Abstract
The binding of neutralizing antibodies 2F5 and 4E10 to human immunodeficiency virus type 1 (HIV-1) gp41 involves both the viral membrane and gp41 membrane proximal external region (MPER) epitopes. In this study, we have used several biophysical tools to examine the secondary structure, orientation, and depth of immersion of gp41 MPER peptides in liposomes and to determine how the orientation of the MPER with lipids affects the binding kinetics of monoclonal antibodies (MAbs) 2F5 and 4E10. The binding of 2F5 and 4E10 both to their respective nominal epitopes and to a biepitope (includes 2F5 and 4E10 epitopes) MPER peptide-liposome conjugate was best described by a two-step encounter-docking model. Analysis of the binding kinetics and the effect of temperature on the binding stability of 2F5 and 4E10 to MPER peptide-liposome conjugates revealed that the docking of 4E10 was relatively slower and thermodynamically less favorable. The results of fluorescence-quenching and fluorescence resonance energy transfer experiments showed that the 2F5 epitope was more solvent exposed, whereas the 4E10 epitope was immersed in the polar-apolar interfacial region of the lipid bilayer. A circular dichroism spectroscopic study demonstrated that the nominal epitope and biepitope MPER peptides adopted ordered structures with differing helical contents when anchored to liposomes. Furthermore, anchoring of MPER peptides to the membrane via a hydrophobic anchor sequence was required for efficient MAb docking. These results support the model that the ability of 2F5 and 4E10 to bind to membrane lipid is required for stable docking to membrane-embedded MPER residues. These data have important implications for the design and use of peptide-liposome conjugates as immunogens for the induction of MPER-neutralizing antibodies.
The two broadly neutralizing human monoclonal antibodies (MAbs) 2F5 and 4E10 target conserved core amino acid residues that lie in the membrane proximal external region (MPER) of human immunodeficiency virus type 1 (HIV-1) gp41 (6, 9, 18, 25, 29). Structural studies of 2F5 and 4E10 in complex with their nominal epitope peptides led to the proposition that the long hydrophobic heavy chain CDR3 (CDR H3) loop might be involved in binding to the virion membrane due to the lack of direct contact of the tip of the CDR H3 loop with their bound epitopes (6, 25). MAbs 2F5 and 4E10 indeed were found to have enhanced binding to gp41 MPER in the presence of membrane (12, 25). Subsequent studies have revealed the lipid reactivity of both the 2F5 and 4E10 MAbs (2, 14, 23, 27), emphasizing the need to understand how MAbs 2F5 and 4E10 recognize their epitopes in the context of a membrane-gp41 MPER interface.
It has been hypothesized that the ability of MAbs 2F5 and 4E10 to interact with membrane lipids is required for binding to the membrane-bound gp41 MPER region and subsequent HIV-1 neutralization (2, 14, 15). The binding of both the 2F5 and 4E10 MAbs to their epitope peptides presented on synthetic liposomes was remarkably different from that of epitope peptides alone and was best described by a two-step “encounter-docking” model (2). In this model, neutralizing MPER MAbs make an initial encounter complex, and such an interaction is associated with faster association and dissociation rates. The formation of the encounter complex induces the formation of the final “docked” complex, which is associated with slower dissociation rates and provides the stability of the overall interaction. A more recent study has also observed the same mode of interaction for MAb 4E10 when it binds to MPER peptide in liposomal form (31). The studies of Sun et al. revealed that critical residues of the 4E10 epitope may be buried in the viral membrane and that interaction of 4E10 with lipids is important in extracting the immersed residues from the lipid bilayer. Although 2F5 binding was not described in the study, the model shows that the N-terminal helix of the “L”-shaped MPER structure projects away from the membrane and that residues K665 and W666 of the core 2F5 epitope (residues DKW) are placed on the surface and in the interfacial region, respectively, of the membrane lipid (31). Thus, as for MAb 4E10, stable docking of 2F5 would also require some level of conformational rearrangement of MPER to release critical residues within the core epitope. This is consistent with binding kinetics data that showed that the final docking of MAbs 2F5 and 4E10 to MPER peptide-lipid conjugates might require conformational rearrangements (2). It is also likely that the CD4 and coreceptor-mediated triggering of HIV-1 Env (10, 28) that leads to the formation of the fusion intermediate conformation might also expose critical residues for MPER MAb binding. Both the 4E10 and 2F5 MAbs bound strongly to a recombinant trimeric gp41 intermediate design and either bound weakly or failed to bind, respectively, to the trimeric gp140 (11) and a putative prefusion-state trimeric MPER (22). However, the orientation of the MPER sequence in a viral-lipid-bound form is not known and, thus, it is possible that in the early stages of the triggered intermediate state, MPER residues may be lying in the plane of the membrane head groups and interaction of MPER MAbs with lipids and extraction of critical residues may be essential for stable docking (31).
In order to gain further understanding of the binding mechanism involved in the interaction of MAbs 2F5 and 4E10 with their epitopes presented in the membrane environment, we have constructed three different novel gp41 MPER peptide-liposome conjugates, including a 2F5 nominal epitope peptide, a 4E10 nominal epitope peptide, and a peptide having sequences of epitopes for both the 2F5 and 4E10 MAbs. Unlike our previously designed constructs (2), the MPER peptides used in the current study were anchored to the liposomes by a hydrophobic sequence (YKRWIILGLNKIVRMYS), named GTH1, placed at their carboxyl termini. Using these second-generation peptide-liposome conjugates, we addressed the following questions. (i) How do MAbs 2F5 and 4E10 bind to the different peptide-liposome conjugates? (ii) How do the kinetics of MAb binding vary with temperature? (iii) How are the peptides oriented in the liposomal membrane in each construct? (iv) How does antibody binding correlate with differences in the membrane orientation of peptides? (v) Is there any difference in the secondary structures adopted by the peptides in the peptide-liposome complex?
Our study of antibody interactions with their membrane-anchored epitope peptides indicates that both the 2F5 and 4E10 MAbs bind to their nominal epitope peptide-liposome conjugates with high affinity. The results of tryptophan fluorescence-quenching and fluorescence resonance energy transfer (FRET) experiments showed that the nominal 2F5 peptide is exposed on the surface of the membrane close to the polar head group, whereas the nominal 4E10 peptide is immersed in the interfacial region of the lipid bilayer. Circular dichroism (CD) spectroscopic studies revealed that the nominal epitope and biepitope peptides adopted ordered structures when anchored to the liposomal membrane. The membrane orientation data and secondary structural features of MPER peptides correlated well with antibody binding characteristics, thus suggesting that membrane-anchored MPER peptide conformations are a physiologic component of the native 2F5 and 4E10 binding epitopes in HIV-1 virions.
MATERIALS AND METHODS
Antibodies.
Anti-HIV-1 gp41 MPER MAbs 2F5 and 4E10 were purchased from Polymun Scientific, Austria.
Peptides.
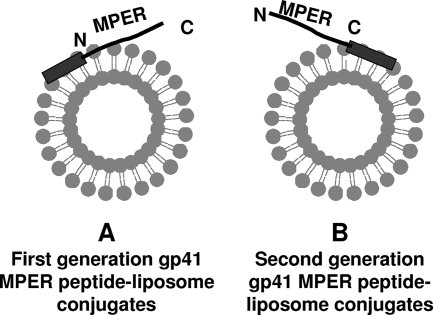
Peptides were synthesized and purified by reverse-phase high-pressure liquid chromatography. The purity of peptides was assessed by high-pressure liquid chromatography to be >95% and was confirmed by mass spectrometric analysis. The peptides used in this study include the 2F5 nominal epitope peptide (QQEKNEQELLELDKWASLWN), the 4E10 nominal epitope peptide (SLWNWFNITNWLWYIK), and a biepitope peptide (NEQELLELDKWASLWNWFNITNWLWYIK). To facilitate membrane anchorage, all of these peptides were synthesized with the GTH1 sequence (YKRWIILGLNKIVRMYS), attached to their carboxyl termini (Fig. 1) instead of the amino-terminal end as described earlier (2).
FIG. 1.
Schematic representation of two different strategies of membrane anchoring of MPER peptides. (A) Nonphysiologic anchoring at the amino terminus of MPER used in our earlier report (2). (B) Physiologic anchoring at the carboxyl terminus of MPER used in this study. The hydrophobic membrane-anchoring sequence is shown as a rectangle and was attached at either the amino or carboxyl terminus of the MPER peptide, which is shown as a black line.
Phospholipids.
Chloroform stocks of phospholipids 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1,2-dimyristoyl-sn-glycero-3-phosphate (DMPA), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(5-dimethylamino-1-naphthalenesulfonyl) (ammonium salt) (DANSYL-PE), 1-palmitoyl-2-stearoyl(6-7)dibromo-sn-glycero-3-phosphocholine (67DBrPC), and 1-palmitoyl-2-stearoyl(9-10)dibromo-sn-glycero-3-phosphocholine (910DBrPC) and cholesterol powder were purchased from Avanti Polar Lipids and were used without further purification.
Preparation of liposomes.
Peptide-liposome conjugates were prepared by using a lipid extrusion method as described earlier (2). Chloroform stocks of POPC, POPE, DMPA and cholesterol were mixed in chloroform-resistant tubes at a molar ratio of 45:25:20:1.33. For preparing peptide-liposomes, an appropriate volume of the relevant peptide stock (made in chloroform-methanol, 7:3 [vol/vol]) solution was added to this mixture to give a final peptide-to-total-lipid (P/L) ratio of 1:420. The mixture was dried in a stream of gaseous nitrogen. Any residual chloroform was removed by placing dried lipid films under a high vacuum overnight. Phosphate-buffered saline (pH 7.4) was added to the dried lipid or lipid-peptide films, and the suspensions were vortexed rigorously and incubated at 37°C for 45 min. The suspensions were then sonicated (3 cycles of 15 s of sonication at 70 W power output with a 30-s resting pulse between cycles) in a bath sonicator (Misonix Sonicator 3000). Liposomes were prepared from these suspensions by serial extrusion through polycarbonate membranes (400-nm and 100-nm pore size) using mini extruders purchased from Avanti Polar Lipids. The stability of peptide-liposomes upon storage was assessed by recording antibody binding. Peptide-liposomes were found to be stable for a week's time if stored at 4°C after capping with nitrogen to prevent oxidation. The surface concentration of peptides was calculated from the concentrations of peptides and lipids used in the experiment and from an aggregation number of 80,047 lipids per 100-nm-diameter liposome and an area per lipid of 0.71 nm2. This calculation yielded ∼190 and 760 peptides per liposome at P/L ratios of 1:400 and 1:100, respectively.
Surface plasmon resonance measurements.
All surface plasmon resonance measurements were made using a BIAcore 3000 instrument, and data analyses were performed using BIAevaluation 4.1 software (BIAcore). Peptide-free synthetic liposomes and peptide-conjugated liposomes were captured on a BIAcore L1 sensor chip which uses an alkyl linker for capturing liposomes. Before capturing liposomes, the L1 chip was conditioned by immobilizing ∼3,000 resonance units of bovine serum albumin in each of the four flow cells using amine coupling chemistry. This step was necessary to minimize the nonspecific binding of MAbs 2F5 and 4E10 to the blank L1 chip surface (2). Prior to the capture of liposomes, the surface of the L1 chip was cleaned with a 60-s injection of 40 mM octyl β-d-glucopyranoside at 100 μl/minute, followed by washes with excess buffer to remove any traces of detergent. Peptide-free synthetic liposomes and peptide-conjugated liposomes were immobilized at a predetermined optimum level of ∼500 resonance units. The MAbs, at a 100-μg/ml concentration, were then injected for 120 s at a 20-μl/min flow rate. After each MAb injection, the surface was cleaned by injecting 40 mM octyl β-d-glucopyranoside at a 100-μl/min flow rate, followed by a 5-s injection of 25 mM NaOH at a 50-μl/min flow rate. Using BIAevaluation 4.1 software, the low levels of nonspecific binding to peptide-free control liposomes were subtracted to obtain the peptide-specific binding of MAbs and were used to analyze the binding of MAbs in a two-step encounter-docking model as described previously (2).
FRET and fluorescence-quenching measurements.
Steady-state fluorescence spectra were recorded by using a Varian Cary Eclipse fluorescence spectrometer. Peptide-conjugated liposomes or control liposomes were diluted as needed into a rectangular fluorescence cuvette with a path length of 0.5 cm. The final concentration of the peptides varied from 10 to 20 μg/ml. Tryptophan fluorescence spectra were recorded by exciting the samples at 280 nm with a slit width of 5 nm. In the case of FRET experiments, tryptophan fluorescence spectra were recorded for peptide-liposomes and for 5 mol% DANSYL-PE label-containing peptide-liposomes and peptide-free liposomes. For acrylamide quenching experiments, the tryptophan fluorescence spectra of peptide-conjugated liposomes were recorded at different concentrations of added acrylamide. The fluorescence spectra from peptide-free liposomes at the same concentrations of added acrylamide were subtracted to obtain fluorescence arising from the peptides only. In the case of DBr quenching experiments, 10 mol% of either 67DBrPC or 910DBrPC was included in the lipid mixture and the POPC fraction reduced accordingly.
CD spectroscopy measurements.
Since liposomes prepared by the extrusion technique lead to excessive light scattering in the CD spectrum, small unilamellar liposomes were prepared by probe sonication. The aqueous suspensions (20 mM phosphate buffer, pH 7.4) of lipid or peptide-lipid films were sonicated for 4 min at a 50% duty cycle using a Misonix 3000 equipped with a titanium probe tip. The liposomes were fractionated (4) by centrifugation at 70,000 rpm for 25 min at 4°C using a Beckman TL-100 ultracentrifuge. CD spectra were measured on an Aviv model 62 DS spectropolarimeter using a 1-mm quartz cuvette. Spectra were obtained at 25°C at 0.5-nm intervals and times averaging from 1 to 2 s. Three scans were averaged to obtain CD spectra. The CD signal from peptide-free synthetic liposome samples was subtracted to remove the contribution to the peptide-liposome CD signal of scattering arising from liposomes. The blank subtracted CD spectra were smoothed using the Savitsky and Golay variation method with a moving window of 10 to 25 points and a polynomial order of 3.
RESULTS
Interaction of MAbs 2F5 and 4E10 with membrane-anchored nominal epitope and biepitope peptides.
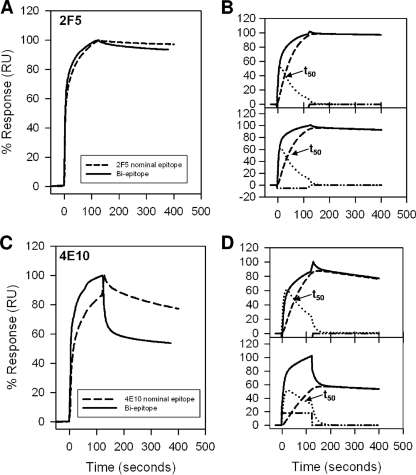
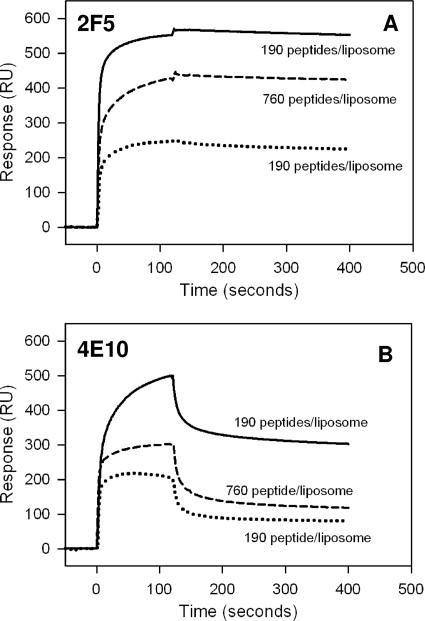
We have previously described the binding kinetics of MAbs 2F5 and 4E10 to first-generation nominal epitope peptides and shown in a two-step encounter-docking model that both antibodies bind to peptide-liposome conjugates (having a nonphysiologic orientation of MPER peptides, i.e., membrane anchored at their N terminus) (Fig. 1A) (2). This mode of binding was not observed with several nonneutralizing antibodies that either failed to bind (2) or bound in a simple model (Langmuir model) (S. M. Alam, M. K. Gorny, S. Zolla-Pazner, and B. F. Haynes, unpublished data). In the current study, in addition to nominal epitope peptide-liposome conjugates, we have designed a longer (28 residues) biepitope peptide-liposome conjugate that includes both 2F5 and 4E10 binding epitopes anchored to the membrane at the C-terminal end (physiologic orientation) of MPER peptides (Fig. 1B). Thus, we first compared the binding kinetics of 2F5 and 4E10 to nominal epitope and biepitope peptide-lipid conjugates and determined whether the binding epitopes are similarly presented on the membrane surface for efficient docking of the MAbs. Figure 2 displays the specific binding of 2F5 (Fig. 2A) and 4E10 (Fig. 2C) MAbs to their nominal epitope and biepitope peptide-liposome conjugates. As described earlier for the nominal epitope conjugates, the peptide-specific binding of both the 2F5 and 4E10 MAbs was also biphasic for the biepitope peptide-lipid conjugates. The results in panels B and D in Fig. 2 show that the biphasic binding pattern of MAbs 2F5 and 4E10 could be best described by a two-step “encounter-docking” model, consistent with our previous report (2) and the work of Sun et al. (31). The association and dissociation rate constants (ka and kd) of the encounter, as well as docking steps, are collected in Table 1. The affinity of MAb 2F5 for both the 2F5 nominal epitope and biepitope peptide-liposomes was higher than that of MAb 4E10 for the corresponding 4E10 nominal epitope and the biepitope by an order of magnitude (1 to 2 nM versus 20 nM). The relatively faster (two- to fourfold) dissociation rates of the docking step in MAb 4E10 binding compared to those of MAb 2F5 primarily accounted for the weaker affinity of 4E10. This relatively lower docking efficiency is also reflected in the longer (∼50 to 80 s for 4E10 versus 28 to 35 s for 2F5) time taken for half of the encounter complexes to be converted to docked complexes (Fig. 2B and D). When the results for their respective nominal epitope peptide-liposome and biepitope peptide-liposome conjugates were compared, the apparent binding constant (Kd) values of 2F5 and 4E10 interaction were similar. However, the docking dissociation rates differed by twofold between the nominal epitope and the biepitope conjugates (Table 1). These results suggested that the orientation of the peptides on the liposomal surface may differ between the shorter nominal epitope and the longer biepitope peptides and that such differences might influence the binding stabilities of the MAbs.
FIG. 2.
Interaction of MAbs 2F5 and 4E10 with MPER peptide-liposome conjugates. (A) Results for binding of MAb 2F5 to 2F5 nominal epitope peptide-liposome conjugate and biepitope peptide-liposome conjugate are shown. RU, resonance unit. (B) The binding of MAb 2F5 to 2F5 nominal epitope (top) and biepitope (bottom) peptide-liposome conjugates follows the two-step conformational change model. (C and D) Binding of MAb 4E10 to the 4E10 nominal epitope and biepitope peptide-liposome conjugates as described for panels A and B. In each of the overlays (B and D), the binding data are shown by solid lines and represent the observed total binding response. The component curves for the encounter (dotted lines) and docked complexes (dashed lines) were simulated from the experimentally determined rate constants (Table 1). t50 is the time required for half of the encounter complex to be converted to docked complex.
TABLE 1.
Rate constants of 2F5 and 4E10 binding to nominal and biepitope peptide-lipid conjugates
| MAb | Peptide-lipid conjugate | Encounter
|
Docking
|
Kd (nM) | ||
|---|---|---|---|---|---|---|
| ka (105M−1 s−1) | kd (10−2 s−1) | ka (10−2 s−1) | kd (10−4 s−1) | |||
| 2F5 | Nominal | 1.79 | 5.72 | 3.06 | 1.05 | 1.09 |
| Biepitope | 2.34 | 5.02 | 2.37 | 2.32 | 2.08 | |
| 4E10 | Nominal | 1.27 | 5.97 | 1.90 | 8.32 | 19.65 |
| Biepitope | 5.72 | 5.99 | 0.92 | 4.34 | 26.95 | |
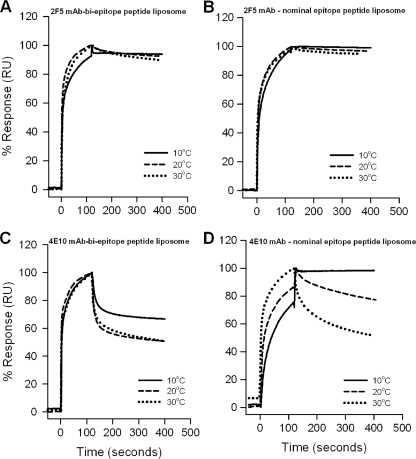
For the purpose of defining the mechanism of binding of MAbs 2F5 and 4E10 to peptide-liposome conjugates and to ascertain differences in the antibody binding stabilities between the nominal epitope and biepitope peptide-liposome conjugates, we measured the temperature dependence of 2F5 and 4E10 binding to the nominal epitope and biepitope peptide-liposome conjugates by recording the binding kinetics at different temperatures (Fig. 3). The rate constants and binding parameters obtained from fitting the time courses to the two-step encounter-docking model and the free-energy changes estimated from them are shown in Tables 2 and 3 for 2F5 and 4E10 binding, respectively. The increase in temperature increased the Kd of 2F5 binding to biepitope peptide-liposomes by 40-fold, because the docking was found to be thermodynamically less favorable at higher temperature (kd2, ΔG2 and %ΔG in Table 2). However, the Kd of 2F5 binding to its nominal epitope peptide-liposomes was relatively less dependent on temperature (Fig. 3B), showing only a threefold difference in Kd values at 10 and 30°C, respectively (Table 2). The free-energy changes of 2F5 binding remained essentially invariant (Table 2). These data showed that 2F5 binding to the biepitope conjugate was relatively more sensitive to temperature than binding to the nominal epitope conjugate.
FIG. 3.
Temperature dependence of 2F5 and 4E10 binding to peptide-liposome conjugates. Binding kinetics of 2F5 (A and B) and 4E10 (C and D) were measured at different temperatures, as indicated, for the biepitope (A and C) and 2F5 and 4E10 nominal epitope (B and D) peptide-lipid conjugates. The specific binding signal was recorded with reference to the signal for peptide-free synthetic liposomes, as described in Materials and Methods. The data recorded at different temperatures were normalized and are presented as the percent response. RU, relative units.
TABLE 2.
Effect of temperature on binding parameters of MAb 2F5 with nominal and biepitope peptide-lipid conjugates
| Parametera | Binding of 2F5 at indicated temp to:
|
|||||
|---|---|---|---|---|---|---|
| Nominal epitope
|
Biepitope
|
|||||
| 10°C | 20°C | 30°C | 10°C | 20°C | 30°C | |
| Rate constant | ||||||
| ka1 (105M−1 s−1) | 0.84 | 2.48 | 2.97 | 2.74 | 3.70 | 3.15 |
| kd1 (10−2 s−1) | 16.6 | 8.30 | 9.80 | 4.66 | 8.81 | 13.6 |
| ka2 (10−2 s−1) | 3.06 | 3.15 | 2.79 | 3.05 | 3.12 | 4.72 |
| kd2 (10−4 s−1) | 5.28 | 1.24 | 1.48 | 0.20 | 2.85 | 4.48 |
| Binding constant | ||||||
| Kd (10−9 M) | 0.65 | 1.30 | 1.75 | 0.11 | 2.16 | 4.07 |
| Kd1 (10−7 M) | 1.21 | 3.34 | 3.30 | 4.63 | 2.38 | 4.31 |
| Kd2 (10−2) | 0.55 | 0.39 | 0.53 | 0.05 | 0.92 | 0.95 |
| Free-energy change | ||||||
| ΔG (kcal) | −12.10 | −11.91 | −12.13 | −12.71 | −11.62 | −11.63 |
| ΔG1 (kcal) | −9.11 | −8.68 | −8.99 | −8.35 | −8.88 | −8.82 |
| ΔG2 (kcal) | −2.98 | −3.22 | −3.16 | −4.35 | −2.73 | −2.80 |
| %ΔG (ΔG2/ΔG) | 25 | 27 | 26 | 34 | 23 | 24 |
ka1, ka2 for association and kd1, kd2 for dissociation processes in encounter and docking steps, respectively. Kd, Kd1, and Kd2 for overall reaction, encounter, and docking steps, respectively. ΔG, ΔG1 and ΔG2 for overall reaction, encounter, and docking steps, respectively.
TABLE 3.
Effect of temperature on binding parameters of MAb 4E10 with nominal and biepitope peptide-lipid conjugates
| Parameterd | Binding of 4E10 at indicated temp to:
|
|||||
|---|---|---|---|---|---|---|
| Nominal epitope
|
Biepitope
|
|||||
| 10°C | 20°C | 30°C | 10°C | 20°C | 30°C | |
| Rate constant | ||||||
| ka1 (105M−1 s−1) | 0.66 | 1.42 | 1.92 | 2.59 | 2.50 | 2.21 |
| kd1 (10−2 s−1) | —a | 5.84 | 8.13 | 10.4 | 9.47 | 12.4 |
| ka2 (10−2 s−1) | 2.77 | 2.02 | 1.20 | 1.15 | 0.80 | 1.07 |
| kd2 (10−4 s−1) | 0.76 | 8.34 | 15.6 | 4.18 | 6.29 | 9.65 |
| Binding constant | ||||||
| Kd (10−9 M) | —b | 16.39 | 50.0 | 14.01 | 27.78 | 45.45 |
| Kd1 (10−7 M) | —c | 4.11 | 4.24 | 4.01 | 3.79 | 5.62 |
| Kd2 (10−2) | 0.28 | 4.13 | 13.0 | 3.63 | 8.0 | 9.0 |
| Free-energy change | ||||||
| ΔG (kcal) | −15.48 | −10.44 | −10.12 | −10.17 | −10.13 | −10.18 |
| ΔG1 (kcal) | −12.16 | −8.56 | −8.83 | −8.28 | −8.61 | −8.66 |
| ΔG2 (kcal) | −3.31 | −4.54 | −4.01 | −4.45 | −4.16 | −4.22 |
| %ΔG (ΔG2/ΔG) | 21 | 43 | 40 | 44 | 41 | 41 |
In contrast, a more dramatic effect was observed with 4E10 binding to nominal epitope peptide-lipid conjugates (Fig. 3D). As observed previously with MAb 2F5, the fitting of these data to a two-step encounter-docking model revealed that the increase in temperature had a greater effect on the docking step than on the encounter step (Table 3). 4E10 docking to the nominal epitope peptide-liposomes showed a 20-fold difference in kd2 values at 10 and 30°C, respectively (Table 3). 4E10 binding to the biepitope peptide liposomes, however, did not exhibit such sensitive temperature dependence, showing only a twofold difference in kd2 values at 10 and 30°C (Fig. 3C and Table 3). Thus, the energetics of 4E10 binding to the biepitope liposomes (Table 3; compare kd2 and Kd2 values for nominal epitope and biepitope liposomes) were more favorable, showing greater stability over the temperature range studied (Fig. 3C andD).
These kinetics and thermodynamics data suggested that the orientations of the nominal epitopes of 2F5 and 4E10 were perhaps different in the two peptide-liposome conjugates. In order to explain these differences in the binding of MAbs 2F5 and 4E10 to epitope peptides, we next determined the differences in the orientation assumed by the peptides in the membrane and the variations in secondary structures adopted by the peptides.
Orientation assumed by HIV-1 gp41 MPER peptides in membranes.
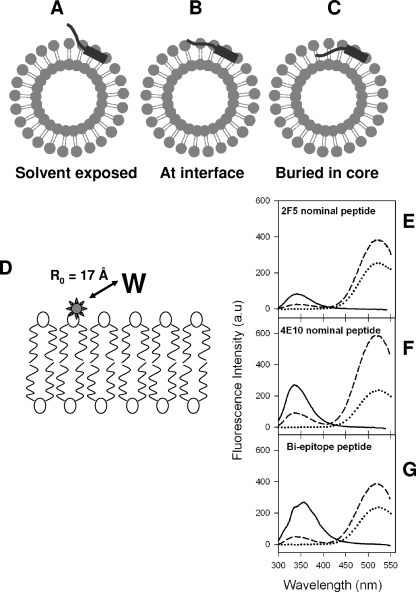
We reasoned that the differences in the docking of MAbs 2F5 and 4E10 to the membrane-anchored nominal epitope and biepitope peptides in our study could be due to the differences in the overall orientation and, more specifically, to the depth of immersion of key core residues of the MPER epitope on the liposomes. Hence, it was necessary to compare the membrane orientation of nominal epitope and biepitope peptides. When anchored into liposomes, the nominal epitope and biepitope peptides could assume one of the three possible orientations: they could project out of the surface of the membrane, lie on the surface or interfacial region of the membrane, or be embedded into the outer monolayer of the membrane (illustrated in Fig. 4A to C). To deduce whether the peptides project out of the surface of the membrane or remain associated with the membrane, we performed FRET experiments (scheme is shown in Fig. 4D). The tryptophan fluorescence spectra of the peptide-liposome conjugates were recorded in the absence and in the presence of fluorescent probe DANSYL attached to the polar head group of the lipid in the liposomal membrane. Figure 4E to G shows the tryptophan fluorescence spectra of MPER peptides conjugated to unlabeled liposomes and to 5 mol% DANSYL-PE-labeled liposomes. The decrease in tryptophan fluorescence emissions for all three peptides, with concomitant increases in DANSYL fluorescence at 520 nm (Fig. 4E to G), can be attributed to an efficient FRET process from tryptophan residues of the peptides to the DANSYL moiety attached to the head group of the phospholipid. Tryptophan and DANSYL are a FRET pair with a Forster distance of 17 Å (32). The observation of strong quenching of tryptophan fluorescence suggests that in all three peptide-liposome conjugates, the majority of tryptophan residues are either proximal to the membrane or remain membrane associated, although one or two residues could be solvent exposed.
FIG. 4.
Possible orientations of MPER peptides and membrane proximity of tryptophan residues of MPER peptides. (A to C) Pictorial representations showing possible orientations that the MPER peptides could assume when conjugated to liposomes. (D) Schematic diagram showing the location of DANSYL label (star) in the lipid bilayer. The Forster distance (R0) for observing 50% FRET efficiency for the DANSYL-tryptophan pair is indicated (32). (E to G) Tryptophan-to-DANSYL FRET for 2F5 nominal epitope (E), 4E10 nominal epitope (F), and biepitope (G) MPER peptide-liposome conjugates. In each panel, the solid curves show the fluorescence spectra of the peptide-liposome conjugates in the absence of DANSYL-PE, the dashed curves show the fluorescence spectra of peptide-liposome conjugates having 5mol% DANSYL-PE, and the dotted curves show the fluorescence of DANSYL-PE liposomes with no peptides.
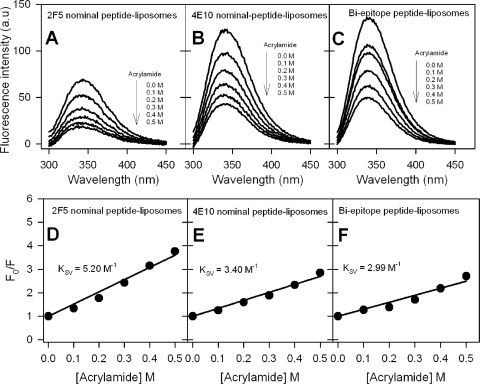
Furthermore, to distinguish whether the peptides remain at the surface of the membrane or at the interfacial region of the membrane, we have examined the quenching of tryptophan fluorescence using aqueous and lipidic quenchers. Fluorescence quenching by acrylamide is a well established method to explore the solvent accessibility of tryptophan residues in proteins and peptides (20) and can be used to infer whether tryptophan residues are exposed or immersed in the lipid bilayer (7). The results for quenching of tryptophan fluorescence of MPER peptide-liposome conjugates by the aqueous quencher acrylamide are shown in Fig. 5A to C. Plots of the ratio of fluorescence intensity in the absence of quencher to that at various concentrations of quencher (Fig. 5D to F) yielded linear responses, indicating the dynamic nature of the quenching process (20), with Stern-Volmer quenching constant (KSV) values of 5.2 M−1, 3.4 M−1, and 3.0 M−1 for the 2F5 nominal epitope, 4E10 nominal epitope, and biepitope peptide-liposomes, respectively. A comparison of KSV values, which are a direct measure of quenching efficiency, shows that the tryptophan residues in the 2F5 nominal epitope peptide are more exposed to aqueous quencher than those of the other two peptides and thus indicates that the C-terminal region of gp41 MPER is more likely to be membrane embedded.
FIG. 5.
Evaluation of exposure of membrane-anchored 2F5 and 4E10 nominal epitope and biepitope peptides to aqueous quencher. (A to C) Tryptophan fluorescence emission spectra of 2F5 nominal epitope (A), 4E10 nominal epitope (B), and biepitope (C) peptide-liposome conjugates at different added acrylamide concentrations. a.u., arbitrary unit. (D to F) Stern-Volmer plots for the quenching experiments whose results are shown in panels A to C. Ratio of tryptophan fluorescence intensity of peptide-liposomes in the absence of acrylamide to that at different acrylamide concentrations (F0/F) as a function of acrylamide concentration.
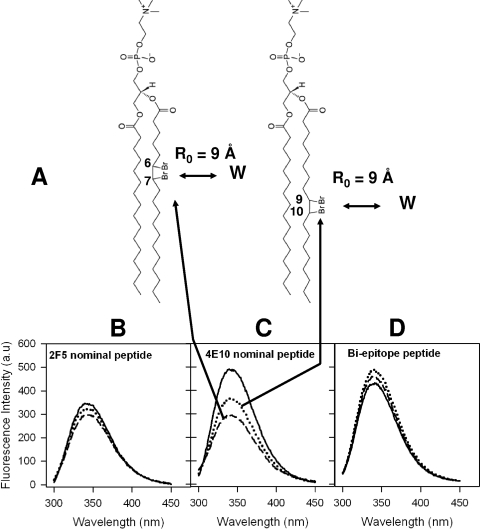
Finally, to ascertain the depth of immersion of the tryptophan residues in gp41 MPER peptide constructs, we recorded the tryptophan fluorescence spectra of peptide-liposome conjugates in the absence and in the presence of brominated phospholipids with the bromine attached at either the 6,7 position or the 9,10 position of the acyl chain of the phospholipid (schematically shown in Fig. 6A) (5, 16). Figure 6B to D displays the tryptophan fluorescence spectra of peptide-liposome conjugates recorded with no DBrPC in the liposomes and with 10 mol% 67DBrPC- and 910DBrPC-labeled liposomes. As can be seen from Fig. 6C, significant quenching was observed for the 4E10 nominal epitope peptide when the DBr label was at the 6,7 position, indicating that one or more tryptophan residues are lying proximal to this DBr label. The absence of any significant quenching for the other two peptide liposome conjugates, the 2F5 nominal epitope (Fig. 6B) and the biepitope (Fig. 6D), suggests that tryptophan residues in these peptides are not proximal to the DBr labels placed at the two different depths of the bilayer (Fig. 6A).
FIG. 6.
Evaluation of membrane insertion of 2F5 and 4E10 nominal epitope and biepitope peptides. (A) Chemical structures of DBr lipids used in the experiment. The positions of DBr labels and Forster distance (R0) requirements for observing 50% quenching efficiency (5) are also shown. (B to D) Tryptophan fluorescence spectra of 2F5 nominal epitope (B), 4E10 nominal epitope (C), and biepitope (D) peptide-liposome conjugates. In each panel, the solid curves show the spectra recorded with no DBr lipids and the dotted and dashed curves show the spectra recorded with 10 mol% 67DBrPC and 910DBrPC liposomes, respectively. a.u., arbitrary unit.
Taken together, these fluorescence-quenching studies indicate that the 2F5 nominal epitope peptide lies proximal to the polar head groups of the phospholipid, with its tryptophan residues more exposed. On the contrary, the 4E10 nominal epitope peptide resides at the interfacial region of the bilayer, with much-reduced exposure of tryptophan residues, and one or more of them are likely to orient into the acyl chain region proximal to the bilayer interface. These findings are at least qualitatively consistent with an earlier report based on membrane immersion depth measurements using electron paramagnetic resonance spectroscopy (31). The biepitope peptide, based on the results, appears to assume an orientation which is intermediate between those of the 2F5 and 4E10 nominal epitope peptides, with the tryptophan residues having relatively more shallow penetration in the membrane bilayer. The data on the depth of immersion of the tryptophan residues are consistent with our binding kinetics data. The efficiency of MAb docking to MPER peptide-lipid conjugates is related to the relative exposure of the binding epitope on the liposomal surface. More efficient docking was observed when the MPER epitope was relatively less immersed in the lipid bilayer. Thus, our data suggest that the ability of both 2F5 and 4E10 to bind to lipids may be required to interact with MPER epitopes embedded into the viral membrane.
Secondary structures adopted by gp41 MPER peptides in the liposomal membrane.
Previously, MPER peptide structures in solution have been defined to be random by CD spectroscopic analysis (21). However, more defined secondary structures have been described for some shorter peptides (D664-K683) both in solution and in micelle- or liposome-bound states in infrared (17, 26, 30) and CD spectroscopic studies (16). More recently, nuclear magnetic resonance spectroscopic studies of micelle-bound peptides corresponding to E662-K683 (31) and S649-K683 (8) revealed that the C-terminal half of MPER consists of two shorter helices separated by a short hinge and that the N-terminal half of MPER is mostly disordered.
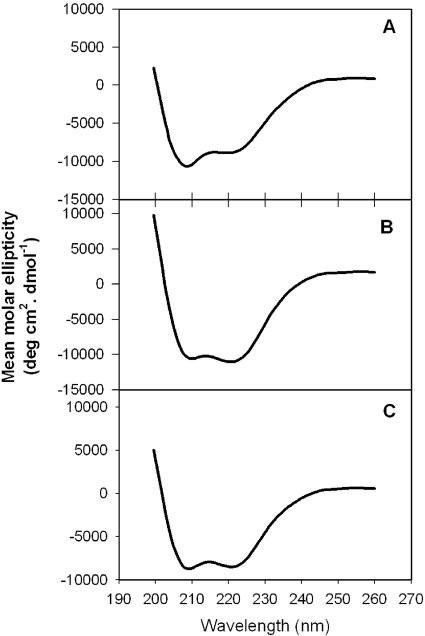
In this study, we used sequences Q652-N671 for the 2F5 nominal epitope peptide, S668-K683 for the 4E10 nominal epitope peptide, and N656-K683 for the biepitope peptide. All these peptides were conjugated to liposomes via the membrane-anchoring GTH1 sequence placed at their C termini (Fig. 1B). Since there are no structural studies of membrane-anchored MPER peptides available, we attempted to define the secondary structures of these MPER peptides on the liposomal surface, to obtain correlation between antibody binding characteristics and the secondary structures adopted by the epitope peptides in the membrane environment, by carrying out CD spectroscopic studies. The CD spectra of the nominal epitope and biepitope peptides conjugated to small unilamellar vesicles (P/L ratio of 1:200) made by probe sonication are shown in Fig. 7. CD spectra collected for samples at a 1:400 P/L ratio were qualitatively similar (not shown) and were not used to deconvolute the different secondary structures due to a low signal-to-noise ratio. As can be seen from the results in Fig. 7, all peptides, especially the 4E10 nominal epitope peptide and biepitope peptide, showed characteristic double minima at 222 and 205 nm, indicating the presence of α-helical structure. In order to quantify the different secondary structures assumed by the peptides, the CD spectra were analyzed by using the K2D program, which utilizes a self-organizing neural network to extract secondary structural features present in the data from a set of CD spectra of proteins with known structures (3); the results are shown in Table 4. The 4E10 nominal epitope peptide assumed a predominantly α-helical (38%) structure with some β-sheet (8%) structure and significant (54%) random structures. The 2F5 peptide adopted a reduced α-helical component (29%) with roughly the same amount of β-sheet structure (29%) along with significant random (42%) structures (Table 4). The biepitope peptide adopted secondary structures that are somewhat intermediate between those assumed by the 2F5 and 4E10 nominal epitope peptides. Since the membrane-anchoring sequence was the same in all peptide-liposome conjugates, it is reasonable to assume that the observed differences directly reflect the differences in MPER peptide structure on the membrane surface.
FIG. 7.
CD spectra of MPER peptides conjugated to small unilamellar liposomes. Smoothed CD spectra (see Materials and Methods) are shown for 2F5 nominal epitope (A), 4E10 nominal epitope (B), and biepitope (C) MPER peptide-liposomes. The P/L ratio was kept at 1:200.
TABLE 4.
Secondary structures adopted by MPER peptides when conjugated to liposomesa
| Peptide | % α-Helix | % β-Sheet | % Randomb |
|---|---|---|---|
| 2F5 nominal epitope peptide | 29.0 | 29.0 | 42.0 |
| 4E10 nominal epitope peptide | 38.0 | 8.0 | 54.0 |
| Biepitope peptide | 33.0 | 17.0 | 50.0 |
Determined by deconvolution of CD spectra of the peptide-lipid conjugates.
Includes beta bends, which are not well estimated by the method employed here (3).
MPER peptide anchorage on the membrane surface facilitates efficient 2F5 and 4E10 binding.
Recent studies that have described the binding of MAbs 2F5 and 4E10 to MPER peptides on liposomes used either no membrane anchor (31) or a lipidic anchor (8) and relied on the inherent membrane affinity of the MPER peptides. The importance of membrane anchoring on peptide orientation and the effect on MAb binding have not been studied. Here and in our previous study (2), we used a membrane-anchoring sequence (GTH1) to facilitate homogeneity in orientation and structure. In order to test the significance of membrane anchoring of MPER peptides for docking of 2F5 and 4E10, we examined their interactions with the biepitope peptide-liposome conjugate that lacked the membrane-anchoring GTH1 sequence used in our peptide-liposome conjugate design. When added to liposomes, the biepitope MPER peptide associated with the membrane and remained at the bilayer interfacial region, as determined by an increase in tryptophan fluorescence, as well as the existence of FRET between tryptophans and membrane head group-located DANSYL-PE (17, 27; our data not shown). We compared the binding of MAbs 2F5 and 4E10 to the biepitope MPER peptide anchored on liposomes via the GTH1 sequence at a P/L ratio of 1:400 (∼190 peptides per liposome) with their binding to the liposome-bound biepitope peptide that lacked the membrane anchor at P/L ratios of 1:400 and 1:100 (∼190 and 760 peptides per liposome). At a surface concentration of 190 peptides per liposome, the steady-state binding of 2F5 to the membrane-bound biepitope peptide construct was ∼2.5-fold lower than its binding to the membrane-anchored biepitope peptide construct (Fig. 8A). An increase in the surface concentration of peptide to ∼760 peptides/liposome decreased this difference significantly (Fig. 8A). A strikingly different behavior for 4E10 binding to differently presented (membrane-anchored versus membrane-bound) biepitope peptide constructs was noted (Fig. 8B). Compared to its binding to the membrane-anchored biepitope peptide construct, the binding of 4E10 to the membrane-bound biepitope peptide construct was lower, and the improvement at the higher concentration was smaller than for 2F5 (Fig. 8B). Table 5 compares the rate constants of the encounter and docking steps for both these constructs. It is evident from the table that the rates of the encounter step remained comparable and the main difference was in the docking step. The relatively inefficient docking of MAb 4E10 to the membrane-bound biepitope peptide construct stems from the relatively slower formation (∼19-fold) and faster dissociation (∼sevenfold) of docked complexes.
FIG. 8.
Comparison of 2F5 and 4E10 binding to biepitope MPER peptide presented differently on liposomes. Binding of 2F5 (A) and 4E10 (B) to membrane-anchored biepitope MPER peptide (solid lines) at a P/L ratio of 1:400 and membrane-bound biepitope MPER peptide at P/L ratios of 1:400 (dotted lines) and 1:100 (dashed lines). The surface concentrations of peptides are also indicated. RU, resonance unit.
TABLE 5.
Comparison of rate constants of 4E10 binding to membrane-anchored and membrane-bound biepitope MPER peptide constructs
| Peptide-lipid conjugate | Encounter
|
Docking
|
Kd (nM) | ||
|---|---|---|---|---|---|
| ka (105M−1 s−1) | kd (10−2 s−1) | ka (10−3 s−1) | kd (10−3 s−1) | ||
| Membrane bound | 1.9 | 6.6 | 0.6 | 3.4 | 20.0 |
| Membrane anchored | 1.0 | 4.7 | 11.1 | 0.5 | 19.4 |
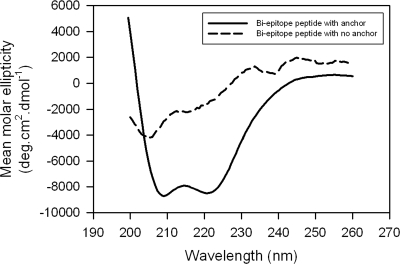
In order to define whether this inefficient conversion of the encounter complex into the docked complex is due to any difference in the structure adopted by the peptide when presented differently on the membrane, we performed a CD spectroscopic comparison of these two constructs (Fig. 9). Interestingly, unlike the membrane-anchored biepitope peptide, which adopted some α-helical structure, as revealed by characteristic double minima at 222 and 205 nm, the membrane-bound biepitope peptide remained largely unstructured. Since the membrane orientation of the membrane-anchored (with GTH1) biepitope peptide described here is similar to that of the membrane-bound (without GTH1) biepitope peptide shown by Sun et al. (31), the difference in secondary structural features seen in Fig. 9 might be due to constraints imposed by the attached GTH1 anchor.
FIG. 9.
CD spectra of biepitope MPER peptides presented differently on small unilamellar liposomes. Smoothed CD spectra of membrane-anchored and membrane-bound biepitope MPER peptides at a P/L ratio of 1:200.
DISCUSSION
In this study, using synthetic peptide-liposome conjugates, we have shown that (i) membrane anchoring is important for the presentation of 2F5 and 4E10 MPER epitope structure; (ii) the N and C termini of gp41 MPER are differentially exposed on the membrane; and (iii) the stable docking of MAbs 2F5 and 4E10 to peptide-liposomes is influenced by the extent of membrane immersion of the binding epitopes. The results from our study, as summarized in Fig. 10, can be used to construct a model that illustrates the presentation of gp41 MPER residues on the liposomal surface and their interactions with MAbs 2F5 and 4E10. We observed that the docking of MAb 2F5 to its nominal epitope-liposome conjugate was more efficient than that of MAb 4E10 and that this difference in MAb binding could be explained by the differences in the level of immersion of the MPER epitopes in the lipid bilayer. Thus, the efficiency of MAb docking correlated well with the more exposed nature of the 2F5 nominal epitope peptide on the membrane surface versus the less exposed orientation of the 4E10 nominal epitope peptide, which had one or more tryptophans immersed into the acyl chain region of the bilayer. In our biepitope peptide-liposome constructs, we observed more efficient docking of both 2F5 and 4E10 and, thus, this construct design could be a potential immunogen for the induction of neutralizing MPER antibodies.
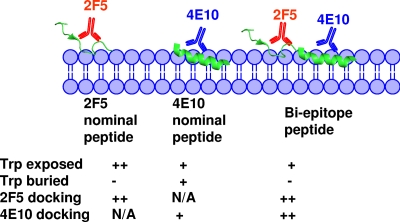
FIG. 10.
Pictorial representation of differential membrane immersion of nominal epitope and biepitope MPER peptides influencing the efficiency of 2F5 and 4E10 docking. The 2F5 and 4E10 binding regions of MPER are rendered from their respective Fab-bound crystal structures (6, 25). The different immersion depths indicated for nominal epitope and biepitope peptides in the lipid bilayer were inferred from the fluorescence-quenching and FRET experimental results. The relative stabilities of 2F5 and 4E10 docking shown were derived from the temperature dependence of antibody binding to the respective nominal epitope and biepitope peptide-liposome conjugates. N/A, not applicable. Trp-exposed row, ++ and + indicate a KSV value of 5.0 and ∼3.0 M−1, respectively, in acrylamide quenching experiment. Trp-burried row, + and − represent a ≥25% and <25% quenching of tryptophan fluorescence, respectively, by dibromo lipids. Rows of 2F5 and 4E10 docking, ++ and + represent no change and a twofold change, respectively, in %ΔG (ΔG2/ΔG) on increasing temperature from 10 to 30°C.
The mechanism that leads to the affinity enhancement at the membrane interface could be due to direct interaction of MAbs 2F5 and 4E10 with membrane lipids via their CDR3 loops (6, 25), thereby increasing the stability of the MAb-epitope complex. It has also been suggested that the lipid binding properties of MAbs 2F5 and 4E10 (14, 27) aid in their gaining proximity to the membrane-bound MPER epitopes and thereby result in a stable MAb-epitope interaction, as the binding of 2F5 and 4E10 was best explained by the two-step encounter-docking model (2). The requirement of lipid binding in HIV-1 neutralization is supported by the results of our recent studies that showed loss of neutralization of HIV-1 by CDR H3 2F5 and 4E10 mutants that were designed to have impaired lipid binding ability without alteration of epitope binding (1; S. M. Alam, presented at HIV Vaccines: Progress and Prospects, Banff, Alberta, Canada, 27 March to 1 April 2008). While these reports demonstrated the role of lipid reactivity in binding to membrane-embedded MPER epitopes and in HIV-1 neutralization, the orientation of the epitopes and its influence on antibody binding remained to be described.
The binding characteristics of both 2F5 and 4E10 to the carboxyl terminus-conjugated nominal epitope MPER peptide-liposomes used in this study, which are more physiologic than the amino terminus-conjugated constructs used previously (2), were quite similar. As observed with the N terminus-conjugated peptide-liposomes, the binding of MAbs 2F5 and 4E10 was biphasic and best described by a two-step encounter-docking model, with the 2F5 MAb forming a more favorable complex than 4E10 (Fig. 2 and Table 1) (2). Striking differences, however, were evident in the binding stability of the MAbs (Table 1 and Fig. 3A and B) (2). The marked binding stability noted for MAb 2F5 in the constructs used here might be due to the more native C-terminal anchoring of peptides, unlike the N-terminal anchoring strategy used before (2). It is likely that the relatively lower 2F5 binding stability observed in the previous study (2) was due to the constraints imposed on the 2F5 epitope region by the membrane upon N-terminal anchoring of peptides. The biepitope peptide-liposome conjugate is a more physiologic mimic, as it consists of the entire MPER region anchored to the liposomes and offers the advantage of using the same construct to examine the binding of both 2F5 and 4E10 (solid lines in Fig. 2A and C). Our results clearly show that the biepitope peptide-liposome construct provides a more favorable MPER epitope orientation for docking of both 2F5 and 4E10 (Fig. 2).
We have used an artificial peptide membrane anchor in our design of peptide-liposome complexes, and therefore, such liposome constructs may not be similar in the membrane orientation of MPER within the context of the envelope trimer. However, recent studies using such artificial systems have provided data to generate models of gp41 MPER membrane orientation (31). The clear correlation that exists between efficient docking of antibodies and the nature of the orientation of epitope peptides in the liposomal membrane is noteworthy. Antibody docking was more efficient when the epitope peptide was solvent exposed (stable docking of 2F5 to its nominal epitope peptide) but was less efficient if the epitope peptide was immersed in the membrane (less efficient docking of 4E10 binding to its nominal epitope). The efficiency of docking to the biepitope peptide of 2F5 and 4E10 remained in between the above two extremes, suggesting that the peptide assumes a somewhat intermediate orientation with probably minor variations in the immersion depth of key residues involved in MAb binding. These data also suggest that the length of the peptide sequence plays a role in determining the membrane immersion depth of key residues. A recent study (17) which calculated the energetics of interaction of different segments of MPER with the membrane interface reported that the free energy for partitioning from water into membrane interfaces was highest for the D664-K683 stretch of MPER and that extending the sequence at the N terminus of this stretch progressively decreased the free energy. Based on these energetics data, one would predict that the S668-K683 stretch of MPER that we used in our 4E10 nominal epitope peptide would be more readily immersed in the membrane than the longer biepitope peptide that contains 12 additional amino acid residues N terminal to it (N656-K683), while the 2F5 nominal epitope peptide (Q652-N671), which contains residues of the N terminus of MPER, would be the least immersed in the membrane. Thus, our membrane immersion data and the energetics of 2F5 and 4E10 binding are in agreement with this prediction.
Besides the differences in the nature of their membrane orientations, any variation in the structure adopted by the epitope peptides could influence the mode of their interaction with MAbs 2F5 and 4E10 differently. Thus, a key question that needed to be addressed was whether the MPER peptides adopted a unique structure in the membrane environment that favored the docking of 2F5 and 4E10. Even though high-resolution structures of MPER peptides of various lengths in micelles or bicelles have been reported (8, 31), no high-resolution structure of membrane-bound MPER peptide is available except for a structure based on the electron paramagnetic resonance-measured membrane immersion depth parameters of MPER amino acid residues (31). The CD spectroscopic experiments showed that the nominal epitope and biepitope peptides adopted ordered structures with different helical contents (Table 4). The 4E10 nominal epitope peptide had the highest helical content, whereas the 2F5 nominal epitope peptide had a lower helical content, with the biepitope peptide adopting a helical content intermediate between these two peptides. These low-resolution structural data provide some insight about the overall secondary structural elements present in the membrane-anchored state but do not give the much-needed information on the conformation of the epitopes.
However, we have managed to address this very important issue for the 4E10 epitope region by exploring the significance of membrane anchorage of MPER peptides. The comparison of the antibody binding characteristics and secondary structures of membrane-anchored and membrane-bound biepitope MPER peptides clearly revealed that membrane anchorage induces helical structures of MPER peptides, resulting in enhanced binding of MAb 4E10. Our finding is consistent with the crystal structure showing 4E10-bound epitope peptide adopting a helical structure (6). Together, these observations indicate that perhaps a transmembrane domain is needed for the 4E10 epitope region of the MPER to adopt a proper helical structure. On the other hand, 2F5 binding does not seem to have a preference for a more ordered structure of biepitope MPER peptide, because an increase in peptide concentration could reduce the difference in 2F5 binding levels between membrane-anchored and membrane-bound biepitope MPER peptides. Thus, it is possible that the 2F5 epitope region of the membrane-displayed biepitope MPER peptide does not have an ordered structure. This interpretation is consistent with recent nuclear magnetic resonance studies of micelles at pH 6.0 that showed that the N-terminal part of MPER did not have a regular secondary structure by itself (8) or in its 2F5-bound state (25). It is likely that the N terminus of gp41 MPER is engaged with some other component of HIV-1 Env protein (19). Such interactions might make the 2F5 epitope inaccessible to gp41 MPER-specific antibodies.
An important implication of our results is that only rare MPER antibodies that can bind to both lipids and MPER epitopes will be able to bind to native gp41 on the viral membrane surface. There are several lines of evidence that favor this hypothesis. Our binding kinetics data suggest that although some gp41 antibodies might be able to form the encounter complex with gp41 MPER, their inability to interact with membrane-embedded residues will make them ineligible to form a stable docked complex. This is particularly true for the 4E10 epitope that remains largely immersed in the membrane interfacial region (31). The results described above also show that MAb docking was enhanced in the biepitope peptide-liposome construct, in which the depth of membrane immersion of the binding epitope was relatively more favorable for antibody binding. Nonneutralizing antibodies, like 13H11, that do not bind to lipids failed to bind peptide-lipid conjugates (2). Other nonneutralizing MAbs, like the cluster II MAbs 126-6 and 167-D, were only able to form the encounter complex, and the resulting weak-affinity interactions with peptide-lipid conjugates (Alam et al., unpublished data) could explain their inability to neutralize HIV-1. The 2F5 epitope, although more solvent exposed than the C-terminal region of MPER, may still have a critical residue immersed in the lipid bilayer. A possible candidate might be the W666 residue in the core 664DKW666 epitope (31). Moreover, the randomness of the N-terminal region suggests that in the prefusion gp41 state, this portion of the MPER may interact with some other component of HIV-1 Env (19) or be occluded in the gp41 trimer (11, 22) and may not be accessible to MAb binding (22). Thus, both the 2F5 epitope and the 4E10 epitope of gp41 MPER may not be exposed in the prefusion state of gp41(11, 22). This is consistent with our recent finding that MPER MAbs, including 2F5 and 4E10, do not bind to native prefusion gp41 on native virion membranes (S. M. Alam, M. Morelli, S. M. Dennison, H. Liao, R. Zhang, S. Xia, S. Rits-Volloch, L. Sun, S. Harrison, B. F. Haynes, and B. Chen, unpublished data) and the designed trimer of Liu et al. (22). Since both 2F5 and 4E10 bound strongly, with slower dissociation rates, to a trimeric gp41 intermediate protein (11), it is likely that the MPER-neutralizing epitopes are only exposed in the fusion intermediate state following coreceptor triggering of HIV-1 Env. It is possible, therefore, that the membrane-embedded residues may be exposed in the intermediate gp41 structure; this is consistent with our binding kinetics data showing more favorable docking of 4E10 to the biepitope construct, in which the C-terminal region was relatively less immersed in the membrane bilayer. Since we observed strong binding of both 2F5 and 4E10 to the MPER peptide-liposome conjugates, we can conclude that the liposomal form of our MPER peptide constructs is not representative of the prefusion gp41 state but is likely to be representative of an advanced intermediate or postfusion state of gp41.
Our findings are very significant in the context of HIV-1 vaccine development. To date, attempts to elicit broadly neutralizing MPER antibodies have not been successful. It is believed that the neutralizing MPER antibodies are not made either due to the difficulty in presenting the correct MPER conformation to the immune system or due to the tolerance mechanism involving the immune system (13, 24). The study described here is an important step forward in designing immunogens that encompass both the membrane and epitope components for the purpose of presenting a near-native MPER structure. We have demonstrated that membrane-anchored MPER peptides adopt structures and/or orientations that favor efficient docking of MPER antibodies. Thus, the MPER peptide-liposome conjugates described here can potentially be used as immunogens for inducing HIV-1-neutralizing MPER antibodies either alone or with appropriate adjuvants incorporated in them.
Acknowledgments
This research was conducted as part of the Collaboration for AIDS Vaccine Discovery (CAVD) with support from the Bill & Melinda Gates Foundation and from the NIAID Center for HIV/AIDS Vaccine Immunology (CHAVI) grant AI067854 from the NIH.
We acknowledge the technical expertise of David Boren in the preparation and surface plasmon resonance characterization of peptide-lipid conjugates. We are grateful to Vann Bennet for providing access to the CD spectrometer and Khadar Abdi for assisting in data collection. We also thank Prabakaran Ponraj for assistance in making Fig. 10.
Footnotes
Published ahead of print on 29 July 2009.
REFERENCES
- 1.Alam, S. M., S. Dennison, M. Morelli, H. Liao, R. Zhang, B. F. Haynes, S. C. Harrison, and B. Chen. 2008. The role of lipid reactivity of HIV-1 gp41 membrane proximal antibodies in neutralization of HIV-1: novel liposome based immunogen design, abstr. OA07 to 06. Abstracts from AIDS Vaccine 2008, Cape Town, South Africa. October 13-16, 2008. AIDS Res. Hum Retrovir. 24(Suppl. 1):23-24. [Google Scholar]
- 2.Alam, S. M., M. McAdams, D. Boren, M. Rak, R. M. Scearce, F. Gao, Z. T. Camacho, D. Gewirth, G. Kelsoe, P. Chen, and B. F. Haynes. 2007. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J. Immunol. 178:4424-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrade, M. A., P. Chacon, J. J. Merelo, and F. Moran. 1993. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 6:383-390. [DOI] [PubMed] [Google Scholar]
- 4.Barenholz, Y., D. Gibbes, B. J. Litman, J. Goll, T. E. Thompson, and R. D. Carlson. 1977. A simple method for the preparation of homogeneous phospholipid vesicles. Biochemistry 16:2806-2810. [DOI] [PubMed] [Google Scholar]
- 5.Bolen, E. J., and P. W. Holloway. 1990. Quenching of tryptophan fluorescence by brominated phospholipid. Biochemistry 29:9638-9643. [DOI] [PubMed] [Google Scholar]
- 6.Cardoso, R. M., M. B. Zwick, R. L. Stanfield, R. Kunert, J. M. Binley, H. Katinger, D. R. Burton, and I. A. Wilson. 2005. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22:163-173. [DOI] [PubMed] [Google Scholar]
- 7.Chang, D. K., S. F. Cheng, E. A. Kantchev, C. H. Lin, and Y. T. Liu. 2008. Membrane interaction and structure of the transmembrane domain of influenza hemagglutinin and its fusion peptide complex. BMC Biol. 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coutant, J., H. Yu, M. J. Clement, A. Alfsen, F. Toma, P. A. Curmi, and M. Bomsel. 2008. Both lipid environment and pH are critical for determining physiological solution structure of 3-D-conserved epitopes of the HIV-1 gp41-MPER peptide P1. FASEB J. 22:4338-4351. [DOI] [PubMed] [Google Scholar]
- 9.D'Souza, M. P., D. Livnat, J. A. Bradac, S. H. Bridges, et al. 1997. Evaluation of monoclonal antibodies to human immunodeficiency virus type 1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. J. Infect. Dis. 175:1056-1062. [DOI] [PubMed] [Google Scholar]
- 10.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Ann. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 11.Frey, G., H. Peng, S. Rits-Volloch, M. Morelli, Y. Cheng, and B. Chen. 2008. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc. Natl. Acad. Sci. USA 105:3739-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grundner, C., T. Mirzabekov, J. Sodroski, and R. Wyatt. 2002. Solid-phase proteoliposomes containing human immunodeficiency virus envelope glycoproteins. J. Virol. 76:3511-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haynes, B. F., and S. M. Alam. 2008. HIV-1 hides an Achilles' heel in virion lipids. Immunity 28:10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haynes, B. F., J. Fleming, E. W. St. Clair, H. Katinger, G. Stiegler, R. Kunert, J. Robinson, R. M. Scearce, K. Plonk, H. F. Staats, T. L. Ortel, H. X. Liao, and S. M. Alam. 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308:1906-1908. [DOI] [PubMed] [Google Scholar]
- 15.Haynes, B. F., and D. C. Montefiori. 2006. Aiming to induce broadly reactive neutralizing antibody responses with HIV-1 vaccine candidates. Exp. Rev. Vaccines 5:347-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huarte, N., M. Lorizate, R. Kunert, and J. L. Nieva. 2008. Lipid modulation of membrane-bound epitope recognition and blocking by HIV-1 neutralizing antibodies. FEBS Lett. 582:3798-3804. [DOI] [PubMed] [Google Scholar]
- 17.Huarte, N., M. Lorizate, R. Maeso, R. Kunert, R. Arranz, J. M. Valpuesta, and J. L. Nieva. 2008. The broadly neutralizing anti-human immunodeficiency virus type 1 4E10 monoclonal antibody is better adapted to membrane-bound epitope recognition and blocking than 2F5. J. Virol. 82:8986-8996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Julien, J. P., S. Bryson, J. L. Nieva, and E. F. Pai. 2008. Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site. J. Mol. Biol. 384:377-392. [DOI] [PubMed] [Google Scholar]
- 19.Kowalski, M., J. Potz, L. Basiripour, T. Dorfman, W. C. Goh, E. Terwilliger, A. Dayton, C. Rosen, W. Haseltine, and J. Sodroski. 1987. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science 237:1351-1355. [DOI] [PubMed] [Google Scholar]
- 20.Lakowicz, J. 2006. Principles of fluorescence spectroscopy, 3rd ed., Springer, New York, NY.
- 21.Lawless, M. K., S. Barney, K. I. Guthrie, T. B. Bucy, S. R. Petteway, Jr., and G. Merutka. 1996. HIV-1 membrane fusion mechanism: structural studies of the interactions between biologically-active peptides from gp41. Biochemistry 35:13697-13708. [DOI] [PubMed] [Google Scholar]
- 22.Liu, J., Y. Deng, A. K. Dey, J. P. Moore, and M. Lu. 2009. Structure of the HIV-1 gp41 membrane-proximal ectodomain region in a putative prefusion conformation. Biochemistry. 48:2915-2923. doi: 10.1021/bi802303b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matyas, G. R., Z. Beck, N. Karasavvas, and C. R. Alving. 2009. Lipid binding properties of 4E10, 2F5, and WR304 monoclonal antibodies that neutralize HIV-1. Biochim. Biophys. Acta 1788:660-665. [DOI] [PubMed] [Google Scholar]
- 24.Montefiori, D., Q. Sattentau, J. Flores, J. Esparza, and J. Mascola. 2007. Antibody-based HIV-1 vaccines: recent developments and future directions. PLoS Med. 4:e348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ofek, G., M. Tang, A. Sambor, H. Katinger, J. R. Mascola, R. Wyatt, and P. D. Kwong. 2004. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 78:10724-10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saez-Cirion, A., J. L. Arrondo, M. J. Gomara, M. Lorizate, I. Iloro, G. Melikyan, and J. L. Nieva. 2003. Structural and functional roles of HIV-1 gp41 pretransmembrane sequence segmentation. Biophys. J. 85:3769-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez-Martinez, S., M. Lorizate, H. Katinger, R. Kunert, and J. L. Nieva. 2006. Membrane association and epitope recognition by HIV-1 neutralizing anti-gp41 2F5 and 4E10 antibodies. AIDS Res. Hum. Retrovir. 22:998-1006. [DOI] [PubMed] [Google Scholar]
- 28.Sattentau, Q. J., and J. P. Moore. 1991. Conformational-changes induced in the human-immunodeficiency-virus envelope glycoprotein by soluble Cd4 binding. J. Exp. Med. 174:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stiegler, G., R. Kunert, M. Purtscher, S. Wolbank, R. Voglauer, F. Steindl, and H. Katinger. 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 17:1757-1765. [DOI] [PubMed] [Google Scholar]
- 30.Suarez, T., S. Nir, F. M. Goni, A. Saez-Cirion, and J. L. Nieva. 2000. The pre-transmembrane region of the human immunodeficiency virus type-1 glycoprotein: a novel fusogenic sequence. FEBS Lett. 477:145-149. [DOI] [PubMed] [Google Scholar]
- 31.Sun, Z. Y., K. J. Oh, M. Kim, J. Yu, V. Brusic, L. Song, Z. Qiao, J. H. Wang, G. Wagner, and E. L. Reinherz. 2008. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity 28:52-63. [DOI] [PubMed] [Google Scholar]
- 32.Vaz, W. L., and G. Schoellmann. 1976. Specific fluorescent derivatives of macromolecules. A fluorescence study of some specifically modified derivatives of chymotrypsin, trypsin and subtilisin. Biochim. Biophys. Acta 439:206-218. [DOI] [PubMed] [Google Scholar]