Abstract
Pandemic influenza A viruses that emerge from animal reservoirs are inevitable. Therefore, rapid genomic analysis and creation of vaccines are vital. We developed a multisegment reverse transcription-PCR (M-RTPCR) approach that simultaneously amplifies eight genomic RNA segments, irrespective of virus subtype. M-RTPCR amplicons can be used for high-throughput sequencing and/or cloned into modified reverse-genetics plasmids via regions of sequence identity. We used these procedures to rescue a contemporary H3N2 virus and a swine origin H1N1 virus directly from human swab specimens. Together, M-RTPCR and the modified reverse-genetics plasmids that we designed streamline the creation of vaccine seed stocks (9 to 12 days).
The influenza A virus genome (∼13.5 kb) is composed of eight negative-sense RNA segments (0.89 to 2.3 kb), encoding up to 11 proteins (15). Genetically diverse influenza A viruses differentiated on the basis of their hemagglutinin (HA) (H1 to H16) and neuraminidase (NA) (N1 to N9) glycoproteins infect humans, marine mammals, horses, pigs, waterfowl, and poultry. H1N1 and H3N2 subtype viruses currently predominate in humans, and antigenic drift variants lead to annual epidemics that affect 500 million people during moderate years (23). Antigenic shift variants emerge from animal reservoirs and cause pandemics that infect the majority of the population and have the potential to kill millions of people (12).
The unfolding pandemic of swine origin H1N1 virus demonstrates the speed with which influenza A virus can disseminate among our population and illustrates the need for technologies that enable high-throughput sequence analysis and accelerate vaccine production via reverse genetics (5, 7, 9, 14). Very few approaches have been developed for single-reaction genomic amplification (1) or for cloning into reverse-genetics plasmids (8). Thus, our goal was to develop robust methods to amplify the entire genome of influenza A virus in a single reaction, regardless of subtype, and use these amplicons for both high-throughput sequencing and direct incorporation into modified reverse-genetics plasmids to rapidly create vaccines.
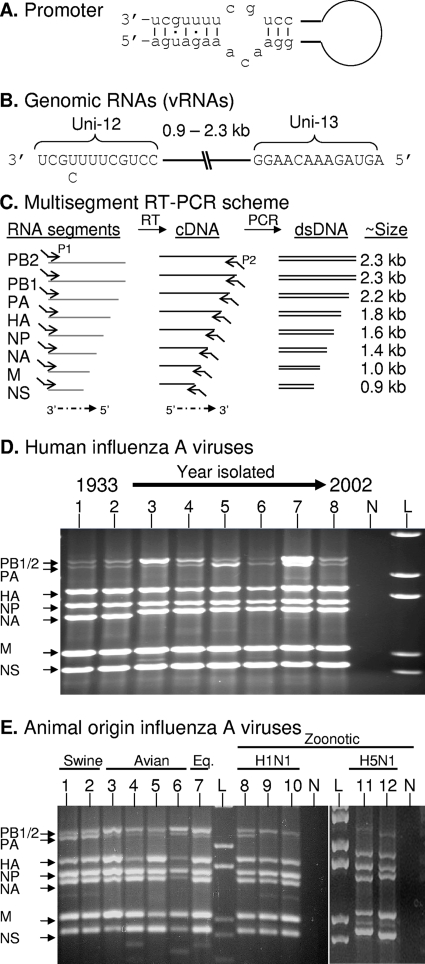
The multisegment reverse transcription-PCR (M-RTPCR) scheme for genomic amplification takes advantage of the fact that the influenza A virus genomic RNAs (vRNAs) each have 12 nucleotides at the 3′ terminus and 13 nucleotides at the 5′ terminus that are conserved and complementary. Base pairing between these conserved residues forms a “panhandle” (Fig. 1A) or “corkscrew” (4); this RNA structure is required for transcription, replication, and packaging of the genome (15). The only known variation (U/C) occurs at position 4 at the 3′ terminus (Fig. 1B, Uni-12), and this variation is involved in differential gene expression (3, 13). In contrast to multiplex RT-PCR strategies that use multiple primer pairs (2), our initial strategy uses one pair of primers to amplify all of the vRNAs. Initially, two primers (MBTuni-12 [5′-ACGCGTGATCAGCAAAAGCAGG] and MBTuni-13 [5′-ACGCGTGATCAGTAGAAACAAGG]) were designed to take advantage of the conserved vRNA termini and were 5′ tailed for efficient PCR amplification (Fig. 1C).
FIG. 1.
M-RTPCR designed to exploit the conserved elements of the influenza A virus promoter. (A) Classical panhandle depiction of base pairing between the 3′ and 5′ termini that occurs in each of the genomic RNA segments. (B) Illustration of genomic RNA segments (vRNAs) of influenza A virus. (C) M-RTPCR scheme illustrating the tailed reverse transcription primer P1, which is complementary to the Uni-12 sequence (e.g., MBTuni-12), and the tailed forward primer P2, which is complementary to the Uni-13 sequence (e.g., MBTuni-13). dsDNA, double-stranded DNA. (D) M-RTPCR successfully amplifies genomes from pandemic human influenza A viruses isolated from 1933 to 2002, regardless of subtype. The influenza A virus strains used were (i) A/WS/1933(H1N1), (ii) A/PR/8/1934(H1N1), (iii) A/Weiss/1947(H1N1), (iv) A/Denver/1/1957(H1N1), (v) A/Japan/305/1957(H2N2), (vi) A/HK/8/1968(H3N2), (vii) A/USSR/1977(H1N1), and (viii) A/NY/1469/2002(H3N2). N, negative control; L DNA ladder (1 kb plus; Invitrogen). (E) Genomic amplification of multiple subtypes of influenza A viruses isolated from swine, avian, equine, and zoonotic viruses that caused fatalities in humans. The influenza A viruses used were (i) A/Sw/IN/1728/1988(H1N1), (ii) A/Sw/WI/1915/1988(H1N1), (iii) A/Dk/Alb/35/1976(H1N1), (iv) A/Mal/WI/944/1982(H5N2), (v) A/R26(Ty/Ont/7732/1966-WSN/1933(H1N9)), (vi) A/Ty/Ont/7732/1966(H5N9), (vii) A/Eq2/Mia/1/1963(H3N8), (viii) A/NJ/8/1976(H1N1), (ix) A/WI/3523/1988(H1N1), (x) A/MD/12/1991(H1N1), (xi) A/HK/156/1997(H5N1), and (xii) A/HK/213/2003(H5N1). N, no-template M-RTPCR control; L, DNA ladder (1 kb plus). For panels D and E, arrows indicating each of the vRNA segments amplified are on the left; note that it is possible to differentiate the PA segment (2.2 kb) from the PB1 and PB2 segments (2.3 kb) for most of the strains. Amplicons were electrophoresed through a 1.5% agarose gel and stained with ethidium bromide.
M-RTPCR efficiently amplifies the genome of any subtype of human or animal lineage influenza A virus.
vRNA isolated from 100 μl of culture supernatant or allantoic fluid served as a template in an RT-PCR with 0.2 μM of MBTuni-12 and MBTuni-13, performed using the SuperScript III one-step RT-PCR system (Invitrogen). The temperature cycle parameters were 42°C for 60 min, 94°C for 2 min, and then 5 cycles (94°C for 30 s, 45°C for 30 s, and 68°C for 3 min), followed by 31 cycles (94°C for 30 s, 57°C for 30 s, and 68°C for 3 min). vRNA segments for polymerase basic 1 (PB1), polymerase basic 2 (PB2), polymerase acidic (PA), HA, nucleoprotein (NP), NA, matrix (M), and nonstructural (NS) were simultaneously amplified from historical and contemporary human influenza A virus isolates (Fig. 1D). The first human influenza A virus isolated [A/WS/33(H1N1)] and subsequent antigenic drift variants isolated in 1934, 1947, and 1957 were all amplified efficiently (Fig. 1D, lanes 1 to 4). M-RTPCR of RNA isolated from A/Japan/305/57(H2N2), A/HK/8/68(H3N2), and A/USSR/77(H1N1) demonstrates that genomes from prototypical antigenic shift viruses that caused the 1957, 1968, and 1977 pandemics were also amplified (Fig. 1D, lanes 5 to 7). M-RTPCR of A/NY/1469/02(H3N2) illustrates that genomes from contemporary viruses are also amplified (Fig. 1D, lane 8). Genomic amplification of (i) classical swine viruses [e.g., A/Sw/IN/1726/88(H1N1)], (ii) avian viruses isolated from waterfowl [A/Dk/Alb/35/76(H1N1)] or created by reassortment [A/R26(turkey/Ontario/7732/1966-WSN/1933(H1N9)], (iii) a highly pathogenic avian virus [A/Ty/Ont/7732/66(H5N9)], and (iv) an equine isolate [A/Eq2/Miami/1/63(H3N8)] further demonstrates that M-RTPCR is both subtype and lineage independent (Fig. 1E, lanes 1 to 7). M-RTPCR successfully amplified fatal zoonotic viruses of swine origin (11, 17, 22) (Fig. 1E, lanes 8 to 10) and prototypical avian H5N1 viruses (19) (Fig. 1E, lanes 11 and 12). The vRNA segments for the RNA polymerase subunits often migrate together or are difficult to visualize (Fig. 1D and E): however, they can be consistently differentiated by restriction endonuclease digestion, nucleotide sequencing, and/or cloning (data not shown).
We have amplified the complete genomes of >450 viruses, comprising nearly all of the known subtypes of human, swine, equine, and avian viruses (data not shown). M-RTPCR amplicons are excellent templates for dideoxynucleotide sequencing, and in collaboration with D. Spiro, we have successfully used M-RTPCR amplicons from 30 avian viruses for high-throughput multiplex 454 sequencing (personal communication).
M-RTPCR and reverse genetics of viruses from human nasopharyngeal (nas) and oropharyngeal (oro) swab specimens.
M-RTPCR of a fraction (1/120) of specimens that were previously identified as positive for influenza A virus by using real-time RT-PCR specific for M vRNA yielded complete genomic amplification of classical H3N2 and H1N1 viruses for all of the primary specimens analyzed (Fig. 2A, lanes 1 to 6), including those with a low level of vRNA (high cycle threshold [CT] value by quantitative RT-PCR for M vRNA). We are often not able to isolate influenza A viruses by inoculation of primary rhesus monkey cells with swab specimens that have CT values of >30. Therefore, M-RTPCR amplifies the complete genomes of specimens from which isolates may not be recovered by conventional virus isolation. M-RTPCR of novel swine origin H1N1 viruses that recently emerged in humans (Fig. 2A, lane 7) is being used to sequence viruses contained in swab specimens (e.g., GenBank accession no. CY039901 to CY039908), thereby avoiding inevitable mutations that occur during laboratory isolation/propagation, which is often required prior to sequencing.
FIG. 2.
Genomic amplification of human influenza A viruses directly from clinical swab specimens. (A). M-RTPCR of influenza A virus rescued directly from human swab specimens. RNA was isolated directly from nas and oro swab specimens collected in New York during the 2005/2006 influenza season (lanes 1 to 5), the 2001/2002 season (lane 6), and the 2008/2009 season that had previously tested positive for influenza A virus. Work with blinded patient swab specimens was done in accordance with Institutional Review Board protocols 07-022 and 09-025. The vRNA was isolated from 100 μl of swab suspension, and one-sixth of the total RNA was amplified by M-RTPCR and subjected to agarose gel electrophoresis. The amplicons shown are from the (i) H3N2 (nas), (ii) H3N2 (oro), (iii) H3N2 (nas or oro), (iv) H3N2 (nas), (v) H3N2 (oro), (vi) H1N1 (nas), and (vii) swine origin H1N1 (nas) subtypes. Where known, the real-time RT-PCR CT corresponding to a particular swab is listed below the lanes. ND, not determined; N, no-template M-RTPCR control; and L, DNA ladder (1 kb plus; Invitrogen). Amplicons were electrophoresed through a 1.5% agarose gel, which was then stained with ethidium bromide. (B) Modification of reverse-genetics plasmid for in-fusion (Clontech) cloning. Shown is a schematic diagram for modified reverse-genetics vector pG26A12 and cloning of M-RTPCR amplicons for generation of vRNA expression plasmids. Twenty-five nucleotides, comprising the 13 nucleotides of the 5′ termini (Uni13) and the 12 nucleotides of the 3′ termini (Uni12) of all influenza A vRNAs, were inserted between the RNA polymerase 1 promoter and terminator. pG26A12 is linearized by StuI digestion, exposing nucleotides identical to those at the termini of M-RTPCR amplicons. The M-RTPCR amplicons have the same termini (Uni13/Uni12 plus some sequence of the Pol I promoter/terminator) as the linearized plasmid for facilitating in-fusion cloning. The clones generated produce authentic vRNA upon transfection into appropriate cells.
The majority of reverse-genetics plasmids used to engineer influenza A viruses take advantage of host RNA polymerase I promoters at the 5′ terminus and RNA polymerase I terminators or ribozymes following the 3′ terminus to generate exact influenza virus-like vRNAs (5, 6, 14). The strict requirement for precise initiation and termination of the vRNA transcripts dramatically limits the restriction enzymes available for cloning into reverse-genetics plasmids. To accelerate the cloning process, we modified pHH21 (14) and introduced an in-fusion (Clontech) cloning site, creating pG26A12 (Fig. 2B). This new plasmid has 25 nucleotides (5′-AGTAGAAACAAGGCCTGCTTTTGCT-3′) between the Pol I promoter and terminator that are identical to the 13 nucleotides conserved in the 5′ termini and the 12 nucleotides conserved in the 3′ termini of influenza A vRNAs and creates a StuI site. We also modified the M-RTPCR primers by replacing the 5′ tails on the MB-Tuni primers with 5′ tails that are identical to the short terminal regions of the RNA polymerase 1 promoter and terminator (Uni12/Inf-1 [5′-GGGGGGAGCAAAAGCAGG-3′] and Uni13/Inf-1 [5′-CGGGTTATTAGTAGAAACAAGG-3′]). These modifications create a cloning strategy that can be accomplished using short regions (18 to 22 nucleotides) of identity between the plasmid and inserts and commercially available enzymes. Eight genomic amplicons that are present after M-RTPCR can then be cloned simultaneously into the StuI-linearized pG26A12 vector in a single reaction by using in-fusion cloning (Fig. 2B).
Rescue of H3N2 and swine origin H1N1 viruses directly from human specimens.
The genome of a contemporary H3N2 virus [A/New York/238/2005(NY238)] was amplified by M-RTPCR of RNA isolated from a clinical specimen. Amplicons were purified and cloned into pG26A12 by using an in-fusion dry-down PCR cloning kit (Clontech). The positive rate of clones containing genomic length segments was ∼70%, of which 45% were clones of the three polymerase vRNAs. M-RTPCR fidelity was very high; of the PB2, PB1, PA, HA, NP, NA, M, and NS clones initially selected, three had synonymous substitutions, and one (NP) contained a nonsynonymous substitution in comparison to the consensus sequence (GenBank accession no. CY006139 to CY006146). We replaced the mutant NY238-NP clone with another sequence-verified NY238-NP clone that was identified during colony screening and rescued the virus, as described previously (7, 14). The average virus titer in rescue experiments was 6.3 × 103 50% tissue culture infectious doses (TCID50)/ml (n = 6; 4 days posttransfection). The reverse genetics NY238 virus (rgNY238) was propagated in MDCK cells and was compared to the wild-type virus (wtNY238), which was isolated by inoculation of primary Rhesus monkey kidney cells with the swab material and propagated once in MDCK cells. The phenotypes of rgNY238 and wtNY238 were similar in both plaque size (Fig. 3A) and replication kinetics (Fig. 3B).
FIG. 3.
Reverse-genetics-rescued A/New York/238/05(H3N2) (rgNY238) has a plaque phenotype similar to and growth kinetics equivalent to those for the cell culture-isolated virus (wtNY238). (A) Plaque phenotypes of the rgNY238 and wtNY238 viruses on MDCK cells with an agarose overlay. Representative pictures were taken at 4 days postinfection. (B) Multistep growth curves of rgNY238 and wtNY238 viruses. Confluent MDCK cells were inoculated at a multiplicity of infection of 0.02 TCID50/cell. At 1 hour postinoculation, cells were washed three times with phosphate-buffered saline, and then maintenance medium with tosylsulfonyl phenylalanyl chloromethyl ketone-treated trypsin was added. Supernatants were harvested at 1, 2, 7, 14, 21, 28, 35, 42, and 49 h postinoculation and titrated by a TCID50 assay. Data shown are averages of results from three experiments.
To help combat and study the emerging swine origin H1N1 influenza A virus, the genome of A/NY/1682/2009(NY1682) was amplified, sequenced, and rescued directly from a patient specimen. M-RTPCR of RNA purified from the NY1682 specimen was performed as described for NY238, except two forward primers (Uni12/Inf-1 and Uni12/Inf-3 [5′-GGGGGGAGCGAAAGCAGG-3′], 0.1 μM of each) were used. Uni12/Inf-3 was added to account for the U/C variation at position 4 of the 3′ terminus, because we found that this modestly improves M-RTPCR amplification. NY1682 amplicons (Fig. 3, lane 7) were cloned into a bidirectional reverse-genetics plasmid (pDZ plasmid [16]), which we modified for in-fusion cloning. The new plasmids, pBZ61A15(A4) and pBZ61A18(G4), contain adenosine and guanosine, respectively, in the fourth position of the complement of the 3′-terminal promoter element of influenza A virus. Clones representing each of the vRNAs were identified, sequenced (GenBank accession no. GQ332640 to GQ332647), and used to create rgA/NY/1682-WC1/2009. We simultaneously generated a 6:2 reassortant [rgPR8(6):NY1682(2)] containing the H1 and N1 vRNAs of the novel swine origin virus and six other vRNAs from A/PR/8/1934, which is commonly used for vaccine seed stock production. The titers of wild-type A/NY/1682/2009 (which was isolated by inoculation of rhesus monkey kidney cells), rgA/NY/1682-WC1/2009, and rgPR8(6):NY1682(2) were very similar (2.8 × 107, 3.2 × 107, and 3.2 × 107 TCID50/ml, respectively).
The recombinant viruses contain nucleotide substitutions that differentiate them from the consensus sequences of wild-type viruses, and the presence of these substitutions was confirmed by nucleotide sequencing. The rescue of rgA/NY/1682-WC1/2009 and the vaccine-like virus was completed in 11 days, demonstrating that this scheme accelerates the rescue of emerging influenza A viruses. To our knowledge, this is the first time recombinant influenza A viruses containing the entire genomes of wild-type viruses were rescued directly from human swab specimens.
In summary, this study provides a system enabling the rapid genomic amplification of influenza A virus, irrespective of lineage, for subsequent sequence analysis and cloning into reverse-genetics plasmids. M-RTPCR is a very robust method for amplification of historic, contemporary, or novel influenza A viruses, regardless of genotype. One primer set amplifies all vRNAs in a single reaction, whereas the best method to date requires eight sets of primers, which are used in separate RT-PCRs (8). M-RTPCR amplification of viruses rescued directly from primary specimens is ideally suited for high-throughput sequencing and DNA array platforms and provides sequence information free of artifacts selected during virus propagation.
An important application of M-RTPCR is to speed the incorporation of cDNAs representing vRNAs of emerging influenza A viruses into reverse-genetics plasmids to produce genetically engineered viruses. Recently, two new strategies for cloning influenza A vRNAs into reverse-genetics plasmids were published (18, 21). One approach modified the reverse-genetics plasmid as we described for pG26A12; however, this approach used recombination in RecA-positive Escherichia coli to clone fragments apparently amplified from four subtypes by using 32 primer pairs (21). Another approach to speeding the cloning of influenza A virus RNA segments used target-primed plasmid amplification, whereby eight RT-PCRs using previously described primers (8) were used to generate inserts that serve as megaprimers for a second round of PCR to incorporate the reverse-genetics plasmid (18). While both of these cloning methods (18, 21) improve influenza A virus reverse-genetics approaches, each has limitations, and neither provides the speed, specificity, fidelity, and efficiency of our M-RTPCR/in-fusion cloning approach.
We can now create recombinant influenza A viruses directly from a primary specimen in 9 to 12 days (Fig. 4). This rescue time frame has important benefits for vaccine production, particularly as we move beyond currently licensed influenza A virus vaccines, which are 6:2 reassortants containing six vRNAs from donor strains that grow well in embryonated chicken eggs (e.g., A/PR/8/1934) or have temperature-sensitive mutations (e.g., A/Ann Arbor/6/1960) and have the HA and NA vRNAs from strains predicted to antigenically match epidemic strains. The procedures presented here accelerate the rational design of novel, and potentially more efficacious, vaccine seed stocks that impart temperature-sensitive phenotypes (10) or NS-1 mutations responsible for interferon sensitivity (20) into the natural gene constellation of influenza viruses that emerge in humans.
FIG. 4.
Illustration of a streamlined scheme for rapid generation of recombinant influenza A viruses that could be used as vaccine seed stocks. The illustration begins with a swab specimen containing any influenza A virus.
Biosafety.
All experiments with infectious virus were performed using procedures and facilities that met or exceeded the requirements set forth by the U.S. Department of Health and Human Services for propagation of influenza A viruses. All experiments involving highly pathogenic influenza A viruses were performed in an enhanced biosafety level 3 containment laboratory approved for such use by the Centers for Disease Control and Prevention and the U.S. Department of Agriculture. Experiments with infectious swine origin H1N1 viruses were conducted using enhanced biosafety level 2 laboratory practices and procedures as described by the Centers for Disease Control and Prevention interim biosafety guidelines.
Nucleotide sequence accession numbers.
New sequences for A/NY/1682-WC1/2009 were deposited in GenBank under accession no. GQ332640 to GQ332647.
Acknowledgments
We thank Virginia Hinshaw for providing many of the unique influenza A virus isolates utilized here and for her continued support and Robert Webster and William Bean for providing some of the viruses analyzed. We also thank Adolfo Garcia-Sastre and Peter Palese for providing the pDZ reverse-genetics plasmid. We are grateful to Yan Li for her technical assistance, the Wadsworth Center's Molecular Genetics Core for nucleotide sequencing, and the Tissue Culture and Media Core for providing some of the cells utilized.
D.E.W. was funded by NIH/NIAID R21AI 057941-02 and the New York State Department of Health. B.Z., M.E.D., and D.T.S. were supported by and most supplies were provided by NIH/NIAID R21AI 057941-02. This work was also supported by National Institute of Allergy and Infectious Diseases Public Health Service research grants, by grants-in-aid from the Ministry of Health, and by ERATO (Japan Science and Technology Agency).
Footnotes
Published ahead of print on 15 July 2009.
REFERENCES
- 1.Adeyefa, C. A., K. Quayle, and J. W. McCauley. 1994. A rapid method for the analysis of influenza virus genes: application to the reassortment of equine influenza virus genes. Virus Res. 32:391-399. [DOI] [PubMed] [Google Scholar]
- 2.Chan, C. H., K. L. Lin, Y. Chan, Y. L. Wang, Y. T. Chi, H. L. Tu, H. K. Shieh, and W. T. Liu. 2006. Amplification of the entire genome of influenza A virus H1N1 and H3N2 subtypes by reverse-transcription polymerase chain reaction. J. Virol. Methods 136:38-43. [DOI] [PubMed] [Google Scholar]
- 3.de Wit, E., T. M. Bestebroer, M. I. Spronken, G. F. Rimmelzwaan, A. D. Osterhaus, and R. A. Fouchier. 2007. Rapid sequencing of the non-coding regions of influenza A virus. J. Virol. Methods 139:85-89. [DOI] [PubMed] [Google Scholar]
- 4.Flick, R., and G. Hobom. 1999. Interaction of influenza virus polymerase with viral RNA in the ‘corkscrew’ conformation. J. Gen. Virol. 80:2565-2572. [DOI] [PubMed] [Google Scholar]
- 5.Fodor, E., L. Devenish, O. G. Engelhardt, P. Palese, G. G. Brownlee, and A. Garcia-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann, E., G. Neumann, G. Hobom, R. G. Webster, and Y. Kawaoka. 2000. “Ambisense” approach for the generation of influenza A virus: vRNA and mRNA synthesis from one template. Virology 267:310-317. [DOI] [PubMed] [Google Scholar]
- 7.Hoffmann, E., G. Neumann, Y. Kawaoka, G. Hobom, and R. G. Webster. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 97:6108-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann, E., J. Stech, Y. Guan, R. G. Webster, and D. R. Perez. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275-2289. [DOI] [PubMed] [Google Scholar]
- 9.Hoffmann, E., and R. G. Webster. 2000. Unidirectional RNA polymerase I-polymerase II transcription system for the generation of influenza A virus from eight plasmids. J. Gen. Virol. 81:2843-2847. [DOI] [PubMed] [Google Scholar]
- 10.Jin, H., H. Zhou, B. Lu, and G. Kemble. 2004. Imparting temperature sensitivity and attenuation in ferrets to A/Puerto Rico/8/34 influenza virus by transferring the genetic signature for temperature sensitivity from cold-adapted A/Ann Arbor/6/60. J. Virol. 78:995-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kendal, A. P., M. Goldfield, G. R. Noble, and W. R. Dowdle. 1977. Identification and preliminary antigenic analysis of swine influenza-like viruses isolated during an influenza outbreak at Fort Dix, New Jersey. J. Infect. Dis. 136(Suppl):S381-S385. [DOI] [PubMed] [Google Scholar]
- 12.Kilbourne, E. D. 2006. Influenza pandemics of the 20th century. Emerg. Infect. Dis. 12:9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, K. H., and B. L. Seong. 1998. The position 4 nucleotide at the 3′ end of the influenza virus neuraminidase vRNA is involved in temporal regulation of transcription and replication of neuraminidase RNAs and affects the repertoire of influenza virus surface antigens. J. Gen. Virol. 79:1923-1934. [DOI] [PubMed] [Google Scholar]
- 14.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 96:9345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palese, P., and M. L. Shaw. 2007. Orthomyxoviridae: The viruses and their replication, p. 1647-1690. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, S. E. Straus, M. A. Martin, and B. Roizman (ed.), Fields virology. Lippincott Williams and Wilkins, Philadelphia, PA.
- 16.Quinlivan, M., D. Zamarin, A. Garcia-Sastre, A. Cullinane, T. Chambers, and P. Palese. 2005. Attenuation of equine influenza viruses through truncations of the NS1 protein. J. Virol. 79:8431-8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rota, P. A., E. P. Rocha, M. W. Harmon, V. S. Hinshaw, M. G. Sheerar, Y. Kawaoka, N. J. Cox, and T. F. Smith. 1989. Laboratory characterization of a swine influenza virus isolated from a fatal case of human influenza. J. Clin. Microbiol. 27:1413-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stech, J., O. Stech, A. Herwig, H. Altmeppen, J. Hundt, S. Gohrbandt, A. Kreibich, S. Weber, H. D. Klenk, and T. C. Mettenleiter. 2008. Rapid and reliable universal cloning of influenza A virus genes by target-primed plasmid amplification. Nucleic Acids Res. 36:e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subbarao, K., A. Klimov, J. Katz, H. Regnery, W. Lim, H. Hall, M. Perdue, D. Swayne, C. Bender, J. Huang, M. Hemphill, T. Rowe, M. Shaw, X. Xu, K. Fukuda, and N. Cox. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279:393-396. [DOI] [PubMed] [Google Scholar]
- 20.Vincent, A. L., W. Ma, K. M. Lager, B. H. Janke, R. J. Webby, A. Garcia-Sastre, and J. A. Richt. 2007. Efficacy of intranasal administration of a truncated NS1 modified live influenza virus vaccine in swine. Vaccine 25:7999-8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang, S., Q. Liu, J. Pu, Y. Li, L. Keleta, Y. W. Hu, J. Liu, and E. G. Brown. 2008. Simplified recombinational approach for influenza A virus reverse genetics. J. Virol. Methods 151:74-78. [DOI] [PubMed] [Google Scholar]
- 22.Wentworth, D. E., B. L. Thompson, X. Xu, H. L. Regnery, A. J. Cooley, M. W. McGregor, N. J. Cox, and V. S. Hinshaw. 1994. An influenza A (H1N1) virus, closely related to swine influenza virus, responsible for a fatal case of human influenza. J. Virol. 68:2051-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright, P. F., and R. G. Webster. 2001. Orthomyxoviruses, p. 1533-1579. In D. M. Knipe and P. M. Howely (ed.), Fields virology. Lippincott Williams and Wilkins, Philadelphia, PA.