Abstract
Hepatitis C virus (HCV) often causes chronic infection and may lead to hepatocellular carcinoma (HCC). We have shown previously that HCV core protein has pleiotropic functions, including transcriptional regulation of a number of cellular genes, although the mechanism for gene regulation remains unclear. In this study, a mammalian two-hybrid screen identified a novel binding partner, HS1-associated protein X-1 (HAX-1), for HCV core protein from a human liver cDNA library. An association between HAX-1 and HCV core protein was further verified by confocal microscopy and coimmunoprecipitation in HepG2 cells expressing HCV core or full-length (FL) gene. Both HCV core protein and a chemotherapeutic agent for HCC, 5-flouorouracil (5-FU), are known to modulate p53. We examined here whether an association between core and HAX-1 has any functional relevance to p53 modulation in 5-FU-treated cells. For this, the role of HAX-1 on 5-FU treatment was examined in HepG2 cells expressing HCV core or FL gene using cell proliferation, p53 expression, and caspase activation analysis. Cells expressing HCV-core or FL gene were more susceptible to 5-FU-induced growth inhibition than control cells, whereas cell survival was enhanced after suppression of HAX-1 by small interfering RNA. Further, 5-FU-mediated p53 expression was reduced with concurrent HAX-1 suppression in core- or polyprotein-expressing cells compared to control HepG2 cells, and caspase-2 and -7 activities were diminished. On the other hand, HCV core protein did not play a detectable role in 5-FU-mediated caspase-7 activation in the absence of functional p53 in Hep3B or Huh-7 cells. These observations underscore an association between HCV core and HAX-1, which promotes 5-FU mediated p53-dependent caspase-7 activation and hepatocyte growth inhibition.
Hepatitis C virus (HCV) core protein has pleiotropic functions, suggesting a complex role in cellular interactions during viral infection (26). Many of the properties suggest that HCV core protein, in concert with cellular factors, may contribute to the pathogenesis during chronic HCV infection. In infected liver, HCV core protein may stimulate cells to escape from replicative senescence, allowing for the rise of selective clonal proliferation (25). We have shown that the inhibition of HCV core protein expression in immortalized human hepatocytes (IHH) results in an increase in p53 expression preceding the onset of apoptosis (1). Apoptosis observed after inhibition of HCV core protein expression by antisense sequences correlates with an upregulation of Apaf-1 and the activation of a caspase-9-related cascade in the absence of cytosolic accumulation of cytochrome c (13, 18, 34). Kao et al. (10) suggested that HCV core protein has the potential to fine tune p53 functions via at least three means: physical interaction, modulation of p53 transcriptional activity, and posttranslational modifications. One or all of these functions may occur even in the cytoplasm (16).
In the present study, we have identified a novel HCV core protein binding partner HS1-associated protein X-1 (HAX-1) by a mammalian two-hybrid screen from a protein fragment complementation assay (28, 29). The HAX-1 protein was first identified by a two-hybrid screen using the hematopoietic lineage cell-specific protein 1 (HS1) as a bait (35). HAX-1 interacts with a variety of structurally unrelated proteins, suggesting its involvement in intracellular signaling and shuttling of various intracellular molecules and in cytoskeletal control (3, 11, 24). The biological function of HAX-1 was primarily divided into three categories: (i) association with viral proteins for involvement in apoptotic regulation processes, (ii) involvement in cell motility processes, and (iii) acting as a cytoplasmic retention factor. HAX-1 mRNA is expressed ubiquitously in different tissues, including liver (17, 19). Several studies have shown that Hax-1 expression is upregulated in different types of tumors (7, 14, 17, 41, 42). HAX-1 is localized mainly in mitochondria but is also found in the endoplasmic reticulum and nuclear envelope in the cells (35). Subcellular localization of HAX-1 may vary among different tissues; depending on its interacting partners, which in turn may modulate the properties of HAX-1 or the interacting protein. Thus, similar to HCV core protein, HAX-1 may have a multifunctional impact on biological processes.
5-Flouorouracil (5-FU) is widely used in the treatment of several cancers. Specifically, it shows a promising effect when used in conjunction with alpha interferon (IFN-α) or PEG-IFN for the treatment of advanced hepatocellular carcinoma (12, 21). Hagiwara et al. (5) reported that 5-FU treatment of tumors generated by subcutaneous injection of HepG2 cells in nude mice was associated with significantly more apoptotic cells than the control tumors. This result supports the fact that 5-FU treatment induces apoptosis in vivo. In general, 5-FU works by altering DNA metabolism (24), thereby causing strand breaks that, in turn, activate p53-dependent apoptosis (4, 8, 23, 40). A functional connection between p53 and caspase-2 is essential for the initiation of 5-FU-induced apoptosis in human colon cancer cells (39).
Previously, HAX-1 was reported to act as an antiapoptotic protein; however, the role of HAX-1 in context to core protein remained to be examined. Recently, Uka et al. (37) reported that ongoing HCV infection is a significant pretreatment predictor with 5-FU and IFN-α for early response and survival of patients with advanced hepatocellular carcinoma (HCC). We predicted that the interaction of HCV core protein with HAX-1 may modulate 5-FU-induced p53-mediated apoptosis. Therefore, the aim of the present study was to investigate the importance of the association between HCV core protein and HAX-1 in the regulation of p53, control on cell proliferation and downstream caspase activation upon 5-FU treatment. We have observed that the induction of growth inhibition and caspase activation in HCV core protein-expressing hepatocytes rely on a pathway that includes the action of p53 and HAX-1.
MATERIALS AND METHODS
Cells and transfections.
HepG2, Huh7, and Hep3B cells were transfected with a mammalian expression vector (pcDNA3) containing HCV core gene under the control of a cytomegalovirus promoter (pcDNA3core) or a mammalian expression vector (pcDNA3) containing HCV full-length (FL) gene, mutated at NS5b region, under the control of a cytomegalovirus promoter (pCI-neo-HCV FL) plasmid DNA using Lipofectamine 2000 (Life Technologies, Inc.), and stable cell colonies were selected using neomycin as previously described (1). Pooled cells transfected with HCV genomic region were used in subsequent studies to avoid artifactual results from clonal variation. Parental HepG2 cells transfected with empty vector were used in parallel as a control. Transfected HepG2 cells were maintained in Dulbecco modified Eagle medium, containing 10% fetal calf serum and a lower dose of the selection antibiotic (400 μg of G418/ml). Cells were transiently transfected with small interfering RNA (siRNA) to HAX-1 (sc-43366, a pool of three target-specific 20- to 25-nucleotide siRNAs designed to knockdown gene expression; Santa Cruz Biotechnology, Inc., CA) using Lipofectamine 2000, whenever necessary, and used for analysis within 2 to 3 days.
IHH were generated and maintained in SABM medium (Lonza, MD) supplemented with 5% heat inactivated fetal calf serum as previously described (25). HCV (genotype 1a, clone H77) were grown in cell culture as recently described (9).
Antibodies.
Commercially available anti-caspase-9 or -7, anti-HA, anti-p53 (Santa Cruz), anti-caspase-2 (R&D Systems), anti-HAX-1 (Covance, CA), and anti-human actin coupled to horseradish peroxidase (Sigma, MO) antibodies were procured.
PCA.
The interaction between cellular proteins and HCV core protein was studied by protein fragment complementation assay (PCA) as described earlier (28, 29). HCV core protein packages the viral RNA genome to form a nucleocapsid. In addition to its function as a structural protein, core protein is involved in the regulation of cellular transcription, virus-induced transformation, and pathogenesis. To gain insights into cellular functions of the core protein, we used a cDNA library screening strategy, based on a PCA using yellow fluorescent protein (YFP) as a reporter. The assay combines a simple cell-based cDNA-screening approach (interactions of a “bait” protein of interest with “prey” cDNA products) with specific functional assays that use the same system and provide initial validation of the cDNA products as being biologically relevant. Human liver cDNA library was inserted upstream of 5′ end of YFP fragment (EYFP1-h-liver library). HCV core gene was subcloned downstream of the 3′ end of the YFP fragment (YFP2-core191). The reagents for cloning of liver cDNA and HCV core gene into YFP fragments were kindly provided by S. W. Michnick (Universite de Montreal, Canada). Briefly, cells were plated in 150-mm dishes 24 h before transfection. Cells were transfected using Lipofectamine 2000 at ca. 90% confluence, with YFP1-cDNA library and YFP2-core. One day after transfection, cells were harvested by gentle pipetting with 0.1 mM EDTA-containing phosphate-buffered saline (PBS), and cells were washed and suspended in DMEM containing 5% fetal bovine serum. Approximately 2 × 107 cells were harvested and subjected to the sorting. COS-7 cells transfected with empty vector and YFP1- and YFP2-leucine zipper expression vectors were used for negative and positive control for adjusting conditions for cell sorter. Fluorescence-activated cell sorting (FACS) analysis of COS cells that were cotransfected with these expression plasmids suggested that ∼0.2% of the total cell number gave rise to YFP fluorescence. Cell sorting was performed to sort the 0.2% fluorescent cells with large-scale transfection and YFP1-cDNA in the YFP-positive population was extracted from sorted cells with DNeasy kit (Qiagen). Extracted plasmid coding cDNA was transformed, and approximately 4 × 104 clones were isolated onto LB-ampicillin plates. The isolated cDNA from some of the clones were used for further verification of YFP fluorescence by FACS analyses in a 96-well format (Guava Co.). The cDNAs were sequenced to identify cellular genes and examined to understand the cellular function of HCV core protein.
Fluorescence and confocal microscopy.
Formaldehyde (4%) fixed cells were incubated with anti-HCV core (monoclonal antibody 017; Virogen, MA) or anti-HAX-1 (Santa Cruz) antibody, and stained with a secondary antibody conjugated with fluorochrome (Molecular Probes, Carlsbad, CA). Cells were stained with MitoTracker for mitochondria, and DAPI (4′,6′-diamidino-2-phenylindole) for nuclear staining (Molecular Probes). After staining, cells were mounted for confocal microscopy (Olympus FV1000). Whenever necessary, the images were merged digitally to monitor colocalization in which two different colors produce a distinct color, whereas physically separate signals retain their individual colors.
In vivo coimmunoprecipitation assay.
HAX-1 cDNA was kindly provided by Venkat S. R. K. Yedavalli (National Institute of Allergy and Infectious Diseases, MD). HepG2 cells were transfected with FLAG-HCV core and/or a HA-HAX-1 plasmid DNA construct using Lipofectamine 2000. Empty vector DNA was used in the place of core plasmid DNA as a control. Cells were lysed after 24 h of transfection with TNTG buffer (30 mM Tris [pH 8.0], 150 mM NaCl, 1% TritonX-100, 10% glycerol) supplemented with a cocktail of protease inhibitors. After sonication, cell debris was removed by centrifugation. Clear cell lysates were mixed with anti-hemagglutinin (anti-HA) antibody (for HA-HAX-1 interaction) and protein G-agarose beads overnight at 4°C. The beads were washed with TNTG buffer and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for Western blotting.
Western blot analysis.
Cells were washed with PBS and lysed in TNTG buffer. After a freeze-thaw, cell debris was removed by centrifugation, and clear lysates were subjected to SDS-PAGE. Proteins were transferred onto nitrocellulose membrane, incubated with specific antibody, and identified by enhanced chemiluminescence (Pierce Chemical Company, IL). Cellular actin was detected similarly for relative quantitation of proteins in each lane.
Real-time PCR.
A quantitative real-time PCR analysis was performed for HAX-1 by using specific primers (5′-AGCCCAAATCCTATTTCA-3′ [sense] and 5′-CATGGCCCCAGTTCACTATT-3′ [antisense]). Isolation of RNA from HepG2 cells was performed with a RNA isolation kit (Purescript; Gentra Systems, MN) according to the manufacturer's instructions. cDNA synthesis was carried out by using random hexamers and Thermoscript II RNase H reverse transcriptase (Invitrogen). The expression of HAX-1 in HepG2 cells in the presence or absence of HAX-1 siRNA (25 μM) was analyzed by quantitative real-time PCR using Power SYBR green PCR Master Mix (Applied Biosystems, CA). HAX-1 mRNA expression was normalized against GAPDH (glyceraldehyde-3-phosphate dehydrogenase), and the expression level was calculated by using the “delta delta threshold cycle” (ΔΔCT) method.
5-FU-induced inhibition of cell proliferation and caspase activation.
5-FU (Sigma) was used at different doses to determine its cell growth inhibitory effect. Approximately 5 × 104 cells were exposed to 5-FU before harvesting. To determine the level of 5-FU-induced growth inhibition, control HepG2 cells or cells stably transfected with the HCV core or FL gene were treated with various doses of 5-FU, and cell viability was determined using the tetrazolium compound MTS (Promega, WI). Briefly, cells (∼3 × 103/well) transfected with or without 25 μM siRNA to HAX-1 was seeded in triplicate wells in a 96-well plate. After 48 h, cells were treated with different doses of 5-FU (50, 250, 750, or 1,250 μM) or left untreated in a final volume of 0.2 ml and incubated for 48 h. Cells were washed with PBS, and 0.1 ml of fresh medium containing 0.02 ml of a 5-ng/ml solution of MTS was added to each well. After 2 h of incubation at 37°C, the quantity of released formazan product, as a function of living cells, was measured at 490 nm by using a 96-well multiscanner autoreader (MR 5000; Dynatech, VA) with the extraction buffer serving as a blank. The apoptotic sensitivity of cells stably transfected with HCV core or FL cDNA to 5-FU was determined from the caspase-9, -2, and -7 status by Western blot analysis with specific antibodies.
RESULTS
HAX-1 interacts with HCV core as determined by PCA.
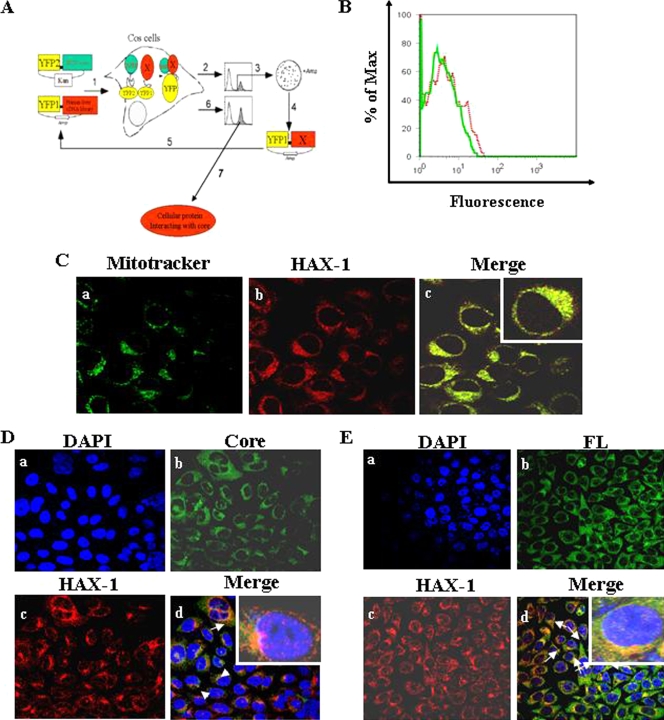
We used cDNA library screening, based on a PCA using YFP as a reporter (Fig. 1A), to further gain insights into the mechanism of cellular functions of the HCV core protein. FACS analysis of cells cotransfected with the expression plasmids was performed to determine positive YFP fluorescence (Fig. 1B). Cell sorting was performed for YFP fluorescent cells with large-scale transfection. We initially identified 34 positive clones. Plasmid DNA was isolated from 17 clones, amplified, and sequenced for confirmation of the positive interaction. On further testing of these clones for interaction, three clones were found to specifically interact with HCV core and not with other heterologous protein baits. All three clones were sequenced and analyzed by the BLAST program. Sequence analysis revealed that these isolates represent an independent overlapping cDNA with homology to HAX-1. The remaining clones identified from the PCA assay are under investigations and will be examined in separate studies. Approximately 4 × 104 cells (0.2% of total cell number) were sorted, and ca. 70% purity of YFP-positive cells was archived. The process was repeated twice to obtain specific interactive proteins. Among a number of other interacting clones identified from PCA, we focused on HAX-1 to understand its interaction with HCV core protein and functional relevance.
FIG. 1.
(A) Schematic diagram of isolation of cellular protein interacting with HCV core by PCA. A human liver cDNA library was fused to fragment 1 of YFP (YFP[1]-cDNA library) and HCV core cDNA to fragment 2 (YFP2-core), in mammalian expression vectors harboring Escherichia coli selection markers ampicillin (Amp) and kanamycin (Kan), respectively. In the first step (step 1) COS-7 cells were cotransfected with core “bait” and human liver cDNA library “prey” fusions, and a physical interaction between the bait and a prey protein induces the folding and reconstitution of YFP from its fragments, generating fluorescence. Positive clones were collected by FACS (step 2) and plasmid that codes human liver cDNA library. The DNA was extracted from the pools and transformed into E. coli grown on ampicillin plates to select only for plasmids harboring cDNA (step 3). Clones were picked up, plasmids were extracted (step 4), and interaction of individual proteins with core was reconfirmed by cotransfecting COS-7 cells with the core fusion and individual cDNA fusions (step 5) and detection by FACS (step 6). Reconfirmed cDNAs were sequenced for identification of the cellular gene and further examined (step 7). (B) The interaction of HAX-1 protein with HCV core is shown from cotransfection of cells with the core and HAX-1 cDNA YFP fusions and detection by FACS. A dotted red line represents the HAX-1 clone, and a solid green line represent results from the negative control. The data were analyzed using FlowJo software (Treestar). The percentage of Max represents the number of events normalized according to the FlowJo algorithm. (C) Localization of HAX-1 into mitochondria. HepG2 control cells stained with MitoTracker green (a), HAX-1-red (b), and merge of immunofluorescence (c) are shown. The inset in the merged immunofluorescence image shows a higher magnification of a single cell. (D) Colocalization of HAX-1 and HCV core protein by confocal microscopy. HAX-1 gene was introduced into HepG2-HCVcore stable transfectants. Cells were stained with DAPI for nuclear staining (a), antibody to core tagged with Alexa green (b), and antibody to HAX-1 tagged with Alexa red (c). (d) Colocalization of merged green (b) and red (c) colors. The inset in the merged immunofluorescence image shows a higher magnification of a single cell. (E) Results from similar experiment in panel D are shown using HAX-1 introduced HepG2-HCV FL stable transfectants. Arrows show colocalization of core and HAX-1 proteins. The inset in the merged immunofluorescence image shows a higher magnification of a single cell.
HAX-1 colocalizes with HCV core protein.
HAX-1 normally localizes into mitochondria (8, 39). We examined the subcellular localization of HAX-1 in HepG2 cells after staining with a MitoTracker dye. Cells were fixed and stained with an antibody to HAX-1 (Fig. 1C). Cells were immunolabeled with MitoTracker (Fig. 1Ca). A speckled localization pattern of HAX-1 was observed when stained with an anti-HAX-1 antibody (Fig. 1Cb). HAX-1 also exhibited mitochondrial localization as the two colors from different fluorochromes merged (Fig. 1Cc). Thus, confocal microscopy confirmed the localization of HAX-1 to mitochondria. HAX-1 expression and localization into mitochondria in HepG2-core stable transfectants was also examined by immunofluorescence (Fig. 1D). Cell nuclei were stained with DAPI (Fig. 1Da). The expression of core and HAX-1 proteins was observed by immunofluorescence with specific antibodies (Fig. 1Db and c, respectively). Merging of these two distinct fluorochrome images further suggested colocalization of HCV core and HAX-1 proteins (Fig. 1Dd). Similarly, HepG2-FL stable transfectants displayed colocalization of core and HAX-1 proteins (Fig. 1Ed).
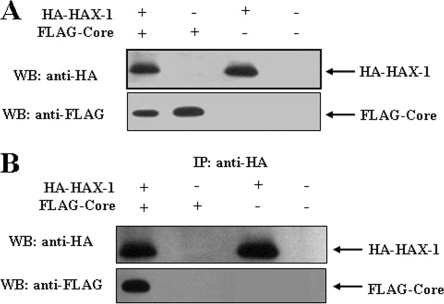
Detection of HAX-1-HCV core complexes in vivo.
HepG2 cells were transiently transfected with HA-HAX-1 and/or FLAG-core, or empty vector as a negative control. The expression of the respective protein(s) was verified in the cell lysates by Western blot analysis with specific antibody (Fig. 2A). A coimmunoprecipitation assay was performed using lysates of HepG2 cells cotransfected with HA-HAX-1 and/or FLAG-core or empty vector as a negative control to verify the in vivo association of HAX-1 and HCV core protein. Cell lysates were immunoprecipitated with a monoclonal antibody to HA and separated by SDS-PAGE. The presence of FLAG-Core protein was detected by immunoblotting with an antibody to FLAG (Fig. 2B). Therefore, coimmunoprecipitation results suggested an association of HAX-1 and HCV core proteins in HepG2 cells.
FIG. 2.
Coimmunoprecipitation of HAX-1 and HCV core proteins. HepG2 cells were transfected with plasmid DNAs encoding HA-HAX1 and/or FLAG-core. Cells were separately transfected with empty vector for use as a negative control. (A) Cell lysates were analyzed for HAX-1 and HCV core protein expression by Western blotting with epitope tag-specific antibodies. (B) Cell lysates were immunoprecipitated with an antibody to HA and probed with FLAG- or HA-specific antibody to detect core or HAX-1. Positions of the protein bands are indicated by arrows on the right.
HAX-1 sensitizes growth inhibition by 5-FU in HCV protein-expressing HepG2 cells.
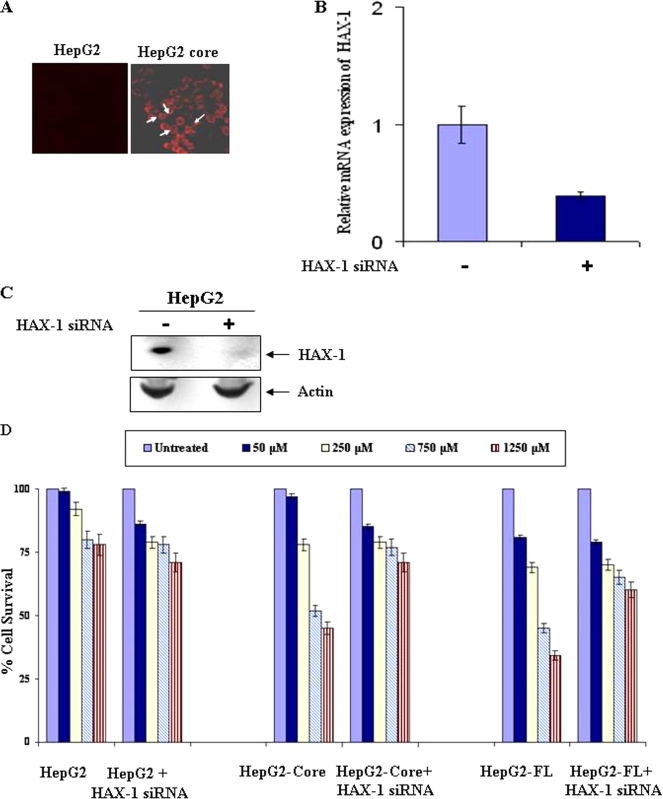
To determine the effect of 5-FU on HepG2 core-expressing cells, we first confirmed the presence of HCV core protein in HepG2 core- or FL gene-expressing cells by immunofluorescence. HepG2 stable transfectants displayed HCV core protein expression by immunofluorescence (Fig. 3A). Similar observations were also made with HepG2-FL stable transfectants (figure not shown). The inhibition of endogenous HAX-1 in HepG2 cells upon introduction of specific or unrelated siRNA was verified by quantitative real-time PCR analysis. A significant level of HAX-1 mRNA (∼60%) inhibition in HepG2 cells was observed after the introduction of siRNA (Fig. 3B). Inhibition of HAX-1 expression at the protein level was also observed in HepG2 cells transfected with HAX-1 siRNA (Fig. 3C).
FIG. 3.
Stable transfectants of HepG2 cells expressing HCV core protein associate with endogenous HAX-1 and sensitize 5-FU-induced growth inhibition. (A) Detection of HCV core protein by immunofluorescence in HepG2 stably transfected with core gene. The results from empty vector-transfected HepG2 cells are shown as a negative control. (B) HAX-1 mRNA expression was analyzed by real-time PCR assay in the presence or absence of siRNA to HAX-1. GAPDH gene expression was used to normalize HAX-1 mRNA expression. (C) Western blot analysis showing the HAX-1 expression status in the presence or absence of HAX-1 siRNAs. (D) 5-FU-mediated growth inhibition of HepG2 cells stably transfected with HCV core or FL upon introduction of siRNA to HAX-1. Cell proliferation after 5-FU treatment was measured by MTS (Promega) assay using the supplier's protocol. The proliferation of untreated cells was arbitrarily considered as 100% and compared to 5-FU-treated cells. The results from three different experiments are shown as bar diagrams, with the standard errors indicated.
Different doses of 5-FU (50, 250, 750, and 1,250 μM) were used to determine the dose-dependent growth inhibition of control HepG2 cells and HepG2 cells stably transfected with HCV core- or FL gene constructs in comparison to untreated cells. HepG2 cells displayed a modest reduction (<25%) in cell viability in the presence of 750 to 1,250 μM 5-FU (Fig. 3C), whereas, under similar conditions, HCV core or FL transfected cells exhibited an enhanced level of 5-FU-induced cytotoxicity (≥50%). Interestingly, the introduction of HAX-1-specific siRNA into HCV core-expressing cells resulted in the level of cell death observed (∼25%) with the 5-FU treatment of control cells. Thus, it appears that HCV core-expressing hepatocytes are sensitized to the induction of 5-FU-mediated cell death via a mechanism that relies upon the cooperative or complementary action of HAX-1 and HCV core protein.
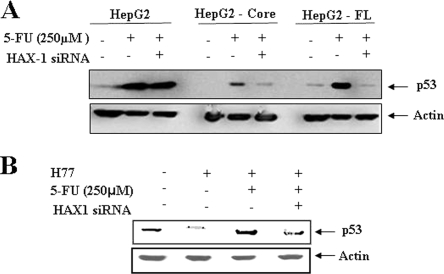
Suppression of HAX-1 inhibits p53 activation in 5-FU-treated cells.
We have shown earlier that HCV core protein represses p53 at the transcriptional level (27). The DNA-damaging drug, 5-FU, functions in a p53-dependent manner (2). When the p53 is activated by DNA damage, it stimulates the transcription of its target genes, which then induce cell cycle arrest and apoptosis. However, little is known about the relationship of p53 activation with HAX-1. To gain insight into this, we examined the interrelationship between p53 and HAX-1 upon 5-FU treatment. HepG2 cells stably transfected with HCV core or FL gene were treated with 5-FU. Parental HepG2 cells were similarly treated for comparison. Alteration of p53 status in cells treated in combination with siRNA to HAX-1 was also analyzed. The results suggested that the status of p53 in parental HepG2 cells was not significantly altered in the presence of siRNA to HAX-1 after 5-FU treatment (Fig. 4A). On the other hand, an increase in p53 was observed in HCV core-expressing hepatocytes in response to 5-FU treatment compared to untreated HepG2-core cells. Interestingly, the reduction of HAX-1 in core-expressing cells by siRNA prior to 5-FU treatment led to the loss of p53 enhancement in response to treatment. Similar observations were also made using HepG2 cells transfected with HCV FL gene. In contrast, the suppression of HAX-1 in parental HepG2 cells by siRNA did not lead to a significant alteration of p53, indicating that HAX-1 itself cannot potentiate the accumulation of p53. IHH is a hepatic cell line derived from primary tissues immortalized in response to the introduction of HCV core plasmid DNA (3, 27). Similar results on p53 status were also observed using HCV (clone H77) grown in IHH (Fig. 4B). Thus, our observations indicated that although HCV core suppresses the basal levels of p53, it can potentiate the activity of 5-FU through the induction of p53, and subsequent cell death, via a mechanism that relies upon the expression of HAX-1.
FIG. 4.
siRNA to HAX-1 inhibits p53 upregulation in 5-FU treated cells. (A) Western blot analysis with specific antibodies for determination of levels of total p53 status in HepG2 cells expressing HCV protein(s) after treatment with siRNA to HAX-1 and/or 5-FU. Cellular actin was used as an internal control for comparison of protein load in each lane. (B) Similar analysis for total p53 level in cell culture grown HCV genotype 1a (clone H77)-infected IHH is shown. The molecular weights of the specific protein bands were verified from the positions of prestained molecular weight markers (Cambrex). Cellular actin was used as an internal control for comparison of the protein load in each lane.
p53 induces the expression of Bax, a proapoptotic member of the Bcl-2 family of proteins, while also acting to downregulate the antiapoptotic Bcl-2 protein. We investigated the potential for HCV core protein in modulating members of the Bcl-2 family (Bax, BclxL) in 5-FU-treated cells by Western blot analysis. Our results did not suggest a significant alteration in the level of Bax expression after 5-FU treatment or the introduction of siRNA to HAX-1 compared to control cells (data not shown). Similarly, the level of BclxL protein under these conditions remained at a similar level. These results indicated that 5-FU treatment does not alter Bax or BclxL, and introduction of siRNA to HAX-1 does not have a significant effect.
Suppression of HAX-1 by siRNA inhibits 5-FU-mediated caspase-2 and -7 cleavages.
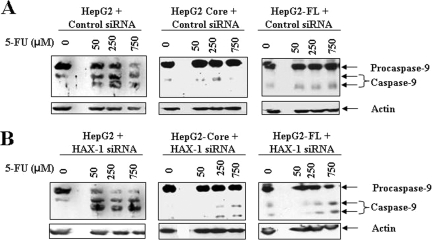
Since caspase-9 interacts with HAX-1 (6, 32) we first determined whether caspase-9 activation was induced by 5-FU in HCV core protein-expressing cells. Our results indicated that procaspase-9 was cleaved in HepG2 control cells when incubated with different doses (50 to 750 μM) of 5-FU (Fig. 5). Suppression of HAX-1 by siRNA did not significantly increase procaspase-9 cleavage in 5-FU-treated hepatocytes compared to cells treated with unrelated siRNA as a negative control. These results suggested that sustained HAX-1 expression does not have a significant effect upon caspase-9 activation in HepG2 cells. However, in HCV core- or FL gene-expressing hepatocytes, 5-FU did not significantly activate caspase-9. Suppression of HAX-1 by siRNA had a minor effect on caspase-9 activation in HepG2 core- or FL gene-expressing cells. These results indicated that caspase-9 may not have a significant contribution toward 5-FU-mediated growth inhibition of HepG2 cells stably transfected with HCV core or FL gene.
FIG. 5.
HAX-1 suppression by siRNA does not activate caspase-9 in 5-FU treated HCV protein-expressing cells. The caspase-9 status after treatment with different doses of 5-FU in parental HepG2, HepG2-core, and HepG2-FL gene transfected cells was determined by Western blot analysis (A). Introduction of siRNA to HAX-1 in control HepG2 cells did not alter cleavage of caspase-9 to its signature polypeptide of 45 kDa compared to inactivation status of caspase-9 in cells expressing HCV proteins (B).
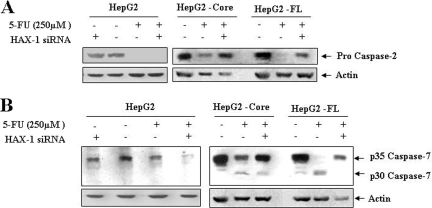
To delineate the involvement of other caspases in 5-FU mediated apoptosis of HCV core- or FL gene-expressing HepG2 cells, further analysis was performed. The results indicated a 5-FU-dependent increase in caspase-2 and -7 activities in HepG2 core- or FL gene-expressing cells, and inhibition of caspase activity was observed for cells transfected with siRNA to HAX-1 (Fig. 6). However, in control HepG2 cells, suppression of HAX-1 was not sufficient to block caspase activation. Activation of caspase-2 was indicated by the disappearance of a 55-kDa procaspase band. Similarly, processing of the caspase-7 was indicated by the disappearance of a 35-kDa procaspase-7 band. Therefore, our findings indicate a potential role for caspase-2 as an effector caspase. The activation of caspase-2 occurs in a high-molecular-weight complex containing RAIDD (for RIP-associated ICH1/CED3 homologous protein with DD) and death domain protein PIDD (for p53-inducible protein with a DD), whose expression is regulated by p53 (15, 17, 36, 41). We do not know whether the caspase-2 regulatory function of p53 primarily occurs during normal or apoptotic conditions, but our study suggests that a p53/caspase-2 apoptotic axis may play an important role for cell death induced by 5-FU in HCV protein-expressing hepatocytes.
FIG. 6.
HAX-1 suppression by siRNA inhibits caspase-2 and -7 activation in 5-FU-treated HCV protein-expressing cells. Caspase-2 status after 5-FU treatment of parental HepG2, HepG2-core, or FL gene transfected cells in the presence or absence of siRNA to HAX-1 (A). Similar analyses were performed for caspase-7 using specific antibodies (B).
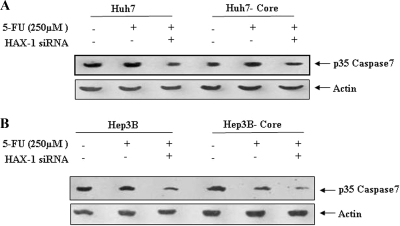
To further investigate the involvement of p53 in caspase-mediated apoptosis of HCV core-expressing cells, we analyzed 5-FU-mediated caspase-7 activation in the presence or absence of HAX-1 siRNA in p53 nonfunctional (Huh7) or null (Hep3B) cells stably transfected with HCV core gene (Fig. 7). The introduction of HAX-1 siRNA in p53 defective cell transfectants marginally enhanced caspase-7 activation, unlike p53 normal HepG2 transfectants, indicating a protective benefit for HAX-1 not associated with p53-mediated cell death. Further, it is apparent that HCV core protein alone cannot mediate 5-FU-associated caspase-7 activation. Therefore, our results suggested that 5-FU-mediated cell death is enhanced in the presence of HCV core and HAX-1 protein and requires the presence of functional p53.
FIG. 7.
HAX-1 suppression by siRNA does not inhibit caspase-7 activation in 5-FU-treated HCV core protein-expressing Huh7 (A) or Hep3B (B) cells. The caspase-7 status after 5-FU treatment of parental or core transfected hepatocytes in the presence or absence of siRNA to HAX-1 (25 μM) is shown.
DISCUSSION
We have identified HAX-1 as a novel HCV core binding protein from a mammalian two-hybrid system. Association of HCV core protein with HAX-1 was verified by confocal microscopy and coimmunoprecipitation experiments. Our study further suggested that 5-FU-induced growth inhibition is enhanced in the presence of HAX-1 in HCV core protein-expressing HepG2 cells. The mechanisms involved p53 and caspase-2 and -7 activations in association with HAX-1 and was reduced upon knockdown of HAX-1.
The tumor suppressor gene p53, known to be activated by DNA damage, is induced by anticancer drugs such as 5-FU. Combination therapy of PEG/IFN-α and 5-FU elicits DNA damage, elevates p53 protein expression, and leads to enhanced apoptosis of hepatocellular carcinoma cells in vivo (2, 22). p53 disruption renders cells strikingly resistant to the effects of the 5-FU. We selected the HepG2 cell line, since its p53 molecule is functional and does not bear mutations. We found that while 5-FU-treated HepG2 cells had a significantly elevated p53 level, HCV core- or FL gene-expressing cells exhibited a reduction of p53 in the presence of HAX-1 siRNA. 5-FU therapy can effectively treat patients with advanced HCC (12, 37). We do not know how HAX-1 contributes to p53 increase upon 5-FU treatment, and this may be addressed by further investigations.
HAX-1 concentrates to mitochondria. Upon interaction with other cellular proteins, its subcellular localization is redistributed, and this may play an important role in modulating different biological activities (19). Vafiadaki et al. (38) reported that an association of HAX-1 with phospholamban, a key regulator of contractibility in the heart, leads to redistribution and colocalization of HAX-1 with phospholamban at the endoplasmic reticulum. This association enhances the protective effects of HAX-1 displayed against hypoxia/reoxygenation-induced cell death. HAX-1 is suggested to avert cardiac myocyte cell death by blocking the biological activation of caspase-9 (6, 32). siRNA directed against HAX-1 increases the sensitivity of cardiac myocytes to death signals by increased caspase-9 activity. However, HAX-1 may not exclusively interact with caspase-9 and may also exert an influence upon members of the Bcl-2 gene family to influence mitochondrial function. HAX-1 may prevent cell death and postmitochondrial defects, leading to caspase-9 activation by preventing the actions of Bax, Bak, Bad, and Bim at the level of the mitochondria or the endoplasmic reticulum. However, our data from HepG2 cells with or without introduction of siRNA to HAX-1 did not display a significant difference in the activation status of caspase-9 upon 5-FU treatment. Further, 5-FU treatment did not significantly change the Bax or BclxL level. We have shown previously that HCV core inhibits caspase-9 activation, which may have implication for inhibition of apoptotic cell death (18). Our study also suggests that HepG2 cells expressing HCV core or FL gene are sensitized to 5-FU-mediated cell death in the absence of caspase-9 activation.
Procaspase-2 is present constitutively in the nucleus (33) and has also been found in the cytosol, in mitochondria, and in the Golgi system (20). Apart from a premitochondrial function leading to cytochrome c release and the processing of caspase-9 (30), caspase-2 may directly interact with the mitochondria, and this interaction occurs independently of its proteolytic activity (31). The importance of p53 for caspase-2 activation was observed in 5-FU-treated human colorectal cancer cells (HCT116) (39). In the present study, reduction of HAX-1 by specific siRNA inhibits 5-FU-mediated caspase-2 and caspase-7 activation. We also have observed that HCV core protein does not play a role in 5-FU-mediated caspase-7 activation in the absence of a functional p53 in Hep3B or Huh-7 cells, further supporting a role of p53 in this process.
The sensitization of hepatocytes to 5-FU-mediated growth inhibition is likely to be linked with the association of HCV core and HAX-1. Uka et al. (37) recently reported that ongoing HCV infection may be a significant pretreatment prediction of early response and survival of patients with advanced HCC when treated with 5-FU and IFN. Our results on the mechanistic role of 5-FU upon HCV core-expressing hepatocytes are in line with the pretreatment predictor. In summary, we have identified an association between HCV core protein and cellular HAX-1 protein. This association inhibited the ability of HCV core protein to modulate p53 expression in the presence of 5-FU treatment, leading to an enhanced level of death in cells expressing HCV proteins by an as-yet-unidentified mechanism. Our work highlights the rationale for 5-FU treatment of HCV-infected hepatocytes. Further elucidation of the mechanisms of hepatocyte growth regulation by 5-FU may provide the mode of action as an important chemotherapeutic agent against HCV-related liver cancer.
Acknowledgments
We thank S. W. Michnick, Universite de Montreal, Montreal, Quebec, Canada, for providing reagents for protein-fragment complementation assays; Venkat S. R. K. Yedavalli, NIAID, Bethesda, MD, for the HAX-1 cDNA; and Lin Cowick for preparation of the manuscript.
This study was supported by research grant DK080812 (R.R.) from the National Institutes of Health and funding from Saint Louis University.
Footnotes
Published ahead of print on 15 July 2009.
REFERENCES
- 1.Basu, A., K. Meyer, R. B. Ray, and R. Ray. 2002. Hepatitis C virus core protein is necessary for the maintenance of immortalized human hepatocyte. Virology 298:53-62. [DOI] [PubMed] [Google Scholar]
- 2.Bunz, F., P. M. Hwang, C. Torrance, T. Waldman, Y. Zhang, L. Dillehay, J. Williams, C., Lengauer, K. W. Kinzler, and B. Vogelstein. 1999. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J. Clin. Investig. 104:263-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gallagher, A. R., A. Cedzich, N. Gretz, S. Somlo, and R. Witzgall. 2000. The polycystic kidney disease protein PKD2 interacts with HAX-1, a protein associated with the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 97:4017-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green, D. R., and J. C. Reed. 1998. Mitochondria and apoptosis. Science 281:1309-1312. [DOI] [PubMed] [Google Scholar]
- 5.Hagiwara, S., M. Kudo, T. Nakatani, Y. Sakaguchi, M. Nagashima, N. Fukuta, M. Kimura, S. Hayakawa, and H. Munakata. 2007. Combination therapy with PEG-IFN-alpha and 5-FU inhibits HepG2 tumour cell growth in nude mice by apoptosis of p53. Br. J. Cancer 97:1532-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han, Y., Y. S. Chen, Z. Liu, N. Bodyak, D. Rigor, E. Bisping, W. T. Pu, and P. M. Kang. 2006. Overexpression of HAX-1 protects cardiac myocytes from apoptosis through caspase-9 inhibition. Circ. Res. 99:415-423. [DOI] [PubMed] [Google Scholar]
- 7.Jiang, Y., W. Zhang, K. Kondo, J. M. Klco, J. M., T. B. St Martin, M. R. Dufault, S. L. Madden, W. G. Kaelin, Jr., and M. Nacht. 2003. Gene expression profiling in a renal cell carcinoma cell line: dissecting VHL and hypoxia-dependent pathways. Mol. Cancer Res. 1:453-462. [PubMed] [Google Scholar]
- 8.Johnson, T. M., Z. X. Yu, V. J. Ferrans, R. A. Lowenstein, and T. Finkel. 1996. Reactive oxygen species are downstream mediators of p53-dependent apoptosis. Proc. Natl. Acad. Sci. USA 93:11848-11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanda, T., A. Basu, R. Steele, T. Wakita, J. S. Ryerse, R. Ray, and R. B. Ray. 2006. Generation of infectious hepatitis C virus in immortalized human hepatocytes. J. Virol. 80:4633-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kao, C.-F., S.-Yi. Chen, J.-Y. Chen. and Y.-H. Wu. Lee. 2004. Modulation of p53 transcription regulatory activity and posttranslational modification by hepatitis C virus core protein. Oncogene 23:2472-2483. [DOI] [PubMed] [Google Scholar]
- 11.Kawaguchi, Y., K. Nakajima, M. Igarashi, T. Morita, M. Tanaka, M. Suzuki, A. Yokoyama, G. Matsuda, K. Kato, M. Kanamori, and K. Hirai. 2000. Interaction of Epstein-Barr virus nuclear antigen leader protein (EBNA-LP) with HS1-associated protein X-1: implication of cytoplasmic function of EBNA-LP. J. Virol. 74:10104-10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurokohchi, K., K. Takaguchi, K. Kita, T. Masaki, and S. Kuriyama. 2005. Successful treatment of advanced hepatocellular carcinoma by combined administration of 5-fluorouracil and pegylated interferon-alpha. World J. Gastroenterol. 11:5401-5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lauber, K., H. A. Appel, S. F. Schlosser, M. Gregor, K. Schulze-Osthoff, and S. Wesselborg. 2001. The adapter protein apoptotic protease-activating factor-1 (Apaf-1) is proteolytically processed during apoptosis. J. Biol. Chem. 276:29772-29781. [DOI] [PubMed] [Google Scholar]
- 14.Li, W. B., J. Feng, S. M. Geng, P. Y. Zhang, X. N. Yan, G. Hu, C. Q. Zhang, and B. J. Shi. 2009. Induction of apoptosis by Hax-1 siRNA in melanoma cells. Cell Biol. Int. 33:548-554. [DOI] [PubMed] [Google Scholar]
- 15.Lin, Y., W. Ma, and S. Benchimol. 2000. Pidd a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nature Genet. 26:122-127. [DOI] [PubMed] [Google Scholar]
- 16.Majumder, M., A. K. Ghosh, R. Steele, R. Ray, and R. B. Ray. 2001. Hepatitis C virus NS5A physically associates with p53 and regulates p21/waf1 gene expression in a p53-dependent manner. J. Virol. 75:1401-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirmohammadsadegh A., U. Tartler, G. Michel, A. Baer, M. Walz, R. Wolf, T. Ruzicka, and U. R. Hengge. 2003. HAX-1, identified by differential display reverse transcription polymerase chain reaction, is overexpressed in lesional psoriasis. J. Investig. Dermatol. 120:1045-1051. [DOI] [PubMed] [Google Scholar]
- 18.Meyer, K., A. Basu, K. Saito, R. B. Ray, and R. Ray. 2005. Inhibition of hepatitis C virus core protein expression in immortalized human hepatocytes induces cytochrome c independent increase in Apaf-1 and caspase-9 activation for cell death. Virology 336:198-207. [DOI] [PubMed] [Google Scholar]
- 19.Oberndorfer, I., D. Schmid, R. Geisberger, G. Achatz-Staussberger, R. Crameri, M. Lamers, and G. Achatz. 2006. HS1-associated protein X-1 interacts with membrane-bound IgE: impact on receptor-mediated internalization. J. Immunol. 177:1139-1145. [DOI] [PubMed] [Google Scholar]
- 20.O'Reilly, L. A., P. Ekert, N. Harvey, V. Marsden, L. Cullen, D. L. Vaux, G. Hacker, C. Magnusson, M. Pakusch, F. Cecconi, K., Kuida, A. Strasser, D. C. Huang, and S. Kumar. 2002. Caspase-2 is not required for thymocyte or neuronal apoptosis even though cleavage of caspase-2 is dependent on both Apaf-1 and caspase-9. Cell Death Differ. 9:832-841. [DOI] [PubMed] [Google Scholar]
- 21.Ota, H., H. Nagano, M. Sakon, H. Eguchi, M. Kondo, T. Yamamoto, M. Nakamura, B. Damdinsuren, H. Wada, S. Marubashi, A. Miyamoto, K. Dono, K. Umeshita, S. Nakamori, K. Wakasa, and M. Monden. 2005. Treatment of hepatocellular carcinoma with major portal vein thrombosis by combined therapy with subcutaneous interferon-alpha and intra-arterial 5-fluorouracil; role of type 1 interferon receptor expression. Br. J. Cancer 93:557-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker, W. B., and Y. C. Cheng. 1990. Metabolism and mechanism of action of 5-fluorouracil. Pharmacol. Ther. 48:381-395. [DOI] [PubMed] [Google Scholar]
- 23.Polyak, K., Y. Xia, J. L. Zweier, K. Polyak, Y. Xia, and Zweier. 1997. A model for p53-induced apoptosis. Nature 389:300-305. [DOI] [PubMed] [Google Scholar]
- 24.Radhika, V., D. Onesime, J. H. Ha, and N. Dhanasekaran. 2004. Gα13 stimulates cell migration through cortactin-interacting protein HAX-1. J. Biol. Chem. 279:49406-49413. [DOI] [PubMed] [Google Scholar]
- 25.Ray, R. B., K. Meyer, and R. Ray. 2007. Hepatitis C virus core protein promotes immortalization of primary human hepatocytes. Virology 271:193-204. [DOI] [PubMed] [Google Scholar]
- 26.Ray, R. B., and R. Ray. 2001. Hepatitis C virus core protein: intriguing properties and functional relevance. FEMS Microbiol. Lett. 202:149-156. [DOI] [PubMed] [Google Scholar]
- 27.Ray, R. B., R. Steele, K. Meyer, and R. Ray. 1997. Transcriptional repression of p53 promoter by hepatitis C virus core protein. J. Biol. Chem. 272:10983-10986. [DOI] [PubMed] [Google Scholar]
- 28.Remy, I., and S. W. Michnick. 2004. A cDNA library functional screening strategy based on fluorescent protein complementation assays to identify novel components of signaling pathways. Methods 32:381-388. [DOI] [PubMed] [Google Scholar]
- 29.Remy, I., and S. W. Michnick. 2004. Mapping biochemical networks with protein-fragment complementation assays. Methods Mol. Biol. 261:411-426. [DOI] [PubMed] [Google Scholar]
- 30.Robertson, J. D., M. Enoksson, M. Suomela, R. Zhivotovsky, and S. Orrenius. 2002. Caspase-2 acts upstream of mitochondria to promote cytochrome c release during etoposide-induced apoptosis. J. Biol. Chem. 277:29803-29809. [DOI] [PubMed] [Google Scholar]
- 31.Robertson, J. D., V. Gogvadze, A. Kropotov, H. Vakifahmetoglu, R. Zhivotovsky, and S. Orrenius. 2004. Processed caspase-2 can induce mitochondria-mediated apoptosis independently of its enzymatic activity. EMBO Rep. 5:643-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaw, J., and L. A. Kirshenbaum. 2006. HAX-1 represses postmitochondrial caspase-9 activation and cell death during hypoxia-reoxygenation. Circ. Res. 99:336-338. [DOI] [PubMed] [Google Scholar]
- 33.Shikama, Y. U. M., T. Miyashita., and M. Yamada. 2001. Comprehensive studies on subcellular localizations and cell death-inducing activities of eight GFP-tagged apoptosis-related caspases. Exp. Cell Res. 264:315-325. [DOI] [PubMed] [Google Scholar]
- 34.Slee, E. A., M. T. Harte, R. M. Kluck, B. B. Wolf, C. A. Casiano, D. D. Newmeyer, H. G. Wang, J. C. Reed, D. W. Nicholson, E. S. Alnemri, D. R. Green, and S. J. Martin. 1999. Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J. Cell Biol. 144:281-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki, Y., C. Demoliere, D. Kitamura, H. Takeshita, U. Deuschle, and T. Watanabe. 1997. HAX-1, a novel intracellular protein, localized on mitochondria, directly associates with HS1, a substrate of Src family tyrosine kinases. J. Immunol. 158:2736-2744. [PubMed] [Google Scholar]
- 36.Tinel, A., and J. Tschopp. 2004. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 304:843-846. [DOI] [PubMed] [Google Scholar]
- 37.Uka, K., H. Aikata, S. Takaki, D. Miki, T. Kawaoka, S. C. Jeong, S. Takahashi, N. Toyota, K. Ito, and K. Chayama. 2007. Pretreatment predictor of response, time to progression, and survival to intraarterial 5-fluorouracil/interferon combination therapy in patients with advanced hepatocellular carcinoma. J. Gastroenterol. 42:845-853. [DOI] [PubMed] [Google Scholar]
- 38.Vafiadaki, E., D. Sanoudou, D. A. Arvanitis, D. H. Catino, E. G. Kranias, and A. Kontrogianni-Konstantopoulos. 2007. Phospholamban interacts with HAX-1, a mitochondrial protein with anti-apoptotic function. J. Mol. Biol. 367:65-79. [DOI] [PubMed] [Google Scholar]
- 39.Vakifahmetoglu, H., M. Olsson, S. Orrenius, and B. Zhivotovsky. 2006. Functional connection between p53 and caspase-2 is essential for apoptosis induced by DNA damage. Oncogene 25:5683-5692. [DOI] [PubMed] [Google Scholar]
- 40.Vayssiere, J. L., P. X. Petit, Y. Risler, and B. Mignotte. 1994. Commitment to apoptosis is associated with changes in mitochondrial biogenesis and activity in cell lines conditionally immortalized with simian virus 40. Proc. Natl. Acad. Sci. USA 91:11752-11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Velculescu, V. E., L. Zhang, B. Vogelstein, and K. W. Kinzler. 1995. Serial analysis of gene expression. Science 270:484-487. [DOI] [PubMed] [Google Scholar]
- 42.Yamaguchi, H., J. Wyckoff, and J. Condeelis. 2005. Cell migration in tumors. Curr. Opin. Cell Biol. 17:559-564. [DOI] [PubMed] [Google Scholar]