Abstract
The molecular basis underlying hepatitis C virus (HCV) core protein maturation and morphogenesis remains elusive. We characterized the concerted events associated with core protein multimerization and interaction with membranes. Analyses of core proteins expressed from a subgenomic system showed that the signal sequence located between the core and envelope glycoprotein E1 is critical for core association with endoplasmic reticula (ER)/late endosomes and the core's envelopment by membranes, which was judged by the core's acquisition of resistance to proteinase K digestion. Despite exerting an inhibitory effect on the core's association with membranes, (Z-LL)2-ketone, a specific inhibitor of signal peptide peptidase (SPP), did not affect core multimeric complex formation, suggesting that oligomeric core complex formation proceeds prior to or upon core attachment to membranes. Protease-resistant core complexes that contained both innate and processed proteins were detected in the presence of (Z-LL)2-ketone, implying that core envelopment occurs after intramembrane cleavage. Mutations of the core that prevent signal peptide cleavage or coexpression with an SPP loss-of-function D219A mutant decreased the core's envelopment, demonstrating that SPP-mediated cleavage is required for core envelopment. Analyses of core mutants with a deletion in domain I revealed that this domain contains sequences crucial for core envelopment. The core proteins expressed by infectious JFH1 and Jc1 RNAs in Huh7 cells also assembled into a multimeric complex, associated with ER/late-endosomal membranes, and were enveloped by membranes. Treatment with (Z-LL)2-ketone or coexpression with D219A mutant SPP interfered with both core envelopment and infectious HCV production, indicating a critical role of core envelopment in HCV morphogenesis. The results provide mechanistic insights into the sequential and coordinated processes during the association of the HCV core protein with membranes in the early phase of virus maturation and morphogenesis.
The hepatitis C virus (HCV), a major cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma, is an enveloped, single-stranded, positive-sense RNA virus in the Hepacivirus genus of the Flaviviridae family, which also includes the flaviviruses and pestiviruses (42; for a review, see reference 5). The HCV genome contains a single open reading frame flanked by nontranslational regions at its 5′ and 3′ ends and encodes for an ca. 3,000-amino-acid precursor polyprotein. The polyprotein is cotranslationally and/or posttranslationally processed by cellular and viral proteases into mature structural proteins including core and envelope (E) glycoproteins E1 and E2, p7, and the nonstructural (NS) proteins of NS2, NS3, NS4A, NS4B, NS5A, and NS5B (for a review, see reference 68). The core protein is a multifunctional molecule that constitutes the viral nucleocapsid and is also involved in the pathogenesis and carcinogenesis of HCV (for reviews, see references 40, 53 and 71). The structural envelope glycoproteins, E1 and E2, mediate attachment of the virus to receptor(s) on host cells and acidic pH-dependent membrane fusion between the viral envelope and endosomal membranes. NS proteins, although not thought to be assembled into virus particles, participate in viral replication.
Based on the hydrophobicity and clustering of basic amino acids, the core protein consists of three distinct predicted domains (Fig. 1A, panel 1): a basic and hydrophilic region covering two-thirds of the N-terminal portion (domain I; residues 1 to 118); a hydrophobic domain (domain II; residues 119 to 173) covering the central one-third; and the last hydrophobic signal sequence of the downstream protein E1 (domain III; residues 174 to 191) (for a review, see reference 40). The mature core is a dimeric, α-helical protein with membrane-associated features (12). Domain I is mainly involved in RNA binding and encapsidation of the viral genome within a nucleocapsid particle and in homotypic interactions necessary for particle formation (for reviews, see references 40 and 45). Domain I also contains three basic residue stretches of Arg- and Lys-rich sequences, which function as nuclear localization signals, to mediate core localization in the nucleus (32, 62, 77). Domain II is not only required for proper folding of domain I but is also critical for the membrane property of the core including core association with endoplasmic reticula (ER) and outer mitochondrial membranes (12, 63). On the other hand, core proteins, via domain II, target lipid storage droplets (3, 23, 41), where they colocalize with apolipoprotein AII and directly associate with triglycerides (3). Two amphipathic α-helices and the intermediate hydrophobic loop, all of which constitute major structural elements within domain II, were shown to be essential for the association with lipid droplets (LDs), which points to an in-plane membrane interaction of the two helices at the lipid bilayer interface (11). This LD targeting event has been implicated in HCV replication and/or pathogenesis (40, 43), and the LD association site in domain II is a critical determinant of the efficiency of HCV assembly (59).
FIG. 1.
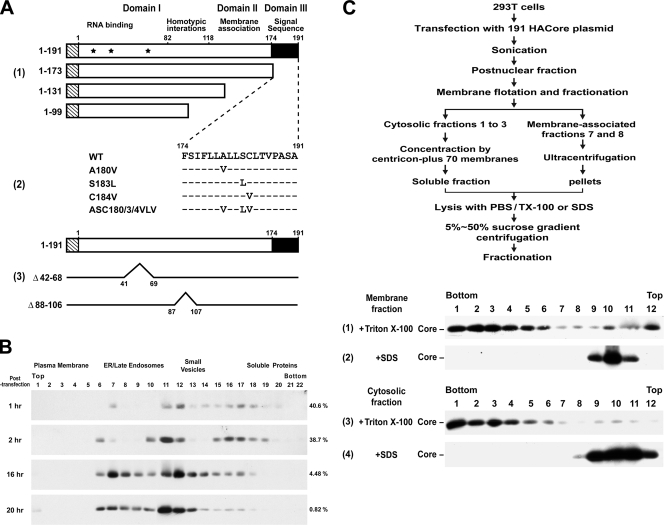
Construction of core plasmids and analyses of membrane-bound and -free core complexes. (A) pCAGGS-based plasmids encoding residues 1 to 191 of the genotype 1a H77 strain core protein and C-terminal-truncated mutants tagged with the influenza virus HA epitope at the N terminus, as shown by the striped rectangles, were constructed as described in Materials and Methods. Domain I, which contains three nuclear localization sequences (located at residues 6 to 13, 38 to 43, and 59 to 71, respectively, and marked with asterisks) and a region involved in core homotypic interaction; domain II, which contains hydrophobic sequences for membrane association; and domain III, which serves as the signal sequence for E1, are also indicated (panel 1). pCAGGS-HACore-based mutant plasmids encoding substitutions in the signal peptide at the residues as indicated were also constructed (panel 2). Dashes indicate that the residue in that position of the mutant core protein is identical to that of the WT core. The Δ88-106 and Δ42-68 mutants, and their corresponding deleted regions are shown in panel 3. (B) 293T cells were transfected with the 191 core-expressing plasmid, and the calcium phosphate-DNA complexes were washed away 4 h posttransfection. Cells were then incubated at 37°C for different times as indicated and harvested, and the postnuclear fractions were subjected to Optiprep subcellular fractionation, followed by Western blot analysis with an HA MAb. The blots were scanned, and the percentages of core proteins distributed into soluble fractions 16 to 22 of the total core populations are indicated on the right of each panel. (C) The steady-state 191 core proteins localized in membrane-associated fractions 7 and 8 after the membrane flotation assay (data not shown) were combined, mixed with an equal volume of cold TE buffer, and then centrifuged in a Beckman SW41 rotor at 36,000 rpm for 2 h. The pelleted membranes were lysed with 0.5 ml of PBS containing 1% Triton X-100 (panel 1) or SDS (panel 2) at 37°C for 10 min prior to 5 to 50% sucrose gradient centrifugation. The combined cytosolic fractions 1 to 3 after the membrane flotation assay (data not shown) were diluted with 6 volumes of cold TE buffer and concentrated by Amicon Centricon plus-70 membranes (Millipore, Bedford, MA). The samples (0.5 ml) were treated with a final concentration of 1% Triton X-100 (panel 3) or SDS (panel 4) before 5 to 50% sucrose gradient centrifugation. The scheme illustrating these procedures is shown at the top.
Maturation of the HCV is thought to proceed through budding of nascent particles into the ER lumen, and then viral particles are transported via a constitutive secretory pathway to the extracellular milieu. The internal signal sequence located between the core and E1 proteins targets the nascent polypeptide to ER membranes and induces translocation of E1 into ER. After membrane insertion, the signal peptide is first cleaved from E1 by a signal peptidase to yield the 191-amino-acid core precursor, p23. This initial cleavage is subsequently followed by a second cleavage catalyzed by an intramembrane-cleaving protease, termed the signal peptide peptidase (SPP) (29, 41, 50), to yield the mature core p21 (41). This mature p21 form predominates in both transfected cell cultures and virus particles from infected sera (77). Once cleaved from the polyprotein, the p21 core is thought to assemble into capsids at the cytoplasmic face of the ER and acquires its envelopes by budding into the ER (6-8, 33, 34), during which time the E1 and E2 proteins are embedded into the viral envelope.
The morphogenesis of the core protein coexpressed with or without other structural proteins has been studied in bacteria (28), yeast (1), insect cells (6), and cell-free systems (25). Many mammalian cell culture systems have also been developed to study the capsid assembly pathway. Although HCV capsids can be detected in many mammalian cells using various expression systems by electron microscopy, core assembly into capsids appears to be inefficient, since no virus-like particles (VLPs) are detected at all (7, 8, 18), presumably due to the instability of core complexes in further isolation, quantification, and biochemical studies. It was reported that the core protein expressed in cells or the purified recombinant core protein self-assembles into nucleocapsid-like particles which require the presence of stem-loop RNA structures, such as those in tRNA (28, 48). Moreover, structural proteins including the core are expressed in various replicon systems using the full-length HCV genome; nonetheless, evidence for virion assembly and release from human liver cell lines was not available (9, 10, 24, 35, 55) until recently when cloned HCV genomes, based on the JFH1 clone isolated from a patient with fulminant hepatitis C, which allowed robust replication of infectious HCV in tissue cultures (HCVcc), were developed (31, 70, 78). The advancement in HCV tissue culture systems has tremendously facilitated our understanding of the molecular mechanisms underlying the HCV replication cycle and virus-cell interactions.
In the present study, we first establish a core subgenomic expression system to dissect several coordinated, key events in the early phase of the HCV core maturation process. The results also demonstrate that association with membranes and SPP-catalyzed cleavage at the C terminus of the core are necessary but not sufficient for core envelopment and that domain I also contains sequences important for core envelopment. We also show that the core protein expressed in the robust HCVcc replication system undergoes several key steps in the early phase of core maturation processes similar to those utilized by the core when expressed alone. In addition, the results from treatment with the SPP-specific inhibitor, (Z-LL)2-ketone, or coexpression with the loss-of-function SPP D219A mutant reveals a critical role of core envelopment in HCV propagation. Our study thus reveals the sequential and concerted processes of core multimerization and interactions with and envelopment by membranes during the HCV assembly and/or budding process. To our best knowledge, this is the first demonstration of this kind.
MATERIALS AND METHODS
Cells, antibodies, and reagents.
293T and Huh7 cells were cultured in Dulbecco modified Eagle medium containing 10% heat-inactivated fetal bovine serum. In particular, nonessential amino acids were added to the Huh7 culture medium. To obtain anti-HCV core antiserum, the coding sequence of residues 2 to 173 of the core of the genotype 1 H77 strain was cloned in a pET21(a) vector (Merck Biosciences-Novagen; Darmstadt, Germany). The fusion protein with a His6 tag attached at the N terminus of the core was purified by BD Talon affinity metal resin (BD Biosciences; San Diego, CA), and the purified protein was used to immunize New Zealand White rabbits. A mouse monoclonal antibody (MAb) for the influenza virus hemagglutinin (HA) tag (HA.11, clone 16B12) was purchased from Covance (Richmond, CA). A MAb specific for HCV core (clone C7-50) was obtained from Affinity BioReagents (Golden, CO). MAbs directed against HCV E1 (clone BD1198) and E2 (clone BD1167) were purchased from Biodesign (Saco, ME). A MAb against HCV E2 (clone HCM-0919-5) was purchased from Austral Biologicals (San Ramon, CA). Rabbit anti-HCV E2 peptide antibody (catalog no. GB10416) was from Genesis Biotech (Xindian, Taipei County, Taiwan). A MAb against DsRed (clone 64-1077) was purchased from BD Biosciences. Rabbit anti-green fluorescent protein (GFP) immunoglobulin G (IgG), goat anti-β-Gal IgG, and goat anti-TIP47 IgG (N-15) were all obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Affinity-purified fluorescein isothiocyanate (FITC)-conjugated secondary antibodies were purchased from Kirkegaard & Perry Laboratories (Gaithersburg, MD). 1,3-Di-(N-carboxybenzoyl-l-leucyl-l-leucyl)amino acetone [(Z-LL)2-ketone] was purchased from Merck Biosciences-Calbiochem (Darmstadt, Germany).
Construction of plasmids.
To construct the pCAGGS-based core plasmids encoding amino acid residues 1 to 191, 1 to 173, 1 to 131, and 1 to 99 with the HA peptide, YPYDVPDYA, tagging the N terminus (Fig. 1A, panel 1), the HACore-coding sequences were amplified from pBRTM-HCV-H1-3011, a full-length HCV genotype 1a clone, through a PCR using HA342f-EcoRI as the sense primer and 914r-EcoRI, 860r-EcoRI, 735r-EcoRI, and 639r-EcoRI, respectively, as the antisense primers. The number in the name of a primer indicates the position of the first nucleotide of the primer in the sequence of the pBRTM-HCV-H1-3011 clone. EcoRI-restricted DNA fragments were then inserted into pCAGGS at the same site. To construct pCAGGS-HACore-A180V, -S183L, -C184V, and -ASC180/3/4VLV core mutant plasmids (Fig. 1A, panel 2), which encoded core variants with a single substitution of Val, Leu, and Val for Ala-180, Ser-183, and Cys-184, respectively, or all three substitutions for the three residues, HA342f-EcoRI was used as the sense primer and A180Vr-EcoRI, S183Lr-EcoRI, C184Vr-EcoRI, and ASC180/3/4VLVr-EcoRI, respectively, were used as the antisense primers in the PCR. The EcoRI-cut DNA fragments were cloned into the pCAGGS vector to generate mutant core plasmids. The pCAGGS-based core plasmids expressing the Δ88-106 and Δ42-68 mutants (Fig. 1A, panel 3) were, respectively, PCR amplified using del88-106f and del88-106r, and del42-68f and del42-68r, as the paired primers. To construct pcDNA3-HACE1E2, the sense and antisense primers in the PCR were HA342f-HindIII and 2579r-XbaI, respectively, and the HindIII- and XbaI-cut DNA fragments were cloned into the pcDNA3 vector (Invitrogen). pcDNA3-HACE1E2 was then cut with HindIII and XbaI and end blunted with T4 DNA polymerase, and the insert was ligated to the end-blunted EcoRI site in pCAGGS to generate pCAGGS-HACE1E2. Kozak-342f-HindIII and 914r-XbaI were used as primers to generate the DNA fragment, which was then cloned into pcDNA3 at the HindIII and XbaI sites to generate pcDNA3-HACore. Kozak-735f-EcoRI and 2579r-XbaI were used as primers and pBRTM-HCV-H1-3011 as the template to generate the E1E2 insert, which was then cloned into the corresponding sites in pcDNA3 to yield pcDNA3-E1E2. The sequences of oligonucleotides used as PCR primers are available upon request.
Plasmid DNA transfection.
Subconfluent 293T cells grown on 6- or 10-cm petri dishes were transfected with 5 or 10 μg of core- or CE1E2-expressing plasmids, respectively, using a standard calcium phosphate coprecipitation method (54). Unless otherwise indicated, the DNA and calcium phosphate complexes were washed away 6 to 8 h after transfection. Huh7 cells were transfected with DNA plasmids by using the Arrest-in transfection method (Openbiosystems, Huntsville, AL) in accordance with the manufacturer's procedures. Unless otherwise indicated, all transfected 293T and Huh7 cells were harvested 48 h after transfection for analyses.
In vitro RNA transcription and RNA transfection.
The JFH1 and chimeric F6/JFH1 (also termed Jc1) RNA genomes were synthesized in vitro as previously described (70). In brief, the JFH1 and Jc1 plasmids were first restricted with XbaI and treated with mung bean nuclease (New England Biolabs; Beverly, MA) to remove the four terminal nucleotides, resulting in the correct 3′ end of the HCV cDNAs. Digested plasmid DNAs were then used as a template for in vitro RNA synthesis by using a MEGAscript T7 kit (Ambion, Austin, TX). Synthesized RNAs were treated with DNase I and purified by using an RNeasy minikit (Qiagen, Valencia, CA). Huh7 cells were transfected with 10 μg of RNA by electroporation using a BTX Model ECM 630 Electro cell manipulator (Holliston, MA) with settings at 240 V and 975 μF. Cells were harvested 72 h posttransfection for various biochemical analyses. Alternatively, 16 h after JFH1 RNA transfection, Huh7 cells were transfected with plasmid DNA by the Arrest-in transfection method described above.
Virus infection and determination of virus titers.
Culture supernatants obtained from Huh7 cells electroporated with HCV RNAs were passed through a filter with a 0.45-μm pore size and fivefold serially diluted with fresh medium supplemented with 20 mM HEPES [4-(2-hydroxyethyl)-1-1-piperazine ethanesulfonic acid] and 8 μg of Polybrene/ml. The supernatants were then used to infect Huh7 cells grown in 96-well plates. The inoculum was removed at 8 h postinfection. At 72 h postinoculation, the cells were processed for indirect immunofluorescence staining. First, cells were fixed with methanol at −20°C, followed by blocking. Cells were then successively incubated with an NS5A (clone 9E10) MAb and FITC-conjugated anti-mouse IgG. The immunostained cells were scored for NS5A foci under an inverted immunofluorescence microscope (Eclipse TE2000-U; Nikon, Tokyo, Japan). The virus titer was expressed as focus-forming units per milliliter determined by the average number of NS5A-positive foci.
SDS-PAGE and Western blot analysis.
Lysates obtained from transfected cells were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), followed by Western blotting with appropriate primary and secondary antibodies and then enhanced chemiluminescent detection. Alternatively, proteins in each fraction after density gradient centrifugation or subcellular fractionation were precipitated with 10% cold trichloroacetic acid prior to SDS-PAGE. An HA MAb was used as the primary antibody to detect core proteins by Western blotting unless otherwise indicated. For quantitation of the core amounts, images within the linear range were scanned by using a Microtek ScanMaker 8700 (Zntennation; Hsinchu, Taiwan) and quantified by using MetaMorph (Molecular Devices; Sunnyvale, CA).
Analysis of core complexes by sucrose linear density gradient centrifugation.
Transfected cells were washed with cold phosphate-buffered saline (PBS) twice, and lysed with PBS containing 1% Nonidet P-40 (NP-40), 1% sodium deoxycholate, and a protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). After centrifugation at 18,000 × g for 30 min at 4°C, cell lysates were loaded onto 5 to 50% (wt/vol) linear sucrose gradients, and the gradients were centrifuged at 100,000 × g for 16 h at 4°C as previously described (2). After centrifugation, samples were fractionated from the bottom of the gradients, and 1 ml per fraction was collected.
Sucrose density gradient-based membrane flotation and Optiprep membrane subcellular fractionation analyses.
For membrane flotation, previously described procedures (15) were followed. In brief, PBS-washed transfected cells were sonicated twice, each time for 15 s, in 0.5 ml of hypotonic TE buffer (10 mM Tris-HCl [pH 7.5] containing 1 mM EDTA and Complete protease inhibitor cocktail) supplemented with 10% (wt/vol) sucrose on ice. After low-speed centrifugation to remove nuclei, the postnuclear supernatants (0.25 ml) were mixed with 1.25 ml of 85.5% sucrose prepared in TE buffer and placed at the bottom of a Beckman SW41 ultracentrifuge tube, and then 7 ml of 65% sucrose and 3.25 ml of 6% sucrose (both prepared in TE buffer) were successively loaded into the tube. The gradients were centrifuged at 100,000 × g for 18 h at 4°C. Samples were then fractionated from the bottom of the gradients, and 1.2 ml per fraction was collected.
For iodixanol (Axis-Shield, Oslo, Norway)-based subcellular fractionation studies, a previously reported procedure (22, 26) was followed. Briefly, transfected cells were sonicated in 1 ml of 10 mM Tris-HCl (pH 7.5) containing 0.25 M sucrose, 1 mM EDTA, and protease inhibitor cocktail. After removal of the nuclei, the postnuclear supernatants were subjected to subcellular fractionation to separate different membrane compartments. First, 0.33 ml of the postnuclear supernatants was mixed with 0.67 ml of 60% Optiprep and then transferred to the bottom of a Beckman SW60 centrifuge tube, which was successively overlaid with 1 ml each of 30%, 20%, and 10% Optiprep. The gradients were centrifuged at 337,000 × g in a Beckman SW60 rotor (50,000 rpm) for 3 h at 4°C. Fractions (180 μl) were collected from the top of the gradient.
In vitro protein synthesis.
In vitro-coupled transcription/translation was performed using the reticulocyte-based TNT quick coupled transcription/translation kit (Promega; Madison, WI) with 1 μg of pcDNA3 plasmids in a 25-μl reaction mixture. For protein synthesis in the presence of membranes, 1.5 μl of a canine pancreatic microsomal membrane (Promega) was added per 25 μl of reaction mixture. After incubation at 30°C for 90 min, a final concentration of 0.1 mg of cycloheximide/ml was added to stop the reaction.
Protease digestion assay.
A membrane protection assay was performed as previously indicated (15). For analysis of proteins expressed in cells, transfected 293T cells were sonicated in 50 mM Tris-HCl (pH 8.0), containing 10 mM CaCl2, 1 mM dithiothreitol, and protease inhibitor cocktail. JFH1 or Jc1 RNA-transfected Huh7 cells were passed through a 27-gauge syringe 30 times. The postnuclear supernatants or samples obtained from in vitro synthesis were divided into three portions. One portion received no treatment. The second portion was treated with 50 μg (for proteins expressed in 293T cells) or 10 μg (for proteins expressed in Huh7 or in vitro) of proteinase K/ml. The third portion was solubilized with a final concentration of 1% Triton X-100 before proteinase K treatment. After incubation on ice for 1 h, samples were supplemented with a final concentration of 5 mM phenylmethylsulfonyl fluoride (dissolved in ethanol), followed by incubation on ice for 10 min. All samples were solubilized with detergents, boiled at 95°C for 10 min, and then analyzed by SDS-PAGE followed by Western blotting.
Reconstituted binding of the core complex to membranes.
In vitro-translated core proteins were incubated with or without 3 μl of microsomal membranes in 250 μl of PBS containing 120 mM potassium acetate, 5 mM magnesium acetate, 1 mM dithiothreitol, and 5 mM ATP at 30°C for 3 h. Alternatively, detergent-lysed cell extracts containing core proteins were first subjected to 5 to 50% linear density sucrose gradient centrifugation. The bottom fractions containing high-ordered, multimeric core complexes were pooled, and the concentrated core complexes were incubated with or without microsomal membranes. The reaction mixtures were then mixed with 1.25 ml of 85.5% sucrose prepared in TE buffer and resolved by a membrane flotation analysis as described above.
Immunoprecipitation of viral proteins.
Transfected 293T cells were sonicated with the proteinase K digestion buffer, and the postnuclear fractions were divided into three parts. The first portion was incubated with 5 μl of rabbit preimmune serum, the second portion was incubated with 5 μl of rabbit anti-core (for 191 and 131 core transfection) or with 1 μg of affinity-purified rabbit anti-E2 (for HACE1E2 transfection), and the third portion was treated with 1% each of NP-40 and sodium deoxycholate prior to incubation with rabbit anti-core or anti-E2. Then, 30 μl of 10% washed protein A-Sepharose beads was added to each mixture, followed by additional incubation. The precipitated antigens were resolved by SDS-PAGE, followed by Western blotting analysis.
Assessment of core envelopment by differential membrane permeabilization methods.
Huh7 cells were transfected with JFH1 RNA, and at 72 h posttransfection the cells were washed with PBS and fixed with 4% paraformaldehyde (prepared in PBS) at room temperature for 30 min. The cells were then incubated with PBS alone or with PBS containing 0.5% each of saponin or Triton X-100 at room temperature for 15 min. After three washes with PBS and blocking with PBS containing 1% fetal bovine serum and 5% bovine serum albumin, the cells were incubated with rabbit preimmune serum or an anti-core, followed by incubation with FITC-conjugated goat anti-rabbit IgG. The immunostained cells were quantitated by fluorescence-activated cell sorter (FACS) analysis using a FACSCalibur (Becton Dickinson Immunocytometry Systems, San Jose, CA).
RESULTS
Formation of high-ordered, multimeric core complexes in cells.
To provide mechanistic insights into the HCV core protein maturation process, we first examined the expression, intracellular localization, and maturation of the full-length HCV H77 strain core protein containing residues 1 to 191, which includes the signal sequence, i.e., residues 174 to 191 at the C terminus, as well as core proteins with a series of truncations from the C terminus (Fig. 1A, panel 1) in 293T cells. To expedite detection of the core protein, all core proteins encoded by the pCAGGS, a cytomegalovirus immediately-early enhancer/chicken β-actin promoter-driven vector (13), were tagged with an HA epitope prefaced by a start codon at their N termini (Fig. 1A, panel 1). The shorter forms of core proteins were effectively expressed as the 1∼191 core (data not shown). In a chemical cross-linking analysis with glutaraldehyde, all of the truncated core proteins, as well as the 191 core exhibited ladderlike, oligomeric forms (data not shown).
When the postnuclear extract containing the 191 core was subjected to 5 to 50% linear density sucrose gradient centrifugation, the 191 core was mainly detected in the bottom one to four fractions (data not shown), which had a buoyant density ranging 1.148 to 1.192 g/ml, similar to those found for VLPs produced by heterologous expression systems (18). The core complexes located in top fractions 6 to 12 might represent membrane-associated multimeric structures, since treatment with 1% each of NP-40 and sodium deoxycholate prior to gradient centrifugation significantly reduced their distribution in these fractions (data not shown). The cell-free human immunodeficiency virus type 1 immature Gag VLPs were located in the bottom fractions 1 to 4, with a peak buoyant density of 1.164 g/ml (data not shown) as previously reported (27, 75). Therefore, the species detected in bottom fractions 1 to 4 represent high-ordered, multimeric core complexes (data not shown). In addition, the shorter core proteins were all distributed in bottom fractions 1 to 4 (data not shown), indicating that the C-terminal segment of core is not critical for core multimerization into high-ordered structural complexes.
In these analyses, we found that the 191 and 173 core proteins comigrated on SDS-PAGE (data not shown). To provide evidence for SPP-mediated conversion of the 1-191 innate p23 to the processed p21, a pulse-chase was performed. With a 15-min pulse time, a fraction of p23 was already processed to p21; nevertheless, the p23 form was still predominant (data not shown). After a 5-min chase, the level of innate p23 had decreased, which was accompanied by an increase in the level of the processed p21, which was at a slightly higher level than that of p23 (data not shown). The instability of the HCV core observed after chasing for 5 min (data not shown) was in line with previous results that degradation of the HCV core protein (64) was mediated by E6AP ubiquitin ligase (60) or the proteasome activator, PA28γ (46). Cotransfection of the 191 core plasmid with or without pcDNA3-SPP(D219A)-HA (50), which encodes an Asp-to-Ala substitution for Asp-219, one of two protease-active sites in SPP, revealed unprocessed p23 and processed p21 forms, respectively (data not shown). These observations show the precursor-product relationship of p23 and p21.
Subcellular fractionation of core proteins.
To determine the intracellular transport of the core complex, a cell fractionation method using iodixanol-based, isopycnic linear density gradient centrifugation (22, 26) was used. As a control, the human immunodeficiency virus type 1 Env glycoproteins gp160, gp120, and gp41 were mainly distributed in the ER/late-endosomal compartments, while a significant fraction of gp41 was also detected in plasma membranes (data not shown), which is consistent with the intracellular transport and maturation process of Env. The Pr55 Gag precursor was recovered from all four fractions (data not shown), in accordance with the maturation process of Gag after its biosynthesis in the cytoplasm and targeting to the plasma membrane for budding. Cellubrevin and TI-VAMP (vesicle-associated membrane protein) are markers for recycled endosomes and late endosomes/secretory vesicles, respectively (17, 37). As previously shown (22), GFP-Cellubrevin was mainly associated with plasma membranes, ER/late endosomes, and small vesicles, whereas GFP-TI-VAMP was predominantly associated with small vesicles and ER/late endosomes (data not shown). Lysosome-associated membrane protein (LAMP) 1, a late endosome marker, was mainly associated with endosomal and plasma membranes (data not shown). Rab11, which was detected in the trans-Golgi network, secretory vesicles, and the pericentriolar recycling endosomes, was recovered from endosomal membranes, small vesicles, and plasma membranes (data not shown). The distribution profiles of LAMP1 and Rab11 were similar to those previously reported (26).
When the postnuclear fraction of the 191 core was analyzed, the processed 191 core was primarily recovered from ER/late endosomes and small vesicles, and a small amount of this protein was associated with plasma membranes (data not shown). Only a small amount of the 191 core was recovered from soluble proteins (data not shown). A similar distribution pattern of the 191 core associated with intracellular membranes was obtained when the core was expressed in COS-1 cells (data not shown). In contrast, the 173, 131, and 99 core proteins were primarily associated with soluble fractions, and a fraction of these proteins was associated with small vesicle membranes (data not shown). Also, a small amount of 173 core protein was transported to plasma membranes (data not shown). Nevertheless, these truncated core proteins were not efficiently associated with ER/late endosomes, indicating that the signal sequence is critical for core association with ER/late endosomes.
Intracellular localization of core complexes.
We then examined the intracellular localization of recombinant core complexes using confocal immunofluorescence microscopy with known intracellular organelle markers. The 191 core protein exhibited a finely reticular perinuclear staining pattern and also formed granules disseminated throughout the cytoplasm (data not shown). Both the perinuclear and the cytoplasmic staining showed “ringlike” and “dot” structures. The 173, 131, and 99 core proteins were primarily localized to the nucleus, and only a small percentage of these proteins formed patches or speckles in the cytoplasm (data not shown), which is consistent with previous findings that removal of the signal sequence between the C and E1 proteins causes the core to detour from the cytoplasm to the nucleus (14, 46, 50, 57, 62). We also observed a considerable degree of colocalization of the 191 core protein with calnexin, an ER marker, GM130, a Golgi marker, and with LAMP3, a late endosome marker (data not shown). Nevertheless, the 191 core protein did not colocalize with EEA1, an early endosome marker (data not shown).
The core protein expressed from subgenomic-expressing systems and in the context of JFH1 RNA replication were shown to target LDs, which exist in virtually all eukaryotic cells and even in some prokaryotes (for reviews, see references 47 and 52). A confocal microscopic analysis showed that a significant fraction of the cytoplasmic 191 core which formed a “ring” structure was localized at the surface of LDs, which were stained with the LD marker, BODIPY 493/503, whereas those core complexes present as “punctates” or “dots” did not seem to be associated with LDs (data not shown). The cytoplasmic 173 core was predominantly localized at the surface of LDs, whereas the cytoplasmic 131 and 99 core proteins were barely detected there (data not shown). The LD association property of 191 and 173 core proteins was also observed when these core proteins were expressed in Huh7 cells (data not shown), a hepatoma cell line which supports HCV replication.
Membrane-associated core proteins form high-ordered, multimeric structures.
To understand the relationship between membrane-free and -bound 191 core proteins, DNA-calcium phosphate coprecipitates were washed away 4 h posttransfection, and the cultures were left to recover in fresh medium for an additional 1, 2, 16, and 20 h. This type of time course study was previously used to monitor intracellular trafficking of the Marburg virus matrix protein VP40 between cellular compartments (26). The percentage of the 191 core distributed in the soluble fraction of the total core population decreased as the cultures were harvested and analyzed at later posttransfection times (Fig. 1B), revealing a progressive shift in the 191 core from soluble to membrane fractions as the posttransfection time proceeded.
To understand whether the structural organization of membrane-free and -associated 191 core proteins in a steady state differs, which thus affects their membrane association properties, a cell-based protein-membrane interaction assay, which separates membrane proteins from cytosolic proteins (16, 61), was used. As previously reported (39), the 191 core could float to membrane fractions 7 and 8 (data not shown), which corresponded to the 6 and 65% sucrose interfaces. When the membrane-associated fractions 7 and 8 and cytosolic fractions 1 to 3 of the 191 core after membrane flotation were separately collected, concentrated, supplemented with a final concentration of 1% Triton X-100, incubated at 37°C for 10 min, and then subjected to a 5 to 50% linear sucrose gradient centrifugation, they all predominantly formed high-ordered, multimeric structures (Fig. 1C, panels 1 and 3, respectively). In contrast, these membrane-free and -associated core proteins were both located in the top fractions of the gradient when treated with 1% SDS prior to sucrose gradient centrifugation, (Fig. 1C, panels 2 and 4, respectively), indicating that SDS not only interferes with core-membrane interactions but also strongly disrupts intersubunit interactions of high-ordered, multimeric core complexes, resulting in monomeric core molecules.
The 191 core expressed in cells, but not synthesized in vitro, is enveloped by membranes.
The core protein translated in vitro provides a ready system to study SPP-mediated signal anchor processing (44, 57, 73, 77). To understand whether the 191 core synthesized in vitro can be enveloped, the 1-191 core translated in vitro was subjected to a standard membrane protection assay. If the core is bound to the inner leaflet of vesicles which are right side out, it will be protected from proteolysis, whereas the cytosolic core and core proteins present on inside-out vesicles are protease sensitive. In an in vitro transcription/translation reaction that synthesized E1 and E2, the E1 translated in the presence of a microsomal membrane was resistant to proteinase K digestion; however, treatment with detergent prior to proteolysis rendered E1 sensitive to proteinase K digestion (Fig. 2A, lanes 1 to 3), indicating that the synthesized E1 had been translocated across the membrane. Although the in vitro-synthesized core was processed by SPP in the presence of membranes (Fig. 2A, compare lanes 4 and 7), it was not enveloped, as judged by its sensitivity to protease digestion (Fig. 2A, compare lanes 7 and 8) (25).
FIG. 2.
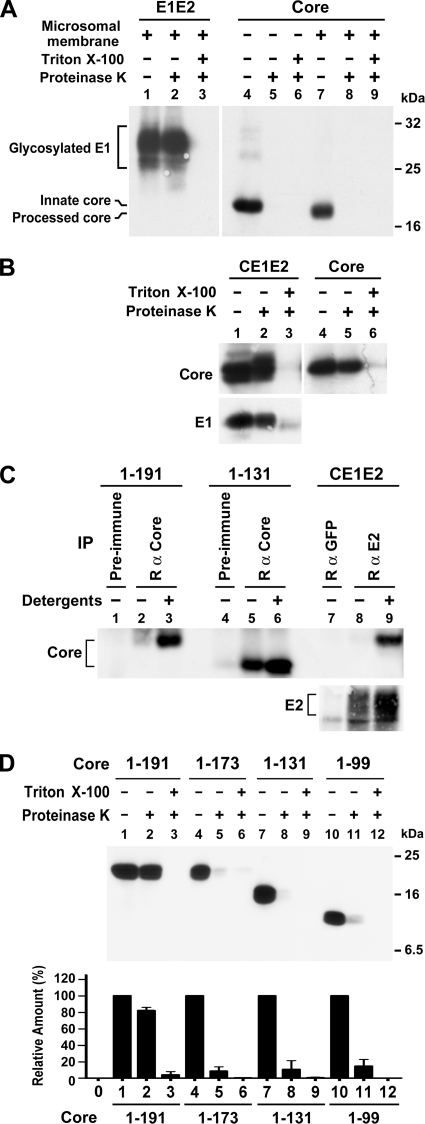
Assessment of core envelopment by membrane protection and immunoprecipitation assays. (A) E1E2 and core proteins translated in vitro from pcDNA3-E1E2 and pcDNA3-HACore, respectively, in the presence or absence of canine pancreatic microsomal membranes were subjected to a membrane protection assay. Proteins were resolved by SDS-PAGE, followed by Western blotting with E1- and HA-specific MAbs, respectively. (B) Equal volumes of postnuclear fractions obtained from 293T cells transfected with pCAGGS-HACE1E2 or pCAGGS-HACore were subjected to a membrane protection assay, and samples were analyzed by Western blotting with HA- and E1-specific MAbs, respectively. (C) Postnuclear fractions obtained from 293T cells expressing core or CE1E2 proteins were left untreated or treated with 1% each of NP-40 and sodium deoxycholate prior to immunoprecipitation with the core or E2-specific antibodies as indicated. Incubation with appropriate rabbit preimmune serum or purified rabbit anti-GFP was used as the control. The precipitated antigens were analyzed by Western blotting with MAbs directed against HA and E2, respectively. (D) Postnuclear samples from cells transfected with each core plasmid were analyzed by a membrane protection assay followed by Western blotting with the HA MAb (top panel). The density of bands corresponding to the HA core proteins in each treatment was scanned and quantified. The percentages of core levels detected in samples treated with proteinases K alone or with detergent and protease relative to that of an untreated sample were determined. The diagram represents results from three independent studies with the standard deviation shown (bottom panel).
We next determined whether the intracellular 191 core complex was enveloped by membranes. Previous studies suggested that HCV capsids acquire an envelope by interacting with E proteins and budding through the ER membrane to form VLPs (7). Thus, the core and E1 proteins encoded by pCAGGS-HACE1E2 were included as a control. The majority of the coexpressed core and E1 proteins and the core protein expressed alone were both resistant to proteinase K treatment in the absence of Triton X-100; however, they all became protease sensitive when treated with detergent before protease digestion (Fig. 2B). To strengthen the viewpoint that the core expressed in cells is enveloped by membranes, the ability of core- and E2-specific antibodies to bind to these two proteins was assessed. The E2 antibody was able to bind E2 and coprecipitate the core if the postnuclear fraction was treated with NP-40 and sodium deoxycholate prior to immunoprecipitation (Fig. 2C, lane 9). However, this antibody bound to E2 at a much smaller degree in the postnuclear fraction without detergent treatment (Fig. 2C, compare lanes 8 and 9). These observations indicate that the majority of E2 and core proteins are enveloped inside membrane vesicles, that E2 is accessible to anti-E2 upon disruption of the membrane by detergents, and that precipitation of the core by anti-E2 occurs via its association with E1 in the E1-E2 heterodimer. Moreover, the anti-core antibody was able to precipitate a considerable amount of the 131 core, but not the 191 core, even when the membrane remained intact (Fig. 2C, compare lanes 2 and 5). Nevertheless, the anti-core was able to bind the 191 core upon disruption of the membranes by detergents (Fig. 2C, lane 3).
When the sensitivities of the 191 and other truncated core proteins to proteinase K were assessed and quantified, 82% of the 191 core molecules were resistant to protease treatment in the absence of Triton X-100 (Fig. 2D, compare lanes 1 and 2), whereas detergent treatment abolished the protease resistance of the processed 191 core (Fig. 2D, lane 3). Although the other three truncated core proteins possibly remained associated with small vesicles (data not shown), they were predominantly associated with cytoplasmic leaflets of right-side-out vesicles, as evidenced by their sensitivity to proteolysis even in the absence of detergent (Fig. 2D, lanes 4 to 12). These results indicate that most of the 191 core molecules coexpressed with E1 and E2 proteins or expressed alone in cells are enveloped by membranes, and also suggest that the signal sequence located at the C terminus of the core is required for core envelopment in cells.
Intracellular cytosolic core complexes isolated from cells bind to membranes but are not enveloped by membranes in vitro.
To determine whether the high-ordered, multimeric core complexes can bind to membranes, the membrane-free multimeric 191 core and cytosolic 173 core complex after treatment with detergents were first isolated by sucrose gradient centrifugation and then incubated with or without microsomal membranes prior to membrane flotation. The cytosolic 191 and 173 complexes could float to the 6 and 65% sucrose interfaces only after incubation with membranes (Fig. 3A, compare panels 2 and 4 to panels 1 and 3, respectively). The 191 core translated in vitro in the presence of membranes was also associated with membranes (Fig. 3A, compare panels 5 and 6). Nevertheless, all of the membrane-free and -bound processed 191 and 173 core complexes were sensitive to protease digestion (Fig. 3B). These results indicate that in spite of their ability to bind to membranes, the preassembled 191 and 173 core complexes were not enveloped by membranes in this reconstituted in vitro membrane binding assay.
FIG. 3.
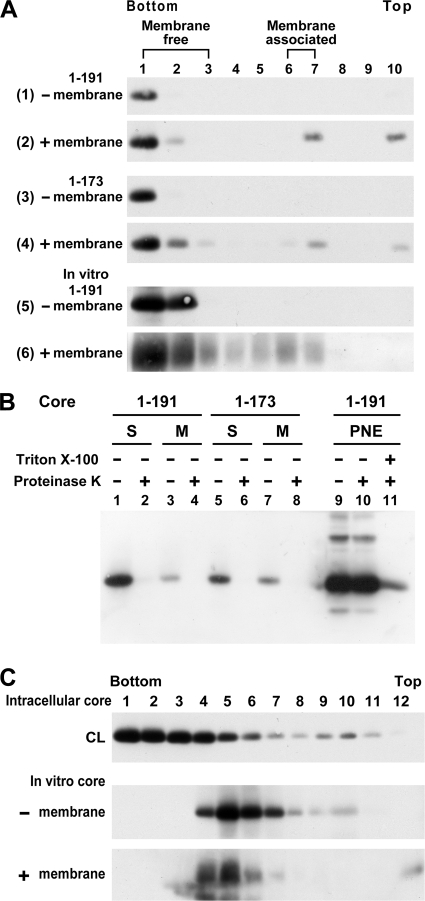
In vitro membrane-binding ability and multimerization of core complexes. (A) 293T cells expressing the 1-191 (panels 1 and 2) and 1-173 (panels 3 and 4) core proteins were lysed with 1% each of NP-40 and sodium deoxycholate and then fractionated by 5 to 50% sucrose gradient centrifugation. The bottom fractions containing the high-ordered, multimeric core complexes were diluted with PBS and ultracentrifuged. After resuspension with PBS, the core complexes were divided into two portions: one left untreated (panels 1 and 3, respectively) and the other incubated with canine pancreatic microsomal membrane (panels 2 and 4, respectively). The mixtures were then analyzed by a membrane flotation assay, followed by Western blotting with the HA MAb. The 191 core protein translated in vitro from pcDNA3-HACore in the absence (panel 5) or presence (panel 6) of microsomal membranes was also resolved by membrane flotation. (B) The pooled membrane-free fractions 1 to 3 and membrane-associated fractions 7 and 8, marked as S and M, respectively, obtained from panels 2 and 4 in panel A were concentrated and treated with or without proteinase K prior to Western blotting (lanes 1 to 8). Postnuclear fractions containing the processed 191 core were simultaneously digested with proteinase K in the presence or absence of detergent (lanes 9 to 11) and were used as the controls. (C) Detergent-treated cell lysates (marked as CL) containing the 191 core and core synthesized in vitro in the absence or presence of microsomal membranes were analyzed by 5 to 50% sucrose gradient centrifugation.
Next, we determined whether the structural organization of the core expressed in cells and in vitro might differ. In contrast to the core complex expressed in cells, which were located in the bottom fractions 1 to 4 of the gradient, the core proteins synthesized in vitro in the presence or absence of microsomal membrane all peaked at fraction 5 (Fig. 3C), indicating that the in vitro-translated core assembles into a complex with a buoyant density lighter than that of the core complex synthesized in cells.
Effects of an SPP inhibitor on the maturation processes of the core.
To understand the role of cleavage of the membrane-anchoring domain in membrane association of the core complex, cells expressing the 191 core were treated with or without (Z-LL)2-ketone, a cysteine protease inhibitor which specifically targets SPP intramembrane cleavage activity (29, 41, 73). Unlike the core expressed without (Z-LL)2-ketone, the innate core expressed in the presence of the SPP inhibitor predominated over the processed form and was primarily recovered from the soluble protein fraction, while only a small fraction of the innate core was coassociated in ER/late endosomes with the processed core (Fig. 4A), the cleavage of which by SPP was apparently incompletely blocked by the inhibitor under these experimental conditions. It was also noted that this SPP inhibitor was not effective at inhibiting SPP activity (76).
FIG. 4.
Characterization of the 191 core synthesized in the presence of (Z-LL)2-ketone. (A) 293T cells expressing the 191 core were treated with or without 100 μM (Z-LL)2-ketone at 37°C for 14 h, and the postnuclear extracts were analyzed by Optiprep-based subcellular fractionation. Migration of the innate p23 and processed p21 core proteins is marked. Percentages of core proteins distributed in soluble fractions 16 to 22 of the total core populations are indicated on the right of each panel. (B) 293T cells expressing the 191 core were treated with or without 20 μM (Z-LL)2-ketone at 37°C for 20 h, and the postnuclear extracts were subjected to a proteinase K digestion assay. The amount of the core synthesized in the presence of the SPP inhibitor was adjusted for better comparison of the protease resistance to that of the core expressed without the SPP inhibitor. (C) Cells expressing the 191 core were treated with 20 μM (Z-LL)2-ketone for 20 h or untreated, and cell lysates were subjected to 5 to 50% sucrose gradient centrifugation.
In the membrane protection assay, a small fraction of the innate and processed core proteins synthesized in the presence of the SPP inhibitor was still resistant to proteolysis despite the level of proteinase K resistance of the innate and processed forms being lower than that without inhibitor treatment (Fig. 4B, compare lanes 4 and 5 to lanes 1 and 2). This result shows that the inhibition of SPP activity decreased the membrane association of the core and the core's subsequent envelopment. Nevertheless, the innate and processed core proteins synthesized in the presence of the inhibitor were also localized in the bottom fractions of the sucrose linear density gradient (Fig. 4C), indicating that treatment with the SPP inhibitor did not affect high-ordered, multimeric core complex formation.
Characterization of core mutants with substitutions in the signal peptide.
To further understand the effects of signal peptide cleavage on core envelopment, core variants with mutations in the signal peptide were examined. Ala-180, Ser-183, and Cys-184 in the signal sequence were individually replaced by Val, Leu, and Val, respectively, or replaced with a combination of these residues (Fig. 1A, panel 2). The triple substitutions were designed to disrupt a critical helix break required for SPP processing (29, 41). The two mutant proteins, A180V and C184V, were fully processed, as judged by their comigration with the processed wild-type (WT) p21 (data not shown). The triple mutant, ASC180/3/4VLV, and the S183L mutant migrated slightly faster than the innate WT p23 but slightly slower than the processed WT p21 on SDS-PAGE (data not shown) (29, 69). Optiprep gradient analyses showed that the A180V and C184V mutants exhibited membrane distribution profiles similar to that of the processed WT core, whereas the S183L and triple mutants exhibited increased distribution to the soluble protein fraction (Fig. 5A). In the membrane protection assay, the A180V and C184V mutants, just like the processed WT core, displayed a higher degree of resistance to protease treatment, while the S183L and triple mutants were more sensitive to proteolysis compared to the WT core (Fig. 5B).
FIG. 5.
Analyses of core proteins with mutations in the signal sequence. (A) Postnuclear fractions of cells expressing the 191 WT core or core mutants were subjected to Optiprep-based subcellular fractionation. Percentages of core proteins partitioned into soluble fractions 16 to 22 of the total core populations are indicated on the right of each panel. (B) Postnuclear fractions obtained from cells expressing various core proteins were assessed by a membrane protection assay (top panel). Blots were scanned for the intensity of the core band. In each case, the relative amounts of the core treated with proteinase K or with detergent and protease relative to that of the core left untreated were assessed. The diagram represents data from three separate experiments with the standard deviation shown (bottom panel).
Coexpression of the 191 core with exogenous SPP.
To confirm that processing of the signal sequence by SPP is required for core envelopment, the 191 core was coexpressed with the WT or D219A mutant SPP, both of which contained the influenza virus HA tag and the ER retrieval signal, KEKK, at the C terminus (50). The WT and mutant SPP proteins were expressed as three forms migrating around a molecular mass of 45 kDa and a dimer migrating at ∼90 kDa was also observed (Fig. 6A, lanes 5 and 6). The top, middle, and bottom bands with an apparent molecular mass of 45 kDa corresponded to SPP containing two, one, and no glycans, respectively (50). The core was predominantly processed when expressed alone or coexpressed with the WT SPP (Fig. 6A, lanes 1, 2, 4, and 5). The core was mainly present as the innate p23 form, and only a small amount of processed p21 was detected when coexpressed with the SPP mutant (Fig. 6A, lanes 3 and 6), indicating that D219A mutant SPP confers dominant-negative interference with endogenous SPP-catalyzed SPP cleavage (50). In addition, a smaller amount of the processed p21 core was detected when the core was coexpressed with the WT SPP compared to that of the processed p21 core expressed alone (Fig. 6A, compare lanes 2 and 5 to lanes 1 and 4, respectively). This result was consistent with a previous observation that SPP-catalyzed cleavage of the core protein may destabilize the viral capsid (69). In the membrane protection assay, the WT and mutant SPP proteins were sensitive to protease digestion (Fig. 6B, top panel, compare lanes 5 and 8 to lanes 4 and 7, respectively); this is because the C terminus of SPP, to which the HA tag was attached, is located in the cytoplasm (72). Coexpression with the WT SPP did not greatly affect the protease resistance of the processed p21 core, whereas coexpression with the mutant SPP rendered the innate p23 core sensitive to proteolysis (Fig. 6B, top and middle panels). Two bands under the core on SDS-PAGE were visible with WT or mutant SPP coexpression (Fig. 6B, top panel, lanes 4, 5, 7, and 8). These two bands were not further processed products of the core but were nonspecific bands due to SPP coexpression. This notion was supported by their appearance in mutant SPP expression alone when a similar proteinase K digestion assay was performed (Fig. 6B, bottom panel, lanes 4 and 5).
FIG. 6.
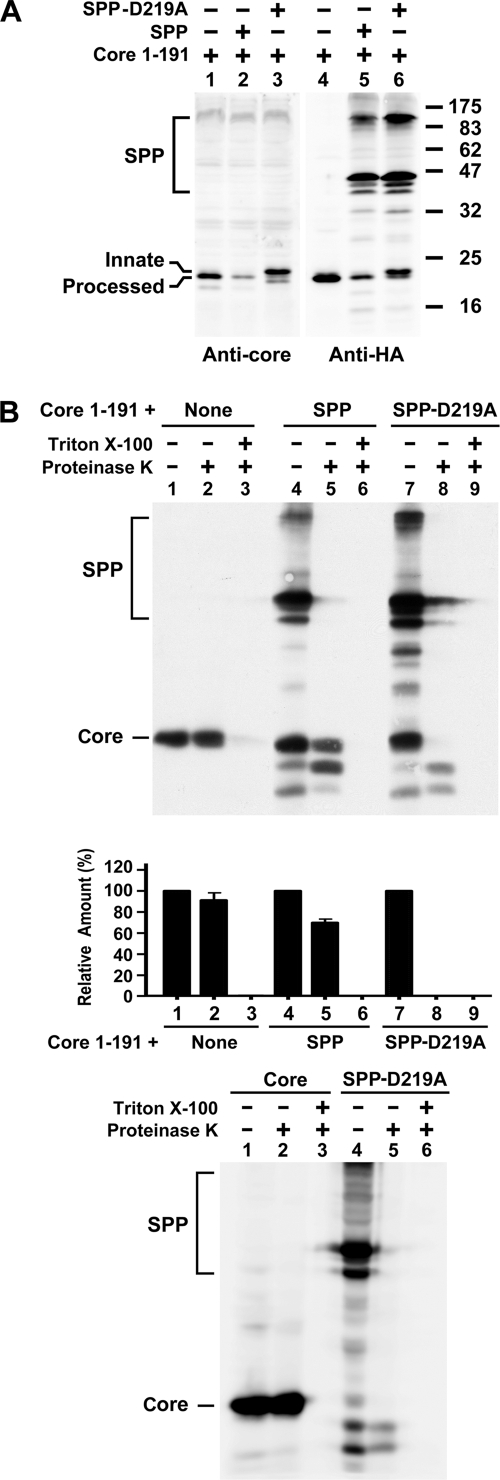
Analyses of the 191 core coexpressed with the WT or D219A mutant SPP. (A) 293T cells were cotransfected with the 191 core plasmid in the presence of pcDNA3, the WT, or D219A mutant SPP plasmid. Cell lysates were analyzed by Western blotting with the rabbit anti-core and HA MAb, respectively. (B) Postnuclear fractions from cells expressing the core alone or coexpressing the core and WT or mutant SPP were subjected to a membrane protection assay, followed by Western blotting with the HA MAb (top panel). The results from three individual studies were quantified, and the average values with the standard deviation are also shown (middle panel). Postnuclear fractions from cells transfected with the 191 core or D219A SPP mutant plasmid were analyzed by a proteinase K digestion assay (bottom panel).
Domain I of the core contains sequences important for core envelopment.
To further understand the molecular basis of core envelopment, the effects of deletions of residues 88 to 106 and residues 42 to 68 in the N-terminal half of the core (Fig. 1A, panel 3) on core envelopment were assessed. These two core mutants still possessed an oligomerization ability, as shown by chemical cross-linking with glutaraldehyde (data not shown), and were able to assemble into high-ordered multimeric complexes, as shown by sucrose linear density gradient centrifugation (Fig. 7A). Since multiple regions in domain I of the core may contribute to core oligomerization (for a review, see reference 65), it is not surprising that these two core N-terminal deletion mutants still formed high-ordered, multimeric complexes. These two mutants also exhibited patterns of association with intracellular membranes similar to those of the 191 core in the Optiprep subcellular fractionation study (Fig. 7B). Coexpression with or without the catalytically inactive D219A mutant SPP showed that these two core mutants were efficiently processed by SPP in cells (Fig. 7C). Nevertheless, a membrane protection assay showed that deletions of these two sequences greatly increased the core's sensitivity to proteinase K digestion (Fig. 7D), indicating that these two mutants were not efficiently enveloped by membranes.
FIG. 7.
Characterization of core mutants with deletions in domain I. (A) Detergent-treated lysates from 293T cells expressing each of the 191 and deletion mutants were subjected to 5 to 50% sucrose linear density gradient centrifugation followed by Western blotting with the HA MAb. (B) Postnuclear fractions obtained from cells expressing each of indicated core proteins were analyzed by Optiprep subcellular fractionation. (C) 293T cells were cotransfected with the core plasmids as indicated in the presence of pcDNA3 or D219A mutant SPP plasmid. Cell lysates were analyzed by Western blotting with the HA MAb. (D) Postnuclear fractions from cells expressing each of the core proteins were subjected to a membrane protection assay, and a representative immunoblot is shown (top panel). The results from three separate analyses were quantified, and the average values with the standard deviation are shown (bottom panel).
Characterization of the core protein expressed by infectious HCV RNAs.
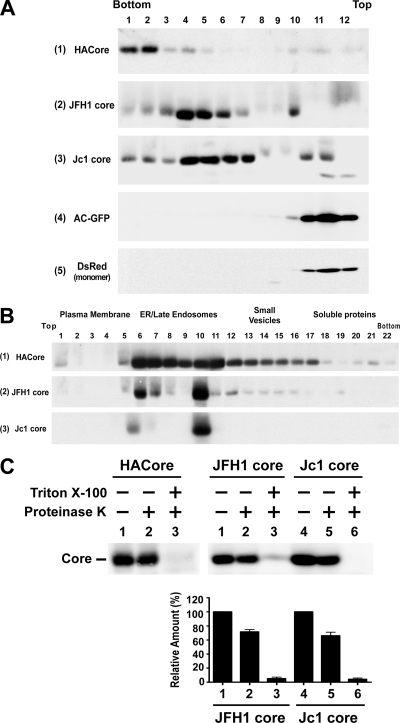
To understand whether core proteins expressed from the HCVcc system also undergo assembly and maturation processes similar to those observed for the 191 recombinant core expressed alone, core proteins expressed from JFH1 and Jc1 RNAs were analyzed in Huh7 cells. The core proteins expressed by the JFH1 and Jc1 clones were mainly located in fractions 4 to 7 (Fig. 8A, panels 2 and 3, respectively), which corresponded to buoyant densities ranging from 1.12 to 1.16 g/ml. These observed densities were close to those of intracellular JFH1 infectious particles reported by other investigators (20) but lighter than that of the recombinant core complex (data not shown). As a control, both Aequorea coerulescens GFP and Discosoma DsRed fluorescence protein, respectively, which are known not to form aggregated proteins, peaked in fraction 11 (Fig. 8A, panels 4 and 5). In the recombinant core expression system, the core may nonselectively enclose cellular RNAs, resulting in a complex with a buoyant density higher than that of authentic HCV particles, in which a single copy of the viral genome is encapsidated. This notion was further supported by the lower buoyant density of 1.13 g/ml of the core complex synthesized in vitro (Fig. 3C), which might incorporate no or a only low level of RNA in the core complex.
FIG. 8.
Characterization of core proteins expressed from infectious HCV RNAs. (A) Huh7 cells were transfected with the 191 pCAGGS-HACore plasmid, JFH1 and Jc1 RNAs, pAcGFP-N1, and pDsRed-monomeric-C1 (Clontech Laboratories, Mountain View, CA), respectively. Transfected cells were lysed with 1% each of NP-40 and sodium deoxycholate, and cell lysates were then subjected to 5 to 50% sucrose density gradient centrifugation, followed by Western blotting with the core MAb (panels 1 to 3), rabbit anti-GFP (panel 4), and MAb for DsRed (panel 5), respectively. (B) Huh7 cells were transfected with the 191 core plasmid, and JFH1 and Jc1 RNAs, respectively, and postnuclear fractions were analyzed by Optiprep-based subcellular fractionation followed by Western blotting with the core MAb. (C) Postnuclear fractions obtained from Huh7 cells transfected with the 191 core plasmid, and JFH1 and Jc1 RNAs, respectively, were analyzed by a membrane protection assay (top panel). The percentages of resistance to proteinase K digestion in the presence or absence of detergent for the JFH1 and Jc1 core proteins were quantified, and the results from four independent studies with the standard deviation are shown (bottom panel).
In the Optiprep subcellular fractionation analysis, the core proteins expressed by the subgenomic system, as well as JFH1 and Jc1 infectious clones, were all predominantly fractionated into the ER/late-endosomal fractions (Fig. 8B). The ability of the HCVcc core complex to float to the lighter ER/late-endosomal membranes indicates that these high-ordered, multimeric complexes are not core aggregates. Unlike the recombinant core which was detected at the plasma membrane, the JFH1 and Jc1 core proteins were only barely detected at the plasma membrane (Fig. 8B). In the membrane protection assay, a significant fraction, ranging between 66 and 72%, of the core synthesized from JFH1 and Jc1 RNAs was still resistant to proteolysis in the absence of detergent, but they were highly sensitive to proteinase K digestion upon detergent treatment (Fig. 8C).
We then examined the intracellular localization of the core expressed by the HCVcc system using markers of intracellular organelles and the endocytic pathway. We did not observe colocalization of the core and calnexin, an ER marker; GM130, a marker of the Golgi apparatus; EEA1, a marker of early endosomes; or LAMP3, a marker of late endosomes (data not shown). Nevertheless, a fraction of the core located in the perinuclear region was associated with the surface of LDs (data not shown).
Assessment of core envelopment by differential membrane permeabilization methods.
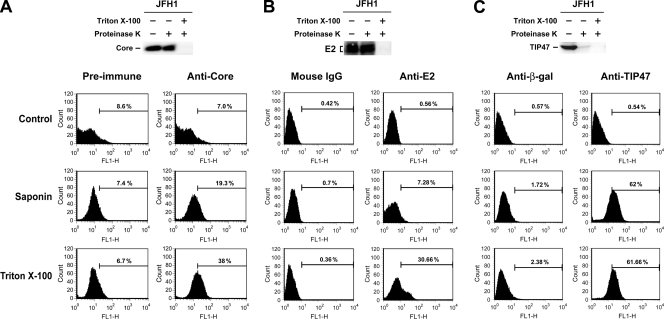
To further address the membrane envelopment property of the HCVcc core, the accessibility of the core's epitopes to a core-specific antibody was assessed in JFH1 RNA-transfected Huh7 cells permeabilized by saponin or Triton X-100, followed by immunostaining and flow cytometry. Permeabilization with saponin moderately increased the accessibility of the core's epitopes to a rabbit anti-core antibody; however, treatment with Triton X-100 greatly exposed the core's epitopes to the anti-core compared to saponin permeabilization (Fig. 9A).
FIG. 9.
Assessment of core envelopment by differential membrane permeabilization. (A) Huh7 cells transfected with JFH1 RNA were incubated with PBS, or permeabilized by saponin or Triton X-100. After blocking, cells from each treatment were separately incubated with 5 μl each of rabbit preimmune or anti-core serum, which was followed by incubation with FITC-conjugated goat anti-rabbit IgG. The immunostained cells were quantified by flow cytometry. The specific levels of anti-core-positive cells revealed by saponin and Triton X-100, designated as S and T values, respectively, were obtained by subtracting the background level with preimmune antibody incubation from those with anti-core incubation. The percentage of core envelopment is defined as [(T − S)]/T × 100%. Two separate analyses were performed with similar results. The data from a representative set are thus shown. The postnuclear fraction from the same experiment was also subjected to the membrane protection assay. (B and C) JFH1 RNA-transfected Huh7 cells were divided into two parts for the studies as shown. In each case, a portion of cells was prepared for postnuclear fractions which were then subjected to the HCV E2 protein (B) and TIP47 (C) membrane protection assays, respectively. Another portion of cells was further divided into three parts for the differential membrane permeabilization assay as described in panel A. Cells from each treatment were then separately incubated with 1 μg each of affinity-purified control mouse IgG or an E2 MAb (B) or with 1 μg each of goat anti-β-Gal or goat anti-TIP47 IgG (C) followed by incubation with FITC-linked rabbit anti-mouse (B) or rabbit anti-goat IgGs (C). The immunostained cells were then analyzed by FACS. In panels B and C, three independent analyses were performed with similar results. Thus, data from a representative set are shown.
We further verified that saponin permeabilized only the plasma membrane, which exposes epitopes on the core that are membrane-free or located at the LD surface, while Triton X-100 permeabilizes plasma membrane and intracellular membrane vesicles, which also exposes epitopes on the membrane-enveloped core as well. To do this, the accessibility of epitopes on HCV E2 and TIP47 to their respective antibodies was assessed. As expected, E2 was largely translocated into the ER lumen, as judged by its resistance to proteinase K digestion (Fig. 9B). Permeabilization with Triton X-100 greatly increased the levels of anti-E2-positive cells compared to permeabilization with saponin (Fig. 9B). TIP47 is known to move from the cytosol to coat nascent LDs during rapid fat storage and thus plays a critical role in packaging fat into LDs (for reviews, see references 19 and 74). TIP47 was completely digested by proteinase K even in the absence of detergent (Fig. 9C). Treatment with saponin and Triton X-100 showed a similar level of anti-TIP47-positive cells (Fig. 9C). These observations are consistent with the nonenveloped nature of TIP47.
Based on these results, we defined the degree of core envelopment using this differential permeabilization approach as the percentage of the differential level of the core that was specifically exposed by Triton X-100, but not by saponin, relative to the total core level exposed by Triton X-100 (also refer to the legend to Fig. 9A for details). Thus, the percentage of core envelopment shown in Fig. 9A was 62%. As controls, results from three independent studies showed that the extents of E2 envelopment as determined by proteinase K digestion and differential membrane permeabilization methods were 82.4% ± 5.1% and 80.3% ± 9.5%, respectively, while those of TIP47 were negligible by both assays.
Concomitant inhibition of core envelopment and infectious virus production by (Z-LL)2-ketone and D219A mutant SPP coexpression.
To provide evidence for the involvement of membrane envelopment in HCV replication, (Z-LL)2-ketone was added to Huh7 cells transfected with JFH1 RNA. This SPP inhibitor reduced the core's resistance to proteolysis to 32.7% compared to 79.8% of the core without inhibitor treatment (Fig. 10A, top and middle panels). When the culture supernatants were titrated to determine viral infectivity, the SPP inhibitor decreased the titer of virus released into the culture medium to 49.7% of that without inhibitor treatment (Fig. 10A, bottom panel).
FIG. 10.
Analysis of the effects of (Z-LL)2-ketone and D219A mutant SPP on core envelopment and infectious virus production. (A) Huh7 cells were first transfected with 10 μg of JFH1 RNA, and at 16 h posttransfection the cell cultures were supplemented with or without 20 μM (Z-LL)2-ketone. At 64 h after the beginning of transfection, the cells were harvested, and postnuclear fractions were subjected to a proteinase K digestion assay. The diagram represents results from three independent studies with the standard deviation shown (top and middle panels). The viral infectivity of cell-free supernatants obtained was determined as focus-forming units (FFU) per milliliter, and the results from three independent studies with the standard deviation are shown (bottom panel). (B) JFH1 RNA-transfected Huh7 cells were treated with or without (Z-LL)2-ketone, and transfected cells were then analyzed by differential membrane permeabilization, followed by FACS. The data from a representative set are shown. The degree of core envelopment in the presence or absence of the SPP inhibitor was calculated as described in Fig. 9A. The diagram represents results from three independent studies with the standard deviation shown. (C) Huh7 cells were first transfected with 10 μg of JFH1 RNA and 16 h posttransfection cells were separately transfected with 10 μg of the vector, marked as “Mock”, or the D219A mutant SPP plasmid. At 64 h after the beginning of transfection, postnuclear fractions were prepared and subjected to a proteinase K digestion assay (top and middle panel), and culture supernatants were titrated for viral infectivity (bottom panel). In each case, the diagram represents results from three independent studies with the standard deviation shown. Differences between samples treated with (Z-LL)2-ketone or cotransfected with D219A plasmid and controls were determined by using the Student t test: P < 0.05 (A and C) and P = 0.0001 (B).
To confirm that treatment with (Z-LL)2-ketone decreases core's envelopment, the accessibility of epitopes to the anti-core for the HCVcc core expressed in the presence or absence of the SPP inhibitor was examined by a differential membrane permeabilization assay. In cells treated with (Z-LL)2-ketone, permeabilization with saponin showed a higher level of anti-core-positive cells compared to cells without the inhibitor; however, permeabilization with Triton X-100 showed similar levels of anti-core-positive cells regardless of the presence or absence of the SPP inhibitor (Fig. 10B). These observations indicated that treatment with (Z-LL)2-ketone increases the percentage of unenveloped core in total core population. The results from three independent studies showed that treatment with the SPP inhibitor reduced core envelopment to 38% as opposed to 63.6% of the core envelopment when core was synthesized without (Z-LL)2-ketone (the diagram shown in Fig. 10B).
Next, we confirmed the role of core envelopment in infectious virus production using the D219A mutant SPP. Coexpression of the D219A mutant SPP not only decreased the core's resistance to proteolysis to 27% compared to 65.7% of the core without mutant SPP coexpression (Fig. 10C, top and middle panels), it also suppressed the production of infectious virus to 33.8% of that of the virus produced in the absence of mutant SPP coexpression (Fig. 10C, bottom panel).
In these studies, the core proteins expressed in the presence or absence of (Z-LL)2-ketone or coexpressed with or without the D219A mutant SPP migrated indistinguishably on SDS-PAGE, suggesting that the SPP inhibitor and D219A mutant SPP may inhibit core processing at a level which is below the detection sensitivity of SDS-PAGE in this infectious virus system. Since HCVcc core proteins with various mutations designed to disrupt the SPP cleavage site in the signal sequence were still efficiently processed and even (Z-LL)2-ketone only partially inhibited the processing of these mutants, Targett-Adams et al. (67) therefore speculated that the HCV core-E1 signal peptide is an extremely efficient substrate for SPP. Okamoto et al. (49) recently showed that treatment with L685,458, another SPP inhibitor, resulted in production of the unprocessed core and the processed form in a Huh7-derived Huh7OK1 cell clone. It is not clear whether different Huh7 clones can cause differences in the efficiency of blocking SPP activity. Because presenilins, which belong to the same aspartic protease family as SPP (72), are able to cleave the β-amyloid precursor protein at multiple sites (51, 58), it is also likely that SPP can cleave the core at multiple sites in the HCVcc system, which thus complicates discrimination of the unprocessed core from the processed one. Alternatively, the possibility that the SDS-PAGE system used may not truly differentiate the unprocessed and processed core proteins expressed by the HCVcc system cannot be completely ruled out despite its ability to differentiate the two forms of the recombinant core.
DISCUSSION
Thus far, mechanistic insights into the consecutive steps of core multimerization and its interaction with membranes during the early phase of HCV morphogenesis are still poorly understood. Several groups have reported a complex intracellular localization of core protein; it predominantly resides in the cytoplasm, either in association with the ER, LDs, or mitochondria or in the nucleus (for reviews, see references 4 and 66). When signal sequence cleavage is inhibited by mutations in the signal sequence or at the signal peptide cleavage site between the core and E1, the innate p23 is retained in ER membranes (29, 41). Nevertheless, we showed that the presence and appropriate processing of the signal anchor by SPP are critical for retention of the recombinant core on ER/late-endosomal membranes (Fig. 5A and data not shown). In line with this notion, it was reported that substitutions of Leu-139, Val-140, and Leu-144 in the core with Ala residues inhibit SPP processing and prevent the core from attaching to ER membranes (50).
In the present study, we provide several lines of evidence to support the notion that the core is assembled into oligomeric complexes before or upon binding to ER/late endosome membranes. This includes (i) cofractionation of the innate 191 core with the processed core to the ER/late endosomes and small vesicles in the presence of (Z-LL)2-ketone (Fig. 4A); (ii) inhibition of the core's association with membranes, but not of multimeric complex formation, by deletion of the signal sequence or inhibition of core processing by an SPP-specific inhibitor (Fig. 4 and data not shown); (iii) the ability of the isolated multimeric 191 and 173 core complexes to bind to membranes in vitro (Fig. 3A); and (iv) formation of a high-ordered, multimeric complex of core translated in vitro (Fig. 3C). On the other hand, proteinase K digestion of the recombinant core synthesized in the presence of (Z-LL)2-ketone exhibited both protease-sensitive and -resistant complexes, both of which contained innate and processed core proteins (Fig. 4B), implying that envelopment of the core occurs after intramembrane cleavage. If intramembrane processing occurred after membrane envelopment, it would be expected that all of the innate p23 core molecules would be sensitive, whereas all of the processed p21 core molecules would be resistant to protease treatment.
It is thought that the HCV particles assemble and bud into the ER lumen and are transported via exocytosis through the Golgi apparatus to the plasma membrane, where the membrane of vesicles is fused with the plasma membrane to release virions into the extracellular milieu. Nevertheless, this proposition has not been verified in the HCVcc system. The recombinant core may ultimately be routed and attach to the plasma membrane without further fission or secretion, resulting in a greater tendency of a recombinant core to be located at plasma membranes than would an HCVcc core (Fig. 8B). Similar to the HCVcc core which targets the surface of LDs (data not shown), where the core recruits viral NS proteins and replication complexes (40, 43), a significant fraction of the recombinant 191 core and the majority of cytoplasmic 173 core were also located on LDs (data not shown). These results indicate a critical role of LDs in the maturation process of the recombinant core as well. Rouillé et al. previously noted that the core expressed in the context of the infectious cycle is not localized in well-defined intracellular compartments such as the ER or the Golgi complex or the intermediate compartment, nor is it associated with markers of the endocytic pathway such as EEA1 or LAMP1 (56). We did not observe colocalization of the HCVcc core with markers of ER, the Golgi complex, or early or late endosomes by confocal microscopy (data not shown) either. These findings are in contrast with the general belief that the HCV core interacts with the ER and that the core protein of the genotype 1b BK strain expressed by stably transfected cells is localized in the ER, intermediate compartment, and cis-Golgi complex region (38). Rouillé et al. proposed that this disparity can be attributed to the rapid transfer of the core from the ER to LDs in infected cells or to the differential levels of LDs in various cell types used, since the latter factor may also affect different extents to which the core is associated with the ER or LDs (56).
Importantly, we provide compelling evidence to show that the recombinant and HCVcc core proteins are indeed enveloped by a membrane. Besides the conventional membrane protection assay that is generally used to assess enclosure of a protein by membranes, we performed coimmunoprecipitation with specific antibodies to detect envelopment of the recombinant core (Fig. 2C). We also established a differential membrane permeabilization assay to differentiate the HCVcc core population that is membrane-enveloped from those that remain membrane-free or are localized at the surface of LDs (Fig. 9A). The fidelity of this method was further validated by examining the HCV E2 protein (Fig. 9B) and an LD-associated protein, TIP47 (Fig. 9C). Moreover, we showed that treatment with the SPP inhibitor decreased membrane envelopment of the HCVcc core using both proteinase K digestion and differential membrane permeabilization assays (Fig. 10A and B). Remarkably, the extent of core envelopment as determined by the differential membrane permeabilization assay was similar to that assessed by the membrane protection assay regardless of the addition of SPP inhibitor or not (compare Fig. 8C and 10A to Fig. 9A and 10B, respectively). All of these results confirmed that a significant portion of the membrane-enveloped core, which is not localized at the LD surface or in the vicinity of LDs (data not shown), is present inside the cytoplasmic membranous compartments or vesicles.
Just like the recombinant core, the core expressed by Huh7 cells from infectious JFH1 and Jc1 RNAs formed a high-ordered, multimeric complex, was mainly associated with ER/late-endosomal membranes, and was enveloped by membranes (Fig. 8). Both the SPP inhibitor and the D219A mutant SPP interfered with the production of infectious HCV production as well as core envelopment (Fig. 10A and C), linking a function of core envelopment to HCV maturation and morphogenesis. This finding is in line with our subgenomic core analyses which showed that processing of the core by SPP is required for core envelopment and with recent results from other researchers that SPP-mediated core processing is required for HCV propagation (49, 67). Our results also underline the more-profound effect of the SPP inhibitor and mutant SPP on inhibition of core envelopment and infectious virus production than its seemingly unnoticed effect on retarding the mobility of the core on SDS-PAGE. These observations collectively imply that during the early phase of core assembly and budding, the recombinant core expressed alone undergoes steps similar to those utilized by the HCVcc core.
The envelopment of HCV nucleocapsids is thought to be achieved by interactions at the ER membrane between the first hydrophobic domain of E1 and the C-terminal hydrophobic domain of the core protein (33, 36). Nevertheless, our subgenomic core and infectious RNA clone studies showed that the HCV core protein, which constitutes the budding apparatus of the virus, can direct the core's assembly into a high-ordered multimeric complex and envelopment by membranes without the help of other viral proteins. It is likely that once it is folded and assembled, the core multimeric complex may possess the intrinsic capacity to assemble laterally into an isometric lattice on ER membranes. The core complex may be sufficient to introduce the membrane curvature needed to form a bud and may trigger the fission reaction required for pinching off membrane vesicles. Deletion of the E1 and E2 open reading frame was found to result in no production of infectious viral particles into the culture medium (70), indicating that E1 and E2 are required for the release of infectious viral particles into the medium. By taking our results and this observation into consideration, we hypothesize that the core itself is sufficient for assembly into VLPs and budding, likely into the lumen of ER. Nevertheless, these VLPs are incapable of being released into the culture fluid without acquiring E1 and E2 proteins on their viral envelope.
On the other hand, we showed that the association with membranes and processing of core signal sequences are required but not sufficient for envelopment of the recombinant core and that sequences in domain I also play crucial roles in core envelopment (Fig. 7). The core protein binds to a variety of cellular proteins and modulate signaling pathways, cell proliferation, cellular and viral gene expressions, cell transformation, apoptosis, and lipid metabolism (for reviews, see references 21, 30, and 68). In particular, the binding sites in the core of many cellular proteins have been mapped to domain I (for a review, see reference 40). It is likely that cellular proteins are involved in the nucleocapsid envelopment process. The differential protease-resistance of core complexes expressed in cells and in vitro (Fig. 2) supports this notion.
The results presented here using a subgenomic core and HCVcc expression systems provide a model to delineate the sequential and coordinated events of core complex formation and interactions with and envelopment by membranes during the core assembly process (Fig. 11). The core is first synthesized in the cytoplasm, and forms oligomeric structures prior to or upon attaching to the cytoplasmic side of ER membranes. On the ER membrane, the core is processed by SPP to yield the mature core. Once the signal sequence is removed, the processed core complex is subsequently enveloped into the lumen of the ER. In particular, core envelopment, a critical step in the infectious virus life cycle, has not previously been identified. An understanding of the mode of core assembly into multimeric complexes and interactions with and envelopment by membranes will not only provide better insights into nucleocapsid maturation and virus assembly and budding but may also reveal potential viral and/or cellular targets with antiviral implications.
FIG. 11.
Model for the concerted events of core interaction with and envelopment by ER membranes. The 1-191 core is synthesized in the cytoplasm and assembled into an oligomeric structure before or upon attachment to the cytoplasmic side of the ER membrane via the core C-terminal signal peptide. The core oligomers may continue to multimerize into a high-ordered, structural complex on the cytoplasmic surface of the ER membrane. The signal peptide is then cleaved off by intramembrane SPP, which is subsequently followed by envelopment of core complex into the lumen of ER membranes. The SPP inhibitor, (Z-LL)2-ketone, interferes with both signal sequence processing and attachment of the core complex to the ER membrane.
Acknowledgments
We thank Yi-Miao Chen for technical assistance. We are also indebted to Charles M. Rice (Rockefeller University, New York, NY) for kindly providing pBRTM-HCV-H1-3011, the Jc1 construct, and the NS5A MAb (clone 9E10) and to Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan) for generously providing pUC-JFH1. pEGFP-C3-derived GFP-Cellubrevin and GFP-TI-VAMP plasmids were generous gifts from Thierry Galli (Institut du Fer-à-Moulin, Paris, France). pcDNA3-SPP-HA and pcDNA3-SPP(D219A)-HA were kindly provided by Kohji Moriishi (Osaka University, Osaka, Japan).
This study was supported by a grant from the National Science Council (NSC95-2320-B-001-032-MY3), a Genomic and Proteomic Research grant (94M001-2), a Foresight Research grant (AS97FP-L15-1 and AS98FP-L15-1), and a Genomic Research grant (Summit Project III) from Academia Sinica, Taipei, Taiwan, R.O.C.
Footnotes
Published ahead of print on 15 July 2009.
REFERENCES
- 1.Acosta-Rivero, N., A. Musacchio, L. Lorenzo, C. Alvarez, and J. Morales. 2002. Processing of the hepatitis C virus precursor protein expressed in the methylotrophic yeast Pichia pastoris. Biochem. Biophys. Res. Commun. 295:81-84. [DOI] [PubMed] [Google Scholar]
- 2.Acosta-Rivero, N., A. Rodriguez, A. Musacchio, V. Falcon, V. M. Suarez, G. Martinez, I. Guerra, D. Paz-Lago, Y. Morera, M. C. de la Rosa, J. Morales-Grillo, and S. Duenas-Carrera. 2004. In vitro assembly into virus-like particles is an intrinsic quality of Pichia pastoris derived HCV core protein. Biochem. Biophys. Res. Commun. 325:68-74. [DOI] [PubMed] [Google Scholar]
- 3.Barba, G., F. Harper, T. Harada, M. Kohara, S. Goulinet, Y. Matsuura, G. Eder, Z. Schaff, M. J. Chapman, T. Miyamura, and C. Brechot. 1997. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc. Natl. Acad. Sci. USA 94:1200-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartenschlager, R., M. Frese, and T. Pietschmann. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63:71-180. [DOI] [PubMed] [Google Scholar]
- 5.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 6.Baumert, T. F., S. Ito, D. T. Wong, and T. J. Liang. 1998. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J. Virol. 72:3827-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanchard, E., D. Brand, S. Trassard, A. Goudeau, and P. Roingeard. 2002. Hepatitis C virus-like particle morphogenesis. J. Virol. 76:4073-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchard, E., C. Hourioux, D. Brand, M. Ait-Goughoulte, A. Moreau, S. Trassard, P. Y. Sizaret, F. Dubois, and P. Roingeard. 2003. Hepatitis C virus-like particle budding: role of the core protein and importance of its Asp111. J. Virol. 77:10131-10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 10.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boulant, S., R. Montserret, R. G. Hope, M. Ratinier, P. Targett-Adams, J. P. Lavergne, F. Penin, and J. McLauchlan. 2006. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J. Biol. Chem. 281:22236-22247. [DOI] [PubMed] [Google Scholar]
- 12.Boulant, S., C. Vanbelle, C. Ebel, F. Penin, and J. P. Lavergne. 2005. Hepatitis C virus core protein is a dimeric alpha-helical protein exhibiting membrane protein features. J. Virol. 79:11353-11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan, W. E., C. K. Chuang, S. H. Yeh, M. S. Chang, and S. S. Chen. 2006. Functional characterization of heptad repeat 1 and 2 mutants of the spike protein of the severe acute respiratory syndrome coronavirus. J. Virol. 80:3225-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang, S. C., J. H. Yen, H. Y. Kang, M. H. Jang, and M. F. Chang. 1994. Nuclear localization signals in the core protein of hepatitis C virus. Biochem. Biophys. Res. Commun. 205:1284-1290. [DOI] [PubMed] [Google Scholar]
- 15.Chen, S. S., S. F. Lee, and C. T. Wang. 2001. Cellular membrane-binding ability of the C-terminal cytoplasmic domain of human immunodeficiency virus type 1 envelope transmembrane protein gp41. J. Virol. 75:9925-9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chong, L. D., and J. K. Rose. 1993. Membrane association of functional vesicular stomatitis virus matrix protein in vivo. J. Virol. 67:407-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daro, E., P. van der Sluijs, T. Galli, and I. Mellman. 1996. Rab4 and cellubrevin define different early endosome populations on the pathway of transferrin receptor recycling. Proc. Natl. Acad. Sci. USA 93:9559-9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ezelle, H. J., D. Markovic, and G. N. Barber. 2002. Generation of hepatitis C virus-like particles by use of a recombinant vesicular stomatitis virus vector. J. Virol. 76:12325-12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujimoto, T., Y. Ohsaki, J. Cheng, M. Suzuki, and Y. Shinohara. 2008. Lipid droplets: a classic organelle with new outfits. Histochem. Cell Biol. 130:263-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gastaminza, P., S. B. Kapadia, and F. V. Chisari. 2006. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J. Virol. 80:11074-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giannini, C., and C. Bréchot. 2003. Hepatitis C virus biology. Cell Death Differ. 10(Suppl. 1):S27-S38. [DOI] [PubMed] [Google Scholar]
- 22.Grigorov, B., F. Arcanger, P. Roingeard, J. L. Darlix, and D. Muriaux. 2006. Assembly of infectious HIV-1 in human epithelial and T-lymphoblastic cell lines. J. Mol. Biol. 359:848-862. [DOI] [PubMed] [Google Scholar]
- 23.Hope, R. G., D. J. Murphy, and J. McLauchlan. 2002. The domains required to direct core proteins of hepatitis C virus and GB virus-B to lipid droplets share common features with plant oleosin proteins. J. Biol. Chem. 277:4261-4270. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein, K. C., S. J. Polyak, and J. R. Lingappa. 2004. Unique features of hepatitis C virus capsid formation revealed by de novo cell-free assembly. J. Virol. 78:9257-9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolesnikova, L., S. Bamberg, B. Berghofer, and S. Becker. 2004. The matrix protein of Marburg virus is transported to the plasma membrane along cellular membranes: exploiting the retrograde late endosomal pathway. J. Virol. 78:2382-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kotov, A., J. Zhou, P. Flicker, and C. Aiken. 1999. Association of Nef with the human immunodeficiency virus type 1 core. J. Virol. 73:8824-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunkel, M., M. Lorinczi, R. Rijnbrand, S. M. Lemon, and S. J. Watowich. 2001. Self-assembly of nucleocapsid-like particles from recombinant hepatitis C virus core protein. J. Virol. 75:2119-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemberg, M. K., and B. Martoglio. 2002. Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol. Cell 10:735-744. [DOI] [PubMed] [Google Scholar]
- 30.Levrero, M. 2006. Viral hepatitis and liver cancer: the case of hepatitis C. Oncogene 25:3834-3847. [DOI] [PubMed] [Google Scholar]
- 31.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 32.Liu, Q., C. Tackney, R. A. Bhat, A. M. Prince, and P. Zhang. 1997. Regulated processing of hepatitis C virus core protein is linked to subcellular localization. J. Virol. 71:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lo, S., M. Selby, and J. Ou. 1996. Interaction between hepatitis C virus core protein and E1 envelope protein. J. Virol. 70:5177-5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo, S. Y., F. Masiarz, S. B. Hwang, M. M. Lai, and J. H. Ou. 1995. Differential subcellular localization of hepatitis C virus core gene products. Virology 213:455-461. [DOI] [PubMed] [Google Scholar]
- 35.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 36.Ma, H. C., C. H. Ke, T. Y. Hsieh, and S. Y. Lo. 2002. The first hydrophobic domain of the hepatitis C virus E1 protein is important for interaction with the capsid protein. J. Gen. Virol. 83:3085-3092. [DOI] [PubMed] [Google Scholar]
- 37.Martinez-Arca, S., R. Rudge, M. Vacca, G. Raposo, J. Camonis, V. Proux-Gillardeaux, L. Daviet, E. Formstecher, A. Hamburger, F. Filippini, M. D'Esposito, and T. Galli. 2003. A dual mechanism controlling the localization and function of exocytic v-SNAREs. Proc. Natl. Acad. Sci. USA 100:9011-9016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martire, G., A. Viola, L. Iodice, L. V. Lotti, R. Gradini, and S. Bonatti. 2001. Hepatitis C virus structural proteins reside in the endoplasmic reticulum as well as in the intermediate compartment/cis-Golgi complex region of stably transfected cells. Virology 280:176-182. [DOI] [PubMed] [Google Scholar]
- 39.Matsumoto, M., S. B. Hwang, K. S. Jeng, N. Zhu, and M. M. Lai. 1996. Homotypic interaction and multimerization of hepatitis C virus core protein. Virology 218:43-51. [DOI] [PubMed] [Google Scholar]
- 40.McLauchlan, J. 2000. Properties of the hepatitis C virus core protein: a structural protein that modulates cellular processes. J. Viral. Hepat. 7:2-14. [DOI] [PubMed] [Google Scholar]
- 41.McLauchlan, J., M. K. Lemberg, G. Hope, and B. Martoglio. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21:3980-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller, R. H., and R. H. Purcell. 1990. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc. Natl. Acad. Sci. USA 87:2057-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089-1097. [DOI] [PubMed] [Google Scholar]
- 44.Moradpour, D., C. Englert, T. Wakita, and J. R. Wands. 1996. Characterization of cell lines allowing tightly regulated expression of hepatitis C virus core protein. Virology 222:51-63. [DOI] [PubMed] [Google Scholar]
- 45.Moradpour, D., F. Penin, and C. M. Rice. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5:453-463. [DOI] [PubMed] [Google Scholar]
- 46.Moriishi, K., T. Okabayashi, K. Nakai, K. Moriya, K. Koike, S. Murata, T. Chiba, K. Tanaka, R. Suzuki, T. Suzuki, T. Miyamura, and Y. Matsuura. 2003. Proteasome activator PA28γ-dependent nuclear retention and degradation of hepatitis C virus core protein. J. Virol. 77:10237-10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murphy, D. J. 2001. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog. Lipid Res. 40:325-438. [DOI] [PubMed] [Google Scholar]
- 48.Nakai, K., T. Okamoto, T. Kimura-Someya, K. Ishii, C. K. Lim, H. Tani, E. Matsuo, T. Abe, Y. Mori, T. Suzuki, T. Miyamura, J. H. Nunberg, K. Moriishi, and Y. Matsuura. 2006. Oligomerization of hepatitis C virus core protein is crucial for interaction with the cytoplasmic domain of E1 envelope protein. J. Virol. 80:11265-11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okamoto, K., Y. Mori, Y. Komoda, T. Okamoto, M. Okochi, M. Takeda, T. Suzuki, K. Moriishi, and Y. Matsuura. 2008. Intramembrane processing by signal peptide peptidase regulates the membrane localization of hepatitis C virus core protein and viral propagation. J. Virol. 82:8349-8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okamoto, K., K. Moriishi, T. Miyamura, and Y. Matsuura. 2004. Intramembrane proteolysis and endoplasmic reticulum retention of hepatitis C virus core protein. J. Virol. 78:6370-6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okochi, M., S. Eimer, A. Bottcher, R. Baumeister, H. Romig, J. Walter, A. Capell, H. Steiner, and C. Haass. 2000. A loss of function mutant of the presenilin homologue SEL-12 undergoes aberrant endoproteolysis in Caenorhabditis elegans and increases Aβ42 generation in human cells. J. Biol. Chem. 275:40925-40932. [DOI] [PubMed] [Google Scholar]
- 52.Packter, N. M., and E. R. Olukoshi. 1995. Ultrastructural studies of neutral lipid localization in Streptomyces. Arch. Microbiol. 164:420-427. [DOI] [PubMed] [Google Scholar]
- 53.Pawlotsky, J. M. 2004. Pathophysiology of hepatitis C virus infection and related liver disease. Trends Microbiol. 12:96-102. [DOI] [PubMed] [Google Scholar]
- 54.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pietschmann, T., V. Lohmann, A. Kaul, N. Krieger, G. Rinck, G. Rutter, D. Strand, and R. Bartenschlager. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol. 76:4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rouille, Y., F. Helle, D. Delgrange, P. Roingeard, C. Voisset, E. Blanchard, S. Belouzard, J. McKeating, A. H. Patel, G. Maertens, T. Wakita, C. Wychowski, and J. Dubuisson. 2006. Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J. Virol. 80:2832-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santolini, E., G. Migliaccio, and N. La Monica. 1994. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J. Virol. 68:3631-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sastre, M., H. Steiner, K. Fuchs, A. Capell, G. Multhaup, M. M. Condron, D. B. Teplow, and C. Haass. 2001. Presenilin-dependent gamma-secretase processing of beta-amyloid precursor protein at a site corresponding to the S3 cleavage of Notch. EMBO Rep. 2:835-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shavinskaya, A., S. Boulant, F. Penin, J. McLauchlan, and R. Bartenschlager. 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 282:37158-37169. [DOI] [PubMed] [Google Scholar]
- 60.Shirakura, M., K. Murakami, T. Ichimura, R. Suzuki, T. Shimoji, K. Fukuda, K. Abe, S. Sato, M. Fukasawa, Y. Yamakawa, M. Nishijima, K. Moriishi, Y. Matsuura, T. Wakita, T. Suzuki, P. M. Howley, T. Miyamura, and I. Shoji. 2007. E6AP ubiquitin ligase mediates ubiquitylation and degradation of hepatitis C virus core protein. J. Virol. 81:1174-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spearman, P., J.-J. Wang, N. Y. Heyden, and L. Ratner. 1994. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J. Virol. 68:3232-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suzuki, R., Y. Matsuura, T. Suzuki, A. Ando, J. Chiba, S. Harada, I. Saito, and T. Miyamura. 1995. Nuclear localization of the truncated hepatitis C virus core protein with its hydrophobic C terminus deleted. J. Gen. Virol. 76:53-61. [DOI] [PubMed] [Google Scholar]
- 63.Suzuki, R., S. Sakamoto, T. Tsutsumi, A. Rikimaru, K. Tanaka, T. Shimoike, K. Moriishi, T. Iwasaki, K. Mizumoto, Y. Matsuura, T. Miyamura, and T. Suzuki. 2005. Molecular determinants for subcellular localization of hepatitis C virus core protein. J. Virol. 79:1271-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suzuki, R., K. Tamura, J. Li, K. Ishii, Y. Matsuura, T. Miyamura, and T. Suzuki. 2001. Ubiquitin-mediated degradation of hepatitis C virus core protein is regulated by processing at its carboxyl terminus. Virology 280:301-309. [DOI] [PubMed] [Google Scholar]
- 65.Suzuki, T., H. Aizaki, K. Murakami, I. Shoji, and T. Wakita. 2007. Molecular biology of hepatitis C virus. J. Gastroenterol. 42:411-423. [DOI] [PubMed] [Google Scholar]
- 66.Suzuki, T., K. Ishii, H. Aizaki, and T. Wakita. 2007. Hepatitis C viral life cycle. Adv. Drug Deliv. Rev. 59:1200-1212. [DOI] [PubMed] [Google Scholar]
- 67.Targett-Adams, P., G. Hope, S. Boulant, and J. McLauchlan. 2008. Maturation of hepatitis C virus core protein by signal peptide peptidase is required for virus production. J. Biol. Chem. 283:16850-16859. [DOI] [PubMed] [Google Scholar]
- 68.Tellinghuisen, T. L., and C. M. Rice. 2002. Interaction between hepatitis C virus proteins and host cell factors. Curr. Opin. Microbiol. 5:419-427. [DOI] [PubMed] [Google Scholar]
- 69.Vauloup-Fellous, C., V. Pene, J. Garaud-Aunis, F. Harper, S. Bardin, Y. Suire, E. Pichard, A. Schmitt, P. Sogni, G. Pierron, P. Briand, and A. R. Rosenberg. 2006. Signal peptide peptidase-catalyzed cleavage of hepatitis C virus core protein is dispensable for virus budding but destabilizes the viral capsid. J. Biol. Chem. 281:27679-27692. [DOI] [PubMed] [Google Scholar]
- 70.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Watashi, K., and K. Shimotohno. 2003. The roles of hepatitis C virus proteins in modulation of cellular functions: a novel action mechanism of the HCV core protein on gene regulation by nuclear hormone receptors. Cancer Sci. 94:937-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weihofen, A., K. Binns, M. K. Lemberg, K. Ashman, and B. Martoglio. 2002. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science 296:2215-2218. [DOI] [PubMed] [Google Scholar]
- 73.Weihofen, A., M. K. Lemberg, H. L. Ploegh, M. Bogyo, and B. Martoglio. 2000. Release of signal peptide fragments into the cytosol requires cleavage in the transmembrane region by a protease activity that is specifically blocked by a novel cysteine protease inhibitor. J. Biol. Chem. 275:30951-30956. [DOI] [PubMed] [Google Scholar]
- 74.Wolins, N. E., D. L. Brasaemle, and P. E. Bickel. 2006. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 580:5484-5491. [DOI] [PubMed] [Google Scholar]
- 75.Wyma, D. J., A. Kotov, and C. Aiken. 2000. Evidence for a stable interaction of gp41 with Pr55Gag in immature human immunodeficiency virus type 1 particles. J. Virol. 74:9381-9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xia, W., and M. S. Wolfe. 2003. Intramembrane proteolysis by presenilin and presenilin-like proteases. J. Cell Sci. 116:2839-2844. [DOI] [PubMed] [Google Scholar]
- 77.Yasui, K., T. Wakita, K. Tsukiyama-Kohara, S. I. Funahashi, M. Ichikawa, T. Kajita, D. Moradpour, J. R. Wands, and M. Kohara. 1998. The native form and maturation process of hepatitis C virus core protein. J. Virol. 72:6048-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]