Abstract
Understanding the correlates of immune protection against human immunodeficiency virus and simian immunodeficiency virus (SIV) will require defining the entire cellular immune response against the viruses. Here, we define two novel translation products from the SIV env mRNA that are targeted by the T-cell response in SIV-infected rhesus macaques. The shorter product is a subset of the larger product, which contains both the first exon of the Rev protein and a translated portion of the rev intron. Our data suggest that the translation of viral alternate reading frames may be an important source of T-cell epitopes, including epitopes normally derived from functional proteins.
The pathway from viral infection to the cellular immune response is not well understood. Despite the importance of T-cell responses in control of AIDS virus replication (1, 3, 8, 22), the sources of the peptides recognized by virus-specific T cells are still being discovered. AIDS virus-specific CD8+ T lymphocytes (CD8-TL) recognize complexes of major histocompatibility complex (MHC) class I and virus-derived epitopes presented on the surface of infected cells. These epitopes can be derived from exogenous viral proteins in the infecting virion (19, 20) or from de novo synthesis of viral proteins (9, 21). Additional sources of epitopes are also being explored (4, 6).
CD8-TL can also recognize epitopes derived from translation of viral alternate reading frames (ARFs). Though CD8-TL specific for ARF-derived epitopes have been detected in human immunodeficiency virus (HIV) (2), they remain a largely unexplored source of epitopes that might elicit potent antiviral cellular immune responses. We recently showed that SIVmac239-infected rhesus macaques that spontaneously controlled viral replication, termed elite controllers, made immunodominant CD8-TL responses against an epitope (RHLAFKCLW, or cRW9) derived from an ARF of the env gene (15). This response selected for viral escape in vivo and suppressed viral replication in an in vitro assay. These findings imply that CD8-TL specific for ARF-derived epitopes might be an important component of the total AIDS virus-specific cellular immune response.
Here, we show that the cRW9 epitope is translated as part of two distinct products that differ in size due to start codon usage. The larger and more frequent product contains both the first 23 amino acids of the Rev protein (exon 1) and 50 amino acids translated from the rev intron. The smaller is produced by translation initiation at a start codon within the rev intron and is a subset of the larger product. Finally, we show that these products are degraded after translation from the mature Env-encoding mRNA.
cRW9 is consistently recognized at 18 h postinfection.
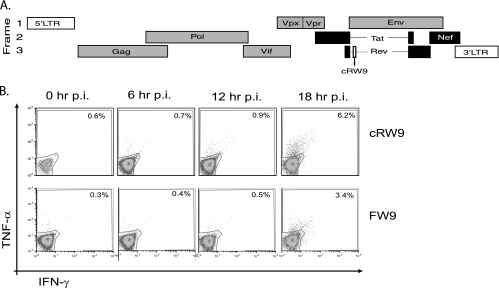
The extent of mRNA splicing is likely a critical determinant of the timing of both viral protein production and epitope recognition. The most rapidly recognized epitopes (excluding those derived from proteins present in the incoming virion) are those derived from proteins encoded by the fully spliced nef and tat (20) and rev (J. B. Sacha, unpublished data) transcripts. In contrast, epitopes derived from translation of partially (or singly) spliced transcripts, such as env, are recognized much later, at 18 h postinfection (19). Consequently, two very different hypotheses emerged as to the origin of cRW9. Since it lies in frame with Rev exon 1 (an early protein), it might be presented early, perhaps within a splice variant form of Rev. However, it also lies in an ARF of Env (a late protein) (Fig. 1A).
FIG. 1.
cRW9 is recognized at 18 h postinfection. cRW9 is located in frame with Rev (early) but in an ARF of Env (late). Early proteins (Tat, Rev, and Nef) are shown in black, while late proteins are shown in gray (A). LTR, long terminal repeat. We synchronously infected CD4+ T cells and used them in a kinetic recognition assay to determine when in the viral life cycle the cRW9 and Env-FW9 epitopes are recognized. Recognition was measured every 6 h as a percentage of cRW9-specific T cells that produce gamma interferon (IFN-γ) and/or tumor necrosis factor alpha (TNF-α) in response to recognition of infected cells. The top row represents recognition of MHC-matched, SIV-infected CD4+ T cells by cRW9-specific CD8-TL, while the bottom row represents recognition by Env-FW9-specific CD8-TL. No recognition was seen of MHC-mismatched cells at any time point. The viral life cycle is approximately 20 to 24 h, and both cRW9 and FW9 are recognized by 18 h after synchronous infection (B). Results are characteristic of at least three distinct experiments. p.i., postinfection.
We used the kinetic intracellular cytokine staining (KICS) assay that we developed previously (19) to examine the timing of cRW9 recognition. We found that cRW9 was consistently recognized late, 18 h after infection, similarly to an Env epitope, FW9 (Fig. 1B). Thus, a partially spliced transcript likely encodes cRW9.
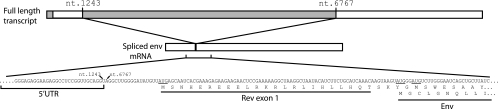
To test this hypothesis, we devised a PCR strategy that selectively amplified nonspliced and partially spliced transcripts. We then synthesized cDNA from simian immunodeficiency virus (SIV)-infected cells and used gene-specific PCR, cloning, and sequencing techniques to determine the presence of a specific mRNA that might uniquely encode the product that contains cRW9. As expected, our PCR strategy identified several partially spliced transcripts (data not shown) but all of the transcripts that we found were previously recognized to encode specific proteins (18, 23). However, closer examination of the Env-encoding mRNA, both from our sequences and from those of others (18, 23), revealed that it was a possible source of the cRW9 epitope (Fig. 2). We excluded the first potential start codon downstream of the splice acceptor site (at the junction of the 5′ untranslated region [UTR] and the coding region) as it would encode only a 2-amino-acid product prior to the ribosome encountering the subsequent stop codon. The second start codon is the rev start codon, while the env start codon lies 74 nucleotides downstream. There is also a potential start codon within the rev intron, which lies just 3 nucleotides downstream of the env start and is in frame with both Rev exon 1 and the cRW9 epitope.
FIG. 2.
The env mRNA is the most likely source of the cRW9 epitope. We cloned and sequenced cDNAs from SIV-infected cells to narrow the search for the SIV mRNA that encodes the cRW9 epitope. We devised a PCR strategy (F_961-980, TGTTCCCATCTCTCCTAGCC; R_6960-6979, AATTGTCGCATTCCTCCAAG) that would selectively amplify nonspliced and partially spliced transcripts. All mRNAs identified had been previously described (18, 23). However, the Env-encoding mRNA contained features consistent with being the source mRNA for the cRW9 epitope. Most importantly, it contained rev exon 1, which is in frame with cRW9. Note that there is an AUG codon upstream of the rev start codon that, if translated, would encode just two amino acids before the next stop codon. The potential significance of this minimal open reading frame is unknown, and it is not included in analyses here. nt., nucleotide.
cRW9 is translated as part of two products from the env mRNA.
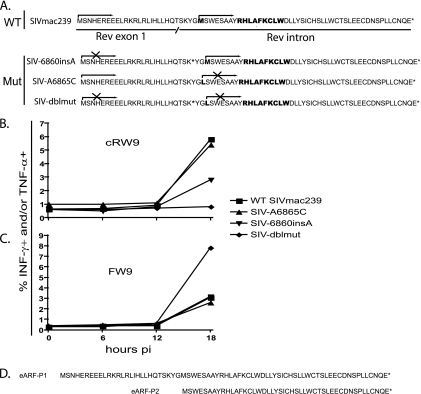
Since cRW9 lies in frame with rev exon 1 but is translated from the rev intron (15), we reasoned that cRW9 might be translated from the env mRNA, using either the rev start codon or the start codon in the rev intron, upstream of the cRW9 epitope (Fig. 2). To test these hypotheses, we used site-directed mutagenesis to create mutant versions of SIVmac239 that either knocked out the cryptic start codon (SIV-A6865C) or prevented translation from Rev exon 1 into the intron, by the introduction of a stop codon (SIV-6860insA) or both (SIV-dblmut). We could not knock out the rev start codon directly as this would likely lead to a completely impotent virus.
Next, we synchronously infected MHC class I-matched CD4+ T cells from SIV-naïve macaques with either wild-type SIVmac239 or the mutant viruses. Using the KICS assay, we tested whether these cells could present the cRW9 epitope to a cRW9-specific CD8-TL clone. Surprisingly, we found that both singly mutated viruses were able to present the cRW9 epitope at 18 h postinfection. However, recognition of the SIV-6860insA virus-infected cells was approximately half of that of wild-type-infected cells, while recognition of the SIV-A6865C virus-infected cells was nearly identical to that of wild-type-infected cells. In contrast, recognition was totally ablated when cells were infected with the SIV-dblmut virus (Fig. 3B). As a positive control, we tested whether cells infected with the various viruses were capable of presenting the Env-FW9 epitope. An Env-FW9-specific T-cell line recognized all cells, indicating that normal viral protein production was not impacted by the introduced mutations (Fig. 3C). Together, these results imply that the cRW9 epitope is derived from two distinct translational products, Env ARF protein 1 (eARF-P1) and eARF-P2, the more frequent of which (eARF-P1) is produced when the ribosome initiates translation at the Rev start codon on the env mRNA and continues into the rev intron. The smaller product is translated when the ribosome initiates translation at the cryptic AUG codon within the intron (Fig. 3d).
FIG. 3.
cRW9 is produced as part of two distinct translation products. We created three mutant viruses to determine the start codon used to translate the cRW9 epitope (A). The first virus, SIV-6860insA (mutagenesis primers: 6860insA-F, GCATCAAACAAGTAAGTAATGGGATGTCTTGGGAATC, and 6860insA-R, GATTCCCAAGACATCCCATTACTTACTTGTTTGATGC), contains an inserted nucleotide which, when translated, introduced a stop codon in frame with cRW9, downstream of the rev splice donor site. This virus was used to test the hypothesis that translation of cRW9 occurred by translation from rev exon 1 into the intron. Another virus, SIV-A6865C (mutagenesis primers: A6865C-F, CAAGTAAGTATGGGCTGTCTTGGGAATCAG, and reverse, A6865C-R, CTGATTCCCAAGACAGCCCATACTTACTTG), mutated a potential methionine start codon in the rev intron to a leucine. This virus was used to test the hypothesis that translation of cRW9 occurred by initiation on this potential start codon. A third virus, SIV-dblmut (mutagenesis primers: dbl-F, GCATCAAACAAGTAAGTAATGGGCTGTCTTGGGAATC, and dbl-R, GATTCCCAAGACAGCCCATTACTTACTTGTTTGATGC), contained both of these mutations. It was used to test the hypothesis that both the rev start codon and the start codon within the rev intron were used to translate the cRW9 epitope. We then performed the KICS assay with CD4+ target cells infected with the different viruses (B and C). Data points represent the percentages of cells that are positive for gamma interferon (INF-γ) and/or tumor necrosis factor alpha (TNF-α) at each time point. SIV-6860insA and SIV-A6865C maintained the ability to present cRW9, but SIV-dblmut was not able to present the epitope at any time point tested. These results indicate that cRW9 is likely translated as part of two products, eARF-P1 and -P2 (D). The results depicted are characteristic of two separate assays. WT, wild type; Mut, mutant; pi, postinfection.
These results strongly suggest that the env mRNA is polycistronic and encodes the Env protein and both eARF-P1 and eARF-P2. To test this hypothesis more directly, we used overlap extension PCR (24) and in vitro transcription to create an env mRNA that contained all of the salient features of the env mRNA produced in infected cells. Specifically, this mRNA contained the same portion of the 5′ UTR present in natural env mRNA, spliced to the coding region in precisely the same way that occurs in the natural env mRNA. Though no particular function has been attributed to this noncoding RNA, it may play an important role in regulation of translation, and so we deemed it necessary to include it in our mRNA.
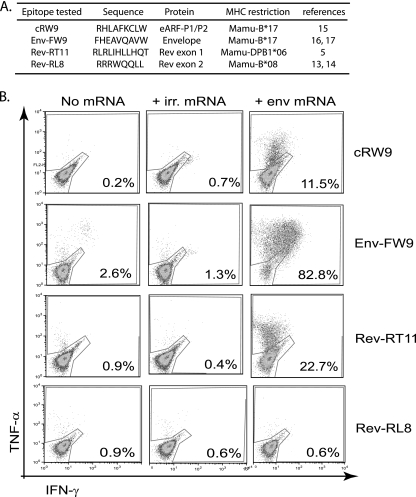
We next cultured dendritic cells (DCs) from monocytes (7). These cells were derived from SIV-naïve macaques that expressed (or did not express) the following MHC molecules: Mamu-B*17, Mamu-B*08, and Mamu-DPB1*06 (Fig. 4A). After 5 days in culture, we transfected these cells with the in vitro-transcribed env mRNA using nucleofection. Twelve hours after transfection, we tested whether epitope-specific T-cell lines or clones could recognize the transfected cells. Epitopes derived from all three sources were presented on cells transfected with the env mRNA (Fig. 4B). A cRW9-specific clone recognized the transfected cells (Fig. 4B), as did an Env-FW9-specific line (Fig. 4B) and a Rev-RT11-specific clone (Fig. 4B), verifying that this single mRNA was translated in both reading frames. As expected, the Rev-RL8 epitope, contained in exon 2, was not recognized (Fig. 4B). Together, these data verify that the env mRNA was the source of the eARF-P1/P2 products. We also tested whether cells transfected with an mRNA lacking the 5′ UTR were recognized by cRW9- and FW9-specific CD8-TL lines. Recognition of these cells was similar to those of cells transfected with mRNA with the UTR, albeit reduced slightly.
FIG. 4.
DCs transfected with the Env-encoding mRNA can present cRW9 as well as epitopes derived from Rev exon 1 and Env but not Rev exon 2. We synthesized the env mRNA in vitro using overlap extension PCR (outside primers, F_CGGAGAGGCTGGCAGATT and R_GGAGAAACCCAGCTGAAGA; overlapping internal primers, R_TCTCCGGAGGCCAACGTCCATCCGAACCCCTA and F_CCGGTTGCAGGTAGGCTTGGGGATATGTTATG; region of overlap is underlined). We cultured DCs from SIV-naïve animals that could present the following epitopes: cRW9 (restricted by Mamu-B*17 [15]), Env-FW9 (restricted by Mamu-B*17 [16, 17]), Rev-RT11 (in exon 1, restricted by Mamu-DPB1*06 [5]), and Rev-RL8 (in exon 2, restricted by Mamu-B*08 [13, 14]) as a negative control, as Rev exon 2 should not be translated from this mRNA (A). We transfected monocyte-derived DCs with the env mRNA and tested whether they were recognized by T-cell lines or clones specific for the epitopes in panel A. Recognition is measured as the percentage of epitope-specific T cells that produce gamma interferon (IFN-γ) and/or tumor necrosis factor alpha (TNF-α) upon recognition of transfected DCs, 12 hours after transfection. Cells either mock transfected (no mRNA) or transfected with irrelevant (irr.) mRNA (green fluorescent protein mRNA) were used as negative controls (B). The cRW9 epitope was translated from this mRNA, as were epitopes derived from Env and Rev exon 1, but not exon 2. The results are characteristic of two distinct assays.
Our data suggest that inefficient ribosomal scanning along the env mRNA might be responsible for translation of eARF-P1/P2. In fact, the rev start codon is a suboptimal Kozak start site (rev start is AUAUGUUAUGA; Kozak consensus is GCC[A/G]CCAUGG [10-12]), which might facilitate ribosomal scanning past the rev start codon to enhance Env translation. These data also provide a scaffold upon which a more-directed search for T-cell epitopes derived from ARFs can be conducted.
Our data do not address the possible functions (or lack thereof) of the eARF-P1/P2 products. They may represent novel viral proteins, or they may be defective ribosomal products without function (25-27). Thus far, attempts to create antibodies against these products have been unsuccessful, likely due to the abundance of hydrophobic amino acids. The SIV-dblmut virus (which knocks out production of these products) replicated as well as did the wild type in vitro (data not shown), indicating that these products are dispensable for in vitro replication. However, it is not known whether this virus can replicate in vivo. Importantly, our data also demonstrate that T-cell epitopes contained within functional proteins can be derived from novel, and possibly aberrant, translation of only a portion of the total protein and from the “wrong” mRNA. Finally, they show that the details of viral gene expression can have unexpected immunological consequences. Understanding these details might enhance our understanding of the total AIDS virus-specific cellular immune response.
Acknowledgments
We are grateful to S. Lank for sequencing, A. T. Bean for help with cell culture, and E. A. Scheef for continued support. Interleukin 2 used in T-cell culture was kindly provided by the NIH AIDS Reference and Reagent Program (Germantown, MD).
This work was supported by NIH grants R01 AI052056, R01 AI049120, and R24 RR015371 to D.I.W. and grant number P51 RR000167 from the National Center for Research Resources (NCRR), a component of the NIH, to the Wisconsin National Primate Research Center, University of Wisconsin—Madison. This research was conducted in part at a facility constructed with support from Research Facilities Improvement Program grant numbers RR15459 01 and RR020141-01.
This publication's contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
Footnotes
Published ahead of print on 15 July 2009.
REFERENCES
- 1.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cardinaud, S., A. Moris, M. Fevrier, P. S. Rohrlich, L. Weiss, P. Langlade-Demoyen, F. A. Lemonnier, O. Schwartz, and A. Habel. 2004. Identification of cryptic MHC I-restricted epitopes encoded by HIV-1 alternative reading frames. J. Exp. Med. 199:1053-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedrich, T. C., L. E. Valentine, L. J. Yant, E. G. Rakasz, S. M. Piaskowski, J. R. Furlott, K. L. Weisgrau, B. Burwitz, G. E. May, E. J. Leon, T. Soma, G. Napoe, S. V. R. Capuano, N. A. Wilson, and D. I. Watkins. 2007. Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication. J. Virol. 81:3465-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garrison, K. E., R. B. Jones, D. A. Meiklejohn, N. Anwar, L. C. Ndhlovu, J. M. Chapman, A. L. Erickson, A. Agrawal, G. Spotts, F. M. Hecht, S. Rakoff-Nahoum, J. Lenz, M. A. Ostrowski, and D. F. Nixon. 2007. T cell responses to human endogenous retroviruses in HIV-1 infection. PLoS Pathog. 3:e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giraldo-Vela, J. P., R. Rudersdorf, C. Chung, Y. Qi, L. T. Wallace, B. Bimber, G. J. Borchardt, D. L. Fisk, C. E. Glidden, J. T. Loffredo, S. M. Piaskowski, J. R. Furlott, J. P. Morales-Martinez, N. A. Wilson, W. M. Rehrauer, J. D. Lifson, M. Carrington, and D. I. Watkins. 2008. The major histocompatibility complex class II alleles Mamu-DRB1*1003 and -DRB1*0306 are enriched in a cohort of simian immunodeficiency virus-infected rhesus macaque elite controllers. J. Virol. 82:859-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hickman, H. D., A. D. Luis, W. Bardet, R. Buchli, C. L. Battson, M. H. Shearer, K. W. Jackson, R. C. Kennedy, and W. H. Hildebrand. 2003. Cutting edge: class I presentation of host peptides following HIV infection. J. Immunol. 171:22-26. [DOI] [PubMed] [Google Scholar]
- 7.Ignatius, R., M. Marovich, E. Mehlhop, L. Villamide, K. Mahnke, W. I. Cox, F. Isdell, S. S. Frankel, J. R. Mascola, R. M. Steinman, and M. Pope. 2000. Canarypox virus-induced maturation of dendritic cells is mediated by apoptotic cell death and tumor necrosis factor alpha secretion. J. Virol. 74:11329-11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin, X., D. E. Bauer, S. E. Tuttleton, S. Lewin, A. Gettie, J. Blanchard, C. E. Irwin, J. T. Safrit, J. Mittler, L. Weinberger, L. G. Kostrikis, L. Zhang, A. S. Perelson, and D. D. Ho. 1999. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189:991-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khan, S., R. de Giuli, G. Schmidtke, M. Bruns, M. Buchmeier, M. van den Broek, and M. Groettrup. 2001. Cutting edge: neosynthesis is required for the presentation of a T cell epitope from a long-lived viral protein. J. Immunol. 167:4801-4804. [DOI] [PubMed] [Google Scholar]
- 10.Kozak, M. 1986. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl. Acad. Sci. USA 83:2850-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozak, M. 1987. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Biol. 196:947-950. [DOI] [PubMed] [Google Scholar]
- 12.Kozak, M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234:187-208. [DOI] [PubMed] [Google Scholar]
- 13.Loffredo, J. T., A. T. Bean, D. R. Beal, E. J. Leon, G. E. May, S. M. Piaskowski, J. R. Furlott, J. Reed, S. K. Musani, E. G. Rakasz, T. C. Friedrich, N. A. Wilson, D. B. Allison, and D. I. Watkins. 2008. Patterns of CD8+ immunodominance may influence the ability of Mamu-B*08-positive macaques to naturally control simian immunodeficiency virus SIVmac239 replication. J. Virol. 82:1723-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loffredo, J. T., T. C. Friedrich, E. J. Leon, J. J. Stephany, D. S. Rodrigues, S. P. Spencer, A. T. Bean, D. R. Beal, B. J. Burwitz, R. A. Rudersdorf, L. T. Wallace, S. M. Piaskowski, G. E. May, J. Sidney, E. Gostick, N. A. Wilson, D. A. Price, E. G. Kallas, H. Piontkivska, A. L. Hughes, A. Sette, and D. I. Watkins. 2007. CD8+ T cells from SIV elite controller macaques recognize Mamu-B*08-bound epitopes and select for widespread viral variation. PLoS One 2:e1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maness, N. J., L. E. Valentine, G. E. May, J. Reed, S. M. Piaskowski, T. Soma, J. Furlott, E. G. Rakasz, T. C. Friedrich, D. A. Price, E. Gostick, A. L. Hughes, J. Sidney, A. Sette, N. A. Wilson, and D. I. Watkins. 2007. AIDS virus specific CD8+ T lymphocytes against an immunodominant cryptic epitope select for viral escape. J. Exp. Med. 204:2505-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maness, N. J., L. J. Yant, C. Chung, J. T. Loffredo, T. C. Friedrich, S. M. Piaskowski, J. Furlott, G. E. May, T. Soma, E. J. Leon, N. A. Wilson, H. Piontkivska, A. L. Hughes, J. Sidney, A. Sette, and D. I. Watkins. 2008. Comprehensive immunological evaluation reveals surprisingly few differences between elite controller and progressor Mamu-B*17-positive simian immunodeficiency virus-infected rhesus macaques. J. Virol. 82:5245-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mothe, B. R., J. Sidney, J. L. Dzuris, M. E. Liebl, S. Fuenger, D. I. Watkins, and A. Sette. 2002. Characterization of the peptide-binding specificity of Mamu-B*17 and identification of Mamu-B*17-restricted epitopes derived from simian immunodeficiency virus proteins. J. Immunol. 169:210-219. [DOI] [PubMed] [Google Scholar]
- 18.Park, I. W., R. Steen, and Y. Li. 1991. Characterization of multiple mRNA species of simian immunodeficiency virus from macaques in a CD4+ lymphoid cell line. J. Virol. 65:2987-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sacha, J. B., C. Chung, E. G. Rakasz, S. P. Spencer, A. K. Jonas, A. T. Bean, W. Lee, B. J. Burwitz, J. J. Stephany, J. T. Loffredo, D. B. Allison, S. Adnan, A. Hoji, N. A. Wilson, T. C. Friedrich, J. D. Lifson, O. O. Yang, and D. I. Watkins. 2007. Gag-specific CD8+ T lymphocytes recognize infected cells before AIDS-virus integration and viral protein expression. J. Immunol. 178:2746-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sacha, J. B., C. Chung, J. Reed, A. K. Jonas, A. T. Bean, S. P. Spencer, W. Lee, L. Vojnov, R. Rudersdorf, T. C. Friedrich, N. A. Wilson, J. D. Lifson, and D. I. Watkins. 2007. Pol-specific CD8+ T cells recognize simian immunodeficiency virus-infected cells prior to Nef-mediated major histocompatibility complex class I downregulation. J. Virol. 81:11703-11712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sacha, J. B., M. R. Reynolds, M. B. Buechler, C. Chung, A. K. Jonas, L. T. Wallace, A. M. Weiler, W. Lee, S. M. Piaskowski, T. Soma, T. C. Friedrich, N. A. Wilson, and D. I. Watkins. 2008. Differential antigen presentation kinetics of CD8+ T-cell epitopes derived from the same viral protein. J. Virol. 82:9293-9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 23.Unger, R. E., M. W. Stout, and P. A. Luciw. 1991. Simian immunodeficiency virus (SIVmac) exhibits complex splicing for tat, rev, and env mRNA. Virology 182:177-185. [DOI] [PubMed] [Google Scholar]
- 24.Warrens, A. N., M. D. Jones, and R. I. Lechler. 1997. Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene 186:29-35. [DOI] [PubMed] [Google Scholar]
- 25.Yewdell, J., L. C. Anton, I. Bacik, U. Schubert, H. L. Snyder, and J. R. Bennink. 1999. Generating MHC class I ligands from viral gene products. Immunol. Rev. 172:97-108. [DOI] [PubMed] [Google Scholar]
- 26.Yewdell, J. W., L. C. Anton, and J. R. Bennink. 1996. Defective ribosomal products (DRiPs): a major source of antigenic peptides for MHC class I molecules? J. Immunol. 157:1823-1826. [PubMed] [Google Scholar]
- 27.Yewdell, J. W., and H. D. Hickman. 2007. New lane in the information highway: alternative reading frame peptides elicit T cells with potent antiretrovirus activity. J. Exp. Med. 204:2501-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]