Abstract
The hepatitis C virus (HCV) core protein is known to modulate apoptosis and contribute to viral replication and pathogenesis. In this study, we have identified a Bcl-2 homology 3 (BH3) domain in the core protein that is essential for its proapoptotic property. Coimmunoprecipitation experiments showed that the core protein interacts specifically with the human myeloid cell factor 1 (Mcl-1), a prosurvival member of the Bcl-2 family, but not with other prosurvival members (Bcl-XL and Bcl-w). Moreover, the overexpression of Mcl-1 protects against core-induced apoptosis. By using peptide mimetics, core was found to release cytochrome c from isolated mitochondria when complemented with Bad. Thus, core is a bona fide BH3-only protein having properties similar to those of Noxa, a BH3-only member of the Bcl-2 family that binds preferentially to Mcl-1. There are three critical hydrophobic residues in the BH3 domain of the core protein, and they are essential for the proapoptotic property of the core protein. Furthermore, the genotype 1b core protein is more effective than the genotype 2a core protein in inducing apoptosis due to a single-amino-acid difference at one of these hydrophobic residues (residue 119). Replacing this residue in the J6/JFH-1 infectious clone (genotype 2a) with the corresponding amino acid in the genotype 1b core protein produced a mutant virus, J6/JFH-1(V119L), which induced significantly higher levels of apoptosis in the infected cells than the parental J6/JFH-1 virus. Furthermore, the core protein of J6/JFH-1(V119L), but not that of J6/JFH-1, interacted with Mcl-1 in virus-infected cells. Taken together, the core protein is a novel BH3-only viral homologue that contributes to the induction of apoptosis during HCV infection.
Hepatitis C virus (HCV), a positive-stranded RNA virus of the family Flaviviridae, is the major cause of non-A, non-B hepatitis worldwide. The HCV genome encodes a precursor polyprotein of ∼3,000 amino acids (aa) that is processed cotranslationally and posttranslationally to give rise to viral structural and nonstructural proteins (2). The core protein is encoded by the N-terminal portion of the HCV precursor polyprotein and cleaved from the polyprotein by cellular signal peptidase to give the immature form of the core protein (aa 1 to 191). This then is further cleaved by membrane-associated signal peptide peptidase to give the mature core protein, whose C terminus is not precisely known but lies between residues 170 and 179 (see reviews in references 33, 42, and 52). The mature core protein is thought to constitute the HCV capsid and is the predominant form detected in virus particles purified from the sera of patients with chronic HCV infection (42, 74). A recent paper also reported that the maturation of the core protein is required for the production of HCV using the JFH-1 infectious clone (65).
Besides its role in the encapsidation of viral RNA, the core protein has been found to interfere with many cellular pathways, including cell signaling, transcriptional activation, lipid metabolism, carcinogenesis, and apoptosis (see reviews in references 33, 42, and 52). As the regulation of apoptosis during viral infection is an important determinant in the struggle between virus and host for survival, many viruses encode viral proteins that can regulate apoptosis in the infected host cells and manipulate this process to their advantage. In the case of HCV, the mechanisms by which the virus maintains viral persistence and promotes hepatocellular carcinoma are not well understood, but several HCV proteins have the ability to modulate apoptosis (see recent reviews in references 20 and 28). In particular, the core protein has been shown to modulate apoptosis, and it seems that the core protein can inhibit as well as promote apoptosis, depending on the death stimuli and types of cells used (3, 9, 13, 25, 36, 40, 49, 53, 54, 57, 60, 76).
In this study, we characterized one of the mechanisms by which the mature form of the core protein from a genotype 1b strain induces apoptosis in Huh7 cells. Following the experimental designs used in previous studies (29, 38, 40, 55, 72), the mature form of the core protein is assumed to be constituted by residues 1 to 173 of the HCV precursor polyprotein, and this shall be referred to as the core protein in this study. Here, we demonstrate that the core protein contains a functional Bcl-2 homology 3 (BH3) domain that is essential for its proapoptotic property and ability to interact with human myeloid cell factor 1 (Mcl-1), a prosurvival member of the Bcl-2 family (31). Detailed molecular analysis and infection studies using the J6/JFH-1 infectious clone showed that the core protein is a bona fide BH3-only protein that contributes to the induction of apoptosis during HCV infection by mimicking Noxa and interfering with the prosurvival function of Mcl-1.
MATERIALS AND METHODS
Construction of plasmids.
Expression plasmids for the wild-type core protein and mutants were generated by PCR using Titanium Taq DNA polymerase (Clontech Laboratories Inc., Palo Alto, CA). Two plasmids containing full-length HCV genomes were used as templates. The first one is a 1b strain cloned in Singapore (59), and the second is the JFH-1 clone, which is a 2a strain (68). All sequences were confirmed by sequencing performed by the core facilities at the Institute of Molecular and Cell Biology, Singapore. The pXJ40flag vector is used so that a flag epitope is fused to the N terminus of the core protein, and this allows the comparison of protein expression levels with an anti-flag antibody.
Transient transfections, CaspACE fluorometric assay, and Western blot analysis.
Transient transfections of Huh7 cells were performed using Lipofectamine reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Approximately 16 h after transfection, the activation of caspase-3 was quantified by using a CaspACE fluorometric assay system from Promega Corporation (Madison, WI) as previously described (63).
Western blot analysis was performed as previously described (64). The primary antibodies (anti-myc monoclonal and anti-myc and anti-Mcl-1 polyclonal [Santa Cruz Biotechnology, Santa Cruz, CA], anti-Mcl-1 monoclonal [Calbiochem, La Jolla, CA], anti-actin monoclonal, anti-Hsp-60 monoclonal, anti-flag monoclonal and polyclonal [Sigma, St. Louis, MO], anti-poly[ADP-ribose] polymerase [PARP] polyclonal [Cell Signaling Technology Inc., Beverly, MA], anti-cytochrome c monoclonal [BD PharMingen, BD Biosciences, San Jose, CA], and anti-Noxa [Imgenex, San Diego, CA]) were purchased. Anti-core protein monoclonal antibody (clone 2H9; a kind gift from T. Wakita, Department of Virology II, National Institute of Infectious Diseases, Tokyo, Japan) was used to detect the core protein of HCV (68).
Coimmunoprecipitation experiments.
For the coimmunoprecipitation experiments, each 6-cm dish of cells was resuspended in 200 μl of immunoprecipitation (IP) buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 0.5% NP-40, 0.5% deoxycholic acid, 0.005% sodium dodecyl sulfate [SDS], and 1 mM phenylmethylsulfonyl fluoride) and subjected to freeze-thawing six times. Anti-flag monoclonal antibody conjugated to Sepharose beads (Sigma) were added to 150 μl of the lysates, and the mixture was subjected to end-over-end mixing at 4°C for 6 h. Beads were washed four times with cold IP buffer, and then 15 μl of Laemmli's SDS buffer was added and the samples were boiled at 100°C for 5 min to release the immunocomplexes. Samples were separated by SDS-polyacrylamide gel electrophoresis and subjected to Western blot analysis.
Alternatively, rabbit anti-Mcl-1 polyclonal antibody was used to immunoprecipitate endogenous Mcl-1 protein. In this case, 7 μg of antibody (either anti-Mcl-1 or anti-hemagglutinin [HA] polyclonal antibody [Santa Cruz Biotechnology]) was added to the lysates obtained from two dishes of cells and allowed to mix for 1 h at room temperature. Protein A agarose beads (Roche, Indianapolis, IN) were added, and the mixture was subjected to end-over-end mixing at 4°C overnight. The coimmunoprecipitated proteins then were detected as described above.
Quantification of autoradiographs.
An imaging densitometer (Bio-Rad, Hercules, CA) was used for the quantification of the intensities of specific bands on autoradiographs.
In vitro cytochrome c release assay.
For the in vitro cytochrome c release assays, mitochondria were isolated from 293T cells as previously described (22). Briefly, 293T cells were suspended in isolation buffer (320 mM sucrose, 1 mM EDTA, 50 mM HEPES [pH 7.5]) and disrupted by 25 expulsions through a 27-gauge needle. The disrupted cells were spun at 1,000 × g for 10 min to remove cell debris and nuclei. The supernatant was centrifuged at 7,000 × g for 10 min, and the pellet was retained as the heavy membrane fraction containing the mitochondria. The mitochondrion-containing pellets then were resuspended in assay buffer (250 mM sucrose, 2 mM KH2PO4, 5 mM sodium succinate, 25 mM EGTA, and 10 mM HEPES [pH 7.5]) at 0.5 mg/ml. Equal amounts of mitochondria were treated with the indicated peptides for 30 min at room temperature, followed by centrifugation. Both the supernatant and pellet then were subjected to SDS-polyacrylamide gel electrophoresis, followed by Western blot analysis to determine the amount of cytochrome c released from the mitochondria. Hsp-60 was used as a loading control for the pellet.
Synthesis of peptides.
A peptide that corresponds to residues 118 to 149 of the genotype 1b core protein (NLGKVIDTLTCGFADLMGYIPLVGAPLGGAAR) was synthesized and purified to 95% purity (Sigma Genosys, Japan). Peptides containing the BH3 domain of Bad (NLWAAQRYGRELRRMSDEFVDSFKK) or Noxa (VPADLKDECAQLRRIGDKVNLRQKL) also were synthesized and purified to 95% purity (Mimotopes, Clayton, Victoria, Australia).
Generation of recombinant HCV.
The pFL-J6/JFH-1 plasmid encoding the entire viral genome of a chimeric strain of HCV genotype 2a, J6/JFH-1 (37), was kindly provided by C. M. Rice, Center for the Study of Hepatitis C, The Rockefeller University. To generate mutant virus possessing a core protein mutation, a nucleotide substitution was introduced into pFL-J6/JFH-1 by site-directed mutagenesis using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). All PCR-amplified DNA fragments were verified extensively using an ABI PRISM 3100-Avant Genetic Analyzer (Applied Biosystems, Foster City, CA). Each of the plasmids was linearized by XbaI digestion and in vitro transcribed by using T7 RiboMAX (Promega) to generate the full-length viral genomic RNA. The in vitro-transcribed RNA (10 μg) was transfected into Huh7.5 cells by means of electroporation (975 μF, 270 V) using a Gene Pulser (Bio-Rad). The cells then were cultured in complete medium, and the supernatant was propagated as a virus stock. Culture supernatants of uninfected cells served as a control (mock preparation). Virus infectivity was measured by indirect immunofluorescence as previously described (17) and expressed as cell-infecting units (CIU) per milliliter.
Proliferation, caspase-3, and DNA fragmentation assays.
Huh7.5 cells were seeded in 96-well plates at a density of 1.0 × 104 cells per well and cultured overnight. The cells then were infected with recombinant HCV at a multiplicity of infection of 0.1 CIU/cell or with a mock preparation. At different time points postinfection (p.i.), cell viabilities were determined by WST-1 proliferation assays (Roche, Mannheim, Germany) as described previously (17, 48).
Caspase-3 and DNA fragmentation assays also were performed on the infected cells as previously described (17).
HCV RNA quantitation.
To measure intracellular HCV RNA replication levels, total RNA was extracted from the cells using an RNeasy Mini kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. One microgram of total RNA was reverse transcribed using a QuantiTect reverse transcription kit (Qiagen) with random primers and was subjected to quantitative real-time PCR analysis using SYBR premix Ex Taq (Takara Bio, Kyoto, Japan) in a MicroAmp 96-well reaction plate and an ABI PRISM 7000 (Applied Biosystems, Foster, CA). The primers used to amplify an NS5A region of the HCV genome were 5′-AGACGTATTGAGGTCCATGC-3′ (sense) and 5′-CCGCAGCGACGGTGCTGATAG-3′ (antisense). As an internal control, human glyceraldehyde-3-phosphate dehydrogenase expression levels were measured using primers 5′-GCCATCAATGACCCCTTCATT-3′ (sense) and 5′-TCTCGCTCCTGGAAGATGG-3′ (antisense).
Statistical analysis.
Either the two-tailed Student's t test or one-way analysis of variance (using SPSS version 16.0) was applied to evaluate the statistical significance of differences measured from the data sets. P < 0.05 was considered statistically significant.
RESULTS
A BH3-like domain is present in the core protein.
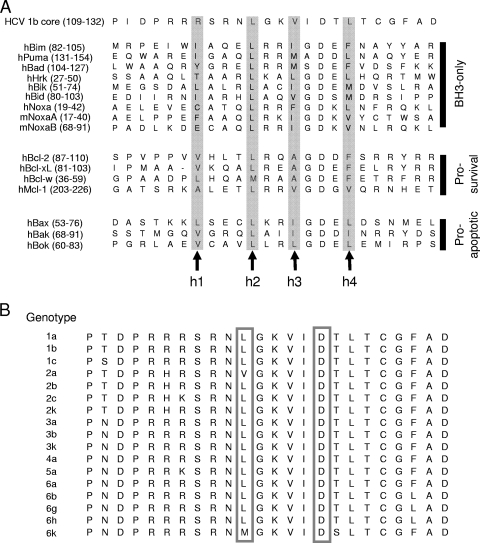
The family of Bcl-2 proteins constitutes one of the biologically important gene products in the regulation of apoptosis (see recent reviews in references 1, 16, 67, and 75). The Bcl-2 proteins may be classified broadly into three classes: prosurvival members containing multiple Bcl-2 homology domains, proapoptotic members containing multiple Bcl-2 homology domains, and proapoptotic members containing the BH3 domain only. The examination of the amino acid sequence of the core protein revealed that there is a BH3-like domain near the C terminus. An alignment of this domain with BH3 domains of the Bcl-2 family of proteins is shown in Fig. 1A. The BH3-like domain of the core protein contains L (residue 119) and D (residue 124) separated by four residues, as in other known BH3 domains. This domain is highly conserved among the major HCV genotypes, with the exception of genotypes 2a and 6k, which have V and M residues at position 119, respectively (Fig. 1B).
FIG. 1.
Identification of a BH3-like domain in the core protein. (A) Alignment of the BH3 domain of the genotype 1b core protein with the BH3 domains of members of the Bcl-2 family. Numbers in parentheses represent the positions of amino acid residues of the respective proteins. The four hydrophobic amino acids that make critical contacts with residues in the BH3 recognition grooves present on the surfaces of the prosurvival Bcl-2 family proteins are indicated as h1 to h4. (B) Alignment of the core protein (residues 109 to 132) of different genotypes. The consensus sequences for these genotypes were obtained from http://hcv.lanl.gov/content/hcv-index. The highly conserved L and D residues, at positions 119 and 124 of the genotype 1b core protein, respectively, in BH3 domains are boxed.
The BH3 domain of the core protein is essential for the induction of apoptosis and its interaction with human Mcl-1.
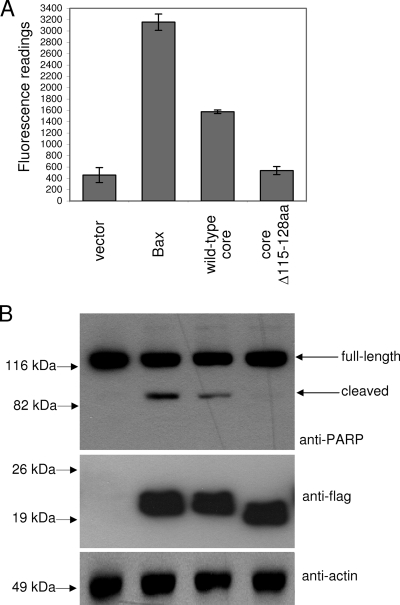
The overexpression of the core protein (with a flag epitope at the N terminus) in Huh7 cells, via the transient transfection of a cDNA expression plasmid containing the genotype 1b core protein gene, induced significant levels of apoptosis as determined by the activation of caspase-3, which is a hallmark of apoptosis (Fig. 2A). The deletion of the BH3 domain in the core protein (designated coreΔ115-128aa) abolished its proapoptotic property, indicating that this domain is essential for the induction of apoptosis. Consistently, the cleavage of endogenous PARP, a substrate of activated caspase-3, was clearly observed in Huh7 cells expressing the wild-type core protein but not in those expressing coreΔ115-128aa (Fig. 2B).
FIG. 2.
Induction of apoptosis by the overexpression of the core protein in Huh7 cells. (A) A CaspACE fluorometric assay system from Promega Corporation (Madison, WI) was used to measure the activation of caspase-3, which is a hallmark of apoptosis, in Huh7 cells that were transfected with vector only, a classical apoptosis inducer (Bax), the wild-type core protein, and a core protein mutant lacking the putative BH3 domain (coreΔ115-128aa). All experiments were performed in triplicate, and the average values with standard deviations are plotted. (B) Western blot analysis also was performed to determine the cleavage of endogenous PARP, which is a substrate of activated caspase-3, from 116 to 83 kDa (top). Similarly, the expression levels of the different proteins were determined using anti-flag antibody (middle). The amounts of total cell lysates loaded were verified by measuring the levels of endogenous actin (bottom).
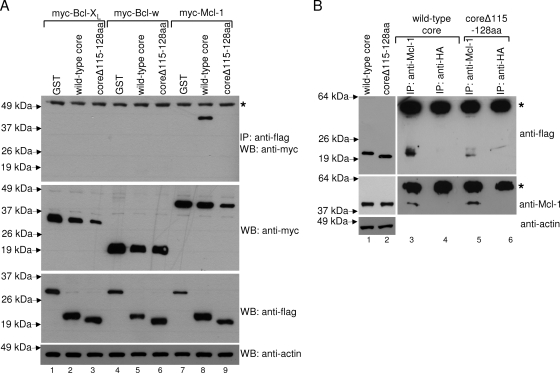
To understand how the core protein modulates the function of the Bcl-2 family of proteins, coimmunoprecipitation experiments were performed to determine if the core protein can interact with representative prosurvival members of the Bcl-2 family. As shown in the top panel of Fig. 3A, Mcl-1 was specifically coimmunoprecipitated by the core protein (lane 8) but not by an irrelevant protein, glutathione S-transferase (GST) (lane 7). The BH3 domain of the core protein is essential for its interaction with Mcl-1, as coreΔ115-128aa failed to coimmunoprecipitate Mcl-1 (lane 9). These results indicate that the core protein induces apoptosis by interfering directly with the prosurvival function of Mcl-1. In contrast, no significant interaction was observed between the core protein and two other prosurvival proteins, Bcl-XL and Bcl-w (lanes 1 to 6).
FIG. 3.
Interaction of the core protein with prosurvival members of the Bcl-2 family determined by coimmunoprecipitation experiments. (A) Huh7 cells were transfected with cDNA constructs for expressing flag-GST (negative control), flag-core, or flag-coreΔ115-128aa, and myc-tagged prosurvival members of the Bcl-2 family (myc-Bcl-XL [lanes 1 to 3], myc-Bcl-w [lanes 4 to 6], and myc-Mcl-1 [lanes 7 to 9]). The cells were harvested at ∼16 h posttransfection, lysed, and subjected to IP with anti-flag monoclonal antibody conjugated to Sepharose beads. The amount of myc-tagged proteins that coimmunoprecipitated (IP) with the flag-tagged proteins was determined by Western blot analysis (WB) with an anti-myc rabbit polyclonal antibody (top). The amounts of myc-tagged and flag-tagged proteins in the lysates before IP were determined by subjecting aliquots of the lysates to Western blot analysis (middle). The protein marked with an asterisk represents the heavy chain of the antibody used for IP (top), and the amounts of total cell lysates loaded were verified by measuring the levels of endogenous actin (bottom). (B) Huh7 cells were transfected with cDNA constructs for expressing flag-core or flag-coreΔ115-128aa. IP then was performed using anti-Mcl-1 or anti-HA rabbit polyclonal antibodies and protein A agarose beads. The amounts of flag-tagged core protein in the lysates before IP (lanes 1 and 2) or coimmunoprecipitated (lanes 3 to 6) were determined by Western blot analysis with an anti-flag monoclonal antibody (top). Similarly, the amounts of endogenous Mcl-1 in these samples were detected using an anti-Mcl-1 monoclonal antibody (middle). The protein marked with an asterisk represents the heavy chain of the antibody used for IP (top), and the amounts of total cell lysates loaded were verified by measuring the levels of endogenous actin (bottom).
The interaction between the core protein and endogenous Mcl-1 was determined by overexpressing the core protein in Huh7 cells. As shown in Fig. 3B, the core protein was coimmunoprecipitated with endogenous Mcl-1 (lane 3). On the other hand, the core protein was not coimmunoprecipitated when an irrelevant antibody (anti-HA) (lane 4) was used for IP. Consistently with the results in Fig. 3A, only a small amount of coreΔ115-128aa was coimmunoprecipitated with endogenous Mcl-1 (lane 5). To estimate the degree of reduction in the binding of coreΔ115-128aa to endogenous Mcl-1, an imaging densitometer was used to quantify the intensities of specific bands on the autoradiographs obtained in three independent coimmunoprecipitation experiments (see Fig. S1 in the supplemental material). The ratios of signals for the expression of flag-tagged core protein, endogenous Mcl-1, and actin (internal control) all are close to 1 (0.94 to 1.14), indicating that the expression levels of these proteins in the two sets of cells (either transfected with cDNA construct for expressing flag-core or flag-coreΔ115-128aa) were similar. From the three independent experiments, the average amount of coreΔ115-128aa coimmunoprecipitated specifically by the Mcl-1 antibody is 14.2% (± 10.7%) of the amount of wild-type core protein coimmunoprecipitated. This implies that the deletion of the BH3 domain does not completely abolish the interaction between the core protein and endogenous Mcl-1 but reduces the interaction greatly.
Overexpression of Mcl-1 or Bcl-XL prevents core protein-induced apoptosis.
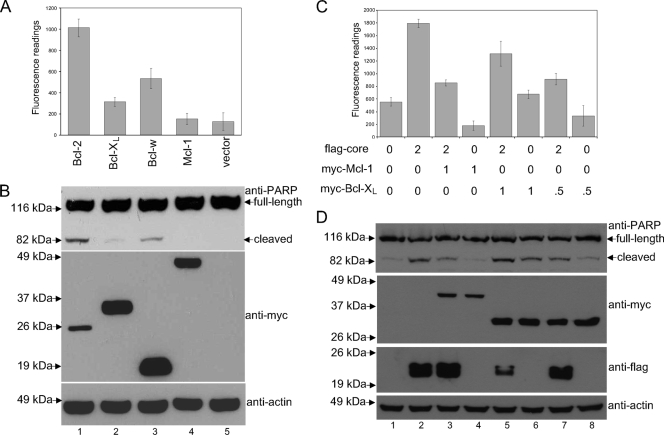
To examine the protective effects of the prosurvival Bcl-2 proteins in Huh7 cells, transfections were performed with plasmids expressing myc-tagged Bcl-2, Bcl-XL, Bcl-w, or Mcl-1 (Fig. 4A and B). Consistent with studies of other cell lines (12, 66), the transient high-level expression of Bcl-2 also caused apoptosis in Huh7 cells (lane 1). Interestingly, the overexpression of Bcl-w also induced a significant level of apoptosis in Huh7 cells (lane 3), and this phenomenon has not been reported previously. Cells overexpressing Bcl-XL, but not Mcl-1, also had a slightly higher level of apoptosis than that of the vector control cells (lanes 2 and 4).
FIG. 4.
Effects of Mcl-1 and Bcl-XL overexpression on the proapoptotic property of the core protein. (A) A CaspACE fluorometric assay system from Promega Corporation (Madison, WI) was used to measure the activation of caspase-3 in Huh7 cells that were transfected with Bcl-2, Bcl-XL, Bcl-w, Mcl-1, or vector only. All experiments were performed in triplicate, and the average values with standard deviations are plotted. (B) Western blot analysis also was performed to determine the cleavage of endogenous PARP (top) and expression levels of the myc-tagged prosurvival members of the Bcl-2 family (middle). The amounts of total cell lysates loaded were verified by measuring the levels of endogenous actin (bottom). (C) A CaspACE fluorometric assay system from Promega Corporation (Madison, WI) was used to measure the activation of caspase-3 in Huh7 cells that were singly transfected with vector, the wild-type core protein, Mcl-1, or Bcl-XL, or that were cotransfected with wild-type core protein and Mcl-1 or Bcl-XL. The amounts of flag-core and myc-Mcl-1 or myc-Bcl-XL DNAs used in each of the transfections are indicated in micrograms. In each transfection, the total amount of DNA was normalized to 3 μg with the addition of empty vector if necessary. All experiments were performed in triplicate, and the average values with standard deviations are plotted. (D) Western blot analysis also was performed to determine the cleavage of endogenous PARP (top) and expression levels of myc-tagged Mcl-1 and Bcl-XL and flag-tagged core protein (middle). The amounts of total cell lysates loaded were verified by measuring the levels of endogenous actin (bottom).
Huh7 cells were cotransfected with plasmids for expressing myc-Mcl-1 and flag-core or myc-Bcl-XL and flag-core. As shown in Fig. 4C and D, the level of apoptosis was significantly reduced in cells expressing both Mcl-1 and the core protein (lane 3) compared to those expressing the core protein only (lane 2). When the same experiment was repeated using Bcl-XL, the level of apoptosis was reduced to a lesser extent (lane 5). However, this may be due to the low level of apoptosis induced by the overexpression of Bcl-XL (lane 6). The level of the core protein expressed in the presence of Bcl-XL also was decreased greatly, but the smaller amount of the core protein expressed still induced a high level of apoptosis (lane 5). However, when a broad caspase inhibitor (z-VAD-fmk) was used, the core protein level in cells coexpressing Bcl-XL increased (see Fig. S2 in the supplemental material), indicating that the transfection efficiencies were similar in the different samples. To resolve this uncertainty, the experiment was repeated with a smaller amount of Bcl-XL plasmid (0.5 μg). Under this condition, the overexpression of Bcl-XL did not induce apoptosis (lane 8), and the level of apoptosis also was reduced in cells expressing both Bcl-XL and the core protein (lane 7) compared to those expressing the core protein only (lane 2). Thus, the results show that the overexpression of either Mcl-1 or Bcl-XL protects against core protein-induced apoptosis.
Bad enhances the ability of the core protein to release cytochrome c from isolated mitochondria.
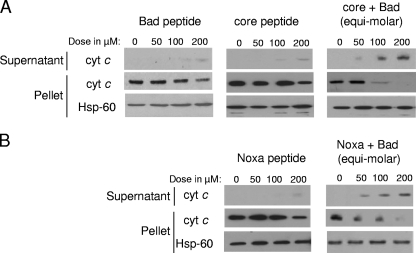
The ability of a core protein peptide, which contains residues 118 to 149 of the genotype 1b core protein, to release cytochrome c from the mitochondria was tested using 293T cells instead of Huh7 cells, as the method for the isolation of mitochondria from 293T cells is well established. The core protein induced apoptosis in 293T cells in the same manner as that in Huh7 cells (see Fig. S3 in the supplemental material). As shown in Fig. 5A, both the Bad and core protein peptides were inefficient in inducing the release of cytochrome c, as only a small amount of cytochrome c was detected in the supernatant from the treated mitochondria when 200 μM of either peptide was used. However, when Bad and core protein peptides were used in combination, the release of cytochrome c was observed at the much lower concentration of 50 μM (consisting of 25 μM Bad peptide and 25 μM core protein peptide). Furthermore, the release of cytochrome c increased in a dose-dependent manner. The amount of cytochrome c left in the treated mitochondria (i.e., pellet) decreased correspondingly, while the amount of control protein, Hsp-60, was not affected.
FIG. 5.
Release of cytochrome c from isolated mitochondria by a combination of the core-BH3 and Bad-BH3 peptides or Noxa-BH3 and Bad-BH3 peptides. (A) Mitochondria isolated from 293T cells were incubated with either Bad peptide, core protein peptide, or a combination of the two peptides in equimolar concentrations. The total amount of peptide used in each experiment is indicated. Following centrifugation, the supernatants and pellets were subjected to Western blot analysis with anti-cytochrome c (cyt c) or anti-Hsp-60 antibodies. (B) The same experiment was repeated with Noxa peptide or a combination of Noxa and Bad peptides in equimolar concentrations.
The same experiment was repeated using the Noxa peptide (Fig. 5B). Consistently with a previous study (11), the Noxa peptide alone was inefficient in inducing the release of cytochrome c, but when it was combined with the Bad peptide, the release was significantly enhanced. The peptide(s) dosage required was similar to the amount required for the core protein and Bad, indicating that the complementation between the core protein and Bad is similar to the complementation between Noxa and Bad. In addition, complementation between the core protein and Bad also was observed when they were coexpressed in Huh7 cells (see Fig. S4 in the supplemental material).
The three hydrophobic residues in the BH3 domain of the core protein are important for apoptosis induction.
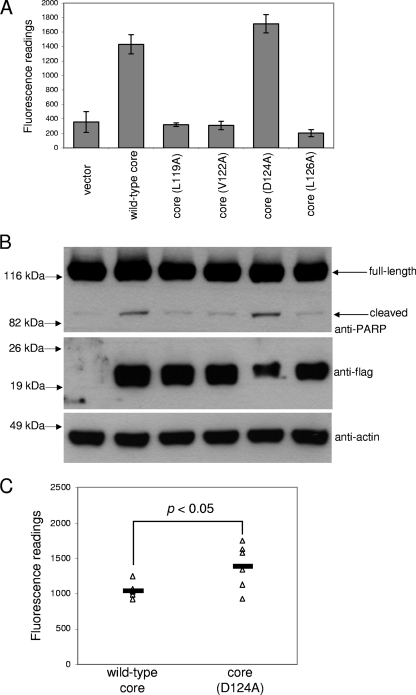
Site-directed mutagenesis and structural studies of the interactions between the prosurvival Bcl-2 proteins and BH3 domains have revealed the mechanism by which BH3 domains are bound to the hydrophobic grooves present on the surfaces of the prosurvival Bcl-2 proteins (see reviews in references 50 and 69). In particular, the BH3 domain usually contains four hydrophobic residues (h1, h2, h3, and h4) (Fig. 1A) that make contacts critical for the stability of the complex. Interestingly, the core protein contains hydrophobic residues at the h2, h3, and h4 positions (Fig. 1A). An alanine substitution experiment was performed to determine if these residues are essential for the proapoptotic property of the core protein (Fig. 6A and B). The results showed that replacement of either L119, V122, or L126 with A completely abolishes the proapoptotic property of the core protein. On the other hand, the replacement of the highly conserved D124 residue with A seems to increase the proapoptotic property of the core protein slightly. The levels of activated caspase-3 induced by the wild-type core protein and the D124A substitution mutant in six independent experiments were compared using the two-tailed Student's t test, and the difference was found to be statistically significant (Fig. 6C). This phenomenon has not been reported for other BH3-only proteins, but there are a few known functional BH3 domains that do not contain D at this position (54, 55).
FIG. 6.
Effects of alanine substitutions on the proapoptotic property of the core protein. (A) A CaspACE fluorometric assay system from Promega Corporation (Madison, WI) was used to measure the activation of caspase-3 in Huh7 cells that were transfected with vector only, wild-type core, or alanine substitution mutants. All experiments were performed in triplicate, and the average values with standard deviations are plotted. (B) Western blot analysis also was performed to determine the cleavage of endogenous PARP (top) and expression levels of the core proteins (middle). The amounts of total cell lysates loaded were verified by measuring the levels of endogenous actin (bottom). (C) The levels of activated caspase-3 induced by the wild-type core protein and the D124A mutant in six independent experiments were compared using the two-tailed Student's t test, and the difference was found to be statistically significant (P < 0.05). The values from each of the experiments are plotted as open triangles, and the average values are plotted as solid lines.
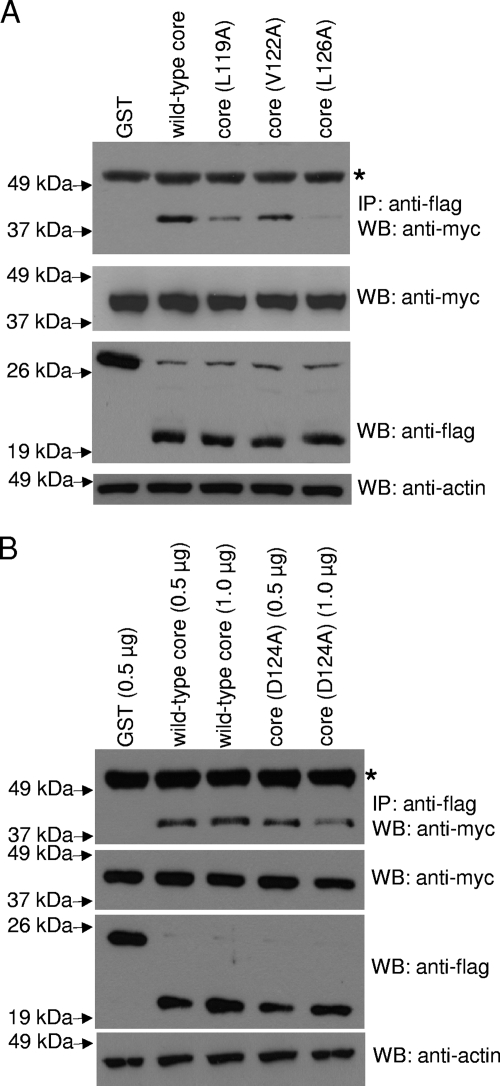
Furthermore, coimmunoprecipitation experiments showed that the L119A, V122A, and L126A substitution mutants have reduced binding to Mcl-1 (Fig. 7A). Similar results were obtained in four independent experiments, and the percentages of binding compared to that of the wild-type core protein were estimated by using an imaging densitometer to measure the intensity of the core protein signals after coimmunoprecipitation. For each experiment, three different autoradiographs (with different exposure times) were used, and the average values are shown in Table S1 in the supplemental material. The average percentages in binding of Mcl-1 to the L119A, V122A, and L126A mutants are 33, 62, and 9% of the binding to the wild-type core protein, respectively. For all three mutants, the reduced interactions with Mcl-1 compared to those of the wild-type core protein are statistically significant (see Table S1 in the supplemental material). As the D124A substitution mutant induced a slightly higher level of apoptosis than the wild-type core protein (Fig. 6), a coimmunoprecipitation experiment also was performed to determine if this mutant can bind Mcl-1. Two different amounts of flag-tagged plasmids (0.5 and 1.0 μg) were used, and the results show that the D124A substitution mutant binds Mcl-1 to an extent similar to that of the wild-type core protein under both conditions (Fig. 7B).
FIG. 7.
Effects of alanine substitutions on the binding of the core protein to Mcl-1. (A) Huh7 cells were transfected with cDNA constructs (1.0 μg) for expressing flag-GST (negative control), flag-tagged wild-type core protein, or single-alanine-substitution mutants (L119A, V122A, and L126A). All cells were cotransfected with myc-tagged Mcl-1 (1.5 μg). (B) Huh7 cells were transfected with cDNA constructs for expressing flag-GST (negative control, 0.5 μg), flag-tagged wild-type core protein (0.5 or 1.0 μg), or single-alanine-substitution mutant D124A (0.5 or 1.0 μg). All cells were cotransfected with myc-tagged Mcl-1 (1.5 μg). Coimmunoprecipitation then was performed as described in the legend to Fig. 3A. The amount of myc-tagged proteins that coimmunoprecipitated (IP) with the flag-tagged proteins was determined by Western blot analysis (WB) with an anti-myc rabbit polyclonal antibody (top). The amounts of myc-tagged and flag-tagged proteins in the lysates before IP were determined by subjecting aliquots of the lysates to Western blot analysis (middle). The amounts of total cell lysates loaded were verified by measuring the levels of endogenous actin (bottom). The protein marked with an asterisk represents the heavy chain of the antibody used for IP (top). Similar results were obtained in four independent experiments, and a representative set of data is presented.
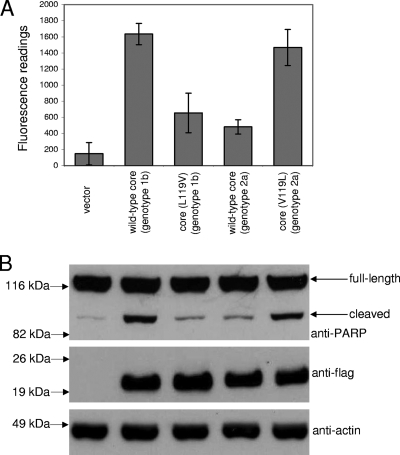
While the V122 and L126 residues, at the h3 and h4 positions, respectively, of the core protein are highly conserved in different genotypes of HCV, the core proteins from genotype 2a strains typically have V instead of L at the h2 position (Fig. 1B). Replacing the L119 of the genotype 1b core protein with V reduced the proapoptotic property of the core protein dramatically (Fig. 8). Interestingly, in all known BH3-only proteins, this position is usually an L residue that is essential for the proapoptotic properties of these proteins (Fig. 1A). The reverse experiment was performed by determining if the core protein of a genotype 2a strain (JFH-1 strain) can induce apoptosis. The overexpression of the genotype 2a core protein induced much less apoptosis than the genotype 1b core protein (Fig. 8). However, replacing the V119 of the genotype 2a core protein with L resulted in a significant increase in apoptosis induction, such that the level was similar to that induced by the genotype 1b core protein (Fig. 8).
FIG. 8.
Comparison of the proapoptotic properties of the core proteins of genotypes 1b and 2a. (A) A CaspACE fluorometric assay system from Promega Corporation (Madison, WI) was used to measure the activation of caspase-3 in Huh7 cells that were transfected with vector only, wild-type core of genotype 1b or 2a, or their substitution mutants. All experiments were performed in triplicate, and the average values with standard deviations are plotted. (B) Western blot analysis also was performed to determine the cleavage of endogenous PARP (top) and expression levels of the core proteins (middle). The amounts of total cell lysates loaded were verified by measuring the levels of endogenous actin (bottom).
A single substitution from V to L at residue 119 in the core protein of the HCV J6/JFH-1 strain is associated with increased abilities to induce apoptosis.
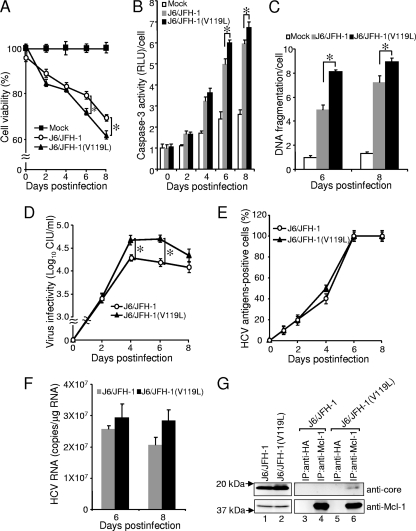
The pFL-J6/JFH-1 plasmid encoding the entire viral genome of a chimeric strain of HCV genotype 2a (J6/JFH-1) can be used to generate infectious HCV (37). In the J6/JFH-1 clone, the core protein contains V at residue 119, just like the JFH-1 clone. A mutant virus, J6/JFH-1(V119L), was generated successfully by replacing the V119 residue with L. Parental J6/JFH-1 and mutant J6/JFH-1(V119L) viruses then were used to infect naïve Huh7.5 cells, and cell viabilities were measured at different time points after infection (Fig. 9A). From day 2 p.i., cells infected by either virus have lower viabilities than mock-infected cells, indicating that the viruses have induced cytopathic effects (CPE). This is consistent with recent observations by us and other researchers (17, 41). Results from days 6 and 8 p.i. show that the J6/JFH-1(V119L) virus induced higher levels of CPE and, therefore, lower levels of cell viability compared to those of the parental J6/JFH-1 virus (Fig. 9A), which is in agreement with the overexpression studies shown in Fig. 8. The CPE is mediated primarily through apoptosis, as indicated by the activation of caspase-3 (Fig. 9B) and DNA fragmentation (Fig. 9C). The production of cell-free infectious virus particles by the J6/JFH-1(V119L) virus also was significantly higher than that produced by the parental J6/JFH-1 virus (Fig. 9D). On the other hand, there was no significant difference in the percentage of HCV-infected cells in the cultures (Fig. 9E) or HCV RNA replication in the cells between the two viruses (Fig. 9F). We next analyzed the possible interaction between endogenous Mcl-1 and the core proteins of either J6/JFH-1 or J6/JFH-1(V119L) in virus-infected cells. As shown in Fig. 9G, the core protein of J6/JFH-1(V119L) was coimmunoprecipitated with Mcl-1 (lane 6). On the other hand, the Mcl-1 interaction of the core protein of J6/JFH-1 was barely detected under the same experimental conditions (lane 4). These results collectively imply the possibility that the V119L mutation of the core protein promotes its interaction with Mcl-1 and is responsible for the increased ability of the virus to induce apoptosis, which favors a higher degree of infectious progeny virus release from the host cell at the late time points of infection compared to that of the parental J6/JFH-1 virus.
FIG. 9.
Comparison of parental J6/JFH-1 and mutant J6/JFH-1(V119L) recombinant viruses. Huh7.5 cells were infected with recombinant HCV at a multiplicity of infection of 0.1 CIU/cell or with a mock preparation, and various assays were performed at different days after infection. (A) Cell viabilities were determined. (B) Caspase-3 activity per cell was determined. (C) The amount of DNA fragmentation per cell was determined. (D) The production of cell-free infectious virus particles was determined. (E) Virus spread in the culture was quantitated. (F) HCV RNA replication was determined by quantitative real-time PCR analysis. (G) Interaction of the core protein with Mcl-1 was determined by coimmunoprecipitation experiments at 3 days p.i. IP was performed using anti-Mcl-1 or anti-HA rabbit polyclonal antibodies and protein A agarose beads. The amounts of the core protein in the lysates before (lanes 1 and 2) and after IP (lanes 3 to 6) were determined by Western blot analysis with an anti-core monoclonal antibody (top). Similarly, the amounts of endogenous Mcl-1 in the samples were determined using an anti-Mcl-1 monoclonal antibody (bottom). Statistical analysis was performed using the one-way analysis of variance to determine if the differences between parental and mutant viruses were statistically significant, and those with P values of <0.05 (marked by asterisks) are considered statistically significant. Data were obtained from three independent experiments, each with triplicate cultures.
DISCUSSION
Besides playing important roles in maintaining homeostasis in healthy cells through the regulation of apoptosis, members of the Bcl-2 family also are involved in viral infections. Indeed, several viruses have been shown to encode homologs of prosurvival Bcl-2 proteins, and these viral proteins act to inhibit apoptosis in infected cells and prevent the premature death of these cells (see reviews in references 14, 26, 51, and 70). Other viral proteins, which can be proapoptotic, prosurvival, or both, do not share any sequence homology with members of the Bcl-2 family but also can modulate apoptosis in the host cells by interfering at different apoptotic checkpoints (see reviews in references 8, 23, 27, and 43).
Unlike the multi-BH domain members, the BH3-only members of the Bcl-2 family contain a single BH3 domain. Although all BH3-only proteins can bind to the hydrophobic grooves on the surfaces of the prosurvival members, recent quantitative measurements have revealed that the affinities of association between different pairs of BH3-only and prosurvival members vary greatly (11, 32). For example, Bim and Puma bind all prosurvival members tested, while Noxa binds strongly only to Mcl-1 and A1. On the other hand, Bad binds much more strongly to Bcl-2, Bcl-XL, and Bcl-w than Mcl-1. Taken together with results from successive studies, it becomes clear that the BH3-only members can be classified into subclasses (see reviews in references 21, 24, 58, and 71). In this study, we demonstrate that the HCV core protein is a BH3-only viral homologue of the Bcl-2 family, and its BH3 domain is essential for the induction of apoptosis (Fig. 1 and 2). In coimmunoprecipitation experiments, the core protein interacted specifically with the prosurvival Mcl-1 protein but not with prosurvival proteins Bcl-XL and Bcl-w (Fig. 3), suggesting that its property is most similar to that of Noxa (11). Consistently, the overexpression of Mcl-1 protects against core protein-induced apoptosis (Fig. 4). However, the overexpression of Bcl-XL also protects against core protein-induced apoptosis (Fig. 4). This may be due to the ability of a high level of Bcl-XL to prevent the complementation between the core protein and endogenous Bad protein, which binds strongly to Bcl-XL (11), as we have observed that a combination of the core protein and Bad peptide mimetics caused efficient cytochrome c release from the mitochondria (Fig. 5). The complementation between Bad and the core protein is similar to that observed between Bad and Noxa, which act in combination to neutralize the two classes of prosurvival proteins, one comprised of Bcl-2, Bcl-XL, and Bcl-w and the other of Mcl-1 and A1 (11). In overexpression studies, the core protein and Noxa also induced comparable levels of apoptosis (see Fig. S3 and S5 in the supplemental material). Taken together, these findings suggest that core can mimic Noxa and interfere directly with the prosurvival function of Mcl-1.
A comparison of the BH3 domain of the core protein to the corresponding domains of other BH3-containing proteins (Fig. 1A) revealed that it contains three out of the four hydrophobic residues that can be accommodated within the hydrophobic pockets of previously described BH3 binding grooves (see reviews in references 50 and 69). Alanine substitution experiments revealed that all three hydrophobic residues in the BH3 domain of the core protein are essential for apoptosis induction (Fig. 6). In coimmunoprecipitation experiments, these alanine substitution mutants also bound Mcl-1 to a lesser extent than the wild-type core protein (Fig. 7A). Since these alanine substitution mutants still can bind Mcl-1, albeit at a lower level than that of the wild-type core protein, it appears that the interactions between these mutants and Mcl-1 are not sufficient to induce apoptosis. In several mutagenesis studies, the interaction between Bcl-2 family members and apoptosis regulation have been observed to be discordant. For example, two mutants of the BH3-only protein Bik, Bik-(43-94) and Bik-(43-120), heterodimerized with prosurvival Bcl-2 and Bcl-XL but were unable to induce efficient cell death (19). A Bad mutant containing an alteration of a critical residue within its BH3 domain, E113 to K, also was found to have significantly reduced apoptotic activity compared to that of wild-type Bad, despite binding to Bcl-2 and/or Bcl-XL to the same extent as wild-type Bad (35). Therefore, the induction of apoptosis by the core protein may be controlled by a critical threshold affinity of binding between the core protein and Mcl-1, or there are contributions from a yet-to-be characterized pathway(s). Two of these residues (V122 and L126) are conserved in the major genotypes of HCV, but residue 119 is a V in genotype 2a (Fig. 1B). When L119 of the genotype 1b core protein was replaced with V, its ability to induce apoptosis was greatly reduced (Fig. 8). Conversely, when V119 of the genotype 2a core protein was replaced with L, its ability to induce apoptosis was greatly enhanced. Thus, the results suggest that the genotype 1b core protein induces apoptosis efficiently via a BH3 domain, while genotype 2a core protein is comparatively less efficient. Another highly conserved residue in the BH3 domain of the core protein is D124. However, the replacement of D124 with A did not reduce the proapoptotic function of the core protein (Fig. 6). Thus far, there are only a few known functional BH3 domains that do not contain D at this position (61, 62). Unlike most BH3-only proteins, the core protein has a charged residue (R115) in the h1 position (Fig. 1A). Interestingly, the second BH3 domain of mouse Noxa (mNoxaB) also has a charged residue (E74) in this position. Indeed, the nuclear magnetic resonance structure of the complex between mouse Mcl-1 and a peptide mimetic of mNoxaB shows that E74 is tolerated at the h1 position because its charged carboxyl group is coordinated by another charged residue, K215, in mouse Mcl-1 (15). However, R115 of the core protein is basic instead of acidic, and how this residue can be accommodated in the hydrophobic groove of Mcl-1 is unclear. Interestingly, replacing the residue at the h1 position (I58) of a novel BimBH3 variant, Bims2A, with A also has little effect on its interaction with Mcl-1 (34). Thus, it appears that the residue in the h1 position is not always involved in the interaction between BH3-only proteins and Mcl-1, but further biophysical and biochemical studies are required to delineate the precise structure-function relationship for the interaction between core and Mcl-1.
To determine if the results from overexpression studies are relevant to the modulation of apoptosis in host cells during HCV infection, the J6/JFH-1-based (genotype 2a) system was used to generate HCV carrying a substitution at residue 119 of the core protein. While the parental wild-type and mutant viruses replicated efficiently in Huh7.5 cells, the J6/JFH-1(V119L) virus (which expresses the core protein with L at the h2 position of the BH3 domain) caused a significantly higher level of apoptosis in the infected cells than the parental J6/JFH-1 virus (which expresses the core protein with V at the h2 position of the BH3 domain) (Fig. 9). This is in good agreement with the overexpression studies and indicates that the BH3 domain of the core protein contributes to the induction of apoptosis in HCV-infected cells. Thus, it appears that core protein-mediated apoptosis during infection by HCV of genotype 2a is less efficient than that of the other genotypes having L at residue 119 of the core protein (Fig. 1B). Coimmunoprecipitation experiments revealed that the core protein of J6/JFH-1(V119L), but not that of J6/JFH-1, interacted with Mcl-l in virus-infected cells (Fig. 9). This result is consistent with the overexpression studies and suggests the possibility that the core protein induces apoptosis, at least partly, through the interaction with Mcl-1 in HCV-infected cells. Interestingly, more progeny virus is released from cells infected with the J6/JFH-1(V119L) virus than by those infected with the parental J6/JFH-1, while there is no difference in the efficiency of infection or amount of HCV replication inside the cells (Fig. 9).
However, it also is apparent that the parental J6/JFH-1 virus still caused a high level of apoptosis in the infected cells, and for the early time points there was no significant difference in the levels of apoptosis induced by the parental J6/JFH-1 virus and the J6/JFH-1(V119L) mutant virus (Fig. 9). This implies that there are other viral factors that contribute to the induction of apoptosis during HCV infection. For example, several nonstructural HCV proteins, like NS3, NS4A, NS5A, and NS5B, can induce apoptosis when they are overexpressed in certain types of cells (see recent reviews in references 20 and 28). In addition, other domains in the core protein have been shown to bind host proteins and may contribute to apoptosis regulation by interfering with different cellular pathways (see reviews in references 33, 42, and 52). For example, the N-terminal domain (aa 1 to 75) of the core protein interacts with Hsp60, leading to the production of reactive oxygen species and enhancement of tumor necrosis factor alpha-mediated apoptosis (30), while a C-terminal domain (aa 153 to 192) facilitates Fas oligomerization and is required for apoptosis induction in Jurkat cells (46). However, the relative contribution of these various factors to apoptosis induction during HCV infection remains to be determined.
We further examined the importance of residue 119 of the core protein in HCV replication. In multiple independent transfection experiments, we observed that the J6/JFH-1 mutant possessing A at position 119 [J6/JFH-1(V119A)] barely replicated in the cells and did not produce any infectious virus particles in the culture supernatants (data not shown). This result suggests the possibility that this single point mutation impairs the interaction of the core protein with other viral and/or cellular protein(s) that is required for HCV RNA replication and infectious virion production. Similarly, the J6/JFH-1 mutants each possessing A at positions 122 [J6/JFH-1(V122A)], 124 [J6/JFH-1(D124A)], or 126 [J6/JFH-1(L126A)] barely replicated in the cells and did not produce any infectious virion in the culture supernatants (data not shown), with the results suggesting an important role(s) for these residues as well as for position 119. In this connection, the essential role for the HCV core protein in infectious virion production recently has been confirmed, and numerous residues required for this role have been identified (47).
By using the JFH-1 infectious clone, recent studies have revealed that the association of the core protein with the lipid droplet (LD) is critical for the production of infectious virus particles (6, 45). Boulant and coworkers reported that there are two amphipathic α-helices in the so-called D2 domain of the core protein (∼118 to 179 aa) (5, 7), and the hydrophobic residues within this domain are critical for the efficient attachment of the core protein to LD (5). Our results showed that residues L119, V122, and L126 of the core protein are essential for the induction of apoptosis, and these residues are found on the hydrophobic face of the first α-helix of the D2 domain. Interestingly, the replacement of L119 with E did not affect LD association, while the replacement of L126 with E significantly reduced LD association (5). The contribution of V122 to LD association was not investigated. Consistently, the J6/JFH-1(V119L) virus, but not the J6/JFH-1(L126A) virus, replicated efficiently to produce infectious virus particles. Since L119 of the genotype 1b core protein, which occupies the crucial h2 position in the BH3 domain, is essential for its proapoptotic property but not for its association with LD, it is clear that the BH3 domain of the core protein is an independent motif that partially overlaps with the LD association domain.
Recently, Makes caterpillars floppy 1 (Mcf1), a bacterial toxin, was reported to contain a BH3-like domain (18). In addition, HBSP, a spliced hepatitis B viral protein, also contains a BH3-like domain (39). Here, we show that the HCV core protein is another BH3-like viral homologue, and it contributes directly to the induction of apoptosis during HCV infection. Our results also reveal that it is a bona fide BH3-only protein that appears to interfere with the prosurvival property of Mcl-1 in a manner similar to that of Noxa. Our observation that the enhanced apoptotic activity of the J6/JFH-1(V119L) virus is correlated with an increase in infectious progeny HCV release seems to be counterintuitive, as many viruses adopted strategies to prevent apoptosis in the infected cells so as to allow viral replication and the packaging of progeny genomes within the cells (14, 26, 51, 70). However, enhanced releases of virus from infected cells that are undergoing apoptosis also have been reported for other viruses, like the infectious bursal disease virus, adenovirus, and Aleutian mink disease parvovirus (4, 44, 73), indicating that apoptosis can be advantageous for viral spreading at the late stages of infection. Future studies to define the precise manner by which the BH3 domain of the core protein modulates apoptosis during infection will provide important insights into HCV replication as well as pathogenesis.
Besides the genotype 1b core protein, the properties of the genotype 1a core protein also have been examined in various studies. The apoptotic property of the genotype 1a core protein has yet to be studied using the JFH-1-based infectious clone system, although previous studies have attributed both prosurvival and proapoptotic properties to it (25, 30, 46, 57). Similar observations also have been described in overexpression studies using the genotype 1b core protein and appear to be dependent on the death stimuli and types of cells used (3, 9, 10, 36, 49, 53, 56, 60, 76). Several studies have identified domains or regions within the core protein that interfere with specific apoptosis pathways. For instance, the interaction of the N-terminal domain (residues 1 to 75) of the genotype 1a core protein with Hsp60 enhanced tumor necrosis factor alpha-mediated apoptosis, while its C-terminal region (residues 153 to 192) is required for Fas ligand-independent apoptosis (30, 46). The genotype 1b core protein (residues 1 to 153) binds to the death domain of FADD, resulting in enhanced apoptosis (76). However, an overlapping domain spanning the first 46 aa of the core protein is involved in ASPP2 interaction, which leads to the inhibition of p53-mediated apoptosis (9). These findings suggest that multiple domains present in the core protein contribute to the modulation of apoptosis via diverse pathways. Therefore, the net apoptotic effect of the core protein may be dependent on the relative strength of its prosurvival and proapoptotic properties. Unlike the genotype 2a core protein, the BH3 domains of the genotype 1b core protein and the genotype 1a core protein share an identical sequence (Fig. 1B) and are expected to function in a similar manner. However, we cannot rule out that there may be differences in the manner in which the core proteins of genotypes 1a and 1b modulate apoptosis during infection. For example, they may be involved in different virus-virus or virus-host interactions. Thus, more studies are needed to understand the contributions of genotype-dependent factors to the regulation of apoptosis during HCV infection.
Supplementary Material
Acknowledgments
We thank T. Wakita for the JFH-1 construct and anti-core monoclonal antibody and C. M. Rice for the J6/JFH-1 construct and Huh7.5 cells.
This work was supported by the Biomedical Research Council of A*STAR (Agency for Science, Technology and Research), Singapore, a Health and Labor Sciences Research Grant from the Ministry of Health, Labor and Welfare, Japan, and the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases, the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Y.-J.T. is an adjunct staff member of the Department of Microbiology at the National University of Singapore.
Footnotes
Published ahead of print on 15 July 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adams, J. M., and S. Cory. 2007. Bcl-2-regulated apoptosis: mechanism and therapeutic potential. Curr. Opin. Immunol. 19:488-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 3.Benali-Furet, N. L., M. Chami, L. Houel, F. De Giorgi, F. Vernejoul, D. Lagorce, L. Buscail, R. Bartenschlager, F. Ichas, R. Rizzuto, and P. Paterlini-Brechot. 2005. Hepatitis C virus core triggers apoptosis in liver cells by inducing ER stress and ER calcium depletion. Oncogene 24:4921-4933. [DOI] [PubMed] [Google Scholar]
- 4.Best, S. M., J. B. Wolfinbarger, and M. E. Bloom. 2002. Caspase activation is required for permissive replication of Aleutian mink disease parvovirus in vitro. Virology 292:224-234. [DOI] [PubMed] [Google Scholar]
- 5.Boulant, S., R. Montserret, R. G. Hope, M. Ratinier, P. Targett-Adams, J. P. Lavergne, F. Penin, and J. McLauchlan. 2006. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J. Biol. Chem. 281:22236-22247. [DOI] [PubMed] [Google Scholar]
- 6.Boulant, S., P. Targett-Adams, and J. McLauchlan. 2007. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J. Gen. Virol. 88:2204-2213. [DOI] [PubMed] [Google Scholar]
- 7.Boulant, S., C. Vanbelle, C. Ebel, F. Penin, and J. P. Lavergne. 2005. Hepatitis C virus core protein is a dimeric alpha-helical protein exhibiting membrane protein features. J. Virol. 79:11353-11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boya, P., A. L. Pauleau, D. Poncet, R. A. Gonzalez-Polo, N. Zamzami, and G. Kroemer. 2004. Viral proteins targeting mitochondria: controlling cell death. Biochim. Biophys. Acta 1659:178-189. [DOI] [PubMed] [Google Scholar]
- 9.Cao, Y., T. Hamada, T. Matsui, T. Date, and K. Iwabuchi. 2004. Hepatitis C virus core protein interacts with p53-binding protein, 53BP2/Bbp/ASPP2, and inhibits p53-mediated apoptosis. Biochem. Biophys. Res. Commun. 315:788-795. [DOI] [PubMed] [Google Scholar]
- 10.Chang, M. L., J. C. Chen, M. Y. Chang, C. T. Yeh, W. P. Lin, C. K. Liang, S. F. Huang, K. N. Dang, C. T. Chiu, and D. Y. Lin. 2008. Acute expression of hepatitis C core protein in adult mouse liver: mitochondrial stress and apoptosis. Scand. J. Gastroenterol. 43:747-755. [DOI] [PubMed] [Google Scholar]
- 11.Chen, L., S. N. Willis, A. Wei, B. J. Smith, J. I. Fletcher, M. G. Hinds, P. M. Colman, C. L. Day, J. M. Adams, and D. C. Huang. 2005. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol. Cell 17:393-403. [DOI] [PubMed] [Google Scholar]
- 12.Cheng, E. H., D. G. Kirsch, R. J. Clem, R. Ravi, M. B. Kastan, A. Bedi, K. Ueno, and J. M. Hardwick. 1997. Conversion of Bcl-2 to a Bax-like death effector by caspases. Science 278:1966-1968. [DOI] [PubMed] [Google Scholar]
- 13.Chou, A. H., H. F. Tsai, Y. Y. Wu, C. Y. Hu, L. H. Hwang, P. I. Hsu, and P. N. Hsu. 2005. Hepatitis C virus core protein modulates TRAIL-mediated apoptosis by enhancing Bid cleavage and activation of mitochondria apoptosis signaling pathway. J. Immunol. 174:2160-2166. [DOI] [PubMed] [Google Scholar]
- 14.Cuconati, A., and E. White. 2002. Viral homologs of BCL-2: role of apoptosis in the regulation of virus infection. Genes Dev. 16:2465-2478. [DOI] [PubMed] [Google Scholar]
- 15.Czabotar, P. E., E. F. Lee, M. F. van Delft, C. L. Day, B. J. Smith, D. C. Huang, W. D. Fairlie, M. G. Hinds, and P. M. Colman. 2007. Structural insights into the degradation of Mcl-1 induced by BH3 domains. Proc. Natl. Acad. Sci. USA 104:6217-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danial, N. N. 2007. BCL-2 family proteins: critical checkpoints of apoptotic cell death. Clin. Cancer Res. 13:7254-7263. [DOI] [PubMed] [Google Scholar]
- 17.Deng, L., T. Adachi, K. Kitayama, Y. Bungyoku, S. Kitazawa, S. Ishido, I. Shoji, and H. Hotta. 2008. Hepatitis C virus infection induces apoptosis through a Bax-triggered, mitochondria-mediated, caspase-3-dependent pathway. J. Virol. 82:10375-10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dowling, A. J., N. R. Waterfield, M. C. Hares, G. Le Goff, C. H. Streuli, and R. H. ffrench-Constant. 2007. The Mcf1 toxin induces apoptosis via the mitochondrial pathway and apoptosis is attenuated by mutation of the BH3-like domain. Cell Microbiol. 9:2470-2484. [DOI] [PubMed] [Google Scholar]
- 19.Elangovan, B., and G. Chinnadurai. 1997. Functional dissection of the pro-apoptotic protein Bik. Heterodimerization with anti-apoptosis proteins is insufficient for induction of cell death. J. Biol. Chem. 272:24494-24498. [DOI] [PubMed] [Google Scholar]
- 20.Fischer, R., T. Baumert, and H. E. Blum. 2007. Hepatitis C virus infection and apoptosis. World J. Gastroenterol. 13:4865-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fletcher, J. I., and D. C. Huang. 2008. Controlling the cell death mediators Bax and Bak: puzzles and conundrums. Cell Cycle 7:39-44. [DOI] [PubMed] [Google Scholar]
- 22.Fu, N. Y., S. K. Sukumaran, and V. C. Yu. 2007. Inhibition of ubiquitin-mediated degradation of MOAP-1 by apoptotic stimuli promotes Bax function in mitochondria. Proc. Natl. Acad. Sci. USA 104:10051-10056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galluzzi, L., C. Brenner, E. Morselli, Z. Touat, and G. Kroemer. 2008. Viral control of mitochondrial apoptosis. PLoS Pathog. 4:e1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Häcker, G., and A. Weber. 2007. BH3-only proteins trigger cytochrome c release, but how? Arch. Biochem. Biophys. 462:150-155. [DOI] [PubMed] [Google Scholar]
- 25.Hahn, C. S., Y. G. Cho, B. S. Kang, I. M. Lester, and Y. S. Hahn. 2000. The HCV core protein acts as a positive regulator of fas-mediated apoptosis in a human lymphoblastoid T cell line. Virology 276:127-137. [DOI] [PubMed] [Google Scholar]
- 26.Hardwick, J. M., and D. S. Bellows. 2003. Viral versus cellular BCL-2 proteins. Cell Death Differ. 10(Suppl. 1):S68-S76. [DOI] [PubMed] [Google Scholar]
- 27.Hay, S., and G. Kannourakis. 2002. A time to kill: viral manipulation of the cell death program. J. Gen. Virol. 83:1547-1564. [DOI] [PubMed] [Google Scholar]
- 28.Herzer, K., M. F. Sprinzl, and P. R. Galle. 2007. Hepatitis viruses: live and let die. Liver Int. 27:293-301. [DOI] [PubMed] [Google Scholar]
- 29.Hope, R. G., and J. McLauchlan. 2000. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J. Gen. Virol. 81:1913-1925. [DOI] [PubMed] [Google Scholar]
- 30.Kang, S. M., S. J. Kim, J. H. Kim, W. Lee, G. W. Kim, K. H. Lee, K. Y. Choi, and J. W. Oh. 2009. Interaction of hepatitis C virus core protein with Hsp60 triggers the production of reactive oxygen species and enhances TNF-alpha-mediated apoptosis. Cancer Lett. 279:230-237. [DOI] [PubMed] [Google Scholar]
- 31.Kozopas, K. M., T. Yang, H. L. Buchan, P. Zhou, and R. W. Craig. 1993. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc. Natl. Acad. Sci. USA 90:3516-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuwana, T., L. Bouchier-Hayes, J. E. Chipuk, C. Bonzon, B. A. Sullivan, D. R. Green, and D. D. Newmeyer. 2005. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol. Cell 17:525-535. [DOI] [PubMed] [Google Scholar]
- 33.Lai, M. M., and C. F. Ware. 2000. Hepatitis C virus core protein: possible roles in viral pathogenesis. Curr. Top. Microbiol. Immunol. 242:117-134. [DOI] [PubMed] [Google Scholar]
- 34.Lee, E. F., P. E. Czabotar, M. F. van Delft, E. M. Michalak, M. J. Boyle, S. N. Willis, H. Puthalakath, P. Bouillet, P. M. Colman, D. C. Huang, and W. D. Fairlie. 2008. A novel BH3 ligand that selectively targets Mcl-1 reveals that apoptosis can proceed without Mcl-1 degradation. J. Cell Biol. 180:341-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, J. W., Y. H. Soung, S. Y. Kim, S. W. Nam, C. J. Kim, Y. G. Cho, J. H. Lee, H. S. Kim, W. S. Park, S. H. Kim, J. Y. Lee, N. J. Yoo, and S. H. Lee. 2004. Inactivating mutations of proapoptotic Bad gene in human colon cancers. Carcinogenesis 25:1371-1376. [DOI] [PubMed] [Google Scholar]
- 36.Lee, S. K., S. O. Park, C. O. Joe, and Y. S. Kim. 2007. Interaction of HCV core protein with 14-3-3epsilon protein releases Bax to activate apoptosis. Biochem. Biophys. Res. Commun. 352:756-762. [DOI] [PubMed] [Google Scholar]
- 37.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 38.Liu, Q., C. Tackney, R. A. Bhat, A. M. Prince, and P. Zhang. 1997. Regulated processing of hepatitis C virus core protein is linked to subcellular localization. J. Virol. 71:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu, Y. W., T. L. Tan, V. Chan, and W. N. Chen. 2006. The HBSP gene is expressed during HBV replication, and its coded BH3-containing spliced viral protein induces apoptosis in HepG2 cells. Biochem. Biophys. Res. Commun. 351:64-70. [DOI] [PubMed] [Google Scholar]
- 40.Marusawa, H., M. Hijikata, T. Chiba, and K. Shimotohno. 1999. Hepatitis C virus core protein inhibits Fas- and tumor necrosis factor alpha-mediated apoptosis via NF-κB activation. J. Virol. 73:4713-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mateu, G., R. O. Donis, T. Wakita, J. Bukh, and A. Grakoui. 2008. Intragenotypic JFH1 based recombinant hepatitis C virus produces high levels of infectious particles but causes increased cell death. Virology 376:397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McLauchlan, J. 2000. Properties of the hepatitis C virus core protein: a structural protein that modulates cellular processes. J. Viral Hepat. 7:2-14. [DOI] [PubMed] [Google Scholar]
- 43.McLean, J. E., A. Ruck, A. Shirazian, F. Pooyaei-Mehr, and Z. F. Zakeri. 2008. Viral manipulation of cell death. Curr. Pharm. Des. 14:198-220. [DOI] [PubMed] [Google Scholar]
- 44.Mi, J., Z. Y. Li, S. Ni, D. Steinwaerder, and A. Lieber. 2001. Induced apoptosis supports spread of adenovirus vectors in tumors. Hum. Gene Ther. 12:1343-1352. [DOI] [PubMed] [Google Scholar]
- 45.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089-1097. [DOI] [PubMed] [Google Scholar]
- 46.Moorman, J. P., D. Prayther, D. McVay, Y. S. Hahn, and C. S. Hahn. 2003. The C-terminal region of hepatitis C core protein is required for Fas-ligand independent apoptosis in Jurkat cells by facilitating Fas oligomerization. Virology 312:320-329. [DOI] [PubMed] [Google Scholar]
- 47.Murray, C. L., C. T. Jones, J. Tassello, and C. M. Rice. 2007. Alanine scanning of the hepatitis C virus core protein reveals numerous residues essential for production of infectious virus. J. Virol. 81:10220-10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nomura-Takigawa, Y., M. Nagano-Fujii, L. Deng, S. Kitazawa, S. Ishido, K. Sada, and H. Hotta. 2006. Non-structural protein 4A of hepatitis C virus accumulates on mitochondria and renders the cells prone to undergoing mitochondria-mediated apoptosis. J. Gen. Virol. 87:1935-1945. [DOI] [PubMed] [Google Scholar]
- 49.Otsuka, M., N. Kato, H. Taniguchi, H. Yoshida, T. Goto, Y. Shiratori, and M. Omata. 2002. Hepatitis C virus core protein inhibits apoptosis via enhanced Bcl-xL expression. Virology 296:84-93. [DOI] [PubMed] [Google Scholar]
- 50.Petros, A. M., E. T. Olejniczak, and S. W. Fesik. 2004. Structural biology of the Bcl-2 family of proteins. Biochim. Biophys. Acta 1644:83-94. [DOI] [PubMed] [Google Scholar]
- 51.Polster, B. M., J. Pevsner, and J. M. Hardwick. 2004. Viral Bcl-2 homologs and their role in virus replication and associated diseases. Biochim. Biophys. Acta 1644:211-227. [DOI] [PubMed] [Google Scholar]
- 52.Ray, R. B., and R. Ray. 2001. Hepatitis C virus core protein: intriguing properties and functional relevance. FEMS Microbiol. Lett. 202:149-156. [DOI] [PubMed] [Google Scholar]
- 53.Realdon, S., M. Gerotto, F. Dal Pero, O. Marin, A. Granato, G. Basso, M. Muraca, and A. Alberti. 2004. Proapoptotic effect of hepatitis C virus CORE protein in transiently transfected cells is enhanced by nuclear localization and is dependent on PKR activation. J. Hepatol. 40:77-85. [DOI] [PubMed] [Google Scholar]
- 54.Ruggieri, A., T. Harada, Y. Matsuura, and T. Miyamura. 1997. Sensitization to Fas-mediated apoptosis by hepatitis C virus core protein. Virology 229:68-76. [DOI] [PubMed] [Google Scholar]
- 55.Sabile, A., G. Perlemuter, F. Bono, K. Kohara, F. Demaugre, M. Kohara, Y. Matsuura, T. Miyamura, C. Brechot, and G. Barba. 1999. Hepatitis C virus core protein binds to apolipoprotein AII and its secretion is modulated by fibrates. Hepatology 30:1064-1076. [DOI] [PubMed] [Google Scholar]
- 56.Sacco, R., T. Tsutsumi, R. Suzuki, M. Otsuka, H. Aizaki, S. Sakamoto, M. Matsuda, N. Seki, Y. Matsuura, T. Miyamura, and T. Suzuki. 2003. Antiapoptotic regulation by hepatitis C virus core protein through up-regulation of inhibitor of caspase-activated DNase. Virology 317:24-35. [DOI] [PubMed] [Google Scholar]
- 57.Saito, K., K. Meyer, R. Warner, A. Basu, R. B. Ray, and R. Ray. 2006. Hepatitis C virus core protein inhibits tumor necrosis factor alpha-mediated apoptosis by a protective effect involving cellular FLICE inhibitory protein. J. Virol. 80:4372-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shibue, T., and T. Taniguchi. 2006. BH3-only proteins: integrated control point of apoptosis. Int. J. Cancer. 119:2036-2043. [DOI] [PubMed] [Google Scholar]
- 59.Soo, H. M., A. Garzino-Demo, W. Hong, Y. H. Tan, Y. J. Tan, P. Y. Goh, S. G. Lim, and S. P. Lim. 2002. Expression of a full-length hepatitis C virus cDNA up-regulates the expression of CC chemokines MCP-1 and RANTES. Virology 303:253-277. [DOI] [PubMed] [Google Scholar]
- 60.Takamatsu, M., T. Fujita, and H. Hotta. 2001. Suppression of serum starvation-induced apoptosis by hepatitis C virus core protein. Kobe J. Med. Sci. 47:97-112. [PubMed] [Google Scholar]
- 61.Tan, K. O., K. M. Tan, S. L. Chan, K. S. Yee, M. Bevort, K. C. Ang, and V. C. Yu. 2001. MAP-1, a novel proapoptotic protein containing a BH3-like motif that associates with Bax through its Bcl-2 homology domains. J. Biol. Chem. 276:2802-2807. [DOI] [PubMed] [Google Scholar]
- 62.Tan, K. O., K. M. Tan, and V. C. Yu. 1999. A novel BH3-like domain in BID is required for intramolecular interaction and autoinhibition of pro-apoptotic activity. J. Biol. Chem. 274:23687-23690. [DOI] [PubMed] [Google Scholar]
- 63.Tan, Y. J., B. C. Fielding, P. Y. Goh, S. Shen, T. H. Tan, S. G. Lim, and W. Hong. 2004. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J. Virol. 78:14043-14047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan, Y. J., E. Teng, S. Shen, T. H. Tan, P. Y. Goh, B. C. Fielding, E. E. Ooi, H. C. Tan, S. G. Lim, and W. Hong. 2004. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 78:6723-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Targett-Adams, P., G. Hope, S. Boulant, and J. McLauchlan. 2008. Maturation of hepatitis C virus core protein by signal peptide peptidase is required for virus production. J. Biol. Chem. 283:16850-16859. [DOI] [PubMed] [Google Scholar]
- 66.Uhlmann, E. J., T. Subramanian, C. A. Vater, R. Lutz, and G. Chinnadurai. 1998. A potent cell death activity associated with transient high level expression of BCL-2. J. Biol. Chem. 273:17926-17932. [DOI] [PubMed] [Google Scholar]
- 67.van Delft, M. F., and D. C. Huang. 2006. How the Bcl-2 family of proteins interact to regulate apoptosis. Cell Res. 16:203-213. [DOI] [PubMed] [Google Scholar]
- 68.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walensky, L. D. 2006. BCL-2 in the crosshairs: tipping the balance of life and death. Cell Death Differ. 13:1339-1350. [DOI] [PubMed] [Google Scholar]
- 70.White, E. 2006. Mechanisms of apoptosis regulation by viral oncogenes in infection and tumorigenesis. Cell Death Differ. 13:1371-1377. [DOI] [PubMed] [Google Scholar]
- 71.Willis, S. N., and J. M. Adams. 2005. Life in the balance: how BH3-only proteins induce apoptosis. Curr. Opin. Cell Biol. 17:617-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yamanaka, T., M. Uchida, and T. Doi. 2002. Innate form of HCV core protein plays an important role in the localization and the function of HCV core protein. Biochem. Biophys. Res. Commun. 294:521-527. [DOI] [PubMed] [Google Scholar]
- 73.Yao, K., and V. N. Vakharia. 2001. Induction of apoptosis in vitro by the 17-kDa nonstructural protein of infectious bursal disease virus: possible role in viral pathogenesis. Virology 285:50-58. [DOI] [PubMed] [Google Scholar]
- 74.Yasui, K., T. Wakita, K. Tsukiyama-Kohara, S. I. Funahashi, M. Ichikawa, T. Kajita, D. Moradpour, J. R. Wands, and M. Kohara. 1998. The native form and maturation process of hepatitis C virus core protein. J. Virol. 72:6048-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Youle, R. J., and A. Strasser. 2008. The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 9:47-59. [DOI] [PubMed] [Google Scholar]
- 76.Zhu, N., C. F. Ware, and M. M. Lai. 2001. Hepatitis C virus core protein enhances FADD-mediated apoptosis and suppresses TRADD signaling of tumor necrosis factor receptor. Virology 283:178-187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.