Abstract
The φX174 DNA pilot protein H contains four predicted C-terminal coiled-coil domains. The region of the gene encoding these structures was cloned, expressed in vivo, and found to strongly inhibit wild-type replication. DNA and protein synthesis was investigated in the absence of de novo H protein synthesis and in wild-type-infected cells expressing the inhibitory proteins (ΔH). The expression of the ΔH proteins interfered with early stages of DNA replication, which did not require de novo H protein synthesis, suggesting that the inhibitory proteins interfere with the wild-type H protein that enters the cell with the penetrating DNA. As transcription and protein synthesis are dependent on DNA replication in positive single-stranded DNA life cycles, viral protein synthesis was also reduced. However, unlike DNA synthesis, efficient viral protein synthesis required de novo H protein synthesis, a novel function for this protein. A single amino acid change in the C terminus of protein H was both necessary and sufficient to confer resistance to the inhibitory ΔH proteins, restoring both DNA and protein synthesis to wild-type levels. ΔH proteins derived from the resistant mutant did not inhibit wild-type or resistant mutant replication. The inhibitory effects of the ΔH proteins were lessened by the coexpression of the internal scaffolding protein, which may suppress H-H protein interactions. While coexpression relieved the block in DNA biosynthesis, viral protein synthesis remained suppressed. These data indicate that protein H's role in DNA replication and stimulating viral protein synthesis can be uncoupled.
Although the atomic structure of protein H remains to be elucidated, the results of bioinformatic analyses predict an N-terminal transmembrane (23) and several coiled-coil domains (16) in the C terminus (Fig. 1). During penetration, H protein is ejected from the capsid along with the penetrating DNA (14). Initially, protein H and the incoming DNA are associated with the outer membrane (13) at sites of cell wall adhesion (2, 3). From there, the single-stranded DNA (ssDNA) is delivered to the cytoplasmic membrane, the site of DNA synthesis (2). The transmembrane domain most likely mediates this DNA-piloting function. Although naked ssDNA can be transfected, replication efficiency is enhanced if H protein is included in the reaction (12). This suggests that the protein may stimulate stage I DNA synthesis: the conversion of the ssDNA genome to a double-stranded replicative-form (RF) molecule, either directly or indirectly by piloting the penetrating DNA to the proper site of replication. Due to the positive polarity of the genome, this occurs without de novo viral protein synthesis. During stage II DNA synthesis, the RF DNA is amplified. This continues until the viral protein C and procapsids accumulate, which signals the switch to stage III DNA synthesis, the concurrent synthesis and packaging of ssDNA genomes.
FIG. 1.
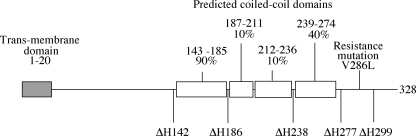
Schematic of the φX174 H protein. The predicted transmembrane and coiled-coil domains are depicted as gray and white boxes, respectively. The probability of each coiled-coil domain prediction (16) is given. The start codons of the N-terminal deletion proteins used in these studies are indicated by ΔH followed by the amino acid number found in the full-length protein. V286L represents the position and amino acid change of a mutation that confers resistance to the expression of the inhibitory ΔH proteins.
While coiled-coil domains are known to mediate protein oligomerization (1, 10, 15) and the purified H protein coiled-coil domains do form oligomers in solutions (J. Nardozzi and G. Cingolani, personal communication), H protein oligomers have yet to be observed during the φX174 life cycle. However, past studies of this low-copy protein have focused on its association with assembly intermediates. The protein appears as a monomer in early assembly intermediates (6), and its incorporation into these intermediates appears to be mediated by the internal scaffolding protein (5, 20). Due to icosahedral averaging, the 10 to 12 copies of the H protein in the virion could not be resolved in the X-ray structure (18, 19). Therefore, it is not known whether H-H protein interactions could occur within the capsid or during DNA penetration.
To determine whether the oligomerizing coiled-coil domains could interact with wild-type H proteins, cloned N-terminal deletion genes (ΔH) were expressed in vivo and assayed for possible effects on wild-type φX174 replication. The ΔH proteins acted as potent inhibitors. The inhibitory mechanism was characterized, and a mutant resistant to the expression of the coiled-coil domains was isolated.
MATERIALS AND METHODS
Phage plating, media, buffers, and stock preparation.
The reagents, media, buffers, and protocols used in this study have been previously described (7). The φX174 mutant resistant to the expression of the inhibitory H proteins, φX174ΔHRV286L, was selected by plating wild-type φX174 on cells expressing the ΔH142 protein.
Escherichia coli C and φX174 strains.
E. coli C strains C122 (supO), BAF8 (supF), and BAF30 (recA) have been previously described (7, 8). The host slyD mutation in E. coli C900 confers resistance to E-protein-mediated lysis (21). The amber H, am(H)Q26, mutant was constructed by oligonucleotide-mediated mutagenesis (9). The mutagenic oligonucleotide was designed to introduce an amber mutation into codon 26 of gene H, which encodes the amino acid glutamine. The mutant was isolated in BAF5 (supE). The genotype of the strain was verified by direct sequence analysis. The am(H)E258 mutant is the am(H)N1 mutant used in earlier studies (22). The name was changed to reflect the location of the amber mutation.
Construction of plasmids expressing H proteins with N-terminal deletions.
To construct the 5′ deletion genes, the φX174 H gene was amplified by PCR. The upstream primers were designed to introduce ATG start codons, contained within NcoI restriction sites, into target codons 142, 186, 238, 277, and 299. The downstream primer was designed to introduce an XhoI restriction site. The PCR products were digested with NcoI and XhoI and cloned into pSE420, digested with the same enzymes. The genes are under lac promoter induction. The plasmid coexpressing the ΔH142 and internal scaffolding proteins (pΔH142+B) was constructed by cloning the internal scaffolding protein with its ribosome binding site directly downstream of the ΔH142 gene. The B gene was amplified by PCR. The upstream and downstream primers were designed to introduce XhoI and HindIII restriction sites, respectively. The PCR product was digested with XhoI and HindIII and cloned into pΔH142, digested with the same enzymes.
Protein and DNA level assays.
Whole-cell lysates were generated by infecting 100-ml cultures of exponential-phase, lysis-resistant cells at a multiplicity of infection of 5.0, incubated for 3 h at 37°C, and concentrated by centrifugation. For protein assays, cells were resuspended in 0.5 ml of a mixture of 100 mM sodium chloride, 5 mM EDTA, 7.2 mM Na2HPO4, and 3.3 mM KH2PO4. Samples were mixed directly with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) running buffer and boiled. After SDS-PAGE, viral proteins were detected by Coomassie staining. Digitized images of protein gels were generated and analyzed with ImageJ software (NIH). Relative protein levels were determined by densitometry and normalization with host cell bands. For DNA assays, RF and plasmid DNAs were extracted as previously described (4). Because the RF and plasmid DNAs are circular and similar in size, the extracted DNA was digested with SspI to distinguish between the two species. Digested DNA was analyzed by agarose gel electrophoresis, followed by ethidium bromide staining. Digitized images were generated and analyzed with ImageJ software (NIH).
RESULTS
Expression of the C terminus of the minor spike protein inhibits wild-type plaque formation.
Using the location of the predicted coiled-coil domains as a guide (Fig. 1), 5′-terminal deletion genes with start codons at positions 142, 186, 238, 277, and 299 were constructed as described in Materials and Methods. The proteins were expressed in vivo and assayed for the ability to inhibit wild-type φX174 plaque formation (Table 1). The expression of the ΔH142, ΔH186, ΔH238, and ΔH277 proteins inhibited wild-type plaque formation. The severity of inhibition appears to be a function of the size of the expressed protein. The expression of ΔH299 had no effect on wild-type plating efficiency.
TABLE 1.
Wild-type and φX174 ΔHRV286La plating efficiencies in cells expressing N-terminal ΔH proteins
| Expressed proteinc | Plating effiiciencyb
|
|
|---|---|---|
| Wild-type φX174 | φX174 ΔHRV286L | |
| None | 1.0 | 1.0 |
| ΔH142 | <10−6 | 0.2 |
| ΔH186 | <10−6 | 0.1 |
| ΔH238 | 10−2 | 0.6 |
| ΔH277 | 10−2 | 1.2 |
| ΔH299 | 1.2 | 1.1 |
| ΔH142+Bd | 0.05 | 1.0 |
| ΔH142V286Le | 0.4 | 1.0 |
A φX174 mutant resistant to the expression of ΔH proteins. The mutation confers a V→L substitution at amino acid 286.
Plating efficiency was determined as the titer on cells expressing ΔH proteins divided by the titer on cells expressing no exogenous proteins.
N-terminal H deletion proteins (ΔH): the number following ΔΗ is the number of amino acids deleted from the N terminus.
The induced plasmid contains two genes, the ΔH142 gene immediately followed by the internal scaffolding protein B gene.
The expressed protein contains the V286L mutation, which confers resistance to the expression of the ΔH proteins.
Effects of de novo H protein synthesis and the expression of the inhibitory ΔH proteins on viral protein synthesis.
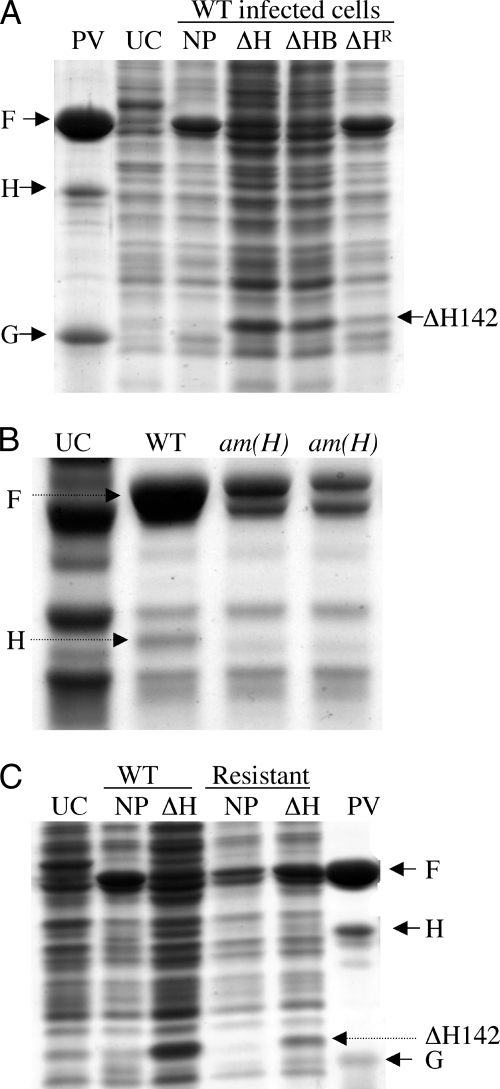
To determine the mechanism by which the C-terminal fragments inhibit wild-type morphogenesis, the infection products synthesized in amber H mutant-infected cells and wild-type-infected cells expressing the inhibitory ΔH142 protein were analyzed (data not shown). However, it was difficult to detect assembly intermediates. The recovery of viral proteins in both the pellet and soluble fractions appeared to be abnormally low compared to that in the wild-type control infection, suggesting a reduction in viral protein synthesis. To determine whether de novo H protein synthesis or the expression of the ΔH proteins affected the overall level of viral protein synthesis, whole-cell lysates were examined by SDS-PAGE. As shown in Fig. 2, the overall viral protein synthesis is decreased in amber H mutant infection cells (Fig. 2B, lanes 3 and 4) and in wild-type-infected cells expressing the ΔH142 protein (Fig. 2A, lane 4) compared to that in the wild-type control infected cells (Fig. 2A, lane 3, and B, lane 2). Using ImageJ software, the relative levels of coat protein F synthesis was determined by comparing its band intensity and area with that of a host cell protein. The level of coat protein synthesis in amber H infections and in cells expressing the inhibitory protein was found to be approximately 20% of that of the wild-type control.
FIG. 2.
(A) Protein expression in wild-type-infected cells expressing the inhibitory ΔH142 protein. Lanes, from the left, contain purified virions (PV), whole-cell extracts of uninfected cells (UC), wild-type (WT)-infected cells with no exogenous protein expression (NP), wild-type-infected cells expressing the ΔH142 protein (ΔH), wild-type-infected cells expressing ΔH142 and internal scaffolding proteins (ΔHB), and wild-type-infected cells expressing the ΔH142V286L protein, which was derived from the resistant mutant (ΔHR). (B) Protein expression in wild-type- and amber H mutant-infected cells. Lanes, from the left, contain whole-cell extracts of uninfected cells (UC), wild-type-infected cells (WT), am(H)Q26 mutant-infected cells, and am(H)E258 mutant-infected cells. The letter and number following am(H) indicate the codon in which the amber mutation is located. (C) Protein expression in wild-type- and φX174ΔHRV286L-infected cells expressing the inhibitory ΔH142 protein. Lanes, from the left, contain whole-cell extracts of uninfected cells (UC), wild-type-infected cells (NP), wild-type-infected cells expressing the ΔH142 protein (ΔH), φX174ΔHRV286L (resistant)-infected cells (NP), φX174ΔHRV286L-infected cells expressing the ΔH142 protein (ΔH), and purified virions (PV).
The ΔH proteins inhibit early stages of DNA synthesis, which does not require de novo H protein synthesis.
There are several mechanisms by which the H protein could lead to elevations in viral protein synthesis. It could stimulate transcription, translation, or viral DNA synthesis. As φX174 is a positive ssDNA virus, transcription is dependent on the synthesis of the negative DNA strand, which occurs during stage I DNA synthesis. Afterwards, the double-stranded RF DNA is amplified, producing more templates for transcription (11).
The effects of de novo H protein synthesis and expression of the ΔH proteins on DNA synthesis were examined in amber H mutant-infected cells and wild-type-infected cells expressing the inhibitory proteins, respectively. In these experiments, plasmid DNA served as an internal standard to which RF DNA levels could be compared. Because both the RF and plasmid DNAs are circular and similar in size, the extracted DNA was digested with SspI. This enzyme cuts RF DNA once, yielding a 5,386-bp linear fragment. There are multiple SspI sites in the plasmid DNA. The large 3,524-bp digestion product was used as an internal standard to assess the efficiency of viral DNA synthesis.
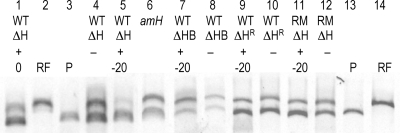
As shown in Fig. 3, the level of RF DNA recovered was significantly lower in cells expressing the ΔH142 protein (lane 5) than in control infected cells, in which the gene for ΔH142 was not induced (lane 4). Using ImageJ software, the relative amounts of RF and plasmid DNAs were determined by comparing band intensities and areas. RF DNA recovered from cells expressing the cloned gene was approximately 5% of that obtained without expression.
FIG. 3.
Relative recovery of plasmid and viral RF DNAs in cells expressing the inhibitory ΔH142 protein. Plasmid and viral RF DNAs were isolated from infected cells and digested with SspI as described in the text. Lanes: 1, wild-type (WT)-infected cells expressing (+) the ΔH142 protein (ΔH) with inducer added at the time of infection (0); 2, digested RF DNA; 3, digested plasmid DNA (P); 4, wild-type-infected cells without induction of the inhibitory gene (−); 5, wild-type-infected cells expressing the ΔH142 protein (+) with inducer added 20 min before infection (−20); 6, amber H-infected cells (amH); 7, wild-type-infected cells coexpressing the internal scaffolding and inhibitory ΔH142 proteins (ΔHB); 8, wild-type-infected cells without induction of the internal scaffolding and inhibitory ΔH142 genes; 9, wild-type-infected cells expressing the ΔH142V286L protein, which was derived from the resistant mutant (ΔHR); 10, wild-type-infected cells without induction of the ΔH142V286L gene; 11, φX174ΔHRV286L (RM, resistant mutant)-infected cells expressing the ΔH142 protein; 12, φX174ΔHRV286L-infected cells without induction of the ΔH142 gene; 13, digested plasmid DNA; 14, digested RF DNA.
The timing of ΔH142 gene induction affected RF recovery. If the inducer was added at the time of infection (lane 1), as opposed to inducing the gene 20 min before infection (lane 5), RF levels were reduced to only 20% of the control level (lane 4). In contrast, RF DNA synthesis does not appear to be inhibited in amber H-infected cells (lane 6). These results indicate that de novo H protein synthesis is not required to stimulate the early stages of DNA synthesis and suggest that the inhibitory proteins interfere with the wild-type protein's DNA-piloting function by interacting with the H protein entering the cell with the penetrating DNA.
A mutation in gene H confers resistance to the expression of the inhibitory fragments.
To further investigate the inhibitory mechanism conferred by the ΔH142 proteins, resistance mutants were selected by plating wild-type φX174 phage on cells expressing the ΔH142 protein. To ensure a wide range of resistant mutant isolation, five independent wild-type φX174 stocks were used in the selection (17); however, only one mutant, φX174ΔHRV286L, was repeatedly recovered. The mutation, which confers a V→L substitution at amino acid 286 of the H protein, elevates plating efficiencies on cells expressing the inhibitory proteins (Table 1). To determine whether the identified mutation was both necessary and sufficient to confer the resistance phenotype, recombination rescue experiments were performed. Wild-type phage was passaged through cells harboring pΔH142 or pΔH142V286L, the construct derived from the V286L resistant mutant. The cloned genes were not induced. Recombination between the wild-type genome and the cloned φX174 DNA sequences would occur in both infections. However, only recombination events that transfer the V286L mutation to the viral genome should produce the resistance phenotype, which was assayed by plating on cells expressing the ΔH142 protein. The resistant progeny frequencies (resistant phage/total progeny) from the pΔH142V286L and pΔH142 passages were 10−4 and 10−7, respectively.
Viral protein and DNA synthesis was investigated in φX174ΔHRV286L-infected cells expressing the inhibitory ΔH142 protein. The presence of the V286L H protein returns viral protein synthesis to nearly wild-type levels (Fig. 2C, lanes 4 and 5) and appears to overcome the block in RF DNA synthesis (Fig. 3, lanes 11 and 12). These data suggest that the V286L H protein either outcompetes the ΔH142 protein for some critical interaction(s) with some host or phage component(s) or is less prone to associate with inhibitory proteins. To distinguish between these two models, the effects of a deletion protein derived from the resistance mutant, ΔH142V286L, on wild-type and φX174ΔHRV286L replication were examined. In a competition-based model, the ΔH142V286L protein should be more inhibitory than the ΔH142 protein. As shown in Table 1, expression of the ΔH142V286L protein does not inhibit wild-type or φX174ΔHRV286L plaque formation. Moreover, the expression of this protein appears to have little or no effect on viral protein and RF DNA synthesis (Fig. 2A, lane 6, and 3, lanes 9 and 10, respectively). These data suggest that the V286L protein is less prone to associate with the inhibitory ΔH proteins.
Coexpression of the internal scaffolding protein lessens the inhibitory effects of the ΔH proteins.
The above data suggest that expression of the ΔH proteins leads to the removal of full-length H into nonfunctional protein complexes. During assembly, H protein appears to be monomeric when it is incorporated into the early 12S* assembly intermediate, in a reaction that is mediated by internal scaffolding protein B (5, 20). In addition to one copy of the H protein, the 12S* intermediate contains five copies of the major coat, spike, and internal scaffolding proteins. As the B protein may prevent H-H protein interactions, the effects of coexpressing the internal scaffolding and ΔH142 proteins were examined. As shown in Table 1, plating efficiency was raised 4 orders of magnitude in cells coexpressing the ΔH142 and B proteins (pΔH142+B). While the coexpression of the B protein did not appear to raise viral protein synthesis to wild-type levels (Fig. 2A, lanes 4 and 5), it did appear to alleviate the block in DNA biosynthesis (Fig. 3, lanes 7 and 8).
DISCUSSION
The results of bioinformatic analyses predict the presence of several coiled-coil domains in the C terminus of the protein (Fig. 1). The ability of coiled-coil domains to mediate protein oligomerization is very well documented (1, 10, 15), and the purified H protein coiled-coil domains do form oligomers in solutions (J. Nardozzi and G. Cingolani, personal communication). To determine whether the expression of the predicted coiled-coil domains could inhibit φX174 replication, a series of N-terminal deletion genes were constructed, expressed in vivo, and assayed for the ability to inhibit φX174 replication. Expression of the proteins containing the predicted coiled-coil domains strongly inhibited φX174 plaque formation. The severity of inhibition correlated with the size of the expressed protein.
At the onset of infection, protein H pilots incoming DNA to the outer membrane (13) at sites of cell wall adhesion (2, 3). This reaction is most likely mediated by the predicted N-terminal transmembrane helix. From there, the ssDNA is delivered to the cytoplasmic membrane, the site of DNA synthesis (2). Microvirus DNA synthesis occurs in three distinct stages (11). During stage I DNA synthesis, the infecting ssDNA is converted into a double-stranded RF molecule. Protein H increases naked ssDNA transfection efficiencies (12, 14), suggesting that the incoming protein may facilitate stage I DNA synthesis. Due to the positive polarity of the genome, this occurs without de novo viral protein synthesis. During stage II DNA synthesis, the RF DNA is amplified. This continues until the viral protein C and procapsids accumulate, which signals the switch to stage III DNA synthesis, the concurrent synthesis and packaging of ssDNA genomes.
The expression of the inhibitory ΔH proteins leads to a reduction in viral DNA synthesis. Two observations suggest that the primary target is stage I DNA synthesis. The recovery of RF DNA is affected by the timing of inhibitory gene induction. Preinduction of the gene results in much lower recovery of RF DNA. In addition, RF DNA recovery is not affected by de novo H protein synthesis. The mechanism of inhibition most likely involves a nonproductive association between the ΔH and incoming wild-type proteins. A single amino acid change in protein H is both necessary and sufficient to confer resistance, restoring viral protein and DNA synthesis to nearly wild-type levels. A ΔH protein derived from this mutant appears to have no inhibitory effects, eliminating models in which the resistant protein outcompetes the inhibitory protein for some critical interaction with a host or viral component. Moreover, these data suggest that the V286L protein is less prone to associate with the ΔH peptides. Finally, the coexpression of the internal scaffolding protein, which may function to keep H protein in a monomeric state for assembly, lessens the inhibitory effects associated with ΔH protein expression.
Protein H may affect several stages in the viral life cycle. While the level of RF production in amber H mutant-infected cells appears to be unaltered, the level of viral protein synthesis is reduced, suggesting that de novo H protein synthesis is required to stimulate viral protein synthesis. The mechanism by which this is accomplished remains to be elucidated. H protein may act directly as a transcription factor. Alternatively, it may direct replicating DNA to locations within the cell where it has access to host cell transcription machinery.
The effects of the inhibitory proteins could well be pleiotropic. Besides inhibiting RF DNA synthesis, they could associate with de novo synthesized wild-type H protein, inhibiting viral protein synthesis or assembly by reducing the pool of H protein monomers. However, in a positive ssDNA viral life cycle, transcription is dependent on DNA synthesis. Thus, these possible secondary effects would be obscured by the reduction of RF DNA. The coexpression of the internal scaffolding protein may somewhat uncouple these pleiotropic effects. While it does not appear to return viral protein synthesis to wild-type levels, it relieves the inhibition of DNA synthesis and allows virion production, which would require the incorporation of the monomeric H protein during assembly.
Acknowledgments
We thank G. Cingolani and J. Nardozzi for communicating unpublished data.
This research was supported by National Science Foundation grant MCB 054297 to B.A.F.
Footnotes
Published ahead of print on 29 July 2009.
REFERENCES
- 1.Alfadhli, A., E. Steel, L. Finlay, H. P. Bachinger, and E. Barklis. 2002. Hantavirus nucleocapsid protein coiled-coil domains. J. Biol. Chem. 277:27103-27108. [DOI] [PubMed] [Google Scholar]
- 2.Azuma, J., J. Morita, and T. Komano. 1980. Process of attachment of φX174 parental DNA to the host cell membrane. J. Biochem. 88:525-532. [DOI] [PubMed] [Google Scholar]
- 3.Bayer, M. E., and T. W. Starkey. 1972. The adsorption of bacteriophage φX174 and its interaction with Escherichia coli: a kinetic and morphological study. Virology 49:236-256. [DOI] [PubMed] [Google Scholar]
- 4.Burch, A. D., J. Ta, and B. A. Fane. 1999. Cross-functional analysis of the Microviridae internal scaffolding protein. J. Mol. Biol. 286:95-104. [DOI] [PubMed] [Google Scholar]
- 5.Chen, M., A. Uchiyama, and B. A. Fane. 2007. Eliminating the requirement of an essential gene product in an already very small virus: scaffolding protein B-free φX174, B-free. J. Mol. Biol. 373:308-314. [DOI] [PubMed] [Google Scholar]
- 6.Cherwa, J. E., Jr., A. Uchiyama, and B. A. Fane. 2008. Scaffolding proteins altered in the ability to perform a conformational switch confer dominant lethal assembly defects. J. Virol. 82:5774-5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fane, B. A., and M. Hayashi. 1991. Second-site suppressors of a cold-sensitive prohead accessory protein of bacteriophage φX174. Genetics 128:663-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fane, B. A., S. Head, and M. Hayashi. 1992. Functional relationship between the J proteins of bacteriophages φX174 and G4 during phage morphogenesis. J. Bacteriol. 174:2717-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fane, B. A., S. Shien, and M. Hayashi. 1993. Second-site suppressors of a cold-sensitive external scaffolding protein of bacteriophage φX174. Genetics 134:1003-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harbury, P. B., T. Zhang, P. S. Kim, and T. Alber. 1993. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262:1401-1407. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi, M., A. Aoyama, D. L. Richardson, and N. M. Hayashi. 1988. Biology of the bacteriophage φX174, p. 1-71. In R. Calendar (ed.), The bacteriophages, vol. 2. Plenum Press, New York, NY. [Google Scholar]
- 12.Jazwinski, S. M., A. A. Lindberg, and A. Kornberg. 1975. The gene H spike protein of bacteriophages φX174 and S13. I. Functions in phage-receptor recognition and in transfection. Virology 66:283-293. [DOI] [PubMed] [Google Scholar]
- 13.Jazwinski, S. M., A. A. Lindberg, and A. Kornberg. 1975. The lipopolysaccharide receptor for bacteriophage φX174 and S13. Virology 66:268-282. [DOI] [PubMed] [Google Scholar]
- 14.Jazwinski, S. M., R. Marco, and A. Kornberg. 1975. The gene H spike protein of bacteriophages φX174 and S13. II. Relation to synthesis of the parenteral replicative form. Virology 66:294-305. [DOI] [PubMed] [Google Scholar]
- 15.Li, N., W. Zhang, S. W. White, and R. W. Kriwacki. 2001. Solution structure of the transcriptional activation domain of the bacteriophage T4 protein, MotA. Biochemistry 40:4293-4302. [DOI] [PubMed] [Google Scholar]
- 16.Lupas, A. 1996. Prediction and analysis of coiled-coil structures. Methods Enzymol. 266:513-525. [DOI] [PubMed] [Google Scholar]
- 17.Luria, S. E., and M. Delbruck. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna, R., L. L. Ilag, and M. G. Rossmann. 1994. Analysis of the single-stranded DNA bacteriophage φX174, refined at a resolution of 3.0 A. J. Mol. Biol. 237:517-543. [DOI] [PubMed] [Google Scholar]
- 19.McKenna, R., D. Xia, P. Willingmann, L. L. Ilag, S. Krishnaswamy, M. G. Rossmann, N. H. Olson, T. S. Baker, and N. L. Incardona. 1992. Atomic structure of single-stranded DNA bacteriophage φX174 and its functional implications. Nature 355:137-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novak, C. R., and B. A. Fane. 2004. The functions of the N terminus of the φX174 internal scaffolding protein, a protein encoded in an overlapping reading frame in a two scaffolding protein system. J. Mol. Biol. 335:383-390. [DOI] [PubMed] [Google Scholar]
- 21.Roof, W. D., H. Q. Fang, K. D. Young, J. Sun, and R. Young. 1997. Mutational analysis of slyD, an Escherichia coli gene encoding a protein of the FKBP immunophilin family. Mol. Microbiol. 25:1031-1046. [DOI] [PubMed] [Google Scholar]
- 22.Spindler, K. R., and M. Hayashi. 1979. DNA synthesis in Escherichia coli cells infected with gene H mutants of bacteriophage φX174. J. Virol. 29:973-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tusnády, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849-850. [DOI] [PubMed] [Google Scholar]