Abstract
CD8 T cells control cytomegalovirus (CMV) infection in bone marrow transplantation recipients and persist in latently infected lungs as effector memory cells for continuous sensing of reactivated viral gene expression. Here we have addressed the question of whether viral immunoevasins, glycoproteins that specifically interfere with antigen presentation to CD8 T cells, have an impact on viral latency in the murine model. The data show that deletion of immunoevasin genes in murine CMV accelerates the clearance of productive infection during hematopoietic reconstitution and leads to a reduced latent viral genome load, reduced latency-associated viral transcription, and a lower incidence of recurrence in lung explants.
Establishment of latency after clearance of acute infection and the potential to reactivate to recurrent infection are key features of herpesvirus pathogenicity (41). Cytomegaloviruses (CMVs) are usually well controlled by the immune system and cause acute as well as recurrent disease mainly in the immunocompromised or immunologically immature host. Recipients (R) of bone marrow transplantation (BMT) are at risk of reactivating intrinsic human CMV (hCMV) or of becoming infected by reactivating donor (D)-derived hCMV transmitted with the transplant or of both (D− R+, D+ R−, and D+ R+ constellations, respectively) (9). Clinical studies (32, 38) and experimental studies of the murine model of infection with murine CMV (mCMV) (18; reviewed in reference 16) have consistently shown that timely endogenous lymphohematopoietic reconstitution of antiviral CD8 T cells is decisive for coping with CMV infection after BMT. Accordingly, preemptive immunotherapy with antiviral CD8 T cells proved to be a promising approach with the murine model (3, 14, 35, 36, 44) and in clinical trials (6, 27, 40).
Both viruses hCMV and mCMV express proteins, so-called immunoevasins, that interfere with the major histocompatibility complex class I pathway of antigen presentation to CD8 T cells (reviewed in reference 33). Whereas many studies have demonstrated the efficacy of immunoevasins in inhibiting the cell surface presentation of antigenic peptides in infected cells in vitro, these molecules do not prevent (11, 19, 26) but rather enhance (4) the priming of viral epitope-specific CD8 T cells, and their role and relevance in viral pathogenesis in vivo are a currently discussed issue (8). Obviously, research on the in vivo function of immunoevasins by using viral immunoevasin gene deletion mutants can be accomplished only using animal models, and the murine model is well established. Although the detailed molecular modes of action differ between the immunoevasins of hCMV and mCMV, the biological outcome in both instances is the inhibition of antigen presentation. Thus, there is good reason to assume that the murine model also gives us valuable predictions for the in vivo role of hCMV immunoevasins.
Three molecules that regulate antigen presentation to CD8 T cells are known for mCMV. The immunoevasins m152/gp40 (7, 47) and m06/gp48 (37) interfere with the vesicular transport of peptide-loaded major histocompatibility complex class I molecules. Although m04/gp34 may cooperate with these two confirmed immunoevasins, more-recent data with a mutant virus expressing m04/gp34 selectively have revealed that it is no CD8 T-cell immunoevasin in its own right (15, 28). A virus lacking all three “viral regulators of antigen presentation” (vRAPs), the deletion mutant mCMV-Δm04+m06+m152 (45), here referred to as mCMV-ΔvRAP, is used to study the immune response and viral pathogenesis in the absence of vRAPs. Importantly, a previous study has shown that deletion of the vRAP genes does not affect viral replicative fitness in immunocompromised mice, as demonstrated by unaltered doubling times in various host tissues (4) compared with results for bacterial artificial chromosome (BAC)-cloned wild-type (WT) virus (46), mCMV-WT.BAC. Therefore, any in vivo growth phenotype of mutant virus mCMV-ΔvRAP in immunocompetent mice or during immunological reconstitution after BMT can be attributed to immunological control.
Previous studies of immunocompetent C57BL/6 and B-cell-deficient μMT mice, both of which are resistant to mCMV due to natural killer (NK) cell activation (1), have suggested that vRAPs have little impact on virus replication, establishment of latency, and virus reactivation upon immunosuppression (12), with the exception of elevated virus titers in salivary glands of mCMV-susceptible BALB/c mice (25). As introduced above, it is a hallmark of CMV biology that infection with WT CMVs is well controlled by the immune system despite the expression of immunoevasins, so in immunocompetent mice, only incremental improvement can be expected from the deletion of immunoevasin genes. An impact of vRAPs might rather be seen in the immunocompromised host, especially in the clinically relevant situation of lymphohematopoietic reconstitution of antiviral CD8 T cells in BMT recipients. Importantly, whereas CD8 T cells can be replaced with other innate and adaptive effector cells in otherwise immunocompetent mice (20), antiviral CD8 T cells are essential for preventing CMV disease in the BMT setting (30, 31). Although deletion of vRAP m152/gp40 can also activate NK cells through expression of the activating NKG2D ligand RAE-1 (for a review, see reference 24), previous work has demonstrated that CD8 T cells outperform NK cells in controlling the in vivo replication of mCMV-ΔvRAP (4). Since CD8 T cells and NK cells are regulated in parallel by m152/gp40, both may contribute to latency in the same direction. Altogether, experimental BMT in the mouse should be a good model for unraveling a potential in vivo role for vRAPs.
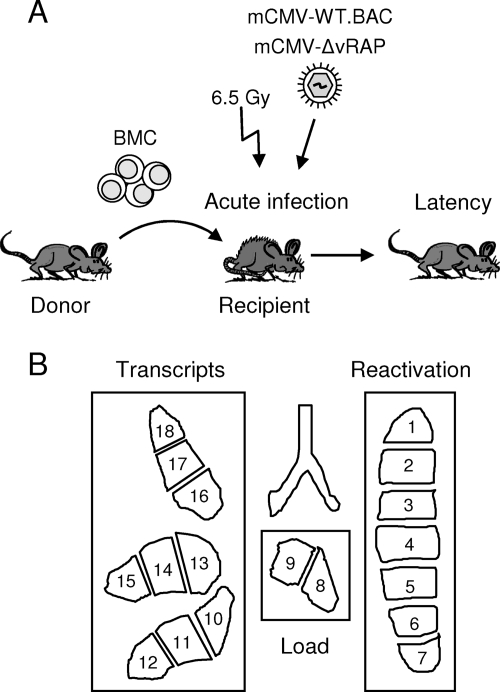
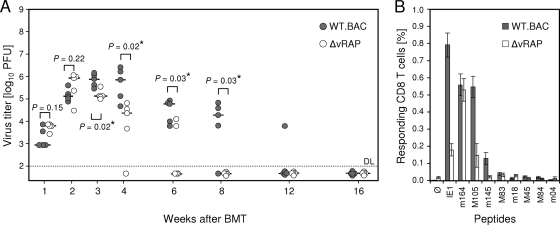
Here we focused on infection of the lungs, since interstitial pneumonia is a very relevant manifestation of CMV disease in BMT recipients (39) and since the lungs are a major organ site of mCMV latency after neonatal infection (2) and after experimental BMT (22, 30). Figure 1 sketches the experimental protocol of syngeneic BMT (Fig. 1A) and the analysis of viral latency (Fig. 1B). As shown in Fig. 2A by the time course of viral replication in the recipients’ lungs, mutant virus replicated like WT virus only in the first 2 weeks, which is consistent with unaltered replicative fitness in the absence of immune cells (4). In contrast, significantly more-efficient control of the mutant virus was found for all later time points, which correlates with the reappearance of virus-specific CD8 T cells in the BMT model (18). As shown previously, selective depletion of CD8 T cells but not of CD4 T cells during lymphohematopoietic reconstitution leads to fatal multiple-organ CMV disease (31), including a fulminant, disseminated viral interstitial pneumonia (30). Beyond 16 weeks, productive infection was resolved for both viruses. Thus, immunoevasins contributed to higher virus peak levels in the third and fourth weeks and to delayed clearance of productive infection. In accordance with previous findings (17), viral epitope-specific CD8 T cells persisted in latently infected lungs, with a particularly high response to the immediate-early epitope IE1, thought to be involved in sensing of early stages in transcriptional reactivation (43). Notably, although in the comparison between the two viruses the replication of mCMV-WT.BAC was higher and prolonged in the acute phase of infection, suggesting a lower level of immune control, CD8 T-cell frequencies in latently infected lungs were actually higher (Fig. 2B).
FIG. 1.
Scheme illustrating the strategy for studying viral latency in the lungs. (A) Protocol of BMT. Syngeneic BMT was performed with 8- to 10-week-old female BALB/c mice as donors and recipients of bone marrow cells (BMC) as described in greater detail previously (29). Briefly, recipients were immunocompromised by total-body γ irradiation with a single dose of 6.5 Gy at 6 h prior to the intravenous transfer of 5 × 106 pooled femoral and tibial BMC and were infected with 105 PFU of either mCMV-WT.BAC or mCMV-ΔvRAP in the left hind footpad at 2 h after BMT. The time course of infection of the lungs was monitored until latency was established. (B) Analysis of latency and reactivation. Shown is the lobular anatomy of the mouse lungs in ventral view. After the lungs were cut into 18 pieces, each representing ∼3 million cells, lung pieces 1 through 7, derived from the left lung, were used for tissue explant cultures to determine the cumulative incidence of virus reactivation and pieces 8 and 9, representing the postcaval lobe, served to determine the latent viral DNA load. Finally, lung pieces 10 through 18, derived from the inferior, middle, and superior lobes of the right lung, were used for the quantitation of IE1 transcripts.
FIG. 2.
Pulmonary infection and immune response after BMT. (A) Time course of viral replication in the lungs of BMT recipients in the presence (gray circles; WT.BAC) or absence (open circles; ΔvRAP) of immunoevasins. Syngeneic BMT (see Fig. 1A), intraplantar infection of the recipients, and determination of virus titers in lung homogenates by virus plaque (PFU) assay were performed as described previously (see reference 29 and references therein). Symbols represent data for individual mice. The median values are marked by short horizontal bars. The statistical significance of differences in virus titers was evaluated by using distribution-free Wilcoxon-Mann-Whitney (rank sum) statistics. Virus titers in two experimental groups differ significantly if the P value (two-tailed test) is <0.05. Calculated P values are indicated for group comparisons of interest. *, significant difference. DL, detection limit of the assay. (B) Frequencies of epitope-specific CD8 T cells in pulmonary infiltrates of latently infected mice. Amino acid sequences of the indicated H-2d-restricted antigenic peptides are listed in reference 14. At 6 months after BMT and infection with WT.BAC (gray bars) or ΔvRAP (open bars), lung infiltrate leukocytes were isolated (18, 29) and CD8 T cells were enriched immunomagnetically (17). Frequencies of gamma interferon-secreting, peptide-sensitized cells were determined by using an enzyme-linked immunospot assay (see reference 4 and references therein). Bars represent most probable numbers calculated by intercept-free linear regression analysis. Error bars indicate the 95% confidence intervals. Ø, no peptide added.
Previous work has revealed a chain of cause and effect, relating virus titers during acute infection to the tissue load of the latent viral genome, latency-associated viral transcription, and the incidence of reactivation to productive infection (23, 34, 44). To verify this here for the role of vRAPs, we used the established strategy of subdividing the lungs into 18 tissue pieces (13, 22). Nine pieces of the three lobes of the right lung were used for the analysis of IE1 transcription, two pieces of the subcaval lobe were used for quantitating latent viral genome, and the seven pieces of the left lung were explanted to determine the cumulative incidence of virus reactivation (Fig. 1B).
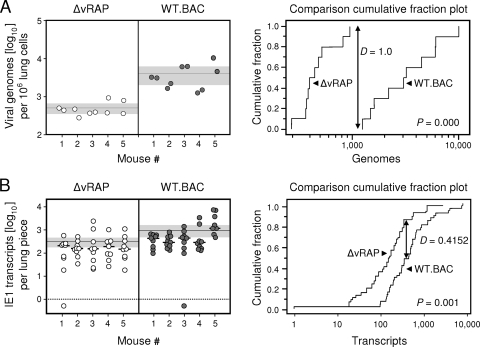
Figure 3A (left panel) shows a much larger viral genome load in lungs latently infected with WT virus, with only modest interindividual and intratissue variance. This load difference turned out to be highly significant, as revealed by Kolmogorov-Smirnov statistics (Fig. 3A, right panel) and by Student's t test. Likewise, IE1 transcription was also elevated in lungs latently infected with WT virus (Fig. 3B, left panel). In agreement with previous data on a Poisson distribution of transcripts (13), the data here were neither normally nor log-normally distributed. Again, the distribution-free Kolmogorov-Smirnov statistics revealed a highly significant difference between the two groups (Fig. 3B, right panel). Thus, apparently, immunoevasins have an impact on latent viral DNA load and on viral transcriptional activity during latency.
FIG. 3.
Comparative molecular characteristics of viral latency established in the lungs. The analyses were performed at 6 months after BMT and infection in the presence (gray-filled circles; WT.BAC) or absence (open circles; ΔvRAP) of immunoevasins. (A) Comparison of latent viral DNA loads determined by M55/gB gene-specific real-time quantitative PCR (qPCR) as described previously (43) and updated (42). Left panel: data. Symbols show the viral DNA content of individually tested lung pieces no. 8 and 9 for five latently infected BMT recipients per group, 10 pieces per group altogether. Data were normalized to 106 lung cells by pthrp gene-specific qPCR. The horizontal line indicates the mean value, and the shaded area shows the 95% confidence interval for the mean. Right panel: statistical analysis. Shown is the “comparison cumulative fraction plot” of the nonparametric and distribution-free Kolmogorov-Smirnov test for comparison of two data sets. In addition to providing a graphical presentation of the data, this test has the advantage of making no assumption about the distribution of data. Instead, it compares the observed empirical distributions, so that it is robust and applicable to all kinds of data sets. In addition, it also tests whether or not the data are normally or log-normally distributed for applying parametric tests such as the Student t test. For a more detailed explanation, go to http://www.physics.csbsju.edu/stats/KS-test.html. Indicated is the maximum difference (D value) between the cumulative fraction functions, also known as “empirical distribution functions,” and the corresponding P value. A high D value and a low P value indicate a high difference between the data sets. The difference is regarded as significant if the P value is <0.05. Note that the cumulative fraction functions for the viral loads are completely separated for the two viruses. Since the viral load data were found to follow a normal distribution, Student's t test was also applied and showed a P value of 0.001, with mean values of 4,091 and 501 for the latent genome loads of viruses WT.BAC and ΔvRAP, respectively. (B) For lung pieces 10 through 18 of the same five mice, for 45 pieces per group altogether, the amounts of IE1 transcripts were determined by reverse transcriptase qPCR as described previously (43), except that total lung tissue RNA was used as a template. Left panel: data, essentially as described above for panel A. In addition, short horizontal bars indicate median values of data from individual mice. Negative data are indicated below the dotted line. Right panel: statistical analysis. Unlike DNA load data, transcript data turned out not to be consistent with a normal or log-normal distribution. Shown are the comparison cumulative fraction plot of the Kolmogorov-Smirnov test and the associated D and P values.
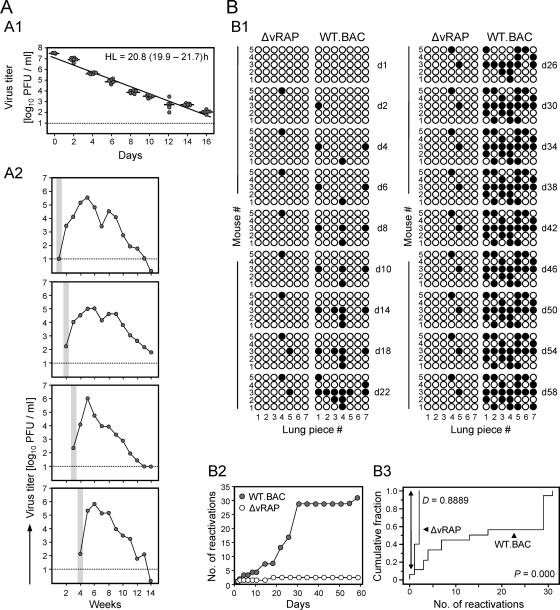
The definition of herpesvirus latency by Roizmann and Sears (41) demands that the viral genomes not only are physically maintained but are reactivatable to productive infection. Since reactivation is a stochastic process, approaches of in vivo reactivation can give only the “point prevalence” of reactivation for the time of analysis (23). We have here therefore chosen the long-established method of tissue explantation (21) to measure the cumulative reactivation incidence over time, which gives a better estimate for the number of reactivatable latent genomes (Fig. 4). Figure 4A introduces the experimental system by showing exemplarily the half-life of infectious mCMV-WT.BAC virions in cell culture medium (Fig. 4A1), as well as differences in the onset of reactivated infection in lung explants (Fig. 4A2). Importantly, the presence of reactivated virus in explant culture supernatants by far exceeded the half-life of virions. Specifically, explanted tissue supported infection for an extended period of ∼2 months until it gradually got exhausted. Accordingly, an explant culture, once positive, remained positive throughout the reactivation assay (Fig. 4B). Figure 4B1 sketches reactivation events as virus-positive cultures observed over time until no further reactivation was observed. At a glance, reactivation occurred frequently from lung pieces carrying latent WT virus but rarely from pieces carrying the latent mutant virus. As shown in the cumulative plot of reactivation events over time, most reactivations occurred between 2 and 4 weeks in culture, which proves that lung pieces were not productively infected at the time of explantation (Fig. 4B2). The difference between the two groups was highly significant as revealed by Kolmogorov-Smirnov statistics (Fig. 4B3). In total, for WT virus, we determined 31 virus reactivations from 35 explants. Since an explant contains ∼3 million cells with a load of ∼4,000 genomes per million cells (recall Fig. 3A), the cumulative reactivation incidence was in the range of 10−4.
FIG. 4.
Virus reactivation in lung tissue explant cultures. (A) Characteristics of the assay. (A1) Functional stability of purified mCMV-WT.BAC virions under standard cell culture conditions. Shown is the time course of activity loss. Symbols represent replicate cultures. The half-life (HL) of infectivity is calculated from the negative slope (a) of the log-linear regression line according to the formula HL = log102/a. Its 95% confidence interval is indicated in parentheses. (A2) Time course of reactivated infection in four selected lung tissue explant cultures. Lung pieces derived from BALB/c mice latently infected with mCMV-WT.BAC were plated directly into 24-well tissue culture plates with 1 ml of medium but with no permissive feeder cells. At the indicated times, 0.1-ml aliquots of the culture supernatants were used for the PFU assay and replaced with fresh medium. The first detection of infectivity is highlighted by gray shading. The dotted lines indicate the detection limit of the assay. (B) Comparative reactivation distributions. Lung pieces no. 1 through no. 7 of the five mice per group, altogether 35 pieces per group, were plated at 6 months after BMT and infection with the respective viruses. (B1) Illustration of the time course of reactivations observed in the explant cultures. Results are shown as binary data. Open circles, no reactivation; black dots, reactivation. (B2) Cumulative reactivation incidence plot. It is important to understand that according to the Poisson distribution of reactivations in each explant, a positive explant culture may reflect more than one reactivation, so that the actual number of reactivations exceeds the observed number of positive cultures. For each time point, the total number of reactivations was calculated from the observed fraction of negative cultures [f(0)], which equals e−λ, according to the formula: Σ F(n) = λ/n × F(n − 1) for all n values of >0, where λ is the Poisson distribution parameter lambda and [F(n)] is the fraction of cultures comprising a number (n) of reactivations. (B3) Statistical analysis. Shown is the comparison cumulative fraction plot of the Kolmogorov-Smirnov test. The maximum difference, D, between the cumulative fraction functions and the corresponding P value is indicated.
The difference in the reactivation incidences most likely reflects the difference in the latent viral DNA load (34). As shown recently, although deletion of the regulatory protein IE1 attenuates mCMV (10), an IE1 deletion mutant was still able to reactivate provided that genome loads of WT and mutant virus were adjusted by using higher doses of mutant virus for acute infection (5).
Notably, the latency parameters load, transcription, and reactivation were here found to be positively correlated with the magnitude of the immune response. This paradox may be explained by the previously described negative feedback regulation of CD8 T-cell stimulation (4). According to this model, the expression of immunoevasins inhibits the recognition of infected cells and thus the control of infection, thereby promoting a sustained antigen supply for cross-priming of the CD8 T-cell response by uninfected antigen-presenting cells. Although such a mechanism was originally proposed for CD8 T-cell priming in the regional lymph node, it may also apply to effector memory cells in latently infected lungs.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft, SFB 490, individual projects E2 (C.K.S. and C.O.S.), E3 (R.H.), and E4 (V.B. and M.J.R.), and Clinical Research Group KFO 183, individual project TP8 (M.J.R.).
Footnotes
Published ahead of print on 15 July 2009.
REFERENCES
- 1.Arase, H., E. S. Mocarski, A. E. Campbell, A. B. Hill, and L. L. Lanier. 2002. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science 296:1323-1326. [DOI] [PubMed] [Google Scholar]
- 2.Balthesen, M., M. Messerle, and M. J. Reddehase. 1993. Lungs are a major organ site of cytomegalovirus latency and recurrence. J. Virol. 67:5360-5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böhm, V., J. Podlech, D. Thomas, P. Deegen, M.-F. Pahl-Seibert, N. A. W. Lemmermann, N. K. A. Grzimek, S. A. Oehrlein-Karpi, M. J. Reddehase, and R. Holtappels. 2008. Epitope-specific in vivo protection against cytomegalovirus disease by CD8 T cells in the murine model of preemptive immunotherapy. Med. Microbiol. Immunol. 197:135-144. [DOI] [PubMed] [Google Scholar]
- 4.Böhm, V., C. O. Simon, J. Podlech, C. K. Seckert, D. Gendig, P. Deegen, D. Gillert-Marien, N. A. W. Lemmermann, R. Holtappels, and M. J. Reddehase. 2008. The immune evasion paradox: immunoevasins of murine cytomegalovirus enhance priming of CD8 T cells by preventing negative feedback regulation. J. Virol. 82:11637-11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Busche, A., A. Marquardt, A. Bleich, P. Ghazal, A. Angulo, and M. Messerle. 2009. The mouse cytomegalovirus immediate-early 1 gene is not required for establishment of latency or for reactivation in the lungs. J. Virol. 83:4030-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cobbold, M., N. Khan, B. Pourgheysari, S. Tauro, D. McDonald, H. Osman, M. Assenmacher, I. Billingham, C. Steward, C. Crawley, E. Olavarria, J. Goldman, R. Chakraverty, P. Mahendra, C. Craddock, and P. A. Moss. 2005. Adoptive transfer of cytomegalovirus-specific CTL to stem cell transplant patients after selection by HLA-peptide tetramers. J. Exp. Med. 202:379-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.del Val, M., H. Hengel, H. Häcker, U. Hartlaub, T. Ruppert, P. Lucin, and U. H. Koszinowski. 1992. Cytomegalovirus prevents antigen presentation by blocking the transport of peptide-loaded major histocompatibility complex class I molecules into the medial-Golgi compartment. J. Exp. Med. 176:729-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doom, C. M., and A. B. Hill. 2008. MHC class I immune evasion in MCMV infection. Med. Microbiol. Immunol. 197:191-204. [DOI] [PubMed] [Google Scholar]
- 9.Emery, V. C. 1998. Relative importance of cytomegalovirus load as a risk factor for cytomegalovirus disease in the immunocompromised host, p. 288-301. In M. Scholz, H. F. Rabenau, H. W. Doerr, and J. Cinatl, Jr. (ed.), CMV-related immunopathology. Monographs in Virology, vol. 21. Karger, Basel, Switzerland. [Google Scholar]
- 10.Ghazal, P., A. E. Visser, M. Gustems, R. Garcia, E. M. Borst, K. Sullivan, M. Messerle, and A. Angulo. 2005. Elimination of ie1 significantly attenuates murine cytomegalovirus virulence but does not alter replicative capacity in cell culture. J. Virol. 79:7182-7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gold, M. C., M. W. Munks, M. Wagner, U. H. Koszinowski, A. B. Hill, and S. P. Fling. 2002. The murine cytomegalovirus immunomodulatory gene m152 prevents recognition of infected cells by M45-specific CTL but does not alter the immunodominance of the M45-specific CD8 T cell response in vivo. J. Immunol. 169:359-365. [DOI] [PubMed] [Google Scholar]
- 12.Gold, M. C., M. W. Munks, M. Wagner, C. W. McMahon, A. Kelly, D. G. Kavanagh, M. K. Slifka, U. H. Koszinowski, D. H. Raulet, and A. B. Hill. 2004. Murine cytomegalovirus interference with antigen presentation has little effect on the size or the effector memory phenotype of the CD8 T cell response. J. Immunol. 172:6944-6953. [DOI] [PubMed] [Google Scholar]
- 13.Grzimek, N. K. A., D. Dreis, S. Schmalz, and M. J. Reddehase. 2001. Random, asynchronous, and asymmetric transcriptional activity of enhancer-flanking major immediate-early genes ie1/3 and ie2 during murine cytomegalovirus latency in the lungs. J. Virol. 75:2692-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holtappels, R., V. Böhm, J. Podlech, and M. J. Reddehase. 2008. CD8 T-cell-based immunotherapy of cytomegalovirus infection: “proof of concept” provided by the murine model. Med. Microbiol. Immunol. 197:125-134. [DOI] [PubMed] [Google Scholar]
- 15.Holtappels, R., D. Gillert-Marien, D. Thomas, J. Podlech, P. Deegen, S. Herter, S. A. Oehrlein-Karpi, D. Strand, M. Wagner, and M. J. Reddehase. 2006. Cytomegalovirus encodes a positive regulator of antigen presentation. J. Virol. 80:7613-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holtappels, R., M. W. Munks, J. Podlech, and M. J. Reddehase. 2006. CD8 T-cell-based immunotherapy of cytomegalovirus disease in the mouse model of the immunocompromised bone marrow transplantation recipient, p. 383-418. In M. J. Reddehase (ed.), Cytomegaloviruses: molecular biology and immunology. Caister Academic Press, Wymondham, Norfolk, United Kingdom.
- 17.Holtappels, R., M.-F. Pahl-Seibert, D. Thomas, and M. J. Reddehase. 2000. Enrichment of immediate-early 1 (m123/pp89) peptide-specific CD8 T cells in a pulmonary CD62Llo memory-effector cell pool during latent murine cytomegalovirus infection of the lungs. J. Virol. 74:11495-11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holtappels, R., J. Podlech, G. Geginat, H.-P. Steffens, D. Thomas, and M. J. Reddehase. 1998. Control of murine cytomegalovirus in the lungs: relative but not absolute immunodominance of the immediate-early 1 nonapeptide during the antiviral cytolytic T-lymphocyte response in pulmonary infiltrates. J. Virol. 72:7201-7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holtappels, R., J. Podlech, M.-F. Pahl-Seibert, M. Juelch, D. Thomas, C. O. Simon, M. Wagner, and M. J. Reddehase. 2004. Cytomegalovirus misleads its host by priming of CD8 T cells specific for an epitope not presented in infected tissues. J. Exp. Med. 199:131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jonjic, S., I. Pavic, P. Lucin, D. Rukavina, and U. H. Koszinowski. 1990. Efficacious control of cytomegalovirus infection after long-term depletion of CD8+ T lymphocytes. J. Virol. 64:5457-5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jordan, M. C., and V. L. Mar. 1982. Spontaneous activation of latent cytomegalovirus from murine spleen explants: role of lymphocytes and macrophages in release and replication of virus. J. Clin. Investig. 70:762-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurz, S. K., M. Rapp, H.-P. Steffens, N. K. A. Grzimek, S. Schmalz, and M. J. Reddehase. 1999. Focal transcriptional activity of murine cytomegalovirus during latency in the lungs. J. Virol. 73:482-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurz, S. K., and M. J. Reddehase. 1999. Patchwork pattern of transcriptional reactivation in the lungs indicates sequential checkpoints in the transition from murine cytomegalovirus latency to recurrence. J. Virol. 73:8612-8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lenac, T., J. Arapovic, L. Traven, A. Krmpotic, and S. Jonjic. 2008. Murine cytomegalovirus regulation of NKG2D ligands. Med. Microbiol. Immunol. 197:159-166. [DOI] [PubMed] [Google Scholar]
- 25.Lu, X., A. K. Pinto, A. M. Kelly, K. S. Cho, and A. B. Hill. 2006. Murine cytomegalovirus interference with antigen presentation contributes to the inability of CD8 T cells to control virus in the salivary gland. J. Virol. 80:4200-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munks, M. W., A. K. Pinto, C. M. Doom, and A. B. Hill. 2007. Viral interference with antigen presentation does not alter acute or chronic CD8 T cell immunodominance in murine cytomegalovirus infection. J. Immunol. 178:7235-7241. [DOI] [PubMed] [Google Scholar]
- 27.Peggs, K. S., S. Verfuerth, A. Pizzey, N. Khan, M. Gulver, P. A. Moss, and S. Mackinnon. 2003. Adoptive cellular therapy for early cytomegalovirus infection after allogeneic stem-cell transplantation with virus-specific T-cell lines. Lancet 362:1375-1377. [DOI] [PubMed] [Google Scholar]
- 28.Pinto, A. K., M. W. Munks, U. H. Koszinowski, and A. B. Hill. 2006. Coordinated function of murine cytomegalovirus genes completely inhibits CTL lysis. J. Immunol. 177:3225-3234. [DOI] [PubMed] [Google Scholar]
- 29.Podlech, J., R. Holtappels, N. K. A. Grzimek, and M. J. Reddehase. 2002. Animal models: murine cytomegalovirus, p. 493-525. In S. H. E. Kaufmann and D. Kabelitz (ed.), Methods in microbiology, vol. 32. Immunology of infection, 2nd ed. Academic Press, London, United Kingdom. [Google Scholar]
- 30.Podlech, J., R. Holtappels, M.-F. Pahl-Seibert, H.-P. Steffens, and M. J. Reddehase. 2000. Murine model of interstitial cytomegalovirus pneumonia in syngeneic bone marrow transplantation: persistence of protective pulmonary CD8-T-cell infiltrates after clearance of acute infection. J. Virol. 74:7496-7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Podlech, J., R. Holtappels, N. Wirtz, H.-P. Steffens, and M. J. Reddehase. 1998. Reconstitution of CD8 T cells is essential for the prevention of multiple-organ cytomegalovirus histopathology after bone marrow transplantation. J. Gen. Virol. 79:2099-2104. [DOI] [PubMed] [Google Scholar]
- 32.Quinnan, G. V., Jr., W. H. Burns, N. Kirmani, A. H. Rook, J. Manischewitz, L. Jackson, G. W. Santos, and R. Sarai. 1984. HLA-restricted cytotoxic T lymphocytes are an early immune response and important defense mechanism in cytomegalovirus infections. Rev. Infect. Dis. 6:156-163. [DOI] [PubMed] [Google Scholar]
- 33.Reddehase, M. J. 2002. Antigens and immunoevasins: opponents in cytomegalovirus immune surveillance. Nat. Rev. Immunol. 2:831-844. [DOI] [PubMed] [Google Scholar]
- 34.Reddehase, M. J., M. Balthesen, M. Rapp, S. Jonjic, I. Pavic, and U. H. Koszinowski. 1994. The conditions of primary infection define the load of latent viral genome in organs and the risk of recurrent cytomegalovirus disease. J. Exp. Med. 179:185-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reddehase, M. J., S. Jonjic, F. Weiland, W. Mutter, and U. H. Koszinowski. 1988. Adoptive immunotherapy of murine cytomegalovirus adrenalitis in the immunocompromised host: CD4-helper-independent antiviral function of CD8-positive memory T lymphocytes derived from latently infected donors. J. Virol. 62:1061-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reddehase, M. J., F. Weiland, K. Münch, S. Jonjic, A. Lüske, and U. H. Koszinowski. 1985. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J. Virol. 55:264-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reusch, U., W. Muranyi, P. Lucin, H. G. Burgert, H. Hengel, and U. H. Koszinowski. 1999. A cytomegalovirus glycoprotein re-routes MHC class I complexes to lysosomes for degradation. EMBO J. 18:1081-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reusser, P., S. R. Riddell, J. D. Meyers, and P. D. Greenberg. 1991. Cytotoxic T-lymphocyte response to cytomegalovirus after human allogeneic bone marrow transplantation: pattern of recovery and correlation with cytomegalovirus infection and disease. Blood 78:1373-1380. [PubMed] [Google Scholar]
- 39.Riddell, S. R. 1995. Pathogenesis of cytomegalovirus pneumonia in immunocompromised hosts. Semin. Respir. Infect. 10:199-208. [PubMed] [Google Scholar]
- 40.Riddell, S. R., K. S. Watanabe, J. M. Goodrich, C. R. Li, M. E. Agha, and P. D. Greenberg. 1992. Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science 257:238-241. [DOI] [PubMed] [Google Scholar]
- 41.Roizmann, N., and A. E. Sears. 1987. An inquiry into the mechanisms of herpes simplex virus latency. Annu. Rev. Microbiol. 41:543-571. [DOI] [PubMed] [Google Scholar]
- 42.Seckert, C. K., A. Renzaho, M. J. Reddehase, and N. K. Grzimek. 2008. Hematopoietic stem cell transplantation with latently infected donors does not transmit virus to immunocompromised recipients in the murine model of cytomegalovirus infection. Med. Microbiol. Immunol. 197:251-259. [DOI] [PubMed] [Google Scholar]
- 43.Simon, C. O., R. Holtappels, H.-M. Tervo, V. Böhm, T. Däubner, S. A. Oehrlein-Karpi, B. Kühnapfel, A. Renzaho, D. Strand, J. Podlech, M. J. Reddehase, and N. K. A. Grzimek. 2006. CD8 T cells control cytomegalovirus latency by epitope-specific sensing of transcriptional reactivation. J. Virol. 80:10436-10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steffens, H.-P., S. Kurz, R. Holtappels, and M. J. Reddehase. 1998. Preemptive CD8-T-cell immunotherapy of acute cytomegalovirus infection prevents lethal disease, limits the burden of latent viral genome, and reduces the risk of virus recurrence. J. Virol. 72:1797-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner, M., A. Gutermann, J. Podlech, M. J. Reddehase, and U. H. Koszinowski. 2002. MHC class I allele-specific cooperative and competitive interactions between immune evasion proteins of cytomegalovirus. J. Exp. Med. 196:805-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner, M., S. Jonjic, U. H. Koszinowski, and M. Messerle. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ziegler, H., R. Thäle, P. Lucin, W. Muranyi, T. Flohr, H. Hengel, H. Farrell, W. Rawlinson, and U. H. Koszinowski. 1997. A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-Golgi compartments. Immunity 6:57-66. [DOI] [PubMed] [Google Scholar]