Abstract
The interactions between retroviruses and their hosts can be of a beneficial or detrimental nature. Some endogenous retroviruses are involved in development, while others cause disease. The Genome Parsing Suite (GPS) is a software tool to track and trace all Retroid agents in any sequenced genome (M. A. McClure et al., Genomics 85:512-523, 2005). Using the GPS, the retroviral content was assessed in four model teleost fish. Eleven new species of fish retroviruses are identified and characterized. The reverse transcriptase protein sequences were used to reconstruct a fish retrovirus phylogeny, thereby, significantly expanding the epsilonretrovirus family. Most of these novel retroviruses encode additional genes, some of which are homologous to cellular genes that would confer viral advantage. Although the fish divergence is much more ancient, retroviruses began infecting fish genomes approximately 4 million years ago.
All genetic entities that encode the reverse transcriptase (RT) enzyme are referred to as retroids (22). The formal retroid classification includes endogenous and exogenous retroviruses, as well as pararetroviruses (large DNA viruses), retrotransposons with long terminal repeats (LTRs), retroposons that lack LTRs, retroplasmids, retrointrons, and retrons (20, 21, 43, 44). This classification is based on the phylogeny of the RT protein sequence, the slowest evolving of the retroid gene components (30). In a global analysis of the retroid content of the genomes of the teleost fish Danio rerio (zebrafish), Oryzias latipes (medaka), Gasterosteus aculeatus (stickleback), and Tetraodon nigroviridis (green spotted pufferfish), we identified several new retroviruses (7). Here, we expand those studies, report the genomic details of 11 new fish retroviral species, and provide a reconstruction of the evolutionary history of sequenced fish retroviruses.
In general, retroviruses are classified as alpha-, beta-, gamma-, delta-, and epsilonretroviruses, lentiviruses, and spumaviruses. The current characterization of known exogenous and endogenous fish retroviruses places them in the epsilon family (http://www.ncbi.nlm.nih.gov/ICTVdb/) or between this group and the gammaretroviruses (36). To date, only three members of the epsilon family have been conclusively identified, i.e., walleye dermal sarcoma virus (WDSV) and the walleye epidermal hyperplasia viruses 1 and 2 (WEHV1 and WEHV2), and tentative members include perch hyperplasia virus and snakehead fish retrovirus (SnRV), (http://www.ncbi.nlm.nih.gov/ICTVdb/). Other fish retroviruses include the Atlantic salmon swim bladder sarcoma virus (SSSV) and zebrafish endogenous retrovirus (ZFERV) from D. rerio; both appear to fall between the gamma and epsilon families (36). Studied in less detail is the ERV_Tet species in T. nigroviridis (15), and the retrovirus-stickleback (RV-stickleback), RV-brook trout, RV-freshwater houting, and RV-pufferfish retroviruses only identified by using PCR (17).
WDSV is associated with the development of seasonal tumors in walleye and, along with WEHV1 and WEHV2, has four accessory genes, two of which are cellular d-type cyclin homologues (orfA and orfB). These cyclin homologues may function to induce cell proliferation to allow viral replication (26). orfC in WDSV appears to alter mitochondrial function, resulting in apoptosis, which may contribute to seasonal tumor regression, and is homologous with genes in WEHV1 and WEHV2 (35). Perch hyperplasia virus is basically indistinguishable from WEHV (37) and is therefore not included in the present study. The SnRV genomic organization, tRNA primer, sequence homology, and transcriptional profile are unique and has prevented this virus from being decisively placed into any retroviral family (16). SSSV is associated with leiomyosarcomas in the swim bladders of Atlantic salmon, and it is not present in healthy genomes, suggesting that it has only recently begun infecting Atlantic salmon. This exogenous virus has very high proviral copy numbers per cell and, along with ZFERV, has not been placed into any known viral families (36).
The fish genomes examined here represent a diversity of sizes and divergence times. The zebrafish genome is the largest with about 1,700 Mbp, followed by the medaka with a genome of approximately 1,000 Mbp and the stickleback with a genome of 675 Mbp, while the pufferfish has the smallest genome (385 Mbp) (45). The lineages containing O. latipes and T. nigroviridis are estimated to have diverged from that of D. rerio approximately 290 million years ago (mya), whereas the ancestors of O. latipes and T. nigroviridis diverged from each other about 195 mya (40). Although G. aculeatus was not included in the study of Steinke et al., it and T. nigroviridis are the most recently diverged from each other (49).
The results reported here are from the Genome Parsing Suite (GPS) software (31) used for identification, classification, and comparison of the retroviral content of the D. rerio, O. latipes, G. aculeatus, and T. nigroviridis genomes. The approach of the GPS is radically different from RepeatMasker, which is used to mask out and count repetitive elements using consensus DNA sequences (RepeatMasker Open-3.0 [http://www.repeatmasker.org]). Repeat Masker and similar methods suffer from the loss of signal due to mutational saturation because DNA is used to query a genome rather than amino acid sequences. DNA sequence libraries are also unable to detect new components, such as the novel LTRs discovered in many of the fish retroviruses described here.
Although the structural genes of retroid agents can be highly divergent, the RT gene is considerably more conserved since it is essential for autonomous transposition and the continuance of an exogenous viral life cycle (30). Although any protein sequence can be used in the GPS, in the present study it is populated with a representative diversity of RT protein sequences that afford a thorough query into the retroid content of the four fish genomes, allowing the discovery of multiple species of new retroviruses. We identify and characterize 11 new species of fish retroviruses, with novel LTRs and genes, thereby significantly expanding the epsilon family.
MATERIALS AND METHODS
GPS.
GPS software identifies retroid signals in any genome database (31). Input queries representative of the diversity of RT-encoding agents, along with a set of host species-specific queries, allows a more in-depth characterization of the identified retroids. In the present study, the GPS was populated with 101 queries encompassing all retroid families to ensure detailed classification; however, only the results of the identified retroviruses are presented here. In addition to the 92 queries presented in our global analysis of retroid agents in the four fish genomes (7), 9 retrovirus queries have been added to ensure a thorough retroviral classification: three new O. latipes endogenous retrovirus (OLERV) and three new G. aculeatus endogenous retrovirus (GAERV) queries, as well as the previously identified, WEHV1 and WEHV2 (AF133051 and AF133052) and SSSV (DQ174103) (Fig. 1).
FIG. 1.
Fish retrovirus query gene component maps. The GPS accesses a database populated by retroid genome sequences. This query library contains all of the genes and noncoding component sequences, which the GPS uses to identify and reconstruct potential retroid agents found in genome databases. All fish retroviruses used as queries are presented in phylogenetic order, with those that are novel indicated in boldface. The complexity of intra- and intergenic, as well as interpolycistronic, variation in accumulation of stop codons (asterisks) and frameshifts (a shift to another line) is illustrated. Stop codons (within separate boxes), with or without a frameshift, are usually found between the gag/pol and pol/env polycistrons. Stop codons that are shown within a gene box are intragenic, while those within a separate box are accompanied by an intragenic frameshift. Note that genes with intragenic frameshifts are labeled multiple times; these do not indicate duplications. If a potential retroid agent encodes all of the genes in a specific query-component library, it is considered full-length. Those gene abbreviations not defined in the text as expected retroviral genes are as follows: CNPase, 2′,3′-cyclic nucleotide 3′-phosphodiesterase homolog; UNK, unknown region; UTR, untranslated region; ORF, open reading frame; CYCD/CYDh, cyclin D homologues; and Macro, the viral homologues of the cellular macro domain.
New species of retroviruses were classified phylogenetically. Fifty of the new full-length RT genes from all four fish genomes cleanly clustered into 11 different groups regardless of phylogenetic method used (data not shown). These 11 species were corroborated by the same analysis using the protease (PRO), RNase H (RH), and integrase (IN) protein sequences with the same results. For each of the 11 species the representative query was based on the following criteria: completeness (the presence of the most gene components), the least number of stop codons and frameshifts, and the presence of the motifs defining each of the enzymatic core proteins. The inclusion of these new queries in the GPS analysis resolved all newly identified retroviruses in the four fish genomes into 1 of these 11 species without ambiguity. In addition, phylogenetic trees were constructed for all homologous protein sequences of all of the full-lengths viruses (Table 1) in each species to ensure that outliers or recombinants were not selected as representative queries (data not shown).
TABLE 1.
New LTR-bound retroviral information in three fish genomesa
| Fish species and virus type | Full/InDel mutant/junk-movers | FS/SC at gag/pol and pol/env | Perfect |
|---|---|---|---|
| D. rerio | |||
| DRERV1 | 4/6/2 | 1/1/0 | 0 |
| DRERV2 | 1/2/2 | 0 | 0 |
| DRERV3 | 2/3/1 | 1/1/0 | 0 |
| DRERV4-gag-env | 6/11/0 | 0 | 0/1*/0 |
| DRERV5 | 1/1/0 | 1/1**/0 | 0 |
| ZFERV | 1/2/0 | 1/1/0 | 0 |
| O. latipes | |||
| OLERV1 | 1/1/7 | 1/0/0 | 0 |
| OLERV3 | 1/1/1 | 1**/0/0 | 0 |
| G. aculeatus | |||
| GAERV1 | 1/2/0 | 1/0/0 | 0 |
| GAERV2-env | 6/19/0 | 1/1/0 | 0 |
| GAERV3 | 1/21/2 | 0 | 0 |
Data are grouped by teleost host. All copies of DRERV4 lack the gag and env genes, whereas GAERV2 lacks the env gene. Full copies have all components of the query present in order (Fig. 1). InDel mutants are insertion/deletion mutants relative to the query that may have internal indels in one or more genes or completely lack a gene relative to the query genome. Junk movers are fragments of retroviral genomes bounded by viable LTRs. Copies of each class that have stop codons and/or frameshifts (FS/SC) at one or both regions of translational control at the gag/pol and pol/env polycistronic boundaries are considered potentially active. “Perfect” indicates all genes present relative to the query without stop codons or frameshifts. The value denoted by an asterisk is perfect except for insertions of 700 and 500 nucleotides into UTR1 and UTR2, relative to the query. The other deletion mutants that could be functional either have deletions in a UTR or in the env gene. **, Only two potentially actives have frameshifts and or stop codons at gene boundaries within a polycistron as described in the text.
Host genomes.
The D. rerio, version Zv7, the G. aculeatus version gasAcu1, and the T. nigroviridis tetNig1 chromosomal genomes are from the University of Santa Cruz Genome Bioinformatics Website (http://genome.ucsc.edu/), and the O. latipes MEDAKA1 genome is from the Ensembl website (19).
LTR analysis.
Since the completion of previous GPS analyses (7, 31), new functionality has been integrated into the GPS. When the GPS finds a sequence that has identity to known retroid components, but no LTRs, and these regions do not pull out any known LTRs when BLAST searches (4) are executed, a test for novel LTRs is performed. To facilitate the identification of new LTRs we compared several available LTR identification methods. LTR_FINDER (48) outperforms all others at identification of de novo LTRs. LTR_FINDER was run at the default parameters, with the “automask high repeat region” option enabled. All LTR analysis presented here is based on LTR_FINDER results, which includes identification of the tRNA priming binding sites (PBS), target site repeats (TSR), and polypurine tracts (PPT).
LTRs are identical at each new integration event due to the complexity of the process of reverse transcription (11). The first new DNA LTR is transcribed from two different regions of each of the RNA LTRs. The high rate of mutation in this process occurs when RNA is copied to DNA. Making an identical copy of the first new DNA LTR creates the second new DNA LTR. The encoding portion of the retrovirus genome is free to accumulate mutations by the RT-process, whereas the LTRs are identical copies of one another, with only the initial copy having undergone reverse transcription from RNA to DNA. Mutational differences between the two DNA LTRs, therefore, only occur after integration during the replication process. Mutations in the rest of the endogenous viral genome are due to a combination of the RT process and host DNA polymerase replication. The accumulated mutations of the once identical DNA LTR pairs are the appropriate and standard data to use to calculate the insertion dates of retroviruses and retrotransposons into host genomes (12). Our datasets only include those samples with LTR pairs at least 80% identical. Due to the observation of variable LTRs for sets of retroviruses with highly conserved enzymatic core proteins (Fig. 2), LTR pairs are considered homologous and in the same set if 3′ and 5′ identities among LTRs are greater than 50%, and size variation is less than 100 nucleotides. Interestingly, these criteria often placed LTR pairs with the same PBS signal into the same set, further supporting homology (Table 2).
FIG. 2.
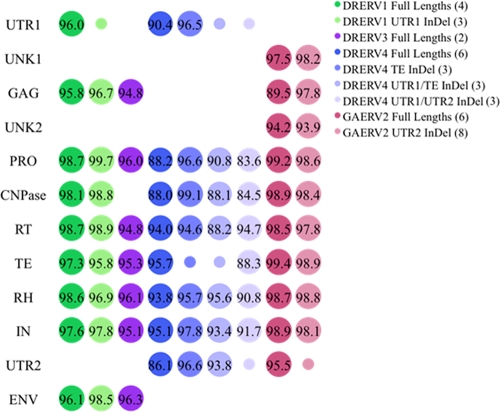
Relationship of gene sequences of full-length and representative indel mutant genomes. The y axis is the retroviral components (as defined in the text and Fig. 1); the x axis is the set of full-length and example indel mutants. Percent nucleotide identities are represented by bubble size and labeled on each bubble. Copy numbers are shown in parentheses next to the retrovirus name. The percent identities are the average pairwise sequence identity over each gene set. Note that not all queries have all components, as shown by blank regions. A deletion or insertion mutation (InDel) in the same region or gene is represented by a smaller bubble. Although most of the deletions overlap the same region, some are in a different position; therefore, no relationship values are given.
TABLE 2.
Identification of novel LTRsa
| Family.set | No. active/ total no. of copies | LTR length (bp) | LTR (% identity) | PBS | PPT | TSR |
|---|---|---|---|---|---|---|
| DRERV1.1 | 1/5 | 434 | 97.90 | Asn | Yes | Yes |
| DRERV1.3 | 1/3 | 553 | 97.10 | Trp | Yes | 2/3 |
| DRERV2 | 0/5 | 455 | 98.56 | Trp | Yes | Yes |
| DRERV3.1 | 1/2 | 329 | 96.95 | Trp | Yes | 1/2 |
| DRERV3.3 | 1/1 | 399 | 100.00 | Pro | Yes | Yes |
| DRERV4.1 | 0/12 | 390 | 99.77 | Leu | Yes | Yes |
| DRERV4.4 | 1/1 | 563 | 99.60 | Null | No | No |
| DRERV5 | 2/2 | 531 | 99.90 | His | No | Yes |
| ZFERV | 2/3 | 617 | 99.90 | Cys | Yes | Yes |
| GAERV1.1 | 1/2 | 962 | 99.95 | Lys | Yes | Yes |
| GAERV2.1 | 2/17 | 499 | 97.90 | Leu | Yes | 5/17 |
| GAERV3.1 | 0/19 | 505 | 98.90 | Ser | 18/19 | 15/19 |
| OLERV1.1 | 1/1 | 800 | 98.50 | Null | Yes | No |
| OLERV3.1 | 1/1 | 510 | 99.80 | Null | Yes | No |
All LTR sets with potentially active copies are presented, in addition to the three sets with the largest number of samples. LTR length and 3′-to-5′ identities are the averages of each set. If the PBS is present, it is indicated by the type of amino acid tRNA that binds to the PBS, e.g., Asn indicates that the PBS is for tRNA-Asn. The PPT and TSR are indicated by “yes” if present in all or as a fraction of the total of each data set. LTRs, PBS, PPT, and TSR were identified by LTR_FINDER (48) and clustered as described in Materials and Methods. All fish retrovirus acronyms are as defined in the text.
The CLUSTAL multiple alignment method from MEGA4 (42) was used to align all LTR pairs. MEGA4 provides a variety of distance estimate methods (P-distance, Jukes-Cantor, Kimura two-parameter, Tajima-Nei, Tamura three-parameter, and LogDet), and they all produce similar distance values (d) within the statistical error (SE) of the data (data not shown). In the data presented here the Kimura two-parameter method was used to calculate the d estimations and the SE for all LTR pairs (42). The rate variation among sites was modeled with a gamma distribution (shape parameter = 8). All positions containing gaps and missing data were eliminated from the data set (complete deletion option). SE estimates were obtained by using the analytical formula option in MEGA4. The insertion times were estimated by using the following equation: t = d/2r. The rate (r) of neutral evolution was estimated for the Tetraodon and Takifugu genuses by using 5,802 orthologues (23) and used for all insertion estimates. For multiple sample sets the maximum and minimum distances were used to display the range of insertion times. The 95% confidence interval (CI) for each insert time was calculated by the SE·1.96/2r.
Multiple alignment and phylogenetic analysis.
All protein sequence multiple alignments used to create trees were made by using mCOFFEE (http://www.igs.cnrs-mrs.fr/Tcoffee/tcoffee_cgi/index.cgi?stage1=1&action=MCOFFEE::Regular). DNA gene alignments were produced by MAFFT at online default parameters (24).
Phylogenetic analyses were conducted with MEGA4 (42) and MrBayes3.1 (38). Neighbor-joining (NJ), minimum evolution (ME), maximum parsimony (MP), and the unweighted pair group method with arithmetic mean (UPGMA) from MEGA4 were used for each data set, and all of these methods produced similar topologies to the MrBayes3.1 result.
The Bayesian method, however, required some parameter settings beyond the initial choosing of a model and evolutionary rate. Preliminary observations of consensus trees generated with a mixed amino acid model and eight category gamma distribution rate produced high posterior probabilities with a number of incorrect internodes even after 100,000's of iterations and apparent convergence. These results are in contrast to the other methods (NJ, ME, MP, and UPGMA), which give biologically supported internodes with lower bootstrap support values. This issue has been attributed to internal branch lengths that are too close for MrBayes3.1 to correctly resolve (1). The MrBayes3.1 documentation suggests the use of topology constraints that allow the incorporation of prior biological knowledge regarding those highly related sequences that should branch together. These constraints provided the ability to generate biologically correct internodes in addition to high posterior probabilities for these nodes (2). In our studies, constraints were designed from previous topologies agreed upon by other phylogentic methods (NJ, ME, MP, and UPGMA), as well as pairwise percent-identity comparison information calculated from the sequences. It should be noted that although the constraint parameter was invoked for the trees, MrBayes3.1 overrides any constraint if the data do not support it. This method, with appropriate constraints, produced trees with higher confidence at each node than did any of the other methods.
RESULTS
There are three classes of GPS-identified, LTR-bound, endogenous retroviral information in fish genomes (Table 1). “Full-length” retroviruses are those that have all gene components of the parent query sequence, although they may have several small internal insertions and/or deletions less than 100 nucleotides, and all motifs of the encoded enzymes are present. As defined here, “indel mutants” have one or more insertions or deletions of 100 nucleotides or larger internal in any gene and/or completely lack a gene. “Junk-movers” are fragments of retroviral genomes. There are 45, 12, and 52 copies of LTR-bounded retroviral information, found in the D. rerio, O. latipes, and G. aculeatus genomes, respectively. Most of these copies are indel mutants (Table 1), and in some cases there are highly related, multiple copies of the same indel mutant genome. The GPS also characterized the previously identified ERV_Tet viruses of T. nigroviridis (data not shown). There is one full-length ERV2_Tet and two indel mutations and only one full-length for ERV3_Tet. Interestingly, for ERV4_Tet, neither the two full-length nor the 19 indel mutant genomes have discernible LTRs, although the sequences of the gene components are well conserved. None of the T. nigroviridis copies fulfill the criteria for being potentially active. The retroviruses of T. nigroviridis are only discussed here in the context of the 11 new retroviral species.
In general, a retrovirus expresses three polycistrons, each encoding multiple proteins: three to five structural proteins from gag; the PRO, the RNA-dependent DNA polymerase (RdDp) comprised of the RT and RH functions, separated by a viable tether region (TE), and the IN from pol; and the two glycoproteins of env. Functional core enzymes encoded by pol are necessary for the autonomous lifestyle of all retroviruses. In some cases, additional enzymatic genes have been acquired from the cell (e.g., the dutp). The analysis for the gag and pol protein sequences encoded by all LTR-bounded samples reveals five D. rerio (DRERV1 to -5), two O. latipes (OLERV1 and -3), and three G. aculeatus (GAERV1 to -3) species of endogenous retroviruses.
The pairwise percent identities for LTR-bound full-length and indel mutation genomes were calculated as the within-set average of all members in each data set to one another from the multiple alignments of nucleotide sequences for each homologous group of protein encoding components (Fig. 2). All protein sequences are highly conserved within each full-length and indel mutant set. In spite of indel mutations, and even the absence of some genes, all of the gene sequences for full-length and indel mutations of a given species (i.e., all DRERV1s) are also highly conserved (data not shown). In addition, some open reading frames of unknown function (UNK1 and -2), and untranslated regions (UTR1 and UTR2) filled with stop codons abutting the LTRs were also identified (Fig. 1 and 2).
Analysis of frameshifts and stop codons.
There is one expected region of regulatory stop codons and frameshifts that controls expression of the pol polycistron. This polycistron is expressed in approximately a 20:1 ratio by readthrough of the stop codon and/or frameshift at the gag/pol boundary. Our analysis indicates that a region between the last gene of pol and the beginning of env has many stop codons and sometimes a frameshift as env is expressed by splicing. In addition to these two expected translationally regulated expression regions, infectious exogenous walleye retroviruses also have frameshifts and/or stop codons at various gene boundaries encoded within the polycistrons (35). We take into consideration these additional criteria to define potentially active retroviruses in all fish genomes (Table 1). Each LTR-bounded potential endogenous virus genome was scanned for the positions and numbers of frameshifts and stop codons.
DRERV1 has a full-length genome and a UTR indel mutant, each with the two expected translational regulatory regions. A third potentially viable copy lacks about 200 nucleotides in the middle of the IN gene. This region of the D. rerio genome has a string of several hundred “N” symbols in it, indicating that an unspecified length has not been sequenced. The discontinuity in the region of IN homology is only 200 nucleotides, however, indicating the actual size of the unsequenced region. DRERV3, DRERV5, and ZFERV each have one full-length mutant and one indel mutant that are potentially viable. All potentially viable indels are mutated in the env gene, TE, or untranslated regions. The only endogenous retrovirus found in any of the four fish genomes that is without mutation leading to stop codons or frameshifts is in the DRERV4 species, all copies of which lack both gag and env genes. The potentially active DRERV4 genome has inserts of 700 and 500 nucleotides in the UTR1 and UTR2 regions, respectively, relative to the query sequence; otherwise, all open reading frames are open to the translational machinery. Within the three O. latipes retrovirus families there are only two full-length gene copies, and both appear viable. OLERV2 lacks LTRs, so viability is not expected. G. aculeatus has the most retroviral copies, with 52 LTR-bounded genomes, of which 8 are full length and 42 are indel mutations. Two of the full-length genomes and one indel mutant genomes are potentially active (Table 1). The LTR-bound copies of retroviral information we call junk-movers are missing most other components and have accumulated many more stop codons and frameshifts than the other two classes.
There are two other sets of retroviral samples that need to be mentioned. Although a dozen or so fragmented copies of different fish retroviruses can be found in each of the four hosts (e.g., ERV4_Tet in G. aculeatus), few of these copies have RTs with greater than 10% identity to the query, and most lack other gene components. These genomes are highly degraded, suggesting there have been multiple infections that are no longer active (data not shown). Only copies of OLERV2 and ERV_Tet have significant sequence conservation to all gene components but lack LTRs. There are also a few copies of some of the other newly described retroviruses that lack any discernible LTRs. Although a few of these copies have highly conserved RTs, the other components are indel mutants, and there is a significant accumulation of stop codons and frameshifts throughout these genomes relative to the copies that have LTRs. The copy number of the LTR minus genomes is proportional to the number and diversity of their LTR-bound relatives, with D. rerio having the most and T. nigroviridis having the least (data not shown).
Novel LTRs.
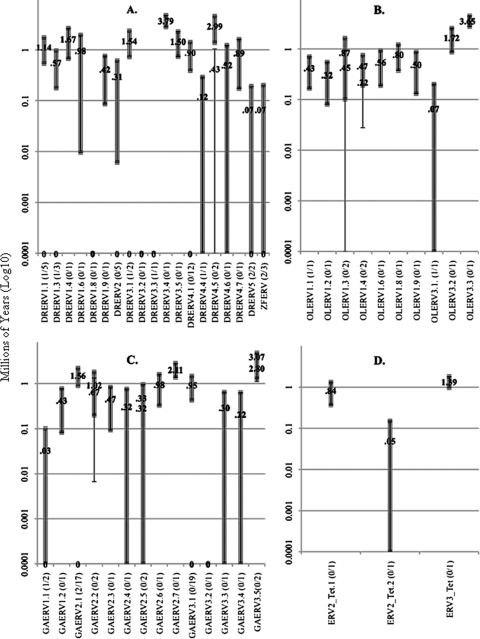
Although the full-length and indel mutant retroviral protein encoding genes are highly conserved (Fig. 2), the LTRs within a given virus species are highly variable in sequence and length (Table 2). When the 5′ and 3′ termini of the newly identified retroviruses were run through a BLAST search, none of them pulled out other LTR sequences, signaling their novelty. Given the sequence and length variability of these LTRs, they were clustered into subsets as described in Materials and Methods, resulting in 46 sets, the largest with 19 samples. The 14 potentially active fish retroviruses have 11 different types of LTRs (Table 2). Among all three classes—full-lengths, indel mutants, and junk-movers—there are LTRs with 100% identity (Fig. 3). While 7 of the 14 potentially active endogenous retrovirus LTR sets are 100% identical, the others inserted between 0.03 and 0.70 mya (Fig. 3). D. rerio has the largest diversity of endogenous retroviruses, with many copies of identical LTRs indicating recent activity, while the oldest LTRs inserted 3.79 mya. T. nigroviridis has the fewest endogenous retroviruses inserting into the genome between 0.05 and 1.39 mya (Fig. 3), all of which have accumulated multiple intragenic stop codons and frameshifts (Fig. 1).
FIG. 3.
Estimates of LTR insertion times. Insertion time estimates are shown for all 46 homologous LTR data sets for D. rerio (A), O. latipes (B), G. aculeatus (C), and T. nigroviridis (D). The y axis shows the years in log scale to display the complete range of insertion times. The x axis indicates the datasets and potentially active copies per total sample size [e.g., DRERV1.1(1/5)]. Sets with single samples are displayed with single values in million of years, while sets with multiple samples have both the maximum and the minimum insertion dates of the set shown. The maximum and minimum 95% CI for multiple sample sets are indicated by thick and thin lines, respectively. The CIs overlap when the maximum and minimum insert dates are close to one another. For single sample sets, the 95% CIs are indicated on the single insertion values by a complete overlap of thick and thin lines. Identical LTRs have no change and are, therefore, the most recent integrations. The distance values, SEs, insertion times, and CIs are calculated as described in Materials and Methods.
Most junk-mover LTRs are not in the same LTR sets as full-length or indel genomes. The LTR insertion estimates from the junk class are 0 to 3.65 mya (Fig. 3, DRERVs 1.8, 1.9, and 3.5; OLERVs 1.3 to 1.9 and 3.3; and GAERV 3.5). In the DRERV2 clade, however, the LTRs of one full-length genome, two copies of a genome with a similar deletion pattern in all three polycistrons, and two junk-movers have highly related LTRs, including conservation of the same PBS (Table 2), although none of the copies are potentially active (Table 1). All other homologous LTRs sets with more than one sample are mixtures of full-length and indel mutant genomes that have variable maximum and minimum insertion dates (Fig. 3).
Phylogenetic reconstruction using RT sequences.
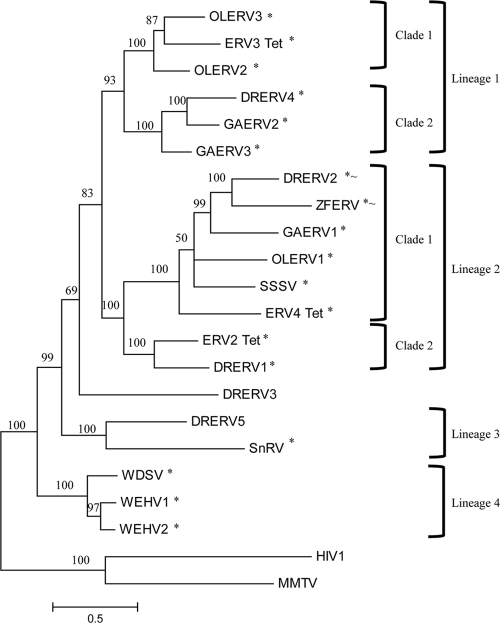
Three of the four lineages of fish retroviruses have representatives from more than one species of fish (Fig. 4). The first lineage contains two clades. The first clade has two O. latipes and one T. nigroviridis retrovirus species. Only OLERV3 has a potentially active copy (Table 1). The estimated insertion of this copy is 0.07 mya, whereas the inactive ERV3_Tet inserted about 1.39 mya (Fig. 3). OLERV2 has no discernible LTRs. Lineage 1, clade 2, has the highest copy numbers, with representatives from D. rerio and G. aculeatus. In this clade, all GAERV3 copies have discernible gag and env polycistrons, while DRERV4 has neither, and GAERV2 lacks the env (Fig. 1). The insertion estimates for the three viable copies of this clade are DRERV4 at 0.12 mya and two GAERV2s—a full-length genome at 0.70 mya and an indel mutation with 100% identical LTRs (Fig. 3).
FIG. 4.
Fish retrovirus phylogeny. RT amino acid sequences from sequenced fish retroviruses were used to reconstruct the evolutionary history of the fish retroviruses. The consensus MrBayes3.1 tree with posterior probabilities at each node and branch lengths scaled to expected substitutions per site is shown. This tree was generated over 80,000 iterations using an eight-category gamma distribution rate with the mixed amino acid model. Six prior constraints were invoked on relationships that have a 100% confidence: (i) OLERV2, OLERV3, and ERV3_Tet; (ii) GAERV3, DRERV4, and GAERV2; (iii) OLERV1, SSSV, ZFERV, DRERV2, GAERV1, and ERV4_Tet; (iv) ERV2_Tet and DRERV1; (v) WDSV, WEHV1, and WEHV2; and (vi) HIV-1 and mouse mammary tumor virus (MMTV). Asterisks indicate queries that encode a CNPase region and tildes show those with macro domains (Fig. 1). All fish retrovirus acronyms are as defined in the text. Mouse mammary tumor virus and HIV-1 serve as outliers.
The second lineage of fish retroviruses contains two clades. The first clade consists of viruses from five different fish genomes. ZFERV has two potentially active copies, one with 100% identical LTRs and the other inserted about 0.05 mya, with a third, degraded copy at 0.07 mya (Fig. 3). DRERV2 has no potentially active copies, although one of the junk-mover LTR pairs are 100% identical, suggesting recent movement while all other copies range back to 0.31 mya. The only potentially viable copy of GAERV1 inserted about 0.03 mya. The LTRs of the potentially viable full-length copy of OLERV1 inserted approximately 0.43 mya, while the only other copy is an indel genome inserting about 0.32 mya (Fig. 3, OLERVs 1.1 and 1.2). All other copies of OLERV1 are junk-movers, all inserting between 0.22 and 0.87 mya. SSSV's LTRs are 100% identical, as is expected of an exogenous virus. ERV4_Tet is the most degraded fish retrovirus, without the gag or env polycistrons or LTRs, and multiple frameshifts and stop codons have accumulated within genes. The second clade consists of only two members: ERV2_Tet and DRERV1. The nonviable ERV2_Tet is highly degraded by frameshifts and stop codons. DRERV1 has two viable copies each with 100% identical LTRs and a variety of types of indel mutants with insertions dating as far back as 1.67 mya. DRERV3 is the outlier to lineages 1 and 2, and there are two copies that are potentially viable (Table 1), each with 100% identical LTRs. One DRERV3 indel mutant is the oldest, dating back to 3.79 mya (Fig. 3, DRERV3.4).
The third lineage of fish retroviruses includes only DRERV5 and the previously identified SnRV from the snakehead fish. There are only two copies of DRERV5, and they are both potentially viable (Table 1), with conserved LTRs that have the same PBS (Table 2). One is a full-length copy with 100% identical LTRs, and the other has a deletion in the TE region that inserted about 0.070 mya. The fourth lineage consists entirely of previously identified exogenous walleye retroviruses, which are closely related with 100% identical LTRs, as expected.
Identification of additional genes. (i) cnp gene.
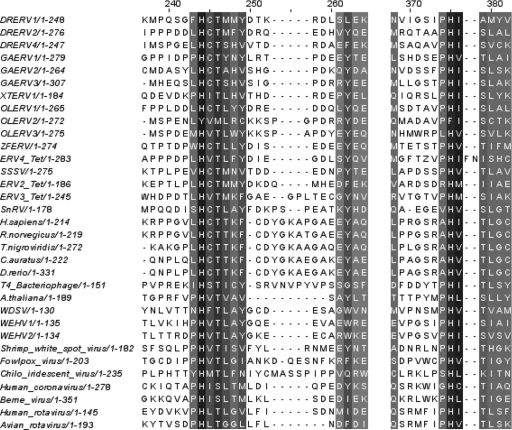
Our use of HHpred analysis (39) reveals that of the 11 new fish retroviral families discovered in the present study, 9 have a homologue to the 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNPase). The HHpred analysis of the aligned nine homologues showed a probability of 96.4% homology to the CNPase family. Only DRERVs 3 and 5 lack the cnp gene. This gene is also present in WDSV, WEHV1 and WEHV2, ZFERV, and SnRV (28). The cnp gene is located in different polycistrons and between different genes in various fish viruses: env and cycd genes in WDSV, WEHV1, WEHV2, PRO, and RT in ZFERV and the RH and IN in SnRV (Fig. 1). We have identified the cnp gene in three of the five virus species of D. rerio and in all species of O. latipes, G. aculeatus, and T. nigroviridis retroviruses (Fig. 1). Each of these cnp genes is found between the PRO and RT, as seen in ZFERV. These retroviral proteins average 242 amino acids and 27% amino acid identity. The cellular copies average 47% amino acid identity. All cellular and viral CNPase sequences share an average of 22.1% amino acid identity. The CNPase active site is composed of two histidine residue motifs, Hh(1)h (h, a hydrophobic residue) (28), which are conserved throughout both the cellular and the viral homologues, with the exception of OLERV2. It is clear from the alignment that although OLERV2 has no longer conserved the H residues, many other features of the motif sites are well conserved. These conserved histidine motifs are at residues 244 and 374 in the 33-sequence alignment (Fig. 5). Although the host genes of goldfish (6) and green puffer fish (23) are in the Pfam database as CNPases, our data show that most of the new endogenous retroviruses encode these proteins. In a recent GPS analysis of the Xenopus tropicalis genome a cnp gene is also found in a novel endogenous retrovirus (XTERV1) between the PRO and the RT (unpublished results).
FIG. 5.
Multiple alignment of the catalytic motifs of the CNPase homologues. Nine of the eleven novel fish retroviruses encode a CNPase homologue. The protein sequence alignment of the two histidine motifs at residues 244 and 374 are shown for the nine fish retrovirus containing a CNPase homolog and other CNPase genes. The alignment was rendered in Jalview and highlighted by conservation. The highest level of conservation is highlighted in black and the lowest in light gray; white indicates no conservation. Accession numbers not found in the text are as follows: Homo sapiens, PDB 1WOJ; Rattus norvegicus, PDB 2ILX; T. nigroviridis, CAG02336; C. auratus, PDB 2I3E; D. rerio, XM_001332834; Arabidopsis thaliana, PDB 1FSI; T4 bacteriophage, 15389; shrimp white spot syndrome virus, 17158251; fowlpox virus, 9634695; Chilo iridescent virus, 15078840; human coronavirus, 608643; Berne virus, 58777; human rotavirus, 6009563; and avian rotavirus, 2754604.
(ii) macro domain.
Cellular and viral macro domain genes form a protein superfamily throughout the three domains of life, which are classified into various families (see macro domain superfamily [http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?hslf=1&uid=cd02749&#seqhrch]). The macro domain between the IN and the ENV of ZFERV was previously identified, and we discovered a copy of this gene in the new species DRERV2 in the same position (Fig. 1) by tBlastn analysis (4). Multiple alignment of ZFERV and DRERV2 to representative members of macro domain proteins show highly conserved regions corresponding to the documented macro domain motifs (14). Phylogenetic reconstruction using MrBayes3.1 puts ZFERV and DRERV2 together, as outliers to all of the cellular macro domains, but within the Pao1p-like family, with a posterior probability of 100 (data not shown).
DISCUSSION
The GPS starts with all unique BLAST query-retrieved RT hits as indicators of potentially new retroviruses and assesses the degree of retained gene components. LTR_FINDER (48) is used to identify new LTRs. This approach allows for the discovery of LTRs not present in any database and provides all instances of retrovirus full-length and deletion mutant genomes, down to the smallest statistically significant RT fragment. Although all of the fish genomes studied here have a variety of nonfish and fish-specific unique RT hits relative to their genomic size (D. rerio, the largest, has more, and T. nigroviridis, the smallest, has the least), most of these are small RT fragments (7). Analysis of the retrovirus query sequence RT hits in the genomes of the model fish species D. rerio, O. latipes, and G. aculeatus, however, reveals 11 new retroviral species.
There are three classes of LTR-bound retroviral information in the fish genomes: full-length genomes, indel mutants, and junk-movers. LTR sets that are 100% identical are found in all three classes, suggesting insertion more recent than 22,000 years since this is the earliest measurable divergence of the fish LTRs (Fig. 3). Regulation of polycistronic gene expression in retroviruses is by way of translational recoding of various stop codons and frameshift at the gag/pol junction. Whereas env is expressed via splicing, avoidance of readthrough at the pol/env junction is assured by a second set of stop codons and frameshifts between pol and env polycistrons. These two regions of translational regulation are present in all of the retroviral full-length and indel mutant genomes analyzed, suggesting that these stop codons and frameshifts are under evolutionary selection to maintain correct polycistronic expression. Only 14 full-length and indel mutant fish retroviral genomes, however, are potentially capable of an autonomous viral life cycle (Table 1), whereas other copies are no doubt involved in nonautonomous mechanisms of replication and movement. The 11 new species of fish retroviruses described here are defined by the phylogeny of their RT protein sequence relationship (Fig. 4).
Cellular homologues in fish retroviruses.
The classical architecture of a retrovirus is: 5′LTR-gag-pol-env-3′LTR. Each of the three polycistrons encode structural, enzymatic, and surface proteins, respectively, and a variety of small, spliced genes in various retroviral families. In addition, some retroviruses encode cellular homologues, e.g., the more recently acquired oncogenes, in contrast to the older ones, such as the dutp gene (29).
Nine of the eleven new fish retroviruses encode a protein homologous to the cellular protein CNPase. Eukaryotes contain two ancient families of 2′,3′-cyclic nucleotide phosphodiesterases involved in RNA processing, transcriptional coactivation, and posttranscriptional gene silencing. One family of phosphodiesterases catalyzes at least two steps in splicing tRNA introns. Domains in a second family of phosphodiesterases, restricted to vertebrates and insects, suggest a role in signal transduction. It is this second family that is also found in some retroviruses and in various other RNA and DNA viruses (Fig. 5), where they may function in capping and processing viral RNAs (14). These retroviral phosphodiesterases were previously identified in WDSV, WEHV1, WEHV2, ZFERV, and SnRV (28).
The cnp genes are found in three different positions in various fish retrovirus genomes. The cnp genes of GAERV1, GAERV2, GAERV3, DRERV1, DRERV2, DRERV4, OLERV1, OLERV2, OLERV3, ERV2, ERV3, ERV4_Tet, SSSV, and ZFERV are located between the PRO and RT, while in SnRV the gene is between the RH and IN genes (Fig. 1). This insertion pattern is similar to that of the mammalian retroviral dutp gene acquisitions (5), suggesting that these regions are susceptible to new gene insertions. In contrast, the walleye CNPase viral genes are located in the env polycistron. The genomic location of this gene will affect its expression. In most cases, the cnp gene would be expressed as part of the pol polycistron (Fig. 1) in low copy numbers relative to the homologues in the walleye viruses found in the env polycistron.
In addition to the cnp gene acquisition, two fish retroviruses have a phosphoesterase of the macro domain superfamily. This gene is found in ZFERV (28), and it is present in the one of the new retroviruses, DRERV2. In both cases it would be expressed as the last protein of the pol polycistron, and these two viruses are nearest neighbors in the RT-based phylogeny (Fig. 4). The retrovirus macro domain clade is grouped within the Pao1p-like cellular macro domains as an outgroup to all cellular versions (data not shown). In cellular organisms, the macro domain is an ADP-ribose binding molecule (13). Given the observation that macro domain proteins do not enter the nucleus, it has been suggested that the cell has a cytoplasmic poly(ADP-ribosylated) messenger that may be intercepted by the viral macro domain. This messenger interception may prevent ATP depletion, maintain the nucleotide pools required for active viral RNA synthesis, and prevent apoptosis (33). Alternatively, the presence of both cnp and macro domain genes may indicate a processing pathway similar to that observed in tRNA splicing (28). The cellular macro domain has also been shown to silence endogenous murine leukemia viruses (MLVs), as evidenced by the distribution of macroH2A1 nucleosomes on endogenous MLVs, the effects of macroH2A1 knockout on proviral DNA methylation, and the enrichment of macroH2A1 nucleosomes on MLV provirus in mouse liver (8). The presence of the macro domain gene in retroviruses may interfere with the histones’ ability to recognize ZFERV and DRERV2 as viral, preventing their repression.
Regardless of the original source of the cnp and macro domain genes in retroviruses or other RNA and DNA viruses, these genes have been acquired multiple times in various viruses. One thing is clear, however, although cellular copies of the cnp and macro domain genes contain introns, all of the RNA and DNA viral copies are intronless. It cannot be determined whether the fish retroviruses acquired these genes as an mRNA or a reverse-transcribed dsDNA copy. As first suggested for the acquisition of the dutp genes in both retroviruses and several DNA viruses (5), it is the RdDp that could mediate these intronless acquisitions via reverse transcription of cellular mRNAs. There is nothing to rule out the acquisition of such auxiliary genes via nonhomologous recombination among viruses. As with a cellular mRNA, a viral cnp or macro domain mRNA could have been acquired directly by the RNA viruses that do not have a DNA stage, whereas the RdDp provides the only mechanism that could produce an intronless dsDNA copy for DNA viruses.
Active or inactive?
In considering the potential autonomous activity of a retrovirus, the partial and complete deletion of genes, the distribution and frequency of stop codons and frameshifts, and the presence of intact LTRs must all be taken into consideration. Of the potentially active fish retroviruses, eight are full-length genomes and six are indel mutant genomes. Active retroviral deletions mutants are known to play a variety of roles. Deletions in the 5′ end of the pro-pol are known to help proviruses escape repression by macroH2A, as evidenced by a high concentration of nucleosomes in this region of MLV (8). The expression of deletion mutants is induced by burn injuries in a variety of mouse tissues (9, 10), demonstrating that deletion mutant expression can increase when the host immune system is under stress. In humans, HERV-H deletion mutants are found in greater abundance and are expressed at higher levels than the intact forms of the virus (46, 47), suggesting that internal sequence deletion may thwart a specific repression mechanism. Viral complementation could then be used to create whole, viable viruses. Exogenous viruses have selective pressure on them to create deletion mutants. Defecting-interfering deletion mutants are those that can compete for viral products provided by nonmutant genomes, thus establishing a cycle in which full-length and deletion mutants coevolve (18). In human immunodeficiency virus type 1 (HIV-1), it has been suggested that formation of deletion mutations may be a mechanism by which HIV-1 acquires heterogeneity, adapts to variable environmental conditions, and escapes the host's defenses, allowing it to cause persistent infection (25), as is well documented for a variety of RNA viruses that do not have a DNA stage. There is no doubt that some deletion mutant genomes in retroviruses are expressed.
The process of endogeneity begins when an exogenous retrovirus accumulates mutations or indels in one or both of the two polycistrons (gag and env), thereby preventing the virus from leaving the cell. Our analysis of the distribution and frequency of stop codons and frameshifts indicates regulatory selection at the gag/pol and pol/env boundaries. In general, there is also a pattern of accumulation of the intrapolycistronic stop codons, frameshifts, and deletions in the gag and env.
There are multiple mechanisms (e.g., pseudoknot formation and selenocysteine insertion) utilized by viruses (http://recode.genetics.utah.edu/) to correct for mutations that cause stop codons and frameshifting, allowing for an autonomous lifestyle. There are examples of retroviruses, retrotransposons, and retroposons that have only one stop codon or frameshift mutation within a gene that are viable. One intriguing study on the mechanism of an active LINEs retroposon with one stop codon between open reading frames indicates that either multiple known mechanisms of readthrough are involved or that there are new mechanisms yet to be discovered that allow translation (3). Given that infectious fish retroviruses can have as many as four frameshifts and five stop codons found at gene boundaries (35), the limit to the variety and position of mutations that can be overcome is unknown. Both translational recoding and/or gene complementation among endogenous retroviruses would allow an autonomous lifestyle of mutated viral genomes. Future plans include adding functionality to the GPS software that would provide predictions of which copies of endogenous viruses could possibly be interacting via complementation to produce viable viruses. Given the criteria described above, the data presented here indicate there are only a few potentially active endogenous fish retroviruses with low copy numbers compared to mammalian retroviruses, suggesting low activity.
Phylogeny.
The expectation of mutational accumulation is that all protein-coding genes are more conserved than the LTRs, with enzymatic genes diverging more slowly than the structural genes (gag is slower than env) (30). The RT region of the RdDp is the slowest of all retroviral genes to evolve, and it is the function that defines classification as a retroid agent (22). Phylogenetic reconstruction of all available fish retroviral RT sequences reveals four major lineages, each containing both exogenous and endogenous viruses (Fig. 4). There are representatives from more than one species of fish in three of the four major lineages, indicating that exogenous retroviruses have infected and, endogenous retroviruses have reinfected, fish hosts multiple times. The results presented significantly increase the number of known viruses in the epsilonretrovirus family. Although the walleye viruses (WDSV, WEHV1, and WEHV2) were the accepted members of the lineage, and the SnRV virus was provisionally included, the phylogenetic reconstruction presented here places all RT sequences available for known fish retroviruses in the epsilon lineage (Fig. 4).
Although D. rerio, O. latipes, and G. aculeatus have accumulated multiple copies of various retroviruses, T. nigroviridis has only three copies of LTR-bound ERV_Tets. Apparently, T. nigroviridis has a rapid deletion rate of repetitive pseudogenes and resistance to large insertions, which may explain why it has the smallest known vertebrate genome (34) and has so few endogenous retroviruses. In these studies, D. rerio is the oldest of the fish with the largest genome, and it has the widest diversity of endogenous retroviruses and more recently active copies, as indicated by identical LTRs (Fig. 3). G. aculeatus has a few recently active endogenous retroviruses, while neither O. latipes nor T. nigroviridis appear to have had any active copies in the last 30,000 to 50,000 years (Fig. 3).
Although human endogenous retroviruses date back to approximately 55 mya (27), most were acquired about 30 to 35 mya, subsequent to the divergence of old and new world monkeys (41). HERV-K expanded during the hominid radiation, 5 to 20 mya (32), while an African great ape lineage specific expansion took place about 3 to 5 mya (50). D. rerio, O. latipes, G. aculeatus, and T. nigroviridis diverged tens of millions of years ago. We estimate that the new retroviruses described here began infecting the teleost fish about 4 mya, which is relatively recent compared to the evolution of retroviruses in primate genomes. Both the antiquity of endogenous retroviruses in mammalian genome and the diversity of roles these viruses play in the reproduction and development of mammals is well documented. Relative to the age of the teleosts studied here, retroviruses are new to the fish genomes and found in low copy numbers, suggesting that they play little if any significant role in the life cycle of these fish.
Acknowledgments
We thank Mensur Dlakic for structural methods advice and Sudhir Kumar for discussion. The D. rerio, O. latipes, G. aculeatus, and T. nigroviridis genomic position data are available for all potentially active retroviruses upon request.
This research was supported by NIH/NIAID grant AI028309-13A2 to M.A.M.
Footnotes
Published ahead of print on 22 July 2009.
REFERENCES
- 1.Alfaro, M. 2003. Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol. Biol. Evol. 20:255-266. [DOI] [PubMed] [Google Scholar]
- 2.Alfaro, M. E., and M. T. Holder. 2006. The posterior and the prior in Bayesian phylogenetics. Annu. Rev. Ecol. Evol. Syst. 37:19-42. [Google Scholar]
- 3.Alisch, R. S., J. L. Garcia-Perez, A. R. Muotri, F. H. Gage, and J. V. Moran. 2006. Unconventional translation of mammalian LINE-1 retrotransposons. Genes Dev. 20:210-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Baldo, A. M., and M. A. McClure. 1999. Evolution and horizontal transfer of dUTPase-encoding genes in viruses and their hosts. J. Virol. 73:7710-7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballestero, R. P., G. R. Wilmot, B. W. Agranoff, and M. D. Uhler. 1997. gRICH68 and gRICH70 are 2*,3*-cyclic-nucleotide 3*-phosphodiesterases induced during goldfish optic nerve regeneration. J. Biol. Chem. 272:11479-11486. [DOI] [PubMed] [Google Scholar]
- 7.Basta, H. A., A. J. Buzak, and M. A. McClure. 2007. Identification of novel retroid agents in Danio rerio, Oryzias latipes, Gasterosteus aculeatus, and Tetraodon nigroviridis. Evol. Bioinform. Online 3:15. [PMC free article] [PubMed] [Google Scholar]
- 8.Changolkar, L. N., G. Singh, and J. R. Pehrson. 2008. macroH2A1-dependent silencing of endogenous murine leukemia viruses. Mol. Cell. Biol. 28:2059-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho, B., J. D. Shaughnessy, Jr., D. A. Largaespada, H. G. Bedigian, A. M. Buchberg, N. A. Jenkins, and N. G. Copeland. 1995. Frequent disruption of the Nf1 gene by a novel murine AIDS virus-related provirus in BXH-2 murine myeloid lymphomas. J. Virol. 69:7138-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho, K., L. K. Adamson, and D. G. Greenhalgh. 2002. Induction of murine AIDS virus-related sequences after burn injury. J. Surg. Res. 104:53-62. [DOI] [PubMed] [Google Scholar]
- 11.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1998. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 12.Du, C., Z. Swigonova, and J. Messing. 2006. Retrotranspositions in orthologous regions of closely related grass species. BMC Evol. Biol. 6:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durkacz, B. W., O. Omidiji, D. A. Gray, and S. Shall. 1980. (ADP-ribose)n participates in DNA excision repair. Nature 283:593-596. [DOI] [PubMed] [Google Scholar]
- 14.Egloff, M. P., H. Malet, A. Putics, M. Heinonen, H. Dutartre, A. Frangeul, A. Gruez, V. Campanacci, C. Cambillau, J. Ziebuhr, et al. 2006. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 80:8493-8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer, C., L. Bouneau, J. P. Coutanceau, and J. Weissenbach., C. Ozouf-Costaz, and J. N. Volff. 2005. Diversity and clustered distribution of retrotransposable elements in the compact genome of the pufferfish Tetraodon nigroviridis. Cytogenet. Genome Res. 110:522-536. [DOI] [PubMed] [Google Scholar]
- 16.Hart, D., G. N. Frerichs, A. Rambaut, and D. E. Onions. 1996. Complete nucleotide sequence and transcriptional analysis of snakehead fish retrovirus. J. Virol. 70:3606-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herniou, E., J. Martin, K. Miller, J. Cook, M. Wilkinson, and M. Tristem. 1998. Retroviral diversity and distribution in vertebrates. J. Virol. 72:5955-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holland, J. H. 1991. Defective viral genomes, p. 151-165. In B. N. Fields and D. M. Knipe (ed.), Fundamental virology, 2nd ed. Raven Press, New York, NY.
- 19.Hubbard, T. J., B. L. Aken, K. Beal, B. Ballester, M. Caccamo, Y. Chen, L. Clarke, G. Coates, F. Cunningham, T. Cutts, et al. 2007. Ensembl 2007. Nucleic Acids Res. 35:D610-D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hull, R. 1999. Classification of reverse transcribing elements: a discussion document. Arch. Virol. 144:209-213. [DOI] [PubMed] [Google Scholar]
- 21.Hull, R. 2001. Classifying reverse transcribing elements: a proposal and a challenge to the International Committee on Taxonomy of Viruses. Arch. Virol. 146:2255-2261. [DOI] [PubMed] [Google Scholar]
- 22.Hull, R., and S. N. Covey. 1983. Does cauliflower mosaic virus replicate by reverse transcription? Trends Biochem. Sci. 8:119-121. [Google Scholar]
- 23.Jaillon, O., J. M. Aury, F. Brunet, J. L. Petit, N. Stange-Thomann, E. Mauceli, L. Bouneau, C. Fischer, C. Ozouf-Costaz, A. Bernot, et al. 2004. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431:946-957. [DOI] [PubMed] [Google Scholar]
- 24.Katoh, K., and H. Toh. 2008. Recent developments in the MAFFT multiple alignment program. Briefings Bioinform. 9:286-298. [DOI] [PubMed] [Google Scholar]
- 25.Kishi, M., Y. H. Zheng, M. K. Bahmani, K. Tokunaga, H. Takahashi, M. Kakinuma, P. K. Lai, M. Nonoyama, R. B. Luftig, and K. Ikuta. 1995. Naturally occurring accessory gene mutations lead to persistent human immunodeficiency virus type 1 infection of CD4-positive T cells. J. Virol. 69:7507-7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaPierre, L. A., J. W. Casey, and D. L. Holzschu. 1998. Walleye retroviruses associated with skin tumors and hyperplasias encode cyclin D homologs. J. Virol. 72:8765-8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavie, L., P. Medstrand, W. Schempp, E. Meese, and J. Mayer. 2004. Human endogenous retrovirus family HERV-K(HML-5): status, evolution, and reconstruction of an ancient betaretrovirus in the human genome. J. Virol. 78:8788-8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazumder, R., L. M. Iyer, S. Vasudevan, and L. Aravind. 2002. Detection of novel members, structure-function analysis and evolutionary classification of the 2H phosphoesterase superfamily. Nucleic Acids Res. 30:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McClure, M. A. 2001. Evolution of the DUT gene: horizontal transfer between host and pathogen in all three domains of life. Curr. Protein Peptide Sci. 2:313-324. [DOI] [PubMed] [Google Scholar]
- 30.McClure, M. A., M. S. Johnson, D. F. Feng, and R. F. Doolittle. 1988. Sequence comparisons of retroviral proteins: relative rates of change and general phylogeny. Proc. Natl. Acad. Sci. USA 85:2469-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McClure, M. A., H. S. Richardson, R. A. Clinton, C. M. Hepp, B. A. Crowther, and E. F. Donaldson. 2005. Automated characterization of potentially active retroid agents in the human genome. Genomics 85:512-523. [DOI] [PubMed] [Google Scholar]
- 32.Medstrand, P., and D. L. Mager. 1998. Human-specific integrations of the HERV-K endogenous retrovirus family. J. Virol. 72:9782-9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nargi-Aizenman, J. L., C. M. Simbulan-Rosenthal, T. A. Kelly, M. E. Smulson, and D. E. Griffin. 2002. Rapid activation of poly(ADP-ribose) polymerase contributes to Sindbis virus and staurosporine-induced apoptotic cell death. Virology 293:164-171. [DOI] [PubMed] [Google Scholar]
- 34.Neafsey, D. E., and S. R. Palumbi. 2003. Genome size evolution in pufferfish: a comparative analysis of diodontid and tetraodontid pufferfish genomes. Genome Res. 13:821-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nudson, W. A., J. Rovnak, M. Buechner, and S. L. Quackenbush. 2003. Walleye dermal sarcoma virus OrfC is targeted to the mitochondria. J. Gen. Virol. 84:375-381. [DOI] [PubMed] [Google Scholar]
- 36.Paul, T. A., S. L. Quackenbush, C. Sutton, R. N. Casey, P. R. Bowser, and J. W. Casey. 2006. Identification and characterization of an exogenous retrovirus from atlantic salmon swim bladder sarcomas. J. Virol. 80:2941-2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quackenbush, S. L., J. Rovnak, R. N. Casey, T. A. Paul, P. R. Bowser, C. Sutton, and J. W. Casey. 2001. Genetic relationship of tumor-associated piscine retroviruses. Mar. Biotechnol. 3:S88-S99. [DOI] [PubMed] [Google Scholar]
- 38.Ronquist, F. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 39.Soding, J., A. Biegert, and A. N. Lupas. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244-W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinke, D., W. Salzburger, and A. Meyer. 2006. Novel relationships among ten fish model species revealed based on a phylogenomic analysis using ESTs. J. Mol. Evol. 62:772-784. [DOI] [PubMed] [Google Scholar]
- 41.Sverdlov, E. D. 2000. Retroviruses and primate evolution. Bioessays 22:161-171. [DOI] [PubMed] [Google Scholar]
- 42.Tamura, K., J. Dubley, M. Nei, and S. Kumar. 2007. molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. and Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 43.Temin, H. M. 1985. Reverse transcription in the eukaryotic genome: retroviruses, pararetroviruses, retrotransposons, and retrotranscripts. Mol. Biol. Evol. 2:455-468. [DOI] [PubMed] [Google Scholar]
- 44.Temin, H. M. 1989. Reverse transcriptases: retrons in bacteria. Nature 339:254-255. [DOI] [PubMed] [Google Scholar]
- 45.Volff, J. N. 2005. Genome evolution and biodiversity in teleost fish. Heredity 94:280-294. [DOI] [PubMed] [Google Scholar]
- 46.Wilkinson, D. A., J. D. Freeman, N. L. Goodchild, C. A. Kelleher, and D. L. Mager. 1990. Autonomous expression of RTVL-H endogenous retroviruslike elements in human cells. J. Virol. 64:2157-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilkinson, D. A., N. L. Goodchild, T. M. Saxton, S. Wood, and D. L. Mager. 1993. Evidence for a functional subclass of the RTVL-H family of human endogenous retrovirus-like sequences. J. Virol. 67:2981-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu, Z., and H. Wang. 2007. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35:W265-W268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamanoue, Y., M. Miya, J. G. Inoue, K. Matsuura, and M. Nishida. 2006. The mitochondrial genome of spotted green pufferfish Tetraodon nigroviridis (Teleostei: Tetraodontiformes) and divergence time estimation among model organisms in fishes. Genes Genet. Syst. 81:29-39. [DOI] [PubMed] [Google Scholar]
- 50.Yohn, C. T., Z. Jiang, S. D. McGrath, K. E. Hayden, P. Khaitovich, M. E. Johnson, M. Y. Eichler, J. D. McPherson, S. Zhao, S. Pääbo, and E. E. Eichler. 2005. Lineage-specific expansions of retroviral insertions within the genomes of African great apes but not humans and orangutans. PLoS Biol. 3:e110. [DOI] [PMC free article] [PubMed] [Google Scholar]