Abstract
Homologs of the essential large tegument protein pUL36 of herpes simplex virus 1 are conserved throughout the Herpesviridae, complex with pUL37, and form part of the capsid-associated “inner” tegument. pUL36 is crucial for transport of the incoming capsid to and docking at the nuclear pore early after infection as well as for virion maturation in the cytoplasm. Its extreme C terminus is essential for pUL36 function interacting with pUL25 on nucleocapsids to start tegumentation (K. Coller, J. Lee, A. Ueda, and G. Smith, J. Virol. 81:11790-11797, 2007). However, controversy exists about the cellular compartment in which pUL36 is added to the nascent virus particle. We generated monospecific rabbit antisera against four different regions spanning most of pUL36 of the alphaherpesvirus pseudorabies virus (PrV). By immunofluorescence and immunoelectron microscopy, we then analyzed the intracellular location of pUL36 after transient expression and during PrV infection. While reactivities of all four sera were comparable, none of them showed specific intranuclear staining during PrV infection. In immunoelectron microscopy, neither of the sera stained primary enveloped virions in the perinuclear cleft, whereas extracellular mature virus particles were extensively labeled. However, transient expression of pUL36 alone resulted in partial localization to the nucleus, presumably mediated by nuclear localization signals (NLS) whose functionality was demonstrated by fusion of the putative NLS to green fluorescent protein (GFP) and GFP-tagged pUL25. Since PrV pUL36 can enter the nucleus when expressed in isolation, the NLS may be masked during infection. Thus, our studies show that during PrV infection pUL36 is not detectable in the nucleus or on primary enveloped virions, correlating with the notion that the tegument of mature virus particles, including pUL36, is acquired in the cytosol.
The herpesvirus virion is composed of an icosahedral nucleocapsid containing the viral genome, an envelope of cellular origin with inserted viral (glyco)proteins, and a tegument which links nucleocapsid and envelope comparable to the matrix of RNA viruses. The herpesvirus tegument contains a multitude of viral and cellular proteins (reviewed in references 45 and 46). Tegument proteins execute various regulatory and structural functions, including activation of viral gene expression (2), modulation of the host cell for virus replication (26, 51, 55), and mediation of posttranslational modification of proteins (10, 27, 50). Numerous interactions have been identified among tegument proteins, between tegument and capsid proteins, and between tegument and envelope proteins (7, 14, 16, 18, 33, 36, 42, 53, 58-61).
The largest tegument proteins found in the herpesviruses are homologs of pUL36 of herpes simplex virus type 1 (HSV-1). Pseudorabies virus (PrV) pUL36 consists of 3,084 amino acids (aa) with a molecular mass of 324 kDa (33). PrV and HSV-1 pUL36 are essential for viral replication (13, 15). In their absence, nonenveloped nucleocapsids accumulate in the cytoplasm. Whereas in several studies nuclear stages like cleavage and packaging of the viral DNA as well as nuclear egress were not found affected (13, 15), another study indicated an effect of pUL36 deletion on PrV nuclear egress (41).
pUL36 homologs complex with another tegument protein, pUL37, as has been shown for HSV-1 (59), PrV (15, 33), and human cytomegalovirus (3, 23), and the interacting region on pUL36 has been delineated for PrV (33) and identified at the amino acid level for HSV-1 (47). Deletion of the pUL37 interaction domain from PrV pUL36 impedes virion formation in the cytosol but does not block it completely, yielding a phenotype similar to that of a pUL37 deletion mutant (31). This indicates an important but nonessential role for pUL37 and the pUL37 interaction domain in pUL36 in virion formation (15). In contrast, absence of pUL37 completely blocks virion formation in HSV-1 (11, 38).
pUL36 is stably attached to the nucleocapsid (39, 43, 56), remains associated with incoming particles during transport along microtubules to the nuclear pore (21, 40, 52), and is required for intracellular nucleocapsid transport during egress (41). In contrast, absence of pUL37 delays nuclear translocation of incoming PrV nucleocapsids but does not abolish it (35). HSV-1 pUL36 is involved not only in transport but also in docking of nucleocapsids to the nuclear pore (9), and proteolytic cleavage of pUL36 appears to be necessary for release of HSV-1 DNA into the nucleus (24).
Immunoelectron microscopical studies of PrV-infected cells showed that pUL36 is added to nucleocapsids prior to the addition of pUL37 (33). Since neither pUL36 nor pUL37 was detected on primary enveloped PrV virions, it was concluded that acquisition of tegument takes place in the cytoplasm (20). However, conflicting data exist whether pUL36 is present in the nucleus, and whether it is already added onto the capsids in this cellular compartment. Indirect immunofluorescence, immunoelectron microscopy and mass spectrometry of intranuclear capsids yielded discrepant results. By immunofluorescence HSV-1 pUL36 was detected both in the cytoplasm and in the nucleus (1, 42, 48). However, whereas one study detected the protein on nuclear C-capsids by Western blotting (6), it was not found by cryo-electron microscopy and mass spectrometry (57). In contrast, the C terminus of PrV pUL36 was suggested to direct pUL36 to capsid assemblons in the nucleus (37) by binding to capsid-associated pUL25 (8), although pUL36 could not be detected in the nucleus during PrV infection (33). These differing results in HSV-1 and between HSV-1 and PrV might be due to the fact that pUL36 could be processed during the replication cycle and that the resulting subdomains may exhibit selective localization patterns (24, 28).
Amino acid sequence analyses of HSV-1 and PrV pUL36 revealed several putative nuclear localization signals (NLS) (1, 4, 5, 49). HSV-1 pUL36 contains four of these NLS motifs (49). Functionality in nuclear localization of a reporter protein was shown for the NLS motif at aa 425 (1). This motif is highly conserved in herpesvirus pUL36 homologs pointing to an important function (1). Besides this conserved NLS (designated in this report as NLS1), two other NLS motifs are predicted in PrV pUL36. One is located adjacent to NLS1 (aa 288 to 296) at aa 315 to 321 (NLS2), and a third putative NLS motif is present in the C-terminal half of the protein (aa 1679 to 1682; NLS3) (4). Whereas this may be indicative for a role for pUL36 inside the nucleus, NLS motifs might also be involved in transport to the nucleus along microtubules (54) and docking at the nuclear pore complex (49).
The discrepancy in pUL36 localization and the putative presence of pUL36 cleavage products with specialized functions and localization prompted us to generate monospecific antisera covering the major part of PrV pUL36 and to study localization of PrV pUL36 by immunofluorescence during viral replication and after transient transfection and by immunoelectron microscopy of infected cells.
MATERIALS AND METHODS
Viruses and cells.
PrV wild-type strain Kaplan (PrV-Ka) (25) was used in all experiments. It was propagated on rabbit kidney cells (RK13) in minimum essential medium supplemented with 10% fetal calf serum. PrV-ΔUL36F lacking almost the complete UL36 coding region was grown in complementing RK13-UL36 cells expressing full-length pUL36 (15) or in cells expressing the truncated, yet fully functional, pUL36Δ2087-2795 (4).
Plasmid constructs.
For generation of NLS-green fluorescent protein (GFP) reporter plasmids, specific primers were used for amplification of fragments containing PrV nucleotides 41256 to 41475 (NLS1/2, primers sb_NLS1_FOR and sb_NLS1_REV1) (Table 1), corresponding to aa 281 to 353 (accession no. BK001744) (30); nucleotides 41384 to 41475 (aa 281 to 310; NLS1, primers sb_NLS1_FOR and sb_NLS1_REV3); nucleotides 41256 to 41376 (aa 314 to 353; NLS2, primers sb_NLS2_FOR and sb_NLS1_REV1); and nucleotides 37043 to 37325 (aa 1664 to 1757; NLS3, primers sb_NLS3_FOR and sb_NLS3_REV) (Table 1). Purified PCR fragments were digested with EcoRI and BamHI, for which restriction sites had been introduced with the primers (Table 1), and ligated into the accordingly cleaved vector pEGFP-N1 (Clontech). Correct cloning was verified by sequencing.
TABLE 1.
Primers used for amplification of putative NLS
| Oligonucleotide | Sequencea |
|---|---|
| sb_NLS1_FOR | 5′-CACAGAATTCATGCCGCCGCGGGTCCAGAAGC-3′ |
| sb_NLS1_REV1 | 5′-CACAGGATCCTCCTCGAGCTCGTCGGCGTGG-3′ |
| sb_NLS1_REV3 | 5′-CACAGGATCCACCGTGTGCGTGCTGCTCAGGTC-3′ |
| sb_NLS2_FOR | 5′-CACAGAATTCATGCGGCCGTCGCGCAAGTTCC-3′ |
| sb_NLS3_REV | 5′-CACAGGATCCATGTACTCGTGCTGCCCGGCCAG-3′ |
| sb_NLS3_FOR | 5′-CACAGAATTCATGGAGAAGGCGCGCGACACGGG-3′ |
Restriction sites for convenient cloning are underlined, and introduced start codons are highlighted in boldface.
Since passive nuclear localization of the ca. 30-kDa NLS-enhanced GFP (EGFP) fusion proteins could not be excluded, additional fusions with the PrV UL25 coding region were created. To this end, pcDNA-UL25 (32) was cleaved with BamHI and XbaI, and the 1.6-kb UL25 insert was cloned into BsrGI/XbaI-digested pEGFP-N1, pNLS1/2-EGFP, and pNLS1-EGFP after Klenow fill-in of the noncompatible ends, giving rise to plasmids pEGFP-UL25, pNLS1/2-EGFP-UL25, and pNLS1-EGFP-UL25, respectively.
For deletion of a fragment comprising the two predicted N-terminal NLS motifs (4), pcDNA-UL36 was digested with SphI (nucleotide 41449) and NcoI (nucleotide 41338) (Fig. 1). After treatment with Klenow and T4 polymerases, the plasmid was religated resulting in pUL36Δ290-326, lacking codons 290 to 326. Sequencing using primer sb_NLS1_REV3 showed correct in-frame cloning.
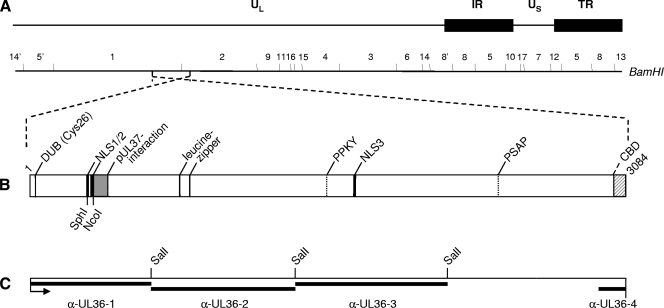
FIG. 1.
Schematic overview of PrV pUL36. (A) Diagram of the PrV genome divided into unique long (UL) and unique short (US) regions by internal repeats (IR) and terminal repeats (TR). The positions of BamHI restriction sites are indicated. (B) Confirmed and putative functional domains in PrV pUL36 are indicated, such as DUB (Cys26), the active-site cysteine of the deubiquitinating activity (28); the pUL37 interacting domain (15, 31); the NLS; the leucine zipper; putative late domain motifs (PPKY and PSAP) (4); and CBD, the capsid binding domain (8). SphI and NcoI sites used for deletion of the two NLS motifs are also indicated. (C) Regions of pUL36 contained in GST-expression proteins used for immunization are marked by black bars above the designations of the resulting antisera. α-, anti-. The SalI restriction sites used for cloning of pGEX-UL36-2 and pGEX-UL36-3 are indicated.
Generation of monospecific antisera.
Monospecific anti-UL36-1 (33) and anti-UL36-2 (4) antisera (Fig. 1) directed against two regions from the N-terminal half of pUL36 have been described previously. For this study, two additional antisera were generated covering parts of the C-terminal region of PrV pUL36. To obtain anti-UL36-3 serum, a SalI-fragment containing codons 1371 to 2158 (Fig. 1) was cloned into pGEX-4T-3 (Amersham Biosciences, Germany), and correct cloning was verified by sequencing. To generate an antiserum directed against the very C terminus of PrV pUL36, several glutathione S-transferase (GST) fusion proteins were tested but only a fragment derived from yeast two-hybrid studies (unpublished data) comprising codons 2946 to 3074 resulted in efficient expression in Escherichia coli after cloning into pGEX-4T-1 (data not shown). The GST fusion proteins were purified and used to immunize rabbits as described previously (33).
Fluorescence analyses.
To analyze intracellular pUL36 location during viral replication, cells were infected with PrV-Ka or PrV-ΔUL36F at a multiplicity of infection (MOI) of 5 and incubated on ice for 1 h. Cells were then fixed either immediately (t = 0 h) or after replacement of the inoculum by prewarmed medium for 1 h (t = 1 h); inactivation of nonpenetrated virus by low-pH treatment (44); and incubation at 37°C for additional 2, 3, 4, 5, 6, 12, and 24 h. Fixation was performed with ice-cold acetone for 20 min at −20°C (for anti-UL36-2, -3, and -4 and anti-UL31) or with 3% paraformaldehyde for 20 min, followed by a 10-min incubation with 3% paraformaldehyde plus 0.3% Triton X-100 (for anti-UL36-1, anti-UL37, and anti-UL48 antisera, which lose their reactivity on acetone-fixed material). Coverslips were then washed with phosphate-buffered saline (PBS), and incubated for 1 h at room temperature with anti-UL36-1 (dilution, 1:1,000), anti-UL36-2 (dilution, 1:1,500), anti-UL36-3 (dilution 1:2,000), or anti-UL36-4 (dilution, 1:2,000) serum, followed by Alexa Fluor 488-conjugated goat anti-rabbit antibodies (Molecular Probes, Invitrogen). For control, parallel coverslips were incubated with anti-UL31 (1:500) (17), anti-UL37 (1:500) (31), or anti-UL48 (1:500) (19) sera and Alexa Fluor 488-conjugated goat anti-rabbit antibodies as secondary antibody (Molecular Probes, Invitrogen) for 1 h at room temperature. For nuclear staining, coverslips were overlaid with mounting medium (a 9:1 mixture of glycerol and PBS containing 25 mg/ml 1,4-diazabicyclooctane) containing 1 μg/ml propidium iodide or were incubated with ToPro3 (1:2,000; Molecular Probes, Invitrogen) for 1 h at room temperature. Images were documented by a confocal laser-scanning microscope (LSM510; Carl Zeiss, Ltd., Oberkochen, Germany).
To study the intracellular localization of the NLS-GFP reporter proteins, RK13 cells were transfected with plasmids pNLS1/2-EGFP, pNLS1-EGFP, pNLS2-EGFP, pNLS3-EGFP, and pEGFP-N1 or pNLS1/2-EGFP-UL25, pNLS1-EGFP-UL25, and pEGFP-UL25. Coverslips were fixed 1 day after transfection with 3% paraformaldehyde for 20 min, followed by a 10-min incubation with 3% paraformaldehyde plus 0.3% Triton X-100. Coverslips were incubated with ToPro3 (1:2,000; Molecular Probes, Invitrogen) for 1 h at room temperature and overlaid with mounting medium. Images were documented by confocal laser-scanning microscopy.
For indirect immunofluorescence after transient expression, RK13 cells were transfected by calcium phosphate coprecipitation with pUL36Δ290-326, lacking both putative N-terminal NLS motifs, or pcDNA-UL36 (4), containing the full-length UL36. In addition, viral DNA of PrV-ΔUL36F was cotransfected with pcDNA-UL36 into RK13 cells. Cells were fixed 1 day after transfection with ice-cold acetone for 20 min at −20°C. pUL36 was detected as described above. Cells (co)transfected with viral DNA were identified by an anti-gC monoclonal antibody (29) and Alexa-Fluor 555-conjugated goat anti-mouse secondary antibody. Representative images were documented by confocal laser-scanning microscopy.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis.
For virus purification, cells were infected at an MOI of 0.1 with PrV-Ka and incubated until a complete cytopathic effect developed. The remaining intact cells were lysed by freezing (−70°C) and thawing (37°C), cellular debris was removed by low-speed centrifugation, and the virus-containing supernatant was cleared by centrifugation through a 35% sucrose cushion. The pellet was resuspended in PBS and layered onto a discontinuous gradient of 30, 40, and 50% sucrose. Virions which accumulated at the boundary between 40 and 50% sucrose were harvested by aspiration, pelleted, and resuspended in PBS. Virion lysates were separated on 6% or 10% polyacrylamide gels containing 1% sodium dodecyl sulfate, electrotransferred onto nitrocellulose membranes, and incubated with anti-UL36-1 (1:20,000) (33), anti-UL36-2 (1:60,000) (4), anti-UL36-3 (1:75,000), and anti-UL36-4 (1:100,000) sera. For control, a parallel blot was probed with antiserum against the tegument protein pUL37 (1:100,000) (31). Binding of peroxidase-conjugated secondary antibody (Dianova, Hamburg, Germany) was detected by chemiluminescence (Super Signal; Pierce, Bonn, Germany) recorded on X-ray film.
Electron microscopy.
RK13 cells were infected with PrV-Ka at an MOI of 1 and incubated for 14 h at 37°C. Fixation and embedding were done essentially as described previously (21). For intracellular labeling of viral proteins, cells were fixed with 0.5% glutaraldehyde in PBS (pH 7.2) for 30 min, embedded in LMP agarose (Biozym), and postfixed with 0.5% glutaraldehyde for another 30 min. Thereafter, samples were blocked with 0.5 M NH4Cl in PBS for 60 min, washed in PBS, stained in 0.5% aqueous uranyl acetate for 15 min, dehydrated in ethanol under progressive lowering of temperature, embedded in the acrylic resin Lowicryl K4M (Lowi, Waldkraiburg, Germany) at −35°C, and polymerized by UV light at a wavelength of 360 nm.
The postembedding labeling of ultrathin sections was performed after blocking of surfaces with 1% cold water fish gelatin, 0.02 M glycine, and 1% bovine serum albumin fraction V (Sigma, Deisenhofen, Germany) in PBS, by either overnight incubation at 4°C or 2 h of incubation at room temperature with anti-pUL36 or anti-pUL31 antibodies diluted in PBS-bovine serum albumin. Diluted gold-tagged goat anti-species antibodies or protein A gold (GAR10 or PAG10; British BioCell, Int., Cambridge, United Kingdom) was added for 60 min at room temperature, and excess antibodies were removed by washing. Specificity of the reaction was controlled on uninfected and infected RK13 cells, by using gold conjugate without primary antibody and by using monospecific anti-pUL31 serum (17). Labeled Lowicryl sections, counterstained with uranyl acetate and lead salts, were examined with an electron microscope (Tecnai 12 electron microscope; Philips, Eindhoven, The Netherlands).
RESULTS
Characterization of anti-pUL36 sera.
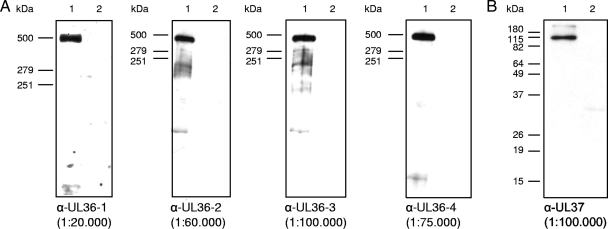
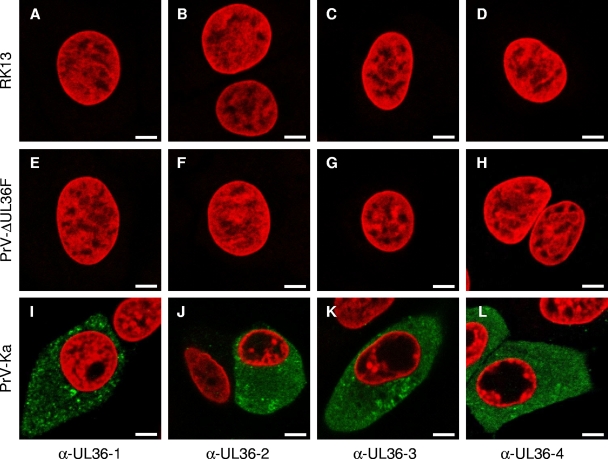
To study the intracellular localization of PrV pUL36 in detail and to reveal presence of putative cleavage products exhibiting different localization patterns, four antisera covering most of the open reading frame were generated. In addition to the previously described antibodies directed against portions from the N-terminal half of PrV pUL36 (4, 33), two additional antisera, anti-UL36-3 and anti-UL36-4, were raised against regions of the C-terminal part (Fig. 1C). The specificity of all sera was tested by Western blotting (Fig. 2) and immunofluorescence (Fig. 3) analyses. In Western blot analyses of lysates from purified PrV virions, the anti-UL36-1, anti-UL36-2, anti-UL36-3, and anti-UL36-4 sera recognized the ∼320-kDa pUL36 (Fig. 2A, lanes 1). Faint smaller bands were sometimes visible with anti-UL36-2 and anti-UL36-3 sera, but varied between different virion preparations (data not shown). The sera did not react with lysates of noninfected RK13 cells (Fig. 2, lanes 2). As a control, a parallel blot was probed with anti-UL37 (Fig. 2B) (31). Moreover, in indirect immunofluorescence analyses, all anti-UL36 sera reacted comparably with PrV-Ka-infected cells (Fig. 3I to L), but not with mock-infected cells (Fig. 3A to D) or cells infected with PrV-ΔUL36F (Fig. 3E to H) demonstrating specificity of the pUL36 antisera.
FIG. 2.
Western blot analyses of purified PrV virions. Purified virions of PrV-Ka (lanes 1) and lysates of noninfected RK13 cells (lanes 2) were separated on 6% (A) or 10% (B) polyacrylamide gels. α-, anti-. After electrotransfer onto nitrocellulose membranes, parallel blots were incubated with the indicated dilutions of the four antisera directed against PrV pUL36 (A). For a control, an antiserum against pUL37 was included (B). Locations of molecular marker proteins (high molecular mass marker; Invitrogen) are indicated to the left of each panel.
FIG. 3.
Indirect immunofluorescence of infected cells. Indirect immunofluorescence of RK13 cells, either infected with PrV-Ka or PrV-ΔUL36F for 24 h or mock infected, with each of the four anti-pUL36 antisera is shown in green. Chromatin was counterstained with propidium iodide or ToPro-3 (shown in red). α-, anti-. Scale bars represent 5 μm.
Intracellular localization of PrV pUL36.
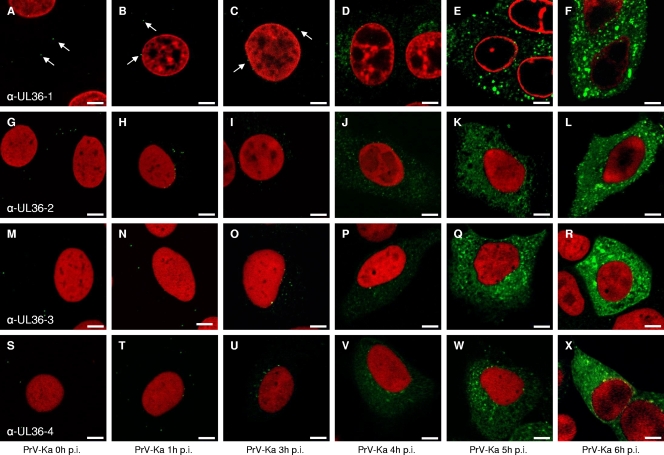
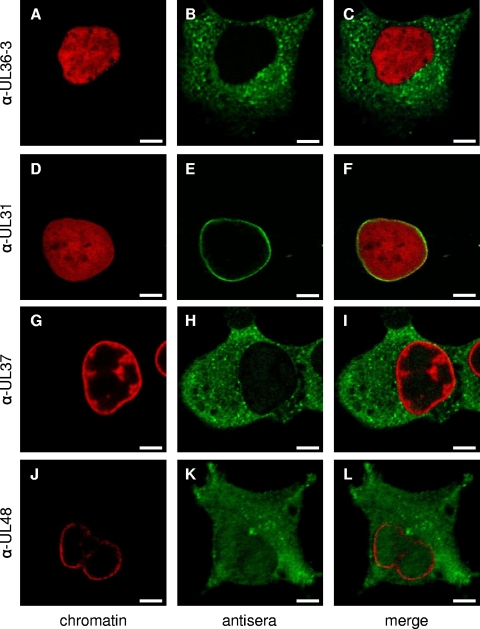
Since there is an ongoing controversy as to the intracellular localization of herpesvirus pUL36 proteins, we used our panel of monospecific anti-pUL36 antisera to investigate intracellular localization of pUL36 during PrV replication (Fig. 4). To this end, cells were infected with PrV-Ka for 1 h on ice to allow virus attachment and either fixed immediately (t = 0) or 1, 3, 4, 5, 6, 12, or 24 h after temperature shift. At t = 0, green fluorescent dots were detected at a distance from the nucleus lining the plasma membrane (as detected by bright-field microscopy), presumably representing input virus particles (Fig. 4A, G, M, and S). After 1 h at 37°C, punctate fluorescence signals were found close to the nucleus, indicating docking of incoming nucleocapsids decorated with pUL36 at the nuclear pore, as has previously been shown also by electron microscopy (Fig. 4B, H, N, and T) (21). Newly expressed pUL36 first became visible in a perinuclear pattern 4 h after infection (Fig. 4D, J, P, and V), subsequently increasing in intensity. At 12 or 24 h after infection, only a few cells remained attached to the coverslip, but pUL36-specific fluorescence was still detectable in the cytoplasm only (data not shown). The reaction patterns of all four tested antisera were comparable. In particular, none of them showed intranuclear labeling at any of the time points analyzed. This is also evident in Fig. 5B, clearly showing that the nucleus is free of specific staining. For controls, pUL31, pUL48, and pUL37, which are known to be associated with the nucleus, were detected in this cellular compartment (Fig. 5D to L). Antiserum directed against the primary tegument protein pUL31 resulted in the typical nuclear rim staining (Fig. 5D to F) (17), whereas antiserum against the pUL48 protein showed a homogeneous cytoplasmic and nuclear staining correlating with its function in immediate-early gene activation in the nucleus and during secondary envelopment (Fig. 5J to L) (19). Antiserum against pUL37, the complex partner of pUL36, showed a predominantly cytosolic but also weak nuclear staining, as described previously (Fig. 5G to I) (31). In summary, none of our anti-pUL36 sera resulted in detection of pUL36 in the nucleus during PrV replication.
FIG. 4.
Intracellular localization of pUL36 during PrV infection. RK13 cells were infected synchronously with PrV-Ka at an MOI of 5 and fixed 0, 1, 3, 4, 5, and 6 h postinfection (p.i.). Immunofluorescence was performed by confocal laser-scanning microscopy using anti-UL36-1 (top row), anti-UL36-2 (second row), anti-UL36-3 (third row), and anti-UL36-4 (bottom row) antisera. Chromatin was counterstained with propidium iodide or ToPro-3 (shown in red). Arrows point to fluorescent dots most likely representing virus particles. α-, anti-. Scale bars represent 5 μm.
FIG. 5.
Indirect immunofluorescence of PrV-infected cells with antisera directed against pUL36, pUL31, pUL37, and pUL48. Cells were infected synchronously with PrV-Ka and fixed after 5 h. Immunofluorescence analysis was performed by confocal laser-scanning microscopy using anti-UL36-3 (top row), anti-UL31 (second row), anti-UL37 (third row), and anti-UL48 (bottom row) antisera and Alexa 488-conjugated secondary antibodies (green). Chromatin was counterstained with ToPro-3 or propidium iodide (shown in red). α-, anti-. Scale bars represent 5 μm.
Nuclear targeting properties of predicted NLS motifs in pUL36.
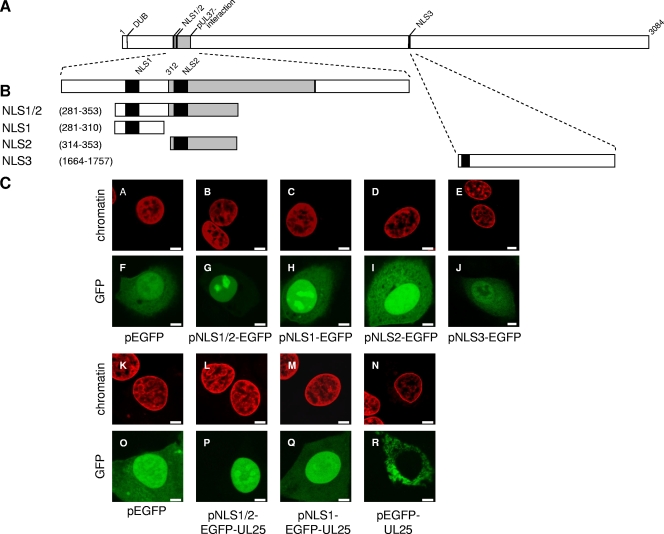
Three putative NLS in pUL36 homologs have been predicted which are conserved in sequence and locations among the Herpesviridae (1, 4). Two of them are located closely adjacent in the N-terminal part of pUL36 (NLS1 and NLS2) (Fig. 1), whereas a third resides in the C-terminal half (NLS3) (Fig. 1). The homologous NLS1 has been shown to be functional in HSV-1 pUL36 (1). Although pUL36 has not been detected in the nucleus during PrV replication (see above), we analyzed functionality of the putative NLS in PrV pUL36 by using GFP as a reporter fused to the predicted NLS sequences. After transient expression in RK13 cells (Fig. 6), NLS1 and NLS2 were able to direct GFP to the nucleus, although significant cytoplasmic fluorescence remained (Fig. 6H and I). The simultaneous presence of both NLS1 and NLS2, however, led to a complete nuclear localization of GFP (Fig. 6G). In contrast, no significant alteration in intracellular GFP distribution was observed after transfection of pNLS3-EGFP (Fig. 6J) compared to pEGFP-N1 (Fig. 6F). Since the molecular mass of these fusion proteins is only ca. 30 kDa, their partial localization to the nucleus by passive diffusion could not be excluded. Thus, to confirm functionality of NLS1 and NLS1/2, reporter constructs were extended by addition of the ∼57-kDa PrV UL25 open reading frame. The resulting NLS-EGFP-UL25 constructs exhibit molecular masses of between 88 and 94 kDa and are excluded from passive diffusion through the nuclear pore. While neither pUL25 alone (data not shown) nor EGFP-UL25 (Fig. 6R) was detected in the nucleus, NLS1/2-EGFP-UL25 (Fig. 6P) showed an exclusively nuclear staining pattern. Fusion to NLS1 alone (Fig. 6Q) paralleled the results of the NLS-EGFP constructs (Fig. 6A to J) resulting in cytoplasmic as well as nuclear localization. Thus, PrV pUL36 carries at least two functional NLS, which, however, do not result in detectable nuclear translocation of pUL36 during infection.
FIG. 6.
Nuclear targeting properties of predicted NLS motifs. (A) Schematic overview of PrV pUL36. Panel B shows enlarged views of the N-terminal region containing the putative NLS motifs NLS1 (aa 288 to 296) and NLS2 (aa 315 to 321) and the C-terminal segment containing NLS3 (aa 1679 to 1682). NLS are shown as black bars, and the pUL37-interacting domain is shaded in gray. Fragments of PrV pUL36 containing putative NLS motifs were fused in frame to the 5′ end of the EGFP- or EGFP-UL25 genes (not shown). Numbers indicate amino acid positions in PrV pUL36. DUB denotes the active-site cysteine of the deubiquitinating activity (28). (C) Intracellular localization of NLS-EGFP or NLS-EGFP-UL25 fusion proteins in RK13 cells. GFP autofluorescence (green) and ToPro-3-stained chromatin (red) were recorded in a confocal laser-scanning microscope. Scale bars represent 5 μm.
Intracellular distribution of pUL36 after transient expression.
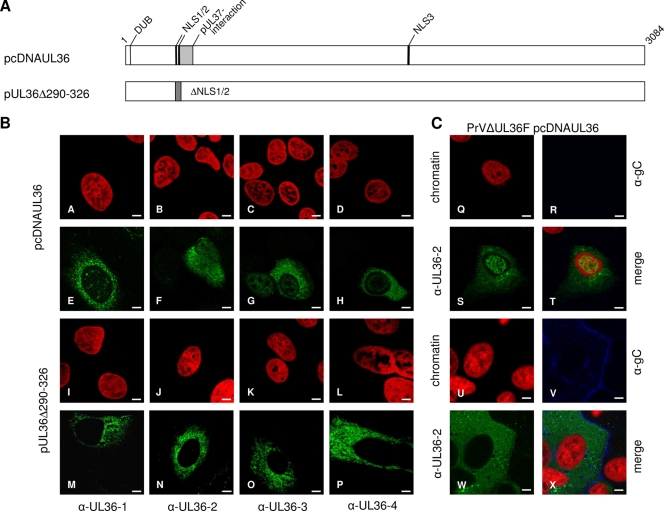
To analyze whether pUL36 is targeted to the nucleus in the absence of other viral proteins, RK13 cells were transfected with pcDNA-UL36 as well as pUL36Δ290-326, lacking both functional NLS. As shown in Fig. 7B, panels A to H, transiently expressed full-length pUL36 showed a predominant cytoplasmic location but was also detected in the nucleus. Deletion of the two functional NLS resulted in abrogation of nuclear localization and exclusive cytoplasmic staining (Fig. 7B, panels I to P). To test whether viral proteins expressed during replication inhibit nuclear localization of pUL36, DNA of PrV-ΔUL36F was cotransfected with pcDNA-UL36 into RK13 cells. As evident in Fig. 7C, panels U to X, in cells cotransfected with PrV-ΔUL36F, which can be identified by expression of gC, pUL36 was found exclusively in the cytoplasm, whereas cells in the same assay which expressed only pUL36 exhibited also nuclear staining (Fig. 7C, panels Q to T). These data indicate that pUL36 is translocated into the nucleus in the absence of other viral proteins but retained in the cytoplasmic compartment during infection.
FIG. 7.
(A) Schematic overview of PrV pUL36 full-length protein (for details see Fig. 1). pUL36Δ290-326 contains a deletion of NLS1 and 2. For immunofluorescence analyses, RK13 cells were transfected with expression plasmids for wild-type pUL36 or pUL36Δ290-326 (B) as well as cotransfected with PrV-ΔUL36F and pcDNA-UL36 (C). PrV-ΔUL36F-transfected cells were detected by indirect immunofluorescence with an anti-gC monoclonal antibody (blue). α-, anti-. Scale bars represent 5 μm.
Ultrastructural analysis.
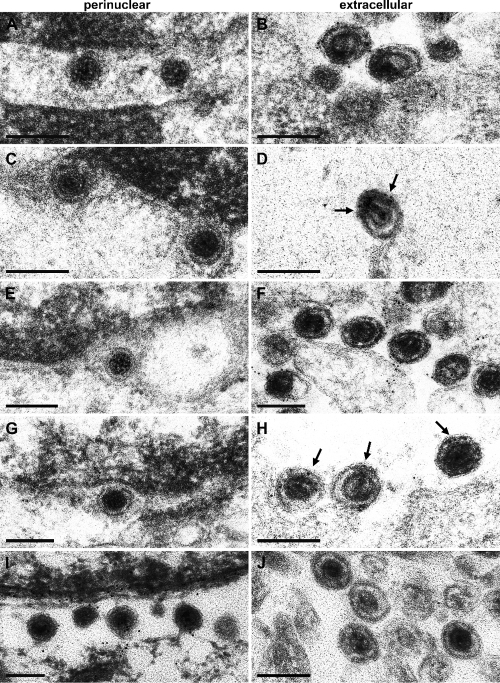
One prominent question in herpesvirus maturation is whether nucleocapsids start acquisition of their final tegument already in the nucleus or whether tegumentation does not commence prior to the arrival of nucleocapsids in the cytosol. The first would imply nuclear egress of partially tegumented nucleocapsids with the possibility that these tegument proteins might execute important functions during nuclear egress. Since pUL36 is thought to represent the innermost layer of the tegument, its presence or absence in primary enveloped virions would give a relevant indication. To assay for the presence of pUL36 on virus particles, RK13 cells were infected with PrV-Ka at an MOI of 1 and analyzed 14 h after infection by immunoelectron microscopy. Ultrathin sections were labeled with the anti-UL36-1, anti-UL36-2, anti-UL36-3, anti-UL36-4, and anti-UL31 sera. Anti-UL36-1, anti-UL36-2, anti-UL36-3, and anti-UL36-4 sera labeled extracellular virus particles intensely (Fig. 8B, D, F, and H), whereas virions in the perinuclear space were not labeled by either serum (Fig. 8A, C, E, and G). In contrast, antiserum against the primary tegument protein pUL31 labeled only virions located in the perinuclear space but not mature particles (Fig. 8I and J), as has been described previously (17).
FIG. 8.
Immunoelectron microscopy. RK13 cells were infected with PrV-Ka at an MOI of 1 and analyzed 14 h after infection. Ultrathin sections were immunogold labeled with anti-UL36-1 (A and B), anti-UL36-2 (C and D), anti-UL36-3 (E and F), anti-UL36-4 (G and H), and anti-UL31 sera (I and J) (17). Whereas no pUL36-specific label was detected on primary enveloped virions in the perinuclear space with any of the antisera (A, C, E, and G), extracellular wild-type virions were heavily labeled, demonstrating the potency of the antisera (B, D, F, and H). For better visibility, arrows in panels D and H highlight the presence of gold particles. For a control, primary enveloped virions in the perinuclear space but no extracellular virions were labeled with antiserum against pUL31. Bars represent 200 nm.
DISCUSSION
In this study, we used a set of four monospecific antisera directed against four different regions of the PrV pUL36 protein to analyze its intracellular distribution during infection and transient expression. The salient findings are as follows. (i) Neither of the anti-pUL36 sera detected pUL36 in the nucleus during PrV infection. (ii) Two putative NLS motifs located in the N-terminal part of PrV pUL36 are functional when fused to EGFP or EGFP-UL25 as a reporter, and a third predicted NLS in the C terminus is not. (iii) After transient expression, PrV pUL36 is detectable also in the nucleus, but nuclear localization is prevented by coexpression of viral proteins. (iv) Deletion of the two functional NLS abrogates nuclear localization of transiently expressed pUL36. (v) Primary enveloped virions in the perinuclear cleft were not labeled by either of the pUL36-specific sera. Thus, pUL36 of PrV has an intrinsic property to enter the nucleus, which, however, does not occur during PrV infection, nor is it incorporated into primary virions.
The studies on intracellular localization of pUL36 published to date rely on several antibodies directed against different fragments of this huge protein revealing different intracellular localizations. Whereas in HSV-1-infected cells pUL36 was detected in the cytoplasm as well as in the nucleus (1, 11, 42, 48), in PrV-infected cells pUL36 was found only in the cytoplasm (33). A possible cleavage of pUL36 during replication (24) could imply specific functions for different parts of the protein. To assay intracellular localization of PrV pUL36 in more detail, we generated monospecific antisera directed against four different regions of pUL36 covering the complete N-terminal half (anti-UL36-1 and anti-UL36-2) as well as substantial portions of the C-terminal part (anti-UL36-3), including the highly conserved and essential extreme C terminus (anti-UL36-4). Using these sera in immunofluorescence analyses of PrV-infected cells, only cytoplasmic fluorescence was observed at different time points after infection from early to late stages. As expected, during early stages the antisera showed a punctate label first at the plasma membrane and then associated with the nuclear rim correlating with attachment and subsequent translocation to the nucleus of pUL36-decorated nucleocapsids (21, 40). During later time points, newly synthesized pUL36 accumulated in the cytoplasm, in part in strongly fluorescent speckles which may represent sites of virus maturation.
The deduced 3,084-aa sequence of PrV pUL36 predicts the presence of three putative NLS, two in the N-terminal half and one in the C-terminal half. The N-terminal NLS1 has been shown to be conserved in herpesvirus pUL36 proteins, and in HSV-1 pUL36, was able to translocate the reporter protein β-galactosidase to the nucleus (1). We used EGFP and EGFP-UL25 constructs for analysis. Fusion of either NLS1 or NLS2 to EGFP-UL25 and/or EGFP resulted in an accumulation of the fusion proteins in the nucleus, although a fraction remained cytoplasmic. In contrast, fusion of both NLS1 and NLS2 to EGFP or EGFP-UL25 resulted in an exclusive nuclear staining pattern demonstrating functionality of these motifs. NLS3 located in the C-terminal half of pUL36 did not enhance autofluorescence in the nucleus compared to EGFP alone. Thus, PrV pUL36 contains two functional NLS which, however, do not result in nuclear translocation of the protein during virus infection.
We hypothesized that during infection other viral proteins may preclude nuclear translocation of pUL36. In support of this assumption, cells transfected with a eukaryotic expression plasmid expressing only pUL36 showed cytoplasmic as well as nuclear localization of pUL36. However, after cotransfection with DNA of a pUL36 deletion mutant, nuclear localization of coexpressed pUL36 was also not observed. Thus, without the context of virus infection PrV pUL36 has the ability to enter the nucleus, most likely mediated by the two N-terminal NLS.
Inactivation or deletion of the functional N-terminal NLS resulted in abrogation of pUL36 function in HSV-1 (1) and PrV (unpublished data). Although indirect effects of these mutations on pUL36 folding cannot be excluded, the data could indicate that the presence of a functional NLS is indeed required for pUL36 function during herpesvirus replication. The apparent discrepancy may be resolved considering a role for pUL36 associated with NLS function but not necessarily translocation into the nucleus. Herpesvirus pUL36 homologs have been implicated in transport of incoming nucleocapsids to and docking at the nuclear pore (9, 21, 40, 52). Both events may be mediated by the interaction of a nucleocapsid-associated NLS with microtubule motor protein adaptors and the nuclear translocation machinery present at the nuclear pore, which involve importin-β. Importin-β was found necessary for docking of nucleocapsids to nuclear pores (49), and importin-β has also been suggested as adaptor protein for movement along microtubules (22) linking both processes. Although the nucleocapsid is too big to be translocated through the nuclear pore, this interaction may trigger subsequent events resulting in opening of the nucleocapsid at the vertex position adjacent to the nuclear pore, followed by genome release into the nucleus. In HSV-1, this process has been shown to require proteolytic processing of pUL36 (24), demonstrating a central role for pUL36 in this step. This correlates with previous observations using the temperature-sensitive HSV-1 mutant tsB7, which exhibits a defect in the release of the viral genome into the nucleus at the nonpermissive temperature (2, 34).
It is unclear which mechanism precludes PrV pUL36 translocation to the nucleus at both early and late times after infection. Presumably, it is retained in the cytoplasm by interaction with other viral proteins or may specifically be targeted to the Golgi complex (12). Herpesvirus pUL36 homologs have been shown to interact with pUL37 (3, 23, 33, 59), the major capsid protein (43), pUL48 (59), and pUL25 (8). pUL25 is intimately capsid associated in a complex with pUL17 (57) and, in PrV, interacts with the C terminus of pUL36, relocating it from the cytosol to the nucleus during transient expression (8). However, it is unclear whether this interaction also occurs in intact pUL36 and during PrV infection.
The question in which cellular compartment tegumentation of nucleocapsids initiates remains. For HSV-1, addition of pUL36, the major component of the “inner,” nucleocapsid-apposed tegument, may already, at least in part, occur in the nucleus (6). However, our immunofluorescence analyses did not show any nuclear localization of PrV pUL36 during infection. Moreover, none of our antisera detected the presence of pUL36 on primary enveloped virus particles in the perinuclear cleft, in line with previous observations (33). In contrast, extracellular virions were heavily labeled. Thus, absence of pUL36 in the nucleus as observed by immunofluorescence analyses correlated well with absence of pUL36 on primary virions, as analyzed by immunoelectron microscopy.
We realize that immunodetection is dependent on a variety of factors. However, with our panel of antisera directed against different portions of the PrV pUL36 protein, we have robust reagents for pUL36 analysis. Although our antisera do not cover the complete length of the 3,084-aa protein, they completely cover the amino-terminal half, which includes important functional domains such as the deubiquitination motif, the two functional NLS, the pUL37 interaction domain, and a putative leucine zipper which may also be relevant for interaction of pUL36 with itself or with other proteins (4, 5). The third antiserum is directed against a region containing a putative late domain as well as a nonfunctional NLS, whereas the fourth antiserum recognizes the essential and highly conserved C terminus of PrV pUL36. Thus, this panel should faithfully detect PrV pUL36 as well as putative proteolytic cleavage products. Nevertheless, we did not find any indication for the presence of pUL36 in the nucleus during PrV infection.
Acknowledgments
This study has been supported by the Deutsche Forschungsgemeinschaft (grant Me 854/8-2).
We thank Diana Werner and Petra Meyer for expert technical assistance and Mandy Jörn for photographic help.
Footnotes
Published ahead of print on 29 July 2009.
REFERENCES
- 1.Abaitua, F., and P. O'Hare. 2008. Identification of a highly conserved, functional nuclear localization signal within the N-terminal region of herpes simplex virus type 1 VP1-2 tegument protein. J. Virol. 82:5234-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batterson, W., D. Furlong, and B. Roizman. 1983. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45:397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bechtel, J. T., and T. Shenk. 2002. Human cytomegalovirus UL47 tegument protein functions after entry and before immediate-early gene expression. J. Virol. 76:1043-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Böttcher, S., B. G. Klupp, H. Granzow, W. Fuchs, K. Michael, and T. C. Mettenleiter. 2006. Identification of a 709-amino-acid internal nonessential region within the essential conserved tegument protein (p)UL36 of pseudorabies virus. J. Virol. 80:9910-9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Böttcher, S., H. Granzow, C. Maresch, B. Möhl, B. G. Klupp, and T. C. Mettenleiter. 2007. Identification of functional domains within the essential large tegument protein pUL36 of pseudorabies virus. J. Virol. 81:13403-13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bucks, M. A., K. J. O'Regan, M. A. Murphy, J. W. Wills, and R. J. Courtney. 2007. Herpes simplex virus type 1 tegument proteins VP1/2 and UL37 are associated with intranuclear capsids. Virology 361:316-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, D. H., H. Jiang, M. Lee, F. Liu, and Z. H. Zhou. 1999. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology 260:10-16. [DOI] [PubMed] [Google Scholar]
- 8.Coller, K. E., J. I.-H. Lee, A. Ueda, and G. A. Smith. 2007. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J. Virol. 81:11790-11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Copeland, A. M., W. W. Newcomb, and J. C. Brown. 2009. Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment. J. Virol. 83:1660-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deruelle, M., K. Geenen, H. J. Nauwynck, and H. W. Favoreel. 2007. A point mutation in the putative ATP binding site of the pseudorabies virus US3 protein kinase prevents Bad phosphorylation and cell survival following apoptosis induction. Virus Res. 128:65-70. [DOI] [PubMed] [Google Scholar]
- 11.Desai, P., G. L. Sexton, J. M. McCaffery, and S. Person. 2001. A null mutation in the gene encoding the herpes simplex virus type 1 UL37 polypeptide abrogates virus maturation. J. Virol. 75:10259-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desai, P., G. L. Sexton, E. Huang, and S. Person. 2008. Localization of herpes simplex virus type 1 UL37 in the Golgi complex requires UL36 but not capsid structures. J. Virol. 82:11354-11361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desai, P. J. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farnsworth, A., T. W. Wisner, and D. C. Johnson. 2007. Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD. J. Virol. 81:319-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J. Virol. 78:11879-11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs, W., H. Granzow, B. G. Klupp, A. Karger, K. Michael, C. Maresch, R. Klopfleisch, and T. C. Mettenleiter. 2007. Relevance of the interaction between alphaherpesvirus UL3.5 and UL48 proteins for virion maturation and neuroinvasion. J. Virol. 81:9307-9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2004. The pseudorabies virus US3 protein is a component of primary and of mature virions. J. Virol. 78:1314-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 79:3200-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harel, A., and D. J. Forbes. 2004. Importin beta: conducting a much larger cellular symphony. Mol. Cell 16:319-330. [DOI] [PubMed] [Google Scholar]
- 23.Harmon, M.-E., and W. Gibson. 1996. High molecular weight virion protein of human cytomegalovirus forms complex with product of adjacent open reading frame, abstr. W35-4, p. 144. Proc. Am. Soc. Virol.
- 24.Jovasevic, V., L. Liang, and B. Roizman. 2008. Proteolytic cleavage of VP1-2 is required for release of herpes simplex virus 1 DNA into the nucleus. J. Virol. 82:3311-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaplan, A. S., and A. E. Vatter. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 7:394-407. [DOI] [PubMed] [Google Scholar]
- 26.Karr, B. M., and G. S. Read. 1999. The virion host shutoff function of herpes simplex virus degrades the 5′ end of a target mRNA before the 3′ end. Virology 264:195-204. [DOI] [PubMed] [Google Scholar]
- 27.Kato, A., M. Yamamoto, T. Ohno, H. Kodaira, Y. Nishiyama, and Y. Kawaguchi. 2005. Identification of proteins phosphorylated directly by the Us3 protein kinase encoded by herpes simplex virus 1. J. Virol. 79:9325-9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kattenhorn, L. M., G. A. Korbel, B. M. Kessler, E. Spooner, and H. L. Ploegh. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19:547-557. [DOI] [PubMed] [Google Scholar]
- 29.Klupp, B. G., and T. C. Mettenleiter. 1999. Glycoprotein gL-independent infectivity of pseudorabies virus is mediated by a gD-gH fusion protein. J. Virol. 73:3014-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klupp, B. G., C. J. Hengartner. T. C. Mettenleiter, and L. W. Enquist. 2004. Complete, annotated sequence of the pseudorabies virus genome. J. Virol. 78:424-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klupp, B. G., H. Granzow, G. M. Keil, and T. C. Mettenleiter. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J. Virol. 80:6235-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knipe, D. M., W. Batterson, C. Nosal, B. Roizman, and A. Buchan. 1981. Molecular genetics of herpes simplex virus. VI. Characterization of a temperature-sensitive mutant defective in the expression of all early viral gene products. J. Virol. 38:539-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krautwald, M., W. Fuchs, B. G. Klupp, and T. C. Mettenleiter. 2009. Translocation of incoming pseudorabies virus capsids to the cell nucleus is delayed in the absence of tegument protein pUL37. J. Virol. 83:3389-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, J. H., V. Vittone, E. Diefenbach, A. L. Cunningham, and R. J. Diefenbach. 2008. Identification of structural protein-protein interactions of herpes simplex virus type 1. Virology 378:347-354. [DOI] [PubMed] [Google Scholar]
- 37.Lee, J. I.-H., G. W. G. Luxton, and G. A. Smith. 2006. Identification of an essential domain in the herpesvirus VP1/2 tegument protein: the carboxy terminus directs incorporation into capsid assemblons. J. Virol. 80:12086-12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leege, T., H. Granzow, W. Fuchs, B. G. Klupp, and T. C. Mettenleiter. 2009. Phenotypic similarities and differences between UL37-deleted pseudorabies virus and herpes simplex virus type 1. J. Gen. Virol. 90:1560-1568. [DOI] [PubMed] [Google Scholar]
- 39.Lemaster, S., and B. Roizman. 1980. Herpes simplex virus phosphoproteins. II. Characterization of the virion protein kinase and of the polypeptides phosphorylated in the virion. J. Virol. 35:798-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luxton, G. W., S. Haverlock, K. E. Coller, S. E. Antinone, A. Pincetic, and G. A. Smith. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. USA 102:5832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luxton, G. W., J.-I.-H. Lee, S. Haverlock-Moyns, J. M. Schober, and G. A. Smith. 2006. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J. Virol. 80:201-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McNabb, D. S., and R. J. Courtney. 1992. Analysis of the UL36 open reading frame encoding the large tegument protein (ICP1/2) of herpes simplex virus type 1. J. Virol. 66:7581-7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McNabb, D. S., and R. J. Courtney. 1992. Characterization of the large tegument protein (ICP1/2) of herpes-simplex virus type-1. Virology 190:221-232. [DOI] [PubMed] [Google Scholar]
- 44.Mettenleiter, T. C. 1989. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology 171:623-625. [DOI] [PubMed] [Google Scholar]
- 45.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2009. Herpesvirus assembly: an update. Virus Res. 143:222-234. [DOI] [PubMed] [Google Scholar]
- 47.Mijatov, B., A. L. Cunningham, and R. J. Diefenbach. 2007. Residues F593 and E596 of HSV-1 tegument protein pUL36 (VP1/2) mediate binding of tegument protein pUL37. Virology 368:26-31. [DOI] [PubMed] [Google Scholar]
- 48.Morrison, E. E., A. J. Stevenson, Y. F. Wang, and D. M. Meredith. 1998. Differences in the intracellular localization and fate of herpes simplex virus tegument proteins early in the infection of Vero cells. J. Gen. Virol. 79:2517-2528. [DOI] [PubMed] [Google Scholar]
- 49.Ojala, P. M., B. Sodeik, M. W. Ebersold, U. Kutay, and A. Helenius. 2000. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol. Cell. Biol. 20:4922-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purves, F. C., W. O. Ogle, and B. Roizman. 1993. Processing of the herpes simplex virus regulatory protein alpha 22 mediated by the UL13 protein kinase determines the accumulation of a subset of alpha and gamma mRNAs and proteins in infected cells. Proc. Natl. Acad. Sci. USA 90:6701-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Read, G. S., B. M. Karr, and K. Knight. 1993. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptide. J. Virol. 67:7149-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts, A. P. E., F. Abaitua, P. O'Hare, D. McNab, F. J. Rixon, and D. Pasdeloup. 2009. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J. Virol. 83:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rozen, R., N. Sathish, Y. Li, and Y. Yuan. 2008. Virion-wide protein interactions of Kaposi's sarcoma-associated herpesvirus. J. Virol. 82:4742-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salman, H., A. Abu-Arish, S. Oliel, A. Loyter, J. Klafter, R. Granek, and M. Elbaum. 2005. Nuclear localization signal peptides induce molecular delivery along microtubules. Biophys. J. 89:2134-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sarma, N., D. Agarwal, L. A. Shiflett, and G. S. Read. 2008. Small interfering RNAs that deplete the cellular translation factor eIF4H impede mRNA degradation by the virion host shutoff protein of herpes simplex virus. J. Virol. 82:6600-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spear, P. G., and B. Roizman. 1972. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J. Virol. 9:143-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trus, B. L., W. W. Newcomb, N. Cheng, G. Cardone, L. Marekov, F. L. Homa, J. C. Brown, and A. C. Steven. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol. Cell 26:479-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uetz, P., Y. A. Dong, C. Zeretzke, C. Atzler, A. Baiker, B. Berger, S. V. Rajagopala, M. Roupelieva, D. Rose, E. Fossum, and J. Haas. 2006. Herpesviral protein networks and their interaction with the human proteome. Science 311:239-242. [DOI] [PubMed] [Google Scholar]
- 59.Vittone, V., E. Diefenbach, D. Triffett, M. W. Douglas, A. L. Cunningham, and R. J. Diefenbach. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79:9566-9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou, Z. H., D. H. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu, W., and R. Courtney. 1994. Chemical cross-linking of virion envelope and tegument proteins of herpes simplex virus type 1. Virology 204:590-599. [DOI] [PubMed] [Google Scholar]