Abstract
Host factor pathways are known to be essential for hepatitis C virus (HCV) infection and replication in human liver cells. To search for novel host factor proteins required for HCV replication, we screened a subgenomic genotype 1b replicon cell line (Luc-1b) with a kinome and druggable collection of 20,779 siRNAs. We identified and validated several enzymes required for HCV replication, including class III phosphatidylinositol 4-kinases (PI4KA and PI4KB), carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase (CAD), and mevalonate (diphospho) decarboxylase. Knockdown of PI4KA could inhibit the replication and/or HCV RNA levels of the two subgenomic genotype 1b clones (SG-1b and Luc-1b), two subgenomic genotype 1a clones (SG-1a and Luc-1a), JFH-1 genotype 2a infectious virus (JFH1-2a), and the genomic genotype 1a (FL-1a) replicon. In contrast, PI4KB knockdown inhibited replication and/or HCV RNA levels of Luc-1b, SG-1b, and Luc-1a replicons. The small molecule inhibitor, PIK93, was found to block subgenomic genotype 1b (Luc-1b), subgenomic genotype 1a (Luc-1a), and genomic genotype 2a (JFH1-2a) infectious virus replication in the nanomolar range. PIK93 was characterized by using quantitative chemical proteomics and in vitro biochemical assays to demonstrate PIK93 is a bone fide PI4KA and PI4KB inhibitor. Our data demonstrate that genetic or pharmacological modulation of PI4KA and PI4KB inhibits multiple genotypes of HCV and represents a novel druggable class of therapeutic targets for HCV infection.
Hepatitis C virus (HCV) causes liver disease in humans, including chronic hepatitis, cirrhosis, and hepatocellular carcinoma (52). The HCV genome is a single-stranded RNA molecule where both the 5′ and the 3′ untranslated region (UTR) contain highly conserved RNA structures necessary for polyprotein translation and genome replication (43). The processed polyprotein yields at least three structural proteins and six nonstructural proteins. The structural proteins include the core, which forms the viral nucleocapsid, and the envelope glycoproteins E1 and E2. The viral proteins processed by signal peptidases form viral particles that assemble at the endoplasmic reticulum (ER) and/or Golgi bodies and are released from the host cell by viral budding. The structural protein coding regions are separated from nonstructural proteins by the short membrane peptide p7, thought to function as an ion channel (43, 53). The nonstructural proteins NS2, NS3/4A, NS5A, and NS5B are involved in coordinating the intracellular processes of the virus life cycle, including polyprotein processing and viral RNA replication (34).
The Luc-1b cell is a human hepatoma cell line (Huh7) that contains a genotype 1b HCV subgenomic replicon, a luciferase reporter, and a neomycin selection marker, allowing HCV replication to be studied both in vitro and in vivo (8, 36). This subgenomic replicon lacks the coding regions for NS2 and the structural proteins but contains the nonstructural proteins in cis, which are required for replication of the viral RNA. Expression of the luciferase gene acts as a surrogate marker for levels of HCV RNA produced in the cell. The goal of the present study was to use this subgenomic HCV replicon to screen siRNA libraries and identify novel host proteins that are involved in HCV replication.
A number of cellular pathways and proteins that play critical roles in HCV replication have recently been described (41, 42, 46). In particular, replication of HCV is tied closely to its localization and transport to various internal membranes and to lipid metabolism (2). Most of the HCV proteins appear to be targeted to the surface of the ER and replication complexes appear to be transported to lipid rafts, where RNA replication can occur (2). Infectious virus particle formation occurs in association with lipid droplets, and this process requires the core and NS5A proteins. In addition, cholesterol pathway production of geranylgeranyl-PP is important to geranylate the FBL2 protein, which serves as a membrane anchor for NS5A (62). The hVAP proteins involved in the localization and trafficking between internal membranous structures are known to be associated with the HCV proteins NS5A and NS5B (59). Thus, host factor lipid metabolism and intracellular protein transport are necessary for HCV replication in cells.
Targeting host factors that are required for viral replication offers a strategy to overcome viral resistance and may allow treatment for more than one genotype of HCV and/or a related Flaviviridae virus such as Dengue, West Nile, or yellow fever virus. The current standard-of-care treatment for the genotype 1 strain of HCV infection is pegylated interferon alpha plus ribavirin over a 6-month time course with more than half of infected patients being refractory to this treatment (57). In addition to genotype 1, there are at least five naturally occurring genotype variants of HCV that can complicate a patient's response to therapy when infected with more than one genotype. As well as the development of mutations, the presence of multiple variants coexisting in patients is thought to contribute to the rapid development of resistance (40). A variety of antiviral therapeutic strategies aim to inhibit viral proteins directly with small molecules or siRNAs (13, 31, 33). Although some small molecule approaches have been successful in preclinical studies, small-molecule strategies directed against the viral targets can still be rendered ineffective due to the development of mutant, treatment-resistant viral strains (13, 40). Thus, combination therapies are a necessary approach to treat the many variants of HCV that exist in the patient population.
In the present study, a set of 779 SMARTpool small interfering RNAs (siRNAs) targeting the kinome and 4 siRNAs targeting 5,000 druggable genes (20,000 siRNAs) were tested for their ability to block replication of the Luc-1b HCV subgenomic replicon. siRNAs targeting CAD (carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase), a tripartite enzyme that catalyzes the first three steps of pyrimidine biosynthesis, inhibited both the Luc-1b replicon and JFH1-2a virus expression. This activity is consistent with the known inhibitor of this enzyme, leflunomide, which has been shown previously to inhibit both respiratory syncytial virus and HCV (12, 54). siRNAs targeting the mevalonate (diphospho) decarboxylase (MVD) enzyme, which catalyzes the formation of mevalonate, were found to inhibit Luc-1b replication (19). Inhibition of the cholesterol biosynthesis pathway and host cell geranylation has been previously reported to inhibit HCV subgenomic replication (3, 24, 51, 62, 67). siRNA-mediated knockdown of the class III phosphatidylinositol 4-kinases PI4KA and PI4KB inhibited luciferase expression not only for the genotype 1b subgenomic replicons (Luc-1a and Luc-1b) but also for the viral RNA levels of SG-1b, Luc-1b, and Luc-1a. PI4KA knockdown also inhibited Renilla expression in the JFH-1 genotype 2a infectious virus (JFH1-2a), genotype 2a subgenomic replicon (SG-1a), and a genomic and subgenomic genotype 1a replicon (FL-1a and SG-1a). Using the small-molecule inhibitor PIK93 in compound affinity competition experiments and in vitro biochemical assays, we demonstrated PIK93 could bind and inhibit both PI4KA and PI4KB enzymatic activity (58). PIK93 could inhibit luciferase expression in the Luc-1b, Luc-1a, and JFH1-2a infectious virus assays in the submicromolar range. Together, our data suggest that PI4KA and PI4KB regulate HCV replication and that pharmacological inhibition of these enzymes represents a new class of antiviral agents for multiple genotypes of HCV. Finally, since PI4KA and PI4KB are known to regulate protein and lipid transport to and from the ER and Golgi bodies, their function may hold clues as to how movement of HCV replication complexes throughout different organelles is regulated.
MATERIALS AND METHODS
HCV replicons and cell culture.
Huh-Luc/neo-ET, a subgenomic genotype 1b (Con1) HCV replicon cell line containing a firefly luciferase gene, was licensed from ReBLikon GmbH and called Luc-1b in the present study (29, 34, 36). The (Con1) subgenomic 1b replicon cell line (clone A or SG-1b) (8), full-length genotype 1b (FL-1b) (9), subgenomic 1a (SG-1a) (10), and genomic genotype 1a (FL-1a) (10) HCV replicon cells were all licensed from Apath, LLC (St. Louis, MO).
The luciferase subgenomic 1a replicon (Luc-1a) was constructed through a multistep subcloning procedure. The backbone plasmid was pH/SG-Neo(L+I), a subgenomic 1a replicon with neoR cassette from Apath LLC (St. Louis, MO) (10). The AgeI/KpnI restriction fragment containing neoR cassette was replaced with the AgeI/KpnI restriction fragment containing the firefly luciferase-ubiquitin-neoR cassette from pFK I389 luc-ubi-neo/NS3-3′/ET, the subgenomic genotype 1b replicon with luciferase reporter from ReBLikon GmbH (30). To enhance replication capacity of the replicon, the nonstructural proteins NS3-NS5A from pH/SG-Neo(L+I) were replaced with the NS3-NS5A coding region from pH77-S (kindly provided by Jang Han, Novartis Vaccines and Diagnostics), a cell culture-adapted genotype 1a virus containing additional adaptive mutations that facilitate higher levels of replication (69). Specifically, this H77-S region contained seven mutations relative to the H77 sequence: Q1067R, E1202G, and V1655I in NS3; K1691R in NS4A; Q1742H in NS4B; and K2040R and S2204I in NS5A. The NS3-NS5A region was replaced with the StuI/BamHI restriction fragment from pH77-S.
The stable luciferase 1a replicon cell line was generated through electroporation of in vitro-transcribed replicon RNA into Huh-Cure cells, followed by selection with 0.5 mg of G418/ml. Replicon RNA was prepared by using a T7 Megascript RNA transcription kit (Ambion) and purified with a MegaClear RNA purification kit (Ambion). HuhCure cells resuspended at 107 cells/ml in OptiMEM-I were combined with 10 μg of in vitro-transcribed replicon RNA in a 0.4-cm gap cuvette and electroporated at 270 V and 960 μF using a Bio-Rad GenePulser II electroporator. Cells were gently transferred to Dulbecco modified Eagle medium containing 10% heat-inactivated fetal bovine serum (FBS), 2 mM l-glutamine, and 1 mM nonessential amino acids. The next day, the medium was replaced with medium containing 0.5 mg of G418/ml for selection of replicon-bearing cells. The medium was replaced every 2 days until large cell clones developed. The colonies were treated with trypsin and pooled, and the cells were passaged an additional five times before assaying for luciferase activity. The luciferase 1a cell line was then validated via testing susceptibility of the replicon to inhibition by NS3 protease inhibitors, NS5B polymerase inhibitors, and IFN-α. Replicon levels were measured both by luciferase activity assay and quantitative reverse transcription-PCR (QRT-PCR) assay to confirm that luciferase activity was representative of replicon RNA levels. In all cases, the 50% inhibitory concentrations (IC50s; that is, the concentration of inhibitor that reduced control reaction rates by 50%) determined via luciferase activity were the same as IC50s determined via QRT-PCR (data not shown). The subgenomic 2a replicon (SG-2a) cell line was produced by using methods previously described (26). The genotype 2a sequences (5′UTR and NS3 though 3′UTR sequences) were taken from the genotype 2a JFH-1 infectious virus. The neoR coding region and encephalomyocarditis virus internal ribosome entry site (IRES) were taken from pCon1/SG-Neo(I) (SG-1b), and the cell line used for clone selection was Huh7.5; both are licensed from Apath LLC (9). All of the cells were maintained in complete medium consisting of Dulbecco modified Eagle medium, 2 mM l-glutamine, 1× nonessential amino acids, 10% heat-inactivated FBS (ΔFBS), and G418 (Invitrogen) at 250 μg/ml of for the Luc-1b cells and 1 mg/ml for the clone A cells and genomic genotype 1a (FL-1a) replicon.
Generation of JFH-1 virus.
A monocistronic JFH-1 construct (pUC-RLF-JFH1) was provided by Jang Han from Novartis Vaccines and Diagnostics and is organized as follows: (5′UTR)-(12 residues of core)-(Renilla luciferase)-(FMDV protein 2a)-(ubiquitin)-(HCV core through NS5B)-(3′UTR). The FMDV 2a and ubiquitin are included to ensure that protease cleavage results in the generation of a Renilla luciferase protein with a correct C terminus sequence and a core protein with a correct N terminus sequence (27). The pUC-RLF-JFH1 plasmid was linearized at the 3′ end of the JFH-1 genome by XbaI digestion and purified by using phenol-chloroform extraction. An RNA transcript was generated (Ambion MEGAscript T7 kit) and purified (Ambion MEGAclear kit) according to the manufacturer's protocols. The JFH-1 RNA was electroporated into Huh7.5 cells (8 μg of RNA, 2 × 106 cells in 200 μl of PBS; Bio-Rad GenePulser, two pulses at 0.35 kV, 10 μF, ∞ resistance), and the cells were plated into a T75 flask. The cells were passaged every 3 to 4 days, and the medium was harvested each time. The production of infectious JFH-1 virus was followed by infection of Huh7.5 cells with the harvested medium (as described below). Medium with a high titer (i.e., 50% tissue culture infective dose of ∼104/ml) was divided into aliquots and frozen at −80°C. Huh7.5 cells to be infected with JFH-1 virus were plated at 1.5 × 103 cells/well in 96-well plates. For reverse transfection siRNA studies, the plates were preloaded with siRNA mix (17). After 24 h of incubation, the medium was removed, the cells were washed with PBS, and 50 μl of JFH-1 virus was added (multiplicity of infection of 0.01 to 0.1). After a 4- to 6-h incubation, 100 μl of medium (containing compound or siRNAs, where appropriate) was added to each well, and the cells were incubated for a further 72 h at 37°C before being assayed. The Renilla luciferase activity was measured by using a Renilla luciferase assay (Promega), and cell viability was measured by using a CellTiter-Glo luminescent cell viability assay (Promega) according to the manufacturer's protocols. The JFH-1 genotype 2a virus construct used in the present study is named JFH1-2a.
siRNA screen.
The kinase siRNA SMARTpool library (Dharmacon, catalog no. H-003500) and the four siRNAs targeting 5,000 druggable genes (Human Druggable Genome siRNA Set V1.0 [Qiagen]) were transfected into the HCV replicon cells as previously described (21, 23, 48). The total number of siRNAs screened was 20,779, of which 20,000 were 4 individual siRNAs targeting 5,000 druggable genes and 779 SMARTpool kinases. The siRNAs were diluted in 4 μl of Opti-MEM (Invitrogen) and stamped out into 384-well tissue culture plates at a concentration of 206 nM. Using 0.07 μl of Oligofectamine transfection reagent (Invitrogen), siRNA dilutions were prepared in 4 μl of Opti-MEM by using a FlexDrop precision reagent dispenser (Perkin-Elmer). The plates were incubated at room temperature for 20 to 30 min to allow siRNA-lipid reagent complexes to form. After the incubation, 1,500 cells in 25 μl of assay medium were added per well using the Multidrop 384 (Thermo Scientific). The cells were assayed for luciferase activity at 72 h posttransfection by using a Bright-Glo luciferase assay (Promega). The siRNA collections were run in sets of n = 2 after screening optimization (11, 70). The screening data was Z-score transformed to pick siRNAs that scored ≤50% below the median (≤0.5 or Z-score of −2) or ≥100% (≥2.0 or Z-score of 2) above the median. The most active siRNAs were validated as three multiple independent duplexes: PI4KA (NM_002650, Dharmacon catalog no. D-006776), PI4KB (NM_002651, Dharmacon catalog no. D-006777), CAD (NM_004341, Dharmacon catalog no. D-009471), and MVD (NM_002641, Dharmacon catalog no. D-006748) (see Table S3 in the supplemental material). The pGL2, GAPDH (glyceraldehyde-3-phosphate dehydrogenase), and Renilla luciferase duplexes were purchased from Dharmacon (pGL2, catalog no. D-001100-01-20; GAPDH, catalog no. D-001140-01-20; Renilla luciferase, catalog no. P-002070-01-20). The validated siRNAs for PI4KA and PI4KB were analyzed in Huh7.5 cells infected with the JFH-1 virus, as described in the previous section. Briefly, Huh7.5 cells were reverse transfected with siRNA and then infected with the JFH-1 virus at a multiplicity of infection of 0.1. Infected cells were incubated for 72 h at 37°C, and Renilla luciferase activity was measured by using the Renilla luciferase assay (Promega), and the cell viability was measured by using the CellTiter-Glo assay. For the siRNA dose experiments, each siRNA was diluted from a 20 μM stock to a working concentration of 250 nM and threefold serial diluted in Opti-MEM before transfection. Cells were assayed for luciferase activity and cell viability 72 h after siRNA transfection. For validation experiments, each siRNA was resuspended in siRNA buffer (Dharmacon catalog no. B-002000-UB-015) to a stock concentration of 20 μM (5× to 1×). Then, 2.5 μl of each stock solution was diluted in 197.5 μl of Dharmacon cell culture reagent (catalog no. B-004500-100) in a 96-well PCR plate (ABgene) to make a working 10× stamp of 250 nM, and then 0.20 μl of DharmaFECT 1 transfection reagent (Dharmacon) was diluted in 10 μl and added to each well of a 96-well tissue culture plate. Next, a 10-μl aliquot of each well in the 10× stamp was added to the 96-well tissue culture plates containing lipid reagent, followed by incubation for 20 min at room temperature to allow the (siRNA/lipid reagent) complexes to form. After the incubation, 6,000 replicon cells in 80 μl of complete medium were plated on top of the siRNA-lipid reagent complexes by using the Multidrop 384. Cells were incubated for 72 h at 37°C and assayed for luciferase activity and cell viability as described above.
Real-time PCR.
Two wells transfected with siRNAs were pooled together, mRNA was isolated by using a TurboCapture 96 mRNA kit (Ambion), and cDNA was generated by using a high-capacity cDNA archive kit (Applied Biosystems). Knockdown of target mRNA was confirmed by using a TaqMan gene expression assay (Applied Biosystems) according to the manufacturer's instructions. The cDNA was quantified using premixed Applied Biosystems probes and primers—PI4KA (NM_002650, catalog no. Hs01021073_m1), PI4KB (NM_002651, catalog no. Hs00356327_m1), CAD (NM_004341, catalog no. Hs00983188_m1), MVD (NM_002641, catalog no. Hs00159403_m1), and GAPDH (NM_002046, catalog no. 4333764F)—using a real-time PCR system (Applied Biosystems 7900HT). RT-PCR was performed on HCV viral RNA in the subgenomic genotype 1b replicon (clone A), Luc-1b, and genomic genotype 1a cells as previously described (37).
shRNAs.
Short hairpin RNAs (shRNAs) targeting PI4KA (NM_002650, Sigma catalog no. SHGLY-NM_002650) and PI4KB (NM_002651, Sigma catalog no. SHGLY-NM_002651) were ordered as five individual shRNA clones (see Table S3 in the supplemental material). shRNAs targeting green fluorescent protein (GFP; Sigma catalog no. TRCN0000072181) were used as negative controls. First, 6,000 cells were plated in 96-well tissue culture plates. The following day, medium was replaced with transduction medium containing Polybrene (Sigma) at a 8-μg/ml final concentration and HEPES (Invitrogen) at a 10 mM final concentration. Then, 1 μl shRNA virus was added per well, and the cells were centrifuged at 2,100 rpm for 90 min at room temperature. The cells were incubated for 24 h and selected by adding puromycin (Sigma) at a 2-μg/ml final concentration. The cells were then incubated for a minimum of 72 h and assayed for luciferase and cell viability as previously described, analyzed by Western blot and RT-PCR, and propagated in culture for long-term knockdown studies.
Western blots.
PI4KB and NS3 protein levels were analyzed by Western blotting using Criterion 10 to 20% Tris-HCl sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels (Bio-Rad) under standard conditions and commercially available antibodies to PI4KB (BD Biosciences) and NS3 (ViroStat) (32).
Compounds.
The compounds PIK93 (4, 5, 58) and NIM811 were synthesized at Novartis (49). HCV replicon and JFH1-2a Huh7.5-infected cells were treated with compound for 48 h prior to Renilla and firefly luciferase, CellTiter-Glo, Western blot, or immunofluorescence analysis as described. An average of five independent experiments, along with the standard deviations, was calculated for each treatment. Calculations of the IC50 and CC50 (the concentration of compound at which 50% of the cells were viable) were determined by using SigmaPlot version 9.0 graphing software.
In vitro biochemistry.
The PI4K cDNA clones were obtained from OriGene, the restriction enzymes and ligases were from New England BioLabs, and the PCR polymerases and primers were from Invitrogen. Bovine liver phosphatidylinositol was purchased from Avanti-Polar Lipids, and ATP was obtained from Amersham Biosciences/GE Healthcare. Expression and purification of PI4KA and PI4KB were performed in Sf9 (Spodoptera frugiperda) cells. Recombinant PI4K proteins were derived from cDNA clones (PI4KA, NM_058004, OriGene catalog no. SC120237; PI4KB, NM_002651, OriGene catalog no. SC118488). Full-length coding regions were subcloned into pFastBac-HT, downstream from the vector-encoded N-terminal His6 tag for the purpose of affinity chromatographic purification. Baculovirus was produced by using a Bac-to-Bac baculovirus expression system (Invitrogen) with Sf9 insect cells. Protein was produced by infecting Sf9 cells with amplified baculovirus. At 68 to 72 h postinfection, the cells were harvested; resuspended in buffer containing 50 Tris (pH 7.5), 10% glycerol, 10 mM imidazole, 300 mM NaCl, 0.2% Triton X-100, 1 mM TCEP [Tris(2-carboxyethyl)phosphine], and EDTA-free protease inhibitor cocktail (Roche); and lysed by using a microfluidizer. The suspension was centrifuged at 21,000 × g for 45 min at 4°C, and the supernatant was mixed with nickel nitrilotriacetic acid resin (Qiagen) for 2 h at 4°C. The resin was washed sequentially with buffer 1 (50 Tris [pH 7.5], 20 mM imidazole, 300 mM NaCl, 0.2% Triton X-100, 1 mM TCEP) and then buffer 2 (50 Tris [pH 7.5], 20 mM imidazole, 0.2% Triton X-100, 1 mM TCEP). His-tagged protein was eluted with 50 Tris (pH 7.5)-250 mM imidazole-0.2% Triton X-100-1 mM TCEP. For PI4KB, protein was further purified via cation-exchange chromatography using HiLoad Q column with buffer A (50 mM Tris [pH 7.5], 1 mM dithiothreitol [DTT], 0.2% Triton X-100) and buffer B (50 mM Tris [pH 7.5], 500 mM NaCl, 1 mM DTT, 0.2% Triton X-100). Pooled fractions for either PI4KA or PI4KB were dialyzed overnight at 4°C against 50 mM Tris (pH 7.5)-150 mM NaCl-1 mM DTT-0.2% Triton X-100-10% glycerol. Purified proteins were then divided into aliquots and stored at −80°C. The PI4K kinase activity was assayed via the use of coupling enzymes pyruvate kinase and lactate dehydrogenase. In this assay system, the PI4K-catalyzed phosphorylation of phosphatidylinositol was coupled to the oxidation of NADH to NAD+. The reaction was monitored as a decrease in NADH fluorescence, with λex = 344 nm and λem = 460 nm. The assay conditions were as follows: 50 mM HEPES (pH 7.3), 10 mM MgCl2, 1 mM DTT, 1 mM EGTA, 1 mM phosphoenolpyruvate, 40 U of pyruvate kinase/ml, 40 U of lactic dehydrogenase/ml, 0.04 mM NADH, 0.2 mM ATP, 0.04 mM bovine liver phosphatidylinositol, and 5 nM PI4KA or 25 nM PI4KB. Inhibitors were serially diluted in dimethyl sulfoxide (DMSO) before dilution into assay buffer, giving a final concentration of 1% DMSO. PI4KA or PI4KB was preincubated with inhibitor dilutions in assay buffer for 10 min at room temperature (25°C) prior to initiating the reaction with phosphatidylinositol and ATP. The percent activity was calculated from the initial rates of the inhibited reactions relative to the uninhibited control. The IC50s were calculated by four-parameter curve fitting of percent control activity at 14 inhibitor concentrations.
Affinity purification.
Compound coupling and affinity purification was performed as described previously (6). All reagents were purchased from Sigma unless otherwise indicated. A PI4KB inhibitor, LCJ217 (see Fig. S4 in the supplemental material), was coupled onto NHS-activated Sepharose 4 beads (Amersham) to be used as the affinity matrix. Huh-7 cells expressing the HCV replicon were homogenized in lysis buffer (50 mM Tris-HCl [pH 7.5], 5% glycerol, 1.5 mM MgCl2, 150 mM NaCl, 20 mM NaF, 1 mM Na3VO4, 1 mM DTT, 5 μM calyculin A, 0.8% Igepal-CA630, and a protease inhibitor cocktail) using a Dounce homogenizer on ice. Lysate was cleared by centrifugation and protein concentration measured by a Bradford assay. PIK93 compound was dissolved in DMSO and added at 10 μM (or DMSO alone) to 50-mg lysate samples for 30 min at 4°C. Then, 100 μl of LCJ217-matrix was added, and incubation resumed at 4°C for another 30 min. After centrifugation, beads were transferred into a column (MoBiTec) and washed. Bound material was eluted with NuPAGE LDS sample buffer (Invitrogen), and the eluates were reduced, alkylated, separated on 4 to 12% NuPAGE gels (Invitrogen) and stained with colloidal Coomassie blue.
Mass spectrometry.
Procedures were performed as previously described (6). Briefly, gel lanes were cut into slices across the separation range, subjected to in-gel tryptic digestion, and monitored by labeling with iTRAQ reagents (Applied Biosystems) as described previously (6). Peptide extract of DMSO vehicle control was labeled with iTRAQ reagent 117 and combined with extract from PIK93-treated sample labeled with iTRAQ reagent 114. Sequencing was performed by liquid chromatography-tandem mass spectrometry on an Eksigent 1D+ high-pressure liquid chromatography system coupled to a LTQ-Orbitrap mass spectrometer (Thermo Scientific). Tandem mass spectra were generated by using pulsed-Q dissociation, enabling the detection of iTRAQ reporter ions. Peptide mass and fragmentation data were used to query an in-house curated version of the IPI database (EBI [http://www.ebi.ac.uk/IDP]). A mass tolerance of 10 ppm was allowed for detection of intact peptide masses and 0.6 Da for fragment ions detected in the linear ion trap. Proteins were quantified by using in-house-developed software as described previously (6). Only peptides unique for identified proteins were used for relative protein quantification. The minimum requirements for acceptance of quantified proteins were at least four quantifiable spectra matching to unique peptides and a total reporter ion intensity of 15,000. Experiments using a decoy database revealed that no false-positive protein identifications are expected in the quantified data set. In order to compensate for variations in the total protein amount yielded from the compound affinity purifications, raw protein fold changes were renormalized by using the median fold change detected between the two experiments. Protein changes had to be at least twofold to be considered significant and were ranked by the number of unique peptides.
RESULTS
Genome-scale HCV replicon siRNA screen.
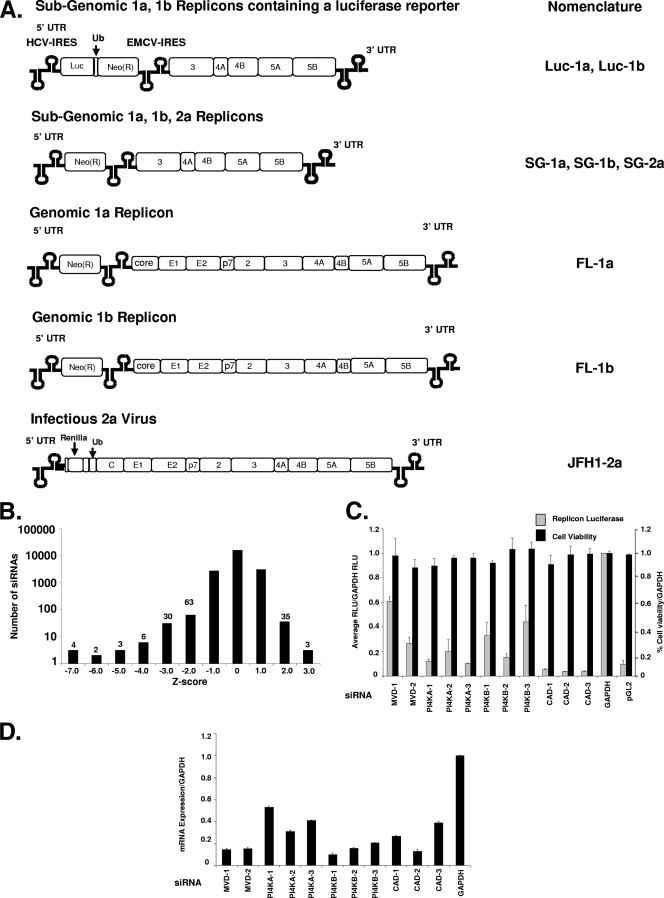
Several self-replicating viral constructs representing three HCV genotypes have been modified to study the HCV replicon activity in vitro (8, 9, 10, 29, 30, 34, 36, 61, 71) (Fig. 1A). Among the salient features of the replicons are the 5′ and 3′ UTRs, which are required for efficient viral RNA replication, translation, and RNA stability. The replicons have been adapted to cell culture growth by selecting for mutations in the genome, including in the NS5A and NS3 proteins (8, 10). For the subgenomic replicons, the structural proteins and NS2 have been replaced with a selectable marker gene neomycin (Luc-1a, Luc-1b, SG-1a, and SG-1b) and two replicons (Luc-1a and Luc1b) contain a firefly luciferase reporter (pGL2) to measure cellular replicon expression. The nonstructural genes are expressed from a second, heterologous encephalomyocarditis virus IRES independent of the HCV IRES that mediates expression of the neomycin and Firefly luciferase gene. The full-length replicons (FL-1a and FL-1b) contain a neomycin selectable marker, followed by the complete HCV genome (9, 10). The JFH1-2a infectious virus contains a Renilla luciferase gene, followed by 12 residues of the core protein. The JFH1-2a viral RNA is made in vitro from a linearized pUC-RLF-JFH1 plasmid prior to electroporation into cells. Viral supernatants are used to infect naive Huh7.5 cells, and Renilla gene expression is the surrogate measure for HCV RNA transcription (see Materials and Methods).
FIG. 1.
Genome-scale siRNA screen in HCV replicon cells. (A) Model of the HCV replicons used in the present study: the subgenomic genotype 1a luciferase replicon (Luc-1a) and the subgenomic genotype 1b luciferase replicon (Luc-1b), both of which contain a Firefly luciferase gene; the subgenomic genotype 1a replicon (SG-1a), the subgenomic genotype 1b replicon (SG-1b), the subgenomic genotype 2a replicon (SG-2a), the genomic genotype 1a replicon (FL-1a), and the genomic genotype 1b replicon (FL-1b), all of which contain a neomycin selectable marker; and the genomic genotype 2a infectious virus (JFH1-2a), which contains a Renilla luciferase gene. (B) The Huh7 stable HCV replicon cells were screened by high-throughput lipid-mediated transfection using a SMARTpool kinome (779 genes) and a druggable genome (four siRNAs targeting 5,000 genes) at 25 nM siRNA per well. The distribution of siRNAs is presented as a Z-score histogram. The frequency of hits at each Z-score is presented above each column. Including the luciferase siRNA, 108 siRNAs scored below a Z-score of −2, whereas 38 siRNAs scored above 2.0. (C) The top-scoring siRNA inhibitors were validated by using multiple independent siRNAs (see Table S3 in the supplemental material for sequences) after a 72-h transfection into the Luc-1b cells. Both luciferase (░⃞) and cell viability (▪) were determined and normalized to the negative control GAPDH siRNA. (D) mRNA levels were quantified in the siRNA-treated cells (in parallel with panel C) using RT-PCR, based on the internal control actin mRNA. The data were normalized to GAPDH siRNA-treated cells to control for transfection effects on total cellular mRNA levels.
Introduction of siRNAs into the Huh7 replicon containing cells was optimized by using a high-throughput siRNA transfection protocol as previously described (11). The HCV replicon cells (Luc-1b) were used to determine antiviral activity by measuring the reduction of luciferase expression in the cell. siRNA SMARTpools targeting 779 kinases and a collection of four independent siRNAs targeting 5,000 druggable genes were transfected into the replicon cells in two independent screens, and luciferase expression was quantified. The screen was analyzed by using a Z-score transformation so the siRNAs with the most significant effects (activation or inhibition of luciferase expression) on replication could be determined (Fig. 1B). The positive control siRNA, pGL2 (targeting firefly luciferase), as well as PI4KA, COPB2, and PLK, all inhibited luciferase levels −7 standard deviations from the median (Table 1) . Additional genes scoring between −5 and −6 standard deviations from the median included MVD, PI4KB, MOS, CAD, and DERMO1 (Table 1). The previously identified host factor regulator of HCV, cyclophilin B, was identified in the screen, scoring −4 standard deviations below the median (63). In addition, we found cyclophilin C siRNAs were as potent as cyclophilin B siRNAs (Z-score of −4, Table 1), although cyclophilin F siRNAs scored below −2 standard deviations (see Table S1 in the supplemental material). Other members of the cyclophilin family tested in the screen, i.e., cyclophilins A, D, and H, did not score below a Z-score of −2 (data not shown). Since the siRNAs in the collection were not prevalidated, it is possible targets would be missed because the siRNAs were not active. An additional 107 genes (excluding pGL2) that scored between −2 and −4 standard deviations from the median are listed in Table S1 in the supplemental material. Of the top-scoring hits, PLK1 and COPB2 siRNAs caused overt cell death in cell viability assays and were not pursued for further study (35; data not shown). Due to the large number of primary hits and the complexity of the signaling pathways they represent, we only validated the top four genes from the primary screen.
TABLE 1.
Summary of the top-scoring siRNAs validated from the replicon screena
| Gene | Description | Accession no. | Z-score |
|---|---|---|---|
| pGL2 | Firefly (Photinus pyralis) luciferase | X65323 | -7 |
| COPB2 | Coatomer protein complex, subunit beta 2 (beta prime) | NM_004766 | -7 |
| PIK4CA | Phosphatidylinositol 4-kinase, catalytic, alpha polypeptide | NM_002650 | -7 |
| PLK1 | Polo-like kinase 1 | NM_005030 | -7 |
| MVD | Mevalonate (diphospho) decarboxylase | NM_002461 | -6 |
| PIK4CB | Phosphatidylinositol 4-kinase, catalytic, beta polypeptide | NM_002651 | -6 |
| MOS | v-mos Moloney murine sarcoma viral oncogene homolog | NM_005372 | -5 |
| CAD | Carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase | NM_004341 | -5 |
| DERMO1 | Likely ortholog of mouse and rat twist-related bHLH protein Dermo-1 | NM_057179 | -5 |
| ITGA7 | Integrin, alpha 7 | NM_002206 | -4 |
| UBB | Ubiquitin B | NM_018955 | -4 |
| PIP5K1A | Phosphatidylinositol-4-phosphate 5-kinase, type I, alpha | NM_003557 | -4 |
| CypC | Peptidylprolyl isomerase C | NM_000943 | -4 |
| SCN1B | Sodium channel, voltage-gated, type I, beta polypeptide | NM_001037 | -4 |
| CypB | Peptidylprolyl isomerase B | NM_000942 | -4 |
siRNAs that inhibited replicon luciferase levels ≥2 standard deviations from the median (Z-score) were picked from the screen (see Table S1 in the supplemental material for a complete list).
In addition to siRNAs that inhibited luciferase expression, there were 38 siRNAs that increased the expression of the luciferase replicon >2 standard deviations from the median (see Table S2 in the supplemental material). The top three hits scoring >3 standard deviations above the median included PDGFRA (NM_006206, for platelet-derived growth factor receptor alpha), SCNN1B (NM_000336, for sodium channel, nonvoltage-gated 1, beta), and RNF26 (NM_032015, for ring finger protein 26). Our attempt to validate the induction of luciferase caused by these siRNAs was not reproducible in the Luc-1b cells (data not shown). Thus, the siRNAs in Table S2 in the supplemental material were not pursued for further study.
In order to confirm the primary screening results and rule out off-target effects, multiple independent gene-specific siRNAs were used in verification experiments to correlate replicon inhibition with efficiency of target mRNA knockdown (for sequences, see Table S3 in the supplemental material). The siRNAs had to meet several criteria to be considered validated: more than one siRNA sequence had to knock down target gene mRNA, mRNA silencing had to correlate with phenotype (luciferase inhibition), and the siRNAs could not affect cell viability. Up to three independent siRNA sequences were repeated in the Luc-1b replicon assay, and the cell viability was determined (Fig. 1C). The siRNAs targeting PI4KA inhibited luciferase activity >5-fold compared to the negative control GAPDH siRNA (scrambled siRNAs performed the same as GAPDH, so all genes were compared to this single negative control throughout this study). All three PI4KB siRNAs inhibited luciferase by 50 to 80%, while two MVD siRNAs inhibited luciferase 40 and 70%, respectively. Each of the CAD siRNAs reduced luciferase expression by >80% similar to the positive control pGL2 siRNA. All of the siRNAs had little or no effect on cell viability as measured by cellular ATP levels (Fig. 1C, black bars). Each siRNA was also analyzed for the ability to knock down its respective target gene as determined by RT-PCR. As shown in Fig. 1D, all of the siRNAs reduced target mRNA levels by >50%, with the exception of PI4KA-1. The MVD siRNAs (MVD-1 and -2) consistently reduced target mRNA levels by >80%. Since the MVD siRNAs could potently reduce mRNA levels but the effects on luciferase expression were modest, more MVD siRNAs need to be analyzed in the Luc-1b cells.
CAD siRNAs silenced target mRNA levels and significantly reduced luciferase expression so the siRNAs were analyzed in substrate rescue experiments. The CAD protein catalyzes the first three reactions of pyrimidine biosynthesis, through three enzyme activities known as carbamoyl phosphate synthetase, aspartate transcarbamoylase, and ornithine transcarbamoylase (25). The requirement of CTP and UTP for HCV replication has been previously identified using antimetabolite compounds (54). Similarly, the addition of exogenous uridine and cytidine should bypass the need for de novo pyrimidine biosyntheses when the CAD complex is silenced, if CAD is indeed the essential enzyme that supplies pyrimidines for HCV replication. After a 24-h siRNA transfection with the validated CAD siRNAs, the addition of 100 μM uridine (as UTP) and cytidine (as CTP) reversed the inhibition of the replicon luciferase levels ∼6-fold (see Fig. S1 in the supplemental material). The addition of exogenous pyrimidines to the Luc-1b cells increased luciferase expression ∼2-fold in both the GAPDH- and the pGL2 siRNA-transfected cells. Although pyrimidine addition stimulates luciferase expression in the Luc-1b cells, knockdown of pGL2 was not able to be rescued, and cell viability was unchanged, suggesting cell growth was equivalent in the treated cell lines (see Fig. S1 in the supplemental material).
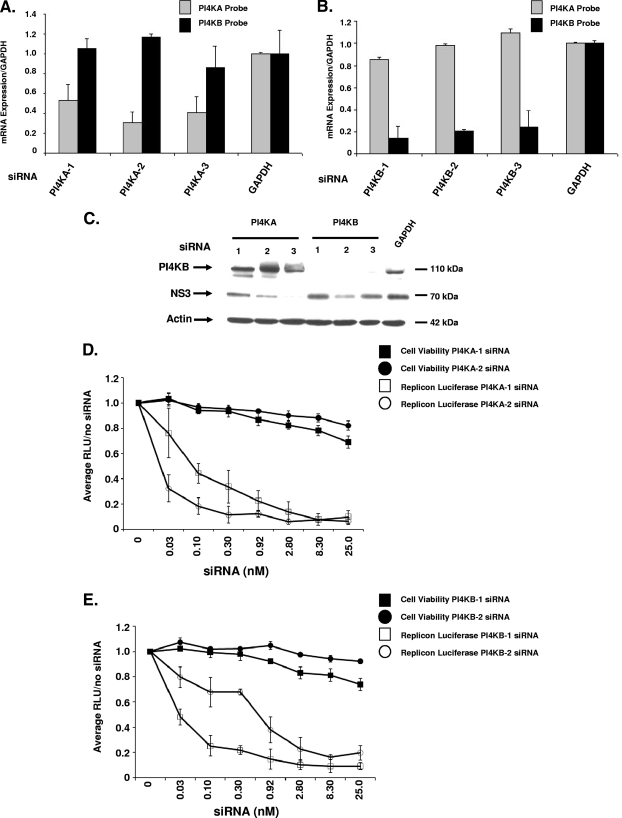
One explanation for the identification of both PI4KA and PI4KB siRNAs is that their respective siRNAs affect the expression of the other isoform. To address the possibility that the siRNAs could cross-react and inhibit both isoforms, we performed both RT-PCR and Western blot analysis. The siRNAs were transfected in Luc-1b cells for 72 h, and primers to each gene were used to quantify mRNA levels in both transfectants. The siRNAs were selective and only reduced the mRNA of their respective isoform (Fig. 2A and B). The three siRNAs were also validated by Western blotting to measure both target protein knockdown and viral protein levels. The siRNAs directed to PI4KA had no significant effect on PI4KB protein levels, while the siRNAs that reduced the mRNA levels of PI4KB-1, -2, and -3 significantly inhibited the PI4KB protein levels (Fig. 2C). PI4KA protein levels were not determined since we were unable to obtain a reliable, specific antibody (data not shown). In addition to PI4KB, we investigated whether NS3 protein levels were altered after PI4KA or PI4KB knockdown. The siRNA-transfected cells had consistently lower levels of NS3 protein compared to GAPDH-transfected cells (Fig. 2C). The most potent siRNAs, as measured by luciferase (PI4KA-1, PI4KA-3, and PI4KB-2, Fig. 1C), had the most significant effect on NS3 protein levels (Fig. 2C). PI4KB-1 and -3 had less of an effect on NS3 protein levels, which correlated with higher luciferase expression (Fig. 1C). There was some variation observed with the mRNA knockdown of PI4KA and PI4KB in the Luc-1b cell line. The variation seen with mRNA or protein knockdown is likely due to the inherent heterogeneity of transient transfection and was not significant when compared across multiple experiments (data not shown).
FIG. 2.
Validation of PI4KA and PI4KB siRNAs. (A and B) Each PI4K siRNA was analyzed by RT-PCR to determine whether the siRNAs were isoform specific. After siRNA transfection with PI4KA (A) and PI4KB (B) siRNAs, the mRNA levels were measured using both a PI4KA and a PI4KB TaqMan probe. (C) PI4KB and NS3 protein levels were measured in replicon cell lysates after a 72-h siRNA transfection with 25 nM siRNA. (D and E) Luc-1b cells were transfected with a dose response of PI4KA-1 (□) and PI4KA-2 (○) (D) or PI4KB-1 (□) and PI4KB-2 (○) (E) siRNAs, luciferase levels (open symbols) and cell viability (solid symbols) were quantified and normalized to the siRNA-free treatment (the zero concentration). RT-PCR was also confirmed across the dose response in Fig. S3A and B in the supplemental material. The RT-PCR data are presented as normalized to GAPDH and are representative of three independent experiments, along with the standard deviations.
To further test the effects on PI4KA and PI4KB siRNAs on Luc-1b expression, we performed a dose titration of both PI4KA and PI4KB siRNAs in the Luc-1b cells. PI4KA-1 and -2 siRNAs had an IC50 in the replicon cells of ∼0.1 nM, while the PI4KB-1 and -2 siRNA IC50 was 0.1 to 0.3 nM on luciferase expression, respectively (Fig. 2D and E). RT-PCR analysis over the dose response confirmed the target mRNA was silenced at or above the IC50 (see Fig. S3A and B in the supplemental material). To determine whether the knockdown of both PI4KA and PI4KB was synergistic to inhibit Luc-1b expression, we dosed PI4KA and PI4KB siRNAs together in a 1:1 concentration and measured luciferase expression after 72 h (see Fig. S2 in the supplemental material). The siRNAs were mixed at a 25 nM:25 nM ratio and added at a final concentration of 25 nM and then added to cells at twofold serial dilutions (see Fig. S2 in the supplemental material). In this way, each siRNA concentration would be half of the total dose added (i.e., at 25 nM, each individual siRNA would be 12.5 nM) so that additive or synergistic effects on luciferase expression could be detected. At an IC50 of 0.80 nM siRNA (0.40 nM PI4KA and 0.40 nM PIK4B), the inhibition of luciferase was twofold greater than for PI4KB-GAPDH siRNA treatment and approximately the same as for the PI4KA-GAPDH siRNA combination. There was no synergy with the combination of PI4KA or PI4KB siRNAs, suggesting that both enzymes play unique but required roles in genotype 1b replication (see Fig. S2 in the supplemental material).
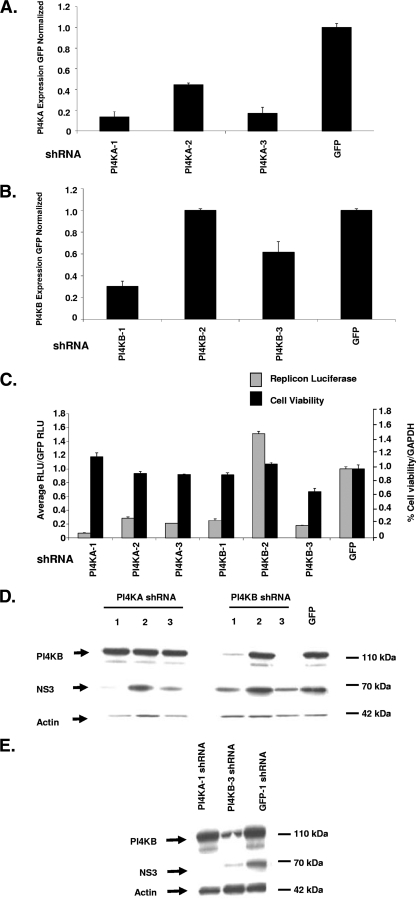
PI4KA and PI4KB shRNA validation.
shRNAs were used to determine whether stable knockdown of PI4KA and PI4KB could maintain inhibition of the Luc-1b cells over time. With the stable knockdown of PI4KA and PI4KB, not only can long-term effects on HCV replication and HCV RNA stability be determined, but the variability of transient siRNA transfection can also be avoided. Three shRNAs were identified that stably reduced mRNA levels for PI4KA (1, 2, and 3) and two for PI4KB (1 and 3) as measured by RT-PCR (Fig. 3A and B). The reduction of mRNA levels also correlated with a reduction in replicon luciferase expression (Fig. 3C, gray bars) while having no significant effect on cell viability (Fig. 3C, black bars), as measured after 2 weeks in cell culture. The two most potent shRNAs targeting PI4KB caused a substantial reduction in protein levels, while all three of the PI4KA shRNAs had no effect on PI4KB protein expression (Fig. 3D). Consistent with the reduced mRNA levels and luciferase activity, the most active (as measured by RT-PCR) shRNAs against PI4KA (1 and 3) and PI4KB (1 and 3) resulted in a reduction of NS3 protein compared to GFP shRNA stable cells (Fig. 3D). Cells transduced with the two most active shRNAs (PI4KA-1 and PI4KB-3) were grown under puromycin selection for 3 weeks. The shRNA-expressing replicon cells showed no significant loss of viability (as measured by actin levels) and NS3 protein levels were reduced in the PI4KB-3 shRNA-expressing cells, while abolished in the PI4KA-1-expressing cells (Fig. 3E). The PI4KB protein was significantly reduced in the PI4KB-3 shRNA-expressing but not in the PI4KA-1-expressing cells. These observations suggest that the stable reduction of PI4KA (measured by RT-PCR) or PI4KB (measured by Western blot) is not overtly toxic to the cells and NS3 levels are reduced, suggesting that these enzymes are required for the maintenance of viral replication.
FIG. 3.
Stable silencing of PI4KA and PI4KB in Luc-1b replicon cells. (A and B) Three shRNAs targeting PI4KA or PI4KB were stably transduced into Luc-1b cells (on puromycin for 2 weeks), and the mRNA levels were quantified by using RT-PCR for PI4KA (A) and PI4KB (B). (C) In parallel, the PI4KA or PI4KB stably transduced cells were analyzed for luciferase signal 96 h after plating (░⃞), and ATP levels were quantified as a surrogate for cell viability (▪). The PI4KB and NS3 protein levels were measured in stable replicon lysates after 96 h (D) or 3 weeks (E) on puromycin selection. All data are presented as normalized to GFP shRNAs and are representative of three independent experiments. The standard deviations are indicated.
PI4KA and -B are required for JFH-1 infectious virus replication.
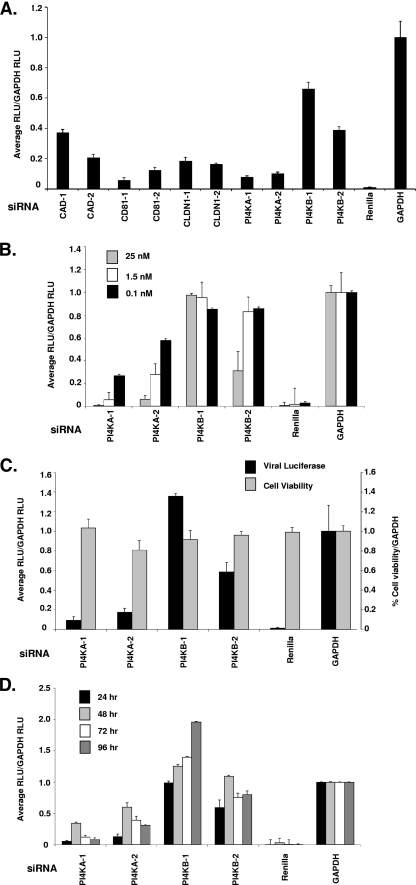
The genotype 1b replicon model system lacks the HCV structural proteins and is unable to form infectious viral particles. It is possible the PI4K enzymes are only required for the adapted subgenomic genotype 1b replicon model and might not be essential for infectious virus replication. We used a JFH1-2a infectious virus to compare the relative roles of the two PI4K isoforms on the complete HCV viral life cycle using the validated siRNAs identified in Fig. 1C and D (56, 66). Along with PI4KA and PI4KB, several positive control siRNAs, CAD, CD81, and Claudin-1 (CLDN1), were transfected into Huh7.5 cells prior to viral infection (Fig. 4A) (14, 28, 44). The control siRNA targeting the Renilla luciferase reporter contained in the JFH1-2a genome could ablate JFH-1 replication in Huh7.5 cells as measured by the Renilla luciferase levels (Fig. 4B). The CD81 and CLDN1 siRNAs inhibited JFH1-2a Renilla expression ≥80% compared to GAPDH siRNA-transfected cells (Fig. 4A). The two validated CAD siRNAs could inhibit JFH1-2a Renilla luciferase ∼60 and 80%, whereas both PI4KA siRNAs blocked luciferase >80%. The PI4KB-1 and -2 siRNAs could inhibit Renilla expression ∼60 and 40%, respectively, which is consistent with the findings presented in Fig. 4B to D.
FIG. 4.
PI4KA and PI4KB siRNAs inhibit JFH-1 infectious virus. (A) A set of SMARTpool siRNAs known to affect HCV entry were transfected into JFH1-2a-infected cells at 25 nM, and the Renilla expression was quantified at 72 h. (B) siRNAs were transfected over a dose response (25, 1.5, and 0.1 nM, as indicated) into Huh7.5 cells for 24 h prior to infection with the JFH-1 virus. After 48 h, the cells were harvested for Renilla luciferase quantification. The mRNA levels were analyzed in the Huh7.5 cells at 72 h after a 25 nM siRNA transfection and viral infection (data not shown). (C) Huh7.5 cells were first infected with JFH-1 for 24 h prior to siRNA transfection for 72 h, and the levels of viral luciferase (▪) and cell viability (░⃞) were measured. (D) Conditioned supernatants from the JFH1-2a-infected Huh 7.5 cells in panel C were collected and added back onto naive Huh7.5 cells. Luciferase was measured from 24 to 96 h (24, 48, 72, and 96 h, as indicated) after supernatant addition to measure reinfection rates. All data are presented as normalized to GAPDH siRNAs and are representative of three independent experiments. The standard deviations are indicated.
To further characterize the PI4KA and PI4KB siRNAs in the JFH1-2a infectious virus assay, a dose of the validated PI4KA and PI4KB siRNAs (25, 1.5, or 0.1 nM) was transfected into Huh7.5 cells 24 h prior to JFH1-2a viral RNA infection. The two independent PI4KA siRNAs potently inhibited viral replication, >80%, at the highest concentration of PI4KA siRNAs tested (25 nM). In contrast, only one of the two PI4KB siRNAs was able to reduce luciferase levels by 50% (Fig. 4B). Despite the difference in luciferase inhibition, knockdown of PI4KA and PI4KB mRNA levels in the Huh 7.5 cells was similar to the Huh7 replicon cells (data not shown).
To measure the effects of the siRNAs on JFH-1 replication (as measured by Renilla luciferase expression) after infectious virus entry has already been initiated, cells were infected first with JFH1-2a virus for 24 h and then transfected with 25 nM concentrations of each siRNA for an additional 72 h (Fig. 4C). The PI4KA siRNAs could potently inhibit replication, while the PI4KB siRNAs had little effect on luciferase levels (PI4KB-2 at 40% inhibition) (Fig. 4C). In addition, virus production was analyzed by removing virus-containing supernatants from the JFH1-2a-infected cells at 24-h intervals after PI4KA and PI4KB siRNA transfection. The virus-containing supernatants were added to naive Huh7.5 cells, and Renilla luciferase expression was determined (Fig. 4D). The percent reinfection was calculated by normalizing pGL2 siRNA by GAPDH siRNA-transfected cells effects on Renilla luciferase levels at each 24-h time point. These results indicate the infectious virus genotype 2a production was selectively sensitive to the loss of PI4KA as early as 24 h after transfection. PI4KA silencing diminished virus production from the cells and inhibited Renilla luciferase when the siRNAs were transfected before and/or after JFH1-2a infection, suggesting PI4KA is a potent inhibitor of JFH1-2a infectious virus replication.
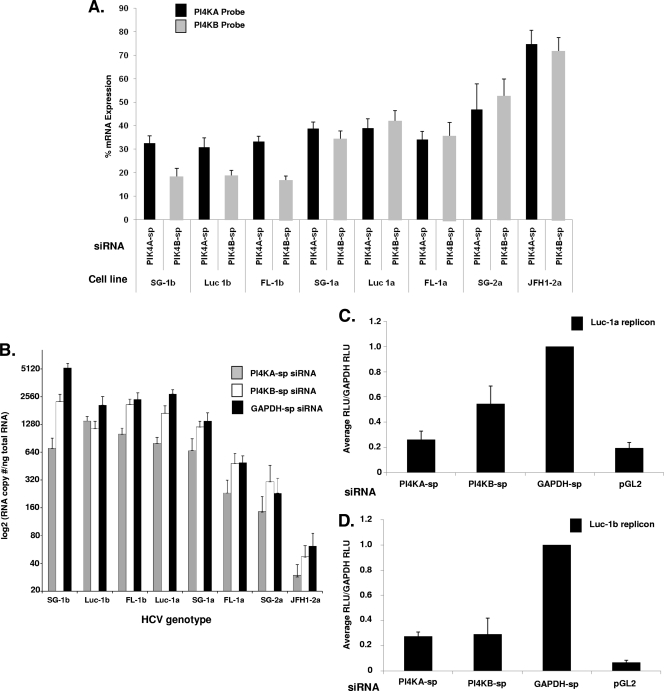
To determine whether PI4KA or PI4KB knockdown could inhibit replication of additional HCV genotypes, we characterized PI4KA-sp, PI4KB-sp, and GAPDH-sp (-sp, SMARTpool, pool of previously validated siRNAs) siRNA silencing in 1a, 1b, and 2a replicons using QRT-PCR as previously described (37). All of the replicon cell lines were transfected with a single 25 nM dose of siRNA for 72 h, and both HCV viral RNA (Fig. 5B) and mRNA knockdown of PI4KA and PI4KB were determined (Fig. 5A). PI4KA and PI4KB mRNA was knocked down ≥58% relative to GAPDH in all of the replicons with the exception of the SG-2a and JFH1-2a transfections, where the knockdown was ≤53%. The copy number of viral RNA [expressed as the viral log 2(RNA/ng of total cellular RNA)] varied in each cell line from >5,000 copies in the SG-1b cells to ∼61 copies in the JFH1-2a cells, each transfected with GAPDH-sp siRNAs. All of the replicons were sensitive to PI4KA-sp siRNAs, and the HCV viral copy number was reduced from 40 to 80% depending on genotype. In contrast, several of the replicons were not significantly inhibited by PI4KB-sp siRNAs, including FL-1b, SG-1a, FL-1a, SG-2a, and JFH1-2a (similar to the results shown in Fig. 4B to D). PI4KA-sp and PI4KB-sp were also transfected into Luc-1a (Fig. 5C) and Luc-1b (Fig. 5D) cells, and the luciferase levels were determined. The PI4KA-sp siRNAs inhibited the Luc-1b and Luc-1a replicon luciferase expression ∼70% compared to GAPDH-transfected cells, while the PI4KB-sp siRNAs inhibited luciferase expression 40% in the Luc-1a and 70% in the Luc-1b cells. The reduction of both HCV viral mRNA and luciferase expression across different HCV genotypes provides independent confirmation that PI4KA and PI4KB are involved in the replication and/or RNA stability of more than one genotype of HCV.
FIG. 5.
QRT-PCR analysis of PI4KA-sp and PI4KB-sp siRNAs across multiple HCV replicons. Eight HCV replicons (SG-1b, Luc-1b, FL-1b, Luc-1a, SG-1a, FL-1a, SG-2a, and JFH1-2a) were transfected with 25 nM concentrations of PI4KA-sp, PI4KB-sp, and GAPDH-sp siRNAs for 72 h, and mRNA was isolated. (A) mRNA knockdown of PI4KA and PI4KB was determined in each transfection by RT-PCR and is shown as the percent mRNA expression for each cell line. (B) The HCV RNA copy number per ng of total RNA was determined as previously described (37). The HCV copy number data was log2 transformed to compare all of the replicons in parallel. The copy number was the average of three independent transfections. The standard deviations are indicated. (C and D) The PI4KA-sp, PI4KB-sp, and GAPDH-sp siRNA-treated (25 nM) Luc-1a cells (C) and Luc-1b cells (D) were analyzed for luciferase levels after a 72-h transfection.
Analysis of pharmacological PI4K inhibitors.
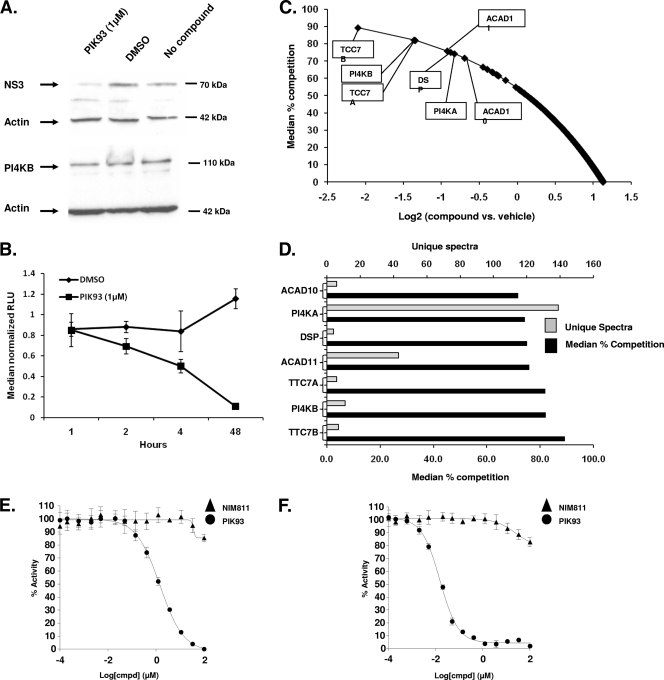
To determine whether the enzymatic activity of PIK4 enzymes was critical for their role in HCV replication, the activity of a known pharmacological PI4KB inhibitor, PIK93, was examined in the Luc-1b, Luc-1a, and JFH1-2a assays (58, 72). The activity of PIK93 was compared to the cyclophilin inhibitor NIM811 (Table 2) (38). The cyclophilin inhibitor NIM811 has been reported to inhibit HCV replication in vitro and is currently in clinical trials as a novel anti-HCV therapy (40). PIK93 inhibited the Luc-1b replicon with a 50 nM IC50, and the Luc-1a replicon with a 98 nM IC50, a concentration ∼100-fold lower than the IC50 for cell viability (10.39 μM). NIM811 had IC50s of 110 and 99 nM in the Luc-1b and Luc-1a replication assays, respectively, values also ∼200-fold less than their effects on cell viability. Thus, PIK93 inhibits the HCV replicon with similar potency as NIM811 in both Luc-1b and Luc-1a replicon cells. We verified that 1 μM PIK93 treatment was not causing a change in the overall PI4KB protein levels (Fig. 6A) and reduces NS3 protein levels consistent with an inhibition of luciferase. Luciferase expression was inhibited in these cells from 2 h up to 48 h (Fig. 6B).
TABLE 2.
IC50 summary of tool compounds in HCV replicon and infectious virusa
| Cell line | Mean IC50 (μM) or CC50 (μM) ± SD
|
|||||
|---|---|---|---|---|---|---|
| HCV
|
Cytotoxicity
|
|||||
| Luc-1b (IC50) | Luc-1a (IC50) | JFH1-2a (IC50) | Luc-1b (CC50) | Luc-1a (CC50) | JFH1-2a (CC50) | |
| PIK93 | 0.05 ± 0.01 | 0.098 ± 0.05 | 0.39 ± 0.04 | 10.39 ± 0.82 | >10 ± 0.14 | 10.8 ± 0.22 |
| NIM811 | 0.11 ± 0.02 | 0.099 ± 0.07 | 0.057 ± 0.003 | >30 ± 0.01 | >10 ± 0.22 | >5 ± 0.73 |
Luc-1b-, Luc-1a-, and JFH-1-infected Huh7.5 cells were treated with an eight-point concentration dose response of compound. Firefly luciferase was determined for the replicon cells, and Renilla luciferase was determined for the JFH-1-infected cells after 48 h of compound treatment (IC50). The cytotoxicity was determined by using CellTiter-Glo. IC50 and CC50 calculations were determined using Sigma plot version 9. The data are representative of three independent experiments, and the standard deviations are shown.
FIG. 6.
Biochemistry of PIK93 in Luc-1b cells. (A) PI4KB, NS3, and β-actin protein levels were analyzed in the Luc-1b cells by Western blot after a 1 μM 48-h incubation with PIK93, DMSO, or without compound. (B) The luciferase activity was also determined in parallel the Luc-1b cells (panel A) after treatment with 1 μM PIK93 over a 48-h time course. (C) Affinity chromatography in-lysate competition experiments were performed in the HCV replicon cells with 10 μM PIK93 and DMSO (affinity matrix supplied in Fig. S4 in the supplemental material). Peptides that competed PIK93 greater than 70% are highlighted in the boxes. All peptides competed from the column are indicated in Table S4 in the supplemental material. (D) Differentially competed proteins are shown as the percent competition verses the DMSO control lysates (▪) and the unique spectra for each protein (░⃞). (E and F) PI4KA (E) or PI4KB (F) protein was preincubated for 10 min at 14 different concentrations of PIK93 (•) or NIM811 (▴) before the addition of ATP and phosphatidylinositol. Depletion of NADH fluorescence was measured for 5 min, and the percent activity was calculated from the initial rates of the inhibited reactions relative to the uninhibited control.
Consistent with the replicon cells, PIK93 inhibited JFH1-2a infectious virus replication at an IC50 of 390 nM, which was >25-fold more potent than cell viability (Table 2). Similarly, NIM811 inhibited JFH1-2a replication at an IC50 of 390 nM and a CC50 of >5 μM. (Table 2). We hypothesized that the activity of PIK93 in the infectious virus assay was likely due to the inhibition of both PI4KA and PI4KB, since the PI4KA siRNAs could inhibit Renilla luciferase in the live virus assay, whereas the PIK4B siRNAs had less of an effect (Fig. 4B and C).
Chemical proteomics of PIK93.
To determine whether PIK93 could bind both PI4KA and PI4KB in the Luc-1b cells, we performed a two-channel iTRAQ (named for isobaric tagging reagents for relative and absolute quantification) quantitative chemical proteomics experiment. To this end, we generated an affinity matrix consisting of a derivatized, bioactive PI4KB inhibitor, LCJ217 (see Fig. S4 in the supplemental material), linked to Sepharose beads to purify and identify cellular and/or viral proteins from the Luc-1b cell lysate. To quantify specific binding to the matrix, we performed an in lysate competition experiment by adding 10 μM soluble PIK93 or DMSO into the replicon cell lysate prior to affinity purification. Using iTRAQ, we multiplexed both samples and quantified binding displacement (percent competition) relative to the vehicle (DMSO) by liquid chromatography-tandem mass spectrometry analysis. A total of 1,275 peptides were quantified by identifying 622 unique proteins (see Table S4 in the supplemental material); among the kinases, five were competed from the column more than 70% of the time. The peptides that were competed from the matrix were plotted as a log ratio of PIK93 versus DMSO by the median percent competition (Fig. 6C). Both class III PI4 kinases, PI4KA and PI4KB, were affinity enriched on the LCJ217 matrix with a high number of unique spectra (139 and 11, respectively; Fig. 6C, gray bars) and were specifically competed (74 and 82% competition, respectively; Fig. 6C, black bars) by PIK93 (Fig. 6D). The number of peptides for PI4KA was greater than PI4KB, suggesting that the competition was more significant, although the percent competition was similar for PI4KA and PI4KB (74 and 82%, respectively), indicating that the cellular binding of the proteins to the matrix was similar.
To validate the chemical proteomic data, we built in vitro biochemical assays for PI4KA and PI4KB. PI4KA or PI4KB were preincubated with PIK93 and NIM811 dilutions in assay buffer for 10 min at room temperature (25°C) prior to initiating the reaction with phosphatidylinositol and ATP. The control compound, NIM811, could not inhibit PI4KA or PI4KB activity as expected, whereas PIK93 could inhibit PI4KB with an IC50 of 0.014 μM and PI4KA with an IC50 of 1.39 μM (Fig. 6E and F).
DISCUSSION
A variety of RNA interference (RNAi) screens using distinct viral replication model systems have been used to identify host factor regulators of HCV (7, 42, 46, 55, 56, 60). In addition to the cyclophilins, the lipid kinase PI4KA, has emerged as an essential host factor in several different HCV genotypes (7, 56, 60). Consistent with these previous studies, our siRNA screens using the Luc-1b replicon identified overlapping targets previously demonstrated to regulate HCV replication, including PI4KA and cyclophilins B and C (7, 56, 41, 60, 63). In addition, we found that PI4KB was essential for genotype 1b replication and that CAD was essential for genotype 1b and 2a replication (54).
The role of the pyrimidine biosynthesis pathway has been previously characterized in HCV and respiratory syncytial virus using the antinucleoside PALA, a transition state inhibitor of the aspartate transcarbamylase activity of CAD (12, 54, 66). We found that CAD knockdown was required for Luc-1b (and JFH1-2a) replication and that the addition of exogenous UTP and CTP was able to restore Luc-1b replication (see Fig. S1 and S4A in the supplemental material). Since the rate of replicon RNA turnover results from a balance between active RNA-dependent RNA polymerases and HCV RNA stability, limited availability of either UTP or CTP could reduce cytoplasmic pyrimidine nucleoside pools and slow viral RNA production. Since viral RNA synthesis competes with endogenous RNA production, a drop in de novo pyrimidine production could be a strategy to inhibit HCV viral replication. However, the observation that PALA is an inhibitor of cell proliferation exposes the liability of targeting this pathway for a chronic disease such as HCV (22, 66).
Cholesterol biosynthesis and lipid droplet formation have been shown to be necessary for HCV replication, including reports on the use of statins and small molecule inhibitors of geranylgeranylation (3, 24, 39, 62, 67). Cholesterol is essential for ER-to-Golgi transport of secretory proteins which is thought to aid in the trafficking of viral proteins through the cell (2, 47). In addition to HCV Luc-1b, we have also shown that silencing of MVD inhibits both a dengue replicon stable cell line and a dengue live virus infectious assay, suggesting that MVD is an important mediator of viral replication in multiple Flaviviridae viruses (50). Although MVD is not predicted to be a rate-limiting enzyme in cholesterol production, its inhibition would likely lead to an accumulation of phosphorylated intermediates (such as mevalonic acid pyrophosphate), which may regulate the phosphorylation state of lipid biogenesis enzymes (19). It is not clear whether MVD knockdown is affecting viral replication through cholesterol modulation or through the accumulation of lipid intermediates.
Several publications have recently identified PI4KA as essential for HCV replication using unbiased siRNA and shRNA screens (7, 56, 60). Distinct from these studies, we identified PI4KB as required for subgenomic 1b HCV replication (Fig. 1C, 2E, and 5B). There are several possible reasons why PI4KB did not emerge as a hit from the published RNAi screens. First, the RNAi screens were not performed using the same cell line model system(s). Thus, the transfection efficiency, the siRNA or shRNA concentration used, and the format and timing of the assay are unique in each approach. The subtle differences in transfection and assay optimization can alter siRNA and/or shRNA potency in transient-transfection assays, as well as contribute to distinct off-target effects (11). This was found to be the case in the Tai et al. study where siRNAs targeting SLC12A5 were found to inhibit HCV replication by an off-target effect (56). Second, the genotype-specific differences (including unique adaptive mutations) of the various replicons could explain why nonoverlapping sets of host factors were identified. Third, the siRNAs or shRNAs analyzed in each screen were not always identical reagents. Both Tai et al. and Berger et al. used Dharmacon SMARTpool siRNAs, whereas Vaillancourt, et al. used an adenovirus-shRNA approach (7, 56, 60).
Both PI4KA and PI4KB siRNAs had a dose-dependent inhibitory effect on Luc-1b replication, and a combination dose response of PI4KA and PI4KB siRNAs could inhibit replication twofold better than PI4KB siRNAs alone (see Fig. S2 in the supplemental material). These results suggest that PI4KA and PI4KB each play unique but required roles in Luc-1b HCV replication. PI4KB overexpression was unable to complement PI4KA shRNA-mediated knockdown in the subgenomic genotype 1b OR6 replicon cells, suggesting that PI4KB is not functionally redundant for PI4KA (56). Our studies demonstrate that that PI4KA knockdown reduces replication and HCV RNA levels in all of the HCV genotypes we tested (Fig. 5B). In complementary studies, a GFP-tagged PI4KA protein was shown to colocalize with HCV double-stranded RNA in Huh7 cells (7). Thus, PI4KA is in a colocalized complex with the HCV double-stranded RNA and might play a role in HCV RNA stability in addition to the formation of membranous web (7).
In a study by Berger et al. (7), PI4KA was shown to play a role in the formation of membrane complexes where HCV replication takes place. Similar to PI4KA, we hypothesize PI4KB can disrupt the ability of HCV proteins to traffic in the cell to sites of the replication complex. PI4KB is necessary for PtdIns4P production in the Golgi which is required to maintain Golgi integrity, bud vesicles from the Golgi membranes, and modulate the production of PtdIns4P (58, 64). If the loss of PI4KB could reduce the local pools of PtdIns4P at the Golgi, it would disrupt the integrity of the Golgi organelle (45). PI4KB might play a role in regulating replication by contributing to a pool of Golgi body-derived membranes that are used in maintaining the web in a stable replicating cell line. PI4KB is known to regulate ceramide and oxysterol transport through the Golgi bodies and to associate with ARF, NCS1, and Rab11, a complex that regulates intra-Golgi body trafficking (58, 18, 64). One of the validated hits in the Tai et al. study was COPZ1, together with the COPI inhibitor brefeldin A, demonstrated that COPI inhibition is required for HCV replication (56). Since PI4KB plays a role in Golgi trafficking, similar to the COPI complex, it suggests that PI4KB could affect membrane-derived compartments necessary for viral replicon assembly. This might explain why PI4KB knockdown is less potent and more variable than PI4KA, because PI4KA is the primary enzyme involved in web formation, whereas PI4KB plays a secondary role by altering the lipid composition of the organelles that make up the membranous web or affects the trafficking of viral proteins.
PI4KB knockdown across various genotypes suggests that it selectively inhibits luciferase-containing or subgenomic replicons (Luc-1a, Luc-1b, and SG-1b). This selectivity could be driven by a unique dependence on the Luc-1b replicon as the cells were grown in culture, such as viral mutations that were selected for expression of the luciferase gene. We have not seen any evidence that adaptive mutations would play a role in this selectivity, and the adaptive mutations in the SG-1b and Luc-1b replicons are similar (data not shown). The presence of the luciferase gene might also create a unique dependency on PI4KB because Golgi trafficking or lipid content of the organelles would be different when luciferase is expressed. Since PI4KB siRNAs did not have an effect on the full-length genotypes we tested, it might be that subgenomic viruses have a specific subset of host factor proteins necessary for their replication. It is also possible that PI4KB knockdown can affect PI4KA activity and/or localization in the cell and would explain why PI4KB knockdown is less potent in several of the HCV replicons. There is a need to determine whether PI4KB is interacting directly with viral proteins or with other host factor proteins, including PI4KA.
The siRNA-mediated knockdown of PI4KA and PI4KB is functionally distinct from compound inhibition. Compound inhibition is expected to rapidly block enzymatic activity, whereas siRNA knockdown reduces mRNA and protein levels. If the enzymatic activity is required for replication, then siRNA knockdown is efficacious only when there is complete ablation of the protein. Even if a fraction of the protein is left present in the cell, it could be sufficient for signaling and PtdIns4P production. If PtdIns4P generation is the essential role of PI4KB in the Luc-1b replicon, then siRNA or shRNA knockdown might not be sufficient to see a consistent phenotype. The reason PI4KA knockdown is absolutely essential for HCV replication is that, in addition to its enzymatic function, it plays an important role as a scaffolding protein by binding to NS5A (1). Since PI4KA and PI4KB have differential subcellular localizations in the cell, it is possible that when the enzymatic activity of PI4KA or PI4KB is inhibited, the local pools of phosphoinositides would be disrupted, reducing the PtdIns4P levels at the Golgi and/or the ER (5, 18, 20). If the PtdIns4P levels drive the ability of NS5A to interact with PI4KA, then this could be a mechanism that regulates membranous web formation. Since the transient siRNA knockdown efficiency of PI4KB in the various replicons is inconsistent, the role of PI4KBs role in HCV replication might be better exploited using selective small molecular inhibitors.
The PIK93 chemoproteomic experiments identified additional proteins that might contribute to the inhibitory activity of PIK93 in the Luc-1a, Luc-1b, and JFH1-2a cells. Some of the notable proteins competed from the affinity matrix included TTC7A, TTC7B, ACAD10, and ACAD11 (Fig. 6C). ACAD10 and ACAD11 are acyl coenzyme A dehydrogenases (ACAD) of a family of flavoproteins that are involved in the beta-oxidation of the fatty acyl coenzyme A derivatives (68). Although ACAD10 or ACAD11 require further validation in the Luc-1b cells, one might speculate these enzymes impinge on fatty acid metabolism and indirectly with cholesterol production in the cell.
The other two related genes identified in the PIK93 competition assay were TTC7A and TTC7B (tetratricopeptide repeat domain 7A and 7B). TTC7B is thought to be involved in iron transport, via an interaction with DMT1, to regulate iron uptake into the cell (65). Iron is known to be a negative regulator of HCV replication by inhibiting the RNA-dependent RNA polymerase (15). Also, subgenomic HCV replication alters liver cell iron metabolism by reducing intracellular iron levels (16). It could be that PIK93 binds to TTC7B modulating its putative interaction with DMT1 and increases iron uptake into the cells. Silencing of the iron homeostasis regulator HAMP was found to inhibit OR6 full-length replication in an independent siRNA screen (56). It was also suggested that HAMP knockdown might disrupt HCV IRES-dependent translation (56). The role of these targets and how the intracellular iron metabolism pathway regulates the HCV life cycle requires further study.
In conclusion, we have identified type III PI4KA and PI4KB as novel cellular proteins that are required for replication of several genotypes of HCV. The identification of novel host factor enzymes required for HCV replication has significant therapeutic implications for the development of new targets in HCV and other related viruses. PI4KA and PI4KB are promising targets for inhibition with low-molecular-weight compounds, as demonstrated using the PI4KA and PIK4B inhibitor PIK93 (Fig. 6 and Table 2). Selective or dual type III PI4K inhibitors should be considered for developing anti-HCV therapies in combination with current standard of care regimens for treating multiple genotypes of HCV.
Supplementary Material
Acknowledgments
We thank Jeff MacKeigan and Christine Chow for critical readings of the manuscript, Tony Orth for stamping and annotating the siRNA collections, and Christina Rau for advice and expertise.
Footnotes
Published ahead of print on 15 July 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ahn, J., K. Chung, D. Kim, M. Won, L. Kim, K. Kim, M. Nam, S. Choi, H. Kim, M. Yoon, S. Chae, and K. Hoe. 2004. Systematic identification of hepatocellular proteins interacting with NS5A of the hepatitis C virus. J. Biochem. Mol. Biol. 37:741-748. [DOI] [PubMed] [Google Scholar]
- 2.Aizaki, H., K. Lee, V. Sung, H. Ishiko, and M. Lai. 2004. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324:450-461. [DOI] [PubMed] [Google Scholar]
- 3.Bader, T., J. Fazili, M. Madhoun, C. Aston, D. Hughes, S. Rizvi, K. Seres, and M. Hasan. 2008. Fluvastatin inhibits hepatitis C replication in humans. Am. J. Gastroenterol. 103:1383-1389. [DOI] [PubMed] [Google Scholar]
- 4.Balla, A., Y. Kim, P. Varnai, Z. Szentpetery, Z. Knight, K. Shokat, and T. Balla. 2008. Maintenance of hormone-sensitive phosphoinositide pools in the plasma membrane requires phosphatidylinositol 4-kinase IIIα. Mol. Biol. Cell 19:711-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balla, A., G. Tuymetova, A. Tsiomenko, P. Várnai, and T. Balla. 2005. A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III alpha: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol. Biol. Cell 16:1282-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bantscheff, M., D. Eberhard, Y. Abraham, S. Bastuck, M. Boesche, S. Hobson, T. Mathieson, J. Perrin, M. Raida, C. Rau, V. Reader, G. Sweetman, A. Bauer, T. Bouwmeester, C. Hopf, U. Kruse, G. Neubauer, N. Ramsden, J. Rick, B. Kuster, and G. Drewes. 2007. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 25:1035-1044. [DOI] [PubMed] [Google Scholar]
- 7.Berger, K. L., J. D. Cooper, N. S. Heaton, R. Yoon, T. E. Oakland, T. X. Jordan, G. Mateu, A. Grakoui, and G. Randall. 2009. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc. Natl. Acad. Sci. USA 106:7577-7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blight, K., A. Kolykhalov, and C. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 9.Blight, K., J. McKeating, and C. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blight, K., J. McKeating, J. Marcotrigiano, and C. Rice. 2003. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J. Virol. 77:3181-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borawski, J., A. Lindeman, F. Buxton, M. Labow, and L. Gaither. 2007. Optimization procedure for small interfering RNA transfection in a 384-well format. J. Biomol. Screen. 13:546-559. [DOI] [PubMed] [Google Scholar]
- 12.Davis, I., E. Lazarowski, J. Hickman-Davis, J. Fortenberry, F. Chen, X. Zhao, E. Sorscher, L. Graves, W. Sullender, and S. Matalon. 2006. Leflunomide prevents alveolar fluid clearance inhibition by respiratory syncytial virus. Am. J. Respir. Crit. Care Med. 173:673-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Clercq, E. 2007. The design of drugs for HIV and HCV. Nat. Rev. Drug Discov. 6:1001-1018. [DOI] [PubMed] [Google Scholar]
- 14.Evans, M., T. von Hahn, D. Tscherne, A. Syder, M. Panis, B. Wölk, T. Hatziioannou, J. McKeating, P. Bieniasz, and C. Rice. 2007. Claudin-1 is a hepatitis C virus coreceptor required for a late step in entry. Nature 446:801-805. [DOI] [PubMed] [Google Scholar]
- 15.Fillebeen, C., A. Rivas-Estilla, M. Bisaillon, P. Ponka, M. Muckenthaler, M. Hentze, A. Koromilas, and K. Pantopoulos. 2005. Iron inactivates the RNA polymerase NS5B and suppresses subgenomic replication of hepatitis C virus. J. Biol. Chem. 280:9049-9057. [DOI] [PubMed] [Google Scholar]
- 16.Fillebeen, C., M. Muckenthaler, B. Andriopoulos, M. Bisaillon, Z. Mounir, M. Hentze, A. Koromilas, and K. Pantopoulos. 2007. Expression of the subgenomic hepatitis C virus replicon alters iron homeostasis in Huh7 cells. J. Hepatol. 47:12-22. [DOI] [PubMed] [Google Scholar]
- 17.Gaither, A., D. Porter, Y. Yao, J. Borawski, G. Yang, J. Donovan, D. Sage, J. Slisz, M. Tran, C. Straub, T. Ramsey, V. Iourgenko, A. Huang, Y. Chen, R. Schlegel, M. Labow, S. Fawell, W. Sellers, and L. Zawel. 2007. A Smac mimetic rescue screen reveals roles for inhibitor of apoptosis proteins in tumor necrosis factor-alpha signaling. Cancer Res. 67:11493-11498. [DOI] [PubMed] [Google Scholar]
- 18.Godi, A., P. Pertile, R. Meyers, P. Marra, G. Di Tullio, C. Iurisci, A. Luini, D. Corda, and M. De Matteis. 1999. ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns4,5P2 on the Golgi complex. Nat. Cell Biol. 1:280-287. [DOI] [PubMed] [Google Scholar]
- 19.Goldstein, J., and M. Brown. 1990. Regulation of the mevalonate pathway. Nature 343:425-430. [DOI] [PubMed] [Google Scholar]
- 20.Haynes, L., G. Thomas, and R. Burgoyne. 2005. Interaction of neuronal calcium sensor-1 and ADP-ribosylation factor 1 allows bidirectional control of phosphatidylinositol 4-kinase beta and trans-Golgi network-plasma membrane traffic. J. Biol. Chem. 280:6047-6054. [DOI] [PubMed] [Google Scholar]
- 21.Hopkins, A., and C. Groom. 2002. The druggable genome. Nat. Rev. Drug Discov. 1:727-730. [DOI] [PubMed] [Google Scholar]
- 22.Huang, M., and L. Graves. 2003. De novo synthesis of pyrimidine nucleotides; emerging interfaces with signal transduction pathways. Cell. Mol. Life Sci. 60:321-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huesken, D., J. Lange, C. Mickanin, J. Weiler, F. Asselbergs, J. Warner, B. Meloon, S. Engel, A. Rosenberg, D. Cohen, M. Labow, M. Reinhardt, F. Natt, and J. Hall. 2005. Design of a genome-wide siRNA library using an artificial neural network. Nat. Biotechnol. 23:995-1001. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda, M., K. Abe, M. Yamada, H. Dansako, K. Naka, and N. Kato. 2006. Different anti-HCV profiles of statins and their potential for combination therapy with interferon. Hepatology 44:117-125. [DOI] [PubMed] [Google Scholar]
- 25.Iwahana, H., M. Fujimura, S. Ii, M. Kondo, M. Moritani, Y. Takahashi, T. Yamaoka, K. Yoshimoto, and M. Itakura. 1996. Molecular cloning of a human cDNA encoding a trifunctional enzyme of carbamoyl-phosphate synthetase-aspartate transcarbamoylase-dihydroorotase in de novo pyrimidine synthesis. Biochem. Biophys. Res. Commun. 219:249-255. [DOI] [PubMed] [Google Scholar]
- 26.Kato, T. T., Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 27.Kato, T., T. Date, M. Miyamoto, M. Sugiyama, Y. Tanaka, E. Orito, T. Ohno, K. Sugihara, I. Hasegawa, K. Fujiwara, K. Ito, A. Ozasa, M. Mizokami, and T. Wakita. 2005. Detection of anti-hepatitis C virus effects of interferon and ribavirin by a sensitive replicon system. J. Clin. Microbiol. 43:5679-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolb, A., J. Maile, A. Heister, and S. Siddell. 1996. Characterization of functional domains in the human coronavirus HCV 229E receptor. J. Gen. Virol. 77:2515-2521. [DOI] [PubMed] [Google Scholar]
- 29.Koutsoudakis, G., A. Kaul, E. Steinmann, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2006. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 80:5308-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krönke, J., R. Kittler, F. Buchholz, M. Windisch, T. Pietschmann, R. Bartenschlager, and M. Frese. 2004. Alternative approaches for efficient inhibition of hepatitis C virus RNA replication by small interfering RNAs. J. Virol. 78:3436-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liebel, U., V. Starkuviene, H. Erfle, J. C. Simpson, A. Poustka, S. Wiemann, and R. Pepperkok. 2003. A microscope-based screening platform for large-scale functional protein analysis in intact cells. FEBS Lett. 20:394-398. [DOI] [PubMed] [Google Scholar]
- 33.Lindenbach, B., and C. Rice. 2005. Unravelling hepatitis C virus replication from genome to function. Nature 436:933-938. [DOI] [PubMed] [Google Scholar]
- 34.Lindenbach, B., M. Evans, A. Syder, B. Wölk, T. Tellinghuisen, C. Liu, T. Maruyama, R. Hynes, D. Burton, J. McKeating, and C. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 35.Liu, X., and R. Erikson. 2003. Polo-like kinase Plk1 depletion induces apoptosis in cancer cells. Proc. Natl. Acad. Sci. USA 100:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 37.Ma, S., J. Boerner, C. TiongYip, B. Weidmann, N. Ryder, M. Cooreman, and K. Lin. 2006. NIM811, a cyclophilin inhibitor, exhibits potent in vitro activity against hepatitis C virus alone or in combination with alpha interferon. Antimicrob. Agents Chemother. 50:2976-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathy, J., S. Ma, T. Compton, and K. Lin. 2008. Combinations of cyclophilin inhibitor NIM811 with hepatitis C virus NS3-4A protease or NS5B polymerase inhibitors enhance antiviral activity and suppress the emergence of resistance. Antimicrob. Agents Chemother. 52:3267-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089-1097. [DOI] [PubMed] [Google Scholar]
- 40.Morishima, C., S. Polyak, R. Ray, M. Doherty, A. Di Bisceglie, P. Malet, H. Bonkovsky, D. Sullivan, D. Gretch, A. Rothman, M. Koziel, K. Lindsay, et al. 2006. Hepatitis C virus-specific immune responses and quasi-species variability at baseline are associated with nonresponse to antiviral therapy during advanced hepatitis C. J. Infect. Dis. 193:931-940. [DOI] [PubMed] [Google Scholar]
- 41.Nakagawa, M., N. Sakamoto, Y. Tanabe, T. Koyama, Y. Itsui, Y. Takeda, C. Chen, S. Kakinuma, S. Oooka, S. Maekawa, N. Enomoto, and M. Watanabe. 2005. Suppression of hepatitis C virus replication by cyclosporin A is mediated by blockade of cyclophilins. Gastroenterology 129:1031-1041. [DOI] [PubMed] [Google Scholar]
- 42.Ng, T., H. Mo, T. Pilot-Matias, Y. He, G. Koev, P. Krishnan, R. Mondal, R. Pithawalla, W. He, T. Dekhtyar, J. Packer, M. Schurdak, and A. Molla. 2007. Identification of host genes involved in hepatitis C virus replication by small interfering RNA technology. Hepatology 45:1413-1421. [DOI] [PubMed] [Google Scholar]
- 43.Penin, F., J. Dubuisson, F. Rey, D. Moradpour, and J. Pawlotsky. 2004. Structural biology of hepatitis C virus. Hepatology 39:5-19. [DOI] [PubMed] [Google Scholar]
- 44.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 45.Rajebhosale, M., S. Greenwood, J. Vidugiriene, A. Jeromin, and S. Hilfiker. 2003. Phosphatidylinositol 4-OH kinase is a downstream target of neuronal calcium sensor-1 in enhancing exocytosis in neuroendocrine cells. J. Biol. Chem. 278:6075-6084. [DOI] [PubMed] [Google Scholar]
- 46.Randall, G., M. Panis, J. Cooper, T. Tellinghuisen, K. Sukhodolets, S. Pfeffer, M. Landthaler, P. Landgraf, S. Kan, B. Lindenbach, M. Chien, D. Weir, J. Russo, J. Ju, M. Brownstein, R. Sheridan, C. Sander, M. Zavolan, T. Tuschl, and C. Rice. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. USA 104:12884-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ridsdale, A., M. Denis, P. Gougeon, J. Ngsee, J. Presley, and X. Zha. 2006. Cholesterol is required for efficient endoplasmic reticulum-to-Golgi transport of secretory membrane proteins. Mol. Biol. Cell 17:1593-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rines, D., M. Gomez-Ferreria, Y. Zhou, P. DeJesus, S. Grob, S. Batalov, M. Labow, D. Huesken, C. Mickanin, J. Hall, M. Reinhardt, F. Natt, J. Lange, D. Sharp, S. Chanda, and J. Caldwell. 2008. Whole genome functional analysis identifies novel components required for mitotic spindle integrity in human cells. Genome Biol. 9:R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosenwirth, B., A. Billich, R. Datema, P. Donatsch, F. Hammerschmid, R. Harrison, P. Hiestand, H. Jaksche, P. Mayer, and P. Peichl. 1994. Inhibition of human immunodeficiency virus type 1 replication by SDZ NIM 811, a nonimmunosuppressive cyclosporine analog. Antimicrob. Agents Chemother. 38:1763-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rothwell, C., A. LeBreton, C. Young Ng, J. Lim, W. Liu, S. Vasudevan, M. Labow, F. Gu, and L. Gaither. 2009. Cholesterol biosynthesis modulation as a novel therapeutic approach to treat Dengue viral infection. Virology 389:8-19. [DOI] [PubMed] [Google Scholar]
- 51.Sagan, S. M., Y. Rouleau, C. Leggiadro, L. Supekova, P. G. Schultz, A. I. Su, and J. P. Pezacki. 2006. The influence of cholesterol and lipid metabolism on host cell structure and hepatitis C virus replication. Biochem. Cell Biol. 84:67-79. [DOI] [PubMed] [Google Scholar]
- 52.Saito, I., T. Miyamura, A. Ohbayashi, H. Harada, T. Katayama, S. Kikuchi, Y. Watanabe, S. Koi, M. Onji, Y. Ohta, Q.-L. Choo, M. Houghton, and G. Kuo. 1990. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 87:6547-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steinmann, E., F. Penin, S. Kallis, A. H. Patel, R. Bartenschlager, and T. Pietschmann. 2007. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 3:e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stuyver, L., T. McBrayer, P. Tharnish, A. Hassan, C. Chu, K. Pankiewicz, K. Watanabe, R. Schinazi, and M. Otto. 2003. Dynamics of subgenomic hepatitis C virus replicon RNA levels in Huh-7 cells after exposure to nucleoside antimetabolites. J. Virol. 77:10689-10694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Supekova, L., F. Supek, J. Lee, S. Chen, N. Gray, J. Pezacki, A. Schlapbach, and P. Schultz. 2008. Identification of human kinases involved in hepatitis C virus replication by small interference RNA library screening. J. Biol. Chem. 283:29-36. [DOI] [PubMed] [Google Scholar]
- 56.Tai, A. W., Y. Benita, L. F. Peng, S. S. Kim, N. Sakamoto, R. J. Xavier, and R. T. Chung. 2009. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5:298-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomson, B., and R. Finch. 2005. Hepatitis C virus infection. Clin. Microbiol. Infect. 11:86-94. [DOI] [PubMed] [Google Scholar]
- 58.Toth, B., A. Balla, H. Ma, Z. Knight, K. Shokat, and T. Balla. 2006. Phosphatidylinositol 4-kinase IIIβ regulates the transport of ceramide between the endoplasmic reticulum and Golgi. J. Biol. Chem. 281:36369-36377. [DOI] [PubMed] [Google Scholar]
- 59.Tu, H., L. Gao, S. Shi, D. Talyor, T. Yang, A. Mircheff, Y. Wen, A. Gorbalenya, S. Hwang, and M. Lai. 1999. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology 263:30-41. [DOI] [PubMed] [Google Scholar]
- 60.Vaillancourt, F. H., L. Pilote, M. Cartier, J. Lippens, M. Liuzzi, R. C. Bethell, M. G. Cordingley, and G. Kukolj. 2009. Identification of a lipid kinase as a host factor involved in hepatitis C virus RNA replication. Virology 387:5-10. [DOI] [PubMed] [Google Scholar]
- 61.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. Krausslich, M. Mizokami, R. Bartenschlager, and T. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, C., M. Gale, B. Keller, H. Huang, M. Brown, J. Goldstein, and J. Ye. 2005. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol. Cell 18:425-434. [DOI] [PubMed] [Google Scholar]
- 63.Watashi, K., N. Ishii, M. Hijikata, D. Inoue, T. Murata, Y. Miyanari, and K. Shimotohno. 2005. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell 19:111-122. [DOI] [PubMed] [Google Scholar]
- 64.Weixel, K., A. Blumental-Perry, S. Watkins, M. Aridor, and O. Weisz. 2005. Distinct Golgi populations of phosphatidylinositol 4-phosphate regulated by phosphatidylinositol 4-kinases. J. Biol. Chem. 280:10501-10508. [DOI] [PubMed] [Google Scholar]
- 65.White, R., S. McNulty, N. Nsumu, L. Boydston, B. Brewer, and K. Shimizu. 2005. Positional cloning of the Ttc7 gene required for normal iron homeostasis and mutated in hea and fsn anemia mice. Genomics 85:330-337. [DOI] [PubMed] [Google Scholar]
- 66.Wyde, P., D. Moore, D. Pimentel, and H. Blough. 1995. Evaluation of the antiviral activity of N-phosphonoacetyl-l-aspartate against paramyxoviruses in tissue culture and against respiratory syncytial virus in cotton rats. Antivir. Res. 27:59-69. [DOI] [PubMed] [Google Scholar]
- 67.Ye, J., C. Wang, R. Sumpter, M. Brown, J. Goldstein, and M. Gale. 2003. Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc. Natl. Acad. Sci. USA 100:15865-15870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ye, X., C. Ji, C. Zhou, L. Zeng, S. Gu, K. Ying, Y. Xie, and Y. Mao. 2004. Cloning and characterization of a human cDNA ACAD10 mapped to chromosome 12q24.1. Mol. Biol. Rep. 31:191-195. [DOI] [PubMed] [Google Scholar]
- 69.Yi, M., R. Villanueva, D. Thomas, T. Wakita, and S. Lemon. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. USA 103:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, J., T. Chung, and K. Oldenburg. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4:67-73. [DOI] [PubMed] [Google Scholar]
- 71.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. Burton, S. Wieland, S. Uprichard, T. Wakita, and F. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zunder, E., Z. Knight, B. Houseman, B. Apsel, and K. Shokat. 2008. Discovery of drug-resistant and drug-sensitizing mutations in the oncogenic PI3K isoform p110 alpha. Cancer Cell 14:180-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.