Abstract
Herpes simplex virus 1 (HSV-1) and HSV-2 cause similar acute infections but differ in their abilities to reactivate from trigeminal and lumbosacral dorsal root ganglia. During latency, HSV-1 and HSV-2 also preferentially express their latency-associated transcripts (LATs) in different sensory neuronal subtypes that are positive for A5 and KH10 markers, respectively. Chimeric virus studies showed that LAT region sequences influence both of these viral species-specific phenotypes. To further map the LAT region sequences responsible for these phenotypes, we constructed the chimeric virus HSV2-LAT-E1, in which exon 1 (from the LAT TATA to the intron splice site) was replaced by the corresponding sequence from HSV-1 LAT. In intravaginally infected guinea pigs, HSV2-LAT-E1 reactivated inefficiently relative to the efficiency of its rescuant and wild-type HSV-2, but it yielded similar levels of viral DNA, LAT, and ICP0 during acute and latent infection. HSV2-LAT-E1 preferentially expressed the LAT in A5+ neurons (as does HSV-1), while the chimeric viruses HSV2-LAT-P1 (LAT promoter swap) and HSV2-LAT-S1 (LAT sequence swap downstream of the promoter) exhibited neuron subtype-specific latent LAT expression phenotypes more similar to that of HSV-2 than that of HSV-1. Rescuant viruses displayed the wild-type HSV-2 phenotypes of efficient reactivation in the guinea pig genital model and a tendency to express LAT in KH10+ neurons. The region that is critical for HSV species-specific differences in latency and reactivation thus lies between the LAT TATA and the intron splice site, and minor differences in the 5′ ends of chimeric sequences in HSV2-LAT-E1 and HSV2-LAT-S1 point to sequences immediately downstream of the LAT TATA.
During an initial infection, herpes simplex virus 1 (HSV-1) and HSV-2 establish latency in the sensory ganglia innervating the peripheral site of inoculation. In response to various stimuli, virus can reactivate from the sensory neurons to cause recurrent disease at or near the original site of inoculation. Although these viruses are very similar, there are notable differences in their abilities to reactivate in a manner specific to each viral species. HSV-1 reactivates most efficiently from the trigeminal ganglia to cause recurrent orofacial herpes, while HSV-2 reactivates more efficiently from the lumbosacral dorsal root ganglia (DRG) to cause recurrent genital herpes (18).
The latency-associated transcript (LAT) is the most abundant latently transcribed RNA in both HSV-1 and HSV-2. Stable LAT introns are spliced from less stable primary transcripts (Fig. 1) (10). HSV-1 and HSV-2 LAT promoter mutants that express no detectable LAT during latency are impaired for reactivation in several animal models (5, 7, 17). The HSV-2 LAT intron expressed transgenically in mice has no influence on HSV infection or the reactivation phenotype (31), and recombinant HSV-1 viruses with deletions in the intron behave like wild-type virus (9). The mutation of the splice branch points destabilizes the accumulation of the HSV-1 LAT intron but does not influence reactivation (22).
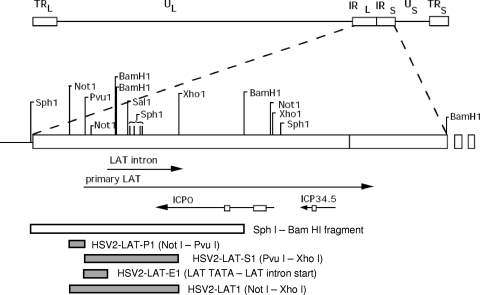
FIG. 1.
Virus construction. Genomic and endonuclease restriction sites are shown relative to HSV-2 strain HG52. The HSV genome consists of unique long and short (UL and US) regions flanked by internal and terminal repeats (IRL/IRS and TRL/TRS). The primary LAT is transcribed from the repeat regions in the direction antisense to ICP0 and ICP34.5 (transcripts are shown here as lines, with arrows indicating the direction of transcription). The primary LAT is spliced into a single stable LAT intron in HSV-2 and two differentially spliced introns in HSV-1. Boxes on the ICP0 and ICP34.5 lines denote introns that are not readily detectable after splicing. Chimeric viruses (HSV2-LAT-P1, HSV2-LAT-S1, HSV2-LAT-E1, and HSV2-LAT1) were constructed by homologous recombination into wild-type virus after cloning HSV-1 sequences corresponding to the shaded areas into the SphI-BamHI clone. The location of the PvuI site relative to the LAT TATAs is illustrated in Fig. 6.
Studies using recombinant viruses with deletions in the LAT region showed that sequences in LAT exon 1 are required for the efficient reactivation of HSV-1 (4, 6, 14, 13, 15, 16, 24, 27, 33) and HSV-2 (3, 13, 33). The LAT exon 1 region also provides enhancer functions for the continued long-term expression of HSV-1 LAT during latency (2), and the corresponding region of HSV-2 also influences LAT expression levels (34). Deletions within HSV-1 LAT exon 1 alter virulence differently in mice and rabbits (23), suggesting that this region responds to cellular factors to exert an effect on virulence. Thus, LAT exon 1 sequences may confer regulatory influence over reactivation and virulence.
Primary sensory neurons are a diverse population of cells that can be classified according to cellular morphology, physiological response properties, and patterns of gene expression. Although all neuronal populations in the trigeminal ganglia and DRG are capable of supporting productive HSV infection, some populations are more likely than others to harbor latent virus, as identified by dual fluorescent staining for HSV LAT and specific surface markers identified by monoclonal antibodies A5 (specific for a population of neurons expressing Galβ1-4GlcNAc-R epitopes) and KH10 (recognizing a different population of neurons that express Galα1-3Galβ1-4Nac-R epitopes). Most A5+ neurons are immunoreactive for the calcitonin gene-related peptide and the high-affinity nerve growth factor receptor (TrkA), whereas KH10+ neurons colabel with the lectin BSL-IB4, identifying them as a population of small-diameter neurons responsive to glial cell-derived neurotrophic factor. Neurons latently infected with HSV-1 that express the LAT are more likely to be identified by monoclonal antibody A5, while LAT expression during HSV-2 latent infection is more associated with neurons identified by monoclonal antibody KH10 (19, 32). In these previous studies, no differences were observed in the ability of A5+ or KH10+ cells to express the HSV-1 or HSV-2 LAT; thus, these findings indicated viral species-specific differences in the ability of the virus to become latent and express LAT at detectable levels in these neuronal subpopulations.
We previously reported on the chimeric HSV-2 viruses HSV2-LAT1, HSV2-LAT-P1, and HSV2-LAT-S1, which contain HSV-1 LAT sequences in place of homologous HSV-2 sequences (Fig. 1). HSV2-LAT1, which contains HSV-1 LAT promoter and downstream sequences homologous to an HSV-2 NotI-XhoI subsequence, displayed an HSV-1 recurrence phenotype in guinea pig genital and rabbit eye models (33) and was found to express LAT in A5+ neurons, also an HSV-1 phenotype (19). HSV2-LAT-P1, containing HSV-1 LAT promoter sequences (in a NotI-PvuI fragment), reactivated efficiently in the guinea pig genital model (an HSV-2 phenotype), while HSV2-LAT-S1, which contained HSV-1 LAT sequences downstream of the promoter (homologous to an HSV-2 PvuI-XhoI fragment), was impaired for reactivation in guinea pigs, implying that sequences within the LAT exon 1 and intron (and not the LAT promoter) were responsible for the efficient reactivation of HSV-2 in the guinea pig genital model (3).
In the present study, to further define the region of LAT that is critical for HSV-2 species-specific latency phenotypes, an additional chimeric virus was constructed and characterized. The region of HSV-2 from the LAT TATA box to the 5′ splice site of the LAT intron (LAT exon 1) was replaced by the corresponding region of HSV-1 and designated HSV2-LAT-E1 (for exon from HSV-1). A rescuant, HSV2-LAT-E1R, also was constructed to restore the native HSV-2 sequences. After the in vitro characterization of HSV2-LAT-E1, the chimeric virus and its rescuant were tested for reactivation in the guinea pig genital model of HSV infection. Tissues also were evaluated for the distribution of viral DNA during acute and latent infection to determine if LAT exon 1 influenced viral spread and the establishment of latency. To evaluate a potential mechanism for any differences, viral transcript expression levels were quantified for LAT and ICP0 (a viral transactivator essential for reactivation). Latently infected LAT-expressing neurons from HSV2-LAT-E1, HSV2-LAT-P1, HSV2-LAT-S1, and their rescuants (where available) also were evaluated for the expression of A5 and KH10 neuronal markers to characterize the subpopulation of neurons in which LAT expression-competent virus established latency.
MATERIALS AND METHODS
Cells and viruses.
Vero cells were obtained from the American Type Culture Collection (ATCC; Rockville, MD) and maintained in minimum essential medium with 10% inactivated fetal bovine serum (FBS) (HyClone) and 1% penicillin-streptomycin-l-glutamine (Quality Biologicals, Gaithersburg, MD). HSV-2 strain 333 originally was obtained from Gary Hayward (Johns Hopkins University, Baltimore, MD). HSV-1 strain 17syn+ originally was obtained from John Hay (SUNY-Buffalo, NY). Virus stocks were produced in Vero cells and titrated by plaque assay in duplicate. To compare one-step growth characteristics of wild-type and mutant viruses, ∼106 Vero cells were inoculated in duplicate at time zero with a multiplicity of approximately 0.1 PFU/cell of each virus. Medium was added after a 2-h adsorption period. At 0, 2, 5, 12, and 20 h postinoculation (p.i.), cells were scraped, freeze-thawed three times, and titrated by plaque assay in duplicate.
Construction of mutant viruses.
Genome locations for HSV-2 are given relative to the sequence of strain HG52 (GenBank accession number NC001798.1) (8), and those for HSV-1 are given relative to the sequence of strain 17syn+ (GenBank accession number NC001806.1) (20). Plasmid clones of an AvrII-AluI fragment of HSV-1 strain 17syn+ (Avr-AluΔXho) and an SphI-BamHI fragment of HSV-2 strain 333 (Sph-Sal-Bam, or SSB) were described previously (33). To construct chimeric virus HSV2-LAT-E1 (Fig. 1), the region from the TATA box to the LAT intron 5′ splice site sequence (CAGGTA) was amplified by PCR from HSV-1 strain 17syn+ using primers GCTGCGTCATCTCAGCCTT and CGACCCTAAACCTACCTGGAAACGC and the following cycle conditions: 95°C for 2 min, 35 cycles of 95°C for 20 s, 50°C for 2 min, and 72°C for 1 min 20 s, and then 72°C for 10 min. The CAGGTA sequence (which is present in both HSV-1 and HSV-2 at the LAT intron 5′ end) is in boldface, and the underlined segment matches the HSV-2 sequence downstream of CAGGTA to facilitate overlap PCR in the subsequent amplification. The HSV-2 strain 333 region from the LAT intron 5′ splice site sequence CAGGTA to the SalI restriction site was amplified using primers GCGTTTCCAGGTAGGTTTAGGGTCG and ACACAACACGACACGACGC and the following cycle conditions: 95°C for 2 min, 35 cycles of 95°C for 20 s, 54°C for 2 min, and 72°C for 1 min 20 s, and then 72°C for 10 min. The CAGGTA sequence is in boldface, and the underlined segment matches the HSV-1 sequence upstream of CAGGTA to facilitate overlap PCR. These two PCR products were amplified together by overlap PCR using primers GCTGCGTCATCTCAGCCTT (HSV-1 5′-end primer) and ACACGACACGACGCGTTTTGC (HSV-2 3′-end primer) and the following cycle conditions: 95°C for 2 min, 5 cycles of 95°C for 15 s, 65°C for 15 s, 72°C for 1 min 20 s, followed by 30 cycles of 95°C for 15 s, 54°C for 45 s, 72°C for 1 min 30 s, followed by 72°C for 5 min. The resulting product joined the sequences at the splice site sequence CAGGTA, yielding a PCR product consisting of HSV-1 sequences upstream of CAGGTA and HSV-2 sequences downstream of CAGGTA. This product was cloned into TOPO vector pCR4 (Invitrogen, Carlsbad, CA), verified by sequencing, and designated pHSV1-HSV2overlap. pHSV1-HSV2overlap was digested with PsiI and SalI restriction endonucleases (Invitrogen) to release the overlapped fragment from the TOPO vector. The released fragment was cloned into HSV-2 plasmid SSB, which also had been digested with PsiI and SalI to release the wild-type HSV-2 sequence. The resulting plasmid, designated SSB-E1, contained HSV-2 sequences upstream of the TATA box, HSV-1 sequences from the TATA box to the 5′ splice site CAGGTA, and HSV-2 sequences downstream of the CAGGTA.
Chimeric virus HSV2-LAT-E1 was constructed by homologous recombination after the cotransfection of SSB-E1 plasmid DNA and parent virus DNA (HSV-2 strain 333) into Vero cells, followed by serial plaque purification until three rounds of analysis showed no evidence of parental virus. The purity and correctness of the chimeric virus were verified by Southern blotting (using restriction endonuclease pairs NotI/SalI and PvuI/SalI) and a radiolabeled probe specific for the mutated sequence. The sequencing of the complete mutated region further validated the mutant virus sequence. A rescuant, HSV2-LAT-E1R, was made from HSV2-LAT-E1 using the same method by cotransfecting mutant viral DNA with the wild-type HSV-2 SSB plasmid DNA to restore the native HSV-2 sequences and was validated by Southern blotting and sequencing.
The chimeric virus and the rescuant were characterized for one-step growth in cell culture to identify inherent growth defects that could account for differences in reactivation as described above. One-step growth kinetics were similar among the wild-type virus, the chimeric virus, and the rescuant.
The construction and characterization of chimeric viruses HSV2-LAT-S1 (3), HSV2-LAT-P1 (3), and HSV2-LAT1 (33) are described elsewhere.
Northern hybridization.
Vero cells were infected at a multiplicity of 1 PFU/cell and harvested 16 h after infection. After RNA was extracted, 5 μg was subjected to electrophoresis through a denaturing formaldehyde-agarose gel as previously described (33). After transfer and immobilization to a Nytran membrane, RNAs were hybridized with a radiolabeled gel-purified SalI-XhoI fragment that overlaps the LAT intron and ICP0 3′ end.
Animal studies.
Female Hartley guinea pigs (Charles River, Wilmington, MA) were inoculated intravaginally with 2 × 105 PFU of each virus. Guinea pigs were monitored and scored daily during acute infection (14 days) for lesion severity (on a scale from 0 to 4; 0 = no disease, 1 = redness/swelling, 2 = 1 to 2 lesions, 3 = 3 to 5 lesions, and 4 = 6 or more lesions or the coalescence of lesions). Recurrences, defined as vesicular lesions, were enumerated during the latent phase from day 15 through day 42 after inoculation and graphed as cumulative recurrences per guinea pig in each group. All observations of guinea pigs were made by two independent investigators masked to the identity of inoculated viruses, and composite scores are presented in the figures. Three separate animal experiments were performed: experiment 1, HSV-2 (n = 8) and HSV2-LAT-E1 (n = 8); experiment 2, HSV-2 (n = 11), HSV2-LAT-E1 (n = 9), and HSV2-LAT-E1R (n = 7); and experiment 3, HSV2-LAT-E1 (n = 8) and HSV2-LAT-E1R (n = 6). Each experiment yielded similar results, and the graphs presented are composites from all three experiments. Animals were housed in facilities approved by the American Association for Accreditation of Laboratory Animal Care and cared for in accordance with institutional guidelines. Lumbosacral DRG and sacral and lumbar spinal cord tissues were collected from each animal immediately after sacrifice and snap-frozen on dry ice. DNA and RNA were isolated from ganglia and spinal cord tissues using the AllPrep DNA/RNA Minikit (Qiagen, Valencia, CA) after homogenization with an Omni rotor-stator homogenizer (Omni International, Marietta, GA).
The ocular infection of mice to study latently infected neuronal subpopulations was performed as previously described (19).
Quantitative real-time PCR and RT-PCR.
The copy number of viral DNA genomes and RNA transcripts in DRG and spinal cord were determined by quantitative real-time PCR (for DNA) and real-time reverse transcription-PCR (RT-PCR) (for RNA) using an ABI 7900 TaqMan PCR system (Applied Biosystems, Foster City, CA). Primers and probes were specific for DNA sequences within HSV-2 glycoprotein D and HSV-1 glycoprotein G and HSV-1 or HSV-2 RNA sequences within LAT and ICP0 (sequence are given in 5′-to-3′ orientation, with final concentrations in parentheses). The HSV-2 gD primers and probe were TCAGAGGATAACCTGGGA (250 nM), GGGAGAGCGTACTTGCAGGA (250 nM), and 6-carboxyfluorescein (FAM)-CCAGTCGTTTTCTTCACTAGCCGCAG-6-carboxytetramethylrhodamine (TAMRA) (200 nM) (30). The HSV-1 gG primers and probe were CTGTTCTCGTTCCTCACTGCCT (1,000 nM), CAAAAACGATAAGGTGTGGATGAC (1,000 nM), and FAM-CCCTGGACACCCTCTTCGTCGTCAG-TAMRA (250 nM). RNA primers and probes for HSV-2 LAT were GTCAACACGGACACACTCTTTTT (1,600 nM), CGAGGCCTGTTGGTCTTTATC (1,600 nM), and FAM-CACCCACCAAGACAGGGAGCCA-TAMRA (200 nM). RNA primers and probes for HSV-1 LAT were ACCCACGTACTCCAAGAAGGC (400 nM), TAAGACCCAAGCATAGAGAGCCA (400 nM), and FAM-TCCCACCCCGCCTGTGTTTTTGT-TAMRA (200 nM). RNA primers and probes for HSV-2 ICP0 were GGTCACGCCCACTATCAGGTA (900 nM), CCTGCACCCCTTCTGCAT (900 nM), and FAM-CAACGGAATCCAGGTCTTCATGCACG-TAMRA (250 nM). RNA primers and probes for HSV-1 ICP0 were GGATGCAATTGCGCAACAC (900 nM), GCGTCACGCCCACTATCAG (900 nM), and FAM-GCTGTGCAACGCCAAGCTGGTGTA-TAMRA (250 nM). The PCRs contained TaqMan Universal PCR Master Mix or TaqMan Universal RT-PCR One-Step Master Mix (Applied Biosystems, Foster City, CA), specific primers and probes, and 50 ng of DNA or 250 ng RNA. Cycle conditions were 50°C for 2 min (48°C for 30 min for RNA for the RT step) and 95°C for 10 min, followed by 40 cycles of 95°C for 20 s and 60°C for 1 min. Standard curves based on 10-fold dilutions of known amounts of viral DNA were used to determine copy numbers. Viral genome copy numbers were normalized to those of the gene for the 18S rRNA, and RNA numbers were normalized to those of 18S rRNA using commercial primers and probe (rRNA control reagents; Applied Biosystems, Foster City, CA). For the detection of viral RNA, all results are presented relative to the quantity of the specific viral RNA found in guinea pigs infected with HSV2-LAT-E1R at day 42 after inoculation. Reverse transcriptase-negative controls were included for all RNA assays to verify that no DNA contamination was present.
Dual fluorescent staining.
Combined fluorescent in situ hybridization and immunofluorescent staining was carried out as previously described using mouse monoclonal antibodies FE-A5 and KH10 (Developmental Studies Hybridoma Bank, Iowa City, IA) and digoxigenin-RNA probes (19, 32).
Statistics.
Statistics were performed using nonparametric analyses with SPSS version 11.5.0, including the Kruskal-Wallis and Mann-Whitney tests (LEAD Technologies, Charlotte, NC). Comparisons of acute infections were based on total areas under the lesion severity curves for days 1 to 14 after inoculation. Comparisons of recurrent infections were based on cumulative recurrences per guinea pig adjusted for the number of days of observation. Log-transformed viral DNA quantities were compared by analysis of variance and post hoc least significant difference analyses. Error bars represent standard errors of the means for each group.
RESULTS
Construction and in vivo characterization of HSV2-LAT-E1.
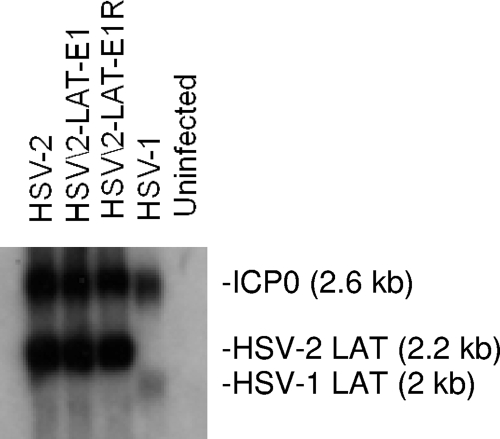
The construction of the chimeric virus HSV2-LAT-E1 and its rescuant were verified by Southern hybridization and the sequencing of the entire mutated region from the LAT promoter through the 5′ splice site of the intron. Figure 1 shows the relevant genomic regions and restriction endonuclease cleavage sites. HSV2-LAT-E1R was indistinguishable from wild-type HSV-2, and each virus yielded the expected bands and sequences (data not shown). One-step growth characteristics of each virus were analyzed in Vero cells, and each of the viruses replicated similarly to wild-type HSV-2 virus in cell culture (data not shown). LAT and ICP0 expression in acutely infected Vero cells was similar among wild-type, mutant, and rescuant viruses by Northern hybridization using a probe that overlaps the HSV-2 LAT intron and the 3′ end of ICP0 and that has some cross-reactivity with these transcripts from HSV-1 (Fig. 2).
FIG. 2.
Northern hybridization of RNA from Vero cells 16 h after infection with HSV-2, HSV2-LAT-E1, HSV2-LAT-E1R, and HSV-1. RNA was probed with an SalI-XhoI probe overlapping the HSV-2 LAT intron, ICP0, with partial homology to these RNAs from HSV-1. Expected sizes of LAT and ICP0 are shown.
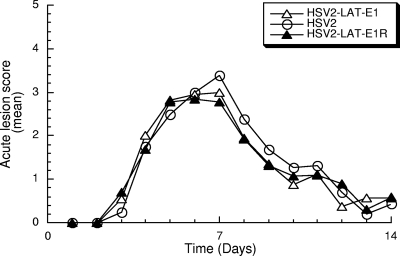
To determine if HSV-2 LAT exon 1 is required for the efficient reactivation of HSV-2, wild-type HSV-2, chimeric virus HSV2-LAT-E1, or its rescuant HSV2-LAT-E1R were inoculated intravaginally (with 2 × 105 PFU of virus) into female guinea pigs. The severity of lesions was analyzed during the acute phase of infection through day 14 p.i. (Fig. 3). The mean lesion scores were similar among HSV2-LAT-E1, its rescuant HSV2-LAT-E1R, and wild-type HSV-2 (P = 0.536 by Kruskal-Wallis test).
FIG. 3.
Acute lesion severity. Lesion severity (as measured on a 4-point scale) is graphed as the mean for each group of guinea pigs on each day of observation from days 1 to 14 p.i. Viruses included HSV-2 (n = 19), HSV2-LAT-E1 (n = 25), and HSV2-LAT-E1R (n = 13). There were no significant differences among the areas under the curve for each virus (P = 0.536 by Kruskal-Wallis test).
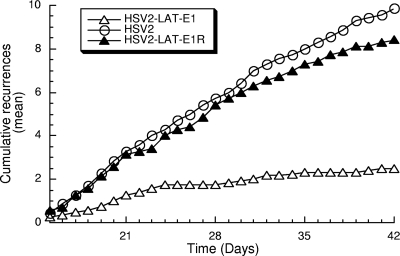
During the latent phase of infection, HSV2-LAT-E1 reactivated inefficiently in the guinea pig genital model compared to the reactivation of wild-type HSV-2 (P < 0.0005) or the rescuant HSV2-LAT-E1R (P = 0.001) (Fig. 4). The rescuant HSV2-LAT-E1R had a wild-type recurrence phenotype (P = 0.221 compared to results for HSV-2). The region substituted in HSV2-LAT-E1 thus provides elements that are essential for efficient recurrence in a genital model of HSV-2 infection.
FIG. 4.
Cumulative recurrences per guinea pig for each group during latent infection. Viruses tested included HSV-2 (n = 7), HSV2-LAT-E1 (n = 13), and HSV2-LAT-E1R (n = 7). Guinea pigs that did not survive acute disease were excluded. P values (using the Mann-Whitney test) for pairwise comparisons of recurrence frequency are the following: for HSV-2 versus HSV2-LAT-E1, P < 0.0005; for HSV2-LAT-E1 versus HSV2-LAT-E1R, P = 0.001; and for HSV-2 versus HSV2-LAT-E1R, P = 0.221.
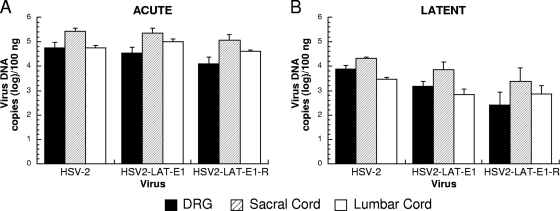
To determine if LAT exon 1 influenced reactivation from these lumbosacral DRG by affecting viral spread and the establishment of latency, tissues from infected guinea pigs were evaluated for viral DNA levels in the lumbosacral DRGs and lumbar and sacral spinal cord regions. Guinea pigs were sacrificed on days 8 and 42 p.i. to evaluate differences between acute and latent viral loads. Viral DNA levels in each of these locations were similar among the viruses (Fig. 5), with no significant differences observed between the chimeric virus HSV2-LAT-E1 and its rescuant. The viral DNA levels in the DRG of the rescuant HSV2-LAT-E1R and the chimeric virus HSV2-LAT-E1 were similar, although both decreased at day 42 compared to the level for wild-type HSV-2 (P = 0.031 and 0.025, respectively), implying that the impaired recurrence phenotype of the chimeric virus cannot be attributed to differences in viral DNA levels when assayed within the whole ganglion.
FIG. 5.
Viral DNA quantities of HSV-2, HSV2-LAT-E1, and HSV2-LAT-E1R in the DRG, sacral cord, and lumbar cord during acute and latent infection. Viral DNA extracted from the lumbosacral DRG, sacral spinal cord, and lumbar spinal cord during acute infection (day 8 p.i.) (A) and latent infection (day 42 p.i.) (B) was quantified by TaqMan PCR assay and normalized to the gene for the 18S rRNA. The day 8 experiment included HSV-2 (n = 10), HSV2-LAT-E1 (n = 11), and HSV2-LAT-E1R (5). The day 42 experiment included HSV-2 (n = 9), HSV2-LAT-E1 (n = 14), and HSV2-LAT-E1R (n = 8).
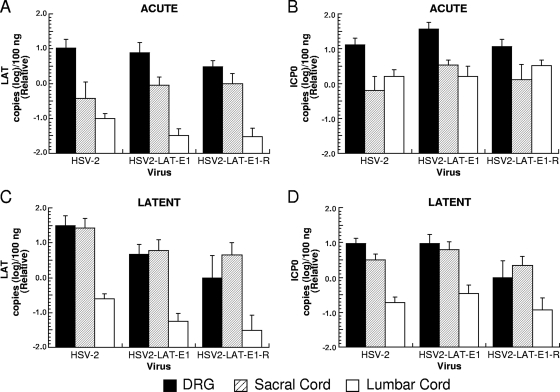
The viral transcripts for LAT and ICP0 were quantified in ganglia from infected guinea pigs by TaqMan quantitative RT-PCR assays. Guinea pigs were sacrificed during acute (day 8 p.i.) and latent infection (day 42 p.i.) to evaluate differences in gene expression in acutely and latently infected animals (Fig. 6). All three viruses expressed less LAT RNA in the lumbar spinal cord than in the DRG or sacral cord during latency (Fig. 6C). During acute infection (Fig. 6A and B) and latency (Fig. 6C and D), LAT and ICP0 RNA expression levels were similar among the viruses, with no statistically significant differences observed. A modest apparent difference in LAT expression during the latency of HSV2-LAT-E1 and its rescuant relative to the level for wild-type HSV-2 did not contribute to the recurrence phenotype, since the rescuant and wild-type HSV-2 had similar recurrence phenotypes. As was the case with viral DNA, differences in recurrence phenotype thus were not correlated with differences in RNA expression as measured in whole ganglia, lumbar spinal cord, or sacral spinal cord, either acutely or latently.
FIG. 6.
LAT and ICP0 transcript expression in DRG, sacral spinal cord, and lumbar spinal cord during acute and latent infection (RNA copies [log]/100 ng total RNA). RNA extracted from the lumbosacral DRG, sacral spinal cord, and lumbar spinal cord during acute infection (day 8 p.i.) (A and B) and latent infection (day 42 p.i.) (C and D) was quantified by TaqMan PCR assay with primers and probes specific for HSV-2 LAT (A and C) and ICP0 (B and D). 18S rRNA also was quantified by TaqMan RT-PCR and used to normalize for RNA loading. Results are shown relative to mean quantities of each transcript that were detected in lumbosacral DRG infected with HSV2-LAT-E1R on day 42. Viruses tested at day 8 included HSV2-LAT-E1 (n = 11), HSV-2 (n = 10), and HSV2-LAT-E1R (n = 5). Viruses tested at day 42 included HSV2-LAT-E1 (n = 14), HSV-2 (n = 9), and HSV2-LAT-E1R (n = 8).
Studies of neuronal subtypes that harbor latent virus.
LAT-positive neurons that are latently infected with HSV-1 or HSV-2 are more likely also to express neuron subtype-specific markers (A5 or KH10, respectively). This phenotype is dependent upon the virus species-specific LAT sequences present in HSV-1 and HSV-2 (19). To investigate the effect of the viral mutations on this phenotype, neuronal subpopulations that harbored latent virus were identified by dual fluorescent in situ hybridization for the LAT and immunohistochemistry for the neuronal subtype markers A5 and KH10 in trigeminal ganglia from mice latently infected with HSV2-LAT-P1, HSV2-LAT-S1, HSV2-LAT-S1R, HSV2-LAT-E1, and HSV2-LAT-E1R. Results are expressed as the proportion of LAT-expressing cells that also express the A5 or KH10 marker (Table 1). During acute infection, A5+ and KH10+ sensory neurons become productively infected in proportion to their representation in uninfected ganglia, but at later time points, cells latently infected with HSV-2 and expressing LAT at detectable levels are more likely to colabel with KH10, and LAT-expressing cells latently infected with HSV-1 (or the chimeric virus HSV2-LAT1, which contains the combined HSV-1 sequences of HSV2-LAT-P1 and HSV-2-LAT-S1) are more likely to colabel with A5 (19).
TABLE 1.
Latent distribution of HSV-1, HSV-2, and chimeric viruses in murine trigeminal ganglia
| Virus | % of colabeled neuronsa (no. of colabeled neurons/total no. of LAT+ neurons)
|
|
|---|---|---|
| A5+ | KH10+ | |
| HSV2-LAT-E1 | 33 (86/257) | 12 (26/214) |
| HSV2-LAT-E1R | 6.8 (19/280) | 49 (170/349) |
| HSV2-LAT-S1 | 8.7 (43/492) | 41 (163/394) |
| HSV2-LAT-S1-R | 3.8 (14/365) | 45 (170/379) |
| HSV2-LAT-P1 | 5.9 (16/271) | 21 (59/285) |
| HSV2-LAT-P1-R | No virus | No virus |
| HSV-1 17syn+ | 39 (133/341) | 3.1 (11/353) |
| HSV-2 333 | 8.2 (17/208) | 47 (124/263) |
Expressed as the percentage of neurons with LAT expression (21 days postinoculation) that colabel with monoclonal antibody A5 (A5+) or KH10 (KH10+).
Twenty-one days postinoculation, 5.9% of LAT-expressing neurons latently infected with HSV2-LAT-P1 were A5+ while 21% were KH10+ (Table 1), similarly to the phenotype observed for HSV-2 (8.2% A5+ and 47% KH10+), although the KH10+ tendency was not as pronounced. Also, at 21 days after inoculation, 8.7% of LAT-expressing neurons in animals infected with HSV2-LAT-S1 were A5+ and 41% colabeled with the KH10 marker, which also is similar to the phenotype observed for its rescuant HSV2-LAT-S1-R (3.8% A5+ and 45% KH10+) and that observed for wild-type HSV-2. Although the combined mutations in HSV2-LAT-P1 and HSV-2-LAT-S1 were equivalent to the sequences substituted in the previously tested chimeric virus HSV2-LAT1, the substitution of neither the LAT promoter sequences nor the sequences downstream of the PvuI site was sufficient alone to confer the A5+ neuronal tendency seen for HSV2-LAT1 (40% A5 and 2% KH10) (19). This finding implied that the A5+ neuronal latency phenotype required HSV-1 sequences on both sides of the PvuI site that separated the chimeric sequences in HSV2-LAT-P1 and HSV2-LAT-S1.
In mice infected with HSV2-LAT-E1, 33% of LAT+ neurons were A5+ and 12% were KH10+, which is similar to the phenotype observed for HSV-1 strain 17syn+ (in which 39% and 3.1% of LAT-expressing neurons were A5+ and KH10+, respectively) and also similar to the proportions exhibited by HSV2-LAT1. The rescuant of HSV2-LAT-E1 displayed a phenotype similar to that of wild-type HSV-2, with 6.8% of LAT-expressing neurons colabeling with A5 and 49% with KH10. Thus, in the context of HSV-2, HSV-1 LAT sequences downstream of the LAT TATA and upstream of the LAT intron splice site were sufficient to confer the HSV-1 A5+ phenotype on HSV-2.
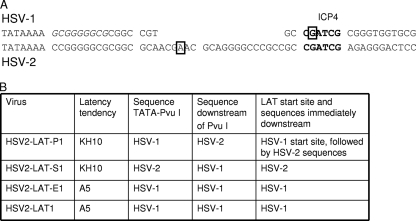
These findings point to the importance of sequences between the TATA and the PvuI site of HSV-1 LAT, which are necessary but not sufficient to confer the HSV-1 neuronal subtype phenotype on HSV-2. The retention of HSV-2 sequences on either side (but not both sides) of the PvuI site in HSV2-LAT-P1 and HSV2-LAT-S1 was sufficient to cause these viruses to retain an HSV-2-like phenotype in specific neuronal subtypes. The sequences in this region were further examined (Fig. 7A). Between the TATA and the PvuI site (18 bp in HSV-1), HSV-1 contains a functional binding site (GCGGGGGCG) for the transcription factor EGR-1 (26) (which is identical to the nerve growth factor inducible A transcription factor) followed by the LAT start site at the PvuI restriction site, which also overlaps an ICP4 binding site (1). HSV-2 contains 38 bp between the TATA and the PvuI site, such that HSV-2 contains a 20-bp insertion relative to HSV-1 and HSV2-LAT-P1; this region in HSV-2 contains two copies (CCGGGGGCG and GCAGGGGCC) of sequences that resemble the single HSV-1 EGR-1 binding site. The HSV-2 LAT start site is at a location relative to the TATA that is similar to that of HSV-1 (29). The location of an ICP4 binding site in HSV-2, if there is one, has not been published.
FIG. 7.
Sequences and phenotypes. (A) Comparison of HSV-1 and HSV-2 sequences between LAT TATAs and PvuI recognition sites. Chimeric viruses HSV2-LAT-P1 and HSV2-LAT-E1 contain HSV-1 sequences in this region, while HSV2-LAT-S1 contains HSV-2 sequences in this region. Potential binding sites for ICP4 and SP1 are shown. An EGR-1 consensus binding sequence (GCGGGGGCG) is italicized in the HSV-1 sequence. The PvuI sites are displayed in boldface. Previously mapped start sites for HSV-1 and HSV-2 primary LATs are shown in boxes. (B) Correlation of sequences near the LAT 5′ end with the chimeric virus latency establishment phenotype.
DISCUSSION
Experiments with HSV2-LAT-E1 show that sequences between the LAT TATA and the LAT intron splice site are required for efficient reactivation in the guinea pig genital model, and that the LAT exon 1 region also regulates the previously described virus species-specific LAT expression in certain neuronal cell subtypes. Experiments with the closely related chimeric viruses HSV2-LAT-S1 and HSV2-LAT-P1 showed that the sequences responsible for this phenotype span the HSV-1 LAT PvuI site, because in these chimeric viruses, HSV-1 sequences on either side of the PvuI site alone did not confer the HSV-1 latency phenotype in A5+ neurons, while their combination in HSV2-LAT1 did (19). The present findings rule out the possible involvement of the LAT promoter region, the stable LAT intron, and the 3′ end of the ICP0 coding region in the previously described HSV species-specific latency phenotypes.
Previous studies validated the use of the LAT expression as a marker for latency in specific neuronal subtypes (19). Because there was no difference in the ability of A5+ and KH10+ neurons to express the LAT, it was concluded that LAT expression is not simply a function of differences in HSV-1 or HSV-2 LAT promoter activity in A5+ or KH10+ cells. Although LAT expression is heterogeneous among ganglionic neurons and some latently infected neurons may accumulate LAT at levels below the level of detection by in situ hybridization, it is unlikely that the distribution of latent infection among A5 and KH10 neurons would differ in neurons that express LAT at levels below this level of detection. Although a significant number of non-LAT-expressing neurons may harbor HSV viral DNA fragments, as detected by in situ PCR (12, 21), it is not known whether these non-LAT-expressing latently infected neurons are reactivation competent. While it also is unknown if viral reactivation is restricted to neurons in which LAT expression can be detected by in situ hybridization, the altered recurrence phenotypes of viruses impaired for LAT expression indicates that it is a biologically relevant marker of HSV latency.
In the context of HSV2-LAT-P1, the HSV-1 region between the TATA and the PvuI site (which, with the exception of a few base pairs, represents a deletion relative to the HSV-2 sequence in the same region [Fig. 7]) did not confer an HSV-1-like latency phenotype in A5+ neurons, but in the context of HSV2-LAT-E1, this region did. Thus, HSV-1 sequences downstream of the PvuI site also are necessary for this phenotype but are insufficient on their own (as was demonstrated by the investigation of HSV2-LAT-S1, which contains HSV-1 sequences downstream but not upstream of the PvuI site). Differences in potential enhancer sequences between HSV-1 and HSV-2 between the LAT TATA and the PvuI site (including a single base pair difference in the EGR-1 binding site directly downstream of the HSV-1 LAT TATA, the potential extra EGR-1 site downstream of the LAT start site in HSV-2, or possible differences in Sp1 and AP2 sites identified in silico [11 and data not shown]) would not by themselves explain the differing phenotypes of HSV2-LAT-E1 and HSV2-LAT-P1, which have the same sequence in this region.
The additional 20 bp in the region between the LAT TATA and the PvuI site of HSV-2 relative to that of HSV-1 implies that there are differences in the likely LAT start site of the chimeric viruses (Fig. 7A). The conservation of the HSV-1 LAT start site and HSV-1 sequences immediately downstream correlated best with the HSV-1 latency phenotype in A5+ neurons (Fig. 7B). Differences in the LAT start site might influence the ability of nearby transcription factor binding sites, including those for ICP4 or neuron-specific transcription factors, to modulate LAT expression.
The quantification of viral DNA and LAT expression suggests that HSV-2 latency is more effectively established in the sacral spinal cord than in the lumbar cord, confirming and extending the previously reported finding that HSV-2 DNA preferentially is found in the sacral spinal cord rather than the lumbar spinal cord (3). One major difference between sacral and lumbar regions of the spinal cord is their different patterns of autonomic innervation, such that parasympathetic neurons synapse in the sacral cord and sympathetic neurons synapse in the lumbar cord. The identification of virus DNA and LAT in the sacral spinal cord is consistent with a tendency of HSV-2 to spread via parasympathetic pathways, although this finding appears not to be influenced by the LAT sequences that were substituted in HSV2-LAT-E1. However, because the mutation in HSV2-LAT-E1 did not cause differences in spread to the spinal cord, differences in spread to the spinal cord do not explain the observed differences in the viral reactivation or neuron subtype preferences during latency.
Although there were mutation-related differences in recurrence phenotype, HSV2-LAT-E1, its rescuant, and wild-type HSV-2 displayed similar viral RNA expression and total viral DNA content in DRG and spinal cord, further supporting the importance of examining neuronal subtype phenotypes in studies of HSV latency. HSV2-LAT-E1 had an HSV-1-like (impaired) recurrence phenotype in the guinea pig genital model and also displayed an HSV-1-like latency neuronal subtype phenotype, with a tendency toward A5+ neurons. The possibility remains that HSV2-LAT-E1 generally is impaired for reactivation not only in the guinea pig genital model but also in other animal species. HSV2-LAT-E1 is not impaired for the establishment of latency in the DRG or spinal cord, since the total viral DNA levels were similar between the chimeric virus and its wild-type parental HSV-2. Since the chimeric virus also demonstrated a preference for A5+ neurons during latency, a phenotype correlated with HSV-1 reactivation competence from the trigeminal ganglion, it will be interesting to determine whether HSV2-LAT-E1 also has an HSV-1-like recurrence phenotype in a trigeminal recurrence model, as was the case with HSV2-LAT1 (in which the HSV-1 LAT promoter, LAT exon 1, and LAT intron sequences were substituted into HSV-2) (33).
The present experimental findings allow the mapping of a region that is critical for HSV-1 and HSV-2 species-specific reactivation in the guinea pig genital model and the finding of LAT expression in specific neuronal subtypes. The sequences necessary for these phenotypes lie directly downstream of the LAT TATA, near the primary LAT start site, spanning a PvuI restriction enzyme site. Because the implicated region includes sequences near the LAT 5′ end, it seems reasonable to hypothesize that the LAT-associated phenotypes controlled by this region are related to the expression, stability, or processing of LAT (including the microRNAs known to be expressed under the LAT promoter (25, 28). We expect that further attention to this region will yield substantial insights into the mechanism by which the LAT region exerts its effect on the HSV-1 and HSV-2 latency and recurrence phenotypes.
Acknowledgments
We thank Kening Wang, Haru Murata, Masahiro Ohashi, Shasta McClenahan, Christine Uhlenhaut, and Shuang Tang for critical readings of the manuscript.
Footnotes
Published ahead of print on 29 July 2009.
REFERENCES
- 1.Batchelor, A. H., K. W. Wilcox, and P. O'Hare. 1994. Binding and repression of the latency-associated promoter of herpes simplex virus by the immediate early 175K protein. J. Gen. Virol. 75:753-767. [DOI] [PubMed] [Google Scholar]
- 2.Berthomme, H., J. Lokensgard, L. Yang, T. Margolis, and L. T. Feldman. 2000. Evidence for a bidirectional element located downstream from the herpes simplex virus type 1 latency-associated promoter that increases its activity during latency. J. Virol. 74:3613-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertke, A. S., A. Patel, and P. R. Krause. 2007. Herpes simplex virus latency-associated transcript sequence downstream of the promoter influences type-specific reactivation and viral neurotropism. J. Virol. 81:6605-6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharjee, P. S., R. K. Tran, M. E. Myles, K. Maruyama, A. Mallakin, D. C. Bloom, and J. M. Hill. 2003. Overlapping subdeletions within a 348-bp in the 5′ exon of the LAT region that facilitates epinephrine-induced reactivation of HSV-1 in the rabbit ocular model do not further define a functional element. Virology 312:151-158. [DOI] [PubMed] [Google Scholar]
- 5.Bloom, D. C., G. B. Devi-Rao, J. M. Hill, J. G. Stevens, and E. K. Wagner. 1994. Molecular analysis of herpes simplex virus type 1 during epinephrine-induced reactivation of latently infected rabbits in vivo. J. Virol. 68:1283-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloom, D. C., J. M. Hill, G. Devi-Rao, E. K. Wagner, L. T. Feldman, and J. G. Stevens. 1996. A 348-base-pair region in the latency-associated transcript facilitates herpes simplex virus type 1 reactivation. J. Virol. 70:2449-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devi-Rao, G. B., D. C. Bloom, J. G. Stevens, and E. K. Wagner. 1994. Herpes simplex virus type 1 DNA replication and gene expression during explant-induced reactivation of latently infected murine sensory ganglia. J. Virol. 68:1271-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolan, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fareed, M. U., and J. G. Spivack. 1994. Two open reading frames (ORF1 and ORF2) within the 2.0-kilobase latency-associated transcript of herpes simplex virus type 1 are not essential for reactivation from latency. J. Virol. 68:8071-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrell, M. J., A. T. Dobson, and L. T. Feldman. 1991. Herpes simplex virus latency-associated transcript is a stable intron. Proc. Natl. Acad. Sci. USA 88:790-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grabe, N. 2002. AliBaba2: context specific identification of transcription factor binding sites. In Silico Biol. 2:S1-S15. [PubMed] [Google Scholar]
- 12.Gressens, P., and J. R. Martin. 1994. HSV-2 DNA persistence in astrocytes of the trigeminal root entry zone: double labeling by in situ PCR and immunohistochemistry. J. Neuropathol. Exp. Neurol. 53:127-135. [DOI] [PubMed] [Google Scholar]
- 13.Hill, J., A. Patel, P. Bhattacharjee, and P. Krause. 2003. An HSV-1 chimeric containing HSV-2 latency associated transcript (LAT) sequences has significantly reduced adrenergic reactivation in the rabbit eye model. Curr. Eye Res. 26:219-224. [DOI] [PubMed] [Google Scholar]
- 14.Hill, J. M., H. H. Garza, Jr., Y. H. Su, R. Meegalla, L. A. Hanna, J. M. Loutsch, H. W. Thompson, E. D. Varnell, D. C. Bloom, and T. M. Block. 1997. A 437-base-pair deletion at the beginning of the latency-associated transcript promoter significantly reduced adrenergically induced herpes simplex virus type 1 ocular reactivation in latently infected rabbits. J. Virol. 71:6555-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarman, R. G., J. M. Loutsch, G. B. Devi-Rao, M. E. Marquart, M. P. Banaszak, X. Zheng, J. M. Hill, E. K. Wagner, and D. C. Bloom. 2002. The region of the HSV-1 latency-associated transcript required for epinephrine-induced reactivation in the rabbit does not include the 2.0-kb intron. Virology 292:59-69. [DOI] [PubMed] [Google Scholar]
- 16.Jin, L., W. Peng, G. C. Perng, D. J. Brick, A. B. Nesburn, C. Jones, and S. L. Wechsler. 2003. Identification of herpes simplex virus type 1 latency-associated transcript sequences that both inhibit apoptosis and enhance the spontaneous reactivation phenotype. J. Virol. 77:6556-6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krause, P. R., L. R. Stanberry, N. Bourne, B. Connelly, J. F. Kurawadwala, A. Patel, and S. E. Straus. 1995. Expression of the herpes simplex virus type 2 latency-associated transcript enhances spontaneous reactivation of genital herpes in latently infected guinea pigs. J. Exp. Med. 181:297-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lafferty, W. E., R. W. Coombs, J. Benedetti, C. Critchlow, and L. Corey. 1987. Recurrences after oral and genital herpes simplex virus infection. Influence of site of infection and viral type. N. Engl. J. Med. 316:1444-1449. [DOI] [PubMed] [Google Scholar]
- 19.Margolis, T. P., Y. Imai, L. Yang, V. Vallas, and P. R. Krause. 2007. Herpes simplex virus type 2 (HSV-2) establishes latent infection in a different population of ganglionic neurons than HSV-1: role of latency-associated transcripts. J. Virol. 81:1872-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 21.Mehta, A., J. Maggioncalda, O. Bagasra, S. Thikkavarapu, P. Saikumari, T. Valyi-Nagy, N. W. Fraser, and T. M. Block. 1995. In situ DNA PCR and RNA hybridization detection of herpes simplex virus sequences in trigeminal ganglia of latently infected mice. Virology 206:633-640. [DOI] [PubMed] [Google Scholar]
- 22.Ng, A. K., T. M. Block, B. Aiamkitsumrit, M. Wang, E. Clementi, T. T. Wu, J. M. Taylor, and Y. H. Su. 2004. Construction of a herpes simplex virus type 1 mutant with only a three-nucleotide change in the branchpoint region of the latency-associated transcript (LAT) and the stability of its two-kilobase LAT intron. J. Virol. 78:12097-12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perng, G. C., D. Esmaili, S. M. Slanina, A. Yukht, H. Ghiasi, N. Osorio, K. R. Mott, B. Maguen, L. Jin, A. B. Nesburn, and S. L. Wechsler. 2001. Three herpes simplex virus type 1 latency-associated transcript mutants with distinct and asymmetric effects on virulence in mice compared with rabbits. J. Virol. 75:9018-9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perng, G. C., S. M. Slanina, A. Yukht, B. S. Drolet, W. Keleher, Jr., H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 1999. A herpes simplex virus type 1 latency-associated transcript mutant with increased virulence and reduced spontaneous reactivation. J. Virol. 73:920-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang, S., A. S. Bertke, A. Patel, K. Wang, J. I. Cohen, and P. R. Krause. 2008. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc. Natl. Acad. Sci. USA 105:10931-10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tatarowicz, W. A., C. E. Martin, A. S. Pekosz, S. L. Madden, F. J. Rauscher III, S. Y. Chiang, T. A. Beerman, and N. W. Fraser. 1997. Repression of the HSV-1 latency-associated transcript (LAT) promoter by the early growth response (EGR) proteins: involvement of a binding site immediately downstream of the TATA. J. Neurovirol. 3:212-224. [DOI] [PubMed] [Google Scholar]
- 27.Trousdale, M. D., I. Steiner, J. G. Spivack, S. L. Deshmane, S. M. Brown, A. R. MacLean, J. H. Subak-Sharpe, and N. W. Fraser. 1991. In vivo and in vitro reactivation impairment of a herpes simplex virus type 1 latency-associated transcript variant in a rabbit eye model. J. Virol. 65:6989-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Umbach, J. L., M. F. Kramer, I. Jurak, H. W. Karnowski, D. M. Coen, and B. R. Cullen. 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nat. Immunol. 454:780-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, K., P. R. Krause, and S. E. Straus. 1995. Analysis of the promoter and cis-acting elements regulating expression of herpes simplex virus type 2 latency-associated transcripts. J. Virol. 69:2873-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, K., G. Mahalingam, Y. Imai, P. L., T. T. Margolis, S. E. Straus, and J. I. Cohen. 2009. Cell type specific accumulation of the major latency-associated transcript (LAT) of herpes simplex virus type 2 in LAT transgenic mice. Virology 386:79-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, K., L. Pesnicak, E. Guancial, P. R. Krause, and S. E. Straus. 2001. The 2.2-kilobase latency-associated transcript of herpes simplex virus type 2 does not modulate viral replication, reactivation, or establishment of latency in transgenic mice. J. Virol. 75:8166-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang, L., C. C. Voytek, and T. P. Margolis. 2000. Immunohistochemical analysis of primary sensory neurons latently infected with herpes simplex virus type 1. J. Virol. 74:209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshikawa, T., J. M. Hill, L. R. Stanberry, N. Bourne, J. F. Kurawadwala, and P. R. Krause. 1996. The characteristic site-specific reactivation phenotypes of HSV-1 and HSV-2 depend upon the latency-associated transcript region. J. Exp. Med. 184:659-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshikawa, T., L. R. Stanberry, N. Bourne, and P. R. Krause. 1996. Downstream regulatory elements increase acute and latent herpes simplex virus type 2 latency-associated transcript expression but do not influence recurrence phenotype or establishment of latency. J. Virol. 70:1535-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]