Abstract
Knowledge of endogenous retroviruses (ERVs) in crocodilians (Crocodylia) is limited, and their distribution among extant species is unclear. Here we analyzed the phylogenetic relationships of these retroelements in 20 species of crocodilians by studying the pro-pol gene. The results showed that crocodilian ERVs (CERVs) cluster into two major clades (CERV 1 and CERV 2). CERV 1 clustered as a sister group of the genus Gammaretrovirus, while CERV 2 clustered distantly with respect to all known ERVs. Interestingly, CERV 1 was found only in crocodiles (Crocodylidae). The data generated here could assist future studies aimed at identifying orthologous and paralogous ERVs among crocodilians.
Endogenous retroviruses (ERVs) are copies or remnants of exogenous viruses derived from past infections of germ cells and subsequent integration into the host genome (7, 8). Most ERVs are defective, having accumulated random inactivating mutations, and are therefore not pathogenic. However, many intact ERVs have been associated with different host diseases (4, 21). Those from pigs are considered potentially hazardous during xenotransplantation due to reactivation or recombination with exogenous retroviruses (18). ERVs have been studied extensively in mammals and birds (9, 10, 16), while knowledge of ERVs in reptiles is limited to a few host species (17, 24). Studies of a few full-length and partial pol gene sequences of some crocodilian species (order Crocodylia) have shown that some ERV sequences clustered distantly from Spumavirus while others clustered closely with Gammaretrovirus-like retroviruses (16, 17). However, their diversity, lineage specificity, and functionality have not been assessed across all extant crocodilian species.
The extant crocodylian lineage consists of 23 species classified in three families, the Alligatoridae (alligators), Crocodylidae (crocodiles), and Gavialidae (gharials). The Crocodylidae and Alligatoridae families are unambiguously recognized, with gharials either lumped with the Crocodylidae family or assigned to a separate family, the Gavialidae (6, 12, 19). It has been estimated, based on DNA and amino acid data, that the Alligatoridae and Crocodylidae lineages diverged from a common ancestor about 97 to 103 million years ago (12, 19).
Here we analyze the distribution, potential to function, and phylogenetic relationships of ERVs in 20 extant crocodilian species by studying the protease-reverse transcriptase (pro-pol) gene. The pro-pol gene (0.8 to 1 kb) was amplified using two sets of degenerate primers (23). The PCR amplicons were gel purified and cloned using standard protocols, and about three ERV inserts were sequenced from each species by the sequencing service at the Australian Genome Research Facility. Sixty-five ERV DNA sequences were generated and translated to putative amino acid sequences using universal translation codes in the Molecular Toolkit interface (http://arbl.cvmbs.colostate.edu/molkit/index.html). Open reading frames and levels of similarity to other available ERV sequences were determined using the blastx tool available through NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Two aligned datasets were generated using the CLUSTALW program (22); the first consisted of 286 predicted amino acids from 65 novel and 9 published (16, 17) crocodilian ERVs (CERVs), and the second comprised 258 predicted amino acids from the novel crocodilian sequences and 73 published endogenous and exogenous retroviral sequences after exclusion of gaps. These known retroviruses included avian leukosis virus (Alpharetrovirus); murine leukemia virus (Gammaretrovirus); mouse mammary tumor virus and Jaagsiekte sheep retrovirus (Betaretrovirus); bovine leukemia virus and human T-lymphotropic virus (Deltaretrovirus); human immunodeficiency virus type 1, equine infectious anemia virus, and visna virus (Lentivirus); human foamy virus (Spumavirus); walleye dermal sarcoma virus (Epsilonretrovirus); 4 avian, 3 reptilian, 2 amphibian, and 14 mammalian ERVs, including human ERVs (13, 14); 3 chicken ERVs (10); 6 avian viruses similar to the alpha/beta group (7); and 22 Gammaretrovirus-like ERVs (9, 16). The best-fit model (JTT matrix model with parameter α = 1.61) for the two data sets was selected by using ProtTest software (1) to implement neighbor-joining (NJ) in the MEGA 4 program (20). In addition, a maximum parsimony analysis was performed. Levels of amino acid similarity between CERV and other ERV sequences were also ascertained in the MEGA 4 program, using the p-distance option.
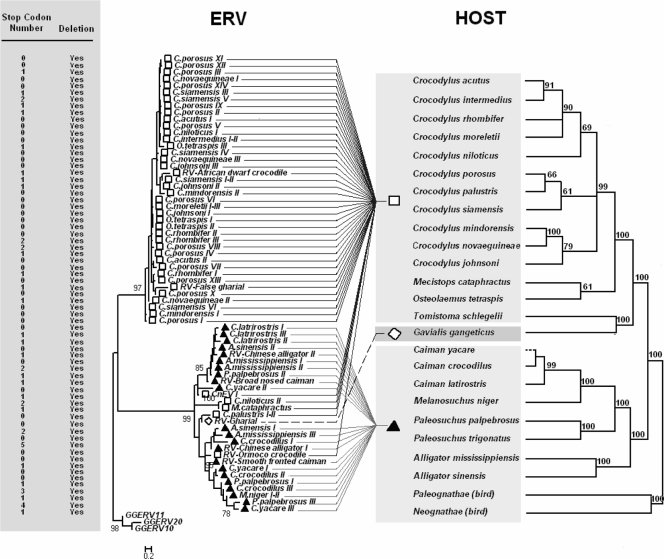
ERVs were found in all crocodilian lineages examined (Fig. 1). All 65 sequences show deletions, and 28 of these contain in-frame stop codons (Fig. 1). These mutations indicate that all CERVs generated in this study are defective and, therefore, nonfunctional, as has been observed in other vertebrate hosts (2, 3, 11, 15). Although only nonfunctional sequences were identified in members of the Alligatoridae and Crocodylidae, our study does not exclude the possibility of functional ERVs in these lineages.
FIG. 1.
NJ tree of 286 putative amino acids of the pol gene from CERV, using chicken ERV sequences as an outgroup (left), and basing the host tree on nuclear, mitochondrial, and morphological data (right), modified with author's permission (5). The column next to the viral tree shows the number of stop codons and deletions observed for the pro-pol reading frame in each CERV. The value on the tree node indicates bootstrap branch support (1,000 replicates). □, Crocodylidae; ▴, Alligatoridae; ⋄, Gavialidae; RV, CERVs from other studies (16, 17); GGERV, chicken ERVs. Branch lengths are proportional to sequence differences among ERVs.
The NJ and maximum parsimony phylogenetic trees were consistent and showed that CERVs cluster into two distinct major clades (Fig. 1), named CERV 1 and 2. CERV 1 consists of 44 sequences from 12 species of Crocodylidae, revealing for the first time the existence of a host lineage-specific ERV clade for crocodiles. CERV 2 consists of 30 sequences representing eight Alligatoridae species and the only Gavialidae species. Four sequences from three Crocodylidae species (Crocodylus niloticus, Crocodylus palustris, and Mecistops cataphractus) also clustered within CERV 2. Interestingly, sequences from both CERV 1 and 2 were found in a single Crocodylidae species (Crocodylus niloticus). Pairwise genetic distances show that the variation within CERV 1 (distance = 0.070 ± 0.006 [mean ± standard deviation]) is very much lower than that within CERV 2 (distance = 0.459 ± 0.020). Analyses also show that all CERV 2 sequences are distinct, revealing additional diversity and new minor clades within this unusual clade (Fig. 1) that have not been reported previously. In contrast, CERV 1 sequences show a high degree of similarity between species, and some of them are identical within species, including those from C. intermedius, C. siamensis, and C. moreletii.
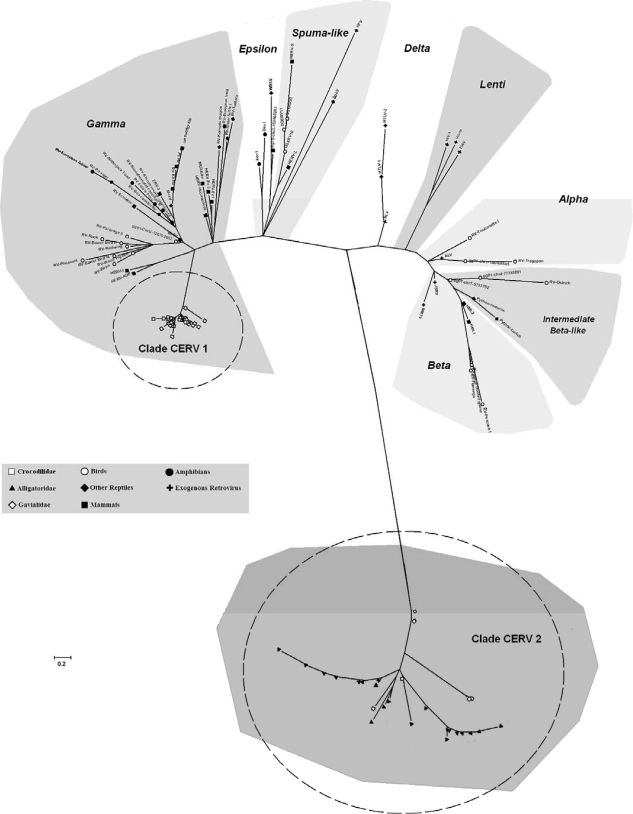
Analysis of phylogenetic relationships with known ERVs showed that CERV 1 clusters closely with two clades of Gammaretrovirus (birds and reptiles/mammals/amphibians/human), showing polytomy and long branch lengths with respect to each other (Fig. 2). Clade CERV 2 clustered distantly from all known ERVs (Fig. 2). These relationships are also supported by the amino acid similarity between a known representative of Gammaretrovirus, murine leukemia virus, and CERV clades. In agreement with the results of previous CERV studies (17), the amino acid similarity was lower for CERV 2 (24%) and higher for CERV 1 (47%) (Table 1).
FIG. 2.
NJ analyses of 258 putative amino acids of the pol gene from CERVs and representatives from seven known ERV genera. Symbols represent the ERV hosts used in this study, as indicated in the key. Dashed circles show the two major CERV clades, CERV 1 and 2. Clustering information within CERVs is provided in Fig. 1. ALV, avian leukosis virus; BaEV, baboon endogenous retrovirus; BLV, bovine leukemia virus; DevI, Dendrobates Ventrimaculatus ERVI; EIAV, equine infectious anemia virus; ERV-3, human ERV3; GGERV, chicken ERV; HERV, human ERV; HFV, human foamy virus; HIV-1, human immunodeficiency virus type 1; HTLV, human T-lymphotropic virus; JSRV, Jaagsiekte sheep retrovirus; MMTV, mouse mammary tumor virus; MuLV, murine leukemia virus; PERV, porcine ERV; Spev, Sphenodon ERV; WDSV, walleye dermal sarcoma virus; Xen1, Xenopus laeris ERV1.
TABLE 1.
Percentages of similarity between CERV 1 and CERV 2 and exogenous members of the Retroviridae
| CERV clade | % Similarity to indicated member of the Retroviradaea
|
||||||
|---|---|---|---|---|---|---|---|
| ALV | MMTV | MuLV | BLV | HIV-1 | HFV | WDSV | |
| 1 | 38 | 30 | 47 | 37 | 20 | 26 | 28 |
| 2 | 25 | 21 | 24 | 28 | 20 | 21 | 21 |
Calculated from alignments of 265 amino acids of the pro-pol gene. Values are based on average similarity observed in CERV 1 and CERV 2 (Pairwise deletion option in the MEGA 4 program). ALV, avian leukosis virus; MMTV, mouse mammary tumor virus; MuLV, murine leukemia virus; BLV, bovine leukemia virus; HIV-1, human immunodeficiency virus type 1; HFV, human foamy virus; WDSV, walleye dermal sarcoma virus.
The comparison of CERV and host species phylogenies (6, 12, 19) showed discordance. While crocodilian host phylogenies based on DNA sequence data are quite well defined, this was not the case within the CERV clades. Both CERV 1 and 2 showed a mixture of internal topologies from symmetric (bushlike), to random, to asymmetric with differential branch lengths, making it difficult to assess any coevolutionary patterns. Given that CERV 1 appeared to be specific to the Crocodylidae family and that its internal branch lengths were considerably shorter than those observed in the crocodile host phylogeny (5) and CERV 2, it is plausible that CERV 1 has a lower mutation rate and represents a relatively recent retroviral infection that occurred after the divergence of the Crocodylidae and Alligatoridae families from the common ancestor.
The current investigation has confirmed the existence of two groups of ERVs and revealed additional distribution and diversity among extant members of the Crocodylia. Interestingly, we found a host lineage-specific clade which could have potential for use in the identification of members of the Crocodylidae at the family level. The data generated here will assist future studies identifying orthologous and paralagous ERVs among crocodilian species to assess the variation, distribution, and taxonomy of these retroelements within crocodilian species and populations.
Nucleotide sequence accession numbers.
GenBank accession numbers for sequences derived in this study are FJ155497 through FJ155561.
Acknowledgments
We thank Travis Glenn, Kent Vliet, Robert Godshalk, Mitch Eaton, and Matthew Shirley who kindly provided us with many of the crocodilian DNA samples included in this investigation. We are also grateful to Porosus Pty. Ltd. for providing the Australian saltwater and Johnston's crocodile samples and to Panya Youngprapakorn of Golden Crocodile Agriculture Co. Ltd., Thailand, and Thanida Haetrakul of Chulalongkorn University, Thailand, for providing us with samples from Siamese crocodile species.
Footnotes
Published ahead of print on 15 July 2009.
REFERENCES
- 1.Abascal, F., R. Zardoya, and D. Posada. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104-2105. [DOI] [PubMed] [Google Scholar]
- 2.Belshaw, R., J. Watson, A. Katzourakis, A. Howe, J. Woolven-Allen, A. Burt, and M. Tristem. 2007. Rate of recombinational deletion among human endogenous retroviruses. J. Virol. 81:9437-9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borysenko, L., V. Stepanets, and A. V. Rynditch. 2008. Molecular characterization of full-length MLV-related endogenous retrovirus ChiRV1 from the chicken, Gallus gallus. Virology 376:199-204. [DOI] [PubMed] [Google Scholar]
- 4.Dolei, A., and H. Perron. 2009. The multiple sclerosis-associated retrovirus and its HERV-W endogenous family: a biological interface between virology, genetics, and immunology in human physiology and disease. J. Neurovirol. 15:4-13. [DOI] [PubMed] [Google Scholar]
- 5.Gatesy, J., and G. Amato. 2008. The rapid accumulation of consistent molecular support for intergeneric crocodylian relationships. Mol. Phylogenet. Evol. 48:1232-1237. [DOI] [PubMed] [Google Scholar]
- 6.Gatesy, J., R. H. Baker, and C. Hayashi. 2004. Inconsistencies in arguments for the supertree approach: supermatrices versus supertrees of Crocodylia. Syst. Biol. 53:342-355. [DOI] [PubMed] [Google Scholar]
- 7.Gifford, R., P. Kabat, J. Martin, C. Lynch, and M. Tristem. 2005. Evolution and distribution of class II-related endogenous retroviruses. J. Virol. 79:6478-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gifford, R., and M. Tristem. 2003. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes 26:291-315. [DOI] [PubMed] [Google Scholar]
- 9.Herniou, E., J. Martin, K. Miller, J. Cook, M. Wilkinson, and M. Tristem. 1998. Retroviral diversity and distribution in vertebrates. J. Virol. 72:5955-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huda, A., N. Polavarapu, I. K. Jordan, and J. F. McDonald. 2008. Endogenous retroviruses of the chicken genome. Biol. Direct. 3:9. doi: 10.1186/1745-6150-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huder, J., J. Boni, J.-P. Hatt, G. Soldati, H. Lutz, and J. Schupbach. 2002. Identification and characterization of two closely related unclassifiable endogenous retroviruses in pythons (Python molurus and Python curtus). J. Virol. 76:7607-7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janke, A., A. Gullberg, S. Hughes, R. K. Aggarwal, and U. Arnason. 2005. Mitogenomic analyses place the gharial (Gavialis gangeticus) on the crocodile tree and provide pre-K/T divergence times for most crocodilians. J. Mol. Evol. 61:620-626. [DOI] [PubMed] [Google Scholar]
- 13.Jern, P., G. O. Sperber, and J. Blomberg. 2005. Use of Endogenous retroviral sequences (ERVs) and structural markers for retroviral phylogenetic inference and taxonomy. Retrovirol. 2:50. doi: 10.1186/1742-4690-2-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kambol, R., P. Kabat, and M. Tristem. 2003. Complete nucleotide sequence of an endogenous retrovirus from the amphibian, Xenopus laevis. Virology 311:1-6. [DOI] [PubMed] [Google Scholar]
- 15.Klymiuk, N. N., M. M. Müller, G. G. Brem, and B. B. Aigner. 2006. Phylogeny, recombination and expression of porcine endogenous retrovirus γ2 nucleotide sequences. J. Gen. Virol. 87:977-986. [DOI] [PubMed] [Google Scholar]
- 16.Martin, J., E. Herniou, J. Cook, R. Waugh O'Neill, and M. Tristem. 1999. Interclass transmission and phyletic host tracking in murine leukaemia virus related retroviruses. J. Virol. 73:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin, J., P. Kabat, E. Herniou, and M. Tristem. 2002. Characterization and complete nucleotide sequence of an unusual reptilian retrovirus recovered from the order Crocodilia. J. Virol. 76:4651-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martina, Y., K. T. Marcucci, S. Cherqui, A. Szabo, T. Drysdale, U. Srinivisan, C. A. Wilson, C. Patience, and D. R. Salomon. 2006. Mice transgenic for a human porcine endogenous retrovirus receptor are susceptible to productive viral infection. J. Virol. 80:3135-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roos, J., R. Aggarwal, and A. Janke. 2007. Extended mitogenomic phylogenetic analyses yield new insight into crocodylian evolution and their survival of the Cretaceous-Tertiary boundary. Mol. Phylogenet. Evol. 45:663-673. [DOI] [PubMed] [Google Scholar]
- 20.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Phylogenet. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 21.Tarlinton, R. E., J. Meers, and P. R. Young. 2008. Biology and evolution of the endogenous koala retrovirus. Cell. Mol. Life Sci. 65:3413-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence-weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tristem, M. 1996. Amplification of divergent retroelements by PCR. BioTechniques 20:608-612. [DOI] [PubMed] [Google Scholar]
- 24.Tristem, M., T. Myles, and F. Hill. 1995. A highly divergent retroviral sequence in the tuatara (Sphenodon). Virology 210:206-211. [DOI] [PubMed] [Google Scholar]