Abstract
A broad Gag-specific CD8+ T-cell response is associated with effective control of adult human immunodeficiency virus (HIV) infection. The association of certain HLA class I molecules, such as HLA-B*57, -B*5801, and -B*8101, with immune control is linked to mutations within Gag epitopes presented by these alleles that allow HIV to evade the immune response but that also reduce viral replicative capacity. Transmission of such viruses containing mutations within Gag epitopes results in lower viral loads in adult recipients. In this study of pediatric infection, we tested the hypothesis that children may tend to progress relatively slowly if either they themselves possess one of the protective HLA-B alleles or the mother possesses one of these alleles, thereby transmitting a low-fitness virus to the child. We analyzed HLA type, CD8+ T-cell responses, and viral sequence changes for 61 mother-child pairs from Durban, South Africa, who were monitored from birth. Slow progression was significantly associated with the mother or child possessing one of the protective HLA-B alleles, and more significantly so when the protective allele was not shared by mother and child (P = 0.007). Slow progressors tended to make CD8+ T-cell responses to Gag epitopes presented by the protective HLA-B alleles, in contrast to progressors expressing the same alleles (P = 0.07; Fisher's exact test). Mothers expressing the protective alleles were significantly more likely to transmit escape variants within the Gag epitopes presented by those alleles than mothers not expressing those alleles (75% versus 21%; P = 0.001). Reversion of transmitted escape mutations was observed in all slow-progressing children whose mothers possessed protective HLA-B alleles. These data show that HLA class I alleles influence disease progression in pediatric as well as adult infection, both as a result of the CD8+ T-cell responses generated in the child and through the transmission of low-fitness viruses by the mother.
Human immunodeficiency virus (HIV)-specific CD8+ T cells play a central role in controlling viral replication (12). It is the specificity of the CD8+ T-cell response, particularly the response to Gag, that is associated with low viral loads in HIV infection (7, 17, 34). Although immune control is undermined by the selection of viral mutations that prevent recognition by the CD8+ T cells, evasion of Gag-specific responses mediated by protective class I HLA-B alleles typically brings a reduction in viral replicative capacity, facilitating subsequent immune control of HIV (2, 20, 21). The same principle has been demonstrated in studies of simian immunodeficiency virus infection (18, 22).
Recent studies showed that the class I HLA-B alleles that protect against disease progression present more Gag-specific CD8+ T-cell epitopes and drive the selection of more Gag-specific escape mutations than those alleles that are associated with high viral loads (23). These protective HLA-B alleles not only are beneficial to infected individuals expressing those alleles but also benefit a recipient following transmission, since the transmitted virus carrying multiple Gag escape mutations may have substantially reduced fitness (3, 4, 8). However, there is no benefit to the recipient if he or she shares the same protective allele as the donor because the transmitted virus carries escape mutations in the Gag epitopes that would otherwise be expected to mediate successful immune control in the recipient (8, 11).
The sharing of HLA alleles between donor and recipient occurs frequently in mother-to-child transmission (MTCT). The risk of MTCT is related to viral load in the mother, and a high viral load is associated with nonprotective alleles, such as HLA-B*18 and -B*5802. This may contribute in two distinct ways to the more rapid progression observed in pediatric HIV infection (24, 26, 27). First, because infected children share 50% or more of their HLA alleles with the transmitting mother, they are less likely than adults to carry protective HLA alleles (16). Thus, infected children as a group carry fewer protective HLA alleles and more nonprotective HLA alleles. Second, even when the child has a protective allele, such as HLA-B*27, this allele does not offer protection if the maternally transmitted virus carries escape mutations within the key Gag epitopes that are presented by the protective allele (11, 19).
However, it is clear that infected children who possess protective alleles, such as HLA-B*27 or HLA-B*57, can achieve durable immune control of HIV infection if the virus transmitted from the mother is not preadapted to those alleles (6, 10). HIV-specific CD8+ T-cell responses are detectable from birth in infected infants (32). Furthermore, as in adult infection (3, 8), HIV-infected children have the potential to benefit from transmission of low-fitness viruses in the situation where the mother possesses protective HLA alleles and the child does not share those protective alleles. MTCT of low-fitness viruses carrying CD8+ T-cell escape mutations was recently documented (28; J. Prado et al., unpublished data).
In this study, undertaken in Durban, South Africa, we set out to test the hypothesis that HIV-infected children are less likely to progress rapidly to disease if either the infected child or the transmitting mother possesses a protective HLA allele that is not shared. The HLA alleles most strongly associated with low viral loads and high CD4 counts in a cohort of >1,200 HIV-infected adults in Durban are HLA-B*57 (-B*5702 and -B*5703), HLA-B*5801, and HLA-B*8101 (16; A. Leslie et al., unpublished data). These four alleles all present Gag-specific CD8+ T-cell epitopes, and in each case the escape mutations selected in these epitopes reduce viral replicative capacity (2-4, 8, 21, 23).
Analyzing a previously described cohort of 61 HIV-infected children in Durban (24, 26, 32), South Africa, who were all monitored from birth, we first addressed the question of whether possession of any of these four alleles by either mother or child is associated with slower disease progression in the child and then determined whether sharing of protective alleles by mother and child affects the ability of the child to make the Gag-specific CD8+ T-cell responses restricted by the shared allele.
MATERIALS AND METHODS
Study subjects.
The pediatric study cohort has been described previously (26, 32). In brief, we enrolled 63 HIV-infected infants born to HIV-infected mothers at St. Mary's and Prince Mshiyeni Hospitals in Durban, South Africa, between 2003 and 2005. The HIV type 1 (HIV-1)-seropositive mothers were recruited in the last trimester of pregnancy and received single-dose nevirapine at the onset of labor and within 48 h of giving birth to the child according to the HIVNET 012 protocol (13, 14). A total of 719 mothers were enrolled and screened, 740 infants were born to the mothers, and 75 were infected. Sixty-three of the 75 infants met the criteria for enrollment into the study. Exclusion criteria comprised prematurity, intrauterine growth restriction, and congenital anomaly.
Forty-three infants received short-term antiretroviral therapy (ART) immediately following diagnosis as part of a separate study investigating the effect of early, short-term ART on disease outcomes of pediatric infection. In this group, ART was discontinued after 12 months of continuous therapy or 18 months of interrupted therapy. The remaining 20 infants did not receive ART immediately but were followed up longitudinally. All infants enrolled in the study received ART once World Health Organization (WHO) clinical or immunological criteria were met (http://www.who.int/hiv/pub/guidelines/WHOpaediatric.pdf). Taking into account that 85% of HIV-infected children progress to CD4 counts of ≤20% by 1 year of age (24), we here define slow progression as a failure to progress to meet WHO criteria to start ART within 12 months in the absence of ART; progressors were those children who required ART within 12 months of life or within 12 months of discontinuing ART.
HLA typing was obtained for 61 of the 63 enrolled mother-child pairs, with two mother-child pairs withdrawing from the study shortly after enrollment. In addition, HLA typing was undertaken antenatally for 236 HIV-infected mothers who did not subsequently transmit the infection.
The mothers gave written informed consent for participation of their children in the study. The study was approved by all of the participating institutional review boards.
HLA typing.
DNA for HLA typing was extracted using a Puregene DNA isolation kit for blood (Gentra Systems, Minneapolis, MN) according to the manufacturer's instructions. HLA class I typing was done by DNA PCR using sequence-specific primers as previously described (16).
Synthetic HIV-1 peptides.
A panel of 410 overlapping peptides (18-mers with a 10-amino-acid overlap) spanning the entire HIV-1 clade C consensus sequence were synthesized on an automated peptide synthesizer (MBS 396; Advanced ChemTech) and used in a matrix system in screening assays (16). Previously defined optimal peptides were also similarly synthesized.
Enzyme-linked immunospot (ELISPOT) assays.
Ex vivo measurement of T cells for gamma interferon (IFN-γ) production was undertaken as previously reported (16, 32). As previously described, specific responses of ≥100 spot-forming cells (SFC)/million peripheral blood mononuclear cells (PBMC) were defined as significant. In the analysis shown in Fig. 2C, responses at less than this level were treated as zero or nonresponsive. In addition, defined optimal peptides were also tested according to the corresponding HLA type of the subject.
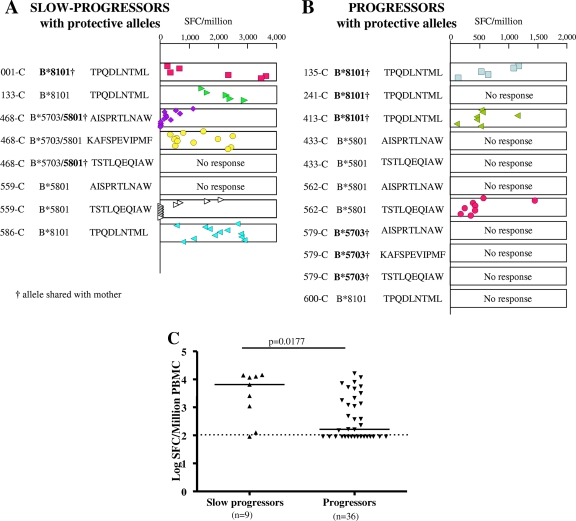
FIG. 2.
CD8+ T-cell responses to Gag epitopes in the first 12 months of life in children not on ART. Gag optimal epitopes presented by HLA-B*57, -5801, and -8101 are shown for slow-progressor (A) and progressor (B) children with protective HLA-B alleles. (C) Gag optimal epitope responses in slow-progressor and progressor children without protective HLA-B alleles. Symbols represent the magnitudes of CD8+ T-cell responses measured by ELISPOT assay at different time points in the first 12 months of life for children who deferred ART or in the first 12 months of cessation of ART for children who received immediate short-term ART. The magnitudes of responses in panel C are median magnitudes of the response for all time points tested. A significant response was defined as a peptide-specific response of ≥100 SFC/million PBMC (see Materials and Methods).
Viral load and CD4 measurement.
Plasma viral loads were measured using the Roche Amplicor Monitor assay (version 1.5), and CD4 counts were determined from fresh whole blood by Tru-Count technology as previously described (32).
Viral sequencing.
Sequencing of HIV proviral DNAs from mothers and children was undertaken as previously described (20). Genomic DNAs were extracted from PBMC pellets; gag sequences were amplified by nested PCR, purified, and directly sequenced.
Statistical analysis.
Fisher's exact test was used to compare proportions defining progressor and slow progressor children and proportions of progressor and slow progressor children with protective and unshared HLA-B alleles. The same test was also used to determine the proportions of progressors and slow progressors with ELISPOT responses to Gag epitopes and escape variants in epitopes presented by protective HLA-B alleles and the proportions of escape variants in shared and unshared HLA-B alleles. The odds ratio test was used to compare the numbers of sequence variations in children who made or did not make responses to the epitopes. The Mann-Whitney test was used to compare differences in the magnitudes of Gag responses in slow progressor and progressor children without protective HLA-B alleles.
RESULTS
Nonprogression in infants is associated with unshared protective HLA-B alleles in the mother or child.
Progression to disease is rapid in HIV-infected children, as 85% of children who did not receive ART immediately following diagnosis met the then WHO criterion to start ART (CD4 count of ≤20%) by 1 year (24). For the purposes of this study, we defined slow progression in these study subjects as a failure to meet WHO treatment criteria within 12 months of birth or within 12 months of cessation of ART for those who were treated with immediate, short-term ART after birth.
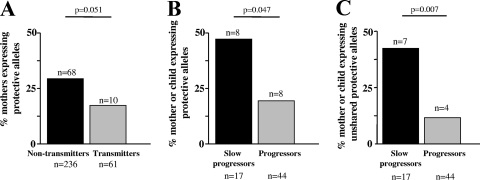
Among the 61 studied mother-child pairs, one or more of the protective HLA alleles, B*5702, B*5703, B*5801, or B*8101, was present in either the mother or child in 16 pairs (Table 1). Ten of 61 transmitting mothers (16%) possessed one of the four protective HLA-B alleles, compared to 68 of 236 (29%) HLA-typed nontransmitting mothers (P = 0.051; Fisher's exact test) (Fig. 1A). Slow progression, as defined above, was observed in 17 children (Table 1).
TABLE 1.
Progressor and slow-progressor mother-child pairs where the child and/or mother expressed one or more of the protective HLA-B alleles (B*5801, B*8101, B*5701, and B*5703)
| Study group | Child no. and alleles | Mother's alleles |
|---|---|---|
| Slow-progressor children (protective allele in 8 of 17 pairs) | ||
| Protective allele in child only | 133-C B*5802/8101 | B*4201/5802 |
| 559-C B*0801/5801 | B*0801/4101 | |
| 586-C B*1510/8101 | B*1510/5802 | |
| Protective allele in mother and child | 001-C B*4201/8101 | B*4403/8101 |
| 468-C B*5703/5801 | B*4403/5801 | |
| Protective allele in mother only | 114-C B*0702/0801 | B*0801/5702 |
| 349-C B*4501/5802 | B*4501/5702 | |
| 568-C B*0702/1801 | B*0702/8101 | |
| Progressor children (protective allele in 8 of 44 pairs) | ||
| Protective allele in child only | 433-C B*1503/5801 | B*1503/1516 |
| 562-C B*1402/5801 | B*1402/1510 | |
| 600-C B*0801/8101 | B*0801/5802 | |
| Protective allele in mother and child | 135-C B*0801/8101 | B*0702/8101 |
| 241-C B*4403/8101 | B*4701/8101 | |
| 413-C B*0701/8101 | B*4403/8101 | |
| 579-C B*4201/5703 | B*1510/5703 | |
| Protective allele in mother only | 284-C B*0801/1501 | B*0801/5801 |
FIG. 1.
Relative contributions of protective HLA-B alleles in mothers and children. Comparisons were performed between protective alleles expressed in transmitting versus nontransmitting mothers (A) and progressor versus slow-progressor children (B). (C) Comparison of protective HLA-B alleles unshared between mother and child.
In the slow-progressor group, a protective allele in either the mother or child was present in 8 of 17 pairs (47%), compared to 8 of the 44 progressor pairs (18%) (P = 0.047; Fisher's exact test) (Fig. 1B). Furthermore, a significantly higher proportion of slow-progressor children had one of the protective HLA-B alleles that were not shared by mother and child (7/17 [41%]). In one pair, although the mother and child shared the protective B*5801 allele, the child (468-C) also carried B*5703, which was not shared with the mother. In contrast, in only 4 of the 44 (9%) progressor mother-child pairs was one of the protective HLA-B alleles unshared by mother and child (P = 0.007; Fisher's exact test) (Fig. 1C). These data therefore support the hypothesis that slow progression of HIV in this cohort of children is influenced by protective HLA-B alleles in either the mother or child, in particular when the protective alleles are not shared by mother and child.
HIV-specific CD8+ T-cell responses in slow-progressor infants tend to target Gag epitopes presented by protective alleles B*57, B*5801, and B*8101.
We next addressed the question of whether the slow-progressor infants possessing one of the four protective HLA-B alleles tended to generate detectable CD8+ T-cell responses to the Gag epitopes presented by these alleles. Among the slow-progressor children expressing one of the protective HLA-B alleles, five of five made IFN-γ ELISPOT responses to one or more of the Gag epitopes presented by these alleles, compared to three of seven progressor children (P = 0.08; Fisher's exact test) (Fig. 2A and B). Overall, the slow-progressor children targeted 6/8 Gag epitopes presented by these alleles, in contrast to the progressor children in this category, who targeted 3/11 Gag epitopes presented by these HLA-B alleles (P = 0.07; Fisher's exact test) (Fig. 2A and B). We additionally analyzed Gag-specific CD8+ T-cell responses of slow-progressor and progressor children without protective HLA-B alleles to optimal epitopes presented by their respective HLA alleles (Fig. 2C). Among the slow-progressor children without protective alleles, eight of nine made IFN-γ ELISPOT responses to one or more Gag epitopes, compared to 20/36 progressor children (P = 0.02; Fisher's exact test). In addition, the magnitude of the Gag-specific response in the slow-progressor children was higher (median, 6,539 SFC/million PBMC; range, 0 to 14,350 SFC/million PBMC) than that for progressor children (median, 164 SFC/million PBMC; range, 0 to 16,428 SFC/million PBMC) (P = 0.0177; Mann-Whitney test) (Fig. 2C). These data suggest that Gag-specific CD8+ T-cell responses tend to slow disease progression in HIV-infected infants, irrespective of the HLA class I alleles presenting the Gag epitopes.
CD8+ T-cell responses and MTCT of escape variants.
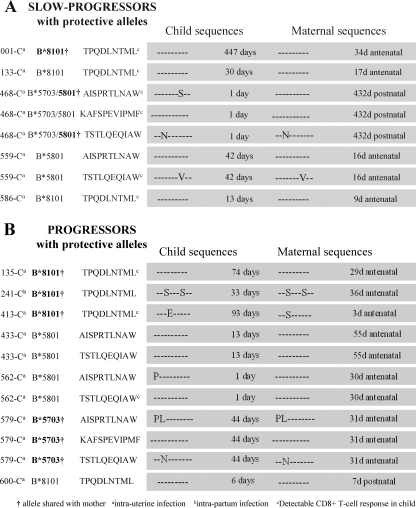
To investigate possible causes for the difference in responses to Gag epitopes presented by the protective HLA-B alleles that were observed in the progressor and slow-progressor groups, we next addressed the question of whether failure to generate detectable CD8+ T-cell responses to the protective epitopes was related to transmission of variants by the mother (Fig. 3). Viruses were sequenced from the mother and child at the time point closest to transmission that was available (Fig. 4). Variants in the children's sequences were observed in 2/9 cases where there were responses to the epitopes detected, compared to 5/10 cases where there were no responses detected (odds ratio, 2.25; P is nonsignificant) (Fig. 3A and B). Thus, whereas responses to epitopes are more likely to be observed if wild-type virus has been transmitted, in 50% of cases no response was detected even when the wild type was transmitted.
FIG. 3.
Mother and child Gag epitope sequence variation in slow-progressor (A) and progressor (B) children.
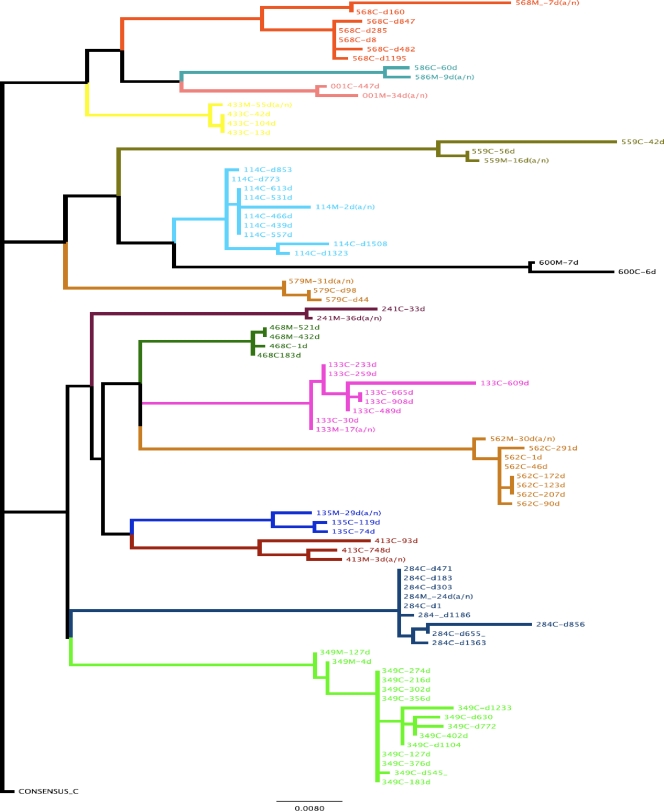
FIG. 4.
Phylogenetic tree of mother and child sequences. Gag p24 nucleotide sequences from mothers (M) before (a/n) or after delivery and from children (C) at different days (d) after infection were edited and aligned with SeqAlv2.0. Maximum likelihood trees were constructed with the GARLI (genetic algorithm for rapid likelihood inference) 0.96 program. Each color represents a mother-child pair, and the scale represents substitutions per site.
Mother-child HLA sharing and MTCT of escape variants.
To investigate further the impact of HLA sharing of protective alleles by the child and the mother, we determined the frequency of transmitted variants within the p24 Gag epitopes (ISW9, KF11, TL9, and TW10) presented by the protective alleles HLA-B*57, -5801, and -8101. Sequences encoding the same epitopes, but in transmitting mothers not expressing the relevant alleles, served as controls. In sequences obtained from the mothers as close to delivery as feasible (median, 20 days prior to delivery; range, 55 days antenatally to 432 days postnatally), 12/16 (75%) epitopes presented by the B*57, -5801, and -8101 alleles and expressed in the transmitting mother contained mutations, compared to 5/24 (21%) epitopes presented by the B*57, -5801, -8101 alleles and not expressed in the transmitting mother (P = 0.001; Fisher's exact test) (Fig. 5A; Table 2). Very similar findings resulted from analysis of sequences obtained from children as close to diagnosis as feasible (median, 31 days of age; range, 1 to 447 days of age; P = 0.0001) (Fig. 5B). Thus, sharing of maternal alleles by the child is likely to contribute to failure of children to make responses to epitopes presented by protective alleles, since in the majority of cases variants will be transmitted.
FIG. 5.
Gag epitope variants in mothers (A) and children (B) in cases where the mother either expressed or did not express the protective HLA-B alleles.
TABLE 2.
Deduced Gag epitope sequences in the 16 mother-child pairs where the mother and/or child possessed one or more of the protective HLA-B alleles
| Child or mother no. | Variant in epitope sequence in allelea
|
|||
|---|---|---|---|---|
| B*5702/5703/5801 (AISPRTLNAW [Gag positions 146 to 155]) | B*5703 (KAFSPEVIPMF [Gag positions 162 to 172]) | B*8101 (TPQDLNTML [Gag positions 181 to 189]) | B*5702/5703/5801 (TSTLQEQIAW [Gag positions 240 to 249]) | |
| Children | ||||
| 001-C | A146S | ▪ | ||
| 114-C | A146P/I147L | T242N | ||
| 133-C | ▪ | |||
| 135-C | ▪ | |||
| 241-C | Q183S/T187S | |||
| 284-C | A146P | T242N | ||
| 349-C | A146P/I147L/S148G | I247L/A248D | ||
| 413-C | D184E | |||
| 433-C | ▪ | ▪ | ||
| 468-C | N153S | ▪ | T242N | |
| 559-C | ▪ | I247V | ||
| 562-C | A146P | ▪ | ||
| 568-C | A146S/I147 M | L185 M/T187S | ||
| 579-C | A146P/I147L | ▪ | T242N | |
| 586-C | A146S | V168I | ▪ | A248G |
| 600-C | A146C/I147L | ▪ | ||
| Mothers | ||||
| 001-M | A146P | ▪ | ||
| 114-M | A146P/I147L | T242N/A248T | ||
| 133-M | ||||
| 135-M | ▪ | |||
| 241-M | Q183S/T187S | A248T | ||
| 284-M | A146P | T242N | ||
| 349-M | A146P/I147L/S148G | I247L/A248D | ||
| 413-M | I147L | Q183S | ||
| 433-M | ||||
| 468-M | ▪ | T242N | ||
| 559-M | T187 M | I247V | ||
| 562-M | ||||
| 568-M | A146S/I147 M | A163G | Q183S/T187S | |
| 579-M | A146P/I147L | ▪ | T242N | |
| 586-M | A146S | V168I | ||
| 600-M | A146C/I147L | |||
Data in bold and filled squares indicate that the mother or child possessed an HLA-B allele presenting the epitope; underlined data indicate that the mother but not the child possessed an HLA-B allele presenting the epitope.
Mothers carrying protective HLA alleles transmit Gag variants reducing viral fitness.
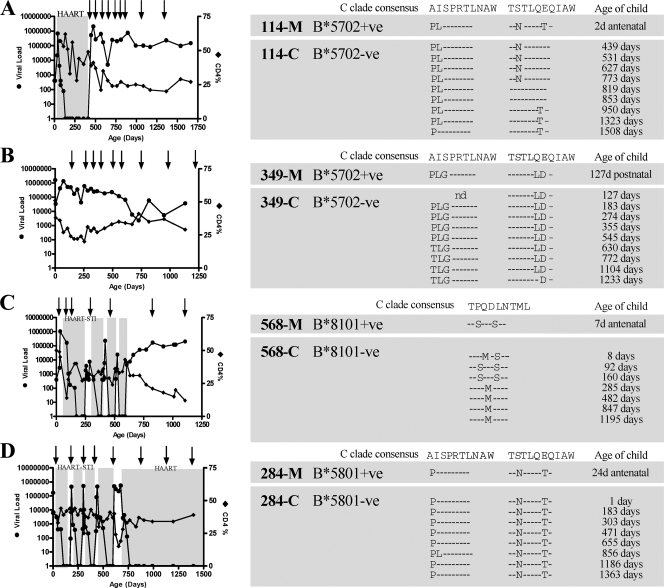
Three of the eight slow-progressor children and one of the eight progressor children had no protective HLA alleles themselves, but their mother possessed one. To address the question of whether the viruses transmitted by these mothers did in fact carry escape mutations in Gag that reduced viral fitness, viruses from these mother-child pairs were sequenced for time points as close as possible to transmission and tracked longitudinally to look for reversion of transmitted escape mutants in the HLA-mismatched children (Fig. 6).
FIG. 6.
Evolution of escape Gag variants in HLA-B*5702- and HLA-B*8101-negative children after transmission from HLA-B*5702-positive and HLA-B*8101-positive mothers. CD4%, viral loads, and epitope sequence assessment are shown for the four cases where mothers expressed protective HLA-B alleles and the child did not, i.e., 114-C (A), 349-C (B), 568-C (C), and 284-C (D).
All of the mothers transmitted escape variants to their children in each epitope presented by the protective maternal alleles. In all three cases where the child was defined as a slow progressor, reverting escape mutations were observed in all of the epitopes, indicating an in vivo fitness cost of T242N and I147L mutations in B*5702-negative child 114-C (Fig. 6A), A146P and I247L mutations in B*5702-negative child 349-C (Fig. 6B), and Q182S and T186S mutations in B*8101-negative child 568-C (Fig. 6C). In the B*5801-negative progressor, child 284-C, even though low-fitness mutants, such as A146P and T242N mutants, were transmitted, reversion did not occur before CD4 decline to the point where the criterion for starting ART was reached rapidly. Thus, from analysis of this small number of mother-child pairs, it is clear that some benefit to the child is likely to accrue from the transmission of Gag escape variants within epitopes presented in the mother by protective HLA alleles, such as B*5702, -5703, -5801, and -8101.
DISCUSSION
While most HIV-infected children progress rapidly to disease following MTCT, a minority of children control the virus for years without the need for ART. The current analyses of 61 HIV-infected children monitored from birth highlight some of the factors associated with slow and rapid progression in children. Slow progression was associated with possession of either one or more of the known protective HLA-B alleles, namely, HLA-B*57, -B*5801, and -B*8101, particularly when these protective HLA alleles were not shared with the mother (P = 0.007). Slow progressors with or without protective HLA-B alleles were more likely than progressors to make HIV-specific CD8+ T-cell responses to Gag epitopes presented by the alleles. Mothers expressing protective alleles were more likely to transmit escape variants within Gag epitopes presented by those alleles than were mothers not expressing them (P = 0.001). Reverting escape mutations were transmitted to and subsequently reverted in all slow-progressor children whose mothers possessed protective HLA-B alleles. These data demonstrate the contributions of protective HLA-B alleles in either the mother or the child to slowing disease progression in pediatric infection.
These data suggest that HIV-infected children can benefit from protective HLA-B alleles in the same way as infected adults, via the targeting of Gag epitopes presented by these alleles (17, 23), and also may benefit from the acquisition of transmitted escape mutations, as described for adult infection (3, 8).
Although the presence of protective HLA-B alleles contributes to outcomes of pediatric HIV infections, these studies illustrate that other factors also operate to influence the outcome. We have previously shown in studies with the same cohort that MTCT is related to maternal viral load (24), and it would not be unexpected for factors other than the protective HLA-B alleles focused on in this study to play a role in determining maternal viral load. Nine of 17 slow-progressor children (53%) did not possess protective HLA-B alleles, nor did their mothers, and of the progressor children, 8 of 44 (18%) carried a protective HLA-B allele, or the mother did. Non-HLA-associated factors likely to play a role would include non-HLA genetic factors (1, 31), the presence of coinfections by pathogens such as hepatitis C virus, cytomegalovirus, and Mycobacterium tuberculosis, and the concomitants of extreme poverty (24).
A further factor reducing the apparent impact of HLA-B on viral fitness may be that mutations reducing viral fitness are transmitted by as many as 21% of mothers not themselves carrying the protective alleles (Table 2). In some cases, it is possible that the father may have transmitted the allele to the child and the variant to the mother, as previously described (25). For example, subject 559-C carried B*5801, which could be expected to select for mutations such as I247V in the TW10 epitope, but the mother carried this mutant without having B*5801 herself. It is striking that a large proportion of the mother-child cohort carried HLA-B*57-associated mutations but did not themselves possess B*57, for example, pair 568-C/M. These data support the notion that these mutations, in spite of having the capacity to revert in the absence of the restricting allele, nonetheless are seen at relatively high frequencies even in HLA-mismatched subjects (15). A potential reason for the persistence of mutations reducing viral fitness is that in some cases they are neutralized by compensatory mutations. For example, in the anecdotal case of child 284-C, the mutations I223N, M228I, A248T, and S252N were observed, which have been shown to contribute to reducing the fitness cost of the T242N mutation (2). In child 114-C, whose mother similarly transmitted a T242N mutant but, unlike child 284-C, in whom reversion did occur subsequent to transmission, the virus consistently, at all four time points prior to reversion (days 439 to 773), showed the wild-type sequence at all of these positions. With such a small number of study subjects in this category, these data are no more than suggestive explanations for the lack of reversion over time seen in child 284-C. A separate report describing the in vitro fitness of viruses from children in this cohort in detail is in preparation (Prado et al., unpublished data).
A potentially important observation is the relative inability of CD8+ T-cell responses to be made by infected children even when they carry the protective HLA allele and the unmutated epitope has been transmitted (Fig. 2 and 3). The reasons for this failure are unclear, but maternal well-being has a strong influence on pediatric disease progression (27). For example, child 600-C, who had HLA-B*8101 and whose virus encoded the unmutated TPQDLTNML epitope, made no detectable response to it (or to any other epitope [data not shown]); the mother, 600-M, had an absolute CD4 count of 4 cells/mm3 (CD4% of 2%).
An additional observation from the data presented in this study is the lack of precise correlation between transmission of escape mutants by the mother and either the ability of the child to make a response to the epitope or the disease outcome for the child. Previous studies (9) focusing on transmission of escape mutants within the HLA-B*27-restricted epitope KK10 (KRWIILGLNK [Gag positions 263 to 272]) suggested that escape variant transmission would result in no response being generated by the child and in more rapid progression in the child. Although the findings from the present study are consistent with the B*27 study, we suggest that the fact that some children can make a detectable response in spite of being infected by an epitope variant and the fact that more rapid progression as a result of MTCT of escape mutants within epitopes restricted by shared alleles is not always observed are the results of two main factors.
First, the impact of transmitted escape mutants on the child's ability to make CD8+ T-cell responses would depend on the particular escape mutants transmitted. In the case of the HLA-B*27-restricted epitope KK10 (KRWIILGLNK [Gag positions 263 to 272]), the transmitted escape mutants referred to above (9) were shown to be nonbinding mutants, which therefore prevented any response being made by a B*27-positive child. However, in cases where the mutant is still capable of binding to the HLA allele, such as a T242N mutant within the B*57/5801 TW10 epitope TSTLQEQIAW (Gag positions 240 to 249), a variant-specific response can be made by the child, as shown previously to be the case for TW10 escape mutants (5). A second possible explanation for the lack of a clear-cut relationship between transmission of escape mutants and disease progression in the infant may relate to the impact on viral fitness of the escape mutants in question. Whereas the fitness costs of B*27-KK10 escape mutants at residue R264 are apparently fully compensated by compensatory mutations elsewhere in p24 Gag (29, 30), this is not the case for the B*57, -5801, and -8101 mutants focused on in this study, as supported by the reversions in vivo shown in Fig. 6. Thus, infants whose ability to generate a CD8+ T-cell response to a particular epitope may be hindered or abrogated by the transmission of an escape mutant may nonetheless benefit from the fact that some of these mutants reduce viral replicative capacity.
One further point in the comparison of maternal viral sequences from samples taken as close to the time of transmission as possible (a median of 2 to 3 weeks prior to delivery) with child sequences, also taken as close to the time of transmission as possible (a median of 31 days of age), is the lack of identity between early child sequences and maternal sequences when variants are involved. Focusing on the four Gag epitopes highlighted in this study and sequences in the 16 mother-child pairs where the mother and/or child possessed one of the protective HLA-B alleles (Table 2), variants were seen in mother or child sequences for 24 of 64 epitopes. However, the variants differ between mother and child in as many as 9/24 instances. In some cases, this may be due to the fact that sequences were not determined from viral RNA but from proviral DNA. However, another possibility is that the most frequent variant present in the mother is not necessarily the variant most likely to be transmitted, a finding previously well described in relation to MTCT (33). Finally, it is possible that very early escape may be selected in the infant, even in utero. In the case of pair 562-C/M, for example, the mother did not have an HLA allele driving variant selection at Gag 146, and only wild-type Gag sequences were observed. The child had HLA-B*5801, however, and at day 1 of life, the child had a virus carrying the B*5801 Gag mutant A146P, and no response to the epitope ISPRTLNAW was detected. In contrast, a strong response was initially observed to the B*5801 epitope TSTLQEQIAW; the escape mutation in this epitope, T242N, was first noted at 10 months of age, coincident with a decline in the size of the CD8+ T-cell response (Fig. 2).
Taken together, these data suggest that infected children have the potential to generate Gag-specific CD8+ T-cell responses and to delay disease progression if they possess protective HLA-B alleles to which the transmitted maternal virus is not preadapted. As in adult infection, the protective HLA class I alleles that drive selection of Gag escape mutations are beneficial not only to the infected mother but also to the infected child not expressing the same alleles. These studies and those reported before could help to identify individuals who qualify for delayed treatment as a result of the advantage of acquisition of a less fit virus. This may be especially relevant to infected children, since the generally observed rapid progression observed in infancy currently dictates that all infants be put on immediate ART following diagnosis.
Acknowledgments
This study was funded by the Doris Duke Charitable Foundation (grant 20011031 to P.J.R.G.), The Wellcome Trust (to P.J.R.G.), and Bristol Myers Squibb Secure the Future (grant RES116/01 to P.J.R.G.). T.N. holds the South African DST/NRF Chair in Systems Biology of HIV/AIDS.
We thank all of the mothers and children for participation in this study, the clinic teams at the St. Mary's Hospital antenatal clinic and Prince Mshiyeni Hospital, and the HIV Pathogenesis Programme (HPP) team (K. Mngwenya, T. Sikhakhane, T. Cele, D. Sindane, N. Linda, T. Mchunu, T. Phahla, M. Mbambo, A. Cengimbo, M. Van der Stok, N. Mlotshwa, E. Govender, T. Moodley, N. Ismail, K. Nair, K. Bishop, L. Maphumulo, and N. Phungula). We are also grateful to M. Brockman and Z. Brumme for their helpful discussions on the manuscript.
Footnotes
Published ahead of print on 15 July 2009.
REFERENCES
- 1.Altfeld, M., and P. Goulder. 2007. ‘Unleashed’ natural killers hinder HIV. Nat. Genet. 39:708-710. [DOI] [PubMed] [Google Scholar]
- 2.Brockman, M. A., A. Schneidewind, M. Lahaie, A. Schmidt, T. Miura, I. Desouza, F. Ryvkin, C. A. Derdeyn, S. Allen, E. Hunter, J. Mulenga, P. A. Goepfert, B. D. Walker, and T. M. Allen. 2007. Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 Gag alter capsid interactions with cyclophilin A. J. Virol. 81:12608-12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chopera, D. R., Z. Woodman, K. Mlisana, M. Mlotshwa, D. P. Martin, C. Seoighe, F. Treurnicht, D. A. de Rosa, W. Hide, S. A. Karim, C. M. Gray, and C. Williamson. 2008. Transmission of HIV-1 CTL escape variants provides HLA-mismatched recipients with a survival advantage. PLoS Pathog. 4:e1000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crawford, H., J. G. Prado, A. Leslie, S. Hue, I. Honeyborne, S. Reddy, M. van der Stok, Z. Mncube, C. Brander, C. Rousseau, J. I. Mullins, R. Kaslow, P. Goepfert, S. Allen, E. Hunter, J. Mulenga, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2007. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J. Virol. 81:8346-8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feeney, M. E., Y. Tang, K. Pfafferott, K. A. Roosevelt, R. Draenert, A. Trocha, X. G. Yu, C. Verrill, T. Allen, C. Moore, S. Mallal, S. Burchett, K. McIntosh, S. I. Pelton, M. A. St. John, R. Hazra, P. Klenerman, M. Altfeld, B. D. Walker, and P. J. Goulder. 2005. HIV-1 viral escape in infancy followed by emergence of a variant-specific CTL response. J. Immunol. 174:7524-7530. [DOI] [PubMed] [Google Scholar]
- 6.Feeney, M. E., Y. Tang, K. A. Roosevelt, A. J. Leslie, K. McIntosh, N. Karthas, B. D. Walker, and P. J. Goulder. 2004. Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an HLA-B27-positive long-term-nonprogressing child. J. Virol. 78:8927-8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geldmacher, C., J. R. Currier, E. Herrmann, A. Haule, E. Kuta, F. McCutchan, L. Njovu, S. Geis, O. Hoffmann, L. Maboko, C. Williamson, D. Birx, A. Meyerhans, J. Cox, and M. Hoelscher. 2007. CD8 T-cell recognition of multiple epitopes within specific Gag regions is associated with maintenance of a low steady-state viremia in human immunodeficiency virus type 1-seropositive patients. J. Virol. 81:2440-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goepfert, P. A., W. Lumm, P. Farmer, P. Matthews, A. Prendergast, J. M. Carlson, C. A. Derdeyn, J. Tang, R. A. Kaslow, A. Bansal, K. Yusim, D. Heckerman, J. Mulenga, S. Allen, P. J. Goulder, and E. Hunter. 2008. Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients. J. Exp. Med. 205:1009-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goulder, P. J., C. Brander, Y. Tang, C. Tremblay, R. A. Colbert, M. M. Addo, E. S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, M. Altfeld, S. He, M. Bunce, R. Funkhouser, S. I. Pelton, S. K. Burchett, K. McIntosh, B. T. Korber, and B. D. Walker. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412:334-338. [DOI] [PubMed] [Google Scholar]
- 10.Goulder, P. J., P. Jeena, G. Tudor-Williams, and S. Burchett. 2001. Paediatric HIV infection: correlates of protective immunity and global perspectives in prevention and management. Br. Med. Bull. 58:89-108. [DOI] [PubMed] [Google Scholar]
- 11.Goulder, P. J., C. Pasquier, E. C. Holmes, B. Liang, Y. Tang, J. Izopet, K. Saune, E. S. Rosenberg, S. K. Burchett, K. McIntosh, M. Barnardo, M. Bunce, B. D. Walker, C. Brander, and R. E. Phillips. 2001. Mother-to-child transmission of HIV infection and CTL escape through HLA-A2-SLYNTVATL epitope sequence variation. Immunol. Lett. 79:109-116. [DOI] [PubMed] [Google Scholar]
- 12.Goulder, P. J., and D. I. Watkins. 2008. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat. Rev. Immunol. 8:619-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guay, L. A., P. Musoke, T. Fleming, D. Bagenda, M. Allen, C. Nakabiito, J. Sherman, P. Bakaki, C. Ducar, M. Deseyve, L. Emel, M. Mirochnick, M. G. Fowler, L. Mofenson, P. Miotti, K. Dransfield, D. Bray, F. Mmiro, and J. B. Jackson. 1999. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: HIVNET 012 randomised trial. Lancet 354:795-802. [DOI] [PubMed] [Google Scholar]
- 14.Jackson, J. B., P. Musoke, T. Fleming, L. A. Guay, D. Bagenda, M. Allen, C. Nakabiito, J. Sherman, P. Bakaki, M. Owor, C. Ducar, M. Deseyve, A. Mwatha, L. Emel, C. Duefield, M. Mirochnick, M. G. Fowler, L. Mofenson, P. Miotti, M. Gigliotti, D. Bray, and F. Mmiro. 2003. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: 18-month follow-up of the HIVNET 012 randomised trial. Lancet 362:859-868. [DOI] [PubMed] [Google Scholar]
- 15.Kawashima, Y., K. Pfafferott, J. Frater, P. Matthews, R. Payne, M. Addo, H. Gatanaga, M. Fujiwara, A. Hachiya, H. Koizumi, N. Kuse, S. Oka, A. Duda, A. Prendergast, H. Crawford, A. Leslie, Z. Brumme, C. Brumme, T. Allen, C. Brander, R. Kaslow, J. Tang, E. Hunter, S. Allen, J. Mulenga, S. Branch, T. Roach, M. John, S. Mallal, A. Ogwu, R. Shapiro, J. G. Prado, S. Fidler, J. Weber, O. G. Pybus, P. Klenerman, T. Ndung'u, R. Phillips, D. Heckerman, P. R. Harrigan, B. D. Walker, M. Takiguchi, and P. Goulder. 2009. Adaptation of HIV-1 to human leukocyte antigen class I. Nature 458:641-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiepiela, P., A. J. Leslie, I. Honeyborne, D. Ramduth, C. Thobakgale, S. Chetty, P. Rathnavalu, C. Moore, K. J. Pfafferott, L. Hilton, P. Zimbwa, S. Moore, T. Allen, C. Brander, M. M. Addo, M. Altfeld, I. James, S. Mallal, M. Bunce, L. D. Barber, J. Szinger, C. Day, P. Klenerman, J. Mullins, B. Korber, H. M. Coovadia, B. D. Walker, and P. J. Goulder. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769-775. [DOI] [PubMed] [Google Scholar]
- 17.Kiepiela, P., K. Ngumbela, C. Thobakgale, D. Ramduth, I. Honeyborne, E. Moodley, S. Reddy, C. de Pierres, Z. Mncube, N. Mkhwanazi, K. Bishop, M. van der Stok, K. Nair, N. Khan, H. Crawford, R. Payne, A. Leslie, J. Prado, A. Prendergast, J. Frater, N. McCarthy, C. Brander, G. H. Learn, D. Nickle, C. Rousseau, H. Coovadia, J. I. Mullins, D. Heckerman, B. D. Walker, and P. Goulder. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13:46-53. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi, M., H. Igarashi, A. Takeda, M. Kato, and T. Matano. 2005. Reversion in vivo after inoculation of a molecular proviral DNA clone of simian immunodeficiency virus with a cytotoxic-T-lymphocyte escape mutation. J. Virol. 79:11529-11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhn, L., E. J. Abrams, P. Palumbo, M. Bulterys, R. Aga, L. Louie, and T. Hodge. 2004. Maternal versus paternal inheritance of HLA class I alleles among HIV-infected children: consequences for clinical disease progression. AIDS 18:1281-1289. [DOI] [PubMed] [Google Scholar]
- 20.Leslie, A. J., K. J. Pfafferott, P. Chetty, R. Draenert, M. M. Addo, M. Feeney, Y. Tang, E. C. Holmes, T. Allen, J. G. Prado, M. Altfeld, C. Brander, C. Dixon, D. Ramduth, P. Jeena, S. A. Thomas, A. St. John, T. A. Roach, B. Kupfer, G. Luzzi, A. Edwards, G. Taylor, H. Lyall, G. Tudor-Williams, V. Novelli, J. Martinez-Picado, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282-289. [DOI] [PubMed] [Google Scholar]
- 21.Martinez-Picado, J., J. G. Prado, E. E. Fry, K. Pfafferott, A. Leslie, S. Chetty, C. Thobakgale, I. Honeyborne, H. Crawford, P. Matthews, T. Pillay, C. Rousseau, J. I. Mullins, C. Brander, B. D. Walker, D. I. Stuart, P. Kiepiela, and P. Goulder. 2006. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 80:3617-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matano, T., M. Kobayashi, H. Igarashi, A. Takeda, H. Nakamura, M. Kano, C. Sugimoto, K. Mori, A. Iida, T. Hirata, M. Hasegawa, T. Yuasa, M. Miyazawa, Y. Takahashi, M. Yasunami, A. Kimura, D. H. O'Connor, D. I. Watkins, and Y. Nagai. 2004. Cytotoxic T lymphocyte-based control of simian immunodeficiency virus replication in a preclinical AIDS vaccine trial. J. Exp. Med. 199:1709-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthews, P. C., A. Prendergast, A. Leslie, H. Crawford, R. Payne, C. Rousseau, M. Rolland, I. Honeyborne, J. Carlson, C. Kadie, C. Brander, K. Bishop, N. Mlotshwa, J. I. Mullins, H. Coovadia, T. Ndung'u, B. D. Walker, D. Heckerman, and P. J. Goulder. 2008. Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J. Virol. 82:8548-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mphatswe, W., N. Blanckenberg, G. Tudor-Williams, A. Prendergast, C. Thobakgale, N. Mkhwanazi, N. McCarthy, B. D. Walker, P. Kiepiela, and P. Goulder. 2007. High frequency of rapid immunological progression in African infants infected in the era of perinatal HIV prophylaxis. AIDS 21:1253-1261. [DOI] [PubMed] [Google Scholar]
- 25.Pillay, T., H. T. Zhang, J. W. Drijfhout, N. Robinson, H. Brown, M. Khan, J. Moodley, M. Adhikari, K. Pfafferott, M. E. Feeney, A. St. John, E. C. Holmes, H. M. Coovadia, P. Klenerman, P. J. Goulder, and R. E. Phillips. 2005. Unique acquisition of cytotoxic T-lymphocyte escape mutants in infant human immunodeficiency virus type 1 infection. J. Virol. 79:12100-12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prendergast, A., W. Mphatswe, G. Tudor-Williams, M. Rakgotho, V. Pillay, C. Thobakgale, N. McCarthy, L. Morris, B. D. Walker, and P. Goulder. 2008. Early virological suppression with three-class antiretroviral therapy in HIV-infected African infants. AIDS 22:1333-1343. [DOI] [PubMed] [Google Scholar]
- 27.Prendergast, A., G. Tudor-Williams, P. Jeena, S. Burchett, and P. Goulder. 2007. International perspectives, progress, and future challenges of paediatric HIV infection. Lancet 370:68-80. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez-Merino, V., M. A. Farrow, F. Brewster, M. Somasundaran, and K. Luzuriaga. 2008. Identification and characterization of HIV-1 CD8+ T cell escape variants with impaired fitness. J. Infect. Dis. 197:300-308. [DOI] [PubMed] [Google Scholar]
- 29.Schneidewind, A., M. A. Brockman, J. Sidney, Y. E. Wang, H. Chen, T. J. Suscovich, B. Li, R. I. Adam, R. L. Allgaier, B. R. Mothe, T. Kuntzen, C. Oniangue-Ndza, A. Trocha, X. G. Yu, C. Brander, A. Sette, B. D. Walker, and T. M. Allen. 2008. Structural and functional constraints limit options for cytotoxic T-lymphocyte escape in the immunodominant HLA-B27-restricted epitope in human immunodeficiency virus type 1 capsid. J. Virol. 82:5594-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneidewind, A., Y. Tang, M. A. Brockman, E. G. Ryland, J. Dunkley-Thompson, J. C. Steel-Duncan, M. A. St. John, J. A. Conrad, S. A. Kalams, F. Noel, T. M. Allen, C. D. Christie, and M. E. Feeney. Maternal transmission of HIV escape mutations subverts HLA-B57 immunodominance but facilitates viral control in the haploidentical infant. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 31.Shostakovich-Koretskaya, L., G. Catano, Z. A. Chykarenko, W. He, G. Gornalusse, S. Mummidi, R. Sanchez, M. J. Dolan, S. S. Ahuja, R. A. Clark, H. Kulkarni, and S. K. Ahuja. 2009. Combinatorial content of CCL3L and CCL4L gene copy numbers influence HIV-AIDS susceptibility in Ukrainian children. AIDS 23:679-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thobakgale, C. F., D. Ramduth, S. Reddy, N. Mkhwanazi, C. de Pierres, E. Moodley, W. Mphatswe, N. Blanckenberg, A. Cengimbo, A. Prendergast, G. Tudor-Williams, K. Dong, P. Jeena, G. Kindra, R. Bobat, H. Coovadia, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2007. Human immunodeficiency virus-specific CD8+ T-cell activity is detectable from birth in the majority of in utero-infected infants. J. Virol. 81:12775-12784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolinsky, S. M., K. J. Kunstman, J. T. Safrit, R. A. Koup, A. U. Neumann, and B. T. Korber. 1996. Response: HIV-1 evolution and disease progression. Science 274:1010-1011. [DOI] [PubMed] [Google Scholar]
- 34.Zuniga, R., A. Lucchetti, P. Galvan, S. Sanchez, C. Sanchez, A. Hernandez, H. Sanchez, N. Frahm, C. H. Linde, H. S. Hewitt, W. Hildebrand, M. Altfeld, T. M. Allen, B. D. Walker, B. T. Korber, T. Leitner, J. Sanchez, and C. Brander. 2006. Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J. Virol. 80:3122-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]