Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) is etiologically linked to Kaposi's sarcoma, primary effusion lymphomas, and multicentric Castleman's disease. Like other herpesviruses, KSHV can exist in either a lytic or a latent phase during its life cycle. We report that the lytic protein encoded by KSHV open reading frame 64 (Orf64) is a viral deubiquitinase (DUB) enzyme capable of deubiquitinating cellular proteins in vitro and in vivo. Orf64 DUB activity is effective against lysine 48 (K48)- and lysine 63 (K63)-linked ubiquitin chains. Thus, KSHV Orf64 is a viral DUB that does not show specificity toward K48 or K63 ubiquitin linkages. Orf64 DUB activity lies within the first 205 residues of the protein, and deubiquitination is dependent on a cysteine at position 29, since mutation of this residue ablated this activity. Cell fractionation studies revealed that the N terminus and the full-length protein localized to both the nuclear and cytoplasmic compartments. The function of Orf64 was tested by short interfering RNA (siRNA) knockdown studies on latently infected cells that were induced into lytic replication. We found that depletion of Orf64 by siRNA resulted in decreased viral lytic transcription and lytic protein expression. These experiments indicate that Orf64 plays a role in KSHV lytic replication.
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8, is the most recently discovered human herpesvirus and the only human rhadinovirus (8). KSHV is the etiological agent of Kaposi's sarcoma (KS), as well as the lymphoproliferative disorders, primary effusion lymphoma (PEL), and multicentric Castleman's disease (MCD) (7, 12, 36). KSHV is consistently found in all clinical forms of KS, PEL, and MCD. Moreover, KS is the most common neoplasm in the human immunodeficiency virus-positive population and is an AIDS-defining illness. Although KS lesions most frequently develop on the external skin, they can also be found on internal organs and in the oral cavity. KS is a highly angiogenic complex lesion that comprises endothelial cell-derived spindle-shaped cells and inflammatory cells that have migrated to the lesion (26).
Like all herpesviruses, KSHV exhibits two distinct phases in its life cycle: a lytic phase and a latent phase. De novo infection of endothelial cells results in temporary lytic replication (11, 30). Lytic replication is characterized by the transcription of the entire complement of viral genes (immediate early, early, and late) in a temporal fashion. Viral replication and virion assembly are followed by the release of infectious progeny from the infected cell. In contrast, during latency, only a few viral proteins are expressed and the viral genome remains in an episomal state. Neither mode of existence is in itself capable of sustaining neoplastic growth, as each state seems to make distinct contributions that are necessary for the development of KSHV-associated malignancies (15). Thus, lytic and latent proteins modulate the environment of the host cell to the advantage of the virus. The role of ubiquitination in KSHV replication, transformation, and pathogenesis has been examined to some extent. During the lytic cycle, KSHV expresses three E3-like ubiquitin ligases (9, 20, 43) encoded by open reading frames K3 (MIR 1), K5 (MIR2), and RTA (Orf50). Both K3 and K5 target major histocompatibility complex class I molecules for degradation in order to hinder presentation of viral antigens. The lytic switch protein RTA targets IRF-7 for degradation in order to preclude induction of the interferon-mediated antiviral state (43).
Ubiquitin (Ub) is a 76-amino-acid molecule that is widely conserved and can be posttranslationally conjugated to specific target proteins through different linkages. The host cell utilizes Ub to control cellular processes such as protein expression, apoptosis, cell cycle regulation, receptor trafficking, DNA repair, and signal transduction (13, 33). Moreover, antigen presentation of pathogen-derived proteins requires Ub-mediated proteosomal processing (42). The Ub pathway consists of a single E1 protein that activates and transfers Ub to one of several conjugating enzymes, known as the E2 adaptor proteins, in an ATP-dependent fashion. The E2 protein subsequently engages an E3 ligase bound to a specific target protein. The Ub moiety is then covalently linked to the target protein through a lysine residue by the E3 Ub ligases (17).
Proteolysis, however, is not the only outcome of ubiquitination. Typically, polyubiquitination through the lysine 48 residue of Ub results in the degradation of the target protein, whereas mono- and diubiquitination through lysine 63 are associated with cellular transport or functional modulation of the target protein (reviewed in reference 19). It has also been demonstrated that polyubiquitination occurs through other lysine residues of Ub such as lysine 29, although these linkages do not appear to be frequently used (3). In addition, monoubiquitination has been shown to be involved in endocytosis, histone regulation, and retrovirus budding (reviewed in reference 18). Thus, Ub modification represents a regulatory mechanism for multiple functions in the cell.
Besides KSHV, other herpesviruses such as herpes simplex virus type 1 (HSV-1) and murine gammaherpesvirus 68 encode E3 Ub ligases (6, 38). In a parallel but opposing fashion, herpesviruses have also pirated deubiquitinase (DUB) enzymes. DUBs cleave Ub molecules from either the E3 ligase or the target protein, thereby preempting Ub-mediated regulation. DUBs have been identified in several herpesviruses (32). The UL36 multifunctional large tegument proteins encoded by HSV-1 (23), Marek's disease virus (21), and pseudorabies virus (PRV) (α subfamily) are DUBs, as is the homologous UL48 encoded by human cytomegalovirus (β subfamily) (39). BPLF-1, BSLF-1, and BXLF-1 of Epstein-Barr virus (EBV) (γ subfamily) (35, 41) are also DUBs.
Although many targets and effects of the herpesviral DUBs are not known, studies of HSV-1 mutants indicate that the HSV DUB plays a role in virion transport through microtubules (34), release of the viral genome into the nucleus (1, 22), tegumentation, and envelopment (10), as well as virion egress (27). DUB activity of HSV UL36 appears to be important, as a DUB-null mutant displayed a 3-log decrease in titers and a 50% reduction in egress of capsids from neurons (24). Null mutations for PRV UL36 are lethal in vitro (25) and delay neuroinvasion in the mouse model (5). Moreover, specifically mutating the DUB catalytic domain reduces titer and plaque size (4). Recently, it was shown that a DUB mutant of Marek's disease virus was severely impaired in its ability to induce lymphomagenesis (21).
We report the first description of a viral DUB, Orf64, encoded by KSHV open reading frame 64. Not much is known about KSHV Orf64 except that it has recently been identified as a lytic protein that is present in the tegument of the virion and appears to act as a scaffold protein during tegumentation (31). We found that KSHV Orf64 is a powerful DUB, as measured by in vitro deubiquitination assays. Additionally, KSHV Orf64 was also able to deubiquitinate cellular proteins when expressed in various cell types. The DUB activity was limited to the N-terminal domain of KSHV Orf64, and, unlike other herpesviral DUBs, which show specificity for K48 Ub linkages (2, 23, 29, 39), KSHV Orf64 appears to deubiquitinate both K48-linked Ub (K48-Ub) and K63-Ub chains. Furthermore, we found that short interfering RNA (siRNA) knockdown of KSHV Orf64 resulted in decreased reactivation from latency and decreased viral replication.
MATERIALS AND METHODS
Cell culture.
HeLa, CV-1, and HEK-293 cell lines were maintained in complete medium: Dulbecco's modified Eagle medium (DMEM) (Cellgro) with 10% fetal bovine serum, l-glutamine, and penicillin plus streptomycin. The rKSHV.219-Vero cells (37) (a kind gift from Jeffrey Vieira) were maintained in complete medium with 5 μg/ml puromycin. KSHV-infected latent HEK-293 cells (KSHV-293 cells) were established by overlaying supernatants from reactivated rKSHV.219-Vero cells on naive HEK-293 cells, followed by puromycin selection at 1 μg/ml. All cells were grown in a 37°C incubator with 5% CO2.
Plasmids.
The N terminus of Orf64 (Orf64N) was constructed by PCR amplification of the first 615 nucleotides of the Orf64 open reading frame with the coding sequence for a C-terminal Flag epitope. This sequence was cloned into the EcoRI and HindIII sites of the pcDEF3 vector with forward primer CGCCGCGAATTCGACATGGCAGCCCAGCCTCTGTACATGGAGGG and reverse primer CGCCGCAAGCTTTCACTTATCGTCGTCATCCTTGTAGTCGTAGTCATGTGGGATAAAGTAAAGGAAGC. The Orf64N fragment coding sequence was also subcloned into pGEX6p-1 (Amersham) to construct a glutathione S-transferase (GST) fusion of Orf64N (GST-Orf64N). Site-directed mutagenesis of Orf64N was conducted to change the cysteine residue at amino acid 29 to a glycine residue (C29G), generating the plasmids encoding Orf64N-C29G and GST-Orf64N-C29G. The coding sequence for the full-length Orf64 (Orf64FL) was PCR amplified and cloned into the pcDEF3 vector with the coding sequence for a C-terminal Flag epitope using the enzymes EcoRI and NotI with primers CGCCGCGAATTCGACATGGCAGCCCAGCCTCTGTACATGGAGGG and CGCGCGGCGGCCGCTCACTTATCGTCGTCATCCTTGTAGTCCAAGTACCACTTCTTTTAACTGTCAACGC, respectively. PCR products were digested with the appropriate enzymes and ligated into pcDEF3. Five microliters of the ligation reaction mixture was transformed into bacteria. The bacteria were grown at 30°C overnight on Luria-Bertani plates with 50 μg/ml ampicillin. Clones were screened by enzymatic digestion and sequencing.
GST-Orf64N protein purification.
BL21(DE3) cells were transformed with plasmid pGEX-6p-1 (control vector), pGEX-6p-Orf64N, or pGEX-6p-Orf64N-C29G. The bacteria were then grown in 2XYT broth (16 g/liter tryptone, 10 g/liter yeast extract, 5 g/liter NaCl) with 50 μg/ml ampicillin and 34 μg/ml chloramphenicol to an optical density at 600 nm of 0.6 in accordance with the manufacturer's directions and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 hours at room temperature. Cells were spun down and resuspended in lysis buffer (200 mM NaCl, 50 mM Tris-HCl, 0.5 mM EDTA, 5% glycerol, 0.5% NP-40 plus Complete protease inhibitor cocktail [Roche]), followed by sonication. Triton X-100 was added to the lysates to a final concentration of 1%, and the samples were rocked for 30 min, followed by centrifugation at 13,000 rpm for 10 min. Supernatants were transferred to new tubes and incubated with a 50% slurry of glutathione-Sepharose beads in 1× phosphate-buffered saline (PBS) for 1 hour. The GST-tagged proteins were washed three times with 1× PBS and used for in vitro deubiquitination assays.
In vitro deubiquitination assays.
In vitro deubiquitination assays were performed in DUB assay buffer containing 50 mM HEPES, 0.01% Brij-35, and 3 mM dithiothreitol (40). Two and one-half micrograms of synthetic lysine 48 (K48)- or lysine 63-linked (K63) Ub chains (Biomol) was added to purified GST-Orf64N or 40 ng of purified isopeptidase T (IsoT; Biomol) (positive control) at 37°C for 3 to 4 h. The sample was subsequently loaded onto a 10% sodium dodecyl sulfate (SDS) denaturing gel, electrophoresed, and transferred to nitrocellulose. Western blotting was performed with an anti-Ub antibody (Sigma). For the in vitro fluorescence deubiquitination assays, 500 nM of the Ub-C-terminal 7-amido-4-methylcoumarin (Ub-AMC) substrate (Biomol) was added to ∼4 μM or 200 nM GST-Orf64N and GST-Orf64N-C29G purified protein in AMC buffer containing 50 mM HEPES, 0.5 mM EDTA, 1 mM dithiothreitol, and 0.1 mg/ml bovine serum albumin. In addition, 200 nM purified proteins was incubated with 1.2 μM Ub-AMC. Some of the samples were then pretreated or mock treated with 1 microgram of the suicide inhibitor Ub aldehyde (Ub-Al) (Santa Cruz). After incubation at room temperature, fluorescence (excitation at 380 nm, emission at 460 nm) was measured using the Fluostar machine (BMG).
In vivo deubiquitination assay.
Deubiquitination assays with transfected cells were also performed. HeLa, HEK-293, CV-1, and rKSHV.219-Vero cells were transfected with 2 μg wild-type Ub (WT-Ub), K48-Ub, or K63-Ub expression plasmids (kind gift from Vishva Dixit [44]) and 8 μg pcDEF3-Orf64N, pcDEF3-Orf64FL, or empty vector pcDEF3. Forty-eight hours posttransfection, cells were lysed with NP-40 lysis buffer and equal amounts of protein lysate were loaded into 10% SDS-polyacrylamide gel electrophoresis (PAGE) gels for Western blot analysis with antihemagglutinin (anti-HA)-horseradish peroxidase (HRP) antibody.
Cell fractionation assay.
HEK-293 cells in 100-mm plates were transfected with pcDEF3-Orf64N or empty vector pcDEF3. Cells were harvested 48 h later by trypsinization for 5 min at 37°C. Complete medium was added to neutralize trypsin, followed by washing twice with cold PBS. The plasma membrane was disrupted with 500 μl cytosolic lysis buffer [5 mM PIPES {piperazine-N,N′-bis(2-ethanesulfonic acid)}, 85 mM KCl, and 0.5% NP-40 with Complete (Roche) protease inhibitors] and incubated on ice for 5 min. Nuclei were spun down, and the cytosolic fraction was transferred to a fresh tube. The nuclei were washed three times with 1× PBS and then lysed in 250 μl radioimmunoprecipitation assay buffer (150 mM NaCl, 1% NP-40, 50 mM Tris [pH 6.8], 0.5% sodium deoxycholate, 0.1% SDS with complete protease inhibitors [Roche]) and freeze-thawed three times. Cell debris was spun down, and the supernatants were quantitated by the Bradford assay (Bio-Rad).
Western blot analysis.
Cells in 100-mm dishes were transfected using Superfect reagent (Qiagen). Cells were harvested 48 h posttransfection, washed twice with PBS, and lysed with 200 μl NP-40 buffer (150 mM NaCl, 1% NP-40, 50 mM Tris [pH 6.8] with Complete [Roche] protease inhibitors). Cell debris was spun down, and the supernatant was quantitated by the Bradford assay (Bio-Rad). Cell lysates were incubated with SDS gel loading buffer at 65°C for 5 min and subjected to SDS-PAGE. Electrophoresed gels were transferred to nitrocellulose membranes (Hybond; Amersham) by wet transfer (20 V for 16 h) and blocked in 5% nonfat dry milk (NFDM), 1% Tween 20, and Tris-buffered saline (TBSt) for 30 min at room temperature. Flag-HRP antibody (Bethyl) was used at 1:10,000 dilution in 3% NFDM-TBSt. HA-HRP antibody (Sigma) was used at 1:10,000 dilution in 1% bovine serum albumin in TBSt. Goat antiactin antibody (Cell Signaling) was used as a control at 1:1,000 dilution in 5% NFDM, and the secondary peroxidase-conjugated anti-goat antibody (Dako) was diluted to 1:2,000 in 5% NFDM. Rabbit anti-Ub (Sigma) was diluted to 1:250 and secondary anti-rabbit peroxidase antibody (Amersham) was diluted to 1:10,000, both in 5% NFDM. GRP-78 antibody (Santa Cruz) was used at 1:500 dilution in 5% NFDM. Antitubulin (Sigma) was diluted to 1:10,000 in NFDM, and the anti-mouse peroxidase-conjugated secondary antibody (Cell Signaling) was used at 1:2,000 dilution also in NFDM. Membranes were washed for 30 min with TBSt, and Western blots were incubated in SuperSignal chemiluminescent substrate (Pierce).
Viral reactivation/replication assay.
KSHV-293 cells in 100-mm plates were either mock induced with 1:1 serum-free DMEM-SF9 medium or induced with 1:1 serum-free DMEM-baculovirus encoding KSHV RTA (BacRTA) in SF9 medium for 1 hour at 37°C. The inoculum was replaced with complete medium in the mock-induced set or complete medium with 1.7 mM sodium butyrate in the induced set. Twenty-four hours postinduction, cells were transfected with siRNAs against luciferase (LUC) or siRNAs against Orf64 in duplicate. Both sets were incubated for 48 h posttransfection, at which point the cells were harvested. The cell sample was split into two fractions to isolate both RNA and protein. Total cell RNA extractions were performed with the RNeasy Plus kit (Qiagen) according to the manufacturer's instructions, followed by RNase-free DNase treatment at 37°C for 1 h. An additional step to remove enzyme and digested DNA was performed with the RNeasy miniprep kit (Qiagen). One microgram total RNA was used in a reverse transcription (RT) reaction (Promega). Each reaction volume was brought up to 120 μl and used as template in PCRs. For cell extracts, cells were lysed with NP-40 lysis buffer as described above and Western blotting was performed as described above with anti-viral interleukin-6 (anti-vIL6) antibody (ABI).
RESULTS
Clustal alignment of KSHV Orf64 homologs in other herpesviruses.
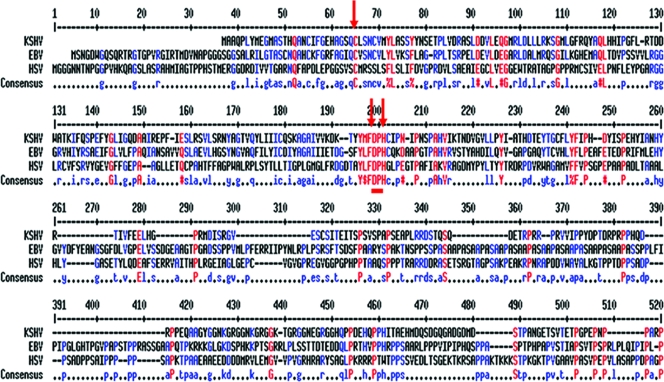
HSV-1 UL36 and EBV BPLF-1 were aligned with KSHV Orf64 using the Clustal W sequence alignment tool. Figure 1 shows the first 500 amino acid residues of these homologs, which display low degrees of identity and similarity. Notably, several short, well-conserved motifs emerge. The cysteine, histidine, and aspartic acid residues in HSV-1 UL36 (amino acid positions 66, 197, and 199) and KSHV (amino acid positions 29, 159, and 161) Orf64 are conserved. These three amino acids represent a catalytic triad that constitutes the catalytic core for DUB activity (2). These residues are conserved in other human herpesvirus homologs as well (23), thus providing strong indication that these residues constitute the catalytic core of the DUB.
FIG. 1.
Sequence analysis predicts a DUB in KSHV Orf64. Clustal W alignment of the amino-terminal 500 amino acid residues of KSHV Orf64 and homologs from EBV (BPLF1) and HSV-1 (UL36) reveal key conserved amino acids, shown in red. Red arrows indicate components of the putative catalytic core of the cysteine protease.
Orf64 protein expression and intracellular localization.
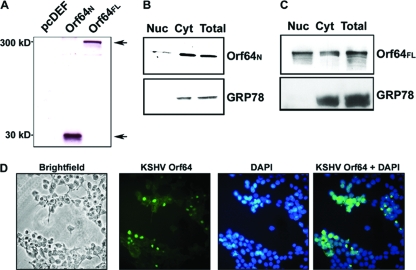
Orf64FL and Orf64N, containing the first 205 amino acids, were each tagged with the Flag epitope, and their coding sequences were cloned into the pcDEF3 (14) mammalian expression vector. In order to determine the molecular sizes of these proteins, the individual plasmids were transfected into HEK-293 cells. Forty-eight hours posttransfection, cells were harvested, lysed, and subjected to Western blot analysis using a Flag-HRP antibody. As depicted in Fig. 2A, the N terminus of Orf64 ran at a mobility of 30 kDa while Orf64FL ran at a much higher mobility of approximately 300 kDa. The predicted size for the full-length, 2,636-amino-acid protein is 279 kDa, which closely correlates with the observed size.
FIG. 2.
Expression and intracellular localization of Orf64. (A) HEK-293 cells were transiently transfected with expression vectors for Flag-tagged Orf64N and Orf64FL. Cells were harvested, and the lysates were subjected to Western blot analysis with an anti-Flag antibody. Orf64N displayed a mobility of ∼30 kDa, and Orf64FL displayed a mobility of ∼300 kDa. (B) Transiently transfected cells expressing Orf64N were subjected to cellular fractionation. The N terminus of Orf64 was present in both the cytoplasm (Cyt) and nucleus (Nuc). Expression of the cytoplasmic marker GRP-78 (bottom) served as a control for the fractionation. (C) Transiently transfected cells expressing Orf64FL were subjected to cellular fractionation. Orf64FL is also detected in both cytoplasmic and nuclear compartments. Expression of the cytoplasmic marker GRP-78 (bottom) served as a control for the fractionation. (D) 293T cells in glass-bottom plates were transiently transfected with a Flag-tagged Orf64FL expression plasmid. Forty-eight hours later, cells were fixed in an acetone-methanol (1:1) mixture. Cells were stained with fluorescein isothiocyanate-conjugated Flag antibody and DAPI nuclear stain. Images were taken on a Zeiss Axiovert 200 inverted microscope using Openlab software. All images shown are at 20×.
To determine the cellular localization of KSHV Orf64, we transfected the pcDEF3-Orf64N and pcDEF3-Orf64FL expression plasmids into HEK-293 cells. Forty-eight hours posttransfection, cells were lysed and either separated into cytosolic and nuclear fractions or harvested as whole-cell lysates. As shown in Fig. 2B and C, the Flag-tagged Orf64N and Orf64FL were detected in both nuclear and cytosolic fractions. The integrity of the cytosol-nuclear separation was confirmed by blotting for the endoplasmic reticulum cytoplasmic protein GRP-78. The signal for GRP-78 was detectable only in the cytoplasmic fraction and the whole-cell lysate, validating the integrity of the fractionation. These results suggest that both Orf64N and Orf64FL may be found in the cytoplasm and nucleus of the cell. Immunofluorescence assays were also performed in 293T cells transiently expressing Orf64FL. KSHV Orf64 was detected with a fluorescein isothiocyanate-conjugated antibody, and the cells were stained with DAPI (4′,6-diamidino-2-phenylindole) to outline the nucleus (Fig. 2D). In agreement with our cell fractionation results, Orf64 is present in both the cytoplasm and the nucleus of the cell. There is an accumulation of Orf64 in the nuclear and perinuclear areas, which reflects the known functions of Orf64 homologs in virion assembly and egress (Fig. 2D). These results also show that Orf64 can locate to the nucleus in the absence of any other viral protein.
In vitro deubiquitination assays.
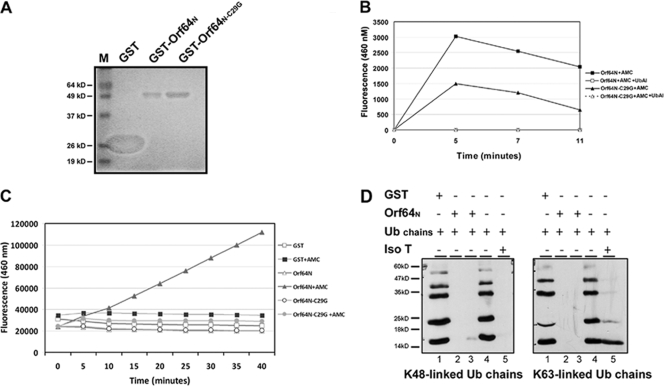
As shown in Fig. 1, KSHV Orf64 appears to contain a conserved DUB domain. In order to determine whether the predicted catalytic core of the Orf64 DUB was functional, we subcloned the Orf64N fragment into pGEX6p-1 to fuse it to GST and express it in vitro. GST-Orf64N was expressed in BL21(DE3) bacteria and purified using a GST column to high purity (Fig. 3A). GST-Orf64N was subsequently used in two different in vitro DUB assays.
FIG. 3.
The N terminus of Orf64 has deubiquitinating activity for both K48- and K63-linked Ub chains in vitro. (A) GST-Orf64N and GST-Orf64N-C29G were expressed in BL21(DE3) cells with the control GST protein (lane GST). After purification, the proteins were electrophoresed by SDS-PAGE and the gel was stained with Coomassie blue dye. (B) Purified GST-Orf64N (4 μM) was incubated with 2 μM of a wild-type Ub fluorogenic substrate, AMC-Ub. Fluorescence generated from the cleaved AMC-Ub substrate was recorded over time at 460 nm. Data from reactions including AMC-Ub with purified GST-Orf64N-C29G, GST-Orf64N plus Ub-Al, or Orf64N-C29G plus Ub-Al are also shown. (C) AMC-Ub reaction mixtures containing 200 nM of the indicated GST-purified proteins and 1.2 μM AMC-Ub substrate were processed as described for panel B. (D) Purified GST-Orf64N was used in an in vitro DUB assay with Ub chains. Lysine 48 (K48)- and lysine 63 (K63)-linked chains are not cleaved by GST (lanes 1) or in the absence of GST-Orf64N (lanes 4). Ub chains containing K48- and K63-linked Ub moieties were completely cleaved by the positive control, IsoT (lanes 5), as well as in the presence of Orf64N (lanes 2 and 3).
The first assay used a WT-Ub substrate. This assay involves the cleavage of the Ub-AMC substrate by a DUB to free a fluorophore capable of excitation and emission at 460 nm. For this assay, wild-type GST-Orf64N or a mutant protein in which the cysteine at position 29 was mutated to a glycine (GST-Orf64N-C29G) was incubated alone, in the presence of the Ub-AMC substrate, or with substrate and a DUB suicide inhibitor, Ub-Al. The samples were then assayed for fluorescence at a wavelength of 460 nm (Fig. 3B). A graph of the fluorescence values obtained at different time points after treatment is shown in Fig. 3B. There is a significant increase in fluorescence in the sample containing GST-Orf64N and Ub-AMC substrate compared to that in the sample containing GST-Orf64N without substrate, GST-Orf64N-C29G with Ub-AMC, or GST-Orf64N/GST-Orf64N-C29G with Ub-AMC substrate and the Ub-Al suicide inhibitor (Fig. 3B). We also performed the same experiment with less protein. Two hundred nanomolar purified GST-Orf64N or GST-Orf64N-C29G was incubated with 1.2 μM Ub-AMC in buffer, and fluorescence was measured at different time points (Fig. 3C). Altogether, these data demonstrate that the purified N terminus of Orf64 is able to cleave synthetic Ub chains in vitro in the absence of other cellular proteins and that the cysteine residue is critical for this activity.
To confirm these findings, a second type of in vitro DUB assay was performed. We incubated synthetic Ub chains, K48-Ub or K63-Ub, with either GST alone or the GST-Orf64N fusion protein (Fig. 3D). The bona fide DUB IsoT (16) was also used as a positive control in this assay. The samples were run on SDS-PAGE gel, and Western blotting with an anti-Ub antibody was performed. Lanes with Ub chains alone or Ub chains plus GST protein displayed a laddered pattern corresponding to Ub chains containing different numbers of Ub linkages (Fig. 3D). The previously characterized DUB, IsoT, cleaved all the chains (either K48 or K63 linkages). Likewise, in the presence of the GST-Orf64N fusion protein, the K48- and K63-Ub chains were also completely cleaved (Fig. 3D). These results indicate that the N terminus of Orf64 has DUB activity and can cleave both types of Ub linkages at least as effectively as the IsoT positive control.
In vivo deubiquitination assays.
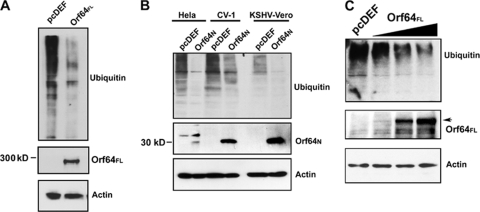
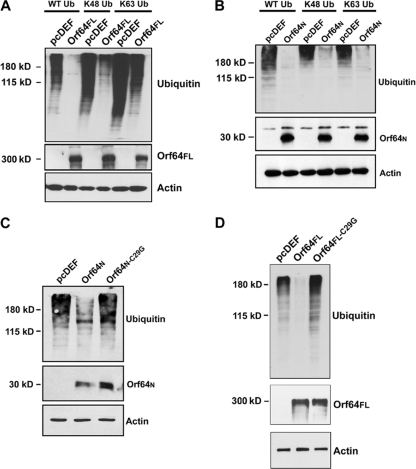
We next determined whether Orf64FL functioned as a DUB in a cell-based assay. HEK-293 cells (2 × 106) were plated on 100-mm dishes and cotransfected with a plasmid encoding a HA-tagged Ub named pcDNA3.1-3XHA-Ub (expressing WT-Ub) (44), the pcDEF3 empty vector, or the pcDEF3-Orf64FL plasmid. Cells were harvested 48 h posttransfection and subjected to Western blot analysis with an anti-HA antibody to detect all ubiquitinated cellular proteins. When pcDEF3-Orf64FL was cotransfected with the WT-Ub expression plasmid, there was very little accumulation of ubiquitinated cellular proteins compared to the control, suggesting that KSHV Orf64FL can cleave Ub moieties off cellular proteins (Fig. 4A). We also performed identical assays with Orf64N in HeLa, CV-1, and the KSHV-positive rKSHV.219-Vero cells and found that Orf64N retained the deubiquitinating activity of Orf64 in all these cell lines, i.e., in the presence or absence of the viral genome (Fig. 4B).
FIG. 4.
Orf64 exerts DUB activity in vivo. (A) Cotransfection of pcDEF3-Orf64FL and HA-tagged Ub expression plasmids in HEK-293 cells. Cells were harvested 48 h posttransfection and subjected to Western blot analysis with an anti-HA antibody. Expression of Orf64FL results in decreased levels of high-molecular-weight ubiquitinated cellular proteins compared to levels seen with the empty vector (pcDEF) control. Western blots for actin show equal loading of protein lysates, and Western blots with anti-Flag antibody show expression of Orf64FL. (B) Cotransfection of pcDEF3-Orf64N and HA-tagged Ub expression plasmids in HeLa, CV-1, and KSHV-positive rKSHV.219-Vero cells (KSHV-Vero). Cells were harvested 48 h posttransfection and subjected to Western blot analysis with anti-HA antibody. Expression of Orf64N results in decreased levels of high-molecular-weight ubiquitinated proteins compared to levels for transfection with the empty vector (pcDEF) control. Thus, the DUB activity is contained within the N-terminal 205 residues of Orf64. Western blots for actin show equal loading of protein lysates, and Western blots with an anti-Flag antibody show expression of Orf64N. (C) Dose-response experiment. 293T cells were transfected with 15 μg of empty vector (lane 1 [left]) or increasing amounts (5, 10, and 15 μg) of Orf64 expression plasmid (lanes 2, 3, and 4, respectively). Cells were harvested 48 hours posttransfection, and lysates were subjected to Western blot analysis with anti-Ub, anti-GAPDH, and antiactin antibodies. There is a decrease in levels of cellular ubiquitinated proteins in the presence of increasing amounts of Orf64FL.
In order to test the ability of different amounts of Orf64 to deubiquitinate cellular proteins, we performed a dose-response experiment by transfecting the empty vector or increasing amounts of Orf64 expression plasmid into 293T cells (Fig. 4C). We found that the degree of deubiquitination depends directly on the amount of transfected Orf64 expression plasmid. These data strongly suggest that Orf64 is directly responsible for the decrease in ubiquitinated cellular proteins.
In order to identify which Ub linkage type is targeted by Orf64 in vivo, we cotransfected HEK-293 cells with pcDEF3-Orf64FL or pcDEF3 and a plasmid encoding HA-tagged WT-Ub, a protein (K48-Ub) in which all lysines in Ub were mutated except for lysine 48, or a protein (K63-Ub) in which only lysine 63 was left intact. The last two constructs produce Ub chains that can be polymerized only through their K48 or K63 lysine residues, respectively. Cells were harvested 48 hours posttransfection and subjected to Western blot analysis with an anti-HA antibody to detect the presence of ubiquitinated proteins in the cell. We found that lysates from cells transfected with the different Ub expression constructs and empty vector (pcDEF) showed accumulation of many ubiquitinated cellular proteins. However, Orf64FL exerted a significant deubiquitination effect on cellular proteins (Fig. 5A) and displayed a DUB preference (but not specificity) for K48-mediated Ub linkages. KSHV Orf64FL was able to deubiquitinate most of the K48-linked ubiquitinated proteins and a significant proportion of the K63-linked ubiquitinated proteins (Fig. 5A). These results do not exclude the possibility that Orf64 may also cleave other, less common types of Ub linkages.
FIG. 5.
Orf64 can effectively cleave both K48 and K63 Ub linkages. (A) pcDEF3 or pcDEF3-Orf64FL expression plasmids were cotransfected with expression plasmids for WT-Ub, K48-Ub, or K63-Ub into HEK-293 cells. Cells were harvested 48 h posttransfection and subjected to Western blot analysis with an anti-HA antibody. Orf64FL was able to deubiquitinate WT-Ub and K48- and K63-linked ubiquitinated cellular proteins. Actin blots show equal loading of protein lysates, and a Western blot with an anti-Flag antibody shows expression of Orf64FL. (B) pcDEF3 or pcDEF3-Orf64N expression plasmids were cotransfected with expression plasmids for WT-Ub, K48-Ub, or K63-Ub into HEK-293 cells. Cells were harvested 48 h posttransfection and subjected to Western blot analysis with an anti-HA antibody. Orf64N was able to deubiquitinate WT-Ub and K48- and K63-linked ubiquitinated cellular proteins. Western blots for actin show equal loading of protein lysates, and a Western blot with an anti-Flag antibody shows expression of Orf64N. (C) Cotransfection of pcDEF3, pcDEF3-Orf64N, and pcDEF3-Orf64N-C29G with WT-Ub expression plasmids in HEK-293 cells. Cells were harvested 48 h posttransfection and subjected to Western blot analysis with an anti-HA antibody. Orf64N can deubiquitinate cellular proteins, but the DUB activity of Orf64N is ablated by the C29G mutation, suggesting that this cysteine is essential for deubiquitination mediated by Orf64. Western blots for actin show equal loading of protein lysates, and a Western blot with an anti-Flag antibody shows expression of both Orf64N and Orf64N-C29G. (D) Cotransfection of pcDEF3, pcDEF3-Orf64FL, and pcDEF3- Orf64FL-C29G with WT-Ub expression plasmids in HEK-293 cells. Cells were harvested 48 h posttransfection and subjected to Western blot analysis with an anti-HA antibody. Orf64FL can deubiquitinate cellular proteins, but the DUB activity of Orf64FL is ablated by the C29G mutation, suggesting that this cysteine is essential for deubiquitination mediated by Orf64. Western blots for actin show equal loading of protein lysates, and a Western blot with an anti-Flag antibody shows expression of both Orf64FL and Orf64FL-C29G.
Next, we determined whether Orf64N displayed the same linkage type specificity in vivo. Notably, both K48- and K63-linked Ub chains were completely cleaved by Orf64N with no apparent preference in the HEK-293 transfected cells (Fig. 5B). We also transfected HEK-293 cells with plasmids expressing WT-Ub and either the wild-type Orf64N or a mutant N-terminal Orf64 (Orf64N-C29G). Cell lysates were harvested 48 h posttransfection and subjected to SDS-PAGE analysis followed by Western blotting with an anti-Ub antibody to detect ubiquitinated proteins. We found that wild-type Orf64N displayed DUB activity against cellular ubiquitinated proteins but that the mutant Orf64N-C29G showed a loss of DUB activity compared to the wild-type Orf64N (Fig. 5C). Finally, we also transfected HEK-293 cells with plasmids expressing WT-Ub and either the wild-type Orf64FL or a mutant full-length Orf64 (Orf64FL-C29G). Cell lysates were harvested 48 h posttransfection and subjected to Western blotting with an anti-Ub antibody to detect ubiquitinated proteins. We found that wild-type Orf64FL displayed DUB activity against cellular ubiquitinated proteins but that the mutant Orf64FL-C29G showed a loss of DUB activity compared to the wild type Orf64FL (Fig. 5D). These results indicate that the predicted N-terminal catalytic core of KSHV Orf64 is functional and the cysteine 29 residue is indeed required for the DUB activity of the full-length and the N-terminal proteins.
Thus, in summary, our data suggest that KSHV Orf64 has DUB activity against ubiquitinated cellular proteins and that both K48- and K63-linked ubiquitinated proteins can be deubiquitinated by KSHV Orf64. Additionally, our data suggest that the first 205 amino acids of KSHV Orf64 are sufficient for this activity.
Orf64 and lytic replication.
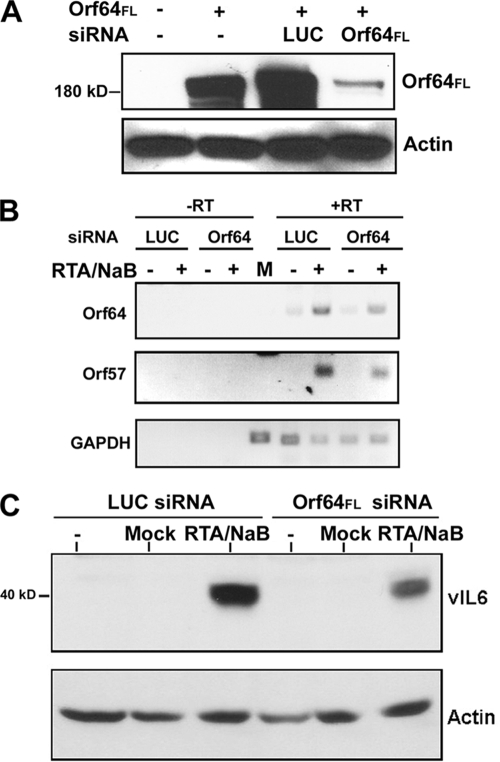
Since Orf64 is present in the tegument of the infecting virion, it is possible that it may play a role during de novo infection. In order to address this possibility, we determined whether Orf64 contributes to the early stages of the KSHV lytic replication cycle. In order to identify the contribution of Orf64 to the lytic cascade, we designed siRNAs targeting Orf64 for degradation via the RNA-induced silencing complex. To test the specificity and effectiveness of Orf64 siRNA knockdown, we transfected HEK-293 cells with the pcDEF3-Orf64FL expression plasmid along with either an irrelevant siRNA directed against LUC or the Orf64-specific siRNA (Orf64FL siRNA) (Dharmacon). Figure 6A shows the extent and specificity of Orf64-directed siRNA knockdown of Orf64FL protein levels as measured by Western blotting to detect the presence of Orf64. Using this Orf64 siRNA we were able to consistently achieve a robust knockdown of the exogenously expressed Orf64FL. Actin was used as a control and showed no difference in protein expression in all four lanes (Fig. 6A).
FIG. 6.
Knockdown of Orf64 affects transcription and protein expression of lytic genes. (A) HEK-293 cells were transfected with pcDEF3- Orf64FL and either siRNA against Orf64 or siRNA against LUC. Cells were harvested 48 h posttransfection and subjected to Western blot analysis with an anti-Flag antibody to detect Orf64FL expression. Knockdown of exogenous Orf64FL in HEK-293 cells was observed when a siRNA specific to Orf64, but not LUC, was cotransfected with the Orf64FL expression vector. Actin blotting was performed to show equal loading of protein lysates. (B) KSHV-293 cells were induced (or mock induced) into lytic replication by adding an RTA-expressing baculovirus (RTA) and sodium butyrate (NaB) and 24 h later were transfected with siRNAs against Orf64 or LUC. Total RNA was harvested 48 h posttransfection and used in RT-PCRs. (Top) Knockdown of the Orf64 transcript; (middle) reduction in the levels of Orf57 transcript; (bottom) GAPDH amplification performed as a control. The reactions performed without reverse transcriptase (−RT lanes) resulted in no detectable product, demonstrating the absence of DNA contaminants in the RT-PCRs. (C) Protein lysates from untreated, mock-induced, and induced cells (RTA/NaB) were analyzed by Western blotting for the levels of the KSHV lytic protein vIL6. Actin blotting was also performed to show equal loading of protein lysates.
KSHV-293 cells were made by infecting HEK-293 cells with the rKSHV.219 virus (37) and selecting with 1 μg/ml puromycin. We subsequently induced the KSHV-293 cells into lytic reactivation by infection with BacRTA and 1.7 mM sodium butyrate. A control mock-induced sample was also included by using medium without BacRTA or sodium butyrate. Twenty-four hours postinduction, we transfected cells with either LUC- or Orf64-directed siRNAs. We achieved 70 to 80% reactivation in these cells, as assayed by red fluorescence, which is driven by the lytic nut-1 promoter (data not shown) (37). Cells were harvested for total RNA analysis as well as protein analysis at 48 h posttransfection. The RNA was used in RT reactions. The resulting cDNAs were then used as templates for PCR amplification with Orf64, Orf57, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) primers to measure viral transcript levels and validate levels of reactivation. Primers for Orf64 were used to assay the level of knockdown of Orf64 transcript, and primers for Orf57, a lytic protein with early kinetics, were used as a marker for lytic reactivation. We also amplified GAPDH as a control. As shown in Fig. 6B, moderate knockdown of the endogenous Orf64 transcript resulted in depressed Orf57 transcription in the reactivated KSHV-293 cells, while GAPDH transcript levels remained unchanged. This suggests that knockdown of Orf64 results in decreased lytic reactivation of KSHV. To confirm this finding, we also performed Western blot analysis with antibody directed against the early lytic protein vIL6 (Fig. 6C). We found that knockdown of Orf64 also resulted in decreased levels of vIL6, while cellular β-actin levels remained unchanged (Fig. 6C).
DISCUSSION
Sequence alignments indicate that all known herpesviruses encode a large tegument protein, which is often encoded by the largest open reading frame in the whole genome. Studies on HSV-1, PRV, and cytomegalovirus have suggested several distinct roles for this herpesviral core protein. These roles include, but are not limited to, delivery of genomic DNA into the nucleus, egress from the nucleus, and scaffolding during tegument assembly (21, 24, 25). Jovasevic et al. have shown that HSV-1 VP1/2 docks at the nuclear pore and mediates delivery of the viral genome into the nucleus after cleavage by a serine-cysteine protease (22). Recently, Shanda and Wilson demonstrated that HSV-1 UL36 (VP1/2) is critical for anterograde transport of enveloped virions along exons using an in vitro microchamber assay (34). Moreover, Lee et al. have determined that the carboxyl terminus of HSV-1 UL36 is sufficient to direct UL36 delivery to nuclear sites of capsid assembly (24).
The study of KSHV Orf64 function in the context of viral infection is complicated by the presence of several virally encoded E3 Ub ligases. KSHV K3 can target several alleles of major histocompatibility complex class I molecules for degradation, while K5 targets ICAM-1 and B7.2 (CD86) (9, 20). The lytic switch protein Orf50 is also an E3 ligase that mediates IRF7 degradation via the proteosome pathway in an effort to preempt the induction of the interferon-mediated antiviral state (43). Hence, the modulation of cellular protein ubiquitination during the lytic life cycle likely involves viral Ub E3 ligases and the viral DUB Orf64.
One unifying role of the Orf64 homologs common to all herpesviruses studied thus far is the Ub-specific protease or DUB activity in the N termini of these proteins. This activity has been found to affect plaque size, titers, and neuroinvasiveness of PRV but has been deemed nonessential in vivo for PRV, yet required in vitro (5, 25). In contrast, in the case of HSV-1, UL36 is indispensable for transport along microtubules. Despite these differences it is clear that UL36 homologs in the alphaherpesviruses play pivotal, if not required, roles in the lytic life cycle.
We are the first to report the functional characterization of the KSHV Orf64 DUB. We found that the full-length Orf64 showed both nuclear and cytoplasmic localization, indicating that Orf64 shuttles between both these cellular compartments. In two different sets of in vitro DUB assays, we found that a GST-Orf64N fusion protein was efficiently able to cleave Ub linkages and did not display preference for K63- or K48-linked Ub chains. Furthermore, expression of both the N terminus of Orf64 and full-length Orf64 in different cell types resulted in a marked reduction of high-molecular-weight ubiquitinated proteins, suggesting that Orf64 can deubiquitinate cellular proteins in vivo. Moreover, an N-terminal mutant Orf64 in which the putative cysteine catalytic residue was mutated to glycine (Orf64N-C29G) was devoid of DUB activity. The most likely explanation for the requirement of this single amino acid is that C29 is part of the catalytic core or catalytic triad that defines the cysteine protease active site, as predicted by sequence analysis (Fig. 1). In sum, our data suggest the following. First, KSHV Orf64 is a robust DUB enzyme. Second, the DUB activity is in the first 205 N-terminal residues. Third, the cysteine residue at position 29 is critical for DUB activity. Fourth, KSHV Orf64 does not display marked specificity for K48- or K63-linked Ub chains, although the deubiquitination of K48-linked Ub chains appears to be slightly favored. The ability of KSHV Orf64 to deubiquitinate both K48 and K63 linkages is unique, since all other alpha- and betaherpesviral DUBs target K48 linkages specifically (2, 23, 29, 39).
It is interesting that KSHV Orf64 can deubiquitinate both types of Ub linkages on proteins. Although exogenous expression of Orf64 appears to deubiquitinate many cellular proteins, in the context of viral infection Orf64 may be localized to a particular region of the cell and its DUB function may be modulated by other viral and/or cellular proteins. Additionally, the targets for Orf64 DUB activity may be cellular and viral. Recently, Whitehurst et al. reported that the EBV homolog of KSHV Orf64 (BPLF-1) negatively regulates EBV ribonucleotide reductase activity.
We also found that Orf64 supports KSHV lytic replication, since siRNA knockdown of Orf64 in KSHV-infected HEK-293 cells induced to enter the lytic cycle resulted in decreased levels of Orf57 lytic transcripts as well as decreased expression of the lytic protein vIL6. This may suggest that Orf64 expression could enhance the KSHV lytic cycle through its deubiquitination function.
The identification of KSHV Orf64 as a robust DUB that can catalyze cleavage of both K48- and K63-linked Ub sets it apart from all its known herpesviral homologs. Since Orf64 is present in the tegument of the incoming virion during de novo infection (31), it may be in a position to exert an effect on KSHV replication upon entry of the virus into the cell. This provides a window of opportunity whence Orf64 could help to modify the cellular environment in order to precipitate the onset of lytic replication, maximize its efficiency, or both.
Acknowledgments
We thank members of the Damania and Dittmer labs for helpful discussions. We thank Stuart Krall for assistance with tissue culture and Aadra Bhatt for critical reading of the manuscript. We thank Jeff Vieira for the recombinant KSHV virus and Vishva Dixit for the Ub plasmids.
B.D. is a Leukemia & Lymphoma Society Scholar and a Burroughs Wellcome Fund Investigator in Infectious Disease. This work was supported by grants CA096500 and DE18281 from the NIH.
Footnotes
Published ahead of print on 29 July 2009.
REFERENCES
- 1.Abaitua, F., and P. O'Hare. 2008. Identification of a highly conserved, functional nuclear localization signal within the N-terminal region of herpes simplex virus type 1 VP1-2 tegument protein. J. Virol. 82:5234-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amerik, A. Y., and M. Hochstrasser. 2004. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta 1695:189-207. [DOI] [PubMed] [Google Scholar]
- 3.Arnason, T., and M. J. Ellison. 1994. Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol. Cell. Biol. 14:7876-7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bottcher, S., H. Granzow, C. Maresch, B. Mohl, B. G. Klupp, and T. C. Mettenleiter. 2007. Identification of functional domains within the essential large tegument protein pUL36 of pseudorabies virus. J. Virol. 81:13403-13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bottcher, S., C. Maresch, H. Granzow, B. G. Klupp, J. P. Teifke, and T. C. Mettenleiter. 2008. Mutagenesis of the active-site cysteine in the ubiquitin-specific protease contained in large tegument protein pUL36 of pseudorabies virus impairs viral replication in vitro and neuroinvasion in vivo. J. Virol. 82:6009-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 8.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 9.Coscoy, L., and D. Ganem. 2000. Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl. Acad. Sci. USA 97:8051-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desai, P. J. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foreman, K. E., J. Friborg, Jr., W. P. Kong, C. Woffendin, P. J. Polverini, B. J. Nickoloff, and G. J. Nabel. 1997. Propagation of a human herpesvirus from AIDS-associated Kaposi's sarcoma. N. Engl. J. Med. 336:163-171. [DOI] [PubMed] [Google Scholar]
- 12.Gessain, A., A. Sudaka, J. Briere, N. Fouchard, M. A. Nicola, B. Rio, M. Arborio, X. Troussard, J. Audouin, J. Diebold, and G. de The. 1996. Kaposi sarcoma-associated herpes-like virus (human herpesvirus type 8) DNA sequences in multicentric Castleman's disease: is there any relevant association in non-human immunodeficiency virus-infected patients? Blood 87:414-416. [PubMed] [Google Scholar]
- 13.Glickman, M. H., and A. Ciechanover. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82:373-428. [DOI] [PubMed] [Google Scholar]
- 14.Goldman, L. A., E. C. Cutrone, S. V. Kotenko, C. D. Krause, and J. A. Langer. 1996. Modifications of vectors pEF-BOS, pcDNA1 and pcDNA3 result in improved convenience and expression. BioTechniques 21:1013-1015. [DOI] [PubMed] [Google Scholar]
- 15.Grundhoff, A., and D. Ganem. 2004. Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis. J. Clin. Investig. 113:124-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hadari, T., J. V. Warms, I. A. Rose, and A. Hershko. 1992. A ubiquitin C-terminal isopeptidase that acts on polyubiquitin chains. Role in protein degradation. J. Biol. Chem. 267:719-727. [PubMed] [Google Scholar]
- 17.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 18.Hicke, L. 2001. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2:195-201. [DOI] [PubMed] [Google Scholar]
- 19.Hicke, L., and R. Dunn. 2003. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19:141-172. [DOI] [PubMed] [Google Scholar]
- 20.Ishido, S., J. K. Choi, B. S. Lee, C. Wang, M. DeMaria, R. P. Johnson, G. B. Cohen, and J. U. Jung. 2000. Inhibition of natural killer cell-mediated cytotoxicity by Kaposi's sarcoma-associated herpesvirus K5 protein. Immunity 13:365-374. [DOI] [PubMed] [Google Scholar]
- 21.Jarosinski, K., L. Kattenhorn, B. Kaufer, H. Ploegh, and N. Osterrieder. 2007. A herpesvirus ubiquitin-specific protease is critical for efficient T cell lymphoma formation. Proc. Natl. Acad. Sci. USA 104:20025-20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jovasevic, V., L. Liang, and B. Roizman. 2008. Proteolytic cleavage of VP1-2 is required for release of herpes simplex virus 1 DNA into the nucleus. J. Virol. 82:3311-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kattenhorn, L. M., G. A. Korbel, B. M. Kessler, E. Spooner, and H. L. Ploegh. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19:547-557. [DOI] [PubMed] [Google Scholar]
- 24.Lee, J. I., G. W. Luxton, and G. A. Smith. 2006. Identification of an essential domain in the herpesvirus VP1/2 tegument protein: the carboxy terminus directs incorporation into capsid assemblons. J. Virol. 80:12086-12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luxton, G. W., J. I. Lee, S. Haverlock-Moyns, J. M. Schober, and G. A. Smith. 2006. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J. Virol. 80:201-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore, P. S., and Y. Chang. 2003. Kaposi's sarcoma-associated herpesvirus immunoevasion and tumorigenesis: two sides of the same coin? Annu. Rev. Microbiol. 57:609-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrison, E. E., A. J. Stevenson, Y. F. Wang, and D. M. Meredith. 1998. Differences in the intracellular localization and fate of herpes simplex virus tegument proteins early in the infection of Vero cells. J. Gen. Virol. 79:2517-2528. [DOI] [PubMed] [Google Scholar]
- 28.Reference deleted.
- 29.Nijman, S. M., M. P. Luna-Vargas, A. Velds, T. R. Brummelkamp, A. M. Dirac, T. K. Sixma, and R. Bernards. 2005. A genomic and functional inventory of deubiquitinating enzymes. Cell 123:773-786. [DOI] [PubMed] [Google Scholar]
- 30.Renne, R., D. Blackbourn, D. Whitby, J. Levy, and D. Ganem. 1998. Limited transmission of Kaposi's sarcoma-associated herpesvirus in cultured cells. J. Virol. 72:5182-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rozen, R., N. Sathish, Y. Li, and Y. Yuan. 2008. Virion-wide protein interactions of Kaposi's sarcoma-associated herpesvirus. J. Virol. 82:4742-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlieker, C., G. A. Korbel, L. M. Kattenhorn, and H. L. Ploegh. 2005. A deubiquitinating activity is conserved in the large tegument protein of the Herpesviridae. J. Virol. 79:15582-15585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schnell, J. D., and L. Hicke. 2003. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J. Biol. Chem. 278:35857-35860. [DOI] [PubMed] [Google Scholar]
- 34.Shanda, S. K., and D. W. Wilson. 2008. UL36p is required for efficient transport of membrane-associated herpes simplex virus type 1 along microtubules. J. Virol. 82:7388-7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sompallae, R., S. Gastaldello, S. Hildebrand, N. Zinin, G. Hassink, K. Lindsten, J. Haas, B. Persson, and M. G. Masucci. 2008. Epstein-Barr virus encodes three bona fide ubiquitin-specific proteases. J. Virol. 82:10477-10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 37.Vieira, J., and P. M. O'Hearn. 2004. Use of the red fluorescent protein as a marker of Kaposi's sarcoma-associated herpesvirus lytic gene expression. Virology 325:225-240. [DOI] [PubMed] [Google Scholar]
- 38.Virgin, H. W., IV, P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, J., A. N. Loveland, L. M. Kattenhorn, H. L. Ploegh, and W. Gibson. 2006. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 80:6003-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wertz, I. E., K. M. O'Rourke, H. Zhou, M. Eby, L. Aravind, S. Seshagiri, P. Wu, C. Wiesmann, R. Baker, D. L. Boone, A. Ma, E. V. Koonin, and V. M. Dixit. 2004. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature 430:694-699. [DOI] [PubMed] [Google Scholar]
- 41.Whitehurst, C. B., S. Ning, G. L. Bentz, F. Dufour, E. Gershburg, J. Shackelford, Y. Langelier, and J. S. Pagano. 2009. The Epstein-Barr virus (EBV) deubiquitinating enzyme BPLF1 reduces EBV ribonucleotide reductase activity. J. Virol. 83:4345-4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.York, I. A., and K. L. Rock. 1996. Antigen processing and presentation by the class I major histocompatibility complex. Annu. Rev. Immunol. 14:369-396. [DOI] [PubMed] [Google Scholar]
- 43.Yu, Y., S. E. Wang, and G. S. Hayward. 2005. The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation. Immunity 22:59-70. [DOI] [PubMed] [Google Scholar]
- 44.Zhou, H., I. Wertz, K. O'Rourke, M. Ultsch, S. Seshagiri, M. Eby, W. Xiao, and V. M. Dixit. 2004. Bcl10 activates the NF-κB pathway through ubiquitination of NEMO. Nature 427:167-171. [DOI] [PubMed] [Google Scholar]