Abstract
An interaction between the helicase domain of the Tobacco mosaic virus (TMV) 126-/183-kDa replicase protein(s) and the Arabidopsis thaliana NAC domain transcription factor ATAF2 was identified via yeast two-hybrid and in planta immunoprecipitation assays. ATAF2 is transcriptionally induced in response to TMV infection, and its overexpression significantly reduces virus accumulation. Proteasome inhibition studies suggest that ATAF2 is targeted for degradation during virus infection. The transcriptional activity of known defense-associated marker genes PR1, PR2, and PDF1.2 significantly increase within transgenic plants overexpressing ATAF2. In contrast, these marker genes have reduced transcript levels in ATAF2 knockout or repressor plant lines. Thus, ATAF2 appears to function in the regulation of host basal defense responses. In response to TMV infections, ATAF2 and PR1 display increased transcript accumulations in inoculated tissues but not in systemically infected tissues. ATAF2 and PR1 transcript levels also increase in response to salicylic acid treatment. However, the salicylic acid treatment of systemically infected tissues did not produce a similar increase in either ATAF2 or PR1 transcripts, suggesting that host defense responses are attenuated during systemic virus invasion. Similarly, noninfected ATAF2 knockout or ATAF2 repressor lines display reduced levels of PR1 transcripts when treated with salicylic acid. Taken together, these findings suggest that the replicase-ATAF2 interaction suppresses basal host defenses as a means to promote systemic virus accumulation.
The suppression of host defenses by invading pathogens represents a key step in the establishment of a successful infection. When associated with plants, pathogens/microbes generally produce sets of molecular components termed pathogen/microbe-associated molecular patterns (PAMPs/MAMPs) (2, 41, 46). Examples of PAMPs include the bacterial elongation factor EF-TU, flagellin, and fungal chitin (26, 43, 49). Plants have evolved to recognize these molecular components and respond with the induction of diverse defense mechanisms (33, 41). PAMP recognition involves the production of reactive oxygen species, increases in cell wall structural components, and the generation of numerous defense proteins and metabolites that function as a first line of defense against the establishment of infection (5, 33, 69). To be successful, pathogens have evolved numerous mechanisms to suppress these PAMP-induced defense responses. This includes targeting the regulatory mechanisms that mediate PAMP-induced responses (66). For example, Pseudomonas syringae secretes effector proteins HopM1 and HopT1-1 to disrupt vesicle trafficking as a means to suppress extracellular cell wall-associated defenses and to suppress cellular microRNA defense pathways, respectively (52, 53). The evidence from these and related studies indicates that pathogens target key cellular pathways to suppress a correlated array of host defenses. Thus, the identification of pathogen-targeted regulatory mechanisms represents an important aspect in understanding pathogenesis and the development of disease.
The dependent nature of viral pathogens on their hosts dictates that their entire genomes potentially function as PAMPs in the induction of host defenses. In addition, transcriptional microarray studies investigating host responses to different plant viruses have established that up to one-third of the host genes induced during susceptible infections are defense related (28, 38, 71). This finding implies that plants are adept at recognizing and responding to virus-derived molecular patterns. The ability of viruses to establish a systemic infection even during host defense activation suggests that viruses possess mechanisms that suppress the efficacy of these defenses. Perhaps the best-studied example of plant virus-mediated defense suppression involves the host recognition of virus-associated RNAs and the subsequent induction of RNA interference (RNAi) defense pathways. To overcome this defense mechanism, viruses encode suppressor proteins that disrupt specific steps in the RNAi pathway (17, 18). The deletion or mutation of a virus's suppressor protein typically compromises virus replication and inhibits infection. Therefore, the suppression of the RNAi defense pathway represents an important, if not essential, anti-defense mechanism. However, the diversity of defense responses induced during virus infections suggests that numerous additional host defense pathways are activated during systemic infections (71). Thus, it seems likely that plant viruses also encode the ability to suppress additional host responses.
In this study, we describe an interaction between the Tobacco mosaic virus (TMV) replicase protein and the NAC domain protein ATAF2 (At5g08790), which is associated with altered host defense responses and changes in virus accumulation. NAC domain proteins represent a large family of plant-specific transcription factors that have been associated with developmental processes, senescence, and defense (55). Members of this protein family contain highly conserved N-terminal domains divided into five subdomains (A through E) and a divergent C-terminal domain involved in transcriptional activation (Fig. 1A) (22, 34, 42). Subdomains D and E appear to comprise a novel DNA binding motif (22, 23, 56), while C and D function in nuclear localization (42). Despite the large number of proteins that comprise this family (∼75 members in rice and ∼105 in Arabidopsis), only a few have been characterized (56). Of the NAC proteins characterized, several, including ATAF2, are induced in response to pathogen invasion (13, 16, 34). Additionally, several NAC family members have been found to associate with specific virus-encoded proteins and can affect virus biology. In wheat, GRAB1 and GRAB2 (for geminivirus RepA-binding) proteins interact with the RepA protein of Wheat dwarf virus (72). Although a direct role for the GRAB-RepA interaction in the virus life cycle is unknown, the overexpression of GRAB proteins severely impairs virus replication in cultured cells (72). Another geminivirus-encoded protein, REn from Tomato leaf curl virus, has been shown to interact with the tomato-encoded NAC protein SlNAC1 (for Solanum lycopersicum NAC1) (64). Interestingly, SlNAC1 is induced in response to infection, and its expression enhances virus accumulation. A third NAC protein, TIP (for TCV-interacting protein), has been found to interact with the coat protein of Turnip crinkle virus (TCV) (61). Interaction with the virus coat protein disrupts TIP localization to the nucleus, while TIP mutation increases the accumulation of both TCV and Cucumber mosaic virus, suggesting that TIP plays a role in basal defense responses (40, 62).
FIG. 1.
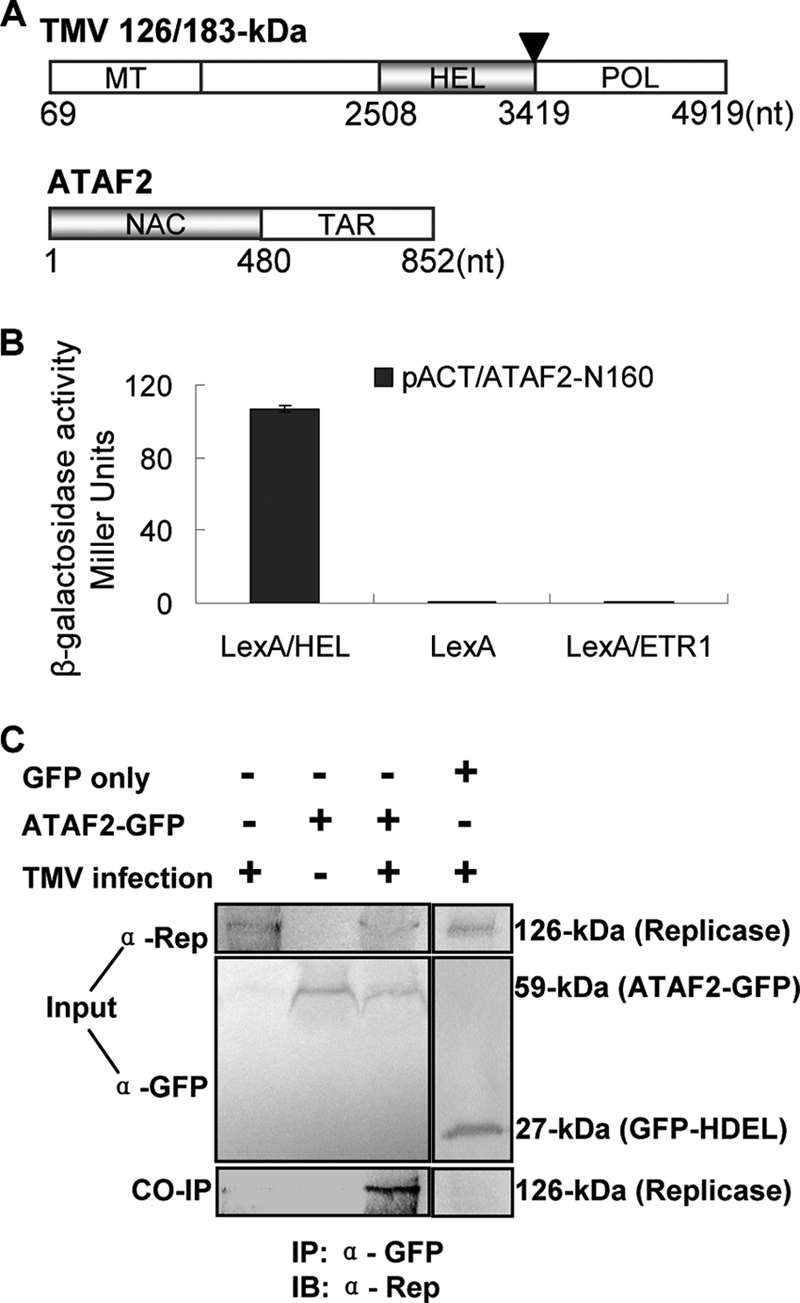
TMV replicase interacts with the NAC domain transcription factor ATAF2. (A) Schematic representation of TMV replicase and ATAF2 ORFs. Gray boxes represent the bait (126-/183-kDa replicase) and prey (ATAF2) interacting regions. MT, methyltransferase; HEL, helicase; POL, polymerase; TAR, transcriptional activation region. (B) Quantitative β-galactosidase assays for interaction between the helicase domain of the TMV replicase protein (nt 2508 to 3419) and the N-terminal 160-amino-acid NAC domain of ATAF2. (C) Coimmunoprecipitation (CO-IP) of ATAF2-GFP fusion protein and the TMV 126-kDa replicase protein within infected and noninfected tissues. α-Rep, anti-Rep antibody; α-GFP, anti-GFP antibody.
The TMV replicase functions as a suppressor of RNAi and is known to associate with several host factors, including TOM1 (for tobamovirus multiplication 1), a putative membrane anchor protein; the tobacco N resistance gene; a chloroplast protein NRIP1 (for N receptor-interacting protein 1) involved in host resistance; and an auxin-responsive transcription factor that affects disease development (7, 8, 19, 57, 67, 73). Thus, in addition to a role in virus replication, the TMV replicase also functions to interface with the host and regulate cellular processes. Consistent with this multifunctional role, our studies demonstrate that interaction with the TMV replicase results in the degradation of ATAF2 and the corresponding disruption of its defense-modulating functions. As a result, ATAF2-induced host defense responses are attenuated during virus infection.
MATERIALS AND METHODS
Plant material, virus infections, and chemical treatment.
Transgenic seedlings first were selected on Murashige and Skoog medium supplemented with 50 μg/ml of kanamycin. After 2 weeks, kanamycin-resistant seedlings were transferred to LC1 potting mix (SunGro Inc., Bellevue, WA). Plants were grown on a 16-h light/8-h dark cycle at 23°C. Leaves of 4-week-old Arabidopsis plants were dusted with carborundum (Fisher Scientific Company, Pittsburgh, PA) and inoculated using a cotton swab with a solution of purified virus (0.1 mg/ml) in 0.1 M sodium phosphate buffer, pH 7.0, or mock inoculated with buffer only.
TMV-infected tissues were collected and homogenized in sample buffer (44). Five micrograms of total protein for each sample, determined by Bradford assay (6), were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred to nitrocellulose paper. Transferred protein blots were probed with a polyclonal rabbit antiserum raised against TMV coat protein (CP), followed by an alkaline phosphatase-conjugated secondary anti-rabbit immunoglobulin G (Sigma, St. Louis, MO). Protein blots were developed by the addition of nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (Fisher Scientific, Pittsburgh, PA). Blots were electronically scanned, and CP levels were compared to known CP standards using AlphaImage software (Alpha Innotech Corp., San Leandro, CA).
Unless specified otherwise, 0.1 mM of salicylic acid (SA) solution (in distilled water) was sprayed onto Arabidopsis leaves prior to inoculation with TMV or real-time quantitative reverse transcription-PCR (qRT-PCR) analysis. For proteasome inhibitor treatment, plant leaves were submerged in 25 μM of MG132 solution (Z-Leu-Leu-Leu-al; Sigma, St. Louis, MO) for 2 h (32). Treated tissues then were subjected to Western immunoblot analysis for the detection of ATAF2-green fluorescent protein (ATAF2-GFP) protein.
Yeast two-hybrid and β-galactosidase assays.
A TMV cDNA fragment (nucleotides [nt] 2508 to 3419) encoding the full-length helicase domain was PCR amplified to carry 5′ SmaI and 3′ SalI restriction sites and cloned in frame into the yeast vector pLexA-NLS to create the bait construct pLexA-HEL (29). This bait construct was cotransformed with the Arabidopsis/GAL4 prey library (CD4-10 from the Arabidopsis Biological Resource Center, Columbus, OH) into Saccharomyces cerevisiae strain L40. Positive transformants for bait-prey interactions were selected on yeast minimal medium lacking uracil, tryptophan, and leucine at 25°C. Positive clones were shuttled into Escherichia coli for sequence determination (70). Interactions were confirmed by retransformation into yeast containing pLexA-HEL as well as control constructs consisting of the empty pLexA-NLS or one encoding the ethylene receptor component ETR1 (11). Positive transformants were assayed quantitatively for β-galactosidase activity in the presence of o-nitrophenyl-β-d-galactopyranoside (ONPG) as previously described (29). Western immunoblots for the detection of Gal4 and LexA fusion proteins were used to confirm the presence of LexA-HEL and Gal4-ATAF2 peptides in yeast (29 and data not shown).
In vivo pull-down assay.
Systemically infected leaf tissue (14 days postinoculation [dpi]) of 35S::ATAF2-GFP transgenic Arabidopsis plants were collected and homogenized in extraction buffer containing 50 mM Tris-HCl at pH 8.0, 150 mM NaCl, 0.5% Triton X-100, 0.2% 2-mercaptoethanol, 5% glycerol, phenylmethylsulfonyl fluoride, and proteinase inhibitor cocktail (Sigma, St. Louis, MO). Tissue extracts were centrifuged for 10 min at 15,000 rpm. The supernatant (1 ml) was incubated with 5 μl of anti-GFP antibody (Sigma, St. Louis, MO) at 4°C overnight with gentle shaking, followed by the addition of 30 μl of protein A agarose (Invitrogen, Carlsbad, CA) to the protein complex for an additional 3 h. Immune complexes then were precipitated by centrifugation and washed three times in 1 ml wash buffer (25 mM Tris-HCl at pH 7.5, 250 mM NaCl, 2 mM EDTA, 0.05% TritonX-100, 1 mM PMSF). The precipitated protein complexes were resuspended in 2× Laemmli sample buffer (44) and separated by SDS-PAGE. Resolved proteins were visualized via Western blotting as described above using GFP-specific antibodies (Clontech, Palo Alto, CA).
ATAF2 transient expression assay.
To create the 35S::ATAF2-GFP fusion construct, the ATAF2 open reading frame (ORF) was RT-PCR amplified from total RNA derived from 4-week-old Shadhara leaves. The PCR-amplified ORF was modified to contain 5′ KpnI and 3′ BsiWI restriction sites for cloning into pCMC-GFP (21). The designated plasmid, pCMC-ATAF2-GFP, utilizes the Cauliflower mosaic virus (CaMV) 35S promoter and the nopaline synthase polyadenylation signal for the expression of the GFP-fused ORF. The transient expression of ATAF2-GFP in Nicotiana benthamiana epidermal cells was achieved using a particle bombardment method as described previously (57). Bombarded tissues were incubated at room temperature for 12 to 14 h and visualized under an LSM510 laser-scanning confocal microscope with dry (magnification, ×10; numerical aperture, 0.8) and wet (with water; magnification, ×63; numerical aperture, 0.8) immersion lenses (Carl Zeiss Inc., Thonwood, NY). Images were further analyzed with Zeiss LSM Imager Examiner software, version 3.0.
Plant transformation and analysis.
pCMC-ATAF2-GFP was used as a template to amplify the ATAF2-GFP ORF using primers containing 5′ KpnI and 3′ PstI restriction sites. The amplified ATAF2-GFP ORF was cloned downstream of the CaMV 35S promoter within the binary transformation vector pBI121 (Clontech, Palo Alto, CA) to produce pBI-ATAF2-GFP. Similarly, GFP-specific primers with sequences for a 5′ KpnI site and 3′ PstI site encoding the endoplasmic reticulum retention signal HDEL was used to amplify and clone pBI-GFP-HDEL.
For promoter analysis, a 2-kb DNA fragment immediately upstream of the ATAF2 coding region was PCR amplified from ecotype Shahdara genomic DNA using primers containing 5′ PstI and 3′ BamHI restriction sites. The resulting DNA fragment was cloned upstream of the β-glucuronidase (GUS) coding sequence found in the binary transformation vector pBI101.1 (Clontech, Palo Alto, CA), resulting in pBI-PATAF2::GUS.
To make the ATAF2 repressor construct, a linker encoding a modified 12-amino-acid (aa) EAR motif repression domain SRDX (amino acids LDLDLELRFGFA) (35) was fused to the 3′ end of the ATAF2 ORF via 5′ BsiWI and 3′ PstI restriction sites. The ATAF2-SRDX fusion construct was cloned downstream of a double CaMV 35S promoter within the vector pPily (27), and the cassette containing the promoter, the ATAF2 repressor fusion, and an NOS terminator was transferred via unique KpnI restriction sites into the binary vector pBin19PLUS (68) to produce pBin-ATAF2-SRDX.
Three binary vector constructs, pBI-ATAF2-GFP, pBI-PATAF2::GUS, and pBin-ATAF2-SRDX, were transformed into Agrobacterium tumefaciens strain GV3101 (37). Transgenic Arabidopsis thaliana ecotype Shahdara plants, including 35S::ATAF2-GFP overexpression lines, PATAF2::GUS promoter lines, and 35S::ATAF2-SRDX repression lines, were created via the floral dip method (12). Genomic DNA from transformed seedlings displaying kanamycin resistance was PCR analyzed for the appropriate construct to confirm transformation. The T1 generations of 35S::ATAF2-GFP and 35S::ATAF2-SRDX were used for virus infection, in vivo pull-down assays, and real-time qRT-PCR analysis. The T2 generation of PATAF2::GUS was used for the promoter study.
Histochemical staining for GUS activity was performed as previously described (39). In summary, plant tissues were vacuum infiltrated in reaction buffer containing 1 mg/ml X-Gluc (5-bromo-4-chloro-3-indolyl p-d-glucuronide), 10 mM EDTA, 100 mM NaH2PO4, pH 7.0, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, and 0.1% vol/vol Triton and incubated at 37°C overnight. Stained tissues then were cleared in 70% ethanol prior to being photographed.
Real-time qRT-PCR.
Total RNA from Arabidopsis plants was purified using the RNeasy RNA extraction kit (Qiagen, Valencia, CA). One microgram of total RNA was treated with RQ1 DNase (Promega, Madison, WI), followed by reverse transcription using a SuperScript first-strand synthesis system (Invitrogen, Carlsbad, CA). SYBR green real-time qRT-PCR was performed in 96-well reaction plates in an ABI Prism 7100 (Applied Biosystems, Foster City, CA). Each 25-μl reaction mixture contains 12.5 μl of SYBR green 2× master mix (ABI, Foster City, CA), 1 μl of forward and reverse primer mix (7.5 μM), 1 μl of cDNA, and 10.5 μl of diethyl pyrocarbonate-treated H2O. Real-time qRT-PCR amplification was carried out for 2 min at 50°C and 10 min at 95°C, and this was followed by 40 amplification cycles of 15 s at 95°C and 1 min at 60°C. The 18S RNA gene was chosen as an internal control for normalization. Total RNA for each real-time qRT-PCR run was pooled from three to five individual test plants. Individual primer sequences were designed using the Primer Express 2.0 software (ABI, Foster City, CA).
RESULTS
The TMV replicase interacts with the NAC domain protein ATAF2.
The helicase domain (nt 2508 to 3419) of the TMV 126- and/or 183-kDa replicase protein(s) was used as the bait in a yeast two-hybrid screen against an Arabidopsis thaliana ecotype Nossen cDNA library (Arabidopsis Biological Resource Center, Columbus, OH). One clone that induced positive β-galactosidase activity contained cDNA (nt 1 to 481) encoding the N-terminal 160-aa NAC domain of the 32-kDa protein ATAF2 (Fig. 1A and B). Subsequently, a cDNA clone encoding the entire ATAF2 ORF was PCR amplified from A. thaliana ecotype Shahdara, a highly susceptible host for TMV (15). Sequence comparisons indicated that ATAF2 cDNA derived from Shahdara and Columbia share 100% identity with the interacting two-hybrid clone.
To examine this interaction in planta, an immuno-pull-down assay was developed using A. thaliana ecotype Shahdara plants transformed with a 35S::ATAF2-GFP fusion construct for the expression of the full-length ATAF2 ORF fused at the C terminus to the GFP. Transgenic plants expressing ATAF2-GFP were mechanically inoculated with TMV, and leaf extracts were subjected to immuno-pull down after 14 dpi using GFP-specific antibodies. The 126-kDa viral replicase protein was significantly enriched in GFP-specific pull-down extracts from TMV-infected leaf tissue expressing ATAF2-GFP (Fig. 1C). In contrast, similar pull-down assays using noninfected ATAF2-GFP or infected tissue from nontransgenic Shahdara plants failed to yield detectable levels of the viral 126-kDa protein (Fig. 1C). An additional control using TMV-infected leaf tissues from transgenic Nicotiana benthamiana plants expressing a 35S::GFP-HDEL fusion construct (30) for GFP localization to the endoplasmic reticulum also failed to pull down detectable levels of 126-kDa protein, indicating that the interaction observed for the ATAF2-GFP construct is specific to the ATAF2 peptide. Taken together, these findings indicate that ATAF2 is capable of interacting with the full-length replicase within the context of a virus infection.
TMV infection interferes in ATAF2-GFP accumulation.
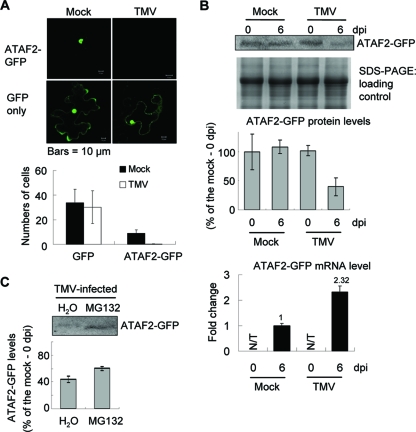
Like other NAC domain family members, ATAF2 contains a predicted nuclear localization signal at aa 68 to 74 (51). Transient expression assays using an ATAF2-GFP fusion construct under the control of the 35S promoter confirmed that ATAF2-GFP localized predominately to the nucleus (Fig. 2A). Similar transient expression assays with TMV-infected tissues, however, showed no GFP-derived fluorescence. In contrast, the transient expression of GFP alone produced similar patterns of localization and displayed no difference in the number of fluorescent cells in either mock- or TMV-infected tissues (Fig. 2A). This observation suggested that during TMV infection, ATAF2 fails to accumulate.
FIG. 2.
TMV-directed degradation of ATAF2. (A) Fluorescent micrographs of transient ATAF2-GFP or GFP expression in mock- or TMV-infected N. benthamiana epidermal cells. Fluorescent images were taken 14 to 16 h after bombardment. The numbers of cells expressing detectable ATAF2-GFP or GFP fluorescent signal in mock- or TMV-infected tissues are shown. Cell numbers were averaged from 10 independent bombardment experiments. (B) The upper panel shows the reduction of ATAF2-GFP levels in either mock- or TMV-infected 35S::ATAF2-GFP transgenic plants at 6 dpi. The middle panel shows the Coomassie blue detection of total proteins loaded in each lane (SDS-PAGE loading control). Results are averaged from three independent experiments ± the standard deviations (SD). The lower panel shows the real-time qRT-PCR analysis of transcript levels for the ATAF2-GFP transgene in mock- and TMV-infected tissues. (C) The upper panel shows that proteasome inhibition via treatment with MG132 restores ATAF2-GFP accumulation in TMV-infected tissues. In the lower panel, results are presented as averages ± SD from three independent experiments. N/T, not tested.
To quantify ATAF2 accumulation during infection, transgenic 35S::ATAF2-GFP Shahdara plants were assayed for ATAF2-GFP accumulation in either mock- or TMV-inoculated tissues at 6 dpi. Pull-down experiments (Fig. 1C) confirmed that the ATAF2-GFP protein maintains its interaction with the virus replicase. Western immunoblot analysis for the detection of ATAF2-GFP averaged from three independent transgenic lines revealed significantly lower accumulations of ATAF2-GFP in TMV-infected leaf tissues (Fig. 2B, upper and middle). SDS-PAGE analysis showed no significant difference in total protein accumulations between infected and noninfected tissues (Fig. 2B). Additionally, real-time qRT-PCR analysis indicated that transcript levels of the ATAF2-GFP transgene were slightly higher within TMV-infected tissues (Fig. 2B, lower), indicating that the virus inhibition of ATAF-GFP transcription was not a factor in observed reductions in ATAF2-GFP protein accumulation. Taken together, these observations are consistent with the virus targeting ATAF2 for degradation, possibly via the proteasome. To test this possibility, the accumulation of ATAF2-GFP was monitored in infected tissues in both the presence and absence of the proteasome inhibitor MG132. Inoculated leaf tissue submerged in 25 μM MG132 accumulated significantly higher levels of ATAF-GFP than did similar tissues submerged in water (Fig. 2C), suggesting that proteasome activity is responsible for TMV-induced perturbations in ATAF2 accumulation. The partial reductions in ATAF2-GFP accumulation observed in infected tissue possibly reflect the nonsynchronous nature of the virus infection. Specifically, only a subset of cells within the infected tissue is actively supporting virus replication at any one time. Since whole-leaf tissue was sampled for these experiments, it is likely that only a portion was supporting active virus replication. Thus, the TMV-targeted degradation of ATAF2 may occur in only a subset of infected cells, perhaps only those supporting virus replication.
ATAF2 overexpression inhibits TMV accumulation.
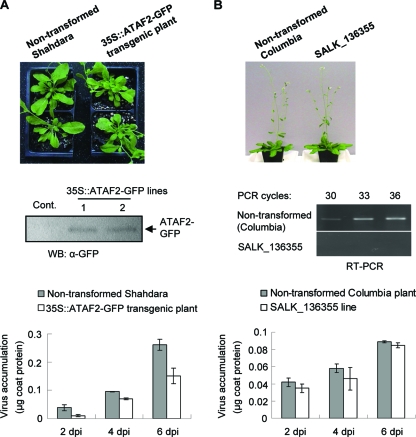
To examine the effect of ATAF2 on TMV accumulation, two independent transgenic lines expressing readily detectable levels of the ATAF2-GFP fusion construct were selected for experimentation. Plants from these two lines displayed similar mild developmental abnormalities that included altered rosette formation and the loss of apical dominance (Fig. 3A). However, plant growth and leaf sizes were not noticeably different from those of the nontransgenic control plants. Fully expanded leaves from 4- to 5-week-old plants were mechanically inoculated with 0.1 mg/ml of purified TMV. An analysis of leaf tissue collected at 2, 4, and 6 dpi demonstrated that TMV accumulations average 40% less in transgenic plants expressing ATAF2-GFP than virus accumulations found in similarly inoculated nontransgenic control plants (Fig. 3A). Based on these findings, we conclude that ATAF2 accumulation prior to infection results in a reduction in overall virus accumulation.
FIG. 3.
Effects of ATAF2 expression on virus accumulation. (A) 35S::ATAF2-GFP transgenic Shahdara plants (4 weeks old) show a moderate developmental phenotype (upper); ATAF2-GFP accumulation was confirmed by Western immunoblot (WB) analysis for the detection of GFP (middle); and the accumulation of TMV CP in 35S::ATAF2-GFP transgenic lines is reduced compared to that of nontransformed control (cont.) plants (lower). The means and standard deviations are averaged from three independent experiments using two different ATAF2 overexpression lines. α-GFP, anti-GFP antibody. (B) Six-week-old ATAF2 T-DNA knockout line Salk_136355 shows no obvious phenotype compared to that of nontransformed Columbia plants (upper); the knockout of ATAF2 was confirmed by RT-PCR analysis (middle); and TMV CP levels are not significantly different between nontransformed and ATAF2 T-DNA knockout lines (lower). Results represent the means ± standard deviations of three independent experiments.
In a similar set of experiments, ATAF2 knockout lines were examined for their effects on virus accumulation. Salk_015750 and Salk_136355 lines, with transposon insertions in an ATAF2 intron and exon, respectively, were analyzed by real-time qRT-PCR for the detection of ATAF2 mRNA (1). Only the Salk_136355 line showed the consistent loss of detectable ATAF2 mRNA, and it was selected for subsequent experiments. As previously reported, plants from the Salk_136355 line displayed no obvious phenotype compared to that of the unmodified A. thaliana ecotype Columbia (Fig. 3B) (16). TMV replicates and travels systemically in ecotype Columbia, although at a reduced level compared to that of ecotype Shahdara (15). TMV accumulations detected in Salk_136355 line-inoculated leaf tissues were similar to those detected in unmodified Columbia plants (Fig. 3B). Thus, the absence of ATAF2 does not dramatically affect TMV accumulation.
ATAF2 is induced in TMV-inoculated but not systemically infected tissues.
The transcriptional control of ATAF2 has been shown to occur in response to wounding and infection by Pseudomonas syringae (16, 31). To examine the expression of ATAF2 in response to TMV infection, a 2-kb region upstream of the ATAF2 start codon was fused to the GUS reporter gene to produce the construct PATAF2::GUS. The T2 generation from three independent Shahdara lines transformed with this construct was examined for GUS reporter activity. Consistent with previous studies, GUS activity was rapidly induced, within 30 min along the margins of cut wounds made in fully expanded leaf tissue (data not shown) (16, 31). However, mechanical inoculation studies demonstrated that mock inoculations using only buffer and carborundum powder did not induce GUS activity (Fig. 4A). In contrast, TMV inoculations (0.1 mg/ml) induced increasing levels of GUS activity during a period of several days (Fig. 4A, upper). The appearance of GUS activity followed a pattern similar to that observed for virus spread in inoculated leaves monitored by immunoblotting for the detection of TMV coat protein (Fig. 4A, lower). The transcriptional induction of endogenous ATAF2 also was monitored using real-time qRT-PCR in response to virus infection. RNA samples taken at 6 dpi displayed a ninefold increase in the detectable level of ATAF2 mRNA in ecotype Shahdara above the levels of similar RNA purified from mock-inoculated leaves (Fig. 4C). Thus, ATAF2 is transcriptionally induced in response to the initial TMV infection.
FIG. 4.
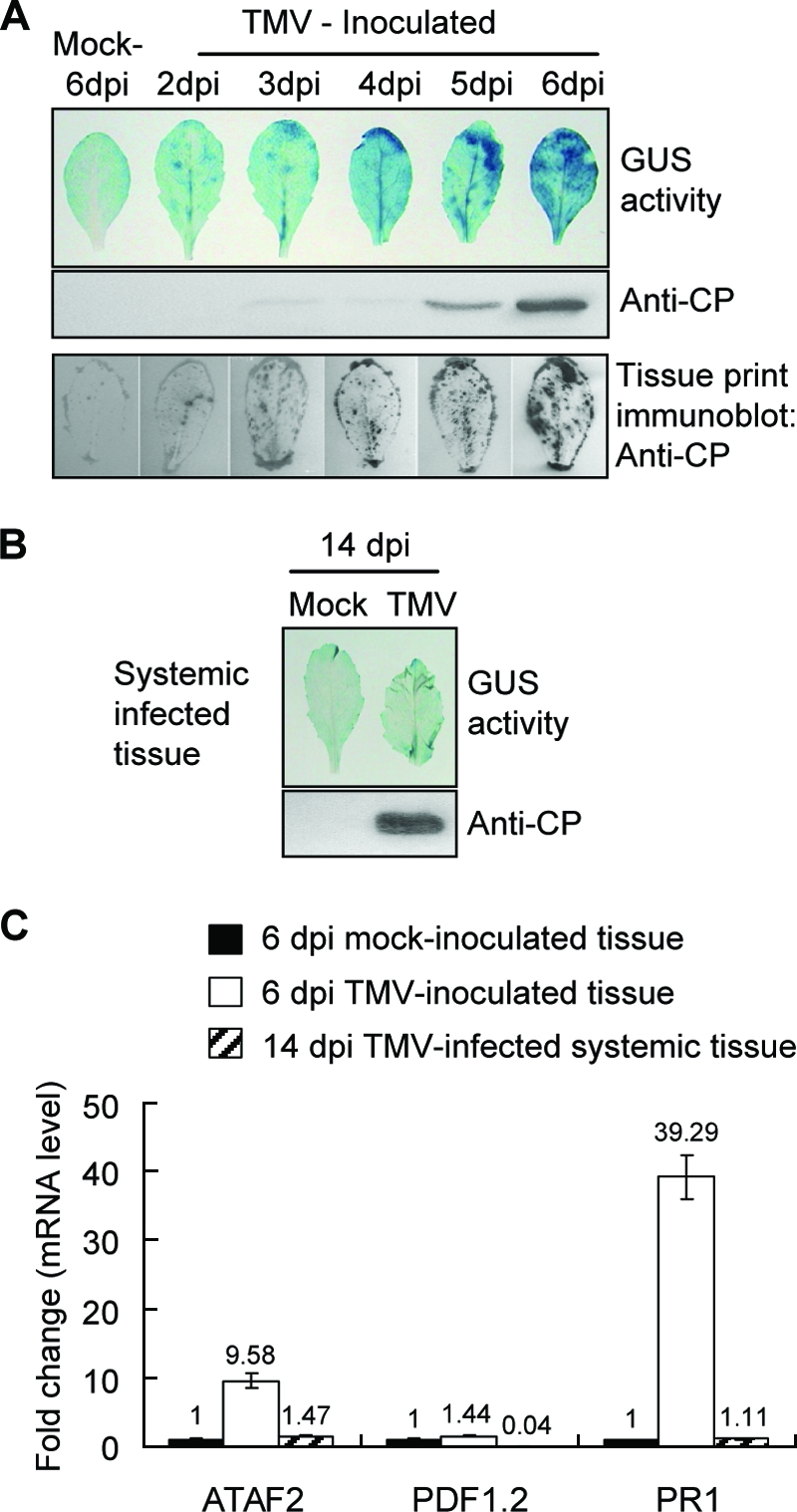
Both ATAF2 and the defense gene PR1 are induced in locally but not systemically infected tissues. (A) The upper panel shows transgenic Shahdara lines carrying the ATAF2 promoter fused to a GUS reporter gene (PATAF2::GUS) that were inoculated with TMV and sampled for GUS activities. For the middle panel, after histochemical staining for GUS activity, individual leaves were analyzed for CP content by Western immunoblotting. The lower panel shows a tissue print immunoblotting method employed to monitor the pattern of virus accumulation in TMV-inoculated leaf tissues. (B) Induction of GUS activity was not observed in systemically infected tissues at 14 dpi. (C) Real-time qRT-PCR analysis monitoring the expression levels of ATAF2 and defense genes PDF1.2 and PR1 in both TMV-inoculated and systemically infected tissues of A. thaliana ecotype Shahdara plants. Total RNA samples were derived from three to five independent test plants. Changes (n-fold) in gene expression are presented as levels relative to those of the mock-inoculated tissue. Data represent averages ± standard deviations of three real-time qRT-PCR replicates.
In contrast to inoculated tissues, ATAF2 was not similarly induced in systemically infected leaves. We first observed this differential response to virus infection in transgenic PATAF2::GUS plants (Fig. 4B). Even though the systemically infected tissues contained high levels of virus, no detectable GUS activity was observed. Additionally, real-time qRT-PCR analysis showed increased accumulations of ATAF2 mRNA within inoculated leaf tissues but not within systemically infected tissues (Fig. 4C). These findings suggest that in response to TMV infection, ATAF2 transcriptional activation is transient and limited primarily to the area of the initial infection.
ATAF2 expression correlates with the transcriptional activation of pathogen-related defense genes.
A previous study examining ATAF2-overexpressing and knockout plants grown in axenic conditions on agar suggested that ATAF2 negatively regulated the expression of basal defense genes (16). In this study, we examined the expression of basal defense-associated marker genes in response to virus infection, ATAF2 overexpression, and ATAF2 knockout in plants grown under nonsterile growth chamber conditions. PR1 and PR2 (for pathogenesis related 1 and 2) were selected as known marker genes for SA-mediated resistance, while PDF1.2 (for plant defensin 1.2) was selected for its association with methyl jasmonate-mediated resistance (48, 65). The real-time qRT-PCR analysis of RNA isolated from either TMV-inoculated or systemically infected leaf tissues did not show substantial increases in the accumulation of PDF1.2 transcripts (Fig. 4C). In contrast, PR1 transcripts showed significant accumulations in inoculated leaf tissues but not in systemically infected tissues (Fig. 4C). Interestingly, the pattern of PR1 accumulation in inoculated but not systemically infected tissues reflected the results observed for the accumulation of ATAF2 transcripts (Fig. 4), demonstrating that these transcripts are similarly regulated in response to a TMV infection.
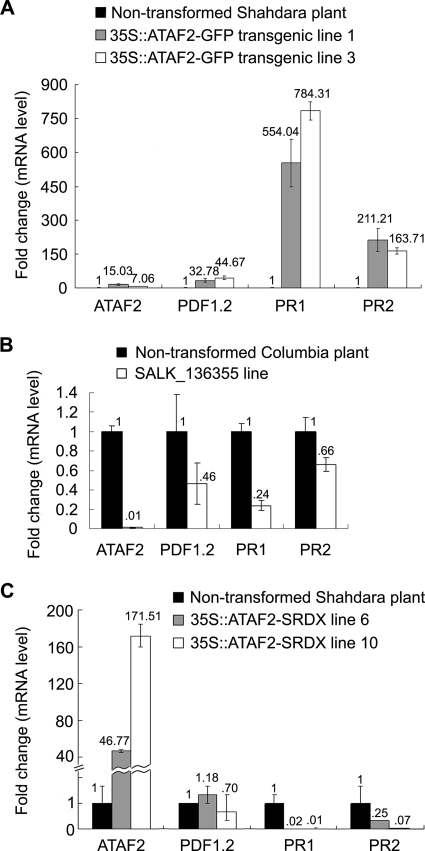
Real-time qRT-PCR analysis of plants transformed with 35S::ATAF2-GFP showed increased levels of ATAF2 mRNA, as expected (Fig. 5A). The analysis of defense-associated genes from these plants demonstrated significant increases in the mRNA accumulation of PR1 and PR2 as well as a lower, although still substantial, increase for PDF1.2 (Fig. 5A). Increased accumulations of these defense genes are consistent with enhanced disease resistance responses that could account for the reduced accumulation of virus observed in these plants (Fig. 3A).
FIG. 5.
Correlated induction of ATAF2 and defense-related marker genes. (A) ATAF2 overexpression stimulates the expression of host basal defense genes PDF1.2, PR1, and PR2. Real-time qRT-PCR analysis obtained from two representative 35S::ATAF2-GFP overexpression lines. (B) Expression of PDF1.2, PR1, and PR2 is reduced in ATAF2 knockout line SALK_136355. (C) Analyses of two representative 35S::ATAF2-SRDX repressor lines show reduced expression levels of PR1 and PR2 but not PDF1.2. Total RNA samples were derived from three to five independent test plants. Changes (n-fold) in gene expression are presented as levels relative to those of nontransformed plants. Data represent averages ± standard deviations of three real-time qRT-PCR replicates.
Similar real-time qRT-PCR studies were done on RNA extracted from the ATAF2 knockout line SALK_136355. As noted above, ATAF2 mRNA levels were undetectable in this knockout line (Fig. 5B). Compared to those of nontransformed plants, PDF1.2, PR1, and PR2 mRNA accumulation levels were reduced in this knockout line by an average of ∼50, ∼75, and ∼30%, respectively (Fig. 5B). This finding correlates with the data described above from the ATAF2-GFP overexpression lines and suggests that ATAF2 plays a role in the positive regulation of basal defense responses.
To further investigate the possible role of ATAF2 in the regulation of defense response genes, we created transgenic Shahdara plants expressing an ATAF2 ORF fused to a 12-aa repressor domain, termed SRDX. This domain is based on the EAR motif repressor from the TFIIIA-type zinc finger transcription repressor SUPERMAN (35, 36). Hiratsu et al. (35) previously demonstrated that the incorporation of this peptide onto the carboxy end of several Arabidopsis transcription factors, including the NAC domain protein CUC1 (for cup-shaped cotyledons 1), produced dominant repressor proteins that repress the transcription of corresponding target genes. In this study, A. thaliana ecotype Shahdara plants transformed with 35S::ATAF2-SRDX were examined for the accumulation of PDF1.2, PR1, and PR2. ATAF2 repressor lines did not show significant reductions in the accumulation of PDF1.2 transcripts; however, accumulations of PR1 and PR2 were greatly reduced compared to those of nontransformed control plants (Fig. 5C). This differential effect on the accumulation of PDF1.2, PR1, and PR2 transcripts suggests that ATAF2 influences the transcriptional activity of SA-mediated defense genes to a greater extent.
Systemic TMV infection interferes in SA-mediated defense responses.
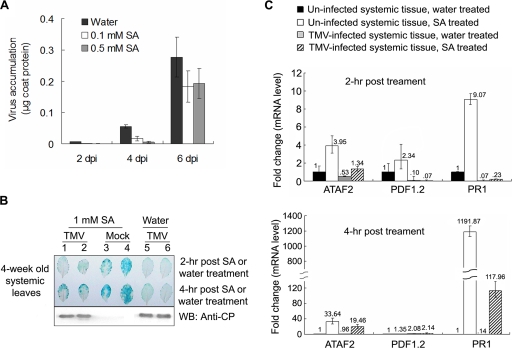
The correlation between ATAF2 expression and the induction of SA-associated defense genes, combined with the lack of ATAF2 transcript accumulation in systemically infected tissues, suggests that host defenses are altered as the virus spreads from the initial site of infection. To examine this possibility, we analyzed ATAF2 promoter-GUS expression in response to SA treatment in the systemic tissues of both TMV- and mock-inoculated plants. Our focus on SA-mediated defense responses was driven in part by studies that show SA treatments inhibit TMV infection in a susceptible host (10, 50). Similarly, for A. thaliana ecotype Shahdara, SA treatments prior to infection produce an average reduction in TMV accumulation of greater than 85% at 2 dpi, 70% at 4 dpi, and 30% at 6 dpi (Fig. 6A). Based on these results, we hypothesized that interaction with ATAF2 provides a mechanism for the virus to counter this host defense process. To test this hypothesis, PATAF2::GUS plants treated with 1 mM SA were monitored for GUS activity at 2 and 4 h posttreatment. In uninfected plants, GUS activity detected by blue coloring displayed notable increases after both 2 and 4 h (Fig. 6B, lanes 3 and 4). This finding is consistent with a previous study and demonstrates that ATAF2 is induced in response to SA (16). However, in leaf tissue systemically infected with TMV, GUS activity was not observed at 2 h after SA treatment and only a moderate level of activity appeared at the 4-h time point (Fig. 6B, lanes 1 and 2). Infected control plants sprayed with water did not display detectable GUS activity in systemically infected tissues (Fig. 6B, lanes 5 and 6). Thus, the induction of the ATAF2 promoter by SA treatment is attenuated in tissues that have been systemically infected prior to SA treatment.
FIG. 6.
TMV infection inhibits SA activation of ATAF2 and the defense marker gene PR1. (A) SA inhibits TMV accumulation. TMV accumulation was monitored in 4-week-old A. thaliana ecotype Shahdara plants sprayed with water, 0.1 mM SA, or 0.5 mM SA 4 h prior to virus inoculation. Data are displayed as the means ± standard deviations of three replicates. (B) Systemic tissues from PATAF2::GUS transgenic plants, either mock or TMV inoculated (14 dpi), were sprayed with H2O (lanes 5 and 6) or 1 mM SA (lanes 1, 2, 3, and 4). Two and 4 h posttreatment the leaves were assayed for GUS activity (upper), and Western immunoblotting (WB) for the detection of the TMV CP was used to confirm systemic infection in the tested tissues (lower). (C) Real-time qRT-PC analysis showing the induction of ATAF2 and PR1 by SA (0.1 mM) in TMV-infected systemic tissue compared to induction for mock-infected tissue at 2 and 4 h posttreatment. Total RNA samples were derived from three to five independent test plants. Changes (n-fold) in gene expression are presented as levels relative to those of uninfected water-treated systemic tissues. Data represent averages ± standard deviations from three real-time qRT-PCR replicates.
To further investigate the attenuation of SA-mediated defenses in response to systemic infection by TMV, the expression of ATAF2, PDF1.2, and PR1 transcripts was analyzed in response to SA treatment in A. thaliana ecotype Shahdara (Fig. 6C). Within uninfected systemic tissues, real-time qRT-PCR results demonstrated inductions of both ATAF2 and PR1 at both 2 and 4 h after SA treatment. In contrast, PDF1.2 did not display induction in response to SA. These findings are consistent with previous studies that show the induction of PR1 functions as a marker for SA-mediated defenses, while PDF1.2 is associated with jasmonate/ethylene-mediated resistance (48, 65). Within systemically infected tissues, the induction of ATAF2 in response to SA was reduced at both 2 and 4 h posttreatment compared to that of uninfected tissues (Fig. 6C). This is consistent with the observed effects of SA on the induction of GUS activity in the above-described ATAF2 promoter plants (Fig. 6B). The accumulation of PR1 in response to SA treatment was notably reduced in systemically infected tissues. At least a 10-fold reduction in PR1 transcript was observed at both 2 and 4 h after SA treatment (Fig. 6C). In contrast, the response of PDF1.2 in systemically infected tissues to SA treatment showed little change at either time point (Fig. 6C). These findings are consistent with the suppression of SA-mediated defense responses within tissues systemically infected with TMV.
ATAF2 functions in the regulation of SA-mediated defense responses.
To further investigate ATAF2's role in SA-mediated defense responses, accumulations of ATAF2, PDF1.2, and PR1 transcripts were determined in response to SA treatment in the Salk_136355 knockout line and the 35S::ATAF2-SRDX repressor lines. We reasoned that if ATAF2 were involved in the regulation of SA-mediated defenses, then its functional absence, through either knockout or a dominant-negative repressor, alters the ability of SA to induce PR1 transcription. As previously shown, nontransformed control plants displayed increased accumulations of ATAF2 and PR1 transcripts in response to SA treatment (Fig. 6C). However, in both the ATAF2 knockout and repressor lines, accumulations of PR1 transcripts in response to SA treatment were noticeably lower, ∼50 and ∼60%, respectively, than those in SA-treated nontransformed plants (Fig. 7A and B). These reduced accumulations of PR1 in response to SA treatment provide additional support for ATAF2 as a regulator of SA-mediated defenses.
FIG. 7.
ATAF2 plays a regulatory role in SA-mediated defense activation. Four-week-old mature leaves were sprayed with 0.1 mM SA, and leaf tissue was collected 4 h after SA treatment. Gene expression is presented relative to the levels observed in water-treated nontransformed plants. (A) ATAF2 knockout line SALK_136355 shows reduced PR1 induction after SA treatment compared to that of nontransformed A. thaliana ecotype Columbia control plants. (B) Two representative ATAF2-SRDX repressor lines show reduced PR1 levels after SA treatment. Total RNA was derived from three to five independent test plants. Data represent the averages ± standard deviations from three real-time qRT-PCR replicates.
DISCUSSION
Plants respond to pathogen attack via distinct signaling pathways that regulate numerous biochemical, metabolic, and molecular defenses. Emerging evidence suggests that in order to establish a successful infection, pathogens employ a variety of countermeasures that disrupt defense-mediated signaling pathways (2, 17, 46, 54). Thus, the ability of a pathogen to suppress host defenses is a key factor in determining the virulence of infection and the severity of the disease. In this study, we identified an interaction between the TMV replicase protein and the NAC domain transcription factor ATAF2. Our interest in this interaction is linked to ATAF2's role in the modulation of basal defense responses and the ability of these responses to limit TMV accumulation within a compatible host. The targeted disruption of this transcription factor via interaction with the virus replicase represents a previously undescribed countermeasure against host basal defense responses and a contributing factor in TMV pathogenicity.
NAC domain proteins encompass a large family of plant-specific transcription factors, of which the functions of only a few have been investigated (55). ATAF2 initially was identified for its ability to trans-activate the CaMV 35S promoter in yeast (72). In plants, the expression of ATAF2 from the 35S promoter results in a mild developmental phenotype that includes curled and cup-shaped leaves and underdeveloped floral organs, demonstrating that ATAF2 expression functionally impacts cellular processes (Fig. 3A). Corresponding ATAF2 T-DNA knockout plants or repressor lines display a normal developmental phenotype (Fig. 3B and data not shown), suggesting the presence of redundancy in the function of this NAC family gene. Links to transcriptional activation and effects on plant development are consistent with a role for ATAF2 in the modulation of plant transcriptional responses.
Considerable evidence indicates that ATAF2 functions in the regulation of defense-associated genes. First, ATAF2 transcription is induced in response to wounding, pathogen infection, treatment with the fungal PAMP chitin, and exposure to SA (16, 31, 45) (Fig. 6C). Second, the overexpression of ATAF2 results in the transcriptional activation of defense-related marker genes PR1, PR2, and PDF1.2, while an ATAF2 T-DNA knockout line produces correspondingly reduced levels of these marker genes (Fig. 5A and B). Finally, transgenic plants expressing an ATAF2 construct fused to a 12-aa transcriptional repressor peptide (35) and designed to function as a dominant repressor of ATAF2 target genes also display reduced levels of selected defense genes (Fig. 5C). These findings indicate that ATAF2 functions in response to pathogen infection and has a role in positively modulating basal defense responses.
Delessert et al. (16) also investigated transcriptional alterations in Arabidopsis in response to the overexpression or knockout of ATAF2. In contrast to the findings presented in this study, Delessert et al. (16) concluded that ATAF2 functions as a negative regulator of pathogen-related host defense genes. This difference in the observed effects ATAF2 has on defense gene expression may be attributed to the different environmental conditions used in the two studies. Delessert et al. (16) analyzed plants grown on agar plates and under sterile conditions. In contrast, plants analyzed in this study were grown in potting mix under nonsterile growth chamber conditions. Previous studies have noted significant differences between the defense responses of plants grown under sterile conditions and those of plants grown in nonsterile conditions. Typically, plants grown under sterile conditions display significant alterations or reductions in the activation of defense-associated transcripts (9, 60). Based on these findings, ATAF2's function in the regulation of the defense response may vary depending upon the environmental conditions of the plant.
It is not known if interaction with the TMV replicase induces ATAF2 transcription in response to infection or if this occurs as a result of another perturbation induced by the pathogen upon infection. The induction of SlNAC1, a related NAC domain protein in tomato, was found to occur in response to the interacting geminivirus REn protein when expressed via Agrobacterium (64). However, within our system, Agrobacterium infiltration alone induced the expression of ATAF2 (data not shown). ATAF2 transcriptional induction also has been shown to occur at sites of severe wounding, primarily along cut edges (16) (data not shown), but not in response to the mechanical damage that is required for TMV infection (Fig. 4A). From these findings, it seems likely that the transcriptional induction of ATAF2 is a general host response to pathogen infection or severe wounding.
In contrast to the transcriptional induction of ATAF2, we observed reduced levels of ATAF2-GFP protein accumulation in TMV-infected tissues (Fig. 2). Both transient and transgene-derived accumulations of ATAF2-GFP were significantly reduced in TMV-infected tissues, suggesting that ATAF2 protein is targeted for degradation during virus infection. Treatments with the proteasome inhibitor MG132 increased ATAF2-GFP accumulations in TMV-infected tissues (Fig. 2D), supporting a proteasome-mediated mechanism for the observed reductions in ATAF2 protein. Only a few examples of virus-directed protein degradation have been identified in plants (14). Of note is the targeted degradation of the catalytic component, Argonaute, of the RNA-induced silencing complex by a polerovirus-encoded suppressor protein (4, 59). Thus, the targeted degradation of host defense factors represents an emerging theme in plant virus-based counterdefense strategies.
SA is a key signaling component in plant defense and regulates the transcriptional activity of numerous defense-associated host genes (3, 24). While SA is an essential signaling component in resistance gene-mediated responses, it also plays a contributing role in PAMP-mediated responses during compatible host-pathogen interactions (10, 38, 66). Pretreatment with SA results in the transcriptional induction of a large group of SA-regulated defense genes, including PR1 and PR2 (47, 63). The regulation of PR genes has been linked directly to the interaction of NPR1 (for nonexpressor of PR1) with TGA transcription factors (25). In addition, WRKY and ERF domain transcription factors also are implicated in PR gene expression (20, 24). Thus, PR gene activation is influenced at multiple levels. The induction of PR1, whether by pathogen or SA treatment, correlates with the induction of ATAF2 in Arabidopsis, suggesting a related mechanism of regulation. Consistently with this possibility is the presence of five WRKY factor binding motifs within the ATAF2 promoter region (58). The mechanism through which ATAF2 affects PR1 gene transcription remains unknown. Presumably ATAF2 plays a supporting role, functioning in conjunction with other factors to enhance PR1 expression. The partial reduction of PR1 transcripts observed in ATAF2 knockout plants treated with SA (Fig. 7) is consistent with such a synergistic role for ATAF2 in the regulation of basal gene defenses.
Based on these studies, we speculate that TMV's interaction with ATAF2 represents a mechanism for the suppression of basal defense pathways during a compatible virus-host interaction. This hypothesis is supported by the reduced ability of SA to transcriptionally activate defense-related genes within tissues systemically infected by TMV. Specifically, host defense responses, such as ATAF2 and PR1, show rapid transcriptional induction in inoculated tissues at the site of infection, but upon the establishment of a TMV infection, the induction of these genes is reduced or inhibited. We conclude that the replication and movement of TMV impacts the transcriptional activation of host defense responses as the virus spreads from the initial site of infection. In support of this, ATAF2 knockout or repressor fusion constructs show reduced defense gene activation in response to SA treatment, similarly to that observed in TMV-infected tissue. Thus, reductions of functional ATAF2 protein accumulation either by virus-mediated degradation, knockout, or transcriptional repression all produce attenuation in the plant's ability to respond to SA treatment. Taken together, these findings are consistent with the TMV replicase-ATAF2 interaction functioning to disrupt host basal defenses.
Acknowledgments
We thank Shunyuan Xiao and members of our laboratory for their critical analysis and input into the writing of the manuscript.
This work was support in part by USDA National Research Initiative Competitive Grants (2005-35319-16099 and 2008-35319-19168).
Footnotes
Published ahead of print on 22 July 2009.
REFERENCES
- 1.Alonso, J. M., A. N. Stepanova, T. J. Leisse, C. J. Kim, H. Chen, P. Shinn, D. K. Stevenson, J. Zimmerman, P. Barajas, R. Cheuk, C. Gadrinab, C. Heller, A. Jeske, E. Koesema, C. C. Meyers, H. Parker, L. Prednis, Y. Ansari, N. Choy, H. Deen, M. Geralt, N. Hazari, E. Hom, M. Karnes, C. Mulholland, R. Ndubaku, I. Schmidt, P. Guzman, L. Aguilar-Henonin, M. Schmid, D. Weigel, D. E. Carter, T. Marchand, E. Risseeuw, D. Brogden, A. Zeko, W. L. Crosby, C. C. Berry, and J. R. Ecker. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301:653-657. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M. 2005. Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 6:973-979. [DOI] [PubMed] [Google Scholar]
- 3.Bari, R., and J. D. Jones. 2009. Role of plant hormones in plant defence responses. Plant Mol. Biol. 69:473-488. [DOI] [PubMed] [Google Scholar]
- 4.Baumberger, N., C. H. Tsai, M. Lie, E. Havecker, and D. C. Baulcombe. 2007. The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 17:1609-1614. [DOI] [PubMed] [Google Scholar]
- 5.Bent, A. F., and D. Mackey. 2007. Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45:399-436. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Burch-Smith, T. M., M. Schiff, J. L. Caplan, J. Tsao, K. Czymmek, and S. P. Dinesh-Kumar. 2007. A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 5:e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caplan, J. L., P. Mamillapalli, T. M. Burch-Smith, K. Czymmek, and S. P. Dinesh-Kumar. 2008. Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 132:449-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cartieaux, F., M. Thibaud, L. Zimmerli, P. Lessard, C. Sarrobert, P. David, A. Gerbaud, C. Robaglia, S. Somerville, and L. Nussaume. 2003. Transcriptome analysis of Arabidopsis colonized by a plant-growth promoting rhizobacterium reveals a general effect on disease resistance. Plant J. 36:177-188. [DOI] [PubMed] [Google Scholar]
- 10.Chivasa, S., A. M. Murphy, M. Naylor, and J. P. Carr. 1997. Salicylic acid interferes with tobacco mosaic virus replication via a novel salicylhydroxamic acid-sensitive mechanism. Plant Cell 9:547-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark, K. L., P. B. Larsen, X. Wang, and C. Chang. 1998. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc. Natl. Acad. Sci. USA 95:5401-5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clough, S. J., and A. Bent. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16:735-743. [DOI] [PubMed] [Google Scholar]
- 13.Collinge, M., and T. Boller. 2001. Differential induction of two potato genes, Stprx2 and StNAC, in response to infection by Phytophthora infestans and to wounding. Plant Mol. Biol. 46:521-529. [DOI] [PubMed] [Google Scholar]
- 14.Culver, J. N., and M. S. Padmanabhan. 2007. Virus-induced disease: altering host physiology one interaction at a time. Annu. Rev. Phytopathol. 45:221-243. [DOI] [PubMed] [Google Scholar]
- 15.Dardick, C. D., S. Golem, and J. N. Culver. 2000. Susceptibility and symptom development in Arabidopsis thaliana to Tobacco mosaic virus is influenced by virus cell-to-cell movement. Mol. Plant-Microbe Interact. 13:1139-1144. [DOI] [PubMed] [Google Scholar]
- 16.Delessert, C., K. Kazan, I. W. Wilson, D. Van Der Straeten, J. Manners, E. S. Dennis, and R. Dolferus. 2005. The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J. 43:745-757. [DOI] [PubMed] [Google Scholar]
- 17.Díaz-Pendón, J. A., and S. W. Ding. 2008. Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu. Rev. Phytopathol. 46:303-326. [DOI] [PubMed] [Google Scholar]
- 18.Ding, S. W., and O. Voinnet. 2007. Antiviral immunity directed by small RNAs. Cell 130:413-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding, X. S., J. Liu, N. H. Cheng, A. Folimonov, Y. M. Hou, Y. Bao, C. Katagi, S. A. Carter, and R. S. Nelson. 2004. The Tobacco mosaic virus 126-kDa protein associated with virus replication and movement suppresses RNA silencing. Mol. Plant-Microbe Interact. 17:583-592. [DOI] [PubMed] [Google Scholar]
- 20.Dong, X. 2004. NPR1, all things considered. Curr. Opin. Plant Biol. 7:547-552. [DOI] [PubMed] [Google Scholar]
- 21.dos Reis Figueira, A., S. Golem, S. P. Goregaoker, and J. N. Culver. 2002. A nuclear localization signal and a membrane association domain contribute to the cellular localization of the Tobacco mosaic virus 126-kDa replicase protein. Virology 301:81-89. [DOI] [PubMed] [Google Scholar]
- 22.Duval, M., T. F. Hsieh, S. Y. Kim, and T. L. Thomas. 2002. Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Mol. Biol. 50:237-248. [DOI] [PubMed] [Google Scholar]
- 23.Ernst, H. A., A. N. Olsen, S. Larsen, and L. Lo Leggio. 2004. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 5:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eulgem, T. 2005. Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 10:71-78. [DOI] [PubMed] [Google Scholar]
- 25.Fan, W., and X. Dong. 2002. In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14:1377-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Felix, G., J. D. Duran, S. Volko, and T. Boller. 1999. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18:265-276. [DOI] [PubMed] [Google Scholar]
- 27.Ferrando, A., R. Farras, J. Jasik, J. Schell, and C. Koncz. 2000. Intron-tagged epitope: a tool for facile detection and purification of proteins expressed in Agrobacterium-transformed plant cells. Plant J. 22:553-560. [DOI] [PubMed] [Google Scholar]
- 28.Golem, S., and J. N. Culver. 2003. Tobacco mosaic virus induced alterations in the gene expression profile of Arabidopsis thaliana. Mol. Plant-Microbe Interact. 16:681-688. [DOI] [PubMed] [Google Scholar]
- 29.Goregaoker, S. P., D. J. Lewandowski, and J. N. Culver. 2001. Identification and functional analysis of an interaction between domains of the 126/183-kDa replicase-associated proteins of tobacco mosaic virus. Virology 282:320-328. [DOI] [PubMed] [Google Scholar]
- 30.Haseloff, J., K. R. Siemering, D. C. Prasher, and S. Hodge. 1997. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94:2122-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hauck, P., R. Thilmony, and S. Y. He. 2003. A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. USA 100:8577-8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He, J. X., J. M. Gendron, Y. Yang, J. Li, and Z. Y. Wang. 2002. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 99:10185-10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He, P., L. Shan, and J. Sheen. 2007. Elicitation and suppression of microbe-associated molecular pattern-triggered immunity in plant-microbe interactions. Cell Microbiol. 9:1385-1396. [DOI] [PubMed] [Google Scholar]
- 34.Hegedus, D., M. Yu, D. Baldwin, M. Gruber, A. Sharpe, I. Parkin, S. Whitwill, and D. Lydiate. 2003. Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol. Biol. 53:383-397. [DOI] [PubMed] [Google Scholar]
- 35.Hiratsu, K., K. Matsui, T. Koyama, and M. Ohme-Takagi. 2003. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34:733-739. [DOI] [PubMed] [Google Scholar]
- 36.Hiratsu, K., M. Ohta, K. Matsui, and M. Ohme-Takagi. 2002. The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett. 514:351-354. [DOI] [PubMed] [Google Scholar]
- 37.Holsters, M., D. de Waele, A. Depicker, E. Messens, M. van Montagu, and J. Schell. 1978. Transfection and transformation of Agrobacterium tumefaciens. Mol. Gen. Genet. 163:181-187. [DOI] [PubMed] [Google Scholar]
- 38.Huang, Z., J. M. Yeakley, E. W. Garcia, J. D. Holdridge, J. B. Fan, and S. A. Whitham. 2005. Salicylic acid-dependent expression of host genes in compatible Arabidopsis-virus interactions. Plant Physiol. 137:1147-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jefferson, R. A., T. A. Kavanagh, and M. W. Bevan. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6:3901-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeong, R. D., A. C. Chandra-Shekara, A. Kachroo, D. F. Klessig, and P. Kachroo. 2008. HRT-mediated hypersensitive response and resistance to Turnip crinkle virus in Arabidopsis does not require the function of TIP, the presumed guardee protein. Mol. Plant-Microbe Interact. 21:1316-1324. [DOI] [PubMed] [Google Scholar]
- 41.Jones, J. D., and J. L. Dangl. 2006. The plant immune system. Nature 444:323-329. [DOI] [PubMed] [Google Scholar]
- 42.Kikuchi, K., M. Ueguchi-Tanaka, K. T. Yoshida, Y. Nagato, M. Matsusoka, and H. Y. Hirano. 2000. Molecular analysis of the NAC gene family in rice. Mol. Gen. Genet. 262:1047-1051. [DOI] [PubMed] [Google Scholar]
- 43.Kunze, G., C. Zipfel, S. Robatzek, K. Niehaus, T. Boller, and G. Felix. 2004. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16:3496-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 45.Libault, M., J. Wan, T. Czechowski, M. Udvardi, and G. Stacey. 2007. Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol. Plant-Microbe Interact. 20:900-911. [DOI] [PubMed] [Google Scholar]
- 46.Mackey, D., and A. J. McFall. 2006. MAMPs and MIMPs: proposed classifications for inducers of innate immunity. Mol. Microbiol. 61:1365-1371. [DOI] [PubMed] [Google Scholar]
- 47.Maleck, K., A. Levine, T. Eulgem, A. Morgan, J. Schmid, K. A. Lawton, J. L. Dangl, and R. A. Dietrich. 2000. The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26:403-410. [DOI] [PubMed] [Google Scholar]
- 48.Manners, J. M., I. A. Penninckx, K. Vermaere, K. Kazan, R. L. Brown, A. Morgan, D. J. Maclean, M. D. Curtis, B. P. Cammue, and W. F. Broekaert. 1998. The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol. Biol. 38:1071-1080. [DOI] [PubMed] [Google Scholar]
- 49.Miya, A., P. Albert, T. Shinya, Y. Desaki, K. Ichimura, K. Shirasu, Y. Narusaka, N. Kawakami, H. Kaku, and N. Shibuya. 2007. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104:19613-19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murphy, A., and J. Carr. 2002. Salicylic acid has cell-specific effects on tobacco mosaic virus replication and cell-to-cell movement. Plant Physiol. 128:552-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakai, K., and M. Kanehisa. 1992. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14:897-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Navarro, L., F. Jay, K. Nomura, S. Y. He, and O. Voinnet. 2008. Suppression of the microRNA pathway by bacterial effector proteins. Science 321:964-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nomura, K., S. Debroy, Y. H. Lee, N. Pumplin, J. Jones, and S. Y. He. 2006. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313:220-223. [DOI] [PubMed] [Google Scholar]
- 54.Nomura, K., M. Melotto, and S. Y. He. 2005. Suppression of host defense in compatible plant-Pseudomonas syringae interactions. Curr. Opin. Plant Biol. 8:361-368. [DOI] [PubMed] [Google Scholar]
- 55.Olsen, A. N., H. A. Ernst, L. L. Leggio, and K. Skriver. 2005. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10:79-87. [DOI] [PubMed] [Google Scholar]
- 56.Ooka, H., K. Satoh, K. Doi, T. Nagata, Y. Otomo, K. Murakami, K. Matsubara, N. Osato, J. Kawai, P. Carninci, Y. Hayashizaki, K. Suzuki, K. Kojima, Y. Takahara, K. Yamamoto, and S. Kikuchi. 2003. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10:239-247. [DOI] [PubMed] [Google Scholar]
- 57.Padmanabhan, M. S., S. P. Goregaoker, S. Golem, H. Shiferaw, and J. N. Culver. 2005. Interaction of the tobacco mosaic virus replicase protein with the Aux/IAA protein PAP1/IAA26 is associated with disease development. J. Virol. 79:2549-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palaniswamy, S. K., S. James, H. Sun, R. S. Lamb, R. V. Davuluri, and E. Grotewold. 2006. AGRIS and AtRegNet. A platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol. 140:818-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pazhouhandeh, M., M. Dieterle, K. Marrocco, E. Lechner, B. Berry, V. Brault, O. Hemmer, T. Kretsch, K. E. Richards, P. Genschik, and V. Ziegler-Graff. 2006. F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc. Natl. Acad. Sci. USA 103:1994-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Penninckx, I. A., B. P. Thomma, A. Buchala, J. P. Metraux, and W. F. Broekaert. 1998. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10:2103-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ren, T., F. Qu, and T. J. Morris. 2000. HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus. Plant Cell 12:1917-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ren, T., F. Qu, and T. J. Morris. 2005. The nuclear localization of the Arabidopsis transcription factor TIP is blocked by its interaction with the coat protein of Turnip crinkle virus. Virology 331:316-324. [DOI] [PubMed] [Google Scholar]
- 63.Ryals, J. A., U. H. Neuenschwander, M. G. Willits, A. Molina, H. Y. Steiner, and M. D. Hunt. 1996. Systemic acquired resistance. Plant Cell 8:1809-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Selth, L. A., S. C. Dogra, M. S. Rasheed, H. Healy, J. W. Randles, and M. A. Rezaian. 2005. A NAC domain protein interacts with tomato leaf curl virus replication accessory protein and enhances viral replication. Plant Cell 17:311-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shah, J. 2003. The salicylic acid loop in plant defense. Curr. Opin. Plant Biol. 6:365-371. [DOI] [PubMed] [Google Scholar]
- 66.Tsuda, K., M. Sato, J. Glazebrook, J. D. Cohen, and F. Katagiri. 2008. Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 53:763-775. [DOI] [PubMed] [Google Scholar]
- 67.Ueda, H., Y. Yamaguchi, and H. Sano. 2006. Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein, N factor during the hypersensitive response in tobacco plants. Plant Mol. Biol. 61:31-45. [DOI] [PubMed] [Google Scholar]
- 68.van Engelen, F. A., J. W. Molthoff, A. J. Conner, J. P. Nap, A. Pereira, and W. J. Stiekema. 1995. pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res. 4:288-290. [DOI] [PubMed] [Google Scholar]
- 69.van Loon, L. C., M. Rep., and C. M. Pieterse. 2006. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44:135-162. [DOI] [PubMed] [Google Scholar]
- 70.Ward, A. C. 1990. Single-step purification of shuttle vectors from yeast for high frequency back-transformation into E. coli. Nucleic Acids Res. 18:5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Whitham, S. A., S. Quan, H. S. Chang, B. Cooper, B. Estes, T. Zhu, X. Wang, and Y. M. Hou. 2003. Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J. 33:271-283. [DOI] [PubMed] [Google Scholar]
- 72.Xie, Q., A. P. Sanz-Burgos, H. Guo, J. A. Garcia, and C. Gutierrez. 1999. GRAB proteins, novel members of the NAC domain family, isolated by their interaction with a geminivirus protein. Plant Mol. Biol. 39:647-656. [DOI] [PubMed] [Google Scholar]
- 73.Yamanaka, T., T. Ohta, M. Takahashi, T. Meshi, R. Schmidt, C. Dean, S. Naito, and M. Ishikawa. 2000. TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc. Natl. Acad. Sci. USA 97:10107-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]