Abstract
The E6 oncoproteins from high-risk mucosotrophic human papillomaviruses (HPVs) target a range of cellular proteins for proteasome-mediated degradation. Apart from the tumor suppressor p53 and proapoptotic Bcl-2 family member Bak, many targets contain class 1 PDZ domains and are involved in cell junction stability and signaling. The targeting mechanism is considered to function by the E6 protein acting as an adaptor molecule linking a cellular ubiquitin ligase to the target protein. In each case, whether the target is the p53 tumor suppressor or a member of the group of PDZ domain-containing targets, this mechanism relies on a direct interaction between E6 and its cellular target. This study focuses on the impact of the HPV type 18 (HPV-18) E6*I protein on the stability of Akt, Dlg, MAGI-1, MAGI-2, and Scribble. We show that HPV-18 E6* expression can downregulate the expression levels of Akt, Dlg, and Scribble in the absence of full-length HPV-18 E6 protein. The reduction in Dlg levels by E6* is independent of transcription and does not require a direct interaction between the two proteins although the proteasome pathway is involved. Further, we provide evidence that activation of certain signal transduction pathways has a profound effect on the targeting of Dlg by E6* and suggest that high-risk HPV E6 oncoproteins can target certain substrates both directly and indirectly through the E6* proteins and may cooperate in their degradation.
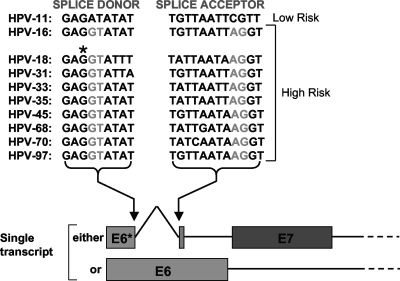
A common feature of the early transcripts of many high-risk mucosotropic human papillomaviruses (HPVs), as opposed to the low-risk types, is the pattern of splicing observed for early transcripts that encode the two principle oncoproteins E6 and E7. A survey of high-risk alpha group HPVs shows that all have a highly conserved splice donor site within the body of the E6 open reading frame (ORF), and most, though not all, also have a conserved splice acceptor site falling within the E6 ORF that lies before the start of E7 and leads to removal of a small intron within the E6 ORF (Fig. 1). Although the significance of these spliced transcripts regarding the translation of these proteins has been controversial, recent evidence suggests that, at least for HPV type 16 (HPV-16) and HPV-18, unspliced mRNA encodes mostly full-length E6 while spliced mRNA encodes both E7 and also the E6* proteins in the context of cell lines derived from cervical tumors (30). Analysis of the early transcripts in these cells has shown that the majority are of this spliced form (25, 28), and, typically, constructs that express full-length HPV-18 or HPV-16 E6 when analyzed after transient transfection into mammalian cells generally express high levels of spliced mRNA.
FIG. 1.
The arrangement of splice donor and acceptor sites within the E6 ORFs of high-risk alpha group HPVs; an asterisk above the splice donor site of HPV-18 E6 indicates the G that is mutated to A to abolish splicing.
The E6* proteins are C-terminally truncated versions of the full-length E6 proteins expressed from a subset of spliced early transcripts. While some papillomavirus types, such as HPV-16, seem to have splicing patterns that allow the expression of up to four E6* species, dependent upon the position of the downstream splice acceptors, HPV-18 appears to transcribe only one mRNA species that is capable of expressing E6*. The polypeptide product of this transcript shares the first 44 amino acids with full-length E6 before the first splice donor site; thereafter, it has 13 unique amino acids that are derived from E6 intronic sequences. Previous studies have shown that HPV-18 E6*, when expressed as a glutathione S-transferase (GST) fusion protein, can bind to in vitro translated full-length HPV-18 and -16 E6 and to the ubiquitin ligase E6-AP but not, significantly, to p53 (21). We subsequently showed that E6* could inhibit the E6-directed degradation of p53 in vitro and also in vivo when all three proteins were overexpressed in p53-null human and mouse cells (21). We also showed that E6* overexpression led both to an increase in p53-mediated transcriptional activity in HPV-positive but not HPV-negative cells (21) and apoptosis in HPV-positive but not HPV-negative cells (22); both of these results suggest that E6* would likely be reactivating p53-dependent growth arrest and apoptotic functions. However, from subsequent studies, we observed that inhibiting E6-directed degradation of p53 is not always sufficient to activate p53-mediated apoptotic pathways in HPV-positive cell lines (15), and more recent studies showing that HPV-16 E6 protein can inhibit the extrinsic death pathway through interaction with tumor necrosis factor receptor 1, FADD, and procaspase 8 strongly suggest other means by which E6* inhibition of E6 function might lead to apoptosis (4, 5, 6).
Studies on the function of the E6 proteins from high-risk mucosotrophic HPVs have revealed a number of cellular proteins whose proteasome-mediated degradation is directed by E6. These include p53, BAK, E6BP, and E6TP1; PDZ domain-containing proteins Dlg, MAGI-1, MAGI-2, MAGI-3, MUPP-1, and hScrib (for a review, see reference 16); and PATJ (29). One of the functions of the E6 oncoproteins is to act as an adaptor, linking a cellular ubiquitin ligase, whose prototype is E6-AP, to the target, allowing it to be ubiquitinated and directed to the proteasome for degradation (12). While the interaction with p53 and Bak is considered to provide a means of evading growth arrest and apoptosis during the E7-induced reactivation or maintenance of host cell S phase, the significance of the E6 interaction with and degradation of its several PDZ domain-containing targets during the normal viral life cycle is incompletely understood. However, given the localization of most of these proteins at cell junctions and the roles they are believed to play in cell polarity, together with the observation that both Dlg and hScrib become aberrantly localized and downregulated during the malignant progression of many cancer types (8, 14), it has been speculated that their targeted degradation by E6 might be involved in the progression and invasiveness of cervical carcinoma. In this study, we show that 293 cells transiently transfected with E6 expression constructs, both when tagged at the amino terminus or untagged, express high levels of spliced transcripts encoding E6* protein, indicating that E6* as well as E7 is likely to play a role in the viral life cycle. Because of this, we considered it important to assess the impact of E6* expression separately from the effect of full-length E6 regarding its known cellular targets. This is especially important in view of the fact that we have previously demonstrated that E6* is able to interact with full-length E6 and regulate some aspects of its cellular localization and function (11).
MATERIALS AND METHODS
Cell lines.
Human 293 cells, HaCaT cells, and Saos-2 cells were maintained in Dulbecco's modified Eagle's medium with fetal bovine serum to a final concentration of 10%.
Plasmid constructs.
Dual hemagglutinin (HA)- and Flag-tagged expression constructs for HPV-18 E6 and HPV-18 E6* were made by PCR cloning of the ORFs of these proteins between the EcoRI and HpaI sites of the in vivo expression vector pCA (34). Splicing-defective mutant pCA18E6 SM (where SM is splice mutant) was constructed using the GeneTailor site-directed mutagenesis system (Invitrogen) introducing a G-to-A substitution at nucleotide 233 in the HPV-18 E6 sequence. Plasmid construct pCDNA3.18E6 has been previously described (19), as has construct pCDNA3.18E6 T156E (7). The splicing-defective mutant pCDNA3.18E6 T156A SM was constructed using the GeneTailor mutagenesis system, introducing the same mutation as for the pCA18E6 SM. Plasmid pGW1 HA-PKB for expression of Akt has previously been described (20), as has plasmid pGW1 DLG for in vivo expression of HA-tagged Dlg (7) and plasmids expressing HA-tagged MAGI-1 (9), MAGI-2 (32), and hScrib (33). Constructs pCDNA3 MKK6glu and pCMV5 MEKK1 (wild type; CMV is cytomegalovirus) were the kind gift of Eric Paulson.
Fluorescence microscopy.
HaCaT cells were plated out at 20 to 30% confluence on coverslips, and after 24 h cells were transfected using a calcium phosphate-mediated transfer system (10). After a further 24 h, cells were washed with phosphate-buffered saline (PBS), fixed for 20 min in 3.7% paraformaldehyde, washed again in PBS, and then permeabilized for 5 min with 0.1% Triton X-100 in PBS. After being washed with PBS, coverslips were incubated for 1 h at 37°C with mouse monoclonal anti-E6 primary antibody 399 (Arbor Vita Corporation), which is directed against an amino-terminal E6 epitope and rabbit anti-Dlg primary antibody (Santa Cruz). Coverslips were washed in PBS and then incubated for 1 h at 37°C with fluorescein-conjugated goat anti-rabbit and rhodamine-conjugated goat anti-mouse secondary antibodies (Alexa Fluor). After a final washing step in PBS, the coverslips were mounted on slides and examined by UV microscopy.
Transfections and Western blot analysis.
Transient transfections were carried out by calcium phosphate-mediated transfer as previously described (10). All transfections were carried out with a LacZ plasmid, which expresses β-galactosidase, in addition to the plasmids described above, as a control for transfection efficiency. Whole-cell lysates were produced by lysing cells for 20 min on ice with a buffer containing 50 mM HEPES, pH 7.0, 250 mM NaCl, 0.1% NP-40, and 20 mg ml−1 aprotinin, with occasional vortexing, and then spinning cells at a relative centrifugal force of 16,000 × g for 1 min, after which supernatants were transferred to fresh tubes. Whole-cell lysates, typically 50 to 100 mg, were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) separation and then electrophoretically transferred to nitrocellulose membranes (Schleicher and Schuell). For probing with antibodies against HA-tagged proteins, p53, or β-galactosidase, membranes were blocked by incubation with 10% milk powder in PBS. Western blots were probed sequentially for expression of residual HA-tagged E6 target proteins using mouse anti-HA monoclonal antibodies (Roche) or, for p53, monoclonal antibody DO-1 (Santa Cruz). Primary antibody probes were amplified with horseradish peroxidase (HRP)-conjugated anti-mouse antibodies (Dako), and then the proteins were visualized by enhanced chemiluminescence (GE Healthcare) according to the manufacturer's instructions. When required for reprobing, membranes were stripped by incubation for 1 h at 65°C in a buffer containing 2% SDS, 60 mM Tris, pH 6.8, and 100 mM β-mercaptoethanol, followed by several washes in distilled water. Membranes were reblocked in milk as before and reprobed with a mouse anti-β-galactosidase monoclonal antibody (Promega) to control for equal plasmid transfection efficiency and loading on gels. To probe for HPV-18 E6 or MAGI-1, membranes were blocked in Tris-buffered saline ([TBS] 25 mM Tris, pH 7.4, 150 mM NaCl, 2.7 mM KCl) with 5% milk and 2% bovine serum albumin (BSA). Anti-E6 monoclonal antibodies 3 and 399 and anti-MAGI-1 monoclonal 236 (Arbor Vita Corporation) were used at 1 mg ml−1 in TBS-0.1% Tween-20 with 0.1% BSA and incubated for 2 h at room temperature. After three washes with TBS-0.1% Tween-20, membranes were probed with HRP-conjugated anti-mouse antibodies (Dako) in TBS-0.1% Tween-20 with 0.1% milk-0.04% BSA for 1 h at room temperature before being visualized as above.
Protease inhibitors.
The following inhibitors were dissolved in dimethyl sulfoxide and used at the final concentrations indicated below: lysosomal inhibitor bafilomycin (Sigma) at 1 mM, broad-spectrum caspase inhibitor Boc-D-FMK [Boc-Asp(OMe)-fluoromethyl ketone] (Calbiochem) at 100 μM, calpain inhibitor II (ALLM [N-acetyl-Leu-Leu-Met-CHO]; Calbiochem) at 100 μM, and proteasome inhibitors MG132 (Sigma) at 100 μM and epoxomicin (Sigma) at 2 μM.
GST pull-downs.
Whole-cell lysates for GST pull-downs were prepared as for the Western blot analyses described above. GST fusion proteins were prepared as previously described (27). Typically, 100 mg of cell extract was incubated with the requisite amount of GST fusion protein bound to glutathione-agarose (Sigma) for 2 h at room temperature, followed by five washes in lysis buffer. Bound proteins were eluted by boiling in loading buffer and then separated by SDS-PAGE prior to transfer onto membranes and probed as for the Western blots described above; 10 mg of whole-cell extract was always run as control for quantification of expressed Akt and Dlg. Membranes were probed with mouse anti-HA monoclonal antibodies and HRP-conjugated secondary antibodies, as described above, before being visualized using enhanced chemiluminescence, as described above. After exposure, membranes were stained with Ponceau Red to quantitate the levels of GST fusion proteins.
Selection of HT1080 colonies stably expressing E6*.
HT1080 cells were seeded onto 10-cm petri dishes and transfected either with the empty vector pCDNA3 or with pCDNA3.18E6* and then grown under G418 selection at a concentration of 600 μg ml−1 for 2 weeks. At this time individual colonies were picked, expanded, and then screened by Western blot analysis for E6* expression.
RT-PCR.
Total RNA was extracted from 10-cm petri dishes of 293 cells, untransfected or transfected with pCA18E6, pCA18E6 SM, pCDNA3.18E6 T156E, or pCDNA3.18E6 T156E SM, using the Tri Reagent system (Sigma) according to the manufacturer's instructions. The purified RNA was treated with RQ1 RNase-free DNase (Promega) according to the manufacturer's instructions to remove any plasmid DNA carried over during the RNA extraction. Two micrograms of purified RNA was then annealed with random decamers, and cDNAs were generated with reverse transcriptase (RT) using a Retroscript kit (Ambion) according to the manufacturer's instructions; to control for plasmid DNA carry-over, a parallel set of random decamer-annealed samples was incubated without RT. The cDNAs generated from the RT step and their control samples were amplified using HPV-18 E6 flanking oligonucleotides and Go-Taq Polymerase (Promega).
Luciferase assays.
Six-centimeter dishes of 293 cells were transiently transfected with pRL-CMV (Promega), which expresses the Renilla luciferase reporter, with and without a 10-fold and 100-fold excess of plasmids pCDNA3.18E6 SM and pCDNA3.18E6* expressing E6 alone and E6*, respectively, together with the LacZ plasmid as a transfection control. After 24 h, cells were harvested, and luciferase assays were performed using a dual luciferase assay system (Promega) according to the manufacturer's instructions, reading on the Renilla channel alone. To control the transfection efficiency, identical quantities of the remaining extracts were subjected to SDS-PAGE and Western blot analysis to visualize levels of β-galactosidase expression. The X-ray film from the Western blot was densitometrically scanned using Adobe Photoshop, and the luciferase data were normalized against the scanned data.
Dlg half-life assays.
293 cells grown on 6-cm petri dishes were transfected with pGW1 DLG in the presence or absence of pCDNA3.18E6*, together with the LacZ plasmid, to assay for plasmid uptake efficiency, as described above. After 24 h, cycloheximide (Sigma) was added to a final concentration of 50 μg ml−1 to one dish expressing Dlg alone and one dish expressing both Dlg and E6*. Thereafter, cycloheximide was added to a pair of dishes at 2-h intervals until the first pair of dishes had been incubated for a total of 10 h. All the dishes were then harvested as described above, the extracts were separated by SDS-PAGE, and residual Dlg levels were quantitated by Western blot analysis using an anti-HA antibody as described above. Plasmid transfection efficiency was tested by stripping the membranes and reprobing with an anti-β-galactosidase antibody as described above.
RESULTS
Constructs expressing tagged and untagged HPV-18 E6 express a majority of spliced transcripts when transfected into 293 cells.
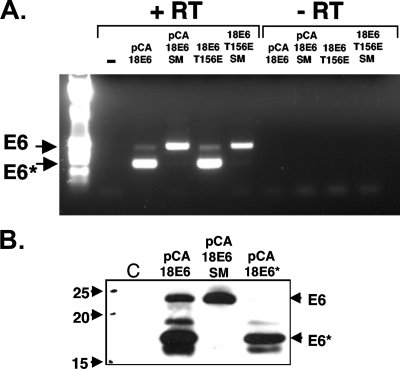
It has been known for many years that the majority of early HPV transcripts in cell lines derived from cervical tumors are spliced within the E6 ORF (25, 28). To obtain an estimate of the ratio of spliced to unspliced transcripts for E6 expression constructs, we carried out semiquantitative RT-PCR on total RNA from 293 cells transfected with four HPV-18 E6 expression constructs. Two constructs expressed amino-terminally HA-tagged HPV-18 E6: wild-type HPV-18 E6 and a mutant with a G-to-A substitution at nucleotide 233 to destroy the splice donor site. The other two constructs expressed untagged HPV-18 E6: pCDNA3.18E6 T156E, which is defective for PDZ domain binding, and pCDNA3.18E6 T156E SM, which also has a mutation at the splice donor site. Figure 2A shows an RT-PCR carried out on total RNA extracted from the cells 24 h posttransfection. Although RT-PCR is only semiquantitative, it can be seen that for both constructs with a wild-type splice donor site, the majority of transcripts are spliced and encode E6*. Mutation of the splice donor site, however, abolishes the overwhelming majority of splicing, and the resulting transcripts express almost exclusively full-length E6; very small amounts of spliced transcript are observable, but it is unclear whether this is due to weak splicing from the mutated donor site or from an adjacent, cryptic splice site. To examine the effects on E6 and E6* protein levels, 293 cells were transfected with the HA-tagged pCA18E6, pCA18E6 SM, and pCA18E6* constructs, and after 24 h whole-cell lysates were made and subjected to immunoblot analysis with an antibody directed against the amino terminus of E6. The results, shown in Fig. 2B, demonstrate that mutation of the splice donor site eliminates expression of E6* protein, allowing expression of only full-length E6. Because the pCA constructs are both Flag and HA tagged at their amino termini, the resulting expressed proteins are substantially larger than their untagged counterparts.
FIG. 2.
The majority of transcripts from both tagged and untagged HPV-18 E6 constructs are spliced. (A) RT-PCR of transcripts from HA-tagged wild-type E6 and PDZ-binding-defective, untagged HPV-18 E6 (18E6) mutant T156E, upon expression in 293 cells before and after introduction of a mutation at the splice donor site. (B) Western blot of pCA18E6 wild type, pCA18E6 SM, and pCA18E6* expressed in 293 cells. C, control; −, without RT; +, with RT.
HPV-18 E6* but not full-length HPV-18 E6 downregulates levels of Akt.
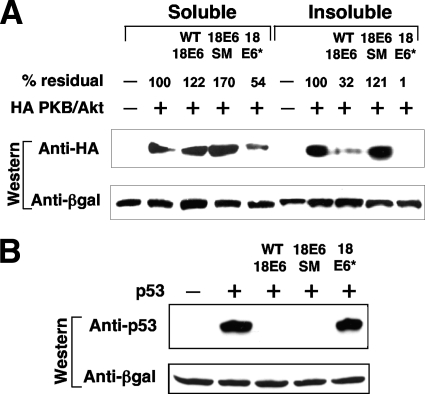
Several groups of investigators have shown that HPV E7 expression increases signal transduction along the phosphatidylinositol 3-kinase/Akt pathway (20, 35). While the majority of this increase may rely on the ability of E7 to bind and inactivate pRb (18), some activity was observed to be independent of upstream signaling from phosphatidylinositol 3-kinase and dependent upon an interaction between E7 and protein phosphatase 2A (20). Because both E6 and E7 are considered to be coexpressed at early stages of the viral life cycle, we were interested to see whether HPV-18 E6 had any effect on Akt activity, and so we transiently transfected constructs expressing wild-type E6, the E6 splice mutant, and E6* into 293 cells along with a construct ectopically expressing HA-tagged Akt. The cells were harvested after 24 h, and both the detergent-soluble and insoluble fractions of the whole-cell lysates were separated by PAGE. After Western transfer, the membranes were probed with anti-HA antibodies to quantify total Akt. The results, shown in Fig. 3A, demonstrate that the wild-type construct expressing both full-length E6 and E6* reduces the levels of Akt protein; the majority of this reduction is observed in the detergent-insoluble fraction. In contrast, the splice mutant construct, expressing only full-length E6, has no effect on Akt levels, whereas the construct expressing only E6* strongly reduces Akt levels, and some effect is also observable in the soluble fraction. While the nature of the Akt localizing to this detergent-insoluble fraction remains unknown, we speculate that E6* is unable to target this fraction. As a formal control to demonstrate the correct functioning of the E6 splice mutant and E6* constructs, we cotransfected constructs expressing wild-type HPV-18 E6, the E6 splice mutant, and E6* into p53-negative Saos-2 cells together with a construct expressing p53. The results, shown in Fig. 3B, indicate that, as expected, full-length E6 but not E6* targets p53 for degradation.
FIG. 3.
Downregulation of protein levels of ectopically expressed Akt, but not p53, by HPV-18 E6*. (A) HA-tagged Akt was overexpressed in 293 cells in the absence (−) and presence (+) of untagged constructs expressing wild-type HPV-18 E6 (WT 18E6), the HPV-18 E6 splice mutant (18E6 SM) which contains a splice donor site mutation, or HPV-18 E6* (18 E6*). Cell extracts were separated by PAGE and then transferred to nitrocellulose and probed with anti-HA antibodies to quantitate residual Akt. After blots were stripped, they were reprobed with anti-β-galactosidase (βgal) antibodies to visualize coexpressed β-galactosidase as a control for transfection and sample loading. (B) Human p53 was ectopically expressed in p53-null Saos-2 cells in the presence or absence of constructs expressing wild-type HPV-18 E6, HPV-18 E6 containing a splice donor site mutation, or HPV-18 E6*. After PAGE separation and transfer to nitrocellulose, residual p53 was visualized by probing with monoclonal antibody DO-1. Transfection efficiency and equal sample loading were controlled for as described above.
HPV-18 E6* targets several PDZ domain-containing targets for degradation, a property that is shared by E6* proteins from other high-risk mucosal HPV types.
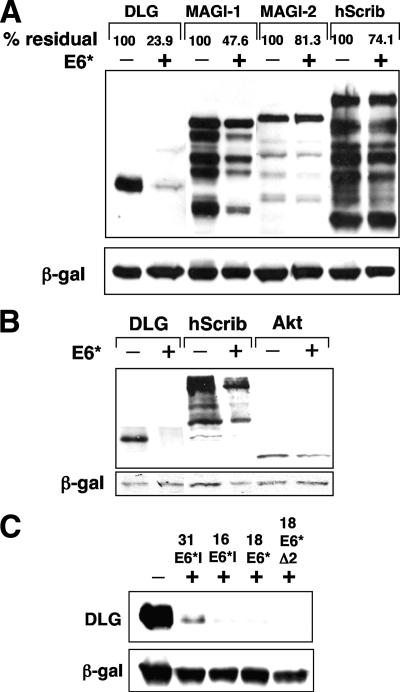
The downregulation of Akt protein levels suggested that E6* can activate a degradation pathway, and since E6*, in addition to E6, has been implicated in the degradation of the PDZ domain-containing junctional protein PATJ (29), we examined a set of other PDZ domain-containing substrates that has previously been shown to be targeted by the full-length E6 proteins from high-risk HPV types. We transfected 293 cells with constructs expressing HA-tagged Dlg, MAGI-1, MAGI-2, and hScrib, together with the E6* expression construct. After 24 h the cells were harvested, and protein levels were analyzed by Western blotting. The X-ray films were subsequently scanned, and then residual protein levels in the presence of E6* were calculated with reference to the levels in the absence of E6*. The results, shown in Fig. 4A, indicate that the levels of Dlg and MAGI-1 are significantly decreased upon E6* expression, but only a very slight decrease was observed for MAGI-2 and hScrib. To eliminate the possibility that degradation of cellular proteins in 293 cells was due to the presence of adenovirus sequences, we performed a similar degradation experiment in Saos-2 cells, examining the levels of ectopically expressed Dlg, hScrib, and Akt both with and without HPV-18 E6* expression. The results, shown in Fig. 4B, confirm that the observed degradation of target proteins is not confined to 293 cells. Similar results were observed for degradation in U2OS cells (data not shown).
FIG. 4.
E6* mediates downregulation of several PDZ domain-containing proteins that are also degraded in the presence of E6. (A) 293 cells were transiently transfected with constructs expressing HA-tagged versions of Dlg, MAGI-1, MAGI-2, and hScrib in the absence (−) and presence (+) of pCDNA3.18E6*. After PAGE separation and transfer to nitrocellulose, residual E6* targets were visualized by probing with anti-HA antibodies. Transfection efficiency and equal sample loading were controlled for as described in Materials and Methods. The X-ray film was scanned, and the percent residual protein levels were calculated and are shown above each lane. (B) Saos-2 cells were transiently transfected with constructs expressing HA-tagged versions of Dlg, hScrib, and Akt in the absence and presence of pCDNA3.18E6*. After PAGE separation and transfer to nitrocellulose, residual E6* targets were visualized by probing with anti-HA antibodies. (C) The ability to induce Dlg degradation is conserved among the E6* proteins from other HPV types. 293 cells were transiently transfected with the construct expressing HA-tagged Dlg, in the absence and presence of pCDNA3.31E6*I, pCDNA3.16E6*I, pCDNA3.18E6*, and deletion mutant pCDNA3.18E6Δ2. Cells were harvested, and Dlg levels were assessed as described above. β-gal, β-galactosidase.
To see if the ability to induce Dlg degradation was conserved among the E6* proteins of other high-risk alpha-group HPVs, we cloned the E6*I open reading frames for HPV-16 and HPV-31 into a mammalian expression vector and examined their effect on Dlg levels. The results, shown in Fig. 4C, demonstrate that these E6* proteins can also induce Dlg degradation. In addition, we expressed HPV-18 E6*Δ2, a deletion mutant that in vitro was previously shown to be defective for binding to E6 and to have a reduced ability to bind to E6-AP as well as to induce apoptosis when overexpressed in HPV-positive cells (22). This mutant was as effective as the wild-type E6* in inducing Dlg degradation.
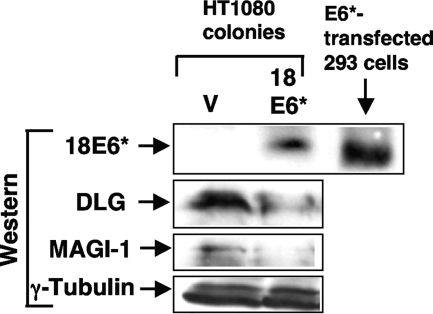
The levels of endogenous Dlg and MAGI-1 are reduced in HT1080 cells stably expressing HPV-18 E6*.
Because of potential differences that might occur between transient and stable expression of E6* in its effects on cellular proteins, we set out to establish cell lines stably expressing HPV-18 E6*. Previous observations indicated that HT1080 cells are able to support E6* overexpression (21), and so these cells were chosen. We transfected HT1080 cells with the empty vector or with the construct expressing untagged HPV-18 E6*, and after 2 weeks under G418 selection, picked and expanded 11 colonies. Of these only one colony expressed high levels of E6* as detected by Western blot analysis. We then compared the levels of endogenous Dlg and MAGI-1 in E6*-expressing cells with the levels in the vector-transfected control cell line; the results, shown in Fig. 5, indicate that in the E6*-expressing line, levels of both proteins are reduced, supporting the hypothesis that E6* expression leads to downregulation of these proteins.
FIG. 5.
An HT1080 cell line that stably overexpresses HPV-18 E6* has reduced levels of endogenous Dlg and MAGI-1. HT1080 cells were transfected with empty pCDNA3 or pCDNA3.18E6* and then placed under G418 selection for 2 weeks. A drug-resistant colony that expressed high levels of HPV-18 E6* (18E6*) as detected using anti-E6 monoclonal antibody 399 (Arbor Vita) was selected, and levels of endogenous Dlg and MAGI-1 were quantitated by Western blot analysis relative to the levels in a vector control-selected cell line (V) by using rabbit anti-Dlg polyclonal antibodies (Santa Cruz) and anti MAGI-1 monoclonal antibody 236 (Arbor Vita).
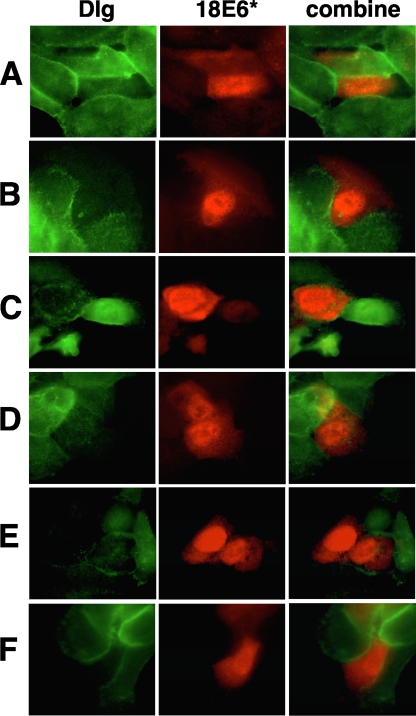
E6* reduces the protein levels of endogenous Dlg when ectopically expressed in HaCaT cells.
Although we had shown a reduction in levels of ectopically expressed Dlg in 293 cells and endogenous Dlg in HT1080 cells, it was clearly also important to demonstrate the same reduction of endogenous Dlg levels in keratinocytes. To do this, HaCaT cells were grown on coverslips and transfected with pCDNA.18E6* which expresses untagged E6* protein. After 24 h, cells were fixed, and the levels of E6* and Dlg were visualized by immunofluorescence. The results are shown in Fig. 6. Because transfection efficiency in HaCaT cells is low, a single field showing multiple cells expressing E6* was not obtained. Nevertheless, in those cells that express E6*, there is invariably reduced Dlg expression, and we did not find any cells expressing E6* with normal Dlg levels. We also observed that the majority of E6* was localized to the nucleus, which is consistent with previous observations for HPV-16 E6* (31) although, in our hands, some cytoplasmic expression of E6* was observed.
FIG. 6.
HPV-18 E6* downregulates endogenous levels of Dlg in HaCaT cells. HaCaT cells were grown on coverslips and then transfected with the pCDNA3.18E6* construct expressing untagged HPV-18 E6*. After 24 h, cells were fixed and permeabilized, and then the ectopically expressed E6* and endogenous Dlg proteins were visualized by performing immunofluorescence with mouse monoclonal anti-E6 399 (Arbor Vita) and rabbit anti-Dlg (Santa Cruz) primary antibodies, followed by rhodamine-conjugated anti-mouse and fluorescein-conjugated anti-rabbit secondary antibodies and UV microscopy. Panels show six typical images from a single experiment of cells expressing HPV-18 E6*; the combined set is the merge of ectopically expressed E6* (red) and endogenous Dlg (green) images to demonstrate that the reduction in endogenous Dlg occurs in the same cells that are expressing ectopic E6*.
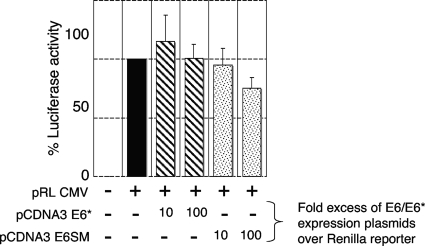
The reduction in levels of ectopically expressed proteins is not due to transcriptional repression of the plasmid construct.
HPV E6 protein has been shown to repress transcription from some promoters (3), and although we did not observe a reduction in levels of ectopically expressed p53 when it was coexpressed with E6*, there remained a possibility that E6* could transcriptionally downregulate the promoters of some expression vectors. Since all the expression constructs used throughout this study are driven by the CMV promoter, we looked for the ability of E6 and E6* to transcriptionally repress a CMV-Renilla luciferase reporter construct. We transiently transfected 293 cells with the plasmid pRL-CMV, which expresses Renilla luciferase (Promega), together with a 10-fold and 100-fold excess of plasmids expressing E6 or E6*. After analyzing the levels of expressed luciferase, we concluded from the results (Fig. 7) that there was a minimal level of transcriptional repression by E6 but no transcriptional repression by E6* even when cells were transfected at an excess of 100 times that of the reporter plasmid. This is consistent with the observation that E6* reduces Dlg, hScrib, and Akt expression at the protein level rather than transcriptionally.
FIG. 7.
HPV-18 E6* does not transcriptionally repress the CMV promoter of expression constructs. The pRL-CMV reporter was transiently transfected into 293 cells with the LacZ-bearing plasmid, without and with a 10- and 100-fold excess of plasmid constructs expressing either E6* or the E6 splice mutant (E6SM). After 24 h cells were harvested, and the extracts were analyzed for luciferase activity. The histogram represents the cumulative data from four experiments with standard deviation bars shown.
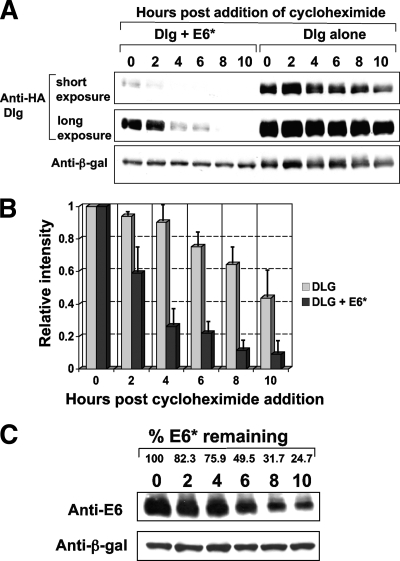
E6* reduces the half-life of ectopically expressed Dlg in 293 cells.
To support further our model that the reduction of Dlg protein levels upon E6* expression occurs at the protein rather than the transcriptional level, we examined the half-life of ectopically expressed Dlg in the absence and presence of E6*. 293 cells were transfected either with a construct expressing HA-tagged Dlg by itself or together with an E6* expression construct. After 24 h, cycloheximide was added to a dish of 293 cells in each transfection set and thereafter to a dish in each set every 2 h until the first dishes had been incubated with cycloheximide for 10 h. At this time all the dishes were harvested, the cells were extracted as previously described, and the levels of Dlg were then monitored by Western blot analysis. The results from a typical half-life experiment are shown in Fig. 8A, and the cumulative data from three separate experiments are shown in a histogram in Fig. 8B. The data show that coexpression of E6* greatly reduces the half-life of Dlg, which is consistent with the idea that the mechanism is degradation rather than transcriptional repression. Because of the formal possibility that the half-life of HPV-18 E6* could be shorter than that of Dlg in this experiment, we examined the half-life of untagged HPV-18 E6* under identical conditions, and the results are shown in Fig. 8C. Much speculation has surrounded the half-life of untagged E6* proteins, with the suggestion that they might be highly unstable. However, our data demonstrate that untagged HPV-18 E6* has a half-life comparable to that previously demonstrated for myc-tagged HPV-18 E6* at around 6 h (29) and, therefore, approximately twice as long as that of Dlg when it is expressed in the presence of HPV-18 E6*.
FIG. 8.
HPV-18 E6* reduces the half-life of ectopically expressed Dlg in 293 cells. (A) 293 cells were transfected with the construct expressing HA-tagged Dlg in the absence and presence of the construct expressing untagged HPV-18 E6*. After 24 h, cycloheximide was added to the dishes at 2-h intervals, and all the dishes were harvested for PAGE at 10 h. Residual Dlg levels were assessed by Western blot analysis. Short and long exposures of the Western blot are included for better comparison of the half-lives. Panel B shows the cumulative data from three experiments, analyzed using the densitometry analysis program in Adobe Photoshop, and then calculated as intensity relative to the time zero levels for Dlg expressed by itself or together with E6*. Standard deviations are indicated as vertical bars. (C) Determination of the half-life of HPV-18 E6*. Untagged HPV-18E6* was expressed in 293 cells, and after 24 h cells were treated with cycloheximide as for the experiments shown in panel A. Residual E6* levels were detected by Western blot analysis using anti-HPV-18 E6 monoclonal antibody 399. β-gal, β-galactosidase.
E6* does not directly interact with either Akt or Dlg while targeting them for degradation.
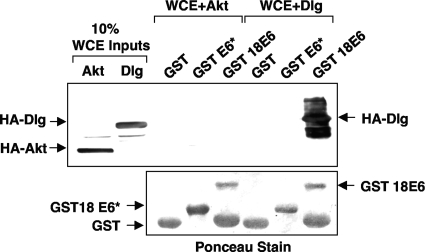
The ability of the full-length E6 proteins from both bovine papillomavirus type 1 and high-risk mucosal HPV types to target cellular proteins for proteasome-mediated degradation relies on E6's binding to the substrate and functioning as an adaptor in order to link a cellular ubiquitin ligase to the target for ubiquitination (12, 13). Additional studies have shown that mutations in E6 that perturb its interaction with a cellular substrate also destroy its ability to direct degradation of that substrate (1, 2, 21, 26). As an example, mutations in the PDZ-binding domain in the extreme C terminus of E6 that eliminate its ability to bind PDZ domain-containing targets also eliminate its ability to degrade them in vitro (7, 9). A recent report has demonstrated the ability of E6* to both interact with and degrade another PDZ domain-containing protein, PATJ (29). Consequently, having shown that E6* could direct the degradation of other cellular targets, we were interested in determining to what degree E6* could also interact with them. We expressed HA-tagged Dlg and Akt in 293 cells and, after harvesting the cells, used the whole-cell lysates as a source of HA-tagged protein to perform pull-downs using full-length HPV-18 E6 and E6* purified as GST fusion proteins. The results are shown in Fig. 9 and demonstrate that although a strong pull-down was observed between GST E6 and Dlg, in agreement with previous studies (7), no pull-down was observed between GST E6 and Akt or between GST E6* and either Akt or Dlg. This result strongly suggests that E6* expression leads to the downregulation of some cellular proteins without directly interacting with them.
FIG. 9.
E6* does not interact with either Akt or Dlg in vitro. 293 cells were transiently transfected with constructs expressing HA-tagged Akt or Dlg. After cells were harvested, 10% of each whole-cell extract (WCE) was retained as a control for input levels of the expressed proteins, and the remainder was used for pull-downs with GST, GST-E6* (GST 18E6*), or GST-E6 (GST 18E6) bound to glutathione resin. After the resin was washed, bound proteins were eluted and visualized by SDS-PAGE and Western blotting with anti-HA antibodies. The membranes were subsequently stained with Ponceau Red to determine input levels of the GST fusion proteins.
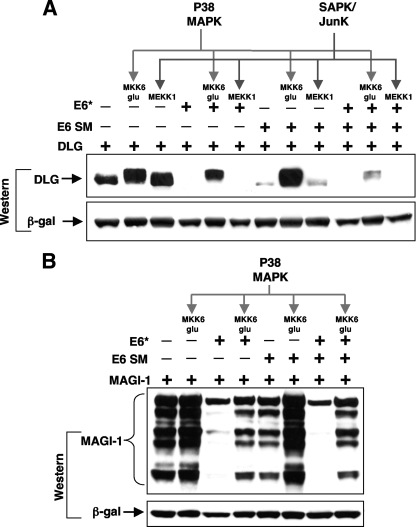
E6* and E6 target different cellular pools of certain substrates and show cooperation in the degradation of Dlg.
Although we have shown that E6* but not E6 can target Akt for degradation and, conversely, that E6 but not E6* can target p53, there remain several targets in common between the two, raising the question as to why more than one protein might have this function. Consequently, we asked whether both proteins target the same cellular pool of Dlg or whether phosphorylation of Dlg along certain kinase pathways might make it differentially susceptible to degradation by one or other of the two viral proteins. To examine this, we induced p38 mitogen-activated protein kinase (MAPK) phosphorylation by ectopic expression of a dominant active MKK6 mutant or stress-activated protein kinase/Jun N-terminal protein kinase (SAPK/JNK) phosphorylation by ectopic expression of wild-type MEKK1 and then analyzed Dlg phosphorylated along these two pathways for susceptibility to E6- or E6*-directed degradation. The results, shown in Fig. 10A, indicated that phosphorylation of Dlg by both p38 MAPK and SAPK/JNK leads to its stabilization and to an increase in Dlg protein levels. It can also be seen that p38 MAPK-phosphorylated Dlg is highly resistant to degradation by E6 and slightly resistant to degradation by E6*, where it can also be seen that E6* preferentially targets the faster-migrating bands of Dlg. In contrast, Dlg phosphorylated by SAPK/JNK is as sensitive to E6- and E6*-directed degradation as unphosphorylated Dlg. Interestingly, when both E6 and E6* are coexpressed, the effects on p38 MAPK-phosphorylated Dlg are greatly increased, suggesting that the two proteins may be acting cooperatively to destabilize Dlg. These findings strongly suggest that E6 and E6* direct the degradation of different cellular pools of Dlg and should be considered in light of recent studies suggesting that JNK- and p38 MAPK-phosphorylated Dlg have different cellular localizations and binding partners (17, 23). When we examined the effect of driving p38 MAPK phosphorylation of MAGI-1 and its susceptibility to degradation by both HPV-18 E6 and E6*, we again saw that MAGI-1 is stabilized against E6-directed degradation and to a lesser degree against E6*-directed degradation. In contrast, however, to the effects observed for Dlg, E6* and E6 expressed together seem to have no greater effect than E6* alone on MAGI-1 degradation, suggesting in this case that although E6* alone can increase the turnover of this protein, it may not cooperate with E6 to do so.
FIG. 10.
E6* and E6 target different cellular pools of Dlg and MAGI-1 when phosphorylated along different kinase pathways. (A) 293 cells were transiently transfected with construct pGW1 HA-Dlg in the absence (−) or presence (+) of pCDNA3.18E6* or pCDNA3.18E6, with or without constructs expressing MEKK1, driving the SAPK/JNK pathway, or MKK6 Glu, driving the p38 MAPK pathway. After 24 h, whole-cell extracts were separated by PAGE, and after Western blotting, the membrane was probed for expressed Dlg using anti-HA antibodies. Transfection efficiency and equal sample loading were controlled for as described in Materials and Methods. (B) The construct expressing HA-MAGI-1 was transfected into 293 cells with pCA18E6* or pCA18E6, as above, together with a construct expressing MKK6 Glu to activate the p38 MAPK pathway. Cells were harvested, and Western blot analysis was carried out to examine levels of MAGI-1, as described above. β-gal, β-galactosidase; E6 SM, E6 splice mutant.
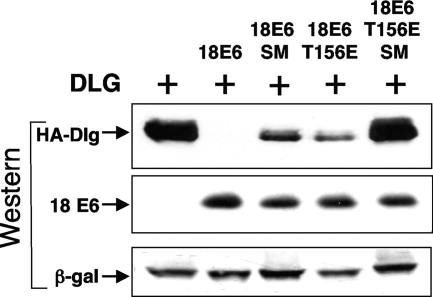
Dlg is degraded by E6* expressed by an E6 mutant that cannot itself bind or degrade PDZ domain-containing proteins.
If E6* can activate degradation of cellular proteins independently of E6, we reasoned that Dlg should be degraded in the presence of a mutant of E6 that cannot bind or degrade Dlg in vitro, since such a construct should still be able to splice and express E6* in vivo. To test this, we ectopically expressed HA-tagged Dlg together with E6 mutant T156E. This mutant of E6 has a threonine-to-glutamic acid substitution at position 156 in the PDZ-binding motif, which destroys its ability to bind or direct the degradation of Dlg in vitro (7). When the mutant is expressed in vivo however, as shown in Fig. 9, we observe that this mutant retains substantial ability to direct the degradation of Dlg, suggesting that, at least in part, it is the E6* protein expressed from this construct that is able to degrade Dlg. To demonstrate that this is true, we introduced a mutation at the splice donor site in this construct to remove its ability to express E6*. Extraction of RNA from cells transfected with this construct and performance of RT-PCR showed that, as for the pCA18E6 SM described above, splicing is almost totally inhibited (Fig. 1B). The results shown in Fig. 11 clearly demonstrate that when this double mutant is expressed in 293 cells and its activity is compared to the single T156E mutant, we observe no degradation of Dlg, as predicted, due to loss of E6* expression; the same mutation introduced at the splice donor site of the wild-type E6 construct reduces but does not inhibit the ability of E6 to direct the degradation of Dlg even though the expression levels of E6, as monitored using an anti-E6 monoclonal antibody, are equivalent.
FIG. 11.
Degradation of Dlg is mediated by both E6 and E6* when a wild-type HPV-18 expression construct is transfected into 293 cells. 293 cells were transiently transfected with construct pGW1 HA-Dlg in the absence or presence of the wild-type construct pCDNA3.18E6 (18E6), pCDNA3.18E6 SM (18E6 SM), which cannot express E6*, and the PDZ-binding-defective mutant construct pCDNA3.18E6 T156E (18E6 T156E) or pCDNA3.18E6 T156E SM (18E6 T156E SM), which also cannot express E6*. After 24 h, whole-cell extracts were prepared and separated by PAGE, and after Western blotting, the membrane was probed for expressed Dlg using anti-HA antibodies and for E6 expression using anti HPV-18 E6 monoclonal antibody 3 (Arbor Vita Corporation). Transfection efficiency and equal sample loading were controlled for as described in Materials and Methods. β-gal, β-galactosidase.
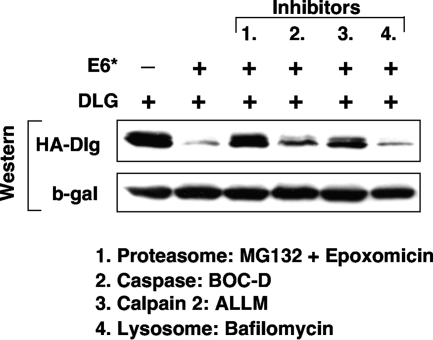
The E6*-directed degradation of Dlg may be proteasome mediated.
To investigate the mechanism of degradation induced by E6* expression, we undertook a degradation experiment with a variety of proteolytic pathway inhibitors. We elected to examine calpain cleavage since this pathway, downstream of calmodulin, is active during cellular differentiation. Similarly, caspase pathways have been observed to be activated during differentiation in an apoptosis-independent manner, and the extrinsic death pathway has also been implicated in E6/E6* interactions (6). We also examined lysosomal degradation. Inhibitors of the proteasome pathway were also included as a comparison since this is the route by which the full-length E6 protein downregulates cellular targets. We transiently transfected 293 cells as before with the construct expressing HA-tagged Dlg together with the E6* expression construct. After 24 h we added the following inhibitors: a cocktail of MG132 and epoxomicin to inhibit proteasome-mediated degradation, the calpain inhibitor II ALLM, and Boc-D FMK, a broad spectrum inhibitor of caspases and bafilomycin to inhibit lysosomal-mediated degradation. The cells were incubated with the inhibitors for 3 h before being harvested, and then the levels of residual Dlg were quantitated by Western blot analysis. The results are shown in Fig. 12, where it is clear that bafilomycin has no effect, suggesting that lysosomal degradation is not involved. In contrast, we observed partial inhibition of degradation with both caspase and calpain inhibitors although specificity may be an issue here since the calpain inhibitor, ALLM, has also been observed to partially inhibit both proteasome-mediated degradation and cathepsin B (24). We also observed that the cocktail of proteasome inhibitors completely inhibited E6*-directed degradation of Dlg, implicating this pathway in E6*-dependent degradation.
FIG. 12.
Degradation of Dlg in the presence of E6* is proteasome dependent. 293 cells were transiently transfected with construct pGW1 HA-Dlg in the absence (−) or presence (+) of pCA18E6* (18E6*).After 24 h, cells were treated with inhibitors against proteasome, caspase, calpain, and lysosomal degradation, as indicated, for 3 h before harvesting. Whole-cell extracts were separated by PAGE, and after Western blotting, the membrane was probed for expressed Dlg using anti-HA antibodies. Transfection efficiency and equal sample loading were controlled for as described in Materials and Methods. β-gal, β-galactosidase.
DISCUSSION
The question of how the transcription of the early regions of high-risk mucosal HPVs relates to expression of the E6 and E7 oncoproteins has been addressed by recent studies, at least in the case of HPV-16- and HPV-18-positive cell lines derived from cervical tumors (30). However, while all of the alpha-group HPVs that we have so far analyzed have a strongly conserved splice donor site early in the E6 ORF, the absence of good consensus downstream splice acceptors before the start of E7 in some alpha-group HPVs such as HPV-59, -52, and -67 suggests that removal of the small intron within E6 may not serve simply to enable E7 translation. Another question that remains unanswered is the function of the E6* proteins. Our previous studies showed an interaction in vitro between HPV-18 E6* and both full-length E6 and the ubiquitin ligase E6-AP and demonstrated that E6* overexpression in HPV-positive but not HPV-negative cell lines led to apoptosis. Despite these observations and subsequent studies from this group concerning the ability of E6* to modulate the functions of the full-length E6 protein (11), there remain many questions regarding the effects of such modulation on the functions of the full-length E6 protein during the normal viral life cycle. Whatever the control of splicing and transcription may be, the effects of E6* need to be considered in relation to both E6 and E7; all three proteins are likely expressed at similar times during the viral life cycle, but there remains a possibility that E6* proteins could be expressed in the absence of full-length E6. This is a question that should be resolved by the recent establishment of productive viral life cycles in raft systems. HPV-16 E7 has been shown to upregulate signaling through the Akt pathway (20, 35), and in this study we have shown that E6* expressed by itself or coexpressed with full-length E6, as would be expected to occur during normal early transcription, leads to degradation of Akt; we have also shown, however, that the degradation is primarily that of Akt localized in a detergent-insoluble fraction of whole-cell lysate. The functional significance of this cellular fraction of Akt and how this relates to the increased activation of Akt in response to E7 expression have yet to be clarified.
In addition to the effects on Akt, we have shown that several of the same targets that are degraded in the presence of full-length E6, such as Dlg and MAGI-1, are also degraded when only E6* is expressed. Although we observed only slight decreases in the levels of MAGI-2 and hScrib when they were ectopically expressed together with HPV-18 E6* in 293 cells, we observed reductions in the protein levels of ectopically expressed Akt, hScrib, and Dlg in Saos-2 cells and clear and reproducible reductions in the levels of endogenous Dlg in HaCaT cells upon expression of E6*. The reductions observed are therefore not restricted to one cell type nor to ectopically expressed Dlg, but clearly the degree to which E6* expression can lead to downregulation of each of these proteins may vary from one cell type to another. Because of potential differences between ectopic expression of E6* and endogenous expression from a stably transfected cell line, we established an HT1080 cell line stably expressing HPV-18 E6* and showed that the endogenous levels of both Dlg and MAGI-1 were reduced in this line. Although the possibility existed that E6* might reduce the cellular levels of these proteins by transcriptional downregulation of their promoters, as has been demonstrated for E6 (3), we considered this highly unlikely for a number of reasons. First, we observed E6*-dependent downregulation of ectopically expressed Dlg and MAGI-1 but not p53 even though all three proteins were expressed from a CMV promoter-driven construct. Second, we observed downregulation of endogenous Dlg in both HaCaT and in HT1080 cells; third, we have demonstrated a clear increase in the rate of Dlg turnover in the presence of E6*. If reduced protein levels had been due to transcriptional repression, the half-life of Dlg would have remained the same. As a final control to show that E6* was not transcriptionally downregulating the promoters of these constructs, we performed a reporter assay to examine the effects of both E6* and full-length E6 on a CMV-Renilla luciferase construct. The results show marginal transcriptional repression by E6, as has previously been reported (3), but not by E6*.
Surprisingly, we observe no interaction between E6* expressed as a GST fusion protein and either HA-tagged Akt or Dlg when these are expressed in cells. This finding is in contrast to recent studies showing that E6* could both interact with novel target PATJ and induce its degradation (29). Although we cannot rule out the possibility that an interaction between E6* and Dlg might require a posttranslational modification of E6*, it seems most likely that the effects of E6* are indirect, possibly through the modulation of signaling pathways that affect Dlg and Akt turnover; studies investigating this possibility are currently ongoing. This may be a property common to all the E6* proteins expressed by high-risk, mucosal, papillomaviruses since we also demonstrate induction of Dlg degradation by the E6* proteins from HPV-16 and HPV-31. Although the question of whether E6-AP is involved has yet to be answered, we note that a mutant of HPV-18 E6* previously shown to be defective for binding to full-length E6 in vitro and reduced for E6-AP binding is as efficient as wild-type E6* for induction of Dlg degradation. Nevertheless we show that the degradation pathway is ultimately proteasome mediated since a cocktail of epoxomicin and MG132 rescues Dlg degradation upon E6* expression. However, we have noted that use of lower concentrations of either MG132 or epoxomicin alone, under conditions that still reproducibly inhibit E6-mediated degradation of Dlg, is barely effective against E6* (data not shown). This observation suggests that the precise characteristics of the degradation pathways, though ultimately linked to the proteasome, are not identical. Interestingly, a recent publication links E6/E6* to the extrinsic caspase pathway (6) and suggests additional complexities to the way in which E6 and E6* interact during the viral life cycle.
Because a subset of cellular targets is degraded in the presence of both E6 and E6*, we asked whether both viral proteins degrade the same pool of cellular targets. We observed increased stability of Dlg when it was phosphorylated by either p38 MAPK or SAPK/JNK and show that while Dlg phosphorylated by p38 MAPK is highly resistant to E6-directed degradation, it can still be degraded in the presence of E6* although exhibiting a slightly increased stability. SAPK/JNK-phosphorylated Dlg shows little change in its susceptibility to degradation by either viral protein. These findings appear at first analysis to be in contrast to previous observations from our group, which showed that osmotic shock-induced phosphorylation of Dlg rendered it more susceptible to E6-directed degradation (17) although in these original studies there was no attempt to separately express and analyze the individual contributions of E6 and E6*. The significance of these observations may relate to cellular localizations of Dlg phosphorylated along different pathways; JNK phosphorylates Dlg in response to osmotic stress, causing accumulation at sites of cell-cell contact (17), and p38 MAPK phosphorylation dissociates Dlg from guanylate kinase domain-associated protein, triggering its release from the cytoskeleton (23). But the most striking finding here is that p38 MAPK-phosphorylated Dlg is efficiently degraded in the presence of E6 and E6* when they are expressed together, suggesting that they function cooperatively and thus explaining our previous findings (17). We also performed a similar experiment to analyze the p38 MAPK phosphorylation of MAGI-1 and its susceptibility to degradation induced when E6 and E6* are expressed separately and together. Although increased turnover of MAGI-1 clearly occurs in the presence of both E6 and E6*, in this case we did not detect cooperation between them, suggesting that they may function independently on this particular target. Finally, we show that expression of an E6 mutant that has been previously demonstrated to be unable to both bind and degrade Dlg in vitro (7) but that, in vivo, would still be able to splice and express E6* also leads to degradation of Dlg in vivo, whereas the same construct with a mutation introduced at the splice donor site is negative for this ability. When an equivalent mutation is introduced in a wild-type E6 construct to inhibit splicing, the ability of the resulting construct to degrade Dlg is reduced but not negative. These data demonstrate that a significant proportion of the ability to degrade Dlg is due to E6* rather than full-length E6 and further confirm that E6* expression can lead to Dlg degradation independently of E6. These observations also suggest ways in which the ratios of E6 to E6* transcripts may have profound effects on the biology of host cells during the normal productive life cycle and during HPV-mediated cancer progression. High E6* protein levels may allow targeting of cellular proteins whose phosphorylation-dependent localization makes them inaccessible to full-length E6. E6* may also inhibit or enhance some of the functions of full-length E6 by interacting directly with it, thus allowing for the possibility of as yet unidentified functions for hetero-oligomers formed by interactions between these two proteins.
Acknowledgments
Anti-HPV-18 E6 monoclonal antibodies 3 and 399 and anti-MAGI-1 monoclonal antibody 236 were generated and kindly provided by the Arbor Vita Corporation, Sunnyvale, CA.
This work was supported, in part, by a research grant from the Associazione Italiana per la Ricerca sul Cancro.
Footnotes
Published ahead of print on 29 July 2009.
REFERENCES
- 1.Chen, J. J., C. E. Reid, V. Band, and E. J. Androphy. 1995. Interaction of papillomavirus E6 oncoproteins with a putative calcium-binding protein. Science 269:529-531. [DOI] [PubMed] [Google Scholar]
- 2.Crook, T., and K. H. Vousden. 1992. Properties of p53 mutations detected in primary and secondary cervical cancers suggest mechanisms of metastasis and involvement of environmental carcinogens. EMBO J. 11:3935-3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Etscheid, B. G., S. A. Foster, and D. A. Galloway. 1994. The E6 protein of human papillomavirus type 16 functions as a transcriptional repressor in a mechanism independent of the tumor suppressor protein, p53. Virology 205:583-585. [DOI] [PubMed] [Google Scholar]
- 4.Filippova, M., H. Song, J. L. Connolly, T. S. Dermody, and P. J. Duerksen-Hughes. 2002. The human papillomavirus 16 E6 protein binds to tumor necrosis factor (TNF) R1 and protects cells from TNF-induced apoptosis. J. Biol. Chem. 277:21730-21739. [DOI] [PubMed] [Google Scholar]
- 5.Filippova, M., L. Parkhurst, and P. J. Duerksen-Hughes. 2004. The human papillomavirus 16 E6 protein binds to Fas-associated death domain and protects cells from Fas-triggered apoptosis. J. Biol. Chem. 279:25729-25744. [DOI] [PubMed] [Google Scholar]
- 6.Filippova, M., M. M. Johnson, M. Bautista, V. Filippov, N. Fodor, S. S. Tungteakkhun, S., K. Williams, and P. J. Duerksen-Hughes. 2007. The large and small isoforms of human papillomavirus type 16 E6 bind to and differentially affect procaspase 8 stability and activity. J. Virol. 81:4116-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardiol, D., C. Kühne, B. Glaunsinger, S. S. Lee, R. Javier, and L. Banks. 1999. Oncogenic human papillomavirus E6 proteins target the discs large tumour suppressor for proteasome-mediated degradation. Oncogene 18:5487-5496. [DOI] [PubMed] [Google Scholar]
- 8.Gardiol, D., A. Zacchi, F. Petrera, G. Stanta, and L. Banks. 2006. Human discs large and scrib are localized at the same regions in colon mucosa and changes in their expression patterns are correlated with loss of tissue architecture during malignant progression. Int. J. Cancer 119:1285-1290. [DOI] [PubMed] [Google Scholar]
- 9.Glaunsinger, B. A., S. S. Lee, M. Thomas, L. Banks, and R. Javier. 2000. Interactions of the PDZ-protein MAGI-1 with adenovirus E4-ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene 19:5270-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 11.Guccione, E., D. Pim, and L. Banks. 2004. HPV-18 E6*I modulates HPV-18 full-length E6 functions in a cell cycle dependent manner. Int. J. Cancer 110:928-933. [DOI] [PubMed] [Google Scholar]
- 12.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1991. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 10:4129-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1993. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol. Cell. Biol. 13:775-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humbert, P., S. Russell, and H. Richardson. 2003. Dlg, Scribble and Lgl in cell polarity, cell proliferation and cancer. Bioessays 25:542-553. [DOI] [PubMed] [Google Scholar]
- 15.Mantovani, F., and L. Banks. 1999. Inhibition of E6 induced degradation of p53 is not sufficient for stabilization of p53 protein in cervical tumour derived cell lines. Oncogene 18:3309-3315. [DOI] [PubMed] [Google Scholar]
- 16.Mantovani, F., and L. Banks. 2001. The human papillomavirus E6 protein and its contribution to malignant progression. Oncogene 20:7874-7887. [DOI] [PubMed] [Google Scholar]
- 17.Massimi, P., N. Narayan, A. Cuenda, and L. Banks. 2006. Phosphorylation of the discs large tumour suppressor protein controls its membrane localization and enhances its susceptibility to HPVE6-induced degradation. Oncogene 25:4276-4285. [DOI] [PubMed] [Google Scholar]
- 18.Menges, C. W., L. A. Baglia, R. Lapoint, and D. J. McCance. 2006. Human papillomavirus type 16 E7 up-regulates AKT activity through the retinoblastoma protein. Cancer Res. 66:5555-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pim, D., A. Storey, M. Thomas, P. Massimi, and L. Banks. 1994. Mutational analysis of HPV-18 E6 identifies domains required for p53 degradation in vitro, abolition of p53 transactivation in vivo and immortalisation of primary BMK cells. Oncogene 9:1869-1876. [PubMed] [Google Scholar]
- 20.Pim, D., P. Massimi, S. M. Dilworth, and L. Banks. 2005. Activation of the protein kinase B pathway by the HPV-16 E7 oncoprotein occurs through a mechanism involving interaction with PP2A. Oncogene 24:7830-7838. [DOI] [PubMed] [Google Scholar]
- 21.Pim, D., P. Massimi, and L. Banks. 1997. Alternatively spliced HPV-18E6* protein inhibits E6 mediated degradation of p53 and suppresses transformed cell growth. Oncogene 15:257-264. [DOI] [PubMed] [Google Scholar]
- 22.Pim, D., and L. Banks. 1999. HPV-18E6* protein modulates the E6-directed degradation of p53 by binding to full-length HPV-18 E6. Oncogene 18:7403-7408. [DOI] [PubMed] [Google Scholar]
- 23.Sabio, G., J. S. Arthur, Y. Kuma, M. Peggie, J. Carr, V. Murray-Tait, F. Centeno, M. Goedert, N. A. Morrice, and A. Cuenda. 2005. p38 gamma regulates the localisation of SAP97 in the cytoskeleton by modulating its interaction with GKAP. EMBO J. 24:1134-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sasaki, T., M. Kishi, M. Saito, T. Tanaka, N. Higuchi, E. Kominami, N. Katunuma, and T. Murachi. 1990. Inhibitory effect of di- and tripeptidyl aldehydes on calpains and cathepsins. J. Enzyme Inhib. 3:195-201. [DOI] [PubMed] [Google Scholar]
- 25.Schneider-Gädicke, A., and E. Schwarz. 1986. Different human cervical carcinoma cell lines show similar transcription patterns of human papillomavirus type 18 early genes. EMBO J. 5:2285-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheffner, M., T. Takahashi, J. M. Huibregtse, J. D. Minna, and P. M. Howley. 1992. Interaction of the human papillomavirus type 16 E6 oncoprotein with wild-type and mutant human p53 proteins. J. Virol. 66:5100-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 28.Smotkin, D., and F. O. Wettstein. 1986. Transcription of human papillomavirus type 16 early genes in a cervical cancer and a cancer-derived cell line and identification of the E7 protein. Proc. Natl. Acad. Sci. USA 83:4680-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storrs, C., and S. Silverstein. 2007. PATJ, a tight junction-associated PDZ protein, is a novel degradation target of high-risk human papillomavirus E6 and the alternatively spliced isoform 18E6*. J. Virol. 8:4080-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang, S., M. Tao, J. P. McCoy, Jr., and Z. M. Zheng. 2006. The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J. Virol. 80:4249-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tao, M., M. Kruhlak, S. Xia, E. Androphy, and Z. M. Zheng. 2003. Signals that dictate nuclear localization of human papillomavirus type 16 oncoprotein E6 in living cells. J. Virol. 77:13232-13247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas, M., R. Laura, K. Hepner, E. Guccione, C. Sawyers, L. Lasky, and L. Banks. 2002. Oncogenic human papillomavirus E6 proteins target the MAGI-2 and MAGI-3 proteins for degradation. Oncogene 21:5088-5096. [DOI] [PubMed] [Google Scholar]
- 33.Thomas, M., P. Massimi, C. Navarro, J. P. Borg, and L. Bank. 2005. The hScrib/Dlg apico-basal control complex is differentially targeted by HPV-16 and HPV-18 E6 proteins. Oncogene 24:6222-6230. [DOI] [PubMed] [Google Scholar]
- 34.Tomaić, V., D. Gardiol, P. Massimi, M. Ozbun, M. Myers, and L. Banks. 2009. Human and primate tumour viruses use PDZ binding as an evolutionarily conserved mechanism of targeting cell polarity regulators. Oncogene 28:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westbrook, T. F., D. X. Nguyen, B. R. Thrash, and D. J. McCance. 2002. E7 abolishes Raf-induced arrest via mislocalization of p21Cip1. Mol. Cell. Biol. 22:7041-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]