Abstract
The severe acute respiratory syndrome coronavirus (SARS-CoV) generates 16 nonstructural proteins (nsp's) through proteolytic cleavage of a large precursor protein. Although several nsp's exhibit catalytic activities that are important for viral replication and transcription, other nsp's have less clearly defined roles during an infection. In order to gain a better understanding of their functions, we attempted to identify host proteins that interact with nsp's during SARS-CoV infections. For nsp2, we identified an interaction with two host proteins, prohibitin 1 (PHB1) and PHB2. Our results suggest that nsp2 may be involved in the disruption of intracellular host signaling during SARS-CoV infections.
Severe acute respiratory syndrome coronavirus (SARS-CoV) is a positive-stranded RNA virus that was first identified in late 2002 as the causative agent for an outbreak of atypical pneumonia originating from southern China (9, 20, 25). SARS-CoV possesses a ∼30-kb genome that consists of two large open reading frames (ORFs), ORF1a and ORF1ab, spanning two-thirds of the genome and 12 additional downstream ORFs (21, 28). The translation of ORF1a and ORF1ab results in the production of two large polyproteins, polyprotein 1a (pp1a) and polyprotein 1ab (pp1ab). Within ppa1a and pp1ab, two viral proteases, a papain-like protease (nsp3) and a 3C-like protease (nsp5), cleave pp1a and pp1ab into 11 and 16 smaller subunits, respectively (33).
Based on findings from studies of SARS-CoV and other, related coronaviruses, several nsp's have been shown to exhibit catalytic activities that are critical at different stages of the viral life cycle. For example, the activities of nsp3, the papain-like protease, and nsp5, the 3C-like protease, are required for cleavage of pp1a and pp1ab into the individual nsp's (33). Likewise, additional nsp's have also been shown to exhibit an RNA helicase activity (nsp13) (15), an exoribonuclease activity (nsp14) (4, 10, 11, 23), an endoribonuclease activity (nsp15) (1-3, 16, 18, 27), and a methyltransferase activity (nsp16) (7, 34). Together, these nsp's are thought to work in concert with the putative viral RNA-dependent RNA polymerase, nsp12, for replication of the full-length genomic RNA and for transcription of a nested set of subgenomic RNAs that encode additional accessory proteins and the structural proteins that are required for virion assembly (5, 30, 33). Other nsp's, however, have less clearly defined activities. For example, the deletion of nsp2 from the SARS-CoV genome results in only a modest reduction in viral titers, and nsp2 is currently thought to be dispensable for replication (13). However, it is unclear whether nsp2 would play a more critical role in the context of an actual infection, in which the virus encounters the selective influence of the immune system.
In order to gain a better understanding of the biology of nsp's, we sought to develop a general strategy for identifying host proteins that interact with viral proteins during an infection. Focusing on nsp1 and nsp2, we purified protein complexes containing C-terminally tagged nsp's and utilized multidimensional protein identification technology (MudPIT) analysis to generate a list of candidate host proteins that potentially interact with these two proteins. Although for nsp1 we were unable to confirm direct interaction with any of the candidate host proteins, for nsp2 we were able to confirm interactions with two proteins, prohibitin 1 (PHB1) and PHB2.
Expression of C-terminally tagged SARS-CoV nonstructural proteins.
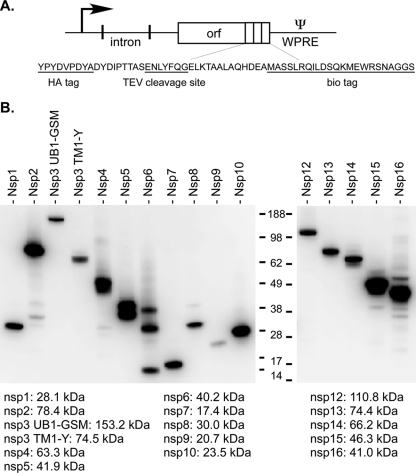
In order to purify nsp-containing protein complexes from SARS-CoV-infected cells, we utilized a 23-amino-acid affinity purification tag that is site-specifically biotinylated by the Escherichia coli biotin ligase BirA (6). All constructs were engineered to contain a C-terminal tag composed of a hemagglutinin (HA) tag followed by a tobacco etch virus protease cleavage site and the biotinylation (bio) tag (Fig. 1A). To test the expression of these constructs, HEK293 cells expressing the SARS-CoV receptor (HEK293/ACE2 cells) were transfected with individual plasmids for tagged nsp's by using FuGENE HD (Roche). After 2 days, protein expression levels were determined by probing Western blots with an anti-HA antibody (Covance). For these constructs, we routinely observed high levels of expression of nsp2, nsp5, nsp10, nsp15, and nsp16 (Fig. 1B). The only construct that failed to show expression was that for full-length nsp3. However, for two shorter nsp3 constructs, an N-terminal fragment spanning the start of nsp3 to the group II-specific marker (GSM) domain located immediately upstream of the first transmembrane domain (nsp3 UB1-GSM) and a C-terminal fragment spanning the first transmembrane domain to the end of nsp3 (nsp3 TM1-Y), we observed moderate levels of expression.
FIG. 1.
Expression of SARS-CoV nonstructural proteins with a C-terminal HA-bio tag. (A) Sequences encoding individual nsp's were cloned into a modified pCAGGS vector that generates transcripts containing the posttranscriptional regulatory element from the Woodchuck hepatitis virus (WPRE). All constructs fuse a C-terminal HA-bio tag to each protein. TEV, tobacco etch virus. (B) Western blotting analysis (with mouse anti-HA [1:1,000; Covance]) to monitor the expression of nsp's. The expected sizes for nsp's with the C-terminal HA-bio tag are listed below the blot. Samples for nsp2, nsp5, nsp7, and nsp10 were diluted 10× prior to loading.
Identification of candidate host proteins copurifying with SARS-CoV nsp2.
SARS-CoV infection has been shown to alter a cell's gene expression profile (8, 19, 32). Therefore, to better simulate the protein expression profile under these conditions, we pursued a purification strategy to isolate nsp-containing complexes from SARS-CoV-infected cells. We initially focused on isolating protein complexes incorporating C-terminally tagged nsp1, nsp2, nsp3 UB1-GSM, nsp3 TM1-Y, and nsp4. As a control to monitor the quality of protein purification, we also included a sample for enhanced yellow fluorescent protein (eYFP) with a C-terminal HA-bio tag. HEK293/ACE2 cells were cotransfected with individual nsp plasmids and a plasmid that expresses BirA. The following day, transfected cells were infected with SARS-CoV. One day following infection, cells were lysed in buffer containing 1% NP-40 and incubated with streptavidin paramagnetic beads (Invitrogen) to isolate protein complexes.
Following single-step purification, we observed multiple bands on Coomassie blue-stained gels for purified samples obtained with C-terminally tagged nsp1, nsp2, and nsp3 UB1-GSM which were absent for samples for nsp3 TM1-Y, nsp4, and eYFP (Fig. 2A). The significance of these multiple bands is presently unclear. In some additional purification runs, we also observed these multiple bands in purified samples for nsp3 TM1-Y and nsp4. However, these bands were never observed in additional purification runs with either eYFP or the nucleocapsid protein for an unrelated virus, the Andes hantavirus (data not shown). Hence, these multiple bands appear to be specifically associated with the expression of SARS-CoV nsp's.
FIG. 2.
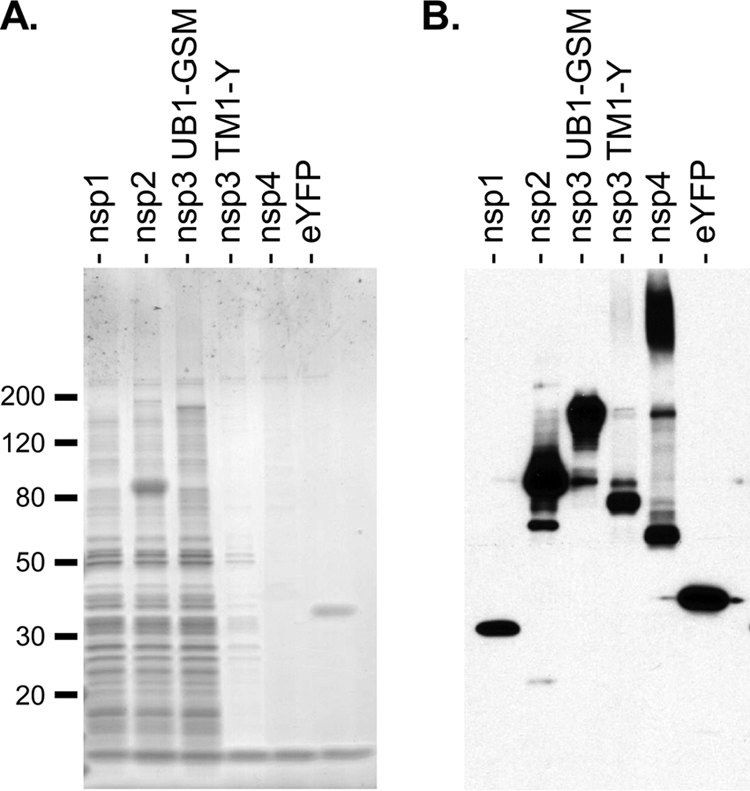
Single-step purification of nsp-containing protein complexes from SARS-CoV-infected cells. (A) Coomassie blue-stained gel of protein samples eluted from streptavidin beads; (B) Western blot analysis (with mouse anti-HA [1:1,000; Covance]) to monitor the amounts of C-terminally tagged nsp's eluted from streptavidin beads. Molecular sizes are given in kilodaltons.
Given the complexity in these purified samples, we performed MudPIT analysis in order to identify potential host proteins that interact with individual nsp's. The MudPIT approach is a sensitive mass spectrometry technique that can be used to identify the protein components in a complex mixture without prior separation by electrophoresis (14, 17, 29, 36). For an initial round of MudPIT analysis, we identified 590 unique host proteins for nsp1, 719 for nsp2, 770 for nsp3 UB1-GSM, 618 for nsp3 TM1-Y, 229 for nsp4, and 240 for eYFP. To reduce the list of candidate host proteins, host proteins that were present in the eYFP sample or were found to be common to all nsp samples were treated as background and subtracted from the list of candidates for each nsp sample. This reduced the total list of candidates to 25 for nsp1, 82 for nsp2, 86 for nsp3 TM1-Y, 40 for nsp3 TM1-Y, and 11 for nsp4. For nsp1 and nsp2, the list of candidate host proteins was further reduced by identifying host proteins that repeatedly associated with either nsp1 or nsp2 in additional rounds of purification and MudPIT analysis. This final reduced list of candidates consisted of 7 host proteins for nsp1 and 11 host proteins for nsp2 (Table 1).
TABLE 1.
Candidate host proteins potentially interacting with nsp1 and nsp2
| nsp | Host protein name (accession no.) | No. of unique peptidesa |
|---|---|---|
| nsp1 | DIDO1 (NP_149072.1) | 3, 2, 5 |
| USP7 (NP_003461.1) | 3, 3 | |
| CKAP5 (NP_001008938.1) | 3, 3 | |
| RFC2 (NP_002905.2) | 4, 6 | |
| CPNE3 (NP_003900.1) | 3, 3 | |
| RPP38 (NP_006405.1) | 3, 3 | |
| SNRNP48 (NP_689764.3) | 3, 3 | |
| nsp2 | GIGYF2 (NP_056390.2) | 16, 22 |
| PHB1 (NP_002625.1) | 9, 10 | |
| VDAC1 (NP_003365.1) | 7, 6 | |
| EIF4E2 (NP_004837.1) | 6, 9 | |
| PHB2 (NP_009204.1) | 6, 9 | |
| PCM1 (NP_006188.2) | 4, 5 | |
| SPEN (NP_055816.2) | 4, 3 | |
| MLL2 (NP_003473.1) | 3, 5 | |
| SLC1A5 (NP_005619.1) | 3, 3 | |
| EIF3M (NP_006351.2) | 3, 3 | |
| STOML2 (NP_038470.1) | 2, 7 |
Data for nsp1 are results from three independent purification and MudPIT runs; data for nsp2 are results from two independent purification and MudPIT runs.
SARS-CoV nsp2 interacts with PHB1 and PHB2.
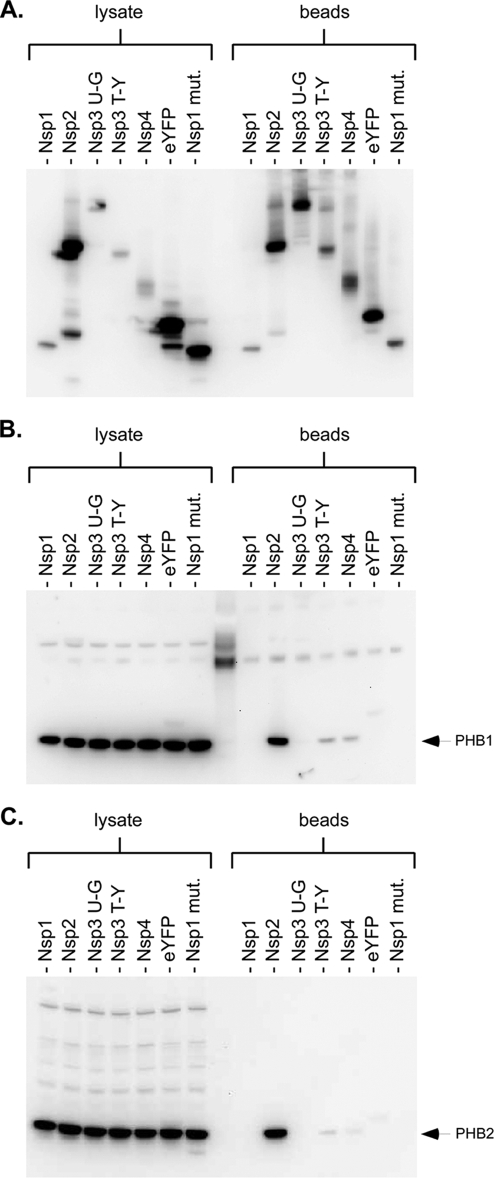
To validate the results from the MudPIT analysis, we again isolated protein complexes containing C-terminally tagged nsp1 and nsp2 and probed for the presence of candidate host proteins through Western blot analysis. For nsp1, we were unable to confirm direct interaction with any of the candidate host proteins identified. For nsp2, GIGYF2 was identified with the highest number of unique peptides in the MudPIT analysis. However, there are no antibodies currently available for this protein. Therefore, we were unable to determine whether this host protein indeed copurifies with nsp2. In Western blot analyses for the next candidate protein, PHB1, we observed a strong signal in purified samples for nsp2 (Fig. 3B). A weak signal could also be detected in purified samples for nsp3 TM1-Y and nsp4. However, no signal for PHB1 was detected in purified samples for nsp1, nsp3 UB1-GSM, eYFP, and an nsp1 mutant that no longer suppresses gene expression (24). We also probed for the presence of the next candidate protein, VDAC1, but did not observe any signal for this protein in any of the purified samples (data not shown). This result indicates that VDAC1 does not specifically interact with nsp2. Finally, because PHB1 is known to form a stable complex with a homologous protein, PHB2, and PHB2 was also identified in the MudPIT analysis, we probed purified samples for the presence of PHB2. Similar to the results with PHB1, the results with PHB2 showed a strong signal in purified samples for nsp2 and faint signals in purified samples for nsp3 TM1-Y and nsp4 (Fig. 3C). No detectable signal for PHB2 was observed in the remaining samples. The significance of the weak signals observed for both PHB1 and PHB2 in purified samples for nsp3 TM1-Y and nsp4 is unclear. Because we typically observed much less purified nsp3 TM1-Y and nsp4 than nsp2, it is possible that both nsp3 TM1-Y and nsp4 do, in fact, interact with PHB1 and PHB2 (Fig. 3A, compare beads for nsp2, nsp3 TM1-Y, and nsp4). Alternatively, because PHB1, PHB2, nsp3 TM1-Y, and nsp4 are integral membrane proteins, the weak signals for PHB1 and PHB2 may simply be the result of nonspecific hydrophobic interactions of transmembrane domains. Nonetheless, for nsp2, the interaction with PHB1 and PHB2 can be confirmed as a specific interaction since both nsp3 UB1-GSM and eYFP were purified at levels comparable to that of nsp2 but were both unable to copurify either PHB1 or PHB2.
FIG. 3.
nsp2 interacts with PHB1 and PHB2. (A) Western blot analysis (with mouse anti-HA [Covance]) to monitor the amount of nsp present in the starting lysate and the amount bound to streptavidin beads; (B) Western blot analysis (with rabbit anti-PHB1 [1:500; Santa Cruz Biotech]) to monitor the amount of PHB1 present in the starting lysate and the amount bound to streptavidin beads; (C) Western blot analysis (with rabbit anti-PHB2 [1:500; Santa Cruz Biotech]) to monitor the amount of PHB2 present in the starting lysate and the amount bound to streptavidin beads. nsp3 U-G, nsp3 UB1-GSM; nsp3 T-Y, nsp3 TM1-Y; mut., mutant.
PHB1 and PHB2 are two evolutionarily conserved proteins that have been implicated in a number of cellular functions. Among these are cell cycle progression (35), cell migration (26), cellular differentiation (31), apoptosis (12), and mitochondrial biogenesis (22). Our finding that nsp2 interacts with PHB1 and PHB2 suggests that nsp2, rather than playing a role in viral replication, may be involved in altering the host cell environment.
Acknowledgments
We thank Jason Botten, Jeremiah Joseph, and Benjamin Neuman for discussions and critical reviews of the manuscript.
This work was supported by National Institutes of Health grants AI059799 (to M.J.B.), P41 RR011823 (to J.R.Y.), and BIMR P30NS057096 (to L.L.) and contract HHSN266200400058C (to P.K.). C.T.C.-T. was supported by grant T32 AI-07354.
Footnotes
Published ahead of print on 29 July 2009.
REFERENCES
- 1.Bhardwaj, K., L. Guarino, and C. C. Kao. 2004. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 78:12218-12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhardwaj, K., S. Palaninathan, J. M. Alcantara, L. L. Yi, L. Guarino, J. C. Sacchettini, and C. C. Kao. 2008. Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. J. Biol. Chem. 283:3655-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhardwaj, K., J. Sun, A. Holzenburg, L. A. Guarino, and C. C. Kao. 2006. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J. Mol. Biol. 361:243-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, P., M. Jiang, T. Hu, Q. Liu, X. S. Chen, and D. Guo. 2007. Biochemical characterization of exoribonuclease encoded by SARS coronavirus. J. Biochem. Mol. Biol. 40:649-655. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, A., W. Zhang, Y. Xie, W. Jiang, E. Arnold, S. G. Sarafianos, and J. Ding. 2005. Expression, purification, and characterization of SARS coronavirus RNA polymerase. Virology 335:165-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Boer, E., P. Rodriguez, E. Bonte, J. Krijgsveld, E. Katsantoni, A. Heck, F. Grosveld, and J. Strouboulis. 2003. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc. Natl. Acad. Sci. USA 100:7480-7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Decroly, E., I. Imbert, B. Coutard, M. Bouvet, B. Selisko, K. Alvarez, A. E. Gorbalenya, E. J. Snijder, and B. Canard. 2008. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2′O)-methyltransferase activity. J. Virol. 82:8071-8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lang, A., T. Baas, T. Teal, L. M. Leijten, B. Rain, A. D. Osterhaus, B. L. Haagmans, and M. G. Katze. 2007. Functional genomics highlights differential induction of antiviral pathways in the lungs of SARS-CoV-infected macaques. PLoS Pathog. 3:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 10.Eckerle, L. D., S. M. Brockway, S. M. Sperry, X. Lu, and M. R. Denison. 2006. Effects of mutagenesis of murine hepatitis virus nsp1 and nsp14 on replication in culture. Adv. Exp. Med. Biol. 581:55-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckerle, L. D., X. Lu, S. M. Sperry, L. Choi, and M. R. Denison. 2007. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 81:12135-12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fusaro, G., P. Dasgupta, S. Rastogi, B. Joshi, and S. Chellappan. 2003. Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. J. Biol. Chem. 278:47853-47861. [DOI] [PubMed] [Google Scholar]
- 13.Graham, R. L., A. C. Sims, S. M. Brockway, R. S. Baric, and M. R. Denison. 2005. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J. Virol. 79:13399-13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guelman, S., T. Suganuma, L. Florens, S. K. Swanson, C. L. Kiesecker, T. Kusch, S. Anderson, J. R. Yates III, M. P. Washburn, S. M. Abmayr, and J. L. Workman. 2006. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Mol. Cell. Biol. 26:871-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivanov, K. A., V. Thiel, J. C. Dobbe, Y. van der Meer, E. J. Snijder, and J. Ziebuhr. 2004. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 78:5619-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joseph, J. S., K. S. Saikatendu, V. Subramanian, B. W. Neuman, M. J. Buchmeier, R. C. Stevens, and P. Kuhn. 2007. Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch. J. Virol. 81:6700-6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaeser, M. D., A. Aslanian, M. Q. Dong, J. R. Yates III, and B. M. Emerson. 2008. BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J. Biol. Chem. 283:32254-32263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang, H., K. Bhardwaj, Y. Li, S. Palaninathan, J. Sacchettini, L. Guarino, J. L. Leibowitz, and C. C. Kao. 2007. Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease. J. Virol. 81:13587-13597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knoops, K., M. Kikkert, S. H. Worm, J. C. Zevenhoven-Dobbe, Y. van der Meer, A. J. Koster, A. M. Mommaas, and E. J. Snijder. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 21.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 22.Merkwirth, C., and T. Langer. 2009. Prohibitin function within mitochondria: essential roles for cell proliferation and cristae morphogenesis. Biochim. Biophys. Acta 1793:27-32. [DOI] [PubMed] [Google Scholar]
- 23.Minskaia, E., T. Hertzig, A. E. Gorbalenya, V. Campanacci, C. Cambillau, B. Canard, and J. Ziebuhr. 2006. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA 103:5108-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narayanan, K., C. Huang, K. Lokugamage, W. Kamitani, T. Ikegami, C. T. Tseng, and S. Makino. 2008. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 82:4471-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poutanen, S. M., D. E. Low, B. Henry, S. Finkelstein, D. Rose, K. Green, R. Tellier, R. Draker, D. Adachi, M. Ayers, A. K. Chan, D. M. Skowronski, I. Salit, A. E. Simor, A. S. Slutsky, P. W. Doyle, M. Krajden, M. Petric, R. C. Brunham, and A. J. McGeer. 2003. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 348:1995-2005. [DOI] [PubMed] [Google Scholar]
- 26.Rajalingam, K., C. Wunder, V. Brinkmann, Y. Churin, M. Hekman, C. Sievers, U. R. Rapp, and T. Rudel. 2005. Prohibitin is required for Ras-induced Raf-MEK-ERK activation and epithelial cell migration. Nat. Cell Biol. 7:837-843. [DOI] [PubMed] [Google Scholar]
- 27.Ricagno, S., M. P. Egloff, R. Ulferts, B. Coutard, D. Nurizzo, V. Campanacci, C. Cambillau, J. Ziebuhr, and B. Canard. 2006. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc. Natl. Acad. Sci. USA 103:11892-11897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 29.Sato, S., C. Tomomori-Sato, T. J. Parmely, L. Florens, B. Zybailov, S. K. Swanson, C. A. Banks, J. Jin, Y. Cai, M. P. Washburn, J. W. Conaway, and R. C. Conaway. 2004. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol. Cell 14:685-691. [DOI] [PubMed] [Google Scholar]
- 30.Sawicki, S. G., D. L. Sawicki, and S. G. Siddell. 2007. A contemporary view of coronavirus transcription. J. Virol. 81:20-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun, L., L. Liu, X. J. Yang, and Z. Wu. 2004. Akt binds prohibitin 2 and relieves its repression of MyoD and muscle differentiation. J. Cell Sci. 117:3021-3029. [DOI] [PubMed] [Google Scholar]
- 32.Tang, B. S., K. H. Chan, V. C. Cheng, P. C. Woo, S. K. Lau, C. C. Lam, T. L. Chan, A. K. Wu, I. F. Hung, S. Y. Leung, and K. Y. Yuen. 2005. Comparative host gene transcription by microarray analysis early after infection of the Huh7 cell line by severe acute respiratory syndrome coronavirus and human coronavirus 229E. J. Virol. 79:6180-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 34.von Grotthuss, M., L. S. Wyrwicz, and L. Rychlewski. 2003. mRNA cap-1 methyltransferase in the SARS genome. Cell 113:701-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, S., N. Nath, M. Adlam, and S. Chellappan. 1999. Prohibitin, a potential tumor suppressor, interacts with RB and regulates E2F function. Oncogene 18:3501-3510. [DOI] [PubMed] [Google Scholar]
- 36.Washburn, M. P., D. Wolters, and J. R. Yates III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242-247. [DOI] [PubMed] [Google Scholar]