreceptor usage by simian and simian/human immunodeficiency viruses and human immunodeficiency virus type 2 in primary cells. J. Virol. 74:6893-6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zingler, K., and D. R. Littman. 1993. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein increases env incorporation into particles and fusogenicity and infectivity. J. Virol. 67:2824-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]